Abstract
Proper specification of germline stem cells (GSCs) in Drosophila ovaries depends on niche derived non-autonomous signaling and cell autonomous components of transcriptional machinery. Stonewall (Stwl), a MADF-BESS family protein, is one of the cell intrinsic transcriptional regulators involved in the establishment and/or maintenance of GSC fate in Drosophila ovaries. Here we report identification and functional characterization of another member of the same protein family, CG3838/ Brickwall (Brwl) with analogous functions. Loss of function alleles of brwl exhibit age dependent progressive degeneration of the developing ovarioles and loss of GSCs. Supporting the conclusion that the structural deterioration of mutant egg chambers is a result of apoptotic cell death, activated caspase levels are considerably elevated in brwl- ovaries. Moreover, as in the case of stwl mutants, on several instances, loss of brwl activity results in fusion of egg chambers and misspecification of the oocyte. Importantly, brwl phenotypes can be partially rescued by germline specific over-expression of stwl arguing for overlapping yet distinct functional capabilities of the two proteins. Taken together with our phylogenetic analysis, these data suggest that brwl and stwl likely share a common MADF-BESS ancestor and they are expressed in overlapping spatiotemporal domains to ensure robust development of the female germline.
Keywords: MADF, BESS, Gene Duplication, Sub-functionalization, Germline Stem Cell, Stonewall, redundancy, robust
Myb/SANT (Swi3, Ada2, N-CoR, TFIIB)-like domain [InterPro, (Finn et al. 2017); IPR001005] protein family is comprised of transcriptional regulators that are involved in a variety of functions in diverse organismal contexts. Consistent with their involvement in gene regulation, a few of these proteins have been shown to bind to naked DNA to modulate transcription of individual genes. Alternatively they can also influence a group of genes simultaneously by regulating chromatin remodeling (Boyer et al. 2004). The Drosophila melanogaster genome consists of fifty-five Myb/SANT like in Adf (MADF) genes (Shukla et al. 2014). In flies, MADF domain containing genes have been implicated in critical biological processes such as hybrid incompatibility (Brideau et al. 2006; Maheshwari et al. 2008), transposition (Casola et al. 2007) and transcriptional regulation (Ratnaparkhi et al. 2008; Cutler et al. 1998; Timmerman et al. 2013). A subgroup of the MADF family is the MADF-BESS family, which consists of sixteen proteins that contain both, a N-terminal MADF and C-terminal BEAF, Su-Var (3–7), Stonewall (BESS) domain (Bhaskar and Courey 2002). The BESS domain, like MADF, may also be a modified SANT domain (Cutler et al. 1998; Bhaskar and Courey 2002). The MADF domain is a DNA binding domain while the BESS domain is thought to be a protein-protein interaction domain (Bhaskar and Courey 2002; Maheshwari et al. 2008).
The MADF-BESS family in Drosophila has been proposed as an example of a gene family that has duplicated and expanded (Lander et al. 2001; Shukla et al. 2014) over 40 million years (My) ago and has in the intervening period sub-functionalized (Shukla et al. 2014). We have been exploring the redundancy and sub-functionalization within this family using molecular genetic analysis (Shukla et al. 2014). In an earlier study, we reported that three members of the family (hinge1, hinge2 and hinge3) show overlapping functions during development of the wing-hinge, primarily by negative regulation of Wingless (Wg) expression (Shukla et al. 2014). Moreover, RNAi mediated knockdowns of all sixteen members in different tissues of the body suggested that different subsets of genes function in distinct spatiotemporal domains. For example, only Dlip3, but not any other MADF-BESS gene, is important for eye development (Duong et al. 2008) while Coop (Song et al. 2010) is involved in patterning of the wing by regulating Wingless signaling.
Drosophila gene stwl (Clark and McKearin 1996) (McKearin 1997) encodes for a heterochromatin-associated protein belonging to the MADF-BESS family (Delattre et al. 2004; Delattre et al. 2000). Aside from being the largest (1037 amino acids) member of this family, it also contains a modified SANT domain (Boyer et al. 2004) and has been implicated in regulating chromatin compaction (Yi et al. 2009). Functional characterization of Stwl protein has suggested its possible involvement during position-effect variegation (PEV) similar to Su(var)3-7. Furthermore, it is also thought that Stwl achieves this by affecting methylation of Lys residues in Histone tails (Yi et al. 2009). As would be expected of a pleiotropic chromatin regulator, mutations in stwl display phenotypes in distinct and seemingly unrelated developmental contexts including wing patterning (Shukla et al. 2014; Brun et al. 2006), DNA damage response (Yi et al. 2009) and, female germline development. (Clark and McKearin 1996) (Akiyama 2002).
stwl function seems to manifest at a number of different stages of ovarian development. Immunostaining analysis with anti-Stwl antibodies indicated that Stwl protein is nuclear protein and is specifically detected in the GSCs and their progeny including cystoblasts and cystocytes. Accordingly, Stwl appears to be required in the germline stem cell (GSC) lineage for GSC maintenance as loss of stwl activity leads to corresponding loss of GSCs (Clark and McKearin 1996; Maines et al. 2007; Akiyama 2002). Interestingly requirement of Stwl appears to be specific to GSCs alone as GSCs undergo precocious differentiation in the absence of stwl, however differentiated population i.e., cystocytes, marked by the presence of fusome, are relatively unaffected. Furthermore, oocyte specification is also defective in stwl ovaries resulting in excess number of cells that retain oocyte determinants such as Orb protein (Clark and McKearin 1996). Moreover, a fraction of stwl cysts undergo an extra round of division (Clark and McKearin 1996). Although many of the phenotypic consequences due to loss of stwl function are readily apparent, they are partially penetrant and are also sensitive to external conditions such as temperature. The variability of stwl phenotypes led Clark & McKearin to speculate that other proteins might partially substitute for stwl function. Thus, they in fact, predicted that ‘Partial functional redundancy for Stwl protein might account for the variable severity of stwl phenotypes’ (Clark and McKearin 1996).
Here we have tested this prediction systematically. The availability of TRiP RNA interference lines (Ni et al. 2009; Ni et al. 2011; Perkins et al. 2015) and germline specific UAS/Gal4 strains (Staller et al. 2013; Rørth 1998) allow for efficient maternal knockdown. Using this strategy, we searched for another member of MADF-BESS family that potentially resembles stwl. Based on female germline specific knockdown and subsequent phenotypic analysis, we have uncovered CG3838, a member of the MADF-BESS family, that has germline-specific functions. Further supporting the claim, transposon insertion mutants spanning the locus are viable but only semi-fertile and fertility of adults deteriorates in an age dependent manner. Moreover, ovaries from mutant females show GSC lineage specific defects. The loss of function phenotypes of CG3838 and stwl thus seem to overlap. Confirming the assertion that the two proteins are partially redundant, Stwl over-expression in the GSC lineage is sufficient to rescue several, but not all, aspects of loss of CG3838 function. Based on our analysis, stwl and CG3838, now christened brickwall (brwl), seem to retain overlapping functions which may reflect the presence of an ancestral gene that existed over 40 My ago.
Materials & Methods
Drosophila husbandry
All flies were raised at 25° in standard corn meal agar. Crosses were set up at 25° and 29°. The females of the F1 progeny were dissected for the phenotypes in all cases.
Fly Lines
For ovarian somatic and maternal knockdowns, VALIUM10, VALIUM20 and VALIUM22 Transgenic RNAi Project lines procured from Bloomington Drosophila Stock Center (BDSC) were used (Ni et al. 2009)(Perkins et al. 2015) in this study. VALIUM lines used have been listed in Table 1. Here, we have used notations V10/V20/V22 to indicate a VALIUM10/VALIUM20 or VALIUM22 line respectively. For example, UAS-CG3838 RNAi (V22) is a VALIUM22 while UAS-CG3838 RNAi (V10) is a VALIUM10 line. Lines expressed in the ovary include (UAS-gene name, (Source)), UAS-CG13204 (DPiM), UAS-CG3838 (FlyORF), UAS-stwl (DPiM), UAS-CG4404 (FlyORF), UAS-CG8359 ( FlyORF), UAS-Coop (FlyORF), UAS-CG3919 (FlyORF). DPiM (Drosophila Protein Interaction Map), indicates use of C-terminal FLAG-HA tagged DNA clones from the DPiM (Guruharsha et al. 2011) project, procured from Drosophila Genomic Resource Centre (DGRC), Indiana and then injected in the Drosophila Transgenic Facilty (http://www.ccamp.res.in/Fly-facility) at NCBS Bangalore. FlyORF lines (https://flyorf.ch/) are from the Zurich FlyORFeome project (Bischof et al. 2013). The expression of Brwl in the GSC lineage was monitored by imaging a CG3838:GFP.FTBP fusion available in BDSC (#56150). The fly line was a Bac construct created as part of the modERN project (Flybase FBrf0225001; Personal Communication by A. Victorsen and K. White 2014). Daughters against Dpp (dad)-lacZ expression (Song et al. 2004) was monitored to mark GSCs. The expression of Stwl was monitored using Stwl:GFP protein trap line procured from FlyTrap (Kelso et al. 2004).
Table 1. Summary of a targeted screen setup to evaluate germline and somatic roles for MADF-BESS genes. RNAi lines from the Harvard in-vivo fly RNAi project (TRiP; https://fgr.hms.harvard.edu/fly-in-vivo-rnai) (Ni et al. 2009; Ni et al. 2011) were used for the experiments. VALIUM10 (V10), VALIUM20 (V20) and VALIUM22 (V22) lines were procured from the Bloomington Drosophila Stock Centre (BDSC) and crossed with nos-Gal4:VP16 (#4937) and c587-Gal4 (#67747). Experiments were done both at 25 °C and 29 °C. Data shown is for the 25 °C cross, which was similar to the results obtained at 29 °C. The data for maternal knockdown using both MTD-Gal4 and nos-Gal4;UAS-Dicer2 has been taken from Suppl. Table of (Yan et al. 2014).
| GENE | Bloomington Line | nos-Gal4 (Germline) | Yan et al. (2014) (Germline) | c587-Gal4 (Somatic) |
|---|---|---|---|---|
| CG8119 | 57761 (V20) | -a | — | Normal |
| CG8119 | 38893 (V22) | Normalb | Normal | — |
| Adf1/ CG15845 | 28680 (V10) | — | Normal | Normal |
| hng2/ CG8539 | 58149 (V20) | Normal | — | Normal |
| CG30403 | 57286 (V20) | Normal | — | Normal |
| CG3919 | 33355 (V20) | Normal | Normal | Normal |
| CG4404 | 62415 (V20) | — | — | Normal |
| CG4404 | 50527 (V22) | Normal | — | — |
| hng1/ CG9437 | 26754 (V10) | — | Normal | Normal |
| hng3/ CG13987 | 29613 (V10) | — | Normal | — |
| hng3/ CG13987 | 42765 (V20) | Normal | — | Normal |
| dlip3/ CG12767 | 27067 (V10) | — | — | — |
| CG11723 | 29349 (V10) | — | Normal | Normal |
| CG11723 | 42514 (V20) | Normal | — | — |
| Coop/ CG1621 | 29350 (V10) | — | Normal | Male pupal lethal |
| CG45071 | — | — | — | — |
| brwl/ CG3838 | 36785 (V22) | Phenotype in ovary seen in flies; 7 days post-eclosion | Normal | Normal |
| brwl/ CG3838 | 31922 (V10) | Normal | — | Normal |
| CG6276 | 62234 (V20) | — | Normal | Normal |
| CG6276 | 38189 (V22) | Normal | Normal | — |
| CG13204 | 31919 (V10) | — | — | Normal |
| CG13204 | 42806 (V22) | Normal | — | — |
| Stwl/ CG3836 | 35415 (V22) | Strong phenotype in ovary; Seen in female flies immediately after eclosion | No Eggs | — |
‘Normal’ indicates a normal ovary without any fusion or degeneration related phenotypes in ovarioles from 10 and 18 day old females. We do see minor follicle phenotypes, such as layering, in some of the crosses but these are distinct from the phenotypes seen in brwl mutants. Also seen are significant variations in number of animals emerging from each cross.
‘-’ indicates that the experiment was not performed. Experimental data in the table represents collated data for 10 and 18 day old females.
Gal4 drivers
nanos(nos)-Gal4:VP16, which expresses in the GSCs and cysts in the germarium (BDSC 4937), was the primary Gal4 used for all the experiments. Other Gal4 drivers used included maternal α-tubulin-Gal4:VP16 (MAT-Gal4, expresses in the germline, but not the GSCs), c587-Gal4 (BDSC 67747, expresses in most somatic cells of the ovary), Traffic Jam (TJ)-Gal4 (Subset of follicle cells), hedgehog (hh)-Gal4 (cap cells) and GR1-Gal4 (BDSC 36287, expresses in the follicle cells) (Hartman et al. 2015; Decotto and Spradling 2005; Jin et al. 2013).
Screening MADF-BESS genes with somatic and maternal drivers
F1 Ovaries were screened for phenotypes using nos-Gal4 as a maternal driver and c587-Gal4 as a somatic driver. For the screen, 5 F1 females were dissected at 3, 10 and 18 days, ovaries stained with Phalloidin and DAPI and visualized under an epifluorescence microscope. ∼150 ovarioles were examined by eye for each time point and representative images were stored. V10 lines were tested with the c587-Gal4, V20 lines tested with both somatic (c587-Gal4) and germline (nos-Gal4) drivers and V22 lines were tested only with nos-Gal4.
Transposon insertion Lines
brwlKG00824 (BDSC 12901) is P-element insertion line in the 5′ of CG3838 and is a part of BDGP Gene Disruption Project collection (Bellen et al. 2004). brwlMI054561 (BDSC 42326) is Minos-element insertion line with the Minos cassette inserted in the 3′ of CG3838 and is the part of transposon Minos-mediated integration cassette (MiMic) collection (Venken et al. 2011). Here, these are referred to in short, as brwlKG0 and brwlMI0.
Immunostaining and Imaging
Adult ovaries were dissected in chilled 1X PBS and fixed with 4% paraformaldehyde in 1X PBS with 0.3% Triton-X for 20 min at room temperature. They were blocked in 2% BSA, and 0.3% Triton for in 1XPBS for 1 hr; incubated with the primary antibody overnight at 4°; washed 4 times for 10 min in 1X PBS containing 0.1% Triton X and incubated with the appropriate fluorescent secondary antibody for 1 hr at room temperature in the dark. The ovaries were then washed and mounted in Antifade (P7481, Themo-Fisher Scientific), DAPI (D1306, Themofisher Scientific) was used to stain the nuclei. Primary antibodies used are mouse anti-alpha spectrin 3A9 (1:50), mouse anti-orb 4H8 (1:10), rat anti-vasa (1:50), anti-hnt 1G9 (1:10), anti B-gal 40-1a (1:10) from DSHB, rabbit anti-phospho-histone S10 (#9701; 1:150), rabbit cleaved Caspase-3 (#9661S; 1:150) from Cell signaling, rabbit anti-cyclin E (1:100) from Cell signaling, rabbit anti-GFP from Sigma and Chicken Anti-GFP (#A10262, 1:500) from Thermo-Fisher Scientific. Mouse anti-C3G (1:500) was kindly provided to us by Prof. Scott Hawley. All secondary antibodies used were from Invitrogen Molecular Probes. Phalloidin 568 and 488 (Invitrogen Molecular Probes) were used to stain actin at a dilution of 1:100. Images were taken on Zeiss 710 LSM confocal microscope and Leica SP8 confocal microscope at 40X and 63X and subsequently processed using Image J software. GraphPad Prism 6 was used for making graphs and for statistical analysis.
Phylogenetic Analysis
The coding sequence of MADF and the BESS domains from brwl and stwl genes in 12 Drosophila species was predicted using NCBI Conserved Domain (Marchler-Bauer et al. 2015). The sequences were aligned using MUSCLE (Edgar 2004) implemented in MEGA7.0 (Kumar et al. 2016). Pairwise raw genetic distances were calculated in MEGA7.0 (Kumar et al. 2016). Sequences were partitioned into three codon positions to create a full partition and then the greedy strategy (Lanfear et al. 2012) implemented in IQ-Tree (Nguyen et al. 2015) was used to find the right partitioning scheme based on minimum Bayesian Information Criterion (BIC) (Nei M 2000) (Schwarz 1978). Pairwise codon substitution pattern (ω = dN/dS) was determined in DnaSP (Librado and Rozas 2009). Each domain was tested for recombination and appropriate codon substitution model in Datamonkey (Delport et al. 2010). Codon wise accumulation of synonymous and non-synonymous codons was analyzed using REL method (Pond et al. 2005) implemented in Datamonkey (Delport et al. 2010).
Data and Reagent availability
Fly lines are available on request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6114977.
Results
A targeted screen to uncover MADF-BESS genes required in the GSC lineage
As previously reported (Clark and McKearin 1996; Akiyama 2002), reducing stwl function in the female germline led to ovarian phenotypes. This phenotype is replicated by driving stwl RNAi (V20) in the germline using nos-Gal4 [See Suppl. Figure 1 and (Yan et al. 2014)]. Consistent with the previous reports the phenotypic consequences due to loss of stwl in adult ovaries were severe resulting in very small ovarioles that degenerated within days. Earlier studies have also demonstrated that Stwl is expressed in the GSCs and cysts in the germarium (Clark and McKearin 1996).
In order to explore the possible germline specific function(s) of other MADF-BESS domain proteins, we sought to compromise the transcript levels of other MADF-BESS family genes in the female germline using a similar RNAi based approach. By employing the UAS-Gal4 system (Brand and Perrimon 1993; Duffy 2002), we tested MADF-BESS genes for which VALIUM20 (V20) and VALIUM22 (V22) RNAi lines (Hu et al. 2017; Perkins et al. 2015) are available (Table 1). nos-Gal4 was employed as the primary Gal4 driver to knockdown the corresponding MADF-BESS genes specifically in the germline (Staller et al. 2013) by using both V20 and V22 lines. As summarized in Table 1 (column #3) compromising the function of only one other gene (CG3838), christened brickwall (brwl), resulted in ovarian phenotypes (Figure 1A-B). Interestingly, Yan et al. (2014) had earlier screened available VALIUM lines using both nos-Gal4:VP16;UAS-Dicer2 and Maternal Triple Driver (MTD)-Gal4. Their studies, also summarized in Table 1 (fourth column), indicated that MADF-BESS genes, with the exception of stwl did not affect oogenesis significantly, after maternal knockdown of transcripts. The study did not uncover CG3838 as a candidate possibly because CG3838/brwl mutant flies are fertile for the first ten days of egg laying, showing age-dependent loss of fertility, as described in the next section.
Figure 1.
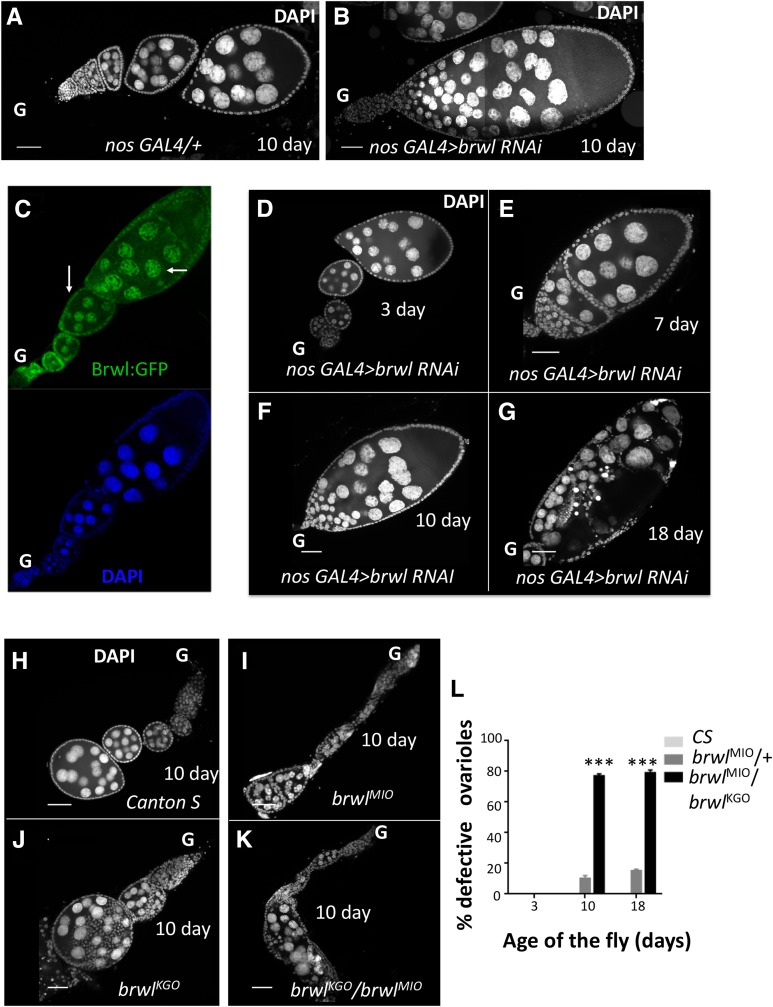
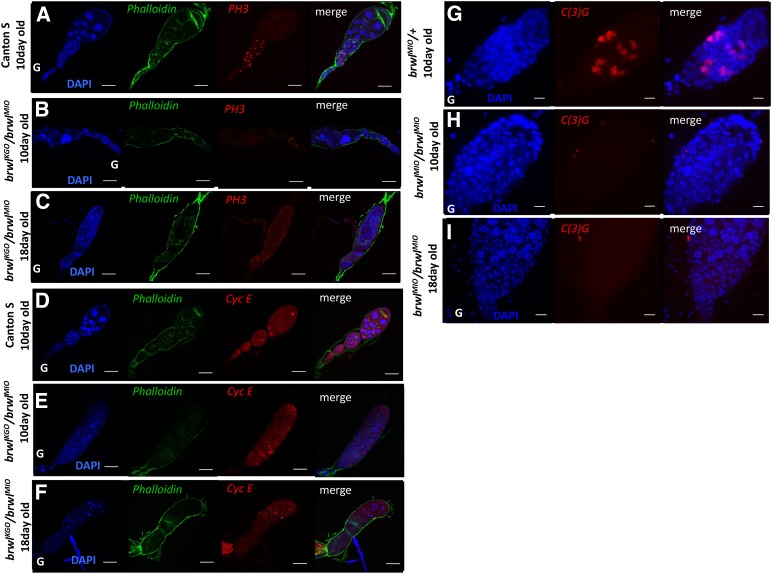
Compromising CG3838/brwl function in the female germline uncovers age dependent ovarian phenotypes. (A-B) Reduction of brwl transcripts in the female germline by expressing brwl TRiP VALIUM22 RNAi line, using nos-Gal4 leads to sterility and a variety of ovarian defects in the adult female. Major morphological defects include fused ovarioles and pycnotic nuclei. (C) Brwl:GFP expresses in the nuclei of both germline and somatic cells. The staining resembles the expression of a Stwl protein trap line (Kelso et al. 2004) (Buszczak et al. 2007). The arrows indicate expression in nuclei of both germline (nurse cells) and the soma (follicle cells). (D-G) Ovarian defects are age dependent and manifested in the adult female 7-10 days (E-F) after of fly eclosion and the phenotypes are enhanced in older flies. 3 day old (D) animals show 100% normal oogenesis and lay fertile eggs while 18 day old (G) animals show ∼100% degenerated egg chambers. Nuclei are marked with DAPI. ‘G’ marks the anterior i.e., germarial end of the egg chamber in all the figures. (H-K) Ovaries from females carrying brwl insertional alleles (brwlKG0, brwlMI0). The ovarioles are dissected from 10 day old females. Canton Special (Canton S) flies are used as controls. The oocytes show phenotypes similar to that of the brwl maternal RNAi knockdowns, with all ovarioles showing degenerating egg chambers by the 18th day. (L) Quantitative data representing the percentage of ovarioles that show the phenotype on third, tenth (n = 80) and eighteenth (n = 25) day after fly eclosion. Ovaries from Canton-special (CS) animals were used as a control for statistical analyses. *, ** and *** denote P < 0.05, P < 0.01, and P < 0.001 respectively for this and all subsequent figures. N: number of adult females = 3-20 and n: number of ovarioles = 25-80.
The ovarian defects induced by the RNAi based knockdown of brwl prompted us to assess the expression of Brwl in the Ovary. Since antibodies against Brwl protein are unavailable, we used a Brwl:GFP fusion (BDSC 56150) line (See Materials and Methods) to monitor the expression of Brwl protein. Brwl:GFP is expressed in the adult ovarioles, where it marks the nuclei of both nurse and follicle cells in developing egg chambers, Stage-1 onwards (Figure 1C). This is similar to the expression of Stwl:GFP (See Figure 2H, Buszczak et al. 2007). We were unable to detect nuclear expression in the GSCs using the Brwl:GFP line. This was also true for the Stwl:GFP line in our hands and in published literature (Rohrbaugh et al. 2013). The evidence for nuclear GSC staining for Stwl is based on the use of an anti-Stwl antibody (Clark and McKearin 1996; Maines et al. 2007).
Figure 2.
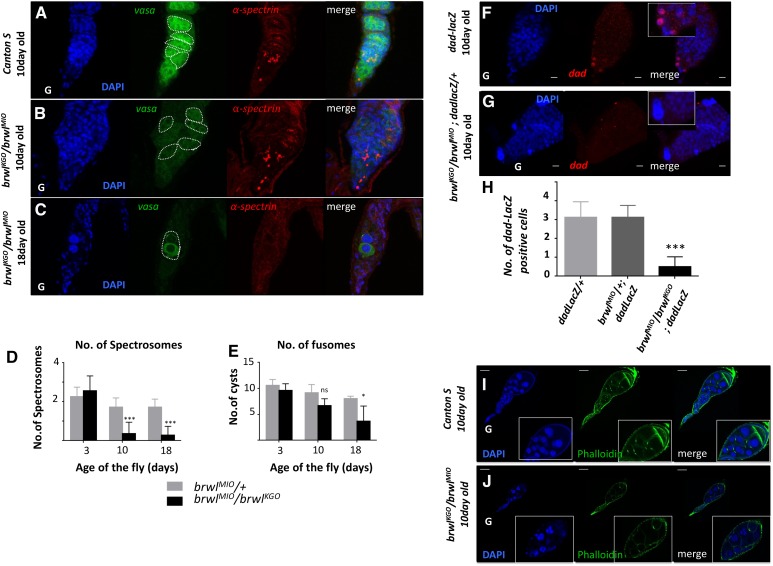
brwl alleles show defects in the cyst architecture and decrease in total number of germline stem cells with age. (A-C) brwlKG0/ brwlMI0 mutants show decrease in Vasa expression (green) by the 10th day PE (B) while α-spectrin staining (red) marks the number of spectrosomes, as measured at day 10 (B) and day 18 (C). N = 5, n = 15-35. Cysts are outlined with dashed lines. (D-E) Quantitative data representing number of spectrosomes (D) and fusomes (E) on third(n = 14), tenth(n = 22) and eighteenth day(n = 11) after fly eclosion. N = 5, n = 15-35. (F-H) brwlKG0/brwlMI0 mutants show decrease in the number of dad-LacZ positive cells (G,H). LacZ levels, as measured by anti-LacZ antibody (red), is highest in germline stem cells of the ovary. N = 5, n = 15-30. Inset here and in following images is a 2X zoom of the main image. dad-LacZ line (F) is used as a control for this experiment. (I-J) brwlKG0 / brwlMI0 mutants show disorganization and decrease in number of ring canals as visualized by phalloidin staining (green). N = 5, n = 15-20.
Unlike the nos-Gal4 driver, MAT-Gal4 does not express in the GSCs (Perkins et al. 2015; Yan et al. 2014) and drives expression in the cystoblasts, Stage-1 onwards. MAT-Gal4; UAS-Brwl RNAi (V22) females (Suppl. Figure 1E) displayed similar age dependent phenotypes as in the case of nos-Gal4. Taken together, the RNAi based inactivation suggests that brwl activity is likely required in the germline from very early stages of development.
Interestingly, Brwl-GFP is also expressed in the somatic (follicular) nuclei (Figure 1C, Suppl. Figure 1B). This expression pattern raises the possibility that brwl function is also required in the soma for proper germline development. We tested this possibility by compromising brwl function in the somatic cells using a variety of somatic drivers, namely GR1-Gal4, c587-Gal4, tj-gal4 and hh-Gal4. The VALIUM10 Brwl RNAi line (BDSC 31922) was used for these experiments. Surprisingly however, none of the resulting F1 females showed any significant ovarian phenotypes, even when the experiments were conducted at an elevated temperature (29°). To confirm these observations, we also tested TJ-Gal4 in combination with UAS-Dicer2 to enhance brwl knockdown and could not detect any role for brwl in the soma. Further, we tested all other MADF-BESS genes, using available V10 and V20 lines in combination with c587-Gal4. The lack of phenotypes (Table 1) suggest at best a relatively minor or insignificant role for brwl, as also for other MADF-BESS genes in the somatic cells of the ovary. Thus, while a role for brwl in the soma is possible, our experiments do not support this possibility.
RNAi mediated germline knockdown of CG3838 uncovers age dependent ovarian phenotypes
To start characterizing the egg chambers compromised for brwl activity, the ovaries were first stained with DAPI, a DNA dye. Readily visible phenotypes upon brwl RNAi included defects in the ovariole architecture; fused egg chambers and pycnotic nuclei (Figure 1B). These defects were highly penetrant as substantial number of the ovarioles (50/69; 72%) displayed a combination of these phenotypes. As indicated in the previous section, the phenotypic consequences appeared to be specific as germline knockdown of other MADF-BESS genes did not display such phenotypes (Table 1) while somatic knockdown did not mimic the maternal phenotypes, when tested in flies 3, 10 and 18 days post-eclosion (PE).
Subsequent careful characterization revealed that the phenotypic consequences upon loss of brwl in the germline were age dependent. At 25 °C, females three days PE showed completely normal ovarian morphology (Figure 1D). Consistently, these females were fertile, and laid eggs that went through normal development post-fertilization (Figure 1D). By the 7th day PE, the abnormalities in the ovarian structure became apparent with fused egg chambers (Figure 1E). This defect became even more penetrant and severe by the 10th day PE (Figure 1F), where multiple instances of a single large ovariole i.e., fused egg chamber with abnormally high number of nurse cells were observed. Moreover, the fused egg chambers showed clear signs of structural abnormalities and the presence of pycnotic nuclei. By the 18th day PE (Figure 1G), all the ovarioles were partially or completely degenerated. As many of these phenotypes are reminiscent of stwl loss of function, stwl and brwl appear to be the only two members of the MADF-BESS family with functions in the GSC lineage during female oogenesis.
Transposon insertion alleles of brwl display similar ovarian phenotypes as germline specific knockdown of brwl
To extend the RNAi dependent phenotypic analysis further, two independent transposon insertion lines in the brwl locus, referred to as brwlKG0 and brwlMI0 (See Materials and Methods) respectively, were tested for phenotypes during ovarian development. Both brwlKG0 and brwlMI0 females show similar range of ovarian defects when homozygous (Figure 1I, J). Notably, these defects are also age dependent and at day 10 PE, appear to be equally drastic as compared to those seen with the brwl-RNAi. brwlMI0 is maintained over a balancer and very few homozygotes emerge while brwlKG0 is a homozygous viable allele. Less than 5% of the total ovarioles display ovarian defects in older heterozygous brwlKG0 /+ and brwlMI0 /+ females. By contrast, significant structural deterioration and egg chamber fusions are observed in the ovaries derived from the trans-heterozygous brwlKG0 / brwlMI0 females (Figure 1K, L). Phenotypic similarities between the germline specific knockdown of brwl function and transposon insertion lines in the brwl locus further supported a germline specific requirement for brwl. We thus sought to analyze this activity further. Both the brwlKG0/brwlMI0 and brwlMIO/brwlMI0 were employed for subsequent characterization of the ovarian defects.
brwl mutants show a range of ovarian defects
The phenotypic consequences due to loss of brwl are highly variable. Thus, to better understand possible function(s) of this novel player, we have undertaken a detailed characterization of the individual phenotypes associated with loss of brwl during different stages of ovarian development. As preliminary data (see above) suggested age dependent progression of the phenotypic severity, we analyzed individual phenotypic traits keeping in mind the age dependence.
Drosophila ovary is made up of 14-16 ovarioles that individually function as an egg production line. The germarium containing GSCs surrounded by the somatic cells is situated at the anterior tip of the ovariole. Typically, the GSCs divide asymmetrically to generate another stem cell and a daughter cell, which undergoes differentiation. Stem cell daughters, or cystoblasts, undergo four rounds of synchronous division to eventually give rise to 16 interconnected cystocytes. While niche derived non-autonomous signaling coordinates GSC specification, cell intrinsic mechanisms contribute to the regulation of both the symmetric as well as asymmetric germline stem cell divisions. The unequal nature of division is reflected in the characteristic morphology of fusomes which are the membranous organelles rich in proteins such as α-Spectrin and, Hu-li tai shao (Hts) (Drosophila homolog of Adducin). Thus, GSCs can be readily identified by the virtue of the presence of a round fusome also referred to as spectrosome, located adjacent to the somatic cap cells. By contrast, the differentiated progeny of GSCs i.e., cystocytes or cystoblasts display diagnostic changes in the shape of the fusome including elongation and bifurcation. Since stwl function is required in the GSCs for their maintenance, we first decided to assess if brwl mutants also show a GSC specific phenotype.
brwl mutants affect cyst architecture and GSC number in the early stages of oogenesis: Preliminary analysis of loss of function mutants in brwl locus as well as germ line specific knock down of brwl function indicated involvement of brwl during several stages of ovarian development. To better understand the function of brwl in the process we decided to take a closer look at the germaria from the brwl mutant females. To this end brwlKG0/brwlMI0 ovaries were first stained with anti-Vasa antibodies and counterstained with DAPI. Vasa is a maternal effect, posterior group gene and vasa RNA as well as Vasa protein are localized in the pole plasm that is assembled and subsequently anchored at the posterior pole of the oocyte. Vasa protein is enriched in a germline specific manner through the course of development. Accordingly, within early germarium Vasa protein is present in germ cells including GSCs and thus has served as a reliable marker of the germline fate (Lasko and Ashburner 1990). In ten day-old brwlKG0/ brwlMI0 ovaries, Vasa protein appears to be considerably less than the wild type (Figure 2B). Additionally, the staining pattern also revealed that Vasa positive cysts appear disorganized and are distributed randomly within the germarium (Figure 2B). By the 18th day, as the number of cysts degenerate, brwlKG0/brwlMI0 ovaries show presence of considerably larger than average sized nuclei (Figure 2C) possibly due to apoptosis (see below). Presence of Vasa around these nuclei also suggests that such cysts might have division defects.
To specifically analyze the influence of Brwl on the GSCs and their progeny, we stained the brwlKG0/brwlMI0 ovaries with antibodies against α-Spectrin and counted the number of spectrosomes and fusomes (Figure 2D-E). Typically, wild type germarium consists of 2-3 GSCs, which can be readily identified by the presence of spectrosome. By contrast, in the ovaries derived from 10 days old brwlKG0/brwlMI0 females, total number of cells with a spectrosome were substantially reduced. At the 18th day, in several germaria, not even a single cell positive for α-Spectrin could be detected (Figure 2C, D), indicating a substantial decrease in the total number of GSCs and cystoblasts. A number of fusomes are present in the wild type is between 9-11. Interestingly in the ovaries from the brwlKG0/brwlMI0 10-day old post-eclosion females, no significant decrease in the number of fusomes was observed. However, this number decreased substantially in the 18th day old flies (3-5 fusomes) (Figure 2E). Moreover, unlike the wild type female ovary samples, brwlKG0/ brwlMI0 ovaries also exhibited disorganized cysts that have lost a stereotypical linear arrangement. Instead of canonical 2, 4, 8, and 16 cell cyst organization, they were unevenly distributed in the mutant germarium.
brwl mutants show decrease in the number of GSCs: In wild type females, total number of GSCs decreases with age. A single ovariole generally consists of 2-4 GSCs which are maintained until late in the lifespan and the GSC count decreases by half only by day 63 post-eclosion (Casanueva and Ferguson 2004; Pan et al. 2007). brwlMI0/ brwlMI0 ovaries show considerable decrease in the number of spectrosome positive cells (Figure 2G,H) between day 10 through 18 post-eclosion. This observation taken together with disorganized cysts with reduced levels of Vasa protein indicated that brwl activity is likely needed for GSC specification and/or maintenance. To confirm this directly a GSC marker dad (daughters against dpp)-lacZ was used to stain the GSCs. brwlKG0/ brwlMI0 ovaries show a 60% reduction (Figure 2H) in the dad-lacZ positive cells 10 days after eclosion.
In a wild type germarium, a cyst consists of 16 cells that are interconnected via cytoplasmic bridges also known as ‘ring canals’. As the egg chambers derived from brwlKG0/ brwlMI0 adult female ovaries appeared to display variety of structural defects, we decided to characterize the cytoskeletal defects by analyzing ring canals, which are actin rich structures. We stained the brwl mutant ovaries with fluorescently labeled phalloidin (Figure 2J), which labels actin. brwlKG0/ brwlMI0 ovaries show significant loss of ring canals (∼70% reduction as compared to Cantos S), critical for oocyte development from day 10 onwards. Fusome, a branched structure made up of continuous endoplasmic reticulum and cytoskeletal components is responsible in determining the mitotic division plane. Because of this influence, eventually after 4 mitotic cell divisions, only two cells end up with four ring canals. One of these two cells eventually would assume the oocyte identity while forcing the other to acquire a nurse cells fate like the remaining cells with fewer number of ring canals. Nurse cells are responsible for the production of cytoplasmic components such as proteins and RNA that are trafficked to the oocyte. As ring canals are the structures through which nurse cells pass the nutrients to the oocyte, reduced number of ring canals raised obvious questions about the oocyte identity in brwlKG0/ brwlMI0 egg chambers.
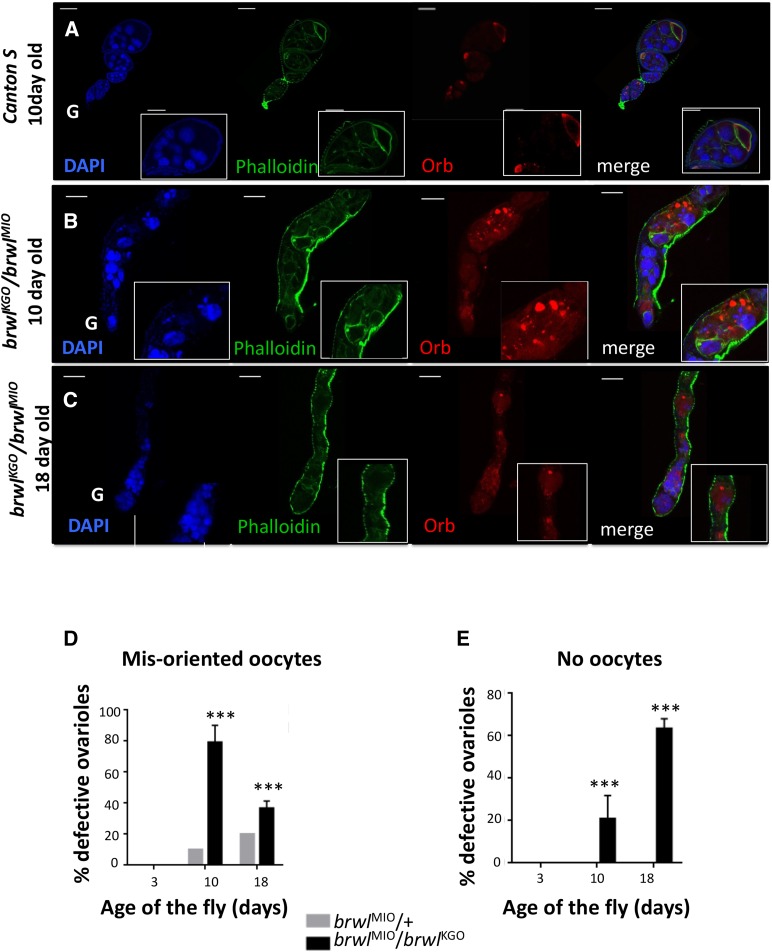
brwl mutants affect oocyte specification: Proteins like Orb, Egl and BicD (Huynh and St Johnston 2000) that accumulate in the prospective oocyte are essential for oocyte specification and axis determination. orb encodes for Drosophila homolog of Cytoplasmic Polyadenylation Element Binding protein, (CPEB) and has been shown to be required at multiple levels during oogenesis including formation of a 16-cell cyst, oocyte specification, and polarity establishment and/or maintenance (Lantz et al. 1994). Orb accumulation is progressively restricted to the presumptive oocyte during early stages of oogenesis. Furthermore, Orb is localized in a crescent shaped pattern at the posterior end of the developing oocyte. To assess if loss of brwl influences oocyte specification, brwlKG0/ brwlMI0 ovaries were stained with anti-Orb antibodies (Figure 3A-C). The staining pattern of Orb protein in mutant egg chambers suggested age dependent loss of oocyte specification. Other phenotypes included mis-positioned oocytes or complete loss of oocyte identity. On several occasions although present, Orb protein was not restricted to a single cell and was instead detected in a group of cells. Consequently, by 10th day, 77.6% of the total ovarioles (n = 45) showed either mis-positioned oocyte and/or several mis-specified i.e., Orb positive cells (Figure 3B). Moreover, 20.83% egg chambers showed absence of a presumptive oocyte as assessed by lack of Orb accumulation in any cell (Suppl. Figure 1D). By the 18th day 63.3% of the ovarioles (n = 32) showed egg chambers with a complete absence of oocyte, while around 36.7% showed mis-positioned oocytes (Figure 3D,E). This demonstrates progressive worsening of the phenotype and deterioration of the tissue from day 10 to day 18.
Figure 3.
Orb is mislocalized or absent in brwl mutants. (A-C) brwlKG0/ brwlMI0 mutants display defective ovarioles. These defects were classified in two major categories, mis-positioned oocytes and no-oocytes, based on actin (green) and Orb (red) staining. Orb related phenotypes enhance with age, with 18 day old flies having a high proportion of ovarioles that have fewer (C) or no oocytes (Suppl. Figure 1). DAPI (blue), F-actin (green) and Orb (red) are used to mark ovarioles. Insets are a 2X Zoom of the images. (D-E) Quantitative data representing the percentage of ovarioles that show the phenotype on third, tenth and eighteenth day after fly eclosion. Ovaries from Canton S 10 day old brwlMIO/brwlKGO animals were used as a control for statistical analyses. N = 3-15, n = 30-50.
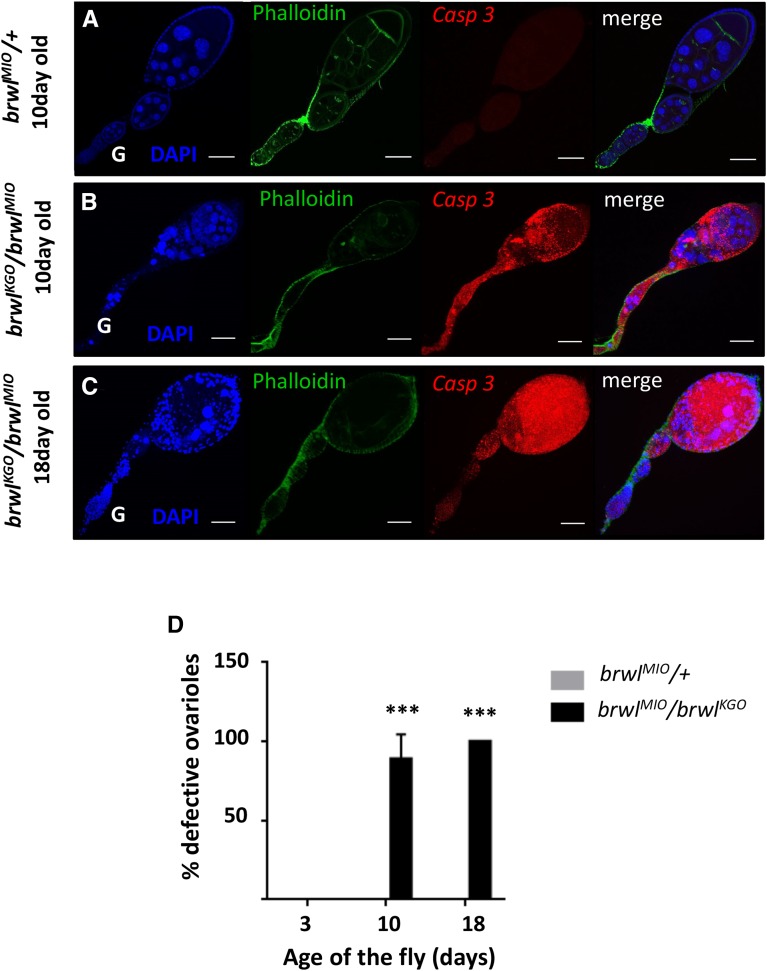
brwl mutants show age dependent Caspase activation: DAPI staining of 10 day brwlKG0/brwlMI0 mutants showed pycnotic nuclei indicative of apoptotic cell death. To test whether this is indeed the case, brwlKG0/brwlMI0 ovaries were stained with cleaved Caspase-3 antibody (Sarkissian et al. 2014), which is a marker for cells undergoing apoptosis. brwl mutant ovaries undergo cell death as shown (Figure 4A-D). 88.88% (n = 39) of ovarioles show extensive apoptosis as assessed by the presence of cleaved Caspase on the 10th day after fly eclosion which deteriorates further by 18th day post-eclosion as all the ovarioles positive for caspase activity (n = 28). This drastic increase in cell death likely results in oogenesis arrest in these mutants (Buszczak and Cooley 2000).
Figure 4.
Caspase is activated in brwl mutants. (A-C) brwlKG0/brwlMI0 mutants are positive for apoptosis as stained by apoptotic marker cleaved caspase-3 (Casp 3). The intensity of staining is enhanced as the fly grows older. 18 day old animals (C) show 100% ovarioles with activated caspase. DAPI (blue), Phalloidin (green) and activated caspase-3 (red) stained ovarioles. (D) Quantitative data representing the percentage of ovarioles that show the Casp 3 staining on third, tenth and eighteenth day after the fly eclosion. Ovaries from three day old brwlMIO/brwlKGO animals were used as a control for statistical analyses. N = 3-15, n = 15-40.
Mitosis and Meiosis is affected in brwl mutants. Mitosis in Drosophila ovaries is tightly regulated at multiple levels by different pathways (Deng et al. 2001). Wild type ovaries when stained for phospho-histone-3 S10 (PH3) show mitotically active cells until stage 6 (Figure 5A-C) after which they enter an endoreplication phase. Interestingly, in brwlKG0/brwlMI0 ovaries this pattern of mitosis is lost. It is difficult to stage the mutant ovary samples owing to degeneration, but it appears that the total number of mitotic nuclei is considerably reduced in brwl egg chambers. In 10 days old ovaries very few, and widely distributed mitotically active cells are observed (Figure 5B) whereas by 18th day old (Figure 5C) unevenly distributed mitotically active cells are seen throughout the ovariole. We confirmed this by staining the ovaries with anti-cyclin E antibody, which also shows an aberrant pattern (Figure 5E-F) and is indicative of cell division defects in brwlKG0/brwlMI0 ovaries.
Figure 5.
brwl mutants affect mitotis and meiosis. (A-C) brwlKG0/brwlMI0 mutants show decrease in the number of mitotically active cells, as stained by a phospho-histone3 S10 (PH3) antibody (red) and counterstained by phalloidin (green), in 10 day old ovaries. This pattern appears abrupt and random by 18th day where mitotic cells are seen randomly situated in the ovariole. N = 5-7, n = 20 = 25. (D-F) brwlKG0 /brwlMI0 mutants show decrease in the number of mitotically active cells, as stained by the Cyclin E antibody (red) in 10 day old ovaries. This pattern is further disrupted by 18th day (C) where randomly scattered mitotic cells are seen in the ovariole. N = 5, n = 15-35. (G-I) brwlMI0/brwlMI0 mutants show complete loss of the formation of synaptonemal complex, as marked by the anti-C(3)G antibody, on the 10th day post eclosion. 100% of ovarioles show this phenotype. N = 5, n = 15-25.
Meiosis initiated in the cysts after 4th mitotic division is essential for oocyte specification in Drosophila ovary. It has been shown that onset of meiosis occurs in 2 pro-oocytes with 4 prospective ring canals after the meiotic markers accumulate in these 2 cells (Carpenter 1975) (Jeffress et al. 2007; Lake and Hawley 2012). Failure of proper cyst formation and accumulation of meiotic markers hinders oocyte specification (Huynh and St Johnston 2000). Since brwl mutants showed defects in oocyte specification we stained the brwl mutants with a meiosis marker C(3)G which marks the synaptonemal complex in a meiotic cell (Jeffress et al. 2007). brwl mutants show complete loss C(3)G staining on 10th day post eclosion (Figure 5G-I). This indicates that none of the cells in cysts form the synaptonemal complex. This is similar to that of BicD null mutants where oocyte specification is not achieved (Huynh and St Johnston 2000).
brwl mutant phenotype is rescued by over-expression of stwl in germ stem cells
We are interested in the sequence homology and its relationship with the functional redundancy among the members of MADF-BESS family of proteins. Detailed phenotypic characterization of brwl mutants revealed remarkable overlap with stwl loss of function. Both the proteins seem to influence similar phenotypic traits including the establishment and/or maintenance of GSC fate, proper cystoblast differentiation and oocyte specification. We thus wondered if the two proteins function in a redundant manner. To directly test this possibility, we sought to assess if brwl mutant phenotypes could be rescued by expression of stwl. As a control, we also decided to test other MADF-BESS domain proteins in an identical experimental set-up. Among the different lines tested UAS-CG13204, UAS-CG3838, UAS-stwl, UAS-CG4404, UAS-CG8359, UAS-Coop and UAS-CG3919 showed expression in the ovary, when driven by nos-Gal4 and as monitored by HA staining. Unfortunately however, flies of only two genotypes survived to adulthood and could be analyzed further: brwlMIO/BrwlKGO; nos-Gal4/UAS-stwl and brwlMIO/brwlKGO; nos-Gal4/UAS-CG3919. All the other genetic combinations failed to eclose making further analysis untenable.
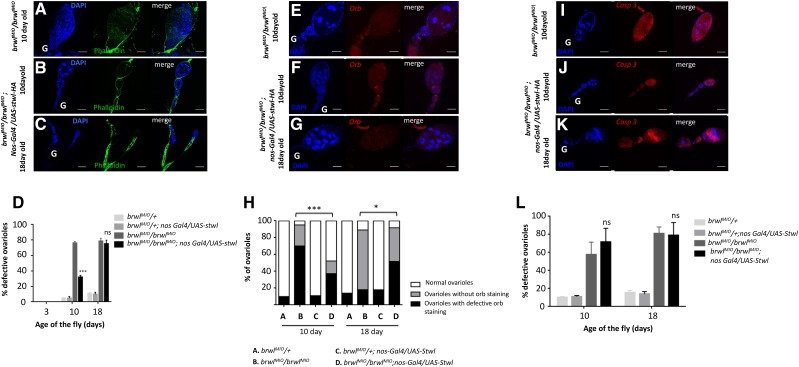
Interestingly we observed a substantial rescue of several phenotypes associated with loss of brwl upon germline specific stwl expression whereas CG3919 expression was unable to achieve rescue. Overexpression of stwl with nos-Gal4 in brwlMI0/ brwlMI0 background led to an amelioration of the ovarian phenotype induced by loss of brwl. For instance, ovarioles from 10 day old female ovaries consisted of normal egg chambers with 15 nurse cells and a properly developing oocyte (Figure 6A-B, E-F). Only 32.8% of total brwlMI0/brwlMI0; nanos-Gal4/UAS-stwl-HA ovarioles (n = 56) show ovarian defects which is substantially lower as compared to brwlMI0/ brwlMI0 ovarioles. Moreover, considerable morphological improvement was also observed as brwlMI0/ brwlMI0; nos-Gal4/UAS-stwl-HA ovarioles display regularly spaced ring canals with in the egg chambers (Figure 6B). Notably however, overexpression of stwl is unable to sustain the rescue levels for the 18-day old ovarioles of brwl mutant females (n = 25) (Figure 6C).
Figure 6.
Ovarian defects due to loss of brwl are ameliorated by over-expression of stwl. (A-C) brwlMI0/brwlMI0 mutants show ovary defects, as measured by DAPI staining (blue) and phalloidin (green) that are rescued in brwlMI0/ brwlMI0; nos-Gal4/UAS-stwl-HA on the 10th (B) and 18th (C) day PE. The rescue is however not as strong on the 18th day PE. (D) Quantitative data representing the number of ovarioles showing the phenotype on 3rd (n = 10), 10th (n = 56) and 18th (n = 25) day after fly eclosion. Ovaries from three day old brwlMIO/brwlKGO animals were used as a control for statistical analyses. (E-G) brwlMI0/brwlMI0; nosGal4/UAS-stwl-HA ovarioles showing reduced oocyte specification defects. Oocyte defects are significantly rescued in brwlMI0/ brwlMI0; nosGal4/ UAS-stwl-HA ovarioles on the 10th day (n = 25). Mild rescue is seen on the 18th day (n = 15) with a decrease in ovarioles without orb staining. (H) Quantitative data representing the percentage of ovarioles that show the phenotype on third, tenth and eighteenth day after fly eclosion. Ovaries from three day old brwlMIO/brwlKGO animals were used as a control for statistical analyses. N = 3-15; n = 25-60. (I-K) brwlMI0/ brwlMI0 mutants show apoptotic egg chambers which is not rescued in 10 day old brwlMI0/ brwlMI0; nanosGal4/ UAS-stwl-HA ovarioles. (L) Quantitative data representing the percentage of ovarioles that show the phenotype on third, tenth (n = 20) and eighteenth (n = 15) day after fly eclosion. Ovaries from three day old brwlMIO/brwlKGO animals were used as a control for statistical analyses. N = 3-10; n = 15-20.
Overexpression of stwl in brwlMI0/ brwlMI0 animals can also partially rescue the oocyte specification defects as assessed by Orb staining (Figure 6F,H). In brwl mutants the percentage of ovarioles with defective oocytes (no oocytes + mis-patterned oocytes) on the 10th day is ∼95%, with <5% normal oocytes. Expression of stwl in brwlMI0/ brwlMI0 leads to a dramatic rescue with >50% of the ovarioles (n = 25) showing normal patterning as compared to <5% in the mutant. The rescue at day 18 is also significant (Figure 6G,H) with egg chambers without Orb staining decreasing from 80% (in brwlMI0/ brwlMI0) to 40% after Stwl overexpression. This clearly indicates that overexpression of stwl in brwl mutants delays the accumulation of brwl associated defects.
Lastly, we examined if the germ cell division pattern is restored by overexpression of stwl in brwl loss of function background. As judged by anti-pH 3 S10 (Suppl. Figure 2A) and anti-CyclinE (Suppl. Figure 2B) stainings the 10-day old brwlMI0/ brwlMI0; nanos-Gal4/UAS-stwl-HA ovarioles show marked improvement. However, consistent with earlier observations mitotic defects persist in the 18th day brwlMI0/ brwlMI0; nanos-Gal4/UAS-stwl-HA ovarioles. Taken together these data argue that bwl activity is continuously required during germline development and Stwl protein can partially substitute loss of brwl albeit in a temporally restricted manner.
Consistent with this limited ability to rescue overexpression of stwl in brwlMI0 / brwlMI0 animals did not decrease the apoptotic cell death that brwlMI0/ brwlMI0 ovarioles show on 10th (n = 20) and 18th day respectively (n = 15) (Figure 6I-L). The temporal rescue of Brwl by Stwl does not appear to extend to blocking the apoptotic program that gets initiated in the absence of brwl function. It is however possible that higher dose of Stwl protein may be sufficient to rescue all the phenotypes due to loss of brwl effectively and additional experiments involving better reagents will be required to examine this possibility.
Negative purifying selection for stwl and brwl
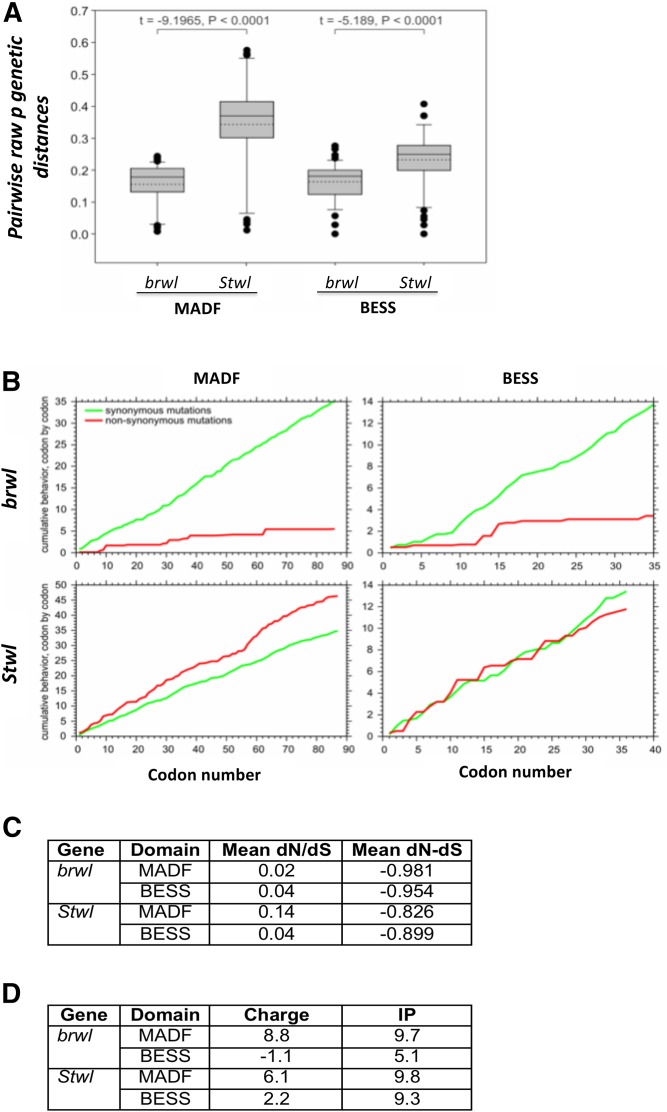
Taken together our analysis of Brwl and Stwl suggests that these two proteins are similar enough but they may have retained independent functions. We decided to explore this possibility using phylogenetic analysis. In an earlier study (Shukla et al. 2014), we found evidence for gene duplication and lineage specific expansion of the entire MADF-BESS family. Since our experimental data suggests that stwl and brwl are functional duplicates in the MADF-BESS family, we analyzed codon substitution patterns for the family with a specific focus on stwl and brwl (See Materials & Methods). We find that the mean pairwise raw genetic distance was two times higher in MADF of stwl than brwl and 1.4 times higher in BESS of stwl than brwl (Figure 7A). This indicates that the rate of evolution of both MADF and BESS domains of stwl was significantly higher than that for brwl. Model selection for nucleotide substitutions suggests that the first two codon positions are evolving by either simple one parameter (Jukes and Cantor 1969) model with equal rates for substitutions of the four nucleotides or with a two parameter (Kimura 1980) model with different rate of transition and tranversion type of nucleotide substitutions (Suppl. Table 1). Third codon position showed a complex model of nucleotide substitution only for MADF domain of brwl, while other domains showed simple substitutions with two parameters (Suppl. Table 1). A cumulative dN and dS plot (Figure 7B) indicates that MADF and BESS domains of stwl accumulated higher number of non-synonymous substitutions as compared to the respective domains of brwl, probably because of their higher rate of mutation. This suggests that brwl is under more stringent-evolutionary control in terms of codon substitutions than stwl. The dN/dS ratio for pairwise comparison between species showed that for both MADF and BESS domains of stwl and brwl was much less than one (Figure 7C), indicating that there is purifying or negative selection. This was also evident from negative dN-dS values for codon-by-codon analysis considering best fit codon substitution model (Figure 7C). Negative or purifying selection indicates selective removal of deleterious alleles, suggesting vital role of the domains and by association for both Brwl and Stwl in cellular functions. Stwl and Brwl have thus maintained their amino acid sequences in spite of changes to the DNA coding sequences, over time. The above analysis thus supports that Brwl and Stwl perform overlapping but distinct functions.
Figure 7.
Evolutionary analyses of changes in nucleotide and amino acid sequences in the MADF-BESS family. (A) Box plot for divergence of nucleotide sequences for stwl and brwl for 12 Drosophila genomes. Solid line in the box is median, dashed line is mean and outliers are shown by solid circles. (B) Cumulative behavior of synonymous and non-synomous substitutions along the coding region of stwl and brwl, showing accumulation of amino-acid substitutions within the MADF or BESS domains for 12 Drosophila genomes. The X-axis represents the position of the codon, and the Y-axis represents the average cumulative number of synonymous (green line) and non-synonymous (red line) codons, estimated at a specific codon position. (C) dN/dS ratio as also mean dN-dS for pairwise comparison between species computed for MADF and BESS coding regions of 12 Drosophila genomes. (D) Predicted charge and Isoelectric point (IP) of MADF and BESS domains in Stwl and Brwl. The values are compared with published values for MADF domain containing proteins (Maheshwari et al. 2008).
(Maheshwari et al. 2008) analyzed the charge and isoelectric point (IP) based on amino-acid sequence of MADF domains in Drosophila and discovered that proteins with multiple MADF domains in a single sequence, for example hybrid male resce (Hmr) and lethal hybrid rescue (Lhr) have distinct Charge and Isoelectric points. They reasoned that MADF domains with a strong positive charge were DNA binding proteins and those with negative charge were primarily binding basic proteins like histones. We find that Stwl and Brwl, having positive Charge of 6.1 and 8.8 respectively (Figure 7D) for their respective MADF domains fall in the category of transcription factors of the type ‘MADF1’, geared for DNA binding (Compare to Table1, (Maheshwari et al. 2008)). In contrast, the BESS domain appears to differ significantly in its charge and IP for Brwl as compared to Stwl, suggesting a possible difference in interactors between the two proteins.
Discussion
The continuity of a species is almost exclusively dependent upon the transmission of genetic information across generations. Multicellular organisms accomplish this task via sexual reproduction, which typically involves fusion of the male and female gametes resulting in the development of a new organism post-fertilization. The progenitors of the male and female gametes, known as GSCs, are thus under constant scrutiny. Consequently, a wealth of information is available regarding the mechanisms underlying the specification and maintenance of the GSC fate. How the specification process follows a defined route to eventually accomplish a reliable outcome namely continuous production of healthy gametes is less clear, however. This is of special significance as GSCs not only self-renew but are also pluripotent and are competent to give rise to daughter progeny that subsequently differentiates into a variety of cell types. Thus, it is important to elucidate how GSCs maintain ‘stemness’ by employing symmetric cell division and also have the capability to generate differentiated daughter cells via asymmetric cell division program.
It is proposed that the process of GSC specification relies upon mechanisms that would ensure both qualitatively and quantitatively robust outcomes (Rudel and Sommer 2003; Félix and Barkoulas 2015; Kitano 2004). To achieve dependable and reproducible outcomes, developmental processes are often reinforced by redundant mechanisms that effectively allow dampening of potential disruptions in the functional gene regulatory network. ‘Gene duplication and sub-functionalization’ is a prominent strategy among the different safeguarding means that have been employed to generate redundancy (Gu 2003; Kitano 2004). Duplicated genes ensure robustness in a system as deleterious mutations on one gene are readily compensated by the activity of the duplicated member. Gene duplicates thus provide functional redundancy, buffering detrimental phenotypes that arise from malfunction of one copy (Gout and Lynch 2015; Shukla et al. 2014). This contributes significantly to the developmental transitions occurring in a sequential and seamless manner from one stage to the next.
In this study, we have presented experimental analysis of two paralogs stwl and brwl, which appear to be partially redundant molecular players that perform similar functions and thus contribute to the robustness to the processes underlying GSC development in Drosophila ovaries. brwl appears to function in the same cell lineage and spatiotemporal domain as stwl, and stwl expression in the germ cell lineage i.e., nos domain can partially rescue/delay brwl phenotypes. Our study finds that similar to stwl, brwl is also required for stem cell maintenance, proper cyst formation and oocyte specification.
First, we examine the implications of the evolutionary data analyzed. Gene duplicates are believed to evolve differently than singletons (Jordan et al. 2004). Post-duplication, the genes could show changes in their rates of evolution with consequences on purifying (negative or positive) selection. Our data, based on the analysis of the MADF and BESS domains suggest that a) stwl and brwl have undergone negative selection, indicating that their ancestral functional state, as defined by the linear sequence of amino acids was critical and had to be maintained and that b) The rate of evolution appears asymmetric with MADF and BESS domains coding regions of stwl and Brwl sequences changing at different rates.
We have compared the stwl phenotypes described in literature with our data as we were unable to collect data for a direct comparison due to unavailability of published Stwl reagents (Clark and McKearin 1996; Akiyama 2002), such as mutant alleles and antibodies. brwl loss of function shows strong age dependent phenotypes that were not reported in the analysis of any stwl mutants. Both stwl and brwl show a range of similar phenotypes, specific to the female germline. The ability of stwl to partially compensate for brwl function indicates that stwl may be a genetic interactor of brwl and that Stwl has function similar to Brwl and can replace, at least partially, Brwl, suggesting redundancy in function. Since both proteins are MADF-BESS domain proteins and predicted paralogs, this leads to a strong possibility that the common function shared by both is an ancestral function retained by both orthologs.
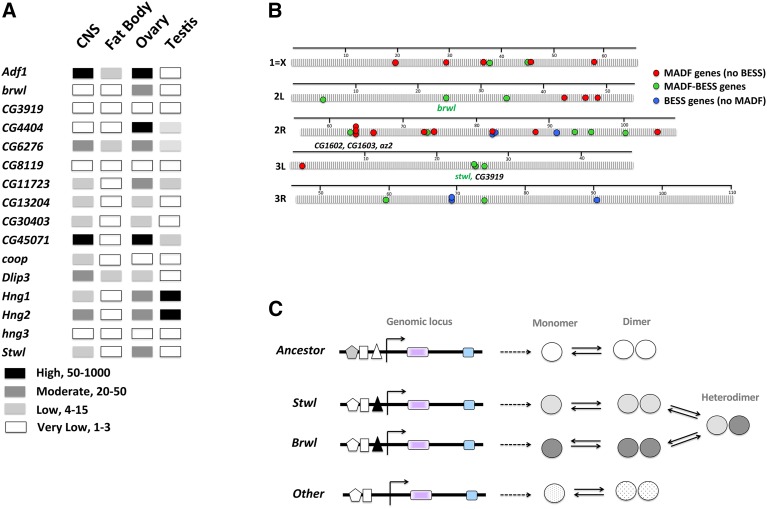
This thesis is further supported by available evidence as summarized below. A comparison of modENCODE data indicates that both brwl and stwl are expressed in the early embryo likely due to maternal deposition. Transcript levels of both the genes are higher in the ovaries as compared to testis (Figure 8A) consistent with the elevated levels in adult females. Some MADF-BESS genes (Hng1, Hng2) show similar expression patterns with others showing varying levels of expression in different tissues. In contrast to the data that shows equivalence for Brwl and Stwl function, there is also evidence for differences between the two indicating gain/loss of function for one member. For example, Loss of stwl in the wing by RNAi shows wing/wing-hinge phenotypes whereas brwl RNAi has no effect (Shukla et al. 2014). Overexpression of stwl perturbs development, both in wings and in the ovarian stem cell lineage. brwl overexpression on the other hand has no such effect. Loss of brwl leads to a decrease in GSC number, with survivability being one of the possible means by which Brwl regulates GSC number.
Figure 8.
A model for Stwl/Brwl function post-duplication and after subsequent Sub-functionalization. (A) MADF-BESS mRNA expression, extrapolated from modENCODE datasets in larval CNS, larval fat body, adult ovary and adult testis of Drosophila melanogaster. These examples highlight the varied expression pattern of these genes indicating that the family members are sub-functionalized in terms of spatiotemporal expression pattern. The levels of transcripts (High, Moderate, Low) are based directly on values from modENCODE. (B) Schematic of genomic organization of MADF-BESS loci. MADF (red circles), MADF-BESS (green circles) and BESS (without MADF, blue circles) domain coding genes are distributed all over the genome. stwl and brwl are on different chromosomes and CG3919, a local duplicate of stwl does not appear to have functional roles in the GSC lineage. The map distances are in centimorgans and genetic distances are based on chromosomal maps, as collated by Flybase. (C) brwl and stwl are expressed in overlapping domains (driven by enhancer, filled triangle) in the GLC lineage and we hypothesize that the functional entity for this overlapping function, which is critical for robust function, is a Stwl/Brwl heterodimer and absence of the Brwl homodimer can be functionally replaced, to a limited extent by a Stwl Homo-dimer.
A genomic map of distribution of MADF-BESS genes reveals interesting features (Figure 8B). The MADF-BESS genes (green circles) are distributed all over the genome, suggesting that the duplication event (∼40 My ago) was transposon based. Local duplication, possibly succeding a transposon based duplication event may also have occurred, for example for MADF genes set CG1602, CG1603 and az2 and possibly for stwl, CG3919 pair are possible local duplicates. Interestingly, CG3919, as tested by RNAi (Table 1), does not appear to retain germ line Cell (GLC) lineage function, nor does it rescue the brwl mutant phenotype. In agreement, ModENCODE data also suggest low expression of CG3919 transcripts in the ovary.
What are the possible mechanisms for redundancy after gene duplication? A possible scenario is a model where MADF-BESS domain proteins are competent fo function as homo as well as heterodimers and and the Stwl-Brwl heterodimer can substitute the Brwl homodimer (Figure 8C). In the female germline, both Stwl and Brwl are expressed, thus retaining ancestral expression and functionality. Although the BESS domains have been suggested to be protein:protein intercation surfaces, there is no experimental evidence for direct molecular interaction between Stwl and Brwl in current interaction databases (Hu et al. 2018).
An open question in the field is the molecular function of MADF and MADF-BESS domain containing proteins. Members appear to modulate diverse processes (Ratnaparkhi et al. 2008; Song et al. 2010; Cutler et al. 1998; Duong et al. 2008; Shukla et al. 2014; Zimmermann et al. 2006; Timmerman et al. 2013; Casola et al. 2007; Barbash et al. 2000; Brideau et al. 2006) with no apparent common theme. The MADF-BESS family members may either act as transcription activators/repressors or as histone modifiers, they may also associate with different subsets of co-factors for diverse functions. Brwl, based on its functional similarity to Stwl may be a DNA binding protein and a putative chromatin regulator (Yi et al. 2009).
In summary, our study uncovers brwl as a novel player in the maintanence of GSCs in the female germline. While our data do not rule out a somatic function conlusively, our phenotypic analysis and expression data strongly suggest a possible germ cell autonomous function of the gene. brwl phenotypes are pleiotropic suggesting that either the protein has an early function with compounding downstream effects or it has multiple roles in the development of the cyst. This pleiotropy is reminiscent of stwl mutants.
Furthermore, our data point to partially redundant functions shared between brwl and stwl in promoting oocyte development in the female germline. We hypothesize that brwl and stwl likely arose as a consequence of a gene duplication event 40 My ago. We further speculate that ancestral functions are retained by both the gene products and are utilized for robust patterning of the germline cells in the developing ovaries.
Acknowledgments
We thank Bloomington Drosophila Stock Center (BDSC), Indiana, supported by NIH grant P40OD018537, for fly stocks; Drosophila Genome Research Centre (DGRC), Indiana supported by NIH grant 2P40OD010949 for vectors and clones; TRiP collection at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing transgenic RNAi fly stocks; Scott Hawley for C(3)G antibodies; IISER Microscopy Facilty; NCBS-CCAMP Bangalore Transgenic Drosophila Facility for DPiM lines; Zurich FlyORFome project for fly lines; Richa Rikhy and LS Shashidhara for useful discussions. This work is funded by a research grant from the Department of Biotechnology, Govt. of India (BT/PR/11935/BRB/10/1316/2014) to GR. ND is a project assistant supported on this grant. VS was a graduate student supported by CSIR Research Fellowship (2010-2015) and a project assistant (2015-2017) in the aforementioned grant. PN is graduate student supported by a UGC Research Fellowship. GD is supported by a grant from NIH (R01GM110015).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.6114977.
Communicating editor: H. Salz
Literature Cited
- Akiyama T., 2002. Mutations of stonewall disrupt the maintenance of female germline stem cells in Drosophila melanogaster. Dev. Growth Differ. 44: 97–102. 10.1046/j.1440-169x.2002.00625.x [DOI] [PubMed] [Google Scholar]
- Barbash D. A., Roote J., Ashburner M., 2000. The Drosophila melanogaster hybrid male rescue gene causes inviability in male and female species hybrids. Genetics 154: 1747–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H. J., Levis R. W., Liao G., He Y., Carlson J. W., et al. , 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167: 761–781. 10.1534/genetics.104.026427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar V., Courey A. J., 2002. The MADF-BESS domain factor Dip3 potentiates synergistic activation by Dorsal and Twist. Gene 299: 173–184. 10.1016/S0378-1119(02)01058-2 [DOI] [PubMed] [Google Scholar]
- Bischof J., Bjorklund M., Furger E., Schertel C., Taipale J., et al. , 2013. A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development 140: 2434–2442. 10.1242/dev.088757 [DOI] [PubMed] [Google Scholar]
- Boyer L. A., Latek R. R., Peterson C. L., 2004. The SANT domain: a unique histone-tail-binding module? Nat. Rev. Mol. Cell Biol. 5: 158–163. 10.1038/nrm1314 [DOI] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Brideau N. J., Flores H. A., Wang J., Maheshwari S., Wang X., et al. , 2006. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314: 1292–1295. 10.1126/science.1133953 [DOI] [PubMed] [Google Scholar]
- Brun S., Rincheval-Arnold A., Colin J., Risler Y., Mignotte B., et al. , 2006. The myb-related gene stonewall induces both hyperplasia and cell death in Drosophila: rescue of fly lethality by coexpression of apoptosis inducers. Cell Death Differ. 13: 1752–1762. 10.1038/sj.cdd.4401861 [DOI] [PubMed] [Google Scholar]
- Buszczak M., Cooley L., 2000. Eggs to die for: cell death during Drosophila oogenesis. Cell Death Differ. 7: 1071–1074. 10.1038/sj.cdd.4400755 [DOI] [PubMed] [Google Scholar]
- Buszczak M., Paterno S., Lighthouse D., Bachman J., Planck J., et al. , 2007. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics 175: 1505–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A. T., 1975. Electron microscopy of meiosis in Drosophila melanogaster females. I. Structure, arrangement, and temporal change of the synaptonemal complex in wild-type. Chromosoma 51: 157–182. 10.1007/BF00319833 [DOI] [PubMed] [Google Scholar]
- Casanueva M. O., Ferguson E. L., 2004. Germline stem cell number in the Drosophila ovary is regulated by redundant mechanisms that control Dpp signaling. Development 131: 1881–1890. 10.1242/dev.01076 [DOI] [PubMed] [Google Scholar]
- Casola C., Lawing A. M., Betran E., Feschotte C., 2007. PIF-like transposons are common in drosophila and have been repeatedly domesticated to generate new host genes. Mol. Biol. Evol. 24: 1872–1888. 10.1093/molbev/msm116 [DOI] [PubMed] [Google Scholar]
- Clark K. A., McKearin D. M., 1996. The Drosophila stonewall gene encodes a putative transcription factor essential for germ cell development. Development 122: 937–950. [DOI] [PubMed] [Google Scholar]
- Cutler G., Perry K. M., Tjian R., 1998. Adf-1 is a nonmodular transcription factor that contains a TAF-binding Myb-like motif. Mol. Cell. Biol. 18: 2252–2261. 10.1128/MCB.18.4.2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decotto E., Spradling A. C., 2005. The Drosophila ovarian and testis stem cell niches: similar somatic stem cells and signals. Dev. Cell 9: 501–510. 10.1016/j.devcel.2005.08.012 [DOI] [PubMed] [Google Scholar]
- Delattre M., Spierer A., Jaquet Y., Spierer P., 2004. Increased expression of Drosophila Su(var)3–7 triggers Su(var)3–9-dependent heterochromatin formation. J. Cell Sci. 117: 6239–6247. 10.1242/jcs.01549 [DOI] [PubMed] [Google Scholar]
- Delattre M., Spierer A., Tonka C. H., Spierer P., 2000. The genomic silencing of position-effect variegation in Drosophila melanogaster: interaction between the heterochromatin-associated proteins Su(var)3–7 and HP1. J. Cell Sci. 113: 4253–4261. [DOI] [PubMed] [Google Scholar]
- Delport W., Poon A. F., Frost S. D., Kosakovsky Pond S. L., 2010. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26: 2455–2457. 10.1093/bioinformatics/btq429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W. M., Althauser C., Ruohola-Baker H., 2001. Notch-Delta signaling induces a transition from mitotic cell cycle to endocycle in Drosophila follicle cells. Development 128: 4737–4746. [DOI] [PubMed] [Google Scholar]
- Duffy J. B., 2002. GAL4 system in Drosophila: a fly geneticist’s Swiss army knife. Genesis 34: 1–15. 10.1002/gene.10150 [DOI] [PubMed] [Google Scholar]
- Duong H. A., Nagaraj R., Wang C. W., Ratnaparkhi G., Sun Y. H., et al. , 2008. Non-cell-autonomous inhibition of photoreceptor development by Dip3. Dev. Biol. 323: 105–113. 10.1016/j.ydbio.2008.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C., 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M. A., Barkoulas M., 2015. Pervasive robustness in biological systems. Nat. Rev. Genet. 16: 483–496. 10.1038/nrg3949 [DOI] [PubMed] [Google Scholar]
- Finn R. D., Attwood T. K., Babbitt P. C., Bateman A., Bork P., et al. , 2017. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 45: D190–D199. 10.1093/nar/gkw1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gout J. F., Lynch M., 2015. Maintenance and Loss of Duplicated Genes by Dosage Subfunctionalization. Mol. Biol. Evol. 32: 2141–2148. 10.1093/molbev/msv095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., 2003. Evolution of duplicate genes vs. genetic robustness against null mutations. Trends Genet. 19: 354–356. 10.1016/S0168-9525(03)00139-2 [DOI] [PubMed] [Google Scholar]
- Guruharsha K. G., Rual J. F., Zhai B., Mintseris J., Vaidya P., et al. , 2011. A protein complex network of Drosophila melanogaster. Cell 147: 690–703. 10.1016/j.cell.2011.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman T. R., Ventresca E. M., Hopkins A., Zinshteyn D., Singh T., et al. , 2015. Novel tools for genetic manipulation of follicle stem cells in the Drosophila ovary reveal an integrin-dependent transition from quiescence to proliferation. Genetics 199: 935–957. 10.1534/genetics.114.173617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Comjean A., Roesel C., Vinayagam A., Flockhart I., et al. , 2017. FlyRNAi.org-the database of the Drosophila RNAi screening center and transgenic RNAi project: 2017 update. Nucleic Acids Res. 45: D672–D678. 10.1093/nar/gkw977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Vinayagam A., Nand A., Comjean A., Chung V., et al. , 2018. Molecular Interaction Search Tool (MIST): an integrated resource for mining gene and protein interaction data. Nucleic Acids Res. 46: D567–D574. 10.1093/nar/gkx1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh J. R., St Johnston D., 2000. The role of BicD, Egl, Orb and the microtubules in the restriction of meiosis to the Drosophila oocyte. Development 127: 2785–2794. [DOI] [PubMed] [Google Scholar]
- Jeffress J. K., Page S. L., Royer S. K., Belden E. D., Blumenstiel J. P., et al. , 2007. The formation of the central element of the synaptonemal complex may occur by multiple mechanisms: the roles of the N- and C-terminal domains of the Drosophila C(3)G protein in mediating synapsis and recombination. Genetics 177: 2445–2456. 10.1534/genetics.107.078717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Flynt A. S., Lai E. C., 2013. Drosophila piwi mutants exhibit germline stem cell tumors that are sustained by elevated Dpp signaling. Curr. Biol. 23: 1442–1448. 10.1016/j.cub.2013.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan I. K., Wolf Y. I., Koonin E. V., 2004. Duplicated genes evolve slower than singletons despite the initial rate increase. BMC Evol. Biol. 4: 22 10.1186/1471-2148-4-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes T. H., Cantor C. R., 1969. Evolution of protein molecules, Academic Press, New York: 10.1016/B978-1-4832-3211-9.50009-7 [DOI] [Google Scholar]
- Kelso R. J., Buszczak M., Quinones A. T., Castiblanco C., Mazzalupo S., et al. , 2004. Flytrap, a database documenting a GFP protein-trap insertion screen in Drosophila melanogaster. Nucleic Acids Res. 32: D418–D420. 10.1093/nar/gkh014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16: 111–120. 10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- Kitano H., 2004. Biological robustness. Nat. Rev. Genet. 5: 826–837. 10.1038/nrg1471 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K., 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33: 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake C. M., Hawley R. S., 2012. The molecular control of meiotic chromosomal behavior: events in early meiotic prophase in Drosophila oocytes. Annu. Rev. Physiol. 74: 425–451. 10.1146/annurev-physiol-020911-153342 [DOI] [PubMed] [Google Scholar]
- Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., et al. , 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Lanfear R., Calcott B., Ho S. Y., Guindon S., 2012. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29: 1695–1701. 10.1093/molbev/mss020 [DOI] [PubMed] [Google Scholar]
- Lantz V., Chang J. S., Horabin J. I., Bopp D., Schedl P., 1994. The Drosophila orb RNA-binding protein is required for the formation of the egg chamber and establishment of polarity. Genes Dev. 8: 598–613. 10.1101/gad.8.5.598 [DOI] [PubMed] [Google Scholar]
- Lasko P. F., Ashburner M., 1990. Posterior localization of vasa protein correlates with, but is not sufficient for, pole cell development. Genes Dev. 4: 905–921. 10.1101/gad.4.6.905 [DOI] [PubMed] [Google Scholar]
- Librado P., Rozas J., 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- Maheshwari S., Wang J., Barbash D. A., 2008. Recurrent positive selection of the Drosophila hybrid incompatibility gene Hmr. Mol. Biol. Evol. 25: 2421–2430. 10.1093/molbev/msn190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines J. Z., Park J. K., Williams M., McKearin D. M., 2007. Stonewalling Drosophila stem cell differentiation by epigenetic controls. Development 134: 1471–1479. 10.1242/dev.02810 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Derbyshire M. K., Gonzales N. R., Lu S., Chitsaz F., et al. , 2015. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43: D222–D226. 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKearin D., 1997. The Drosophila fusome, organelle biogenesis and germ cell differentiation: if you build it. BioEssays 19: 147–152. 10.1002/bies.950190209 [DOI] [PubMed] [Google Scholar]
- Nei M. K. S., 2000. Molecular evolution and phylogenetics, Oxford University Press, Oxford. [Google Scholar]
- Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q., 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32: 268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J. Q., Liu L. P., Binari R., Hardy R., Shim H. S., et al. , 2009. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics 182: 1089–1100. 10.1534/genetics.109.103630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J. Q., Zhou R., Czech B., Liu L. P., Holderbaum L., et al. , 2011. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods 8: 405–407. 10.1038/nmeth.1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L., Chen S., Weng C., Call G., Zhu D., et al. , 2007. Stem cell aging is controlled both intrinsically and extrinsically in the Drosophila ovary. Cell Stem Cell 1: 458–469. 10.1016/j.stem.2007.09.010 [DOI] [PubMed] [Google Scholar]
- Perkins L. A., Holderbaum L., Tao R., Hu Y., Sopko R., et al. , 2015. The Transgenic RNAi Project at Harvard Medical School: Resources and Validation. Genetics 201: 843–852. 10.1534/genetics.115.180208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond S. L., Frost S. D., Muse S. V., 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21: 676–679. 10.1093/bioinformatics/bti079 [DOI] [PubMed] [Google Scholar]
- Ratnaparkhi G. S., Duong H. A., Courey A. J., 2008. Dorsal interacting protein 3 potentiates activation by Drosophila Rel homology domain proteins. Dev. Comp. Immunol. 32: 1290–1300. 10.1016/j.dci.2008.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrbaugh M., Clore A., Davis J., Johnson S., Jones B., et al. , 2013. Identification and characterization of proteins involved in nuclear organization using Drosophila GFP protein trap lines. PLoS One 8: e53091 10.1371/journal.pone.0053091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rørth P., 1998. Gal4 in the Drosophila female germline. Mech. Dev. 78: 113–118. 10.1016/S0925-4773(98)00157-9 [DOI] [PubMed] [Google Scholar]
- Rudel D., Sommer R. J., 2003. The evolution of developmental mechanisms. Dev. Biol. 264: 15–37. 10.1016/S0012-1606(03)00353-1 [DOI] [PubMed] [Google Scholar]
- Sarkissian T., Timmons A., Arya R., Abdelwahid E., White K., 2014. Detecting apoptosis in Drosophila tissues and cells. Methods 68: 89–96. 10.1016/j.ymeth.2014.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz G., 1978. Estimating the dimension of a model. Ann Stat. 1978;6:461–464. Ann. Stat. 6: 461–464. 10.1214/aos/1176344136 [DOI] [Google Scholar]
- Shukla V., Habib F., Kulkarni A., Ratnaparkhi G. S., 2014. Gene duplication, lineage-specific expansion, and subfunctionalization in the MADF-BESS family patterns the Drosophila wing hinge. Genetics 196: 481–496. 10.1534/genetics.113.160531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Goetze S., Bischof J., Spichiger-Haeusermann C., Kuster M., et al. , 2010. Coop functions as a corepressor of Pangolin and antagonizes Wingless signaling. Genes Dev. 24: 881–886. 10.1101/gad.561310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Wong M. D., Kawase E., Xi R., Ding B. C., et al. , 2004. Bmp signals from niche cells directly repress transcription of a differentiation-promoting gene, bag of marbles, in germline stem cells in the Drosophila ovary. Development 131: 1353–1364. 10.1242/dev.01026 [DOI] [PubMed] [Google Scholar]
- Staller M. V., Yan D., Randklev S., Bragdon M. D., Wunderlich Z. B., et al. , 2013. Depleting gene activities in early Drosophila embryos with the “maternal-Gal4-shRNA” system. Genetics 193: 51–61. 10.1534/genetics.112.144915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman C., Suppiah S., Gurudatta B. V., Yang J., Banerjee C., et al. , 2013. The Drosophila transcription factor Adf-1 (nalyot) regulates dendrite growth by controlling FasII and Staufen expression downstream of CaMKII and neural activity. J. Neurosci. 33: 11916–11931. 10.1523/JNEUROSCI.1760-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K. J., Schulze K. L., Haelterman N. A., Pan H., He Y., et al. , 2011. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods 8: 737–743. 10.1038/nmeth.1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D., Neumuller R. A., Buckner M., Ayers K., Li H., et al. , 2014. A regulatory network of Drosophila germline stem cell self-renewal. Dev. Cell 28: 459–473. 10.1016/j.devcel.2014.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X., de Vries H. I., Siudeja K., Rana A., Lemstra W., et al. , 2009. Stwl modifies chromatin compaction and is required to maintain DNA integrity in the presence of perturbed DNA replication. Mol. Biol. Cell 20: 983–994. 10.1091/mbc.E08-06-0639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann G., Furlong E. E., Suyama K., Scott M. P., 2006. Mes2, a MADF-containing transcription factor essential for Drosophila development. Dev. Dyn. 235: 3387–3395. 10.1002/dvdy.20970 [DOI] [PubMed] [Google Scholar]