Abstract
Winter wheat cultivar ‘Jagger’ was recently found to have an alien chromosomal segment 2NS that has Lr37, a gene conferring resistance against leaf rust caused by Puccinia triticina. The objective of this study was to map and characterize the gene(s) for seedling leaf rust resistance in Jagger. The recombinant inbred line (RIL) population of Jagger × ‘2174’ was inoculated with leaf rust pathogen THBJG and BBBDB, and evaluated for infection type (IT) response. A major quantitative trait locus (QTL) for THBJG and BBBDB was coincidently mapped to chromosome arm 2AS, and the QTL accounted for 56.6–66.2% of total phenotypic variation in infection type (IT) response to THBJG, and 72.1–86.9% to BBBDB. The causal gene for resistance to these rust races was mapped to the 2NS segment in Jagger. The 2NS segment was located in a region of approximately 27.8 Mb starting from the telomere of chromosome arm 2AS, based on the sequences of the A genome in tetraploid wheat. The Lr17a gene on chromosome arm 2AS was delimited to 3.1 Mb in the genomic region, which was orthologous to the 2NS segment. Therefore, the Lr37 gene in the 2NS segment can be pyramided with other effective resistance genes, rather than Lr17a in wheat, to improve resistance to rust diseases.
Keywords: leaf rust, Lr37, Lr17, quantitative trait loci (QTL), wheat
The hard red winter wheat variety Jagger (KS84W85/Stephens) (Triticum aestivum, 2n = 6×=42, AABBDD) has been widely grown in the Great Plains region of the U.S. since its release in 1996. Jagger was highly resistant to leaf rust caused by Puccinia triticina Erikss. when initially released, however within a few years races of P. triticina with virulence to Lr17a increased rapidly and the resistance in Jagger was much less effective (Long et al. 1998). Jagger is currently still used extensively as a parent due to the presence of many elite resistance genes for abiotic and biotic stresses, and many current hard red winter wheat cultivars in the U.S. have Jagger in their pedigrees (Fang et al. 2011; Liu et al. 2015, 2016; Turner et al. 2016).
Jagger was initially hypothesized to have leaf rust resistance gene Lr17a, based on its high similarity with the Thatcher near-isogenic line with Lr17a for infection types to different leaf rust races (Zhang et al. 2008; Kolmer 2017). Jagger was later determined to have the 2NS/2AS alien translocation (Fang et al. 2011) derived from Aegilops ventricosa Tausch (2n = 4×=28, DDNN) (Maia 1967) that was originally transferred to the wheat line VPM1 (Bariana and McIntosh 1993; Dyck and Lukow 1988). The 2NS/2AS translocation in VPM1 carries Lr37 for leaf rust resistance, Sr38 for resistance to stem rust caused by P. graminis tritici Eriks. & Henn., and Yr17 for resistance to stripe rust, P. striiformis Westend. (McIntosh et al. 1995). A recombinant inbred line (RIL) population of Jagger × 2174 (IL71-5662/PL145) was tested for stripe rust resistance in different field locations, and the population segregated for Yr17 and effective resistance to common stripe rust pathogen in Washington, Kansas and China (Fang et al. 2011). Jagger also has Sr38 on the 2NS segment, in addition to Sr7a that conditions resistance to stem rust race TTTTF, which is the most virulent race found in North America and and is virulent to many winter wheat cultivars grown in the southern Great Plains (Turner et al. 2016). However, the effectiveness of the resistance genes on the 2NS segment in Jagger is not well known.
Leaf rust resistance conditioned by Lr37 is best expressed in adult plants (McIntosh et al. 2016), however seedlings of the Thatcher line with Lr37 have infection types (IT) that are usually mesothetic, with a mixture of medium to large uredinia with varying amounts of necrosis and chlorosis to avirulent races (Kolmer 1997). In a recent study Kolmer (2017) determined that seedling leaf rust resistance in the hard red wheat cultivar Santa Fe (G1878/Jagger) was conditioned by Lr37 on the 2NS translocation.
The objectives of this study were to use the Jagger × 2174 RIL population to map the seedling leaf rust resistance genes in Jagger wheat, and to characterize the 2NS translocation with additional molecular markers. The Jagger × 2174 RIL population was recently genotyped with the Infinium iSelect 90K wheat bead chip array described in Cavanagh et al. (2013), which has facilitated discovery of additional resistance genes. The common wheat genome sequences recently released by the International Wheat Genome Sequencing Consortium (IWGSC) (https://urgi.versailles.inra.fr/blast_iwgsc/blast.php) have helped to develop markers for the translocated 2NS region, and the assembled genome sequences of the A genome in tetraploid emmer wheat (Avni et al. 2017) have helped to physically locate the resistance genes in common wheat.
Materials and Methods
The mapping population and inoculation
The parental lines Jagger and 2174, 141 F7 RILs derived from the parental lines, and a near-isogenic line of Thatcher wheat with Lr17a, RL6008 were phenotyped with leaf rust races THBJG and BBBDB in 2012 and 2017. The RIL population was arranged in six trays of 24 lines each, and a seventh tray of the remaining RIL and control lines. All entries were arranged as a randomized design. Each line was evaluated for reaction to leaf rust races. These materials were inoculated in the seedling stage with leaf rust races THBJG and BBBDB (Long and Kolmer 1989). Both races are avirulent to seedling plants with Lr17a and adult plants with Lr37 (Helguera et al. 2003; Kolmer et al. 2005, 2006). Race THBJG is virulent to Lr1, Lr2a, Lr2c, Lr3, Lr16, Lr26, Lr10, Lr14a, and Lr28. In contrast, race BBBDB is virulent only to Lr14a and can be used to detect many seedling leaf rust resistance genes. Inoculation was conducted as previously described (Kolmer 2017). The parental lines and the RILs were grown in segmented trays with 8-10 seed of a single wheat line in each segment in a greenhouse at 20-23° and fertilized with 20-20-20 NPK solution 7 days after planting. At the two-leaf stage, the seedlings were inoculated with a single race of P. triticina using a suspension of urediniospores in Soltrol 170 (Phillips Petroleum, Bartlesville, OK) mineral oil. After inoculation the plants were dried for 1 hr, and then placed in a dew chamber overnight at 18°. The plants were then returned to a greenhouse bench at 20-23° with supplemental metal halide lighting. Leaf rust infection types were evaluated 10 days after inoculation.
Infection type scores
Infection types (IT) of seedlings were scored as described by Long and Kolmer (1989) on a 0 to 4 scale: 0 = no visible sign of infection; ; = hypersensitive flecks; 1 = small uredinia with necrosis, 2 = moderate size pustules with chlorosis; 3 = moderate-large size uredinia without necrosis or chlorosis; 4 = large uredinia lacking necrosis or chlorosis. Mixtures of infection types were described with the lowest infection type first. Symbols “–” and “+” denote smaller or larger uredinia. The original phenotypic data were converted to a 0–9 linear disease scale for QTL analysis. IT ;, ;1-, ;1, ;1+, ;2-, 2, 2+, 3-, 3, and 3+ were coded as 0, 1, 2, 3, 4, 5, 6, 7, 8 and 9, respectively. For plants in the same RIL that showed heterogeneous infection types, only the most prevalent IT was used.
Genotypes
The 90K Illumina wheat SNP chip was used to genotype the Jagger × 2174 population of 141 RILs (Li et al. 2015), and an 8-digit SNP code for the mapping population was converted to the reference number for each SNP, i.e., IWA0000 or IWB0000 as published (Cavanagh et al. 2013). However, the sequences of three SNP markers, SNP19777336, SNP30671452 and SNP48765306, were not found to have a reference number, and their sequences for mapping in this study were provided in supplementary information. The SNP markers mapped to chromosome 2A were incorporated with SSR markers and gene markers reported in previous studies (Fang et al. 2011; Tan et al. 2013). Those lines that contained multiple crossovers within a small chromosomal region were excluded for further analyses. Final genotypic data and phenotypic data were integrated in 129 RILs to analyze QTL. QTL were found using MapQTL 6.0. Only those SNP markers that covered a significant QTL were used to further analyze genetic distance using MapMaker 3.0, with the Kosambi mapping function to estimate the map distance. WinQTLCart 2.5 (North Carolina State University, Raleigh) was used to conduct analyses using Composite Interval Mapping (CIM), and a QTL was declared when the logarithm of the odds (LOD) score exceeded the LOD threshold that was determined at 300 permutations and significance level at 0.05.
Development of sequence tagged site (STS) markers
PCR markers for three genes, TaVrga-A1 and cMWG682 containing fragments encoding conserved domains of resistance genes (Fang et al. 2011), and TaOPR2 for a 12-oxo-phytodienoic acid reductase (OPR) gene (Tan et al. 2013), were mapped to the 2NS segment in Jagger. To verify the genotype of the major QTL for IT response to leaf rust races THBJG and BBBDB, 13 STS markers specific to chromosome 2A were developed, based on seven SNP markers, two SSR markers, and five putative genes. The primers for these STS markers and the expected sizes of PCR products are provided in Table 1.
Table 1. Primer sequences of the markers used in this study.
| Markers | Forward primer (5′-3′) | Reverse primer (5′-3′) | PCR (bp) | Tm (°C) |
|---|---|---|---|---|
| Traes_2AS_3381A39C6 | GTTCCACACCCAAGATGGTTAC | TGAAAGAAACACTAAGCTGAGGAG | 503 | 55 |
| SNP19777336 | TGAATATATAGCAGTATGGGTGGCAG | GTACCAAACGCCATGAAAGGG | 1075 | 55 |
| IWB58709 | GGTTGAGTTTCCGAGTTTCGTG | GTTCTTGTTCTCCCGTTTGTGC | 1505 | 58 |
| GWM636-M | TGCGGTAGTTTTTAGCAAAGCG | TTTGTCTGGATAATACGACCCCC | 738 | 56 |
| IWB64569 | AGCCCTTGCGGAATTCATGGCAAA | GCTACAGCAGTATGTACACAAAAGCCTG | 1351 | 55 |
| Traes_2AS_5CAF7A367 | TCCATCCACAACACCTACCC | GCAGCGTAATAATACTTCCTATGAT | 1725 | 55 |
| IWB72490 | CTCCACCGCTCATCTTCCTG | TCCCCCAACCGACATTATCC | 1559 | 55 |
| WMC407-M | GCATCTTGTGTTGGTGTTGCC | TCAAGGGTGCGTTTTTCACTTC | 423 | 53 |
| Traes_2AS_D11AD107B | GTTCGCATCTCCGCCTTTAGG | TGGTCACAGGGTAGAAGTATTTCGG | 1617 | 55 |
| Traes_2AS_27D2CB02A | CAAGACACACAGACCTGCTTTTT | CACTGTAATAAAGTGGTAAAGTGAAGC | 1675 | 53 |
| IWB11136 | GTCTACATGAAAATAAGCTGCTGAGG | GATGGAGTTTTGGGTTTAAGTTCTAG | 365 | 52 |
| SNP30671452 | TGGAGGAAGACGATGGAGACTG | GCAAACCACCAAAGAAGAACCG | 581 | 61 |
| IWB47995 | GCTCCCCAAGGACAACATCA | AGAGAGCCAAGAGCAAGCATACC | 497 | 60 |
Seven SNP markers and two SSR markers (Xgwm636 and Xwmc407) were converted to 2A-specific PCR markers based on the 100 bp SNP probe sequences and the genome sequence harboring the target SSR. The sequences were used as queries to search the IWGSC Chinese Spring wheat scaffold sequences of 2A, 2B and 2D (https://urgi.versailles.inra.fr/blast_iwgsc/blast.php). The BLASTN search was limited to the top hit with an E-value cut off of at least 1E-10. The resulting scaffolds were used to design primers that were specific to chromosome 2A to amplify the fragment covering the SNP or SSR marker sequence region. The amplified PCR products were confirmed to be on chromosome 2A by sequencing PCR products. Five putative T. aestivum genes residing within the QTL region, Traes_2AS_3381A39C6 encoding a hypothetical protein, Traes_2AS_5CAF7A367 encoding a NB-ARC domain-containing disease resistance protein, Traes_2AS_67EFE0FAE encoding a heat shock protein 90 (HSP90), Traes_2AS_D11AD107B encoding a hypothetical protein, and Traes_2AS_27D2CB02A encoding a mortality factor 4-like protein 1, were chosen as representatives to distinguish between the 2NS segment and the orthologous region of chromosome 2A in hexaploid wheat.
The PCR was performed by using a Taq DNA polymerase (New England BioLabs) and 35 thermal cycles after denaturing at 95° for 3 min, with each cycle consisting of 94° for 30 sec, 55-61° for 40 sec, and 72° for 1-2 min, and a final extension at 72° for 10 min. The annealing temperature and extension time of PCRs were determined depending on primers and product size. All of the developed STS markers but IWB47995 showed a dominant pattern for chromosome arm 2AS with presence in 2174 but absence in Jagger. The STS marker for IWB47995 is co-dominant, and the 497 bp fragment was digested with restriction enzyme Hpa II that produced 328 bp and 169 bp fragments for the allele from Jagger and 345 bp and 152 bp fragments for the allele from 2174.
Physical mapping and sequence annotation
To determine the physical position of markers on chromosome arm 2AS, the sequences corresponding to the markers were used as BLASTN queries against the assembled pseudomolecules of chromosome 2A (https://urgi.versailles.inra.fr/download/iwgsc/IWGSC-WGA/) with an E-value cut off of at least 1E-10. The physical distance between Xgwm636-M and Xwmc407-M was predicted based on the assembled genome sequences of the A genome in tetraploid emmer wheat (http://z-data.interomics.eu) (Avni et al. 2017). The genes residing in the region between Xgwm636-M and Xwmc407-M on chromosome 2A were predicted according to the IWGSC Survey sequence annotations (https://wheat-urgi.versailles.inra.fr/Seq-Repository/Genes-annotations). Their putative functions were determined, via BLASTX searches, against the National Centre for Biotechnology Information (NCBI) non-redundant protein sequence database using an E-value cut-off of 1E-5.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6122270.
Results
Infection type response of the Jagger × 2174 RIL population to THBJG and BBBDB
Seedlings of Jagger had low IT (;1-) to THBJG, but seedlings of 2174 had high IT (3+) to THBJG (Table 2), indicating that race THBJG can be used to detect genes in the Jagger × 2174 RIL population. Both Jagger (;1-) and 2174 (;) had low IT to BBBDB. The control line RL6008 carrying the Lr17a gene showed the same IT (;1-) as Jagger to both THBJG and BBBDB. The Thatcher line (RL6081) with Lr37 on the 2NS translocation had IT of:22- to BBBDB, and 2+3 to THBJG. The lower IT of Jagger indicated that Jagger has additional resistance to both races compared to RL6081.
Table 2. IT response to leaf rust races THBJG and BBBDB in parental lines, controls and RILs.
| Race | Parents | Controls | Population | |||
|---|---|---|---|---|---|---|
| Jagger | 2174 | RL6008-Lr17a | RL6081-Lr37 | Mean | Min–Max | |
| THBJG | 1a (;1-)b | 9 (3+) | 1 (;1-) | 6.5 (2+3) | 6.2 | 0–9 |
| BBBDB | 1 (;1-) | 0 (;) | 1 (;1-) | (5.5) ;22- | 4.7 | 0–9 |
converted IT used for QTL analysis.
original IT.
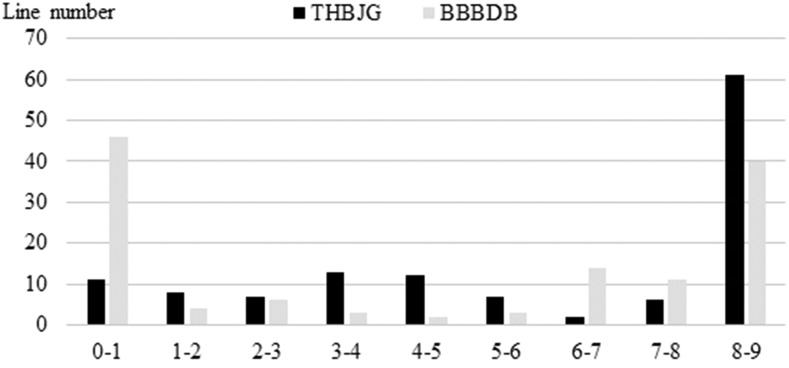
The ITs of 129 Jagger × 2174 RILs inoculated with race THBJG and race BBBDB individually as seedlings in 2012 and 2017 were analyzed in this study. The correlation of IT between the two tests was highly significant for THBJG (r = 0.829, P < 0.001) and for BBBDB (r = 0.829, P < 0.001). The average converted IT of the two tests for the whole population was 6.2 for THBJG, whereas the average converted IT for both tests was 4.7 for BBBDB (Table 2), indicating THBJG had higher virulence than BBBDB to the Jagger × 2174 population. To race BBBDB 60 RILs had IT of 3+ (at 9 in scale) in both tests, 45.7% of the total RILs. To race THBJG 59 RILs had IT of 3+ in both tests, 46.5% of the total RILs. These results suggested that the segregation of the 129 RILs to both races fit a single major gene ratio of 1:1 (P > 0.05). The converted ITs of the RILs to both races ranged from 0 to 9 (Figure 1), The RILs with the lowest ITs likely have some additional minor genes with small effect on IT, in addition to the single major resistance gene that segregated.
Figure 1.
Distribution of the Jagger × 2174 RILs infection type to THBJG and BBBDB. Frequency distribution of the Jagger × 2174 RILs for converted IT using the 0–9 linear disease scale. Y axis indicates the number of lines with different ITs in the population of 129 RILs. The converted ITs were averaged from two tests in 2012 and 2017.
A major QTL for resistance to THBJG and BBBDB was mapped to the 2NS segment
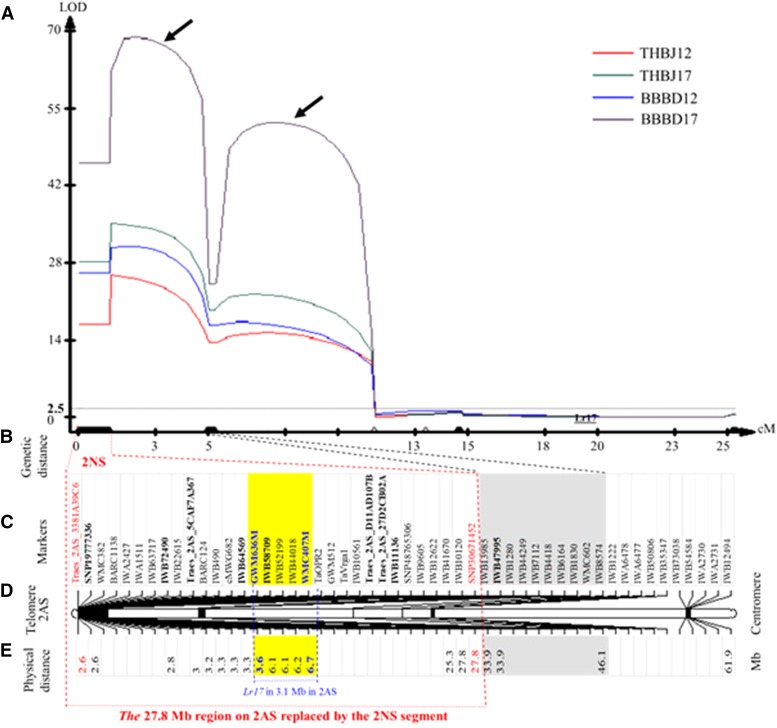
A major QTL for reaction to THBJG and BBBDB was coincidently mapped to the 2NS region (Figure 2A). The LOD value at the peak position of the QTL for THBJG was 25.9 and 35.4, and accounted for 56.5% and 62.2% of total phenotypic variation, in 2012 and 2017 respectively. The LOD value at the peak position of the QTL for BBBDB was 31.1 and 69.1, and accounted for 72.2% and 86.9% of total phenotypic variation, in 2012 and 2017 respectively.
Figure 2.
Genetic and physical relationship between Lr17a and the 2NS segment containing Lr37. A). A major QTL for reaction to THBJG and BBBDB. The genetic map was constructed using MapMaker 3.0 and QTL graphs were constructed using WinQTL Cart 2.5. The vertical dotted line indicates the logarithm of the odds (LOD) significance threshold of 2.5. Arrows point to peaks, under which no marker was mapped, and the peaks should be false. B). Genetic distance of the QTL for resistance to THBJG and BBBDB. C). Markers in the linkage group spanning the QTL. Markers that were converted to STS markers for mapping of the QTL are shown in bold. Markers that were used to determine the physical distance spanning the QTL are shown in red. The markers on chromosome arm 2AS that showed recombination with the 2NS segment are highlighted with gray shaded box. D). A diagram of genetic locations of the mapped markers on chromosome arm 2AS. E). Physical locations of the 3.1 Mb region including Lr17a on chromosome arm 2AS. Markers that were used to determine the physical distance of the Lr17a region are shown in yellow.
A total of 52 markers was assembled to a linkage group spanning approximately 26 cM, with a marker density of 0.53 cM per marker (Figure 2B). The linkage group included 38 SNP markers, seven SSR markers, and seven gene markers (Figure 2C). The peak of the QTL was covered by a cluster of 32 markers from Traes_2AS_3381A39C6 closest to the telomere (Figure 2D), throughout the five markers Xwmc382, Xbarc1138, Xbarc124, Xgwm512 and TaVrga1, which were previously mapped to 2NS (Fang et al. 2011), to SNP30671452 which is the 33rd marker in the linkage group. These clustered markers showed no crossover (Table S1).
The physical distance of the 2A chromosomal region replaced by the 2NS segment
Sequences of the SNP and STS markers were used to search the recently released the emmer wheat genomic sequences, which allowed a physical distance covering the clustered markers on chromosome 2A to be determined. The SNP markers were identified in scaffolds of chromosome 2A (Table S1). On the side closest to the telomere, the first clustered marker for the QTL peak was Traes_2AS_3381A39C6 located at position 2.6 Mb of chromosome arm 2AS (Figure 2E), and the last marker in the cluster was SNP30671452 which was at position 27.8 Mb of chromosome arm 2AS; therefore, the clustered markers for the QTL peak were predicted to cover 27.8 Mb regardless of the distance to the centromere.
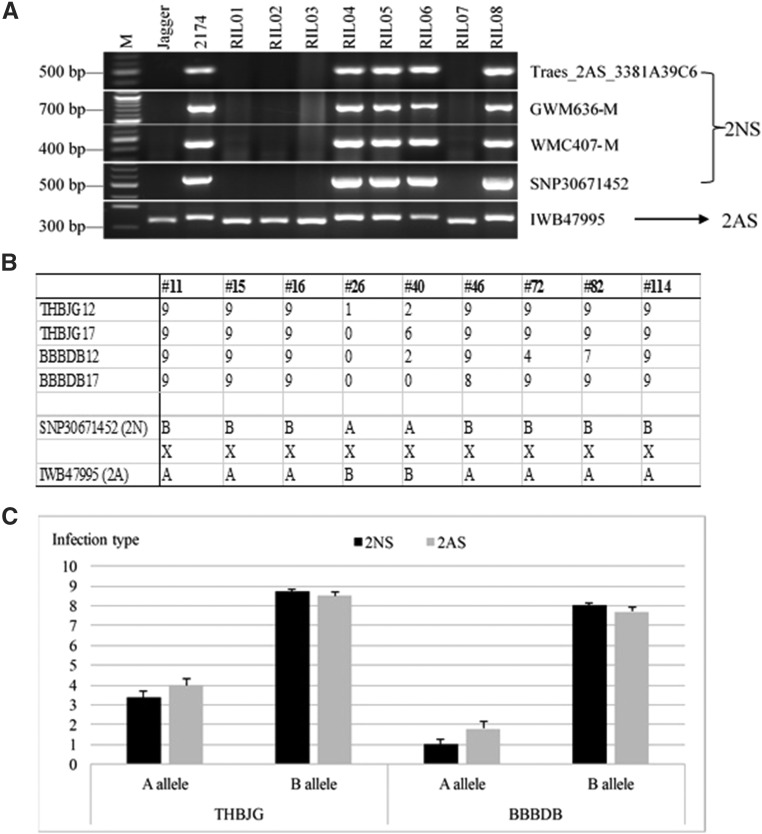
The 2A-specific primers for 13 markers included in the cluster (Table 1) were confirmed to amplify PCR products from chromosome 2A but failed to amplify any products from the 2NS segment, and all of these markers had dominant inheritance, showing their absence was associated with the A allele in Jagger and their presence was associated with the B allele in 2174. Four of the STS markers for 2NS/2AS are shown in Figure 3A.
Figure 3.
Development and utilization of STS markers for the 2NS segment. A). Mapping of five STS markers in eight random lines. M: 100-bp ladder. The numbers to the left indicate the molecular size in bp. The first four markers showed the presence/absence polymorphism, and the last marker IWB47995 shows the polymorphic band pattern after the PCR products were digested with Hpa II. B). Genotypes and phenotypes of nine critical recombinant lines. ‘A’ represents the Jagger allele, ‘B’ represents the 2174 allele, and ‘x’ indicates a crossover between two markers. The phenotype is infection type scored in the 0-9 scale. C). Genetic effects of the 2NS segment to THBJG and BBBDB. The converted ITs were averaged from two tests in 2012 and 2017, and bar indicates standard error (± SE). ‘A’ indicates the Jagger allele in the 2NS region including SNP30671452 and the 2AS region including IWB47995, and ‘B’ indicates the 2174 allele in the 2NS region and the 2AS region.
On the 2AS segment closest to the 2NS segment, the SNP marker IWB47995 which showed recombination with the clustered markers for the 2NS segment, was converted to a cleaved amplified polymorphic sequence (CAPS) marker. In the sequenced 497 bp fragment, three nucleotides were found to differ between Jagger and 2174 alleles, and two of the SNPs were located within the recognition site of restriction enzyme Hpa II. The CAPS marker for IWB47995 was mapped as a co-dominant polymorphism between the two alleles (Figure 3A). IWB47995 was assigned to position 33.9 Mb from the telomere of chromosome arm 2AS in emmer wheat (Figure 2E).
Jagger had the same sequence in IWB47995 as Chinese Spring; therefore, the genome region from IWB47995 to the centromere in Jagger should be chromosome arm 2AS from hexaploid wheat (Figure 2D). Since 50 SNP markers in the 27.8-33.9 Mb region of chromosome arm 2AS (Table S2) were monomorphic between the two alleles and these markers had identical sequences to Chinese Spring, the 27.8-33.9 Mb region in Jagger should also be chromosome arm 2AS. Therefore, the 2AS region replaced by the translocated 2NS segment was approximately 27.8 Mb (Figure 2E), according to the physical distance of assembled genomic sequences of chromosome 2A in emmer wheat.
Nine lines were found to have a crossover between SNP30671452 representing the 2NS segment and IWB47995 representing chromosome arm 2AS (Figure 3B). Among seven of these critical recombinant lines that carried the B allele from chromosome arm 2AS, all had highest converted IT to THBJG, and all of them had high or intermediate susceptibility to BBBDB (Figure 3B). Two critical recombinant lines (RIL26 and RIL40) that carried the A allele on the 2NS segment had very low or intermediate converted IT to THBJG and BBBDB. The tight linkage of 2NS with the resistance of the nine critical recombinant lines indicated that the seedling resistance genes detected in the Jagger × 2174 RILs was present in the 2NS segment.
At the population level as shown in Figure 3C, in the region of SNP30671452 representing the 2NS segment, the average converted IT of those lines carrying the A allele was 1 for BBBDB and 3.4 for THBJG, whereas the average IT of those lines carrying the B allele was 8 for BBBDB and 8.7 for THBJG. However, in the IWB47995 region representing chromosome arm 2AS, the average IT of those lines carrying the A allele was 1.8 for BBBDB and 4 for THBJG, whereas the average IT of those lines carrying the B allele was 7.7 for BBBDB and 8.7 for THBJG.
The physical location of Lr17a on chromosome arm 2AS
The wheat Lr17a gene was mapped close to the distal end of the short arm of chromosome 2A, at 2.7-4.0 cM to Xgwm636 and at 2.5 cM to Xwmc407 (Bremenkamp-Barrett et al. 2008; Zhang et al. 2008). GWM636 and WMC407 were converted to 2A-specific STS markers (Table 1) that were mapped in the population of Jagger × 2174 RILs. The dominant band patterns of the modified PCR markers for the SSR markers demonstrated that they were linked with the 2NS segment (Figure 3A).
Sequences of GWM636-M and WMC407-M were used to search the emmer wheat genome sequence, and GWM636-M was found on scaffold33664 at position 3.6 Mb, and WMC407-M was found on scaffold98927 at position 6.7 Mb. Based on these results, the physical distance between Xgwm636-M and Xwmc407-M was at least 3.1 Mb (Figure 2E), according to the emmer wheat genome sequences.
Discussion
Seedling leaf rust resistance in Jagger wheat mapped to the 2NS translocation derived from A. ventricosa. The 2NS translocation in Jagger was determined to span 27.8 Mb on the distal region of chromosome arm 2AS as delineated by molecular markers. Both races THBJG and BBBD produced a very low IT that mapped to the 2NS translocation. Lr37 has been regarded as an adult plant resistance gene, since lines with the 2NS translocation optimally express resistance in adult plants, and low IT can be difficult to obtain in seedlings (McIntosh et al. 1995). However, wheat populations segregating for the 2NS translocation have also segregated for seedling leaf rust resistance (Dyck and Lukow 1988; Kolmer 2017). At normal greenhouse temperatures of 20°, seedlings of the Thatcher line with Lr37 condition a mesothetic response of necrosis, chlorosis, and various sized uredinia (IT X+) (Kolmer 1997). Temperature also affects expression of Lr37. At 17° wheat lines with the 2NS translocation expressed low IT 12 compared to IT 3 at 20° (Bariana and McIntosh 1994).
Previously Lr17a was postulated to be present in Jagger (Zhang et al. 2008; Kolmer 2017) and many other hard red winter wheat cultivars in the southern Great Plains region of the U.S that were derived from Jagger. This was reinforced by the loss of resistance in Jagger and related cultivars due to the rapid increase of races with Lr17a virulence in North America after the release of Jagger in 1996. Our study placed the Lr17a locus on 2AS within a 3.1 Mb region, which is orthologous to the 2NS translocation, thus eliminating the possibility of Lr17a in Jagger. Bariana and McIntosh (1993) previously determined that the 2NS translocation and Lr17a were in linkage repulsion since they did not recover any F2 plants with both in a cross between a wheat line with the 2NS translocation and the Thatcher line with Lr17a. The P. triticina races with virulence to Lr17a that have been prevalent in North America since the mid 1990s are also virulent to Lr37 (Kolmer et al. 2005). The Lr17 locus has two resistance alleles, Lr17a originally found in wheat cultivars Rafaela and EAP 26127 (Dyck and Kerber 1977) and Lr17b in the Australian cultivar Harrier (Singh et al. 2001). Lr17a and Lr17b alleles also have similar IT to the same races, depending on the temperature and leaf rust race (Singh et al. 2001). Since Lr17a, Lr17b and Lr37 are found in the same region on 2AS and 2NS respectively, these genes may be allelic at the same corresponding loci in the different genomes. Genes Lr2a, Lr2b, and Lr2c are allelic and are highly related for IT to avirulent leaf rust pathogen (Dyck and Samborski 1974). Since the 2NS translocation and the Lr17 locus are linked in repulsion it is not possible to conduct classical allelism tests. Further sequencing and characterization of the corresponding regions on 2NS and 2AS may determine the relationship between Lr37 and the Lr17 genes. In addition, Jagger carrying Lr37 on 2NS can be crossed with a cultivar/line that carries Lr17 on 2AS. This population would segregate for both Lr37 and Lr17, therefore, the two genes could be compared for resistance in the same genetic background.
The wheat Lr17 gene was physically delimited into the 3.1 Mb region between Xgwm636-M and Xwmc407-M. All of the resistance genes residing in the targeted region on chromosome arm 2AS should be candidates for Lr17a in wheat. Four genes were predicted to be related with known resistance genes in the 3.1 Mb region between Xgwm636-M and Xwmc407-M, including Traes_2AS_5CAF7A367 (encoding a NB-ARC domain-containing disease resistance protein), Traes_2AS_089875521 (encoding a disease resistance RPP13-like protein 1), Traes_2AS_6E1B0E6EF (encoding a resistance gene analog 2 (RGA2), and Traes_2AS_AFEB22E37 (encoding a disease resistance RPP13-like protein 1). In addition, another two genes are located in this region, including Traes_2AS_D0C21ADB5 that was predicted to encode a WRKY transcription factor, and Traes_2AS_7B74BE6D6 that was predicted to encode a putative leucine-rich-repeat (LRR) receptor-like serine/threonine-protein kinase. Further study is needed to test the functions of these candidate genes in wheat.
An unresolved issue in this study was that the resistance to BBBD in 2174 was not mapped. Recombinant inbred lines with resistance alleles to BBBD derived from 2174 were not identified. 2174 was also resistant to stripe rust when the same population was tested in China (Fang et al. 2011), but this resistance was also not mapped. Although the Jagger × 2174 RIL population has been genotyped using the wheat 90K SNP assay, not all genomic regions may have been covered, and the uncharacterized part of the genome may have additional stripe and leaf rust resistance genes.
In summary, the seedling resistance in Jagger was mapped to the 2NS translocation. The 2NS translocation in Jagger was determined to be 24 Mb in length, on the distal region of 2A. The leaf rust resistance gene locus Lr17 on 2AS, mapped in a 3.1 Mb region, which is orthologous to the 2NS translocation. Lr37 on the 2NS translocation may be allelic with the Lr17 locus on 2AS. The population in North America has high level of virulence to Lr17a and Lr37, however the three minor QTL may be useful in combinations with other effective resistance genes in development of leaf rust resistance cultivars.
Acknowledgments
This work was supported by USDA-NIFA T-CAP grant No. 2011-68002-30029, the Oklahoma Center of Advanced Science and Technology (OCAST AR131-033), and the Oklahoma Wheat Research Foundation. The authors also thank the Malone Family Land Preservation Foundation and The Land Institute for support of studies on application of alien genes from perennial wheat to bread wheat. The National Natural Science Foundation of China program (31271711) and the Natural Science Foundation of Jiangsu Province of China program (BK20131316) provided research grants to S. Xue.
Author contributions: S. Xue, S. Wang and L. Yan conducted marker development, genotyping, and mapping; J. Kolmer conducted phenotyping. All authors analyzed data and reviewed the manuscript. S. Xue, J. Kolmer and L. Yan wrote the manuscript. The authors declare that they have no conflict of interest.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.6122270.
Communicating editor: E. Akhunov
Literature Cited
- Avni R., Nave M., Barad O., Baruch K., Twardziok S. O., et al. , 2017. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357: 93–97. 10.1126/science.aan0032 [DOI] [PubMed] [Google Scholar]
- Bariana H. S., McIntosh R. A., 1993. Cytogenetic studies in wheat XV. Location of rust resistance genes in VPM1 and their genetic linkage with other disease resistance genes in chromosome 2A. Genome 36: 476–482. 10.1139/g93-065 [DOI] [PubMed] [Google Scholar]
- Bariana H. S., McIntosh R. A., 1994. Characterisation and origin of rust and powdery mildew resistance genes in VPM1 wheat. Euphytica 76: 53–61. 10.1007/BF00024020 [DOI] [Google Scholar]
- Bremenkamp-Barrett B., Faris J. D., Fellers J. P., 2008. Molecular mapping of the leaf rust resistance gene Lr17a in wheat. Crop Sci. 48: 1124–1128. 10.2135/cropsci2007.07.0379 [DOI] [Google Scholar]
- Cavanagh C. R., Chao S., Wang S., Huang B. E., Stephen S., et al. , 2013. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl. Acad. Sci. USA 110: 8057–8062. 10.1073/pnas.1217133110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyck P. L., Kerber E. R., 1977. Inheritance of leaf rust resistance in wheat cultivars Rafaela and EAP 26127 and chromosome location of gene Lr17. Can. J. Genet. Cytol. 19: 355–358. 10.1139/g77-038 [DOI] [Google Scholar]
- Dyck P. L., Lukow O. M., 1988. The genetic analysis of two interspecific sources of leaf rust resistance and their effect on the quality of common wheat. Can. J. Plant Sci. 68: 633–639. 10.4141/cjps88-076 [DOI] [Google Scholar]
- Dyck P. L., Samborski D. J., 1974. Inheritance of virulence in Puccinia recondita on alleles at the Lr2 locus for resistance in wheat. Can. J. Genet. Cytol. 16: 323–332. 10.1139/g74-036 [DOI] [Google Scholar]
- Fang T., Campbell K. G., Liu Z., Chen X., Wan A., et al. , 2011. Stripe rust resistance in the wheat cultivar Jagger is due to Yr17 and a novel resistance gene. Crop Sci. 51: 2455–2465. 10.2135/cropsci2011.03.0161 [DOI] [Google Scholar]
- Helguera M., Khan I. A., Kolmer J., Lijavetzky D., Li Z., et al. , 2003. PCR Assays for the Lr37-Yr17-Sr38 cluster of rust resistance genes and their use to develop isogenic hard red spring wheat lines. Crop Sci. 43: 1839–1847. 10.2135/cropsci2003.1839 [DOI] [Google Scholar]
- Kolmer J. A., 1997. Virulence in Puccinia recondita f. sp. tritici isolates from Canada to genes for adult-plant resistance to wheat leaf rust. Plant Dis. 81: 267–271. 10.1094/PDIS.1997.81.3.267 [DOI] [PubMed] [Google Scholar]
- Kolmer J. A., 2017. Genetics of leaf rust resistance in the hard red winter wheat cultivars Santa Fe and Duster. Crop Sci. 57: 2500–2505. 10.2135/cropsci2016.12.1029 [DOI] [Google Scholar]
- Kolmer J. A., Long D. L., Hughes M. E., 2005. Physiologic specialization of Puccinia triticina on wheat in the United States in 2003. Plant Dis. 89: 1201–1206. 10.1094/PD-89-1201 [DOI] [PubMed] [Google Scholar]
- Kolmer J. A., Long D. L., Hughes M. E., 2006. Physiologic specialization of Puccinia triticina on wheat in the United States in 2004. Plant Dis. 90: 1219–1224. 10.1094/PD-90-1219 [DOI] [PubMed] [Google Scholar]
- Li G., Wang Y., Chen M., Edae E., Poland J., et al. , 2015. Precisely mapping a major gene conferring resistance to Hessian fly in bread wheat using genotyping-by-sequencing. BMC Genomics 16: 108 10.1186/s12864-015-1297-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Lei L., Powers C., Liu Z., Campbell K. G., et al. , 2016. TaXA21-A1 on chromosome 5AL is associated with resistance to multiple pests in wheat. Theor. Appl. Genet. 129: 345–355. 10.1007/s00122-015-2631-9 [DOI] [PubMed] [Google Scholar]
- Liu M., Yu M., Li G., Carver B. F., Yan L., 2015. Genetic characterization of aluminum tolerance in winter wheat. Mol. Breed. 35: 205 10.1007/s11032-015-0398-y [DOI] [Google Scholar]
- Long D. L., Kolmer J. A., 1989. A North American system of nomenclature for Puccinia recondita f. sp. tritici. Phytopathology 79: 525–529. 10.1094/Phyto-79-525 [DOI] [Google Scholar]
- Long D. L., Leonard K. J., Roberts J. J., 1998. Virulence and diversity of wheat leaf rust in the United States in 1993 to 1995. Plant Dis. 82: 1391–1400. 10.1094/PDIS.1998.82.12.1391 [DOI] [PubMed] [Google Scholar]
- Maia N. 1967. Obtention des blés tendres resistants au pietin-verse par croisements interspecifiques blés X Aegilops. C R Acad. Agric. Fr. 53: 149–154. [Google Scholar]
- McIntosh, R.A., J. Dubcovsky, W.J. Rogers, C. Morris, R. Appels et al., 2016 Catalogue of gene symbols for wheat: 2015–2016 supplement. KOMUGI Integrated Wheat Science Database.
- McIntosh R. A., Friebe B., Jiang J., The D., Gill B. S., 1995. Cytogenetical studies in wheat XVI. Chromosome location of a new gene for resistance to leaf rust in a Japanese wheat-rye translocation line. Euphytica 82: 141–147. 10.1007/BF00027060 [DOI] [Google Scholar]
- Singh D., Park R. F., Bariana H. S., Mcintosh R. A., 2001. Cytogenetic studies in wheat XIX. Chromosome location and linkage studies of a gene for leaf rust resistance in the Australian cultivar ‘Harrier’. Plant Breed. 120: 7–12. 10.1046/j.1439-0523.2001.00544.x [DOI] [Google Scholar]
- Tan C. T., Carver B. F., Chen M., Gu Y., Yan L., 2013. Genetic association of OPR genes with resistance to Hessian fly in hexaploid wheat. BMC Genomics 14: 369 10.1186/1471-2164-14-369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M. K., Jin Y., Rouse M. N., Anderson J. A., 2016. Stem rust resistance in ‘Jagger’ winter wheat. Crop Sci. 56: 1719–1725. 10.2135/cropsci2015.11.0683 [DOI] [Google Scholar]
- Zhang J. X., Singh R. P., Kolmer J. A., Huerta-Espino J., Jin Y., et al. , 2008. Inheritance of leaf rust resistance in the CIMMYT wheat Weebill. Crop Sci. 48: 1037–1047. 10.2135/cropsci2007.08.0455 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Supplemental material available at Figshare: https://doi.org/10.25387/g3.6122270.