Abstract
Mre11 is a key player for DNA double strand break repair. Previous studies have shown that mammalian Mre11 is methylated at multiple arginines in its C-terminal Glycine-Arginine-Rich motif (GAR) by protein arginine methyltransferase PRMT1. Here, we found that the Drosophila Mre11 is methylated at arginines 559, 563, 565, and 569 in the GAR motif by DART1, the Drosophila homolog of PRMT1. Mre11 interacts with DART1 in S2 cells, and this interaction does not require the GAR motif. Arginines methylated Mre11 localizes exclusively in the nucleus as soluble nuclear protein or chromatin-binding protein. To study the in vivo functions of methylation, we generated the single Arg-Ala and all Arginines mutated flies. We found these mutants were sensitive to ionizing radiation. Furthermore, Arg-Ala mutated flies had no irradiation induced G2/M checkpoint defect in wing disc and eye disc. Thus, we provided evidence that arginines in Drosophila Mre11 are methylated by DART1 methytransferase and flies loss of arginine methylation are sensitive to irradiation.
Keywords: Mre11, arginine methylation, DART1, Glycine-Arginine-Rich motif, ionizing radiation
Mre11 plays a pivotal role at DNA double strand break repair (haber 1998). It forms a complex with Rad50 and Nbs1 (MRN), and is highly conserved in eukaryotes. The MRN complex executes the sensor function to recognize DNA double strand breaks (DSBs) (petrini and stracker 2003), and regulates DNA damage repair through both homologous recombination and non-homologous end joining pathways (haber 1998). It is widely recognized that post-translational modifications, such as phosphorylation, acetylation, methylation etc, are critical for protein functions (Blanc and Richard 2017). This is also true to Mre11, previous studies have shown that multiple arginines in the C-terminal Glycine-Arginine-Rich motif (GAR) of Mre11 are methylated by protein arginine methyltransferase 1 (PRMT1) in HeLa cells (boisvert et al. 2005a; boisvert et al. 2005b; Dery et al. 2008).
Arginine methylation is catalyzed by the nine-member protein arginine methyltransferase (PRMT) family in mammals (Blanc and Richard 2017). Based on their substrate specificity, PRMTs are classified into three types. Type I and type II PRMTs catalyze the formation of ω-NG-monomethylarginine (MMA), which is the intermediate for production of the ω-N′G, NG-asymmetric dimethylarginine (aDMA) and ω-NG, NG-symmetric dimethylarginine (sDMA). Type III PRMT catalyzes the formation of MMA only on histones. Among PRMTs, PRMT1 is the major arginine methyltransferase and contributes to more than 90% of type I PRMT activity in human cells (tang et al. 2000). The arginines methylated by PRMT1 are usually surrounded by glycines, they together form a GAR motif, which is also called the Arginine- and Glycine-Rich motif (RGG) (thandapani et al. 2013). Study in mouse embryonic fibroblasts (MEFs) has shown that loss of PRMT1 causes genome instability, and delayed cell cycle progression (yu et al. 2009). PRMT1 performs its functions in response to DNA damage through methylation of DNA repair factors, such as Mre11 (boisvert et al. 2005a; Dery et al. 2008), p53BP1 (boisvert et al. 2005c), p53 (scoumanne and chen 2008), DNA polymerase beta (pol β) (el-andaloussi et al. 2007; el-andaloussi et al. 2006), HMGA1 (zou et al. 2007) etc.
Study in mammalian cells has shown that Mre11 interacts with PRMT1, and mutation of arginines in the GAR motif causes intra-S-phase checkpoint defect and loss of exonuclease activity (boisvert et al. 2005a). Although arginine methylation is not required for MRN complex formation, it is essential for its binding with chromatin (boisvert et al. 2005a; Dery et al. 2008; yu et al. 2012). Inhibition of arginine methylation suppresses Mre11 localization to DNA damage sites (8), and reduces γ-H2AX foci formation (boisvert et al. 2005b) as well as Rad51 foci formation upon DNA damage (Dery et al. 2008; yu et al. 2012). Studies in mre11RK/RK knock-in mice and MEFs have shown that arginines substituted with lysines causes hypersensitivity to gamma ray radiation, and MEF cells display significant increase of aberrant chromosomes, defects in G2/M cell cycle checkpoint and ATR/CHK1 signaling activation, and reduced RPA and RAD51 foci after irradiation (yu et al. 2012).
The Drosophila arginine methyltransferase (DART) is identified by sequence homology with mammalian protein arginine methyltransferase (boulanger et al. 2004). DART1 is the Drosophila homolog of PRMT1, it can methylate mammalian PRMT substrates and arginine rich Drosophila substrates in vitro (boulanger et al. 2004). The Drosophila Mre11 is highly conserved, we and others have shown that the major defects in mre11 mutated flies are telomere-telomere associations, aneuploidy (bi et al. 2004; ciapponi et al. 2004) and G2/M checkpoint defect induced by low dose irradiation (bi et al. 2005). While mammalian PRMT1 interacts with Mre11 and catalyzes its methylation, whether highly conserved Drosophila Mre11 is methylated remains unknown.
In this study, we identified four arginine residues methylated by DART1 in the GAR motif of Drosophila Mre11, and knock-down of dart1 reduced the level of arginine methylation in vitro. Methylated Mre11 localizes exclusively in the nucleus as chromatin-binding protein and nucleoplasmic protein. Moreover, Drosophila Mre11 interacts with DART1 in S2 cells, and this interaction does not require the GAR motif. To study the in vivo functions of Mre11 arginine methylation, we generated knock-in flies of Arg-Ala (RA) point mutations and 4 Arg-Ala (4RA) mutation by ends-in approach (rong et al. 2002; rong and golic 2000). The mre11RA and mre114RA flies are sensitive to ionizing radiation. However, the mre11RA and mre114RA flies do not show DNA damage induced G2/M checkpoint defect in the wing disc and eye disc.
Materials and Methods
Plasmids construction
Constructs for in vitro methylation assay:
The full length cDNA of dart1 was PCR amplified with primers dart1-F and dart1-R (Table S1), and cloned into XhoI/EcoRI sites of pRSETb vector (Invitrogen, Grand Island, NY, USA). The full length cDNA of mre11 was amplified with mre11-F and mre11-R, mre11N206 was amplified with mre11-N-F and mre11-N-R, mre11M167 was amplified with mre11-M-F and mre11-M-R, and mre11C247 was amplified with mre11-C-F and mre11-C-R (Table S1). Mre11 was cloned into EcoRI/SalI sites, and mre11C247 was cloned into BamHI/EcoRI sites of pSJ8 vector (pET21a derivative, a gift from Dr. Yuhui Dong). Mre11N206 and mre11M167 were cloned into KpnI/EcoRI sites of pRSETb vector.
Constructs for co-immunoprecipitation:
The full length cDNA of dart1 was PCR amplified with primers V5-dart1-F and V5-dart1-R, and cloned into EcoRV/NotI sites of pAc5.1/V5-His B vector (Invitrogen, Grand Island, NY, USA). Mre11 was amplified with Myc-mre11-F1 and Myc-mre11-R, and cloned into EcoRI/NotI sites of pCMV-Myc vector (Clontech, Mountain View, CA, USA) to generate pCMV-Myc-mre11. Then, the correct pCMV-Myc-mre11 plasmid was used as a template and amplified with Myc-mre11-F2 and Myc-mre11-R, and cloned into EcoRV/NotI sites of pAc5.1/V5-His B vector (Invitrogen, Grand Island, NY, USA) to generate pAc5.1-Myc-mre11. Mre11GARΔ was constructed by two-step PCR. In the first PCR, pAc5.1-Myc-mre11 was used as template, primer pairs Myc-mre11-F2 and mre11GARΔ-overlapPCR-R, mre11-GARΔ-overlapPCR-F and Myc-mre11-R were used separately to generate two fragments. In the second PCR, the products from first PCR were mixed together and used as template, primer pair was Myc-mre11-F2 and Myc-mre11-R (Table S1) and the PCR product was cloned into EcoRV/NotI sites of pAc5.1/V5-His B vector.
Protein expression and purification
DART1, Mre11, Mre11N206, Mre11M167 and Mre11C247 were expressed in BL21. DART1, Mre11N206 and Mre11M167 protein were purified with HIS-Select Nickel Magnetic Agarose Beads (Sigma, St. Louis, MO, USA), Mre11 and Mre11C247 protein were purified with Amylose Resin (NEB, Ipswich, MA, USA) following the manufacture′s instructions.
Methylated arginine antibody production
Antibodies recognizing single methylated arginine at Mre11-R559, R563, R565, R569, R571 or R579 sites were generated by GL Biochem (Shanghai, China). The antigenicity of Drosophila Mre11 was analyzed using DNAStar software, and two peptides with better epitope segments were selected, their sequences were 555-PATGR*GAAR*GR*GT-567, and 568-AR*TR*AGATAATR*GKG-582 (methylated arginines were labeled with *). Peptides with single aDMA modification at specific arginine site were synthesized and used to immunize rabbits. Methylated arginine antibodies (M-Ab) were precleared with non-arginine methylation peptide and affinity purified with a site specific arginine methylated peptide. Specificity was verified by a dot-blot assay. And non-methylated antibodies (NM-Ab1 and NM-Ab2) were generated with two non-arginine methylation peptides respectively.
In vitro methylation assay
In vitro methylation assay was performed as described (boisvert et al. 2005a). Purified Mre11, Mre11N206, Mre11M167 and Mre11C247 were incubated with His-DART1 separately, in 50 mM Tris-HCl pH 8.5, with 0.1 µM of [methyl-3H] S-adenosyl methionine (Sigma, St. Louis, MO, USA), to a final volume of 30 μl for 1 h at 30°. The reaction was stopped by adding 2 × SDS-PAGE sample buffer and boiled for 10 min. Samples were subjected to electrophoresis and visualized by fluorography.
Generation of mre11 mutants
A 6-kb genomic fragment of mre11 with an I-SceI cut site constructed into pCR2.1 (Gao et al. 2009) was used as a template. The R559A, R563A, R565A, R569A, or 4RA change was made by site directed mutagenesis using primers: mre11 R559A-F and mre11 R559A-R, mre11 R563A-F and mre11 R563A-R, mre11 R565A-F and mre11 R565A-R, mre11 R569A-F and mre11 R569A-R, mre11 4RA-F and mre11 4RA-R, respectfully (Table S1). The full length of genomic constructs was sequenced to verify the specified RA change as the only mutation. The 6-kb NotI fragment was excised and constructed into the targeting vector pTV2. Targeting crosses and the subsequent reduction crosses were performed as previously described (Gao et al. 2009; Gong et al. 2005).
The overall integrity of the mre11 genomic region from homozygotes of intermediate flies and mutants was assayed by Southern blot analysis (Gong et al. 2005), using genomic DNA digested with BglII and probe amplified from mre11 gene region with primers Probe-F and Probe-R (Table S1). The mutants were further verified by cDNA sequencing and genomic DNA sequencing.
Fly genetics
All flies were maintained at 25° on standard corn meal unless specified. The fly lines used in this study are: w1118; mre11Δ35K1/TSTL Tb (Gong et al. 2005); mre11R559A1-1T10, mre11R563A3-1D5, mre11R565A2-7M5, mre11R569A6-34H1, and mre114RA2-3P3 flies were generated in this study, kept as homozygote and used in the assay.
Embryo fractionation
Embryos of w1118 wild-type flies were collected at 0 ∼4 h and fractionated following the manufacture′s instructions (Thermo Scientific). Briefly, embryos were dechorionated in 3% NaOCl for 2 min, washed thoroughly with tap water, then grounded gently in 300 μL pre-cooled 1 × phosphate-buffered saline (PBS) with a dounce homogenizer, centrifuged at 6,000 g for 5 min at 4°, and supernatant was carefully removed. The pellet was resuspended with 200 μL of CERI and put on ice for 10 min, 11 μL of CERII was added and treated for 1 min to lysate cell membrane. The sample was centrifuged with maximum speed (∼16,000 × g) at 4° for 5 min, and supernatant (cytoplasm) was immediately transfered to a clean pre-chilled tube. The pellet was washed in l mL precooled 1 × PBS three times, resuspended with 200 μL ice-cold NER to lysate nuclei, then vortexed 15 sec in every 10 min for 4 times. The sample was centrifuged at ∼16,000 × g for 10 min, and supernatant (nucleoplasm) was transfered to a clean pre-chilled tube. The pellet was resuspended in 200 μL ice-cold 1 × PBS and sonicated to get the chromatin binding protein.
RNA interference
S2 cells were seeded into 25 cm2 flasks at 1×106 cells/mL the day before experiments. Five million cells were transfected with dart1 siRNA, 5′-GCAGCGAGGAUACAUACAATT-3′, at final concentration of 40 pmol/L using Lipofectamine RNAiMAX Reagent (Invitrogen, Grand Island, NY, USA). The scrambled sequence of siRNA was used as a negative control (NC). Forty eight hours after transfection, medium was changed and let cells recover for 24 h.
Reverse transcription PCR
Total RNA was prepared from S2 cells using an RNeasy Plus Mini Kit (Qiagen, Hilden, NW, Germany), and reverse transcribed with Oligo(dT)15 primer and M-MLV reverse transcriptase following the manufacturer’s instructions (Promega, Fitchburg, WI, USA). The PCR primers used were dart1-RT-F and dart1-RT-R to detect transcripts of dart1, and gapdh1-RT-F and gapdh1-RT-R to detect gapdh1 as internal control (Table S1). The PCR products were visualized by 2% agarose gel electrophoresis.
Western blot
Protein samples were subjected to SDS-PAGE electrophoresis and transferred to PVDF membrane. Samples were detected with primary antibodies against Mre11 (1:6,000, rabbit), β-actin (1:10,000, abcam ab8224, Cambridge, MA, USA), Histone H3 (1:15,000, abcam ab1791, Cambridge, MA, USA), MeMre11Arg559 (1:1,000), MeArg563 (1:800), MeArg565 (1:1,000), MeArg569 (1:1,500), MeArg571 (1:1,500), MeArg579 (1:1,500). Secondary antibodies were HRP-conjugated goat anti-rabbit (1:5,000, Jackson, West Grove, PA, USA) and goat anti-mouse (1:10,000, MultiSciences Biotech, Hangzhou, China).
Co-immunoprecipitation
DART1-V5 and Myc-Mre11 or Myc-Mre11GARΔ constructs were co-transfected into S2 cells with Effectene (Qiagen, Hilden, Germany) for 24 h at 25°. Cells were lyzed in IP lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100), then pre-cleared with Protein-G agarose beads (Roche, Indianapolis, IN, USA) and incubated with V5 antibody (abcam, Cambridge, MA, USA) or Myc antibody (CST, Beverly, MA, USA) for 3 h at 4°. Protein-G dynabeads (Invitrogen, Grand Island, NY, USA) were added and incubated overnight at 4°. They were then washed 4 times with ELB buffer (50 mM HEPES, pH 7.3, 250 mM NaCl, 0.1% NP40).
Viability assay
Actively crawling third instar larvae of w1118, mre11R559A1-1T10, mre11R563A3-1D5, mre11R565A2-7M5, mre11R569A6-34H1, mre114RA2-3P3 were collected, and irradiated at a rate of 1.3 Gy/min with dosage of 0, 10, 20, 30, 40 Gy in a X-Ray Biological Irradiator (X-RAD 320iX, Precision X-ray Inc), respectively. Let the irradiated flies grow to adult on fly medium, the number of viable flies was counted and survival percentage was calculated as viable flies divided by larvae irradiated. At least 100 larvae were treated at each dosage for each genotype, and experiments were repeated three times.
G2/M checkpoint assay
The G2/M checkpoint assay was performed as described (bi et al. 2005). Briefly, third instar larvae were irradiated with a dosage of 10 Gy. Let the irradiated larvae recovered at 25° for 1.5 h. Wing discs and eye discs were dissected in 1 × phosphate-buffered saline (PBS), fixed in 1 × PBS containing 4% formaldehyde at room temperature (RT) for 25 min. Samples were washed three times with 1 × PBS, blocked with 0.3% PBST containing 0.5% Tween-20, 1% BSA, 2% sheep serum, then stained with primary antibody of rabbit anti-histone H3pS10 (1:500, Millipore, 06-570) overnight at 4°. The secondary antibody used was anti-rabbit IgG (1:400, Cell Singnaling Technology, 4412). At least 6 wing discs or eye discs were examined for each genotype and experiments were repeated three times. The samples were analyzed on a Leica SPE5 confocal laser-scanning microscope.
Quantification by Image J software
The intensity of signals on the film was quantified using Image J software (National Institutes of Health, USA). Protein bands were measured with Uncalibrated OD mode, and the percentage of each fraction is the gray value of each band divided by total value. The confocal images of G2/M cell cycle checkpoint were qualified using Multi-point tool to count the positive cells.
Statistics
All statistical comparisons were performed using SPSS. P-values were calculated by two-tailed student′s t-tests. Data were presented as: *, P ≤ 0.05, **, P ≤ 0.01, ***, P ≤ 0.0001.
Data availability
Strains and plasmids are available upon request. All supplementary materials necessary for confirming the conclusions have been uploaded to figshare and the DOI is 10.6084/m9.figshare.6157718. Figure S1 contains the SDS-PAGE of purified proteins of Mre11, Mre11N206, Mre11M167 and Mre11C247 with Coomassie staining. Figure S2 contains the dot-blotting detection of methylation antibodies. Figure S3 contains methylated arginines detection of Mre11 in S2 cells. Figure S4 shows the ends-in scheme that was used to generate mutant flies. Figure S5 contains the southern-blot detection to verify the intermediate flies and knock-in alleles. Figure S6 contains G2/M cell cycle checkpoint detection after ionizing radiation in eye disc. Sequences of primers used in this study are listed in Table S1. File S1 contains detailed descriptions of all supplemental files. Supplemental material available at Figshare: https://doi.org/10.6084/m9.figshare.6157718.
Results
Drosophila Mre11 is methylated by DART1
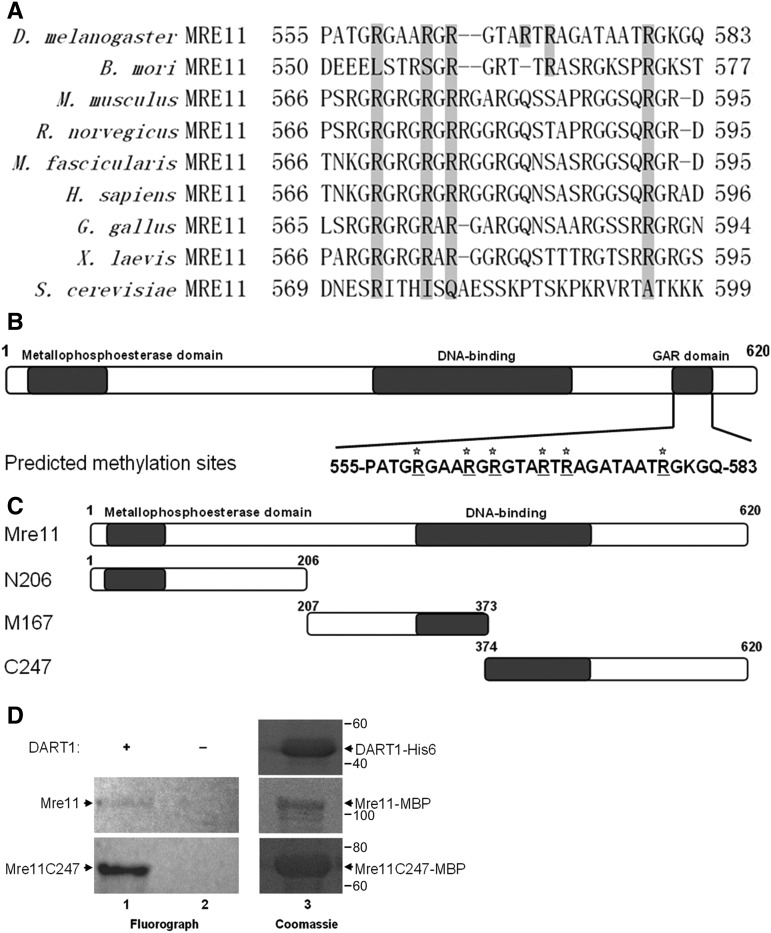
The functions of Mre11 are highly conserved in Drosophila (bi et al. 2005; bi et al. 2004; ciapponi et al. 2004). To investigate whether highly conserved Drosophila Mre11 has a GAR motif and arginines in this motif are methylated, we aligned protein sequence of Drosophila Mre11 with that of B. mori, M. musculus, R. norvegicus, M. fascicularis, H. sapiens, G. gallus, X. laevis and S. cerevisiae using Clustal W2 software (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The Drosophila Mre11 has a GAR motif at its C-terminus between Aa 555-583 (Figure 1A), and there are six arginines in the GAR motif. The following analysis using the MEMO software (http://www.bioinfo.tsinghua.edu.cn/∼tigerchen/memo.html) (chen et al. 2006) suggested that all six arginines are potential methylation sites of protein arginine methyltransferase (Figure 1B).
Figure 1.
Drosophila Mre11 is methylated by DART1. A. Sequence alignment of Mre11 from D. melanogaster, B. mori, M. musculus, R. norvegicus, M. fascicularis, H. sapiens, G. gallus, X. laevis and S. cerevisiae. Drosophila Mre11 has a Glycine-Arginine-Rich(GAR)motif at its C-terminus. B. Predicted arginine methylation in the GAR motif. There are six potential methylated arginines in the GAR motif. C. Diagram of construction of Mre11 and truncated Mre11 proteins. Mre11 protein is expressed as full length, N-terminal Aa 206 (N206), middle part (M167), and C-terminal Aa 247 (C247). D. In vitro methylation assay. Purified full length Mre11 and truncated Mre11 proteins were incubated with 3H-labeled S-adenosylmethionine (SAM) in the presence or absence of DART1. Methylated proteins were subjected to SDS-PAGE and detected by autoradiography. One representative result is shown here, at least three independent experiments were performed. 1. Fluorograph of Mre11 and Mre11C247 methylated by DART1. 2. Fluorograph of Mre11 and Mre11C247 un-methylated without DART1. 3. Coomassie staining of purified DART1, Mre11 and Mre11C247.
To prove that Drosophila Mre11 is methylated by DART1, the Drosophila homolog of PRMT1, full length of DART1, and full length, N-terminus (N206), central region (M167), C-terminus (C247) of Mre11 were expressed in E.coli and purified (Figure 1C and Figure S1). We found that full length Mre11 protein and Mre11C247 protein were methylated by DART1 using in vitro methylation assay (Figure 1D), whereas Mre11N206 and Mre11M167 were not (data not shown), indicating that Mre11 is methylated at its C-terminus by DART1, probably in the GAR motif.
Endogenous Mre11 is methylated at multiple arginines in the GAR motif
To localize the methylated arginines, peptides with single aDMA modified arginine at the Aa 559, 563, 565, 569, 571 or 579 sites were prepared and used separately to generate specific antibodies. All antibodies were incubated with peptide without modification, then affinity purified with specific arginine methylated peptide, and the specificity was verified with a dot-blot assay (Figure S2). Previous studies have shown that methylation is essential for Mre11 binding to chromatin (boisvert et al. 2005b; Dery et al. 2008), suggesting that methylated Drosophila Mre11 might localize specifically in the nucleus and perform its functions.
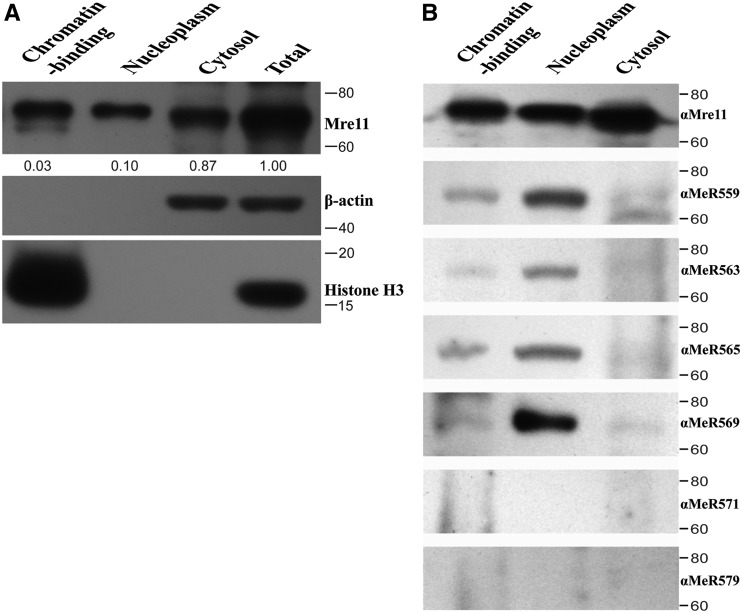
The Drosophila embryos of w1118 flies were fractionated into chromatin-binding protein, soluble nucleoplasmic protein and cytoplasmic protein. We found that Mre11 protein was not equally allocated into each fraction, 87% of Mre11 protein is cytoplasmic, 10% is soluble nucleoplasmic, and only 3% of Mre11 protein binds to chromatin, respectively (Figure 2A).
Figure 2.
Endogenous Mre11 is methylated at multiple arginines in the GAR motif. A. Subcellular localization of Mre11. Embryos of w1118 wild-type flies were fractionated and detected with anti-Mre11 antibody. Mre11 protein is not equally localized in cytoplasm, nucleocytoplasm and chromatin-binding fraction. To visualize Mre11 protein, nucleocytoplasmic fraction and chromatin-binding fraction were concentrated 2.fivefold and 6.25-fold, respectively, and quantified with Image J software. One representative result is shown here, at least three independent experiments were performed. B. Endogenous Mre11 has multiple methylated arginines recognized by specific arginine methylated antibodies, which are at Aa 559, 563, 565 and 569.
To investigate the methylation status of Mre11, we adjusted each fraction loaded on the gel to get as close amount of Mre11 protein in each fraction as possible. Arginine methylated antibodies specifically recognize R559, R563, R565 and R569 sites of Mre11, whereas antibodies of the other two arginines at R571 and R579 could not detect Mre11 protein (Figure 2B). Moreover, only soluble nucleoplasmic and chromatin-binding Mre11 protein can be detected by arginine methylated antibodies, indicating that methylated Mre11 localizes exclusively in the nucleus as the chromatin-binding protein and soluble nucleoplasmic protein. These four methylated arginine sites were also detected using the S2 cells and their nuclear localization was confirmed (Figure S3).
Knock-Down of dart1 reduces level of arginine methylated Mre11 in S2 cells
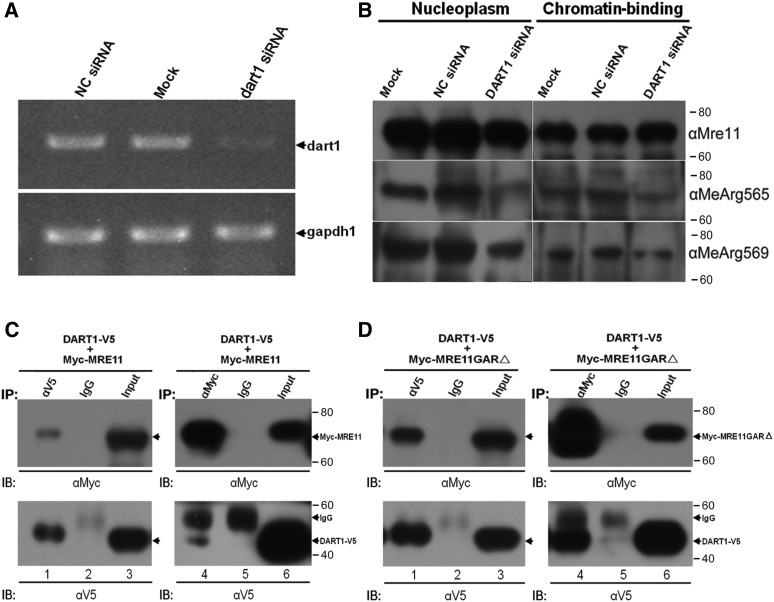
As DART1 catalyzes arginine methylation of Drosophila Mre11 in vitro, we wish to know whether knock-down of dart1 expression influences the methylation of arginines of Mre11. The S2 cells were treated with dart1 siRNA or mock treated for 48 h (Figure 3A), let the cells recover without siRNA for 24 h, and then detected with R565 and R569 specific methylation antibodies. We found that knock-down of dart1 reduced level of methylated Mre11 in S2 cells (Figure 3B).
Figure 3.
Mre11 interacts with DART1 in S2 cells which does not depend on the GAR motif. A. RT-PCR detection of dart1 expression after knock-down by siRNA in S2 cells, gapdh1 was used as the internal control. B. Arginine methylation of Mre11 was reduced when dart1 was knock-down. Arginine methylation of Mre11 in S2 cells was detected with arginine methylation antibodies specific for Arg565 (αMeArg565) and Arg569 (αMeArg569). C. Co-immunoprecipitation was performed by DART1-V5 and MRE11-Myc in S2 cells. Mre11 protein and DART1 protein were detected by Myc antibody and V5 antibody, respectively. D. Mre11GARΔ protein interacts with DART1 shown by co-immunoprecipitation.
Mre11 interacts with DART1 in S2 cells which does not require the GAR motif
Mammalian Mre11 is methylated by PRMT1, and they interact with each other (boisvert et al. 2005a; Dery et al. 2008). Our data indicated that Drosophila Mre11 is methylated by DART1 (Figure 1D), and knock-down of dart1 decreased methylated Mre11. We further investigated whether they interact with each other by performing the co-immunoprecipitation assay. Mre11 and DART1 were co-expressed in Drosophila S2 cells as Myc-Mre11 and DART1-V5 fusion proteins. Using anti-V5 antibody or anti-Myc antibody, we detected Mre11 protein or DART1 protein, respectively, whereas Mre11 protein or DART1 protein was not pulled-down by a control IgG (Figure 3C), suggesting that the Drosophila Mre11 physically interacts with DART1 in S2 cells.
We further investigated whether the GAR motif of the Drosophila Mre11 is required for its interaction with DART1. The GAR motif was removed from Mre11 (Mre11GARΔ) by mutagenesis. The Mre11GARΔ and DART1 were co-expressed in Drosophila S2 cells as Myc-Mre11GARΔ and DART1-V5 fusion proteins. Using anti-V5 antibody or anti-Myc antibody, we can still detect Mre11GARΔ protein or DART1 protein (Figure 3D). Mre11 lacking a GAR motif retains the ability to interact with DART1, indicating that the GAR motif is not required for Mre11-DART1 interaction in S2 cells.
Arginine mutated flies are sensitive to ionizing radiation
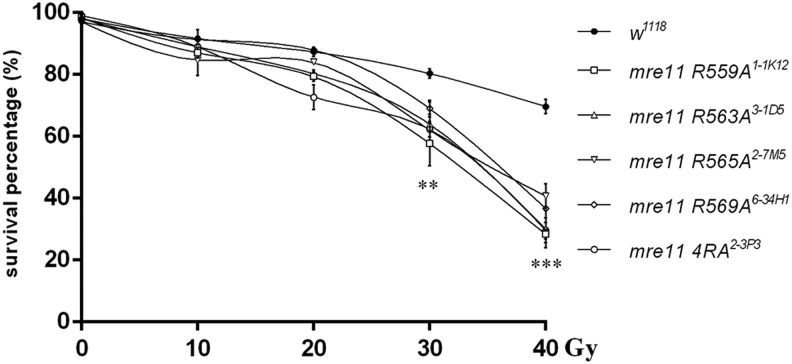
To explore the in vivo functions of arginine methylation of Mre11 in Drosophila, we generated knock-in flies of Arg-Ala (RA) point mutation and 4 Arg-Ala (4RA) mutation of the four methylated arginines, by using ends-in strategy as previously described (Figure S4) (rong et al. 2002; rong and golic 2000). The single RA and 4RA mutated mre11 flies were verified by southern-blot, cDNA sequencing and genomic DNA sequencing (Figure S5 and data not shown). All of mre11RA and mre114RA flies were viable and fertile, indicating the arginine methylation of Mre11 is not essential for fly survival. We collected the late third instar larvae of homozygote of mre11RA and mre114RA flies, treated with ionizing radiation at dosage of 0, 10, 20, 30, and 40 Gy, respectively. Let the irradiated larvae grow, counted the survival adults, and calculated the survival percentage. All mre11RA and mre114RA flies were sensitive to irradiation, however they showed different sensitivity. They were sensitive to 30 and 40 Gy (P ≤ 0.05), while mre11R563A flies were also sensitive to 20 Gy (P = 0.003), and mre11R559A and mre114RA flies were sensitive to 10 Gy (Figure 4, P ≤ 0.05). Moreover, we did not observe the additive effect in mre114RA mutant, suggesting that methylated arginines play the redundant role at DNA damage response.
Figure 4.
Flies with mre11 Arg-Ala mutation are sensitive to ionizing radiation. Late third instar larvae of w1118, mre11R559A1-1T10, mre11R563A3-1D5, mre11R565A2-7M5, mre11R569A6-34H1, and mre114RA2-3P3 were irradiated with dosage of 0, 10, 20, 30, and 40 Gy, respectively. Survival adults were counted one week after irradiation, and survival percentage was calculated as number of viable adult flies divided by number of irradiated larvae. At least 100 larvae were treated at each dosage for each genotype, and three independent experiments were performed. ** P ≤ 0.01, *** P ≤ 0.001; error bars indicate SEM.
Arginine methylation is not required for DNA damage induced G2/M cell cycle checkpoint in wing disc and eye disc
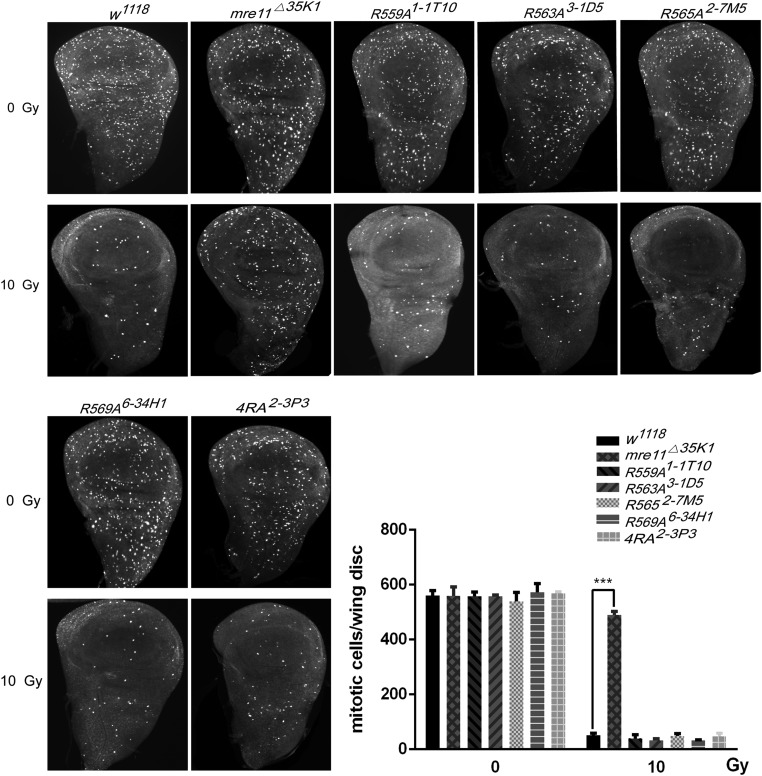
We have previously shown that mre11 knock-out flies have the DNA damage induced G2/M cell cycle checkpoint defect when treated with low-dose ionizing radiation (bi et al. 2005), we wish to know whether arginine methylation is required for DNA damage induced G2/M checkpoint control. We treated the third instar larvae with 10 Gy ionizing radiation as before (bi et al. 2005), dissected wing discs and eye discs, and stained with anti-histone H3pS10 antibody. The mre11 knock-out flies lost the G2/M cell cycle checkpoint, while all mre11RA and mre114RA flies had the normal G2/M cell cycle checkpoint as wild-type flies (Figure 5 and Figure S6), indicating that arginines in Mre11 GAR motif are not required for G2/M cell cycle checkpoint function in wing disc and eye disc.
Figure 5.
Mre11 arginines methylation is not required for irradiation induced G2/M cell cycle checkpoint. A. Photographs of wing discs of w1118, mre11Δ35K1, mre11R559A1-1T10, mre11R563A3-1D5, mre11R565A2-7M5, mre11R569A6-34H1, and mre114RA2-3P3, treated with 0 or 10 Gy, and stained with anti-histone pH 3S10 antibody. B. Quantification of mitotic cells in different genotypes. After ionizing radiation, mitosis was almost completely blocked in w1118 flies, while mre11Δ35K1 flies exhibited a strong G2/M checkpoint defect, and mitosis in mre11R559A1-1T10, mre11R563A3-1D5, mre11R565A2-7M5, mre11R569A6-34H1, and mre114RA2-3P3 flies were almost completely blocked as in w1118 flies. n ≥ 6; *** P ≤ 0.001; error bars indicate SEM.
Discussion
The mammalian Mre11 is methylated at multiple arginines in its GAR motif by protein arginine methyltransferase PRMT1, and arginine methylation is critical for its exonuclease activity (boisvert et al. 2005a; Dery et al. 2008; yu et al. 2012), irradiation induced intra-S phase checkpoint (boisvert et al. 2005a) and G2/M checkpoint (yu et al. 2012), and Mre11 foci formation (boisvert et al. 2005b; Dery et al. 2008). As Mre11 is highly conserved in eukaryotes, whether Mre11 protein in the typical model organism Drosophila has arginine methylated is still an unanswered question.
In this study, we proved that the arginine methylation of Mre11 is conserved in Drosophila. We identified 4 arginines in the GAR motif that are methylated by DART1, the Drosophila homolog of PRMT1, and methylated Mre11 localized exclusively in nucleus as nucleoplasmic protein and chromatin-binding protein, suggesting the dynamic regulation of Mre11 methylation. As the GAR motif is required for Mre11 binding to DNA damage sites in mammals, and inhibition of PRMT1 activity with the pan inhibitor MTA/ADOX abrogates the Mre11-PRMT1 interaction (Dery et al. 2008), we investigated whether the GAR motif of Drosophila Mre11 is required for its interaction with DART1. Surprisingly, the Drosophila Mre11 lacking the GAR motif retains ability to interact with DART1, suggesting that the GAR motif is not required for their interaction in Drosophila. It is possible that the pan inhibitor MTA/ADOX has effects besides inhibition of Mre11 methylation, and further study is required to identify the domain that Drosophila Mre11 uses to interact with DART1.
Similar as knock-in mre11RK/RK mice (yu et al. 2012), all of the mre11RA and mre114RA flies were viable, and showed sensitivity to ionizing radiation, which suggest that the methylation of arginines of Mre11 are not essential for viability both in mice and flies. The mre11RA and mre114RA flies did not show irradiation induced G2/M cell cycle checkpoint defect, which is contrary to that of mammalian Mre11 functions. Therefore, the physiological significance of arginine methylation of Drosophila Mre11 and the underlying mechanism of its effect on sensitivity to ionizing radiation needs to be further investigated.
Acknowledgments
We thank Dr. Yuhui Dong at Institute of High Energy Physics, Chinese Academy of Sciences for pSJ8 plasmid DNA, members of the Bi laboratory for advice and discussions. This work was supported by grants from the National Natural Science Foundation of China (Grant No. 30871388, 31771437, 31700874), Hundred Talent Program of Chinese Academy of Sciences, Pandeng Scholarship of Liaoning Province and Qizhen Gongcheng of Dalian Medical University to X.B. The authors declare that they have no conflict of interest.
Author Contributions
X. Bi designed research; Q. Yuan, R. Tian, H. Zhao and L. Li performed research; Q. Yuan, R. Tian and X. Bi analyzed data; Q. Yuan and X. Bi wrote the paper.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.6084/m9.figshare.6157718.
Communicating editor: C. Gonzalez
Literature Cited
- Bi X., Gong M., Srikanta D., Rong Y. S., 2005. Drosophila ATM and Mre11 are essential for the G2/M checkpoint induced by low-dose irradiation. Genetics 171: 845–847. 10.1534/genetics.105.047720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X., Wei S. C., Rong Y. S., 2004. Telomere protection without a telomerase; the role of ATM and Mre11 in Drosophila telomere maintenance. Curr. Biol. 14: 1348–1353. 10.1016/j.cub.2004.06.063 [DOI] [PubMed] [Google Scholar]
- Blanc R. S., Richard S., 2017. Arginine Methylation: The Coming of Age. Mol. Cell 65: 8–24. 10.1016/j.molcel.2016.11.003 [DOI] [PubMed] [Google Scholar]
- Boisvert F. M., Dery U., Masson J. Y., Richard S., 2005a Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev. 19: 671–676. 10.1101/gad.1279805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert F. M., Hendzel M. J., Masson J. Y., Richard S., 2005b Methylation of MRE11 regulates its nuclear compartmentalization. Cell Cycle 4: 981–989. 10.4161/cc.4.7.1830 [DOI] [PubMed] [Google Scholar]
- Boisvert F. M., Rhie A., Richard S., Doherty A. J., 2005c The GAR motif of 53BP1 is arginine methylated by PRMT1 and is necessary for 53BP1 DNA binding activity. Cell Cycle 4: 1834–1841. 10.4161/cc.4.12.2250 [DOI] [PubMed] [Google Scholar]
- Boulanger M. C., Miranda T. B., Clarke S., di Fruscio M., Suter B., et al. , 2004. Characterization of the Drosophila protein arginine methyltransferases DART1 and DART4. Biochem. J. 379: 283–289. 10.1042/bj20031176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Xue Y., Huang N., Yao X. B., Sun Z. R., 2006. MeMo: a web tool for prediction of protein methylation modifications. Nucleic Acids Res. 34: W249–W253. 10.1093/nar/gkl233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciapponi L., Cenci G., Ducau J., Flores C., Johnson-Schlitz D., et al. , 2004. The Drosophila Mre11/Rad50 complex is required to prevent both telomeric fusion and chromosome breakage. Curr. Biol. 14: 1360–1366. 10.1016/j.cub.2004.07.019 [DOI] [PubMed] [Google Scholar]
- Dery U., Coulombe Y., Rodrigue A., Stasiak A., Richard S., et al. , 2008. A glycine-arginine domain in control of the human MRE11 DNA repair protein. Mol. Cell. Biol. 28: 3058–3069. 10.1128/MCB.02025-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Andaloussi N., Valovka T., Toueille M., Hassa P. O., Gehrig P., et al. , 2007. Methylation of DNA polymerase beta by protein arginine methyltransferase 1 regulates its binding to proliferating cell nuclear antigen. FASEB J. 21: 26–34. 10.1096/fj.06-6194com [DOI] [PubMed] [Google Scholar]
- El-Andaloussi N., Valovka T., Toueille M., Steinacher R., Focke F., et al. , 2006. Arginine methylation regulates DNA polymerase beta. Mol. Cell 22: 51–62. 10.1016/j.molcel.2006.02.013 [DOI] [PubMed] [Google Scholar]
- Gao G., Bi X., Chen J., Srikanta D., Rong Y. S., 2009. Mre11-Rad50-Nbs complex is required to cap telomeres during Drosophila embryogenesis. Proc. Natl. Acad. Sci. USA 106: 10728–10733. 10.1073/pnas.0902707106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong M., Bi X., Rong Y. S., 2005. Targeted mutagenesis of Drosophila atm and mre11 genes. Drosoph. Inf. Serv. 88: 79–83. [Google Scholar]
- Haber J. E., 1998. The many interfaces of Mre11. Cell 95: 583–586. 10.1016/S0092-8674(00)81626-8 [DOI] [PubMed] [Google Scholar]
- Petrini J. H., Stracker T. H., 2003. The cellular response to DNA double-strand breaks: defining the sensors and mediators. Trends Cell Biol. 13: 458–462. 10.1016/S0962-8924(03)00170-3 [DOI] [PubMed] [Google Scholar]
- Rong Y. K. S., Titen S. W., Xie H. B., Golic M. M., Bastiani M., et al. , 2002. Targeted mutagenesis by homologous recombination in D-melanogaster. Genes Dev. 16: 1568–1581. 10.1101/gad.986602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y. S., Golic K. G., 2000. Gene targeting by homologous recombination in Drosophila. Science 288: 2013–2018. 10.1126/science.288.5473.2013 [DOI] [PubMed] [Google Scholar]
- Scoumanne A., Chen X., 2008. Protein methylation: a new mechanism of p53 tumor suppressor regulation. Histol. Histopathol. 23: 1143–1149. 10.14670/HH-23.1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Kao P. N., Herschman H. R., 2000. Protein-arginine methyltransferase I, the predominant protein-arginine methyltransferase in cells, interacts with and is regulated by interleukin enhancer-binding factor 3. J. Biol. Chem. 275: 19866–19876. 10.1074/jbc.M000023200 [DOI] [PubMed] [Google Scholar]
- Thandapani P., O’Connor T. R., Bailey T. L., Richard S., 2013. Defining the RGG/RG motif. Mol. Cell 50: 613–623. 10.1016/j.molcel.2013.05.021 [DOI] [PubMed] [Google Scholar]
- Yu Z. B., Chen T. P., Hebert J., Li E., Richard S., 2009. A Mouse PRMT1 Null Allele Defines an Essential Role for Arginine Methylation in Genome Maintenance and Cell Proliferation. Mol. Cell. Biol. 29: 2982–2996 (erratum: Mol. Cell. Biol. 37: e00298-17) 10.1128/MCB.00042-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z. B., Vogel G., Coulombe Y., Dubeau D., Spehalski E., et al. , 2012. The MRE11 GAR motif regulates DNA double-strand break processing and ATR activation. Cell Res. 22: 305–320. 10.1038/cr.2011.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y., Webb K., Perna A. D., Zhang Q., Clarke S., et al. , 2007. A mass spectrometric study on the in vitro methylation of HMGA1a and HMGA1b proteins by PRMTs: methylation specificity, the effect of binding to AT-rich duplex DNA, and the effect of C-terminal phosphorylation. Biochemistry 46: 7896–7906. 10.1021/bi6024897 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. All supplementary materials necessary for confirming the conclusions have been uploaded to figshare and the DOI is 10.6084/m9.figshare.6157718. Figure S1 contains the SDS-PAGE of purified proteins of Mre11, Mre11N206, Mre11M167 and Mre11C247 with Coomassie staining. Figure S2 contains the dot-blotting detection of methylation antibodies. Figure S3 contains methylated arginines detection of Mre11 in S2 cells. Figure S4 shows the ends-in scheme that was used to generate mutant flies. Figure S5 contains the southern-blot detection to verify the intermediate flies and knock-in alleles. Figure S6 contains G2/M cell cycle checkpoint detection after ionizing radiation in eye disc. Sequences of primers used in this study are listed in Table S1. File S1 contains detailed descriptions of all supplemental files. Supplemental material available at Figshare: https://doi.org/10.6084/m9.figshare.6157718.