Abstract
Histidyl-tRNA synthetase (HARS) ligates histidine to cognate tRNA molecules, which is required for protein translation. Mutations in HARS cause the dominant axonal peripheral neuropathy Charcot Marie-Tooth disease type 2W (CMT2W); however, the precise molecular mechanism remains undefined. Here, we investigated three HARS missense mutations associated with CMT2W (p.Tyr330Cys, p.Ser356Asn, and p.Val155Gly). The three mutations localize to the HARS catalytic domain and failed to complement deletion of the yeast ortholog (HTS1). Enzyme kinetics, differential scanning fluorimetry (DSF), and analytical ultra centrifugation (AUC) were employed to assess the effect of these substitutions on primary aminoacylation function and overall dimeric structure. Notably, the p.Tyr330Cys, p.Ser356Asn, and p.Val155Gly HARS substitutions all led to reduced aminoacylation, providing a direct connection between CMT2W-linked HARS mutations and loss of canonical ARS function. While DSF assays revealed that only one of the variants (p.Val155Gly) was less thermally stable relative to WT, all three HARS mutants formed stable dimers, as measured by AUC. Our work represents the first biochemical analysis of CMT-associated HARS mutations and underscores how loss of the primary aminoacylation function can contribute to disease pathology.
Keywords: aminoacyl-tRNA synthetase, histidyl-tRNA synthetase, Charcot-Marie-Tooth disease type 2W, hereditary motor and sensory neuropathy
INTRODUCTION
With a prevalence of one in 2,500 individuals, Charcot Marie Tooth disease [CMT; also referred to as hereditary motor and sensory neuropathy (HMSN)] represents the most common inherited neuromuscular disorder. CMT disease is a heterogeneous group of peripheral neuropathies mainly characterized by muscle weakness and sensory loss in the distal extremities (Skre, 1974). Several subtypes of CMT have been described, including one form (CMT1) with primary Schwann cell pathology resulting in demyelination and decreased motor nerve conduction velocities (MNCVs), a second form (CMT2) with normal MNCVs but reduced amplitudes of muscle action potentials arising from axonal degeneration, and an intermediate form that has characteristics of both CMT1 and CMT2 (Dyck and Lambert, 1968; Pareyson and Marchesi, 2009a; Pareyson and Marchesi, 2009b; Pareyson, et al., 2009). CMT typically presents in an axon-length dependent manner with the structures innervated by longer axons affected first. CMT specifically affects the extremities, causing progressive muscle weakening and wasting, decreased sensation, and skeletal deformities. The onset and penetrance of CMT is variable, with many patients experiencing symptoms in the first two decades of life. At the current time, there are no effective treatments for CMT.
Over 80 genes have been linked to CMT, distributed among diverse functional categories (Timmerman, et al., 2014). Four genes account for the majority of CMT cases, consisting of peripheral myelin protein 22 (PMP22), myelin protein zero (MPZ), and gap junction beta-1 (GJB1), which are linked to CMT1, and mitofusin 2 (MFN2), which is linked to CMT2. In addition to myelin assembly, the diverse cellular processes affected by CMT include cytoskeleton/axonal transport, protein synthesis and quality control, endosomal sorting, mitochondrial function, channel abnormalities, and mRNA/processing transcription (Bird, 1993; Jerath and Shy, 2015). While the end result of CMT-associated mutations involves impaired Schwann cell or axon dysfunction, the precise mechanisms by which alterations of individual genes bring about these phenotypes is, for many genes, not well understood (Stum, et al., 2011; Timmerman, et al., 2014).
Among the genes linked to CMT are the aminoacyl-tRNA synthetases (ARSs), which attach amino acids to cognate tRNAs in the first step of translation (Antonellis and Green, 2008; Ibba and Soll, 2000). As essential protein synthesis factors, ARSs are present in all kingdoms and are ubiquitously expressed in all tissues. The human nuclear genome contains 37 ARS genes, of which 17 encode for a cytoplasmic synthetase, 17 encode for a mitochondrial synthetase, and three encode for bi-functional enzymes that charge tRNA in both cellular compartments. Interestingly, 31 ARS genes have been implicated in myriad dominant and recessive human disease phenotypes (Meyer-Schuman and Antonellis, 2017; Oprescu, et al., 2017). Many of the diseases linked to ARS mutations are severe, recessive developmental syndromes that affect multiple organs and tissues, including the central nervous system (Frohlich, et al., 2017; Simons, et al., 2015; Zhang, et al., 2014). The majority of the disease-associated mutations have a clear loss-of-function effect, which is supported by the associated patient phenotype. For example, many ARS-associated recessive diseases are specific to mitochondrial ARS enzymes and lead to clear defects in the synthesis of electron transport chain protein synthesis and thus mitochondrial ATP output (Abbott, et al., 2014).
To date, five genes encoding a cytoplasmic or bi-functional ARS enzyme have been implicated in autosomal dominant CMT disease: glycyl-(GARS; MIM# 600287), tyrosyl-(YARS; MIM# 603623), alanyl-(AARS; MIM# 601065), histidyl-(HARS; MIM# 142810), and tryptophanyl-tRNA synthetase (WARS; MIM# 191050) (Jordanova, et al., 2006; Latour, et al., 2010; Safka Brozkova, et al., 2015; Tsai, et al., 2017; Vester, et al., 2013). Methionyl-tRNA synthetase (MARS) variants have been identified in patients with peripheral neuropathy (Gonzalez, et al., 2013)(Hirano, et al., 2016), (Hyun, et al., 2014)(Nam, et al., 2016). However, these variants were identified in single individuals or small families deeming the findings inconclusive. The first ARS implicated in CMT disease was GARS, where mutations were implicated in axonal CMT disease via positional cloning in families with an atypical disease presentation (Antonellis, et al., 2003; Sivakumar, et al., 2005). Notably, the majority of patients presented with an upper limb predominant neuropathy that affected the intrinsic muscles of the hand (Antonellis, et al., 2003; Sivakumar, et al., 2005). Subsequently, YARS, AARS, HARS, and WARS mutations were identified in families with a more typical presentation of CMT disease (Jordanova, et al., 2006; Latour, et al., 2010; Safka Brozkova, et al., 2015; Tsai, et al., 2017; Vester, et al., 2013).
Histidyl-tRNA synthetase (HARS) is unusual among the ARS in that different mutations in the gene encoding the cytoplasmic enzyme lead to two distinct inherited neurological syndromes. Initially, a single HARS mutation (P.Tyr454Ser) in the homozygous state was linked to recessive Usher Syndrome Type 3B in a small number of Amish children presenting with early onset loss of auditory and visual function (Puffenberger, et al., 2012) (Abbott, et al., 2017). Initial characterization of the mutant enzyme did not reveal a significant loss of aminoacylation, change in expression level, or change in intracellular localization. Subsequently, a candidate gene screen of 363 patients with CMT disease and no other previously acknowledged existing disease-causing alleles identified a histidyl-tRNA synthetase (HARS) missense mutation (R137Q) in a single patient with peripheral neuropathy (Vester, et al., 2013). More definitive evidence linking HARS mutations to CMT disease was provided by the identification of multi-generational pedigrees with inherited peripheral neuropathy that segregated with HARS missense mutations (T132I, P134H, D175E, and D364Y) (Safka Brozkova, et al., 2015). While previous studies demonstrated consequences of HARS mutations consistent with pathogenicity, biochemical analyses on the primary function of HARS have yet to be performed.
Here, we report detailed biochemical analyses of HARS missense mutations identified in three families with autosomal dominant CMT disease. As part of our study, each mutant protein was expressed, purified, and characterized biochemically. The results indicate that each of the three mutations leads to a clear reduction in HARS catalytic activity. These findings provide the first indication that a substantial decrease in HARS catalytic activity is associated with CMT disease. The results are discussed in light of current models linking altered ARS activity to CMT pathophysiology.
MATERIALS AND METHODS
Clinical and Genetic analysis of patients with peripheral neuropathy
Family 1
We applied whole-exome sequencing to a cohort of index patients from 82 families with genetically undefined distal hereditary motor neuropathy for which mutations in the known pathogenic genes had previously been excluded. The Ethical Review Boards of the participating institutions approved this study. All patients or their legal representatives signed informed consent prior to enrolment. Blood sampling and DNA extraction was performed according to standard methods. The Nextera Rapid Capture Expanded Exome kit (62Mb) (Illumina) was used for exome enrichment. Subsequently, the libraries were sequenced on a HiSeq 2500 platform (Illumina). The Burrows-Wheeler Aligner (BWA) tool was used to perform the sequence alignment to the reference genome (hg19, UCSC Genome Browser). Variant calling was done with Genome Analysis Toolkit (GATK) Unified Genotyper. For the annotation and filtering we used the Clinical Sequence Analyzer and Miner (Wuxi NextCODE). For further filtering of the data following criteria were applied: no occurrence or a frequency ≤ 0.5% of the variants in public exome variant repositories (Exome Aggregation Consortium, 1000 Genomes Project, Exome Variant Server, in-house data); variants with impact on the encoded protein (missense, nonsense, frame shift, inframe indels and splice site variants); read depth ≥ 7; minimal heterozygous call percentage ≥ 20%, minimal homozygous call percentage ≥ 66%. In addition, all data were imported and re-annotated into the GENESIS (gem.app) platform, a web-based tool for next generation sequencing data analysis (http://thegenesisprojectfoundation.org/) (Gonzalez, et al., 2015). Patients were evaluated by means of details clinical examination in combination with nerve conduction studies according to standard methods.
Family 2
All family members were studied at Bellvitge’s University Hospital. Detailed neurological evaluation was performed on patients and unaffected subjects. Electrophysiological studies were performed using standard methods in all subjects. We classified the severity of neuropathy symptoms according to the CMT neuropathy score (CMTNS). Blood samples were obtained from all family members once informed consent had been given. Genomic DNA was extracted from peripheral blood leucocytes. PMP22, MPZ, MFN, and GJB1 mutations were previously excluded in the index case. Subsequently, a CMT gene panel screen was employed to identify candidate disease-causing variants.
Family 3
The family was evaluated in the University of Iowa CMT Clinic. Informed consent was obtained from all individuals and the study obtained the approval of the Institutional Review Board at the University of Iowa. Neurological examination and neurophysiological studies were performed. The second version of the CMT Neuropathy Score (CMTNSv2) and CMT Exam Score (CMTESv2) [1] were employed to evaluate neuropathy. The SureSelect Human All Exon 50 MB Kit (Agilent) was used for in-solution enrichment, and the HiSeq 2500 instrument (Illumina) was used to produce 100 bp paired-end sequence reads. The Burrows-Wheeler aligner, Picard, and the Genome Analysis Tool-kit were used to align sequence reads and call variants. These data were imported into GENESIS (GEM.app) (Gonzalez, et al., 2015) for further analysis and filtering of variants.
The following HARS variants were identified in each of the three families: c.464T>G; p.Val155Gly (Family 1); c.989A>G; p.Tyr330Cys (Family 2); and c.1067G>A; p.Ser356Asn (Family 3). HARS variant nomenclature for nucleotide and amino acid changes are based on accession numbers NM_002109.5 and NP_002100.2, respectively. The three HARS variants were submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) under the accession numbers SCV000611612 (p.Val155Gly), SCV000611613 (p.Tyr330Cys), and SCV000611614 (p.Ser356Asn).
Computational assessment of HARS mutations
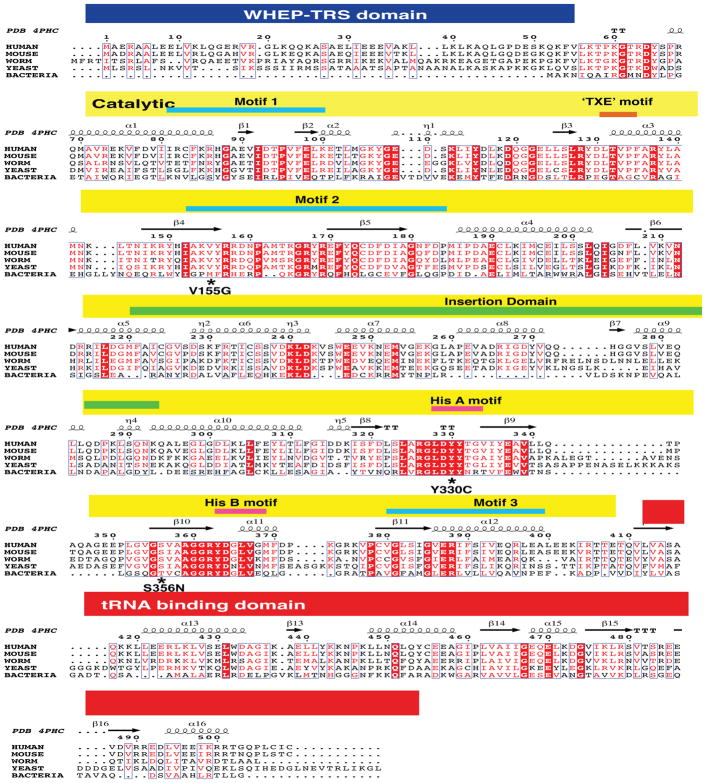
HARS protein sequences were collected from the NCBI Protein Database (http://www.ncbi.nlm.nih.gov/protein/) for the indicated species using the following accession numbers: human (Homo sapiens, NP_002100.2), mouse (Mus musculus, NP_032240.3), worm (Caenorhabditis elegans, NP_001023373.1), yeast (Saccharomyces cerevisiae, EDN61168.1), and bacteria (Escherichia coli, NP_289067.1). Multiple-species amino-acid sequence alignments were generated using Clustal Omega software (Larkin, et al., 2007) and annotated with quaternary structural information by ENDscript (Robert and Gouet, 2014). Rendering of human HARS 3D protein structures and neuropathy-associated substitutions was performed by use of PYMOL using the coordinates corresponding to the structures of apo and histidine bound human cytosolic HARS (PDB IDs: 4PHC and 4X5O). The modeling of ATP into the active site of the human HARS-histidine complex was performed by aligning this structure to the E. coli HARS-ATP complex (PDB 1KMN) using the Ce_align subroutine in PyMol. PyMol sessions of the aligned structures are available upon request. The PDB IDs used in this study include: 4PHC (Fig. 2); 4X5O (Fig. 8A); 4X5O and 1KMN; (Fig. 8B); 4X5O and 1KMN (Fig. 8C); 4X5O and 4PHC (Fig. 8D); 4X5O (Fig. 8E); and 4X5O and 1KMN (Fig. 8F).
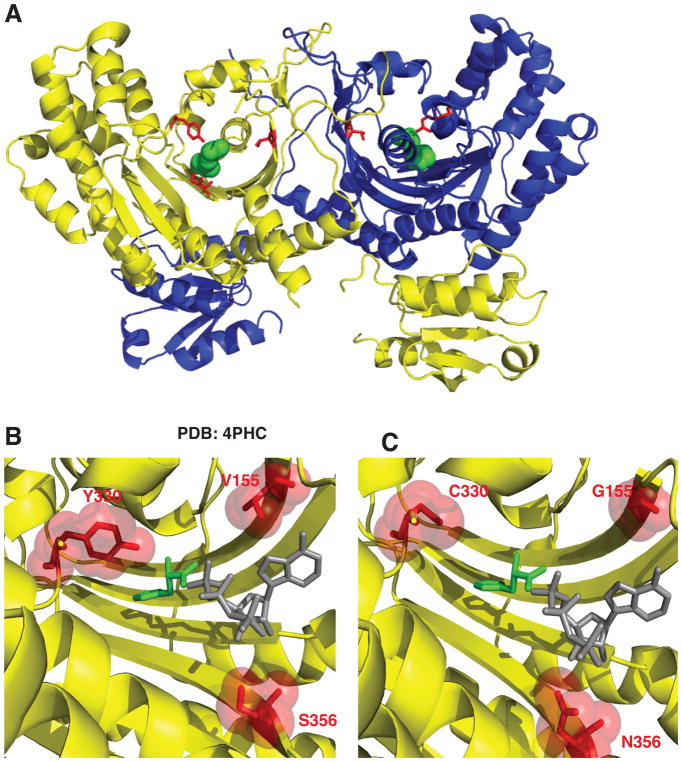
Figure 2. Histidyl-tRNA synthetase CMT variants are found in the active site of the dimeric enzyme.
(A) Histidyl-tRNA synthetase forms a homodimer (first monomer yellow and second monomer blue). The neuropathy-associated HARS residues are shown in the 3D structure as red sticks of the HARS dimer bound to histidine (green spheres) (PDB 4PHC). (B) The HARS active site pocket (yellow) reveals that neuropathy-associated mutations face into the active site (shown as red sticks and spheres). (C) Modeling in HARS neuropathy-associated substitutions indicates that interference with substrate binding (histidine green sticks and ATP gray sticks) can occur. ATP was modeled into the active site of the human HisRS bound to histidine (PDB 4PHC) by aligning the HisRS E. coli structure bound to ATP (PDB 1KMN).
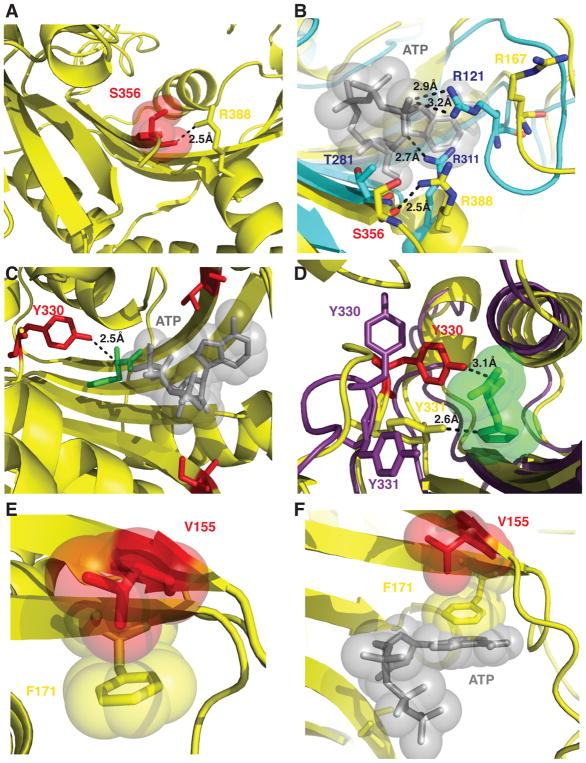
Figure 8. Molecular interactions of HARS neuropathy-associated amino acid residues in the active site.
(A) S356 is positioned in the active site to coordinate a critical arginine residue R388 in motif 3. (B) R388 is highly conserved and corresponds to R311 in E. coli HisRS structure (cyan) that is responsible for positioning the γ phosphate of ATP adjacent R121. (C) Y330 is situated 2.5 Å from the alpha carbon of histidine (green) but makes no immediate interactions with modeled ATP (gray sticks and spheres). (D) Y330 and Y331 facilitate hydrogen bonding interactions with histidine. Y330 in the apo enzyme (shown as purple sticks) can flip away from the active site and swing 8 Å upon histidine binding (shown as red sticks) to facilitate hydrogen bonding interactions. (E) Phe 171 forms a stacking interaction with the ribose ring of ATP (as modeled in the active site as grey sticks and spheres). (F) p.Val155Gly is buried in the active site and does not mediate any direct interactions with either substrate histidine or ATP, but in the apo enzyme forms a CH-Pi interaction with the aromatic ring of Phe 171 within the active site.
Yeast complementation assays
Yeast complementation assays to study the functional consequences of HARS mutations were generated and performed as previously described (Vester, et al., 2013). Each indicated missense variant was modeled in the yeast HARS ortholog HTS1 or in the human HARS open-reading frame (primers available upon request). After mutagenesis, wild-type or mutant HARS or HTS1 were cloned into pRS316 or pYY1 (Pierce, et al., 2011), respectively, using the QuickChange II XL Site-Directed Mutagenesis Kit (Stratagene). Resulting expression clones were purified and fully sequenced to confirm successful mutagenesis and rule out PCR-induced errors. A haploid ΔHTS1 strain (harboring a maintenance vector to express wild-type HTS1 and URA3) was transformed with an empty vector (‘Empty’ in Fig. 4A and B) or the appropriate wild-type or mutant HTS1 or HARS in a LEU2-bearing vector and selected on medium lacking uracil and leucine (Teknova). For each transformation, at least two independent plasmid preparations were used and at least two colonies from each plasmid were selected for additional analysis and grown to saturation for 2 days at 37°C in liquid media lacking uracil and leucine. A 10 μl aliquot of each culture was spotted undiluted or diluted 1:10 or 1:100 in H2O onto plates containing 0.1% 5-FOA (Teknova) or medium lacking uracil and leucine and incubated at 30°C for 48–72 hours. Survival was determined by visual inspection of growth.
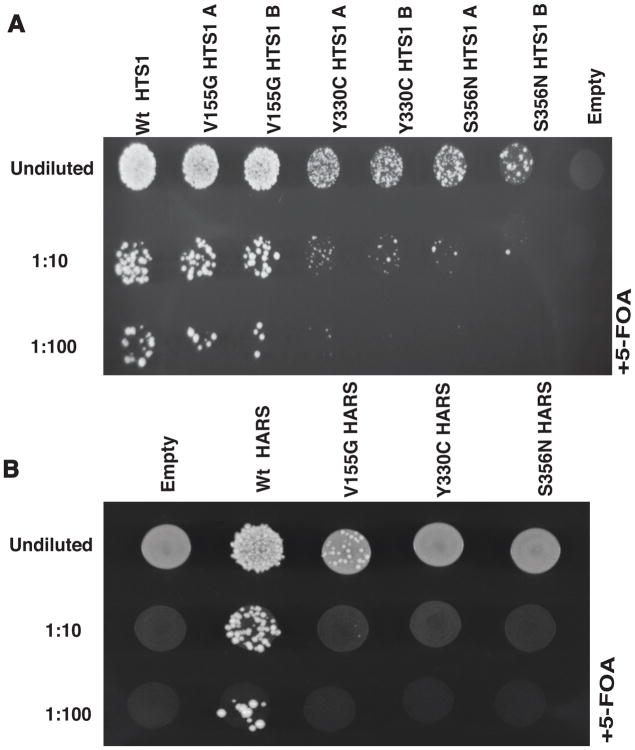
Figure 4. Neuropathy-associated HARS mutations result in loss-of-function in vivo.
(A) Yeast complementation analysis of HARS variants. Haploid ΔHTS1 yeast strains were transformed with a vector containing no insert (‘Empty’) or an insert to express wild-type, p.Val155Gly, p.Tyr330Cys, or p.Ser356Asn HTS1. Two colonies (indicated by ‘A’ and ‘B’) from transformations with p.Val155Gly, p.Tyr330Cys, or p.Ser356Asn HTS1 are shown. Resulting colonies (undiluted, diluted 1:10, or diluted 1:100) were grown on agar plates containing complete media with 0.1% 5-FOA. Note the severe depletion of growth associated with p.Tyr330Cys and p.Ser356Asn HTS1 in the 1:10 and 1:100 dilutions. (B) Similar yeast complementation assays as describe in A using the human HARS open-reading frame. Haploid ΔHTS1 yeast strains were transformed with a vector containing no insert (‘Empty’) or an insert to express wild-type, p.Val155Gly, p.Tyr330Cys, or p.Ser356Asn HARS. After transformations, colonies (undiluted, diluted 1:10, or diluted 1:100) were grown on agar plates containing complete media with 0.1% 5-FOA. Note the severe depletion of growth associated with p.Val155Gly HARS in the undiluted sample and the lack of growth in the 1:10 and 1:100 dilutions.
Expression and purification of human HARS enzymes
Neuropathy-associated HARS mutations p.Ser356Asn, p.Tyr330Cys and p.Val155Gly HARS containing plasmids were generated by QuickChange II Site-Directed Mutagensis (Qiagen) using the (pCAG/FLAG/RFC/A) plasmid with CAG promoter and FLAG-tag from GateWay vector Reading Frame Cassette A as a template (a generous gift from Dr. Robert Jinks) containing the gene for human WT N-terminal FLAG-tagged HARS. The following forward and reverse primers were used in the mutagenesis reactions for p.Ser356Asn, forward 5′-CCAGCAGCCACATTGCCC ACACCCAGG-3′ and reverse 5′-CCTGGGTGTGGGCAATGTGGCTGCTGG-3′, for p.Tyr330Cys forward 5′-TCACCCCAGTGTAGCAATCCAGCCCTCGAG-3′and reverse 5′-CTCGAGGGCTGGATTG CTACACTGGGGTGA-3′ reverse, for p.Val155Gly 5-GGTTATCCCGCCGATATCC CTTTGCTATGTGGTAG-3′ and reverse 5′-CTACCACATAGCAAAGGGATATCGG CGGGATAACC-3′. Successful mutagenesis of the HARS gene was analyzed by the Advanced Genome Technology Core sequencing facility at the University of Vermont and results were validated using Sequencher (Gene Codes Corporation). HEK293 cells were transiently transfected with plasmids expressing N-terminal FLAG-conjugated HARS gene for either WT or neuropathy-associated HARS mutants. Enzymes were purified by affinity and ion exchange chromatography as previously described (Abbott, et al., 2017). Purified proteins were visualized by on a 10% SDS PAGE gel.
Multiple turnover aminoacylation kinetics
Multiple turnover aminoacylation assays were performed using a modified version of the Uhlenbeck-Wolfson assay as previously described (Abbott, et al., 2017; Wolfson, et al., 1998). Multiple turnover experiments were conducted in a buffer composed of (50 mM HEPES pH 7.5, 150 KCl, 10 mM MgCl2, 5 mM β-ME, 2 U/ml PPiase and 32P-labeled tRNAHis) at a fixed concentration of enzyme 5 nM for WT and (20 nM) for CMT variants with saturating concentrations of two of the three substrates. The saturating concentrations of tRNAHis, ATP, and histidine were 5 μM, 10 mM, and 5 mM, respectively. The variable concentrations of these substrates were 100 nM −15 μM, 25 μM – 5 mM, and 1 μM – 5 mM, for tRNAHis, ATP and histidine, respectively. The concentration ranges tested for the mutant proteins typically employed a 2–3 fold higher final concentration for the variable substrate relative to the ranges employed for the wild type protein. Initial rates were typically sampled over the first minute of the reaction. Reaction products were detected by radioisotopic imaging on a Phosphor Imaging screen (Bio-Rad Molecular Imager FX™). The concentration of aminoacylated tRNAHis was quantified by comparing the ratio of the relative amount of aminoacylated A76 (aa) (determined as counts * mm2) to total radiolabeled product (aa + A76) according to equation (1):
Differential scanning fluorimetry
The DSF experiments were carried out as previously described (Abbott, et al., 2017). Briefly purified HARS proteins were incubated with 6X SYPRO orange dye and DSF buffer (25 mM HEPES pH 7.5, 50 mM KCl) at an enzyme concentration of 10 μM in a 96-well microplate (Costar) to a final volume of 20 μL. Tm values for apo enzymes were determined for two biological preparations of enzyme in triplicate for a final of six replicates. Enzymes were also incubated with either 10 mM histidine (n=5), 5 mM ATP (n=2), or 10 μM tRNAHis for WT and 20 μM tRNAHis (n=3) for peripheral neuropathy-associated HARS mutations. Scan rates were initiated at 26 °C and heated at increments of 1 °C per minute, to 95 °C while fluorescence intensity was measured every 1 °C. We fit the fluorescence data with GraphPad Prism 7 software as previously described (Abbott, et al., 2017) to determine melting temperatures of HARS neuropathy-associated enzyme mutations and substrate complexes.
The melting temperature (Tm), defined as the midpoint of the protein unfolding transition curve, was determined using the Boltzman model as previously described (Niesen, et al., 2007). GraphPad Prism 7 software was used to fit the fluorescence data (excluding data after maximal fluorescence intensity) to the equation (3):
Analytical centrifugation sedimentation velocity experiments and analysis
Analytical centrifugation experiments were performed as previously described (Abbott, et al., 2017). Briefly, purified enzymes (WT, p.Tyr330Cys, p.Val155Gly and p.Ser356Asn) were dialyzed into a buffer containing 10 mM potassium phosphate buffer, 50 mM KCl, and no reducing agents. All protein samples were concentrated to 0.3 OD units/mL at 230 nm (0.7 μM). Sedimentation velocity experiments were performed by the Center for Analytical Ultracentrifugation of Macromolecular Assemblies at the University of Texas Health Science Center at San Antonio, and conducted at 20 °C, 35 K rpm, in a Beckman Optima XLI analytical ultracentrifuge using an An60Ti rotor and standard 2-channel epon centerpieces (Beckman-Coulter) and measured by UV intensity.
All data were examined with UltraScan-III ver. 3.5, release 2170 (Brookes E., 2006; Demeler, 2016) and hydrodynamic corrections for buffer density and viscosity was estimated to be 1.0019 g/ml and 0.998 cP, respectively. The partial specific volume of HARS (0.745 ml/g) was estimated by UltraScan from protein sequence analogous using methods outlined in Laue et al. (Laue, et al., 1992). Experimental sedimentation data were pre-processed by 2-dimensional spectrum analysis (2DSA) (Brookes, et al., 2010; Demeler, 2010; Schuck, 1999) and fitted by the parametrically constrained spectrum analysis (PCSA), using a straight-line parameterization coupled with a Monte Carlo approach (Demeler, 2008; Gorbet, et al., 2014). The calculations (Brookes, 2008) were performed on the Lonestar cluster at the Texas Advanced Computing Center at the University of Texas at Austin and on Comet and Gordon at San Diego Supercomputing Center.
RESULTS
Clinical and genetic analysis of patients with peripheral neuropathy
Family 1
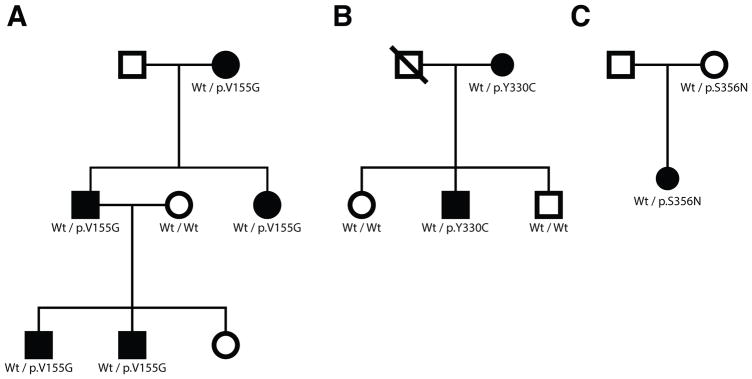
A three-generation family of Persian-Jewish descent (living in Israel) was identified with peripheral neuropathy (Fig. 1A). Five affected individuals are present and the family displays an autosomal dominant inheritance pattern with male-to-male transmission. The age of onset was in the second decade of life for the two youngest individuals, but the age of onset was less clear for the older individuals. In all cases, the affected status was clear upon examination. All individuals display a motor predominant to pure motor phenotype with clear pyramidal features as assessed by brisk reflexes and ankle clonus/Babinski signs. With one exception, all patients had normal sensory exams. At least two of the elder individuals have severe weakness necessitating the use of a cane or a wheelchair; however, the younger individuals are ambulatory. Nerve conduction studies are consistent with a motor predominant axonal neuropathy. There is some co-morbidity, as the female patient from the second generation has systemic lupus and one of the males in the third generation has uveitis, hematuria, and proteinuria.
Figure 1. Histidyl-tRNA synthetase mutations identified in three pedigrees with peripheral neuropathy.
(A–C) Genotyping was performed to determine if HARS variants segregate with disease status. The pedigree structures of Family 1 (A), Family 2 (B), and Family 3 (C) are shown. Circles represent female individuals and squares represent male individuals. Shaded symbols represent affected individuals and non-shaded symbols represent unaffected individuals. Diagonal lines indicate deceased individuals. Genotypes are indicated under each individual where DNA was available.
Whole-exome sequencing was performed on one affected individual, which revealed common or non-segregating variants in SLC5A7, SETX, and LAMA2 (the variant in LAMA2 was not consistent with a dominant neuropathy). This analysis also revealed p.Val155Gly HARS, which fully segregated with the disease phenotype upon PCR and Sanger sequencing of additional individuals. Unfortunately, DNA was not available for the unaffected individual in the third generation. p.Val155Gly HARS affects an amino-acid residue found in the catalytic domain of the HARS enzyme (Fig. 2A–C) and that is conserved from human to yeast (Figure 3). p.Val155Gly HARS has not been previously reported nor has it been detected in the gnomAD database (Table 1) (Lek, et al., 2016).
Figure 3. Neuropathy-associated HARS mutations are highly conserved.
(A) Multiple-species protein alignments were generated to assess the conservation of each affected amino-acid position. Complete amino acid sequence alignment of human, mouse, worm, yeast, and bacterial HARS were generated with Clustal Omega and annotated with quaternary structural information with ENDscript (Robert and Gouet, 2014). The position of known functional domains of the HARS protein indicated in blue (WHEP-TRS domain of unknown function), red (catalytic core), and yellow (tRNA binding domain) and motifs critical for substrate binding are indicated along the top. For each of the three variants, the affected amino acid is indicated by an * in the HARS protein sequence in multiple, evolutionarily diverse species. Note that strictly conserved residues are highlighted in red with white lettering while slightly conserved residues across multiple but not all species are red.
Table 1.
HARS variants identified in patients with peripheral neuropathy
| Amino acid changea | Detection in gnomADb | dbSNP accession no. | Position in yeast HTS1c | Reference |
|---|---|---|---|---|
| p.Thr132Ile | Not detected | None | p.Thr131Ile | (Safka Brozkova, et al., 2015) |
| p.Pro134His | Not detected | None | p.Pro133His | (Safka Brozkova, et al., 2015) |
| p.Arg137Gln | 20 / 246,154 | rs191391414 | p.Arg136Gln | (Vester, et al., 2013) |
| p.Val155Gly | Not detected | None | p.Val154Gly | This study |
| p.Asp175Glu | Not detected | None | p.Asp174Glu | (Safka Brozkova, et al., 2015) |
| p.Tyr330Cys | Not detected | None | p.Tyr330Cys | This study |
| p.Ser356Asn | 5 / 277,222 | rs144322728 | p.Ser370Asn | This study |
| p.Asp364Tyr | Not detected | None | p.Asp378Tyr | (Safka Brozkova, et al., 2015) |
Human amino acid positions are relative to GenBank Accession number NP_002100.2
Yeast amino acid coordinates correspond to GenBank accession number EDN61168.1
Family 2
A two-generation pedigree was identified with peripheral neuropathy (Fig. 1B). The age of neuropathy onset for both patients was during childhood. Both individuals show a motor predominant phenotype, with distal motor deficit and atrophy, hammer toes, and pes cavus. Both affected patients have mild sensory symptoms in the toes, with reduced vibration sense at the knees. Tendon reflexes were brisk in both patients. The mother (patient 1) is restricted to a wheelchair while her son (patient 2) walks with difficulty but without aid. The CMT Neuropathy Score (CMTNS) for patient 1 was 22 and 9 for patient 2. The phenotype of patient 1 is likely exacerbated by concomitant diseases (diabetes, chemotherapy for melanoma, radiculitis secondary to herpes zoster and Parkinson’s disease). Nerve conduction studies show axonal motor-predominant neuropathy in both patients.
Targeted gene panel screening in Family 2 (Fig. 1B) revealed two missense HARS variants in cis: p.Ser227Ala and p.Tyr330Cys. These variants were previously reported as part of a larger study on CMT disease (Lupo, et al., 2016) but the functional consequences of the mutations were not assessed. Both mutations segregate with the disease phenotype; however, p.Ser227Alawas deprioritized due to the number of alleles detected in gnomAD (37 alleles in 282,464 alleles total) and the lack of conservation of the affected residue (see Fig. 3). p.Tyr330Cys HARS affects an amino-acid residue in the catalytic core of the HARS enzyme (Fig. 2B) that is conserved from human to bacteria (Fig. 3). p.Tyr330Cys has not been previously reported nor has it been detected in the gnomAD database (Table 1) (Lek, et al., 2016).
Family 3
A simplex pedigree was identified with two unaffected parents and a single daughter affected with peripheral neuropathy (Fig. 1C). The affected daughter, at 15 years of age, was seen in consultation for difficulty with walking. Her early milestones were normal including walking with a normal gait at 12 months. She was physically active as a child and able to keep up with her peers until the age of 10. At 12 years of age she noticed she could no longer walk on her heels. At ~13 years of age she developed difficulties with walking. This progressed and she had problems with other activities such as jumping and running by the age of 15. She had ankle weakness, a foot drop, and right knee pain. She began to wear foot orthotics bilaterally. No problems with hand function were reported, but a slight hand tremor was reported and observed. Her sensory exam was normal to pinprick, light touch, and joint position sense in all four extremities. Vibration sensation was also normal with the exception of a slight reduction at her toes. Her feet turned in bilaterally but could be brought to a neutral position. She had high arches and atrophy noted in her hands and distal forearms. Her CMT Neuropathy Score (CMTNS) (Burns, et al., 2012) was a 10, which is in the high end of the mild range. Her CMT Pediatric Score (CMTPeds) (Shy, et al., 2005) was a 33, which is in the moderately impaired range.
Whole-exome sequencing was performed on the affected daughter in Family 3 (Fig. 1C), which revealed p.Ser356Asn HARS as the only candidate disease-associated variant. Sanger sequencing revealed that this variant was inherited from the unaffected mother suggesting decreased penetrance or that this is a non-pathogenic variant. p.Ser356Asn HARS is also found in the active site of the HARS enzyme (Fig. 2B, C) and that is conserved from human to yeast (Fig. 3). p.Ser356Asn HARS has not been previously reported; however, it has been detected in the gnomAD database (5 alleles in 277,222 chromosomes; Table 1) (Lek, et al., 2016).
The identified HARS mutations affect viability in yeast complementation studies
To test the functional consequences of the HARS missense variants described above (p.Val155Gly, p.Tyr330Cys, and p.Ser356Asn), each variant was introduced into the yeast ortholog HTS1 to model the effect of the mutation in the context of a eukaryotic cell (Table 1). Yeast complementation assays were then performed to independently test each missense change for the ability to support yeast cell growth compared to wild-type HTS1 or an empty vector. A haploid yeast strain (with the endogenous HTS1 locus deleted and a maintenance vector to express wild-type HTS1 and URA3) was transformed with either a pRS315 vector with no insert (‘Empty’), a pRS315 vector harboring wild-type HTS1, or a mutant version encoding one of the three missense mutations (p.Val155Gly, p.Tyr330Cys, or p.Ser356Asn). Yeast cells were then selected on media containing 5-FOA, which is toxic to yeast carrying the URA3-bearing maintenance vector (Boeke, et al., 1984). Thus, only yeast cells expressing a functional HTS1 allele from the pRS315 vector will grow in this assay.
Yeast transformed with a wild-type HTS1 expression vector demonstrated significant growth, while those transformed with an empty vector did not (Fig. 4A), consistent with HTS1 being an essential gene (Vester, et al., 2013). Yeast expressing p.Tyr330Cys or p.Ser356Asn HTS1 displayed severely depleted, but not ablated, yeast cell growth (Fig. 4A), indicating that these are hypomorphic alleles. In contrast, p.Val155Gly HTS1 supported yeast cell growth in a manner similar to wild-type HTS1 (Fig. 4A). Growth at 37 OC did not affect the cellular phenotype associated with the above mutations (data not shown). In summary, our previously employed in vivo yeast complementation assay demonstrated that p.Val155Gly HTS1 supports growth similar to wild-type HTS1, and that S365N and p.Tyr330Cys HTS1 are hypomorphic alleles in this assay.
The lack of a cellular growth defect associated with p.Val155Gly HTS1 was surprising given that this mutation segregates with disease in a three-generation family. One possible explanation for this discrepancy may be differential effects of p.Val155Gly on the function of yeast HTS1 compared to human HARS. To explore this possibility, we attempted to rescue deletion of yeast HTS1 with the full-length human HARS protein. In yeast complementation assays similar to those described above, expressing human HARS rescued yeast cell growth while a vector with no HARS insert did not (Fig. 4B).This confirms a similar finding that was recently reported (Lee, et al., 2017). We next mutated the HARS expression construct to harbor the three missense mutations (p.Val155Gly, p.Tyr330Cys, or p.Ser356Asn) and found that p.Tyr330Cys and p.Ser356Asn HARS did not support any yeast cell growth indicating that they are functional null alleles (Fig. 4B). Interestingly, p.Val155Gly HARS supported growth, but in a manner that is severely reduced compared to wild-type HARS (Fig. 4B). These data indicate that p.Val155Gly HARS is a hypomorphic allele. Combined, our in vivo functional analyses indicate that p.Val155Gly, p.Tyr330Cys, and p.Ser356Asn HARS are loss-of-function alleles.
Multiple-turnover kinetics identify specific catalytic deficiencies of HARS mutations
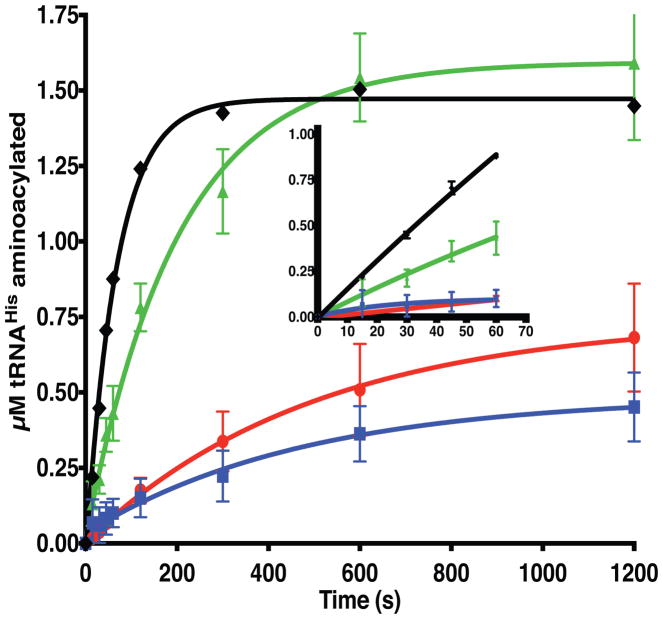
The results of the yeast complementation assays and the location of these neuropathy-associated HARS mutations in the catalytic domain raised the possibility that these substitutions compromise catalytic function. As an initial characterization of catalytic function, the production of histidyl-tRNA by each mutant and a wild type control was monitored over a ten-minute time course in the presence of saturating concentrations of all three cognate substrates. In excellent agreement with the yeast complementation results, the two non-complementing mutants (p.Ser356Asn and p.Tyr330Cys HARS) generated about 20% of the amount of charged tRNAHis generated by wild-type and p.Val155Gly HARS in ten minutes (Fig. 5). Despite its near wild type level of product accumulation after ten minutes, the initial rate of product formation by the p.Val155Gly mutant was approximately 50% of the wild type protein, indicating that this mutant is at least partially compromised for aminoacylation (Fig. 5, inset).
Figure 5. Neuropathy-associated HARS mutations reduce aminoacylation function.
Progress curves for aminoacylation of tRNAHis substrates with histidine by wild type or mutant HARS enzymes under conditions of excess substrates and limiting enzyme. Reaction conditions utilized saturating substrates (10 mM ATP, 10 mM histidine, and 10 μM tRNAHis) with either wild type or mutant HARS enzymes. The progress of the reaction was followed by detection of aminoacylated 32P radiolabeled over time, as described in “Materials and Methods.” The progress curves are color coded as follows: wild-type HARS (black diamonds, ◆); p.Val155Gly HARS (green triangles, ▲); p.Tyr330Cys HARS (red circles, ●), and p.Ser356Asn HARS (blue squares, ■). Each point represents the mean of three independent experiments, and error bars indicate the standard error. Inset, progress curves over the first sixty seconds.
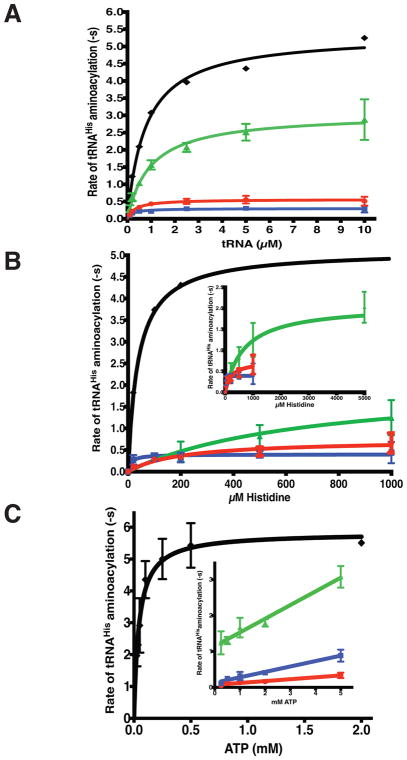
In order to assess the effect of the mutant substitutions on the recognition of individual substrates, the 32P-tRNA aminoacylation experiments were repeated under multiple turnover conditions employing fixed nanomolar concentrations of enzyme and varying substrate concentrations. In the first set of kinetic experiments, aminoacylation was monitored in presence of varying concentrations of tRNA and saturating concentrations of histidine and ATP. This set of experiments showed that, while none of the mutants exhibited an elevated KM for tRNAHis, p.Tyr330Cys and p.Ser356Asn HARS exhibited 9-fold and 18-fold reductions (0.6 s−1 and 0.3 s−1, versus 5.4 s−1 for WT) in the steady state kcat for aminoacylation, relative to wild type HARS (Fig. 6, Table 2). By comparison, the p.Val155Gly HARS mutant exhibited a modest two-fold reduction in kcat (Fig. 6, Table 2). These results confirm and extend the previous yeast complementation and product formation assays.
Figure 6. Identification of specific catalytic defects in neuropathy associated HARS CMT mutants by steady state kinetics.
Multiple turnover aminoacylation reactions were performed under Michaelis-Menten conditions of excess substrates and limiting enzyme as described in “Materials and Methods.” Initial velocity of product formation was plotted against substrate concentration for A, tRNA; B, Histidine, and C, ATP. Fits to the Michaelis-Menten equation returned kcat and KM values, which are reported in Table 2. The plots are color coded as in Figure 5: wild-type HARS (black diamonds, ◆); p.Val155Gly HARS (green triangles, ▲); p.Tyr330Cys HARS (red circles, ●), and p.Ser356Asn HARS (blue squares, ■). When ATP was the variable substrate, none of the mutants reached a saturating velocity at the highest ATP concentration (5 mM). The inset shows linear fits of the V/S data for the mutants to allow an estimation of kcat/KM. Each point represents the mean of three independent experiments, and error bars indicate the standard error.
Table 2.
Steady state kinetics of tRNAHis aminoacylation by human and neuropathy-associated mutations of histidyl-tRNA synthetases
| Variable Substrate | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| tRNA | histidine | ATP | ||||||||
| Km (μM) | kcat (s−1) | kcat/Km (μM−1 s−1) | Km (μM) | kcat (s−1) | kcat/Km (μM−1 s−1) | Km (μM) | kcat (s−1) | kcat/Km (μM−1 s−1) | References | |
| WT HARS | 0.782 ± 0.101 | 5.4 ± 0.2 | 6.9 | 8.0 ± 4.0 | 4.1 ± 0.4 | 0.5 ± 0.4 | 44.2 ± 5.5 | 5.8 ± 0.2 | 0.13 | ATP data this work |
| p.Ser356Asn HARS | 0.199 ± 0.059 | 0.30 ± 0.02 | 1.5 | 10.8 ± 9.1 | 0.39 ± 0.04 | 0.04 | ND | ND | 1.5x10−4 ± 1.2x10−5 | This work |
| p.Tyr330Cys HARS | 0.330 ±0.072 | 0.56 ± 0.03 | 1.7 | 202.9 ± 83.23 | 0.74 ± 0.10 | 0.004 | 1,763 ± 544 | 0.48 | 0.000272 | This work |
| p.Val155Gly HARS | 0.979±0.159 | 3.05 ± 0.14 | 3.1 | 687.2 ± 200.0 | 2.08 ± 0.16 | 0.003 | ND | ND | 3.8x10−4 ± 2.7x10−5 | This work |
| E. coli HisRS | 0.34 ±0.05 | 1.71 ± 0.06 | 5.0 | 35.4 ± 3.7§ | 133 ± 2.2§ | 3.8§ | 380 | ND | ND | (Fahoum and Yang, 1987) |
Values reported are the mean ± standard error of three independent experiments.
Data are for histidine in the pyrophosphate exchange reaction.
When histidine was the variable substrate and tRNA and ATP were the fixed concentration substrates, the turnover numbers for each of the mutants were similar to the values obtained in experiments where tRNA was the variable substrate (Fig. 6, Table 2). The most significant kinetic differences were seen with the p.Tyr330Cys and p.Val155Gly HARS mutants. The KM for histidine was elevated 25-fold for p.Tyr330Cys HARS, and at least 86-fold for p.Val155Gly HARS (Fig. 6, and Table 2). These results illustrate that, despite the fact that p.Tyr330Cys and p.Val155Gly HARS both exhibit deficits in histidine binding, the activity of the p.Val155Gly HARS mutant alone can be rescued by elevated concentrations of histidine. Additionally, these results suggest that the diminished kcat associated with p.Ser356Asn HARS appears not be linked to a decrease in binding to histidine.
The final set of steady state kinetic experiments investigated the kinetics of aminoacylation under conditions of variable ATP concentrations and saturating histidine (10 mM) and tRNAHis (5 μM). Kinetic experiments for the wild type enzyme indicate that, under these substrate conditions, the KM value for ATP in the aminoacylation reaction is 44.2 μM (Table 2). This is comparable to the value of 140 μM determined for the S. typhiumurium WT HisRS catalyzed aminoacylation reaction (De Lorenzo, et al., 1972). Initial pilot experiments conducted with the mutants indicated that substantially higher titration ranges had to be employed in order to accurately determine parameters. Even under these conditions, saturating velocities were not obtained with any of the mutants. However, plotting the velocities against the concentration of [ATP] yielded straight lines from which estimates of the second order rate constant V/ KM could be obtained (Fig. 6C). Relative to wild type HARS, all three mutants showed significant decreases in V/ KM for ATP, corresponding to 342-fold for p.Val155Gly HARS, 866-fold for p.Ser356Asn HARS, and 2549-fold for p.Tyr330Cys HARS (Table 2). Based on these results, we conclude that, for all three mutants, a major component of the attenuated catalytic activity originates from a significant increase in KM, for ATP. It is noteworthy that, while we were unable to achieve saturation at 5 mM ATP, we were able to reproduce kcat results with 10 mM ATP in both the tRNA and histidine titrations (Table 2). In summary, we conclude that the HARS neuropathy-associated mutations lead to significant decreases in catalytic activity, typically reflected in decreases in kca and increases in the Km for ATP binding.
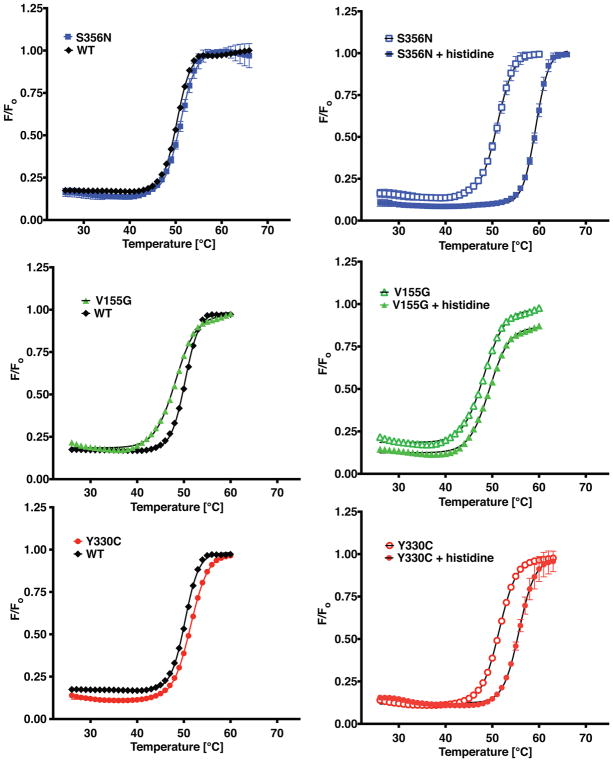
Differential scanning fluorimetry reveals one out of three HARS mutations is unstable and two mutations have substrate binding defects
Differential scanning fluorimetry (DSF) is a useful technique that can be used to assess the melting temperature stabilizing effect of a substrate with respect to an enzyme, as well as aid in the evaluation of the effect of a potential pathogenic missense variant on protein structure (Abbott, et al., 2017). Previously, incubation of HARS with saturating concentrations of the histidine amino acid ligand raised the thermal shift transition temperature by seven degrees (Abbott, et al., 2017). On the basis of the kinetic results indicating weaker binding of both histidine and ATP, we predicted that one or more the mutants would show altered thermal shift behavior. Thermal shift assays were performed for each combination of mutant enzyme and substrate. Consistent with the previous kinetic data, there was an inverse relationship between the KM for histidine of a given mutant protein and the extent of temperature stability provided by incubation with histidine in the assay. As seen in Fig. 7 and Table 3, p.Ser356Asn HARS exhibited the same extent of stabilization by histidine (8.3 vs 7.03°) as WT HARS, consistent with its essentially equivalent KM in aminoacylation. By contrast, p.Tyr330Cys HARS and p.Val155Gly HARS exhibited smaller histidine mediated temperature shifts (4.2 and 1.3°, respectively) consistent with their substantially increased Michaelis constants for histidine (202.9 μM and 687.2 μM fold, respectively).
Figure 7. Differential scanning fluorimetry of neuropathy associated HARS enzymes and their interactions with substrates.
A–C, Melting temperature of apo wild type or mutant HARS enzymes determined at a concentration of 10 mM WT enzyme (black diamonds, ◆); p.Ser356Asn HARS (blue squares, ■); p.Val155Gly (green triangles, ▲); and p.Tyr330Cys HARS (red circles, ●). D–F, Stabilization by histidine was investigated by incubation of mutant HARS enzymes with 5 mM histidine. Empty symbols represent apo enzymes, while filled symbols represent histidine bound complexes of p.Ser356Asn HARS (D); p.Val155Gly HARS (E); and p.Tyr330Cys HARS (F). Thermal shift curves are representations of neuropathy-associated mutants measured in duplicate.
Table 3.
Thermal stability of neuropathy-associated HARS variants and substrate complexes
| Enzyme | Tm °C | Δ Tm °C # | Δ Tm °C ## |
|---|---|---|---|
| WT | 51.72 ± 0.44*** | - | - |
| WT + histidine | 58.75 ± 0.41 | 7.03 | - |
| WT + ATP | 53.76 ± 0.21 | 2.04 | - |
| WT + tRNAHis | 53.43 ± 0.41 | 1.71 | - |
| p.Ser356Asn | 50.87 ± 0.33 | - | −0.85 |
| p.Ser356Asn + histidine | 59.17 ± 0.31 | 8.30 | 0.42 |
| p.Ser356Asn + ATP | 53.12 ± 0.11 | 2.25 | 0.64 |
| p.Ser356Asn + tRNAHis | 55.16 ± 0.20 | 4.29 | NA |
| p.Tyr330Cys | 51.72 ± 0.36 | - | 0 |
| p.Tyr330Cys + histidine | 55.95 ± 0.32 | 4.23 | −2.8 |
| p.Tyr330Cys + ATP | 53.85 ± 0.12 | 2.13 | 0.09 |
| p.Tyr330Cys + tRNAHis | 55.88 ± 0.29 | 4.16 | NA |
| p.Val155Gly | 48.05 ± 0.39*** | - | −3.67 |
| p.Val155Gly + histidine | 49.35 ± 0.67 | 1.30 | −9.4 |
| p.Val155Gly + ATP | 51.09 ± 0.16 | 3.04 | −2.67 |
| p.Val155Gly + tRNAHis | 53.91 ± 0.26 | 5.86 | NA |
Bold values were previously determined (Abbott, et al., 2017).
Relative to the apo form of the same enzyme.
Relative to the same form of WT HARS enzyme. Values reported are the mean ± standard error of 2 independent experiments in triplicate. Values that are significantly different WT to Val155Gly are indicated as
p<0.0001 (extra sum-of-squares F test).
For wild type HARS, the ATP and tRNA mediated temperature shifts are much smaller (2.04 and 1.7°, respectively) than that provided by histidine (Fig. 7 and Table 3). In the presence of saturating ATP (5 mM) all three neuropathy-associated mutations had slightly increased ΔTm values, corresponding to least 2 °C for p.Tyr330Cys and p.Ser356Asn, and 3 °C for p.Val155Gly (Table 3). In the presence of 20 μM tRNAHis, all three mutants exhibited 1 °C in ΔTm increases in stability (Table 3). Notably, none of the three mutants showed a significantly altered melting temperature in the presence of either ATP or tRNA, despite the fact that all three show substantially reduced apparent second order rate constants associated with ATP binding (Table 3).
Analytical ultracentrifugation confirms that HARS neuropathy-associated mutations do not disrupt enzyme dimerization
Previous studies investigating mutations in the GARS gene linked to CMT have reported that a subset of the mutant substitutions lead to weaker tertiary interactions and thus reduced dimer stability (Nangle, et al., 2007). Given the location of the HARS mutants in the active site, we considered the possibility that the decreased activity of these mutants might be a consequence of reduced dimer stability. To address this hypothesis, we subjected highly purified preparations of each of the mutant proteins and a wild type control to sedimentation velocity experiments at various loading concentrations. In these experiments, we monitored protein absorbance at 230 nm, allowing the experiments to be performed at concentrations that would be in the range of the equilibrium dissociation constant (Kd) for the dimer. As shown in Supplemental Figure 1, the molar mass distributions of the wild type and mutant proteins were closely similar. For each mutant, a major species was observed that had a molar mass that was in excellent agreement with the dimer form of HARS (Table 4, Supp. Figure S1). In none of the mutants was there a detectable amount of material corresponding to the monomeric form of the enzyme. On the basis of these results, we conclude that the pathophysiology of these mutants is unlikely to be the result of a weakened dimeric interface.
Table 4.
PCSA – Monte Carlo results (straight-line parameterization) from Sedimentation Velocity experiment.
| Species: | Molar mass (kDa) | D (x 10−7 cm/sec2) | s (x 10−13 sec) | f/f0 | RMSD |
|---|---|---|---|---|---|
| WT | 118.5 (111.6, 125.5) | 4.54 (4.43, 4.66) | 5.67 (5.48, 5.85) | 1.44 (1.43, 1.45) | 0.0025 |
| p.Val155Gly | 122.0 (110.6, 133.5) | 4.43 (4.20, 4.65) | 5.68 (5.42, 5.94) | 1.46 (1.44, 1.49) | 0.0032 |
| p.Tyr330Cys | 120.6 (116.3, 124.9) | 4.52 (4.50, 4.55) | 5.74 (5.51, 5.97) | 1.44 (1.42, 1.46) | 0.0028 |
| p.Ser356Asn | 120.0 (110.3, 129.6) | 4.41 (4.34, 4.49) | 5.57 (5.21, 5.93) | 1.48 (1.46, 1.49) | 0.0034 |
Values in parenthesis are 95% confidence intervals from the Monte Carlo analysis. The theoretical molar mass of HARS WT dimer is 114.9 kDa, indicating the observed species is a dimer. The residual mean square deviation (RMSD) of the fit is shown in units of absorbance at 230 nm.
DISCUSSION
Mutations in five ARS genes were previously implicated in CMT disease (Jordanova, et al., 2006; Latour, et al., 2010; Safka Brozkova, et al., 2015; Storkebaum, 2016; Tsai, et al., 2017; Vester, et al., 2013). A central question emerging from those studies is the degree to which the CMT disease is a direct consequence of a loss of canonical ARS function, or is instead linked to one or more potential secondary functions. In this paper, we describe detailed functional analyses of three HARS mutations linked to CMT. The characterization of these mutants showed that they all displayed losses in catalytic activity when assessed in vivo and in vitro. Notably, the extent of decrease in catalytic activity correlated with the loss of ability to support yeast growth. The most active of the three mutants (p.Val155Gly HARS) was the only one of the three mutants that retained some ability to support yeast growth when studied in the human HARS open-reading frame. In contrast to previous work analyzing ARS mutants linked to CMT, each mutant was fully characterized with respect to defects associated with recognition of each substrate; indeed, biochemical analysis of CMT-associated HARS mutations has not been previously reported. The novel finding emerging from our analysis is that all three mutants displayed substantially elevated Michaelis constants for ATP, and two of the three (p.Tyr330Cys and p.Val155Gly) displayed an elevated KM for histidine. Notably, none of the three exhibited an altered KM for tRNA. Collectively, these mutants represent some of the best-characterized examples of ARSs where specific defects in substrate recognition that produce reduced aminoacylation function are specifically linked to CMT. It would therefore be valuable to know if other CMT-associated ARS mutations lead to similar alterations in substrate interactions, as such information would help develop comprehensive models to explain the gene-disease relationships.
Strength of the evidence implicating the newly identified HARS mutations in neuropathy
Implicating ARS mutations in peripheral neuropathy requires data showing that the variants segregate with disease in large pedigrees and are absent at high frequencies in the general population. Indeed, the majority of previously implicated ARS mutations meet these criteria (Oprescu, et al., 2017). When genetic data are impossible to obtain, functional studies can be employed to predict pathogenicity, provided that the functional assays are informative. With respect to ARS mutations, enzyme kinetic and yeast complementation assays are informative for predicting mutational pathogenicity (Oprescu, et al., 2017). Several independent lines of evidence allow us to conclude that p.Val155Gly and p.Tyr330Cys HARS are the causal mutations in Family 1 and Family 2, respectively. Notably, the mutations segregate with the CMT phenotype in pedigrees (albeit in a small family for p.Tyr330Cys), occur at highly conserved amino acids, are absent from variant databases, and result in loss-of-function effects similar to other CMT-associated ARS variants. By contrast, the evidence for the role of p.Ser356Asn in the CMT phenotype in Family 3 is less convincing. Specifically, the patient’s unaffected mother is heterozygous for p.Ser356Asn HARS, and this variant is present (albeit at a very low frequency) in the gnomAD variant database. However, the fact that p.Ser356Asn affects a highly conserved amino-acid residue and displays impaired function similar to validated, disease-associated ARS mutations strongly supports the conclusion that it may be a pathogenic variant (Safka Brozkova, et al., 2015; Vester, et al., 2013). One possibility is that p.Ser356Asn HARS has reduced penetrance similar to certain GARS mutations (Sivakumar, et al., 2005). This proposal is supported by the low frequency of validated, CMT-associated ARS mutations in the gnomAD variant database (e.g., p.Gly240Arg GARS; http://gnomad.broadinstitute.org/variant/7-30649345-G-C; Gly240Arg is equivalent to p.Gly294Arg on a longer protein isoform). Conversely, it is possible that p.Ser356Asn is not the disease-causing mutation in Family 3 despite the deleterious effect on enzyme function. Thus, we conclude that p.Ser356Asn HARS is an excellent candidate for pathogenicity in Family 3, but that further genetic and phenotypic evidence is required to implicate this variant in CMT disease.
Structural justification for activity defects
All three of the amino acid substitutions analyzed here alter amino acids located in the catalytic domain, consistent with their pronounced effect on aminoacylation. Visualization of these substitutions in the context of available X-ray structure of human HARS provides insights into potential structural and functional consequences. While Ser356 does not appear to be involved directly in substrate binding interactions, it may be important for positioning other amino acid residues within the active site that are critical for binding of the ATP substrate. ATP binding in the active site of HARS is dependent on magnesium ions that are coordinated to the non-bridging oxygens of the β and γ phosphates of ATP, and the strictly conserved glutamate in Motif 2. In the human HARS histidine complex (no ATP), Ser356 is within 2.5 Å of Arg 388 (Fig. 8A), the conserved motif 3 arginine that (based on the E. coli complex) is predicted to interact with the gamma phosphate of ATP (Fig. 8B). Important caveats regarding the likely effects of p.Ser356Asn are that, firstly, a complex with ATP is not available, and secondly, that much of the loop between strands B9 and B10 in which Ser356 residue resides is disordered in the various human HARS structures, and has different conformations in the different subunits of the asymmetric unit (Koh, et al., 2014). Hence, it is difficult to make definitive conclusions about the potential role of Ser356 in stabilizing interactions in the bound complex, and about the consequences of substitution with asparagine. In any event, the elevated KM for ATP seen with p.Ser356Asn could be seen as a likely outcome of a local disruption of contacts to the gamma phosphate.
The one mutant that would be predicted to have the most severe consequence for HARS function is p.Tyr330Cys, which substitutes a highly conserved tyrosine in the His A motif that constitutes part of the binding site for histidine (Fig. 3). Notably, this residue has different conformations in the apo versus histidine bound human cytoplasmic HARS complex. In the apo complex without substrates (4X5O), Y330 adopts a conformation directed away from the active site, pointing towards solvent and donating a hydrogen bond to the main chain carbonyl of Gly108 (Fig. 8C). In the complex of HARS with its amino acid substrate, Y330 moves some 8 Å to approach within 3.1 of the alpha-carbon of histidine (Fig. 8D). In this bound conformation, Y330 may further stabilize the histidine binding pocket by engaging in a stacking interaction with the polar group of the neighboring tyrosine in position 331.
A similar effect may underlie the effect of p.Val155Gly on HARS function. Notably, this mutant exhibited a lower melting temperature than the other mutants, but the substitution had no effect on the association constant for the dimer. Analysis of the human HARS-histidine complex and comparison to the E. coli HisRS-histidinol ATP complex suggests that p.Val155Gly is unlikely to substitute a residue that interacts directly with ATP (Fig. 8E). However, Val155 is in the region of the active site near where ATP binds, and the valine can potentially make a CH-π interaction with the neighboring hydrophobic Phe171 in the active site (Fig. 8E). Phe171 in human HARS corresponds to Phe125 in the E. coli enzyme (Arnez, et al., 1995), which makes a critical stacking interaction with the adenine ring (Fig. 8F) The substitution of glycine for valine at position 155 may allow greater conformational flexibility on the part of Phe171, reducing its ability to generate stable binding pocket for ATP. The effects of this disruption are apparent in the elevated KM value for ATP observed for p.Val155Gly. Additionally, a glycine substitution would permit greater conformational freedom in the accessible phi-psi angles within the peptide backbone of the beta sheet, potentially accounting for the lower melting temperature in the absence of substrate. Furthermore, a less constrained beta sheet created by the glycine substitution could directly impact the helix 3, which contains the ‘TXE’ motif for histidine binding. This provides a structural rationale for the decreased Tm of the p.Val155Gly apo enzyme, the dramatically reduced Tm in the presence of histidine (Table 3), and the elevated KM for histidine (Table 2).
The vast majority of ARS mutations impair enzyme activity
Previous studies examining the links between mutations in ARS genes and CMT have addressed the question of whether mutations lead to a significant loss of aminoacylation activity, or a change in an as yet undetermined secondary function. The biochemical analyses in this study indicate that CMT-linked HARS mutations confer significant reductions in aminoacylation function, in accordance with what has been described for AARS mutations (McLaughlin, et al., 2012). These AARS mutations, like the described HARS mutations, also fail to complement yeast growth over a background where the endogenous yeast gene has been disrupted (Oprescu, et al., 2017). Thus, for AARS and HARS mutations, the link to CMT appears to be directly associated with a loss of aminoacylation function.
The link between CMT and reduced ARS function is slightly less clear for two other aminoacyl-tRNA synthetase genes, GARS and YARS. In these cases, multiple mutations are linked to the disease, some of which have clear reductions in aminoacylation, and some of which appear to retain significant activity. In GARS, twelve GARS mutations have been associated with CMT disease, the majority of which are deleterious to enzyme function (Oprescu, et al., 2017). In the case of YARS, two mutants are reported to have significantly reduced activity (Jordanova, et al., 2006) but one (p.Glu196Lys) appears to be much more active, or even unaffected (Froelich and First, 2011). However, the p.Glu196Lys mutant and another CMT-associated mutation at the same codon (p.Glu196Gln) were unable to fully support yeast cell growth in complementation studies (Gonzaga-Jauregui, et al., 2015; Jordanova, et al., 2006). Furthermore, studies employing a D. melanogaster model of CMT show that, despite its near wild type activity in vitro, p.Glu196Lys YARS leads to decreased protein synthesis rates in flies (Niehues, et al., 2015). These and other results suggest that, in the context of specialized cell types, the consequences of a particular human pathogenic ARS mutation may be difficult to predict a priori. Mutations that lead to decreased activity may have limited phenotypic impact owing to the fact that, under normal conditions, protein synthesis can be met by less than 100% activity of a given tRNA synthetase. Conversely, a mutation that has limited impact on aminoacylation in vitro may exert more deleterious impact in vivo, owing to folding defects that promote proteotoxic stress. Discrepancies may arise as a consequence of additional interactions with other proteins and/or RNAs that are necessary to execute all phases of protein synthesis. Moving forward, a critical question to be addressed for each CMT-related ARS allele is if protein synthesis is affected as a direct downstream consequence of impaired aminoacylation.
Proposed gain-of-function effects of ARS mutations
Clearly more research is needed to resolve the effect of CMT-associated ARS mutations on enzyme function. As an alternative to loss-of-function mechanisms, multiple gain-of-function hypotheses have been proposed, specifically for GARS mutations (Motley, et al., 2010). For example, a mechanism in which mutant monomeric GARS binds to the VEGF receptor neuropilin-1 and alters neuronal signaling has been proposed in the context of a mouse model (He, et al., 2015). As yet, the relevance of this model to human disease-associated mutations is not fully clear. However, three human mutant GARS proteins (p.Glu71Gly, p.Leu129Pro, and p.Gly240Arg) were shown to bind to neuropilin-1 in vitro. Additionally, it was argued that the propensity of mutant GARS to bind to neuropilin-1 was linked to weakened dimer formation (Xie, et al., 2007). While the findings reported here do not explicitly address the neuropilin-1 hypothesis, our characterization of HARS mutant proteins indicates that, despite a loss of canonical catalytic function, there is no evidence for weakened dimerization (Supplemental Fig. 1). It should also be noted that, despite the linkage of over 80 loci to CMT disease, neither mutations in neuropilin-1 nor in any of the ligands for this receptor have been identified in patients with CMT disease.
Closing thoughts
Here we present data that for the first time link impaired enzyme activity to HARS-associated peripheral neuropathy. This observation dictates considerations of how altered tRNA charging might affect peripheral nerve function. Human peripheral nerve axons can extend more than a meter in length, and transport of essential proteins to the cell periphery is essential for maintaining axon function (Giuditta, et al., 1968). There is increasing evidence of local protein translation along axons and at synapses, but the role of ARSs in this process is still not well understood (Holt and Schuman, 2013). Moving forward, it will be important to directly test if CMT-associated ARS mutations alter specific aspects of protein translation. In summary, additional studies are required to determine if CMT-associated ARS mutations act via a loss-of-function or gain-of-function mechanism; we suggest that these two mechanisms are not mutually exclusive. Our study presents data that further supports a loss-of-function molecular pathology and provides key data, newly applied methods, and novel reagents for studying the pathological mechanism of ARS-associated human disease.
Supplementary Material
Acknowledgments
GRANT SUPPORT
J.A.A. is supported by NHLBI Training Grant (T32 HL 007594-30). R.M. is supported by the Michigan Pre-doctoral Training in Genetics Program (GM007544). I.M. is supported by a Ph.D. fellowship of the agency for Innovation by Science and Technology (IWT). L.B.G. is supported by the NIH Cellular and Molecular Biology Training Grant (GM007315), the NIH Medical Scientist Training Grant (GM07863), and an NIH F30 NRSA (NS092238). S.Z is supported by the National Institute of Neurological Diseases and Stroke (NS075764 and NS065712). C.E. is supported by the Miguel Servet Program (CPII14/00002). J.B. is supported by a Senior Clinical Researcher mandate of the Research Fund - Flanders (FWO). A.A. is supported by funding from the National Institute of General Medical Sciences (GM118647) and the Muscular Dystrophy Association (MDA294479). C.S.F. is supported by funding from the National Institute of General Medical Sciences (GM54899). This work was also supported by the Association Belge contre les Maladies Neuromusculaire (ABMM) - Aide à la Recherche ASBL and the EU FP7/2007-2013 under grant agreement number 2012-305121 (NEUROMICS). B.D. acknowledges San Antonio Cancer Institute Grant P30 CA054174 for support of the Cetner for Analytical Ultrancentrifugation of Macromolecular Assemblies at the University of Texas Health Science Center at San Antonio.
We are grateful to the patients and their families who participated in this study. The authors declare no conflicts of interest.
References
- Abbott JA, Francklyn CS, Robey-Bond SM. Transfer RNA and human disease. Front Genet. 2014;5:158. doi: 10.3389/fgene.2014.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott JA, Guth E, Kim C, Regan C, Siu VM, Rupar CA, Demeler B, Francklyn CS, Robey-Bond SM. The Usher Syndrome Type IIIB Histidyl-tRNA Synthetase Mutation Confers Temperature Sensitivity. Biochemistry. 2017 doi: 10.1021/acs.biochem.7b00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonellis A, Ellsworth RE, Sambuughin N, Puls I, Abel A, Lee-Lin SQ, Jordanova A, Kremensky I, Christodoulou K, Middleton LT, et al. Glycyl tRNA Synthetase Mutations in Charcot-Marie-Tooth Disease Type 2D and Distal Spinal Muscular Atrophy Type V. Am J Hum Genet. 2003;72(5):1293–9. doi: 10.1086/375039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonellis A, Green ED. The role of aminoacyl-tRNA synthetases in genetic diseases. Annu Rev Genomics Hum Genet. 2008;9:87–107. doi: 10.1146/annurev.genom.9.081307.164204. [DOI] [PubMed] [Google Scholar]
- Arnez JG, Harris DC, Mitschler A, Rees B, Francklyn CS, Moras D. Crystal structure of histidyl-tRNA synthetase from Escherichia coli complexed with histidyl-adenylate. EMBO J. 1995;14(17):4143–55. doi: 10.1002/j.1460-2075.1995.tb00088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird TD. Charcot-Marie-Tooth Hereditary Neuropathy Overview. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Ledbetter N, Mefford HC, Smith RJH, et al., editors. GeneReviews(R) Seattle (WA): 1993. [PubMed] [Google Scholar]
- Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–6. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Brookes E, Cao W, Demeler B. A two-dimensional spectrum analysis for sedimentation velocity experiments of mixtures with heterogeneity in molecular weight and shape. Eur Biophys J. 2010;39(3):405–14. doi: 10.1007/s00249-009-0413-5. [DOI] [PubMed] [Google Scholar]
- Brookes EDB. In Analytical Ultracentrifugation Data Analysis with UltraScan-III. Analytical Ultracentrifugation: Instrumentation, Software, and Applications. In: Wandrey CCH, editor. Analytical Ultracentrifugation VIII. Progress in Colloid and Polymer Science. Berlin, Heidelberg: Springer; 2006. [Google Scholar]
- Brookes EH, Demeler B. Parallel computational techniques for the analysis of sedimentation velocity experiments in UltraScan. Colloid Polym Sci. 2008;286:138–148. [Google Scholar]; Colloid and Polymer Science. 286(2):139–148. doi: 10.1007/s00396-007-1815-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns J, Ouvrier R, Estilow T, Shy R, Laura M, Pallant JF, Lek M, Muntoni F, Reilly MM, Pareyson D, et al. Validation of the Charcot-Marie-Tooth disease pediatric scale as an outcome measure of disability. Ann Neurol. 2012;71(5):642–52. doi: 10.1002/ana.23572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lorenzo F, Straus DS, Ames BN. Histidine regulation in Salmonella typhimurium. X. Kinetic studies of mutant histidyl transfer ribonucleic acid synthetases. J Biol Chem. 1972;247(8):2302–7. [PubMed] [Google Scholar]
- Demeler B. Methods for the design and analysis of sedimentation velocity and sedimentation equilibrium experiments with proteins. Curr Protoc Protein Sci. 2010;Chapter 7(Unit 7):13. doi: 10.1002/0471140864.ps0713s60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeler B, Brookes E. Monte Carlo analysis of sedimentation experiments. Colloid and Polymer Science. 2008;286(2):129–137. [Google Scholar]
- Demeler B, Gorbet G, Zollars D, Dubbs B, Brookes E, Cao W. UltraScan-III version 3.5: A comprehensive data analysis software package for analytical ultracentrifugation experiments. 2016 http://www.ultrascan3.uthscsa.edu/
- Dyck PJ, Lambert EH. Lower motor and primary sensory neuron diseases with peroneal muscular atrophy. II. Neurologic, genetic, and electrophysiologic findings in various neuronal degenerations. Arch Neurol. 1968;18(6):619–25. doi: 10.1001/archneur.1968.00470360041003. [DOI] [PubMed] [Google Scholar]
- Fahoum SK, Yang DC. Purification of mammalian histidyl-tRNA synthetase and its interaction with myositis-specific anti-Jo-1 antibodies. Biochemistry. 1987;26(18):5871–7. doi: 10.1021/bi00392a044. [DOI] [PubMed] [Google Scholar]
- Froelich CA, First EA. Dominant Intermediate Charcot-Marie-Tooth disorder is not due to a catalytic defect in tyrosyl-tRNA synthetase. Biochemistry. 2011;50(33):7132–45. doi: 10.1021/bi200989h. [DOI] [PubMed] [Google Scholar]
- Frohlich D, Suchowerska AK, Spencer ZH, von Jonquieres G, Klugmann CB, Bongers A, Delerue F, Stefen H, Ittner LM, Fath T, et al. In vivocharacterization of the aspartyl-tRNA synthetase DARS: Homing in on the leukodystrophy HBSL. Neurobiol Dis. 2017;97(Pt A):24–35. doi: 10.1016/j.nbd.2016.10.008. [DOI] [PubMed] [Google Scholar]
- Giuditta A, Dettbarn WD, Brzin M. Protein synthesis in the isolated giant axon of the squid. Proc Natl Acad Sci U S A. 1968;59(4):1284–7. doi: 10.1073/pnas.59.4.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzaga-Jauregui C, Harel T, Gambin T, Kousi M, Griffin LB, Francescatto L, Ozes B, Karaca E, Jhangiani SN, Bainbridge MN, et al. Exome Sequence Analysis Suggests that Genetic Burden Contributes to Phenotypic Variability and Complex Neuropathy. Cell Rep. 2015;12(7):1169–83. doi: 10.1016/j.celrep.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M, Falk MJ, Gai X, Postrel R, Schule R, Zuchner S. Innovative genomic collaboration using the GENESIS (GEM.app) platform. Hum Mutat. 2015;36(10):950–6. doi: 10.1002/humu.22836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M, McLaughlin H, Houlden H, Guo M, Yo-Tsen L, Hadjivassilious M, Speziani F, Yang XL, Antonellis A, Reilly MM, et al. Exome sequencing identifies a significant variant in methionyl-tRNA synthetase (MARS) in a family with late-onset CMT2. J Neurol Neurosurg Psychiatry. 2013;84(11):1247–9. doi: 10.1136/jnnp-2013-305049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbet G, Devlin T, Hernandez Uribe BI, Demeler AK, Lindsey ZL, Ganji S, Breton S, Weise-Cross L, Lafer EM, Brookes EH, et al. A parametrically constrained optimization method for fitting sedimentation velocity experiments. Biophys J. 2014;106(8):1741–50. doi: 10.1016/j.bpj.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Bai G, Zhou H, Wei N, White NM, Lauer J, Liu H, Shi Y, Dumitru CD, Lettieri K, et al. CMT2D neuropathy is linked to the neomorphic binding activity of glycyl-tRNA synthetase. Nature. 2015;526(7575):710–4. doi: 10.1038/nature15510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Oka N, Hashiguchi A, Ueno S, Sakamoto H, Takashima H, Higuchi Y, Kusunoki S, Nakamura Y. Histopathological features of a patient with Charcot-Marie-Tooth disease type 2U/AD-CMTax-MARS. J Peripher Nerv Syst. 2016;21(4):370–374. doi: 10.1111/jns.12193. [DOI] [PubMed] [Google Scholar]
- Holt CE, Schuman EM. The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron. 2013;80(3):648–57. doi: 10.1016/j.neuron.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun YS, Park HJ, Heo SH, Yoon BR, Nam SH, Kim SB, Park CI, Choi BO, Chung KW. Rare variants in methionyl- and tyrosyl-tRNA synthetase genes in late-onset autosomal dominant Charcot-Marie-Tooth neuropathy. Clin Genet. 2014;86(6):592–4. doi: 10.1111/cge.12327. [DOI] [PubMed] [Google Scholar]
- Ibba M, Soll D. Aminoacyl-tRNA synthesis. Ann Rev Biochem. 2000;69:617–50. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- Jerath NU, Shy ME. Hereditary motor and sensory neuropathies: Understanding molecular pathogenesis could lead to future treatment strategies. Biochim Biophys Acta. 2015;1852(4):667–78. doi: 10.1016/j.bbadis.2014.07.031. [DOI] [PubMed] [Google Scholar]
- Jordanova A, Irobi J, Thomas FP, Van Dijck P, Meerschaert K, Dewil M, Dierick I, Jacobs A, De Vriendt E, Guergueltcheva V, et al. Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat Genet. 2006;38(2):197–202. doi: 10.1038/ng1727. [DOI] [PubMed] [Google Scholar]
- Koh CY, Kim JE, Wetzel AB, de van der Schueren WJ, Shibata S, Ranade RM, Liu J, Zhang Z, Gillespie JR, Buckner FS, et al. Structures of Trypanosoma brucei methionyl-tRNA synthetase with urea-based inhibitors provide guidance for drug design against sleeping sickness. PLoS Negl Trop Dis. 2014;8(4):e2775. doi: 10.1371/journal.pntd.0002775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Latour P, Thauvin-Robinet C, Baudelet-Mery C, Soichot P, Cusin V, Faivre L, Locatelli MC, Mayencon M, Sarcey A, Broussolle E, et al. A major determinant for binding and aminoacylation of tRNA(Ala) in cytoplasmic Alanyl-tRNA synthetase is mutated in dominant axonal Charcot-Marie-Tooth disease. Am J Hum Genet. 2010;86(1):77–82. doi: 10.1016/j.ajhg.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laue TM, Shah BD, Ridgeway TM, Pelletier SL. In: Analytical Ultracentrifugation in Biochemistry and Polymer Science. Harding S, Rowe A, editors. Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- Lee YH, Chang CP, Cheng YJ, Kuo YY, Lin YS, Wang CC. Evolutionary gain of highly divergent tRNA specificities by two isoforms of human histidyl-tRNA synthetase. Cell Mol Life Sci. 2017;74(14):2663–2677. doi: 10.1007/s00018-017-2491-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupo V, Garcia-Garcia F, Sancho P, Tello C, Garcia-Romero M, Villarreal L, Alberti A, Sivera R, Dopazo J, Pascual-Pascual SI, et al. Assessment of Targeted Next-Generation Sequencing as a Tool for the Diagnosis of Charcot-Marie-Tooth Disease and Hereditary Motor Neuropathy. J Mol Diagn. 2016;18(2):225–34. doi: 10.1016/j.jmoldx.2015.10.005. [DOI] [PubMed] [Google Scholar]
- Meyer-Schuman R, Antonellis A. Emerging Mechanisms of Aminoacyl-tRNA Synthetase Mutations in Recessive and Dominant Human Disease. Hum Mol Genet. 2017 doi: 10.1093/hmg/ddx231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley WW, Talbot K, Fischbeck KH. GARS axonopathy: not every neuron’s cup of tRNA. Trends Neurosci. 2010;33(2):59–66. doi: 10.1016/j.tins.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam SH, Hong YB, Hyun YS, Nam da E, Kwak G, Hwang SH, Choi BO, Chung KW. Identification of Genetic Causes of Inherited Peripheral Neuropathies by Targeted Gene Panel Sequencing. Mol Cells. 2016;39(5):382–8. doi: 10.14348/molcells.2016.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangle LA, Zhang W, Xie W, Yang XL, Schimmel P. Charcot-Marie-Tooth disease-associated mutant tRNA synthetases linked to altered dimer interface and neurite distribution defect. Proc Natl Acad Sci U S A. 2007;104(27):11239–44. doi: 10.1073/pnas.0705055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehues S, Bussmann J, Steffes G, Erdmann I, Kohrer C, Sun L, Wagner M, Schafer K, Wang G, Koerdt SN, et al. Impaired protein translation in Drosophila models for Charcot-Marie-Tooth neuropathy caused by mutant tRNA synthetases. Nat Commun. 2015;6:7520. doi: 10.1038/ncomms8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2(9):2212–21. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- Oprescu SN, Griffin LB, Beg AA, Antonellis A. Predicting the pathogenicity of aminoacyl-tRNA synthetase mutations. Methods. 2017;113:139–151. doi: 10.1016/j.ymeth.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pareyson D, Marchesi C. Diagnosis, natural history, and management of Charcot-Marie-Tooth disease. Lancet Neurol. 2009a;8(7):654–67. doi: 10.1016/S1474-4422(09)70110-3. [DOI] [PubMed] [Google Scholar]
- Pareyson D, Marchesi C. Natural history and treatment of peripheral inherited neuropathies. Adv Exp Med Biol. 2009b;652:207–24. doi: 10.1007/978-90-481-2813-6_14. [DOI] [PubMed] [Google Scholar]
- Pareyson D, Marchesi C, Salsano E. Hereditary predominantly motor neuropathies. Curr Opin Neurol. 2009;22(5):451–9. doi: 10.1097/WCO.0b013e3283311dfd. [DOI] [PubMed] [Google Scholar]
- Pierce SB, Chisholm KM, Lynch ED, Lee MK, Walsh T, Opitz JM, Li W, Klevit RE, King MC. Mutations in mitochondrial histidyl tRNA synthetase HARS2 cause ovarian dysgenesis and sensorineural hearing loss of Perrault syndrome. Proc Natl Acad Sci U S A. 2011;108(16):6543–8. doi: 10.1073/pnas.1103471108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puffenberger EG, Jinks RN, Sougnez C, Cibulskis K, Willert RA, Achilly NP, Cassidy RP, Fiorentini CJ, Heiken KF, Lawrence JJ, et al. Genetic mapping and exome sequencing identify variants associated with five novel diseases. PLoS One. 2012;7(1):e28936. doi: 10.1371/journal.pone.0028936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42(Web Server issue):W320–4. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safka Brozkova D, Deconinck T, Griffin LB, Ferbert A, Haberlova J, Mazanec R, Lassuthova P, Roth C, Pilunthanakul T, Rautenstrauss B, et al. Loss of function mutations in HARS cause a spectrum of inherited peripheral neuropathies. Brain. 2015;138(Pt 8):2161–72. doi: 10.1093/brain/awv158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. Sedimentation equilibrium analysis of interference optical data by systematic noise decomposition. Anal Biochem. 1999;272(2):199–208. doi: 10.1006/abio.1999.4172. [DOI] [PubMed] [Google Scholar]
- Shy ME, Blake J, Krajewski K, Fuerst DR, Laura M, Hahn AF, Li J, Lewis RA, Reilly M. Reliability and validity of the CMT neuropathy score as a measure of disability. Neurology. 2005;64(7):1209–14. doi: 10.1212/01.WNL.0000156517.00615.A3. [DOI] [PubMed] [Google Scholar]
- Simons C, Griffin LB, Helman G, Golas G, Pizzino A, Bloom M, Murphy JL, Crawford J, Evans SH, Topper S, et al. Loss-of-function alanyl-tRNA synthetase mutations cause an autosomal-recessive early-onset epileptic encephalopathy with persistent myelination defect. Am J Hum Genet. 2015;96(4):675–81. doi: 10.1016/j.ajhg.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar K, Kyriakides T, Puls I, Nicholson GA, Funalot B, Antonellis A, Sambuughin N, Christodoulou K, Beggs JL, Zamba-Papanicolaou E, et al. Phenotypic spectrum of disorders associated with glycyl-tRNA synthetase mutations. Brain. 2005;128(Pt 10):2304–14. doi: 10.1093/brain/awh590. [DOI] [PubMed] [Google Scholar]
- Skre H. Genetic and clinical aspects of Charcot-Marie-Tooth’s disease. Clin Genet. 1974;6(2):98–118. doi: 10.1111/j.1399-0004.1974.tb00638.x. [DOI] [PubMed] [Google Scholar]
- Storkebaum E. Peripheral neuropathy via mutant tRNA synthetases: Inhibition of protein translation provides a possible explanation. Bioessays. 2016;38(9):818–29. doi: 10.1002/bies.201600052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stum M, McLaughlin HM, Kleinbrink EL, Miers KE, Ackerman SL, Seburn KL, Antonellis A, Burgess RW. An assessment of mechanisms underlying peripheral axonal degeneration caused by aminoacyl-tRNA synthetase mutations. Mol Cell Neurosci. 2011;46(2):432–43. doi: 10.1016/j.mcn.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman V, Strickland AV, Zuchner S. Genetics of Charcot-Marie-Tooth (CMT) Disease within the Frame of the Human Genome Project Success. Genes (Basel) 2014;5(1):13–32. doi: 10.3390/genes5010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai PC, Soong BW, Mademan I, Huang YH, Liu CR, Hsiao CT, Wu HT, Liu TT, Liu YT, Tseng YT, et al. A recurrent WARS mutation is a novel cause of autosomal dominant distal hereditary motor neuropathy. Brain. 2017 doi: 10.1093/brain/awx058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vester A, Velez-Ruiz G, McLaughlin HM, Lupski JR, Talbot K, Vance JM, Zuchner S, Roda RH, Fischbeck KH, Biesecker LG, et al. A loss-of-function variant in the human histidyl-tRNA synthetase (HARS) gene is neurotoxic in vivo. Hum Mutat. 2013;34(1):191–9. doi: 10.1002/humu.22210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson AD, Pleiss JA, Uhlenbeck OC. A new assay for tRNA aminoacylation kinetics. RNA. 1998;4(8):1019–23. doi: 10.1017/s1355838298980700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Nangle LA, Zhang W, Schimmel P, Yang XL. Long-range structural effects of a Charcot-Marie-Tooth disease-causing mutation in human glycyl-tRNA synthetase. Proc Natl Acad Sci U S A. 2007;104(24):9976–81. doi: 10.1073/pnas.0703908104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ling J, Barcia G, Jing L, Wu J, Barry BJ, Mochida GH, Hill RS, Weimer JM, Stein Q, et al. Mutations in QARS, encoding glutaminyl-tRNA synthetase, cause progressive microcephaly, cerebral-cerebellar atrophy, and intractable seizures. Am J Hum Genet. 2014;94(4):547–58. doi: 10.1016/j.ajhg.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.