Abstract
Liquid biopsies focusing on the analysis of cell‐free circulating tumor DNA (ctDNA) may have important clinical implications for personalized medicine, including early detection of cancer, therapeutic guidance, and monitoring of recurrence. Mutations in the oncogene, PIK3CA, are frequently observed in breast cancer and have been suggested as a predictive biomarker for PI3K‐selective inhibitor treatment. In this study, we analyzed the presence of PIK3CA mutations in formalin‐fixed, paraffin‐embedded, metastatic tissue and corresponding ctDNA from serum of patients with advanced breast cancer using a highly sensitive, optimized droplet digital PCR (ddPCR) assay. We found 83% of patients with PIK3CA mutation in the metastatic tumor tissue also had detectable PIK3CA mutations in serum ctDNA. Patients lacking the PIK3CA mutation in corresponding serum ctDNA all had nonvisceral metastatic disease. Four patients with detectable PIK3CA‐mutated ctDNA were followed with an additional serum sample during oncological treatment. In all cases, changes in PIK3CA ctDNA level correlated with treatment response. Our results showed high concordance between detection of PIK3CA mutations in tumor tissue and in corresponding serum ctDNA and suggest that serum samples from patients with advanced breast cancer and ddPCR may be used for PIK3CA mutation status assessment to complement imaging techniques as an early marker of treatment response.
Keywords: ddPCR, liquid biopsy, metastatic breast cancer, PIK3CA
Abbreviations
- bp
base pairs
- CT
computed tomography
- ctDNA
circulating tumor DNA
- ddPCR
droplet digital polymerase chain reaction
- FFPE
formalin‐fixed and paraffin‐embedded
- HE
hematoxylin–eosin
- HER2
human epidermal growth factor receptor 2
- PET
positron emission tomography
- PTEN
phosphatase and tensin homolog
- SOP
standard operating procedure
1. Introduction
Cell‐free DNA is released from both normal and cancer cells into the circulation (Choi et al., 2005; Stroun et al., 2000). In contrast to benign tumors and other noncancerous conditions, a rapid turnover of tumor cells is thought to result in a consistently increased release of circulating tumor DNA (ctDNA; Diehl et al., 2005). This enables clinical use of repeated and noninvasive blood samples as a ‘liquid biopsy’ to monitor the dynamic evolution of human cancers (Siravegna et al., 2017) and response to treatment, which could accompany the well‐established RECIST 1.1 and PERCIST 1.0 criteria evaluative of changes in tumor burden during cancer treatment (Eisenhauer et al., 2009).
In some cancers, certain driver mutations are found at high frequencies, such as KRAS in colorectal cancer (Hao et al., 2017). In other cancers, including breast cancer, the pattern of driver mutations is more diverse. The most frequent driver mutation observed in breast cancer other than TP53 is PIK3CA, an oncogene encoding the p110α component of the phosphoinositide 3‐kinase (PI3K). Genetic alterations in PI3K pathway constituents, for example, PI3K‐activating mutations, loss of the antagonistic tumor suppressor phosphatase, and tensin homolog or via the transduction of aberrant receptor tyrosine kinase signals, result in upregulation of the PI3K pathway (Saal et al., 2005; Stemke‐Hale et al., 2008). Abnormal activation of the PI3K pathway is a common finding in a variety of tumor types, including breast cancer, and the recent BELLE‐2 study of the PI3K inhibitor, Buparlisib, showed promising results in progression‐free survival in combination with endocrine therapy, especially in patients with tumors exhibiting PIK3CA mutations (Baselga et al., 2017). Additional PI3K inhibitors are currently under clinical development (Zhao et al., 2017).
PIK3CA mutations are present in approximately 30–40% of all breast cancers (Arsenic et al., 2014; Buttitta et al., 2006; Campbell et al., 2004; Jensen et al., 2011; Koboldt et al., 2012; Li et al., 2006; Maruyama et al., 2007; Saal et al., 2005), with the highest frequency in estrogen receptor (ER)‐positive and human epidermal growth factor receptor (HER)2‐positive breast cancers (Koboldt et al., 2012; Saal et al., 2005). The four most frequent ‘hotspot’ PIK3CA mutations are located within two exons (exon 9: E545K and E542K and exon 20: H1047R and H1047L) and account for 80–90% of all PIK3CA mutations in human malignancies (Kalinsky et al., 2009; Karakas et al., 2006). We (Kodahl et al., 2015) and others (Oshiro et al., 2015) have previously reported disputed points concerning the use of ctDNA containing PIK3CA mutations as a biomarker in early‐stage breast cancer patients. The ctDNA amount is influenced by the extent of the disease, including tumor burden, and generally, very little tumor DNA is present in the circulation in early‐stage breast cancers compared to advanced stage (Diehl et al., 2005). Increasing the blood sample volume to be analyzed as well as technological improvements that increase sensitivity to detection of ctDNA in low blood concentrations is necessary to achieve better assay results (Han et al., 2017). However, ctDNA containing PIK3CA mutations has been suggested to be a promising biomarker of cancer patients with advanced disease both for early detection of recurrence and to monitor treatment response (Board et al., 2010; Dawson et al., 2013; Higgins et al., 2012), and studies reveal a correlation of PIK3CA mutation level and tumor burden in advanced breast cancer (Garcia‐Saenz et al., 2017; Higgins et al., 2012).
This study aimed to investigate whether PIK3CA mutations identified in archived formalin‐fixed, paraffin‐embedded (FFPE), metastatic tissue samples of breast cancer patients could also be detected in their corresponding ctDNA from serum samples using an optimized droplet digital PCR (ddPCR) assay (Kodahl et al., 2015). Furthermore, we sought to investigate whether the level of PIK3CA‐mutated ctDNA could be used as an early marker for monitoring treatment response and supporting clinicians in guiding treatment decisions.
2. Materials and methods
2.1. Patient samples
Metastatic tumor biopsies and corresponding serum samples were obtained from 66 patients who were part of a prospective study, in which patients from 2007 to 2013 were offered a biopsy from the metastasis, when diagnosed with recurrent breast cancer at the Department of Oncology at Odense University Hospital. Sixteen of the included 66 patients had previously had their metastatic tumor tissue analyzed for PIK3CA mutations using pyrosequencing (Jensen et al., 2011). In this study, the tumor biopsies and corresponding serum samples from these 16 patients, in addition to the other 50 patients, were analyzed for PIK3CA mutations using ddPCR (Section 3.3).
Blood was drawn into BD Vacutainer™, SST™ Serum Separation Tubes with polymer gel/silica activator. According to standard operating procedure, serum was prepared within one hour of sample collection after centrifugation (2000G, 1800 g; 10 min at 20 °C) and immediately stored at −80 °C.
Hematoxylin–eosin (HE) sections were prepared from FFPE, tissue biopsies from metastatic sites and reviewed by a skilled breast pathologist to confirm the presence and amount of tumor tissue. Cancer cell content in all cases was > 30%. For DNA extraction, 2 × 10 μm sections of FFPE samples from each tumor sample were used. Follow‐up data, including patient outcome, were available for all patients, and observations on follow‐up were censored at date of data withdrawal (March 2018). Overall survival is defined as time to death. An overall survival curve was generated by Kaplan–Meier survival analysis using stata statistical software (StataCorp LLC, College Station, TX, USA). The experiments were undertaken with the understanding and written consent of each patient. The study methodologies conformed to the standards set by the Declaration of Helsinki. The study was approved by The Regional Scientific Ethical Committee for Southern Denmark (ID: S‐20070003) and the Danish Data Protection Agency.
2.2. Droplet digital PCR of FFPE samples
Genomic DNA was extracted using a Maxwell 16 FFPE Tissue LEV DNA Purification Kit (Promega, Madison, WI, USA) according to the manufacturer's recommendations. Purified DNA was eluted in 100 μL and analyzed by ddPCR in a QX100 droplet digital system (Bio‐Rad, Copenhagen, Denmark) using PrimePCR™ ddPCR™ Mutation Assays (Bio‐Rad) for wild‐type PIK3CA as well as the four most common mutations in the PIK3CA gene: E542K (c.1624G>A) Assay ID PIK3CA:dHsaCP2000073 (mut) and dHsaCP2000074 (wt), chromosome location 3:178936050–178936172, amplicon length 80 bp, E545K (c.1633G>A) Assay ID PIK3CA:dHsaCP2000075 (mut) and dHsaCP2000076 (wt), chromosome location 3:178936068–178936190, amplicon length 80 bp, H1047L (c.3140A>T) Assay ID PIK3CA:dHsaCP2000123 (mut) and dHsaCP2000124 (wt), chromosome location 3:178952024‐178952146, amplicon length 74 bp and H1047R (c.3140A>G) Assay ID PIK3CA:dHsaCP2000077 (mut) and dHsaCP2000078 (wt), chromosome location 3:178952065‐178952187, amplicon length 80 bp. They were all performed in duplicates, as described by the manufacturer. Five microlitre sample DNA was combined with 1 μL 20× wild‐type primers/probe mix (HEX‐labeled) and 1 μL 20× target primers/probe mix (FAM‐labeled) along with 10 μL 2× ddPCR supermix for probes (Bio‐Rad) and 3 μL DNase‐/RNase‐free sterile water in a 20 μL reaction volume. Droplets were generated from the 20 μL reaction mix and 70 μL droplet generator oil for probes (Bio‐Rad) in a droplet generator DG8 cartridge (Bio‐Rad), and the droplets were transferred into a microtiter plate and subjected to thermal cycling, as recommended by the manufacturer. Thermal cycle parameters were as follows: enzyme activation 95 °C for 10 min, followed by 40 cycles of denaturation 94 °C for 30 s and annealing/extension 55 °C for 60 s with a ramp rate of 2 °C/s, and a final enzyme inactivation step 98 °C for 10 min. After PCR thermocycling, the emulsions were enumerated by fluorescence measurement using a QX100 (Bio‐Rad) droplet reader. Mutant populations were identified, and the fractional abundance calculation of mutant to total PIK3CA molecules was calculated for each sample using Poisson statistics from the qx100 software (Bio‐Rad).
2.3. Droplet digital PCR of serum samples
DNA was purified from 1 mL of serum using a MagNA Pure LC Total Nucleic Acid Isolation Kit – large volume (Roche) according to the manufacturer's instructions. Water was added to serum samples with inadequate volumes to 1 mL prior to purification. Also prior to purification, 5 μL non‐human exogenous internal control DNA spike‐in fragment of 191 bp was added to each serum sample. The control fragment was part of the soya bean CPP1 gene and generated by PCR (Pallisgaard et al., 2015). Purified DNA was eluted in 100 μL in the supplied elution buffer. Twenty‐five microlitre of the purified DNA was used to control for loss during purification by measuring the amount of the added spike‐in fragment by qPCR, contamination of lymphocyte DNA by qPCR analysis of the immunoglobulin gene rearrangements of B cells and number of genomic DNA alleles per ml of serum, as previously described (Pallisgaard et al., 2015). To increase the sensitivity of the analysis, the remaining 75 μL serum DNA was subjected to 12 cycles of PCR pre‐amplification with Q5 High‐Fidelity DNA Polymerase (New England BioLabs, Ipswich, MA, USA) using a multiplex PIK3CA primer mix. The PIK3CA ddPCR assay has been extensively validated as reported in Kodahl et al. (2015). In short, serum DNA was up‐concentrated by centrifugation 10 min 12 000 g using a Amicon Ultra‐0.5 Centrifugal Filter Unit (Merck Millipore, Burlington, MA, USA) and volume was adjusted to 20 μL with DNase‐/RNase‐free sterile water. A multiplex pre‐amplification step was incorporated prior to ddPCR using the PIK3CA E542K and H1047L PrimePCR™ ddPCR™ Mutation Assays (Bio‐Rad) primers, diluted 1 : 9. Pre‐amplification was performed in 50 μL reactions using 20 μL of up‐concentrated serum DNA, 5 μL multiplex primer mix (100 nm final concentration of each primer), and 25 μL Q5® Hot Start High‐Fidelity 2X Master Mix (New England BioLabs). Thermal cycling was performed by enzyme activation 98 °C for 10 min, followed by 12 cycles of denaturation 98 °C for 15 s, annealing 55 °C for 60 s and extension 72 °C for 60 s, and a final enzyme inactivation step 99.9 °C for 10 min. The Q5 High‐Fidelity DNA Polymerase (New England BioLabs) and TaqMan® PreAmp Master Mix (Life Technologies, Carlsbad, CA, USA) were tested for potential incorporation of technical mutations due to misincorporation of the Taq polymerase during the pre‐amplification step by comparing the mutant allele fraction in samples from healthy donors before and after pre‐amplification (Fig. 1). It should be noted that as the linearity of the pre‐amplification step was not experimentally verified there may be a slight risk of a quantification bias due to nonperfect linear pre‐amplification. However, as the wild‐type and mutant amplicon only differs by one nucleotide in the middle of the amplicon, it is very unlikely that the mutant/wild‐type ratio will be affected during the pre‐amplification step. The pre‐amplified products were then diluted 50‐fold, and ddPCR was performed in duplicates for PIK3CA E542K, E545K, H1047L, and H1047R mutation detection, according to the manufacturer's protocol (Bio‐Rad). The results were reported as a fractional abundance of mutant DNA alleles to total (mutant plus wild‐type) DNA alleles. Positive control DNA fragments for the four PIK3CA mutations were constructed and spiked into DNA from healthy donors and used as positive controls, as previously described (Spindler et al., 2012).
Figure 1.
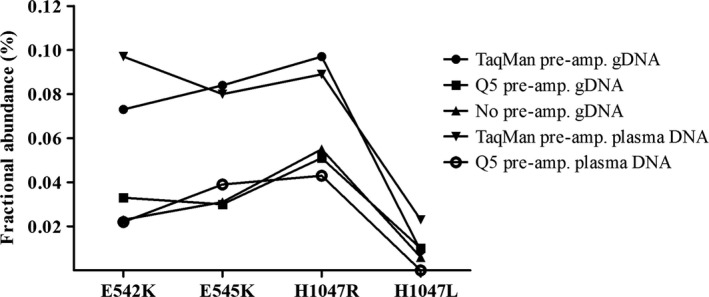
Assay specificity for PIK3CA mutations. Comparison of mutant PIK3CA detection in human control genomic DNA (gDNA) and cell‐free plasma DNA following pre‐amplification or no pre‐amplification using Q5 High‐Fidelity DNA Polymerase (New England BioLabs) or TaqMan® PreAmp Master Mix (Life Technologies). The level of blank (LoB) was 0.04% for the mutation E545K and for H1047R, 0.02% for E542K, and less than 0.01% for H1047L as shown by the graphs.
From the nonmutated alleles, the level of blank (LoB) for each of the four PIK3CA mutation assays was determined using pre‐amplified cell‐free plasma DNA and the Q5 High‐Fidelity DNA Polymerase (New England BioLabs). The LoB % for each sample was calculated by the formula: ((mut allele)/((mut allele) + (wt allele))) × 100. The standard deviation of the average LoB % was calculated, and the level of detection (LoD, with 95% confidence) was determined by (LoB + (2 × Std.dev of LoB) (Table S1). LoD of the ddPCR assay allowed detection of a mutant allele fraction of ≥ 0.08% (Table S1) corresponding to one mutant molecule in a background of 1200 wild‐type molecules.
2.4. Controls
Four 180‐bp DNA fragments containing one of the four PIK3CA mutations analyzed in this study were generated by site‐directed PCR mutagenesis, as previously described (Spindler et al., 2012). The mutated PCR fragments were spiked into normal donor DNA and used as positive controls throughout the study. In all ddPCR and qPCR, positive and negative (water) and wild‐type (normal donor DNA) controls were performed in parallel.
3. Results
3.1. Patient characteristics
Archived formalin‐fixed, paraffin‐embedded metastatic tumor biopsies and corresponding serum samples were available from 66 patients with metastatic disease who were part of a prospective study (2007–2013) at the Department of Oncology, Odense University Hospital, Denmark (Fig. 2). Metastatic tissue samples from 16 patients had, as part of a previous study (Jensen et al., 2011), been tested for the presence of PIK3CA mutations by pyrosequencing, 15 of which were found to be PIK3CA‐mutation positive and one wild‐type. In this study, 50 additional patients were included to a total of 66 patients. Of these 66 patients, four samples were excluded due to less than 10% detectable tumor cells in the FFPE tumor tissue, and two patients were excluded due to missing serum samples, resulting in a total of 60 matched samples. Serum samples from 24 patients with a PIK3CA mutation in their metastatic tissue and five patients with PIK3CA wild‐type metastatic tissue (randomly selected) were analyzed for circulating cell‐free PIK3CA mutations using ddPCR (Fig. 2). The survival curve from Kaplan–Meier estimates of the 29 metastatic breast cancer patients included for further analysis showed a median overall survival of 20 months (Fig. S1).
Figure 2.
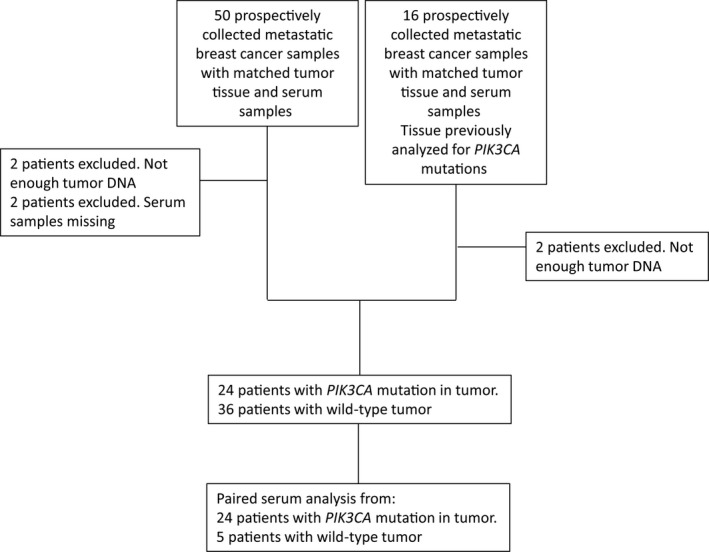
Consort diagram. A total of 66 metastatic tumor biopsies and corresponding serum samples were prospectively collected. Of these, 24 metastasis with PIK3CA mutation and five wild‐type as determined by ddPCR were selected for further analysis.
3.2. PIK3CA mutation analysis of metastatic breast cancer samples
PIK3CA mutations were observed in 40% of the total metastatic tissue samples (24/60), with 33% (8/24) having exon 9 mutations (E542K and E545K) and 67% (16/24) having exon 20 mutations (H1047L and H1047R), as determined by the optimized ddPCR method. However, the inclusion of the preselected samples might account for the high overall observed frequency of PIK3CA mutations in tumor tissue compared to the unselected samples (24%; 11/46).
Of the 24 breast cancer patients with PIK3CA mutation‐positive metastasis, 92% were ER‐positive (n = 22), 8% were ER‐negative (n = 2), and 8% were HER2‐amplified (n = 2) (Table 1). The first blood sample from each patient was taken an average of 1.5 days after diagnostic biopsy (range: 26 days before to 43 days after diagnosis) (Table 2). Of the 24 patients with detectable PIK3CA mutations in the metastatic tumor tissue, 20 (83%) had similar mutations detectable in corresponding ctDNA from serum. Four patients with PIK3CA mutations in metastatic tumor tissue had no detectable mutations in their corresponding serum samples, and interestingly, all had nonvisceral metastatic disease. No patients with PIK3CA wild‐type tumor biopsies (n = 5) exhibited PIK3CA mutations in their corresponding serum samples.
Table 1.
Clinical characteristics of breast cancer patients with metastatic tissue and corresponding ctDNA from serum analyzed for PIK3CA mutation
| Patient no. | ER status | HER2 status | Site of metastatic disease | Site of biopsy |
|---|---|---|---|---|
| 1 | + | + | Non‐visceral | Lymph nodes |
| 2 | + | − | Non‐visceral | Bone |
| 3 | + | − | Non‐visceral | Bone |
| 4 | + | − | Non‐visceral | Lymph nodes |
| 5 | − | − | Non‐visceral | Subcutis, chest wall |
| 6 | + | − | Visceral, Non‐visceral | Liver |
| 7 | + | − | Non‐visceral | Bone |
| 8 | + | − | Visceral | Pleural effusion |
| 9 | − | − | Visceral, Non‐visceral | Lymph nodes |
| 10 | + | − | Non‐visceral | Bone |
| 11 | + | NA | Visceral, Non‐visceral | Lymph nodes |
| 12 | + | − | Visceral, Non‐visceral | Bone |
| 13 | + | − | Visceral, Non‐visceral | Liver |
| 14 | + | − | Non‐visceral | Bone |
| 15 | + | − | Non‐visceral | Bone |
| 16 | + | − | Visceral, Non‐visceral | Liver |
| 17 | + | − | Visceral, Non‐visceral | Liver |
| 18 | + | − | Non‐visceral | Subcutis, chest wall |
| 19 | + | − | Visceral, Non‐visceral | Liver |
| 20 | + | − | Non‐visceral | Lymph nodes |
| 21 | + | − | Visceral, Non‐visceral | Bone |
| 22 | + | − | Visceral, Non‐visceral | Liver |
| 23 | + | − | Visceral, Non‐visceral | Liver |
| 24 | + | + | Non‐visceral | Skin |
| 25 | + | − | Non‐visceral | Bone |
| 26 | − | − | Visceral, Non‐visceral | Liver |
| 27 | + | − | Visceral, Non‐visceral | Bone |
| 28 | − | − | Visceral, Non‐visceral | Subcutis |
| 29 | + | − | Visceral, Non‐visceral | Lymph nodes |
ER+ cutoff was ≥1% positive tumor cells. Visceral metastases are metastases in the lung, liver, brain and/or peritoneum. Non‐visceral localizations are the skin, lymph nodes, soft tissue and/or bone. NA, not available.
Table 2.
PIK3CA mutational status and treatment response of breast cancer patients with metastatic tissue and corresponding ctDNA from serum analyzed for PIK3CA mutation
| Patient no. | Tumor tissuemutation | Serum ctDNAmutation | Previousregisteredmutation | Serum 1(days) | Serum 1ctDNA(copies/μl) | Serum 2(days) | Serum 2ctDNA(copies/μl) | Serum 3(days) | Serum 3ctDNA(copies/μl) | Treatmenttype | Treatment start (days) | Status(days) | Imagingtype | Response |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | E542K | E542K | −4 | 1.6 | ||||||||||
| 2 | E545K | E545K | −6 | 49 | ||||||||||
| 3 | H1047R | H1047R | 0 | 13.6 | ||||||||||
| 4 | WT | No | H1047R | −3 | ||||||||||
| 5 | E542K | No | −14 | |||||||||||
| 6 | E542K | E542K/ E545K | E542K | −2 | 21/230 | |||||||||
| 7 | H1047L | H1047L | −2 | 761 | ||||||||||
| 8 | WT | No | E545K | 18 | ||||||||||
| 9 | WT | No | −10 | |||||||||||
| 10 | H1047R | H1047R | H1047R | 0 | 3.1 | |||||||||
| 11 | H1047L | H1047L | 36 | 5 | ||||||||||
| 12 | E545K | E545K | E545K/ H1047R | −12 | 125.1 | |||||||||
| 13 | H1047R | H1047R | −2 | 1170 | ||||||||||
| 14 | E542K | No | 43 | |||||||||||
| 15 | H1047R | No | −1 | |||||||||||
| 16 | H1047R | H1047R | −5 | 17.4 | ||||||||||
| 17 | H1047R | H1047R | −1 | 9500 | ||||||||||
| 18 | H1047R | No | −13 | |||||||||||
| 19 | E545K | E545K | −3 | 694 | ||||||||||
| 20 | H1047R | H1047R | −10 | 1.8 | ||||||||||
| 21 | H1047L | H1047L | −3 | 21.8 | ||||||||||
| 22 | E545K | E545K | −14 | 114.2 | ||||||||||
| 23 | H1047R | H1047R | H1047R | −2 | 15.4 | |||||||||
| 24 | H1047R | H1047R | 15 | 26.5 | 36 | 3.9 | Docetaxel | 15 | 76 | CT | PR | |||
| 25 | H1047R | H1047R | −26 | 22.5 | 2 | 3 | AI | −26 | 96 | CT | PR | |||
| 26 | H1047R | H1047R | 0 | 566 | 90 | 2.8 | Epirubicin, Cyclophosphamide | 12 | 74 | PET | PR | |||
| 27 | H1047R | H1047R | −22 | 343 | 33 | 41.3 | 47 | 9.2 | Paclitaxel | 13 | 67 | PET | PR | |
| 28 | WT | No | 16 | 53 | Capecitabine | 33 | 83 | CT | PD | |||||
| 29 | WT | No | 16 | 127 | AI | 8 | 86 | CT | NC |
Day 0 refers to the day of diagnostic biopsy from a metastatic lesion. Serum samples and treatment start are given in days after biopsy. No serum ctDNA mutation refers to none measurable PIK3CA mutation in serum. Medical imaging (computed tomography (CT) or positron emission tomography (PET)) was used to measure the response and assessed according to RECIST 1.1 and PERCIST 1.0 criteria: PR, partial response; PD, progressive disease; NC, no change; AI, aromatase inhibitor; WT, wild‐type.
3.3. PIK3CA‐mutated ctDNA as an early marker for treatment response
Six patients had serial serum samples collected during oncological treatment (four patients with detectable ctDNA containing PIK3CA mutations and two wild‐type that remained wild‐type) (Table 2). Response evaluation was performed using computed tomography (CT) or positron emission tomography (PET) imaging according to RECIST 1.1 and PERCIST 1.0 (Eisenhauer et al., 2009) an average of 11.5 weeks after diagnostic biopsy (range: 9.5–15 weeks) (Table 2). Clinicopathological patient characteristics is provided in Table 1 and the results of the response evaluations is provided in Table 2. Correlation between ctDNA PIK3CA mutation level, treatment, and disease history for the four patients is shown in Fig. 3. Serum samples from all four patients (patient nos 24, 25, 26, and 27) showed a decreased level of PIK3CA‐mutated ctDNA in the second and, for one, third serum sample, suggesting tumor shrinkage during treatment. This was verified by CT or PET imaging showing partial response in all four patients during treatment within this follow‐up period (Table 2 and Fig. 3). Unfortunately, no additional serum samples were collected from these patients.
Figure 3.
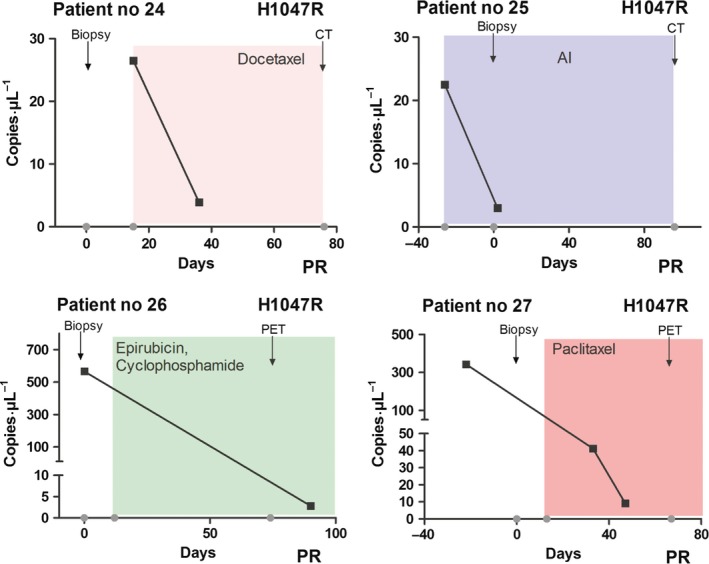
Serial monitoring of PIK3CA point mutation levels and correlation with treatment response in patients with advanced breast cancer. Measurements of serial fractional abundance of PIK3CA‐mutated ctDNA (H1047R) and evaluation of treatment response by CT or PET imaging in four patients (patient nos 24, 25, 26, and 27). Blood samples were collected two to three times per patient and the level of PIK3CA‐mutated ctDNA in serum reported as copies·μL−1 (■). Details regarding type of treatment (docetaxel, aromatase inhibitor (AI), epirubicin, cyclophosphamide, or paclitaxel) and treatment schedule are indicated by colored shading. Time for biopsy and CT or PET imaging is indicated by arrows. All patients showed partial response (PR) after treatment.
4. Discussion
Tissue biopsy is an invasive procedure that may cause distress as well as complications for the patient. Moreover, due to intratumoral heterogeneity, the use of tumor tissue to obtain an accurate genomic landscape of breast cancer could be challenging (Gerlinger et al., 2012). Accurate determination of the genomic landscape of breast tumors is important to identify driver mutations that may make them susceptible to targeted antitumor agents, but also to determine whether subclones within the metastasis subsequently acquire additional mutations that render the lesion drug‐resistant, leading to disease progression (Diaz et al., 2012). Metastatic lesions are, unlike primary tumors, generally not surgically accessible and must be treated with systemic therapies. To make decisions regarding targeted cancer treatment, liquid biopsies, including detection of mutant tumor DNA in the circulation, may be the future method of choice to determine optimal treatment strategy, monitor treatment response, and characterize escaping resistant subclones that may cause metastases.
In this study, we show that PIK3CA mutations can be identified in ctDNA of serum samples from patients with PIK3CA‐mutated metastatic breast cancer and that the circulating PIK3CA mutation level might be used to follow treatment response in these patients. Using an optimized, highly sensitive, ddPCR assay, we found that 83% of patients with PIK3CA mutation in the metastatic tumor tissue had detectable circulating PIK3CA mutations in their corresponding serum sample, which underlines that the method is highly useful to obtain information regarding tumor characteristics and might be favored over the invasive and time consuming solid biopsies. Moreover, our study implies that ctDNA is a resource for therapeutic guidance in patients who may benefit from PI3K‐selective inhibitor treatment (Baselga et al., 2017).
Response evaluation during treatment of metastatic breast cancer is currently assessed primarily by visualization of imaging data and/or clinical examination of the patient at monthly intervals. Liquid biopsies of ctDNA may provide an additional method to monitor tumor response to treatment. Although only four patients with PIK3CA mutation in metastatic tumor tissue were followed with serial serum samples, all showed changes in PIK3CA ctDNA levels that correlated with treatment response according to imaging assessments. Similar results have been reported by others (Garcia‐Saenz et al., 2017), although no correlation between PIK3CA mutation levels and treatment response was observed in two of eight advanced breast cancer patients (Garcia‐Saenz et al., 2017). Cases with discordance between PIK3CA mutation level and treatment response could be explained by the technology used (Kodahl et al., 2015) or the disease stage (early‐stage versus advanced breast cancer; Garcia‐Saenz et al., 2017; Kodahl et al., 2015), or may reflect tumor evolution and heterogeneity within advanced breast cancer disease. However, liquid biopsies have previously been suggested to reflect the global (primary and metastatic sites) molecular status of cancer in terms of tumor heterogeneity better than a solid tumor biopsy (Crowley et al., 2013). While the four patients followed with serial serum samples all had H1047R mutations, we would expect similar results for patients with other hotspot PIK3CA mutations. Our results from analysis of the serial serum samples suggest that the level of circulating PIK3CA mutations could be a marker of disease burden in patients with PIK3CA mutant tumors. However, having a PIK3CA mutation does not necessarily mean that the only effective treatment is PI3K‐targeted and, in the case of the patients we studied, tumor reduction was caused by chemo or antihormonal therapies.
Similar to our study, good concordance between PIK3CA mutation status in formalin‐fixed, paraffin‐embedded, biopsies and ctDNA of patients with metastatic breast cancer has been observed by others (Garcia‐Saenz et al., 2017; Higgins et al., 2012). Interestingly, the patients with PIK3CA mutation identified in the metastatic tissue but in whom similar mutations could not be detected in corresponding serum ctDNA all had nonvisceral metastatic disease, suggesting that the location of the metastasis, in addition to size, may influence the amount of tumor DNA shed into the circulation. On the other hand, the sensitivity of the ddPCR assay allowed detection of a mutant allele fraction of > 0.084%, corresponding to one mutant molecule in a background of 1200 wild‐type molecules, making it highly sensitive. A higher concordance between PIK3CA mutation status in formalin‐fixed, paraffin‐embedded, biopsies and ctDNA might have been achieved if we had used plasma instead of serum, as serum DNA can be contaminated by blood cell DNA (Jen et al., 2000). However, the risk of contamination was limited as serum was isolated within one hour from blood drawing and subsequently immediately stored at −80 °C. Further, all DNA samples were tested for potential blood cell DNA contamination by a novel qPCR assay utilizing the immunoglobulin gene rearrangements in B cells (Pallisgaard et al., 2015).
5. Conclusions
In conclusion, our results show high concordance between tumor tissue and ctDNA mutation status and suggest that serum samples from advanced breast cancer patients and ddPCR may be used for PIK3CA mutation status assessment to complement imaging techniques as a tool for monitoring treatment response. However, as not all metastatic breast cancers are PIK3CA‐positive, additional biomarkers in the metastatic setting are needed.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
ARK, SE, ASK, and HJD participated in the study design. ARK and SE coordinated the project, performed data interpretation, and wrote the first draft of the manuscript. NP performed molecular analysis. AMBJ, JDJ, and AVL provided samples and clinical and paraclinical information. NP and HD assisted in writing the manuscript, and all authors have read and approved the final manuscript.
Supporting information
Fig. S1. Overall survival analysis of patients analyzed for circulating cell‐free PIK3CA mutations in serum.
Table S1. Level of detection and level of blank of the ddPCR assay.
Acknowledgements
The authors thank M. K. Occhipinti for editorial assistance. The public and private foundations that supported the study had no role in the design and conduct of this study; in the collection, management, analysis, and interpretations of the data; or in the preparation, review, or approval of the manuscript. The authors had full access to all the data in the study and had the final responsibility for the decision to submit the manuscript for publication. This work was supported by the Danish Cancer Society (H.J. Ditzel) and the Academy of Geriatric Cancer Research (AgeCare), Odense University Hospital (H.J. Ditzel).
Contributor Information
Annette R. Kodahl, Email: annette.kodahl@rsyd.dk
Sidse Ehmsen, Email: sehmsen@health.sdu.dk.
Henrik J. Ditzel, Email: hditzel@health.sdu.dk.
References
- Arsenic R, Lehmann A, Budczies J, Koch I, Prinzler J, Kleine‐Tebbe A, Schewe C, Loibl S, Dietel M and Denkert C (2014) Analysis of PIK3CA mutations in breast cancer subtypes. Appl Immunohistochem Mol Morphol 22, 50–56. [DOI] [PubMed] [Google Scholar]
- Baselga J, Im SA, Iwata H, Cortes J, De Laurentiis M, Jiang Z, Arteaga CL, Jonat W, Clemons M, Ito Y et al (2017) Buparlisib plus fulvestrant versus placebo plus fulvestrant in postmenopausal, hormone receptor‐positive, HER2‐negative, advanced breast cancer (BELLE‐2): a randomised, double‐blind, placebo‐controlled, phase 3 trial. Lancet Oncol 18, 904–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Board RE, Wardley AM, Dixon JM, Armstrong AC, Howell S, Renshaw L, Donald E, Greystoke A, Ranson M, Hughes A et al (2010) Detection of PIK3CA mutations in circulating free DNA in patients with breast cancer. Breast Cancer Res Treat 120, 461–467. [DOI] [PubMed] [Google Scholar]
- Buttitta F, Felicioni L, Barassi F, Martella C, Paolizzi D, Fresu G, Salvatore S, Cuccurullo F, Mezzetti A, Campani D et al (2006) PIK3CA mutation and histological type in breast carcinoma: high frequency of mutations in lobular carcinoma. J Pathol 208, 350–355. [DOI] [PubMed] [Google Scholar]
- Campbell IG, Russell SE, Choong DYH, Montgomery KG, Ciavarella ML, Hooi CS, Cristiano BE, Pearson RB and Phillips WB (2004) Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res 64, 7678–7681. [DOI] [PubMed] [Google Scholar]
- Choi JJ, Reich CF 3rd and Pisetsky DS (2005) The role of macrophages in the in vitro generation of extracellular DNA from apoptotic and necrotic cells. Immunology 115, 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley E, Di Nicolantonio F, Loupakis F and Bardelli A (2013) Liquid biopsy: monitoring cancer‐genetics in the blood. Nat Rev Clin Oncol 10, 472–484. [DOI] [PubMed] [Google Scholar]
- Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler‐Araujo B et al (2013) Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 368, 1199–1209. [DOI] [PubMed] [Google Scholar]
- Diaz LA Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, Allen B, Bozic I, Reiter JG, Nowak MA et al (2012) The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 486, 537–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, Diaz LA Jr, Goodman SN, David KA, Juhl H et al (2005) Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A 102, 16368–16373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M et al (2009) New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 45, 228–247. [DOI] [PubMed] [Google Scholar]
- Garcia‐Saenz JA, Ayllon P, Laig M, Acosta‐Eyzaguirre D, Garcia‐Esquinas M, Montes M, Sanz J, Barquín M, Moreno F, Garcia‐Barberan V et al (2017) Tumor burden monitoring using cell‐free tumor DNA could be limited by tumor heterogeneity in advanced breast cancer and should be evaluated together with radiographic imaging. BMC Cancer 17, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A et al (2012) Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366, 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Wang J and Sun Y (2017) Circulating tumor DNA as biomarkers for cancer detection. Genomics Proteomics Bioinformatics 15, 59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao YX, Fu Q, Guo YY, Ye M, Zhao HX, Wang Q, Peng XM, Li QW, Wang RL and Xiao WH (2017) Effectiveness of circulating tumor DNA for detection of KRAS gene mutations in colorectal cancer patients: a meta‐analysis. Onco Targets Ther 10, 945–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins MJ, Jelovac D, Barnathan E, Blair B, Slater S, Powers P, Zorzi J, Jeter SC, Oliver GR, Fetting J et al (2012) Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clin Cancer Res 18, 3462–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jen J, Wu L and Sidransky D (2000) An overview on the isolation and analysis of circulating tumor DNA in plasma and serum. Ann N Y Acad Sci 906, 8–12. [DOI] [PubMed] [Google Scholar]
- Jensen JD, Laenkholm AV, Knoop A, Ewertz M, Bandaru R, Liu WH, Hackl W, Barrett JC and Gardner H (2011) PIK3CA mutations may be discordant between primary and corresponding metastatic disease in breast cancer. Clin Cancer Res 17, 667–677. [DOI] [PubMed] [Google Scholar]
- Kalinsky K, Jacks LM, Heguy A, Patil S, Drobnjak M, Bhanot UK, Hedvat CV, Traina TA, Solit D, Gerald W et al (2009) PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res 15, 5049–5059. [DOI] [PubMed] [Google Scholar]
- Karakas B, Bachman KE and Park BH (2006) Mutation of the PIK3CA oncogene in human cancers. Br J Cancer 94, 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Fulton RS, McLellan MD, Schmidt H, Kalicki‐Veizer J, McMichael JF, Fulton LL, Dooling DJ, Ding L, Mardis ER et al (2012) Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodahl AR, Pallisgaard N, Jylling AMB, Cold S, Knoop AS and Ditzel HJ (2015) Detecting plasma tumor DNA in early‐stage breast cancer‐Letter. Clin Cancer Res 21, 3569. [DOI] [PubMed] [Google Scholar]
- Li SY, Rong MN, Grieu F and Iacopetta B (2006) PIK3CA mutations in breast cancer are associated with poor outcome. Breast Cancer Res Treat 96, 91–95. [DOI] [PubMed] [Google Scholar]
- Maruyama N, Miyoshi Y, Taguchi T, Tamaki Y, Monden M and Noguchi S (2007) Clinicopathologic analysis of breast cancers with PIK3CA mutations in Japanese women. Clin Cancer Res 13, 408–414. [DOI] [PubMed] [Google Scholar]
- Oshiro C, Kagara N, Naoi Y, Shimoda M, Shimomura A, Maruyama N, Shimazu K, Kim SJ and Noguchi S (2015) PIK3CA mutations in serum DNA are predictive of recurrence in primary breast cancer patients. Breast Cancer Res Treat 150, 299–307. [DOI] [PubMed] [Google Scholar]
- Pallisgaard N, Spindler KL, Andersen RF, Brandslund I and Jakobsen A (2015) Controls to validate plasma samples for cell free DNA quantification. Clin Chim Acta 446, 141–146. [DOI] [PubMed] [Google Scholar]
- Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, Yu JS, Malmström PO, Mansukhani M, Enoksson J et al (2005) PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res 65, 2554–2559. [DOI] [PubMed] [Google Scholar]
- Siravegna G, Marsoni S, Siena S and Bardelli A (2017) Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 14, 531–548. [DOI] [PubMed] [Google Scholar]
- Spindler KL, Pallisgaard N, Vogelius I and Jakobsen A (2012) Quantitative cell‐free DNA KRAS and BRAF mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clin Cancer Res 18, 1177–1185. [DOI] [PubMed] [Google Scholar]
- Stemke‐Hale K, Gonzalez‐Angulo AM, Lluch A, Neve RM, Kuo WL, Davies M, Carey M, Hu Z, Guan Y, Sahin A et al (2008) An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res 68, 6084–6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroun M, Maurice P, Vasioukhin V, Lyautey J, Lederrey C, Lefort F, Rossier A, Chen XQ and Anker P (2000) The origin and mechanism of circulating DNA. Ann N Y Acad Sci 906, 161–168. [DOI] [PubMed] [Google Scholar]
- Zhao W, Qiu Y and Kong D (2017) Class I phosphatidylinositol 3‐kinase inhibitors for cancer therapy. Acta Pharm Sin B 7, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Overall survival analysis of patients analyzed for circulating cell‐free PIK3CA mutations in serum.
Table S1. Level of detection and level of blank of the ddPCR assay.


