Abstract
The MiT‐TFE family of basic helix‐loop‐helix leucine‐zipper transcription factors includes four members: TFEB, TFE3, TFEC, and MITF. Originally described as oncogenes, these factors play a major role as regulators of lysosome biogenesis, cellular energy homeostasis, and autophagy. An important mechanism by which these transcription factors are regulated involves their shuttling between the surface of lysosomes, the cytoplasm, and the nucleus. Such dynamic changes in subcellular localization occur in response to nutrient fluctuations and various forms of cell stress and are mediated by changes in the phosphorylation of multiple conserved amino acids. Major kinases responsible for MiT‐TFE protein phosphorylation include mTOR, ERK, GSK3, and AKT. In addition, calcineurin de‐phosphorylates MiT‐TFE proteins in response to lysosomal calcium release. Thus, through changes in the phosphorylation state of MiT‐TFE proteins, lysosome function is coordinated with the cellular metabolic state and cellular demands. This review summarizes the evidence supporting MiT‐TFE regulation by phosphorylation at multiple key sites. Elucidation of such regulatory mechanisms is of fundamental importance to understand how these transcription factors contribute to both health and disease.
Keywords: autophagy, lysosome, mTOR, nucleo‐cytoplasmic shuttling, TFEB
Subject Categories: Autophagy & Cell Death, Metabolism, Signal Transduction
Introduction
Transcriptional regulation plays a crucial role in the adaptation of cell homeostasis to environmental cues. Nucleo‐cytoplasmic shuttling of transcription factors is used by the cell to control gene expression programs in response to the environment. Transcription factors exert their function primarily in the nucleus; however, under some conditions, they may be localized in the cytoplasm. The import of transcription factors from the cytoplasm to the nucleus is facilitated by several mechanisms, importin‐mediated and importin‐independent (Xu & Massague, 2004). For example, the NF‐κB and NFAT transcription factors are imported to the nucleus through a nuclear localization signal (NLS), a cluster of basic amino acids that is recognized by importin‐β, which in turn binds importin‐α to promote nuclear import. In contrast, nuclear translocation of SMAD transcription factors is importin‐independent and requires their direct binding to the nuclear pore (Beg et al, 1992; Henkel et al, 1992; Zhu et al, 1998; Hill, 2009). This process is dependent on the activity of the small GTPase Ran, which mediates nuclear import and export of transcription factors (Gorlich & Kutay, 1999; Xu & Massague, 2004).
Extracellular signals may affect nucleo‐cytoplasmic shuttling of transcription factors in several ways. A common mechanism used by the cell to link signaling pathways to the control of gene expression is the phosphorylation of transcription factors (Nardozzi et al, 2010). One mode of regulation by phosphorylation involves controlling protein subcellular localization. Phosphorylation may promote nuclear import by enhancing the binding affinity for importins or by unmasking an NLS, but may also inhibit nuclear import by acting on a component of the nuclear transport machinery or by disrupting the NLS (Nardozzi et al, 2010). In the case of transcription factors, the control of their subcellular localization is an important way to modulate gene expression programs with profound effects on the metabolic adaptation to environmental cues.
In this review article, we focus on how phosphorylation‐mediated signaling pathways regulate the subcellular localization and function of transcription factor EB (TFEB) and the other members of the MiT‐TFE family. Such regulation is of importance for coordinating expression of lysosomal‐autophagic pathway and cell metabolism genes and thereby allowing cells to adapt to changing environmental cues.
Transcription factor EB and the MiT‐TFE family
The MiT‐TFE family
The MiT‐TFE family of transcription factors includes four members: MITF, TFEB, TFE3, and TFEC (Steingrimsson et al, 2004). These are helix‐loop‐helix (HLH) leucine‐zipper transcription factors that share high sequence similarities and activate expression of their target genes by binding DNA either as homo‐ or hetero‐dimers. Like other members of the larger family of HLH leucine‐zipper transcription factors, MiT‐TFE proteins bind a palindromic DNA sequence (CACGTG) located in the proximal promoter of target genes (Fisher et al, 1991; Hemesath et al, 1994). This sequence, referred to as an E‐box, conforms to the CANNTG motif that is recognized by other members HLH/leucine‐zipper family transcription factors (Hemesath et al, 1994). However, specificity for DNA binding in the HLH‐LZ family is influenced by sequences immediately flanking the E‐box such that the MiT‐TFE proteins prefer the GTCACGTGAC consensus sequence that is known as a CLEAR motif (Sardiello et al, 2009; Palmieri et al, 2011).
Structural and biochemical data suggest that MiT‐TFE proteins may heterodimerize with one another but not with other members of the HLH/leucine‐zipper family (Pogenberg et al, 2012). MiT‐TFE genes are present in all metazoan organisms. However, commonly studied invertebrates such as Drosophila and Caenorhabditis elegans only have a single member of the family, named Mitf and HLH‐30, respectively (Rehli et al, 1999; Hallsson et al, 2004), whose function and regulation appear to be similar to TFEB (Lapierre et al, 2013; O'Rourke & Ruvkun, 2013; Settembre et al, 2013b; Zhang et al, 2015; Bouche et al, 2016; Tognon et al, 2016). Proteins of the MiT‐TFE family have a large degree of overlap in their function and regulatory mechanisms. In general, they are ubiquitously expressed but their expression levels in different tissues vary considerably. One exception is an MITF splice variant (MITF‐M) that exhibits constitutive nuclear localization and is expressed almost exclusively in melanocytes (Yasumoto et al, 1998).
TFEB and TFE3 as master regulators of lysosomal function and autophagy
While the function of MITF as regulator of melanoblast survival and differentiation, melanosome biogenesis, and eye development has been known for some time due to the striking coat color and eye development defects detected in mice and rats harboring spontaneous MITF mutations (Hodgkinson et al, 1993; Steingrimsson et al, 1994, 2004; Opdecamp et al, 1997), the function of the other members of the MiT‐TFE family has been more elusive. Notably, while TFEB knock‐out mice are embryonic lethal (Steingrimsson et al, 1998), there was an apparent lack of overt phenotypes in both TFE3 and TFEC knock‐out mice (Steingrimsson et al, 2002).
Important insights into the function of TFEB, and subsequently of its closely related paralogue TFE3, came from a systems biology study aimed at testing the hypothesis that lysosomal function was globally regulated at the transcriptional level. This study led to the identification of a transcriptional network of genes involved in lysosomal biogenesis, named coordinated lysosomal expression and regulation (CLEAR) network, and of TFEB as its master regulator (Sardiello et al, 2009). Subsequent studies demonstrated that TFEB is also able to regulate multiple aspects of lysosome function such as autophagy (Settembre et al, 2011) and lysosomal exocytosis (Medina et al, 2011). Interestingly, TFE3 was also found to control lysosomal biogenesis and autophagy by regulating a gene network that largely overlaps with the one regulated by TFEB (Martina et al, 2014).
The role of TFEB in the control of lysosomal biogenesis, autophagy, and lysosomal exocytosis may be exploited to promote cellular clearance in a number of disease conditions (Medina et al, 2011). This approach was tested in several cellular and mouse models of human diseases resulting from the accumulation of undegraded substances, such as lysosomal storage diseases (LSD; Medina et al, 2011; Song et al, 2013; Spampanato et al, 2013; Rega et al, 2016), Parkinson's disease (Dehay et al, 2010; Decressac et al, 2013; Kilpatrick et al, 2015), Alzheimer's disease (Polito et al, 2014; Xiao et al, 2014; Chauhan et al, 2015), and diet‐induced obesity (Settembre et al, 2013a), among others. Like TFEB, TFE3 was shown to promote cellular clearance in a mouse model of Pompe disease (Martina et al, 2014).
Physiological roles of TFEB and TFE3
Loss‐of‐function approaches based on knock‐out (KO) mice have been used to explore the physiological roles of MiT‐TFE genes. Single and double KO mice were used to investigate redundancy and cooperation between these transcription factors. Interestingly, double KO mice for MITF and TFE3 show osteoclast defects that were not observed in either of the respective single KO strains (Steingrimsson et al, 2002). Due to the embryonic lethality of global TFEB KO mice, which arises due to a defect in placental vascularization (Steingrimsson et al, 1998), conditional KO mice were generated in which TFEB was deleted in specific tissues. Table 1 lists the TFEB full KO line and tissue‐specific conditional KO and conditional overexpressor lines that were generated to explore the physiological role of TFEB in specific organs and tissues. TFEB liver‐specific conditional KO mice displayed severe abnormalities of lipid metabolism that are significantly enhanced by a high‐fat diet, resulting in severe obesity and diabetes (Settembre et al, 2013a; Pastore et al, 2017). This effect of TFEB on lipid metabolism is mediated by genes involved in lipid degradation pathways, such as fatty acid oxidation and lipophagy (Settembre et al, 2013b).
Table 1.
TFEB full KO line and tissue‐specific conditional KO and conditional overexpression lines that were generated to explore the physiological role of TFEB in specific organs and tissues
| Mouse line | Tissue | Phenotype | References |
|---|---|---|---|
| Loss of function | |||
| Full KO | Ubiquitous | Embryonic lethality (E9.5) | Steingrimsson et al (1998) |
| Conditional KO | Bone (CtsK‐Cre) | Defective bone resorption | Ferron et al (2013) |
| Conditional KO | Liver (Alb‐Cre) | Impaired liver metabolism, metabolic imbalance, exacerbated obesity | Settembre et al (2013a) and Pastore et al (2017) |
| Conditional KO | Macrophages (Lys2‐Cre) | Impaired inflammatory and immune response | Pastore et al (2016) |
| Conditional KO | Muscle (Mlc1f‐Cre) | Impaired mitochondrial function and glucose homeostasis | Mansueto et al (2017) |
| Conditional KO | Intestinal epithelium (Villin‐Cre) | Exacerbated colitis | Murano et al (2017) |
| Conditional KO | Endothelial cells (VE‐Cad‐Cre) | Decreased angiogenesis and attenuated blood flow recovery after ischemic injury | Fan et al (2018) |
| Gain of function | |||
| Transgenic | Brain (Thy1 promoter) | Clearance of toxic aggregates in an AD model | Wang et al (2016) |
| Conditional transgenic | Muscle (HSA‐Cre‐Er) | Increased glucose and lipid metabolism | Mansueto et al (2017) |
| Conditional transgenic | Kidney (Cdh16‐Cre; Cdh16‐Cre‐Ert2) | Renal cystic pathology and papillary carcinoma | Calcagni et al 16) |
| Conditional transgenic | Macrophages (LyzM‐Cre) | Enhanced degradative capacity, reduced atherosclerosis in ApoE null mice | Sergin et al (2016) |
| Transgenic | Endothelial cells (Tie2 promoter) | Increased angiogenesis and improved blood flow recovery after ischemic injury | Fan et al (2018) |
Loss of TFEB in the muscle has a major impact on cellular energy metabolism. TFEB muscle‐specific conditional KO mice showed impaired glucose homeostasis and mitochondrial biogenesis with decreased fatty acid oxidation and oxidative phosphorylation. In addition, these mice have reduced metabolic flexibility during physical exercise, which makes them unable to exploit its beneficial effects on metabolism. Transcriptomic analysis of muscle‐specific TFEB KO mice showed that in muscle, TFEB regulates the expression of genes encoding glucose transporters and glycolytic enzymes as well as genes involved in mitochondrial biogenesis (Mansueto et al, 2017).
Transcription factor EB was also shown to play a broad role in the regulation of the innate immune response. A key signaling pathway is the cGAS‐STING pathway, which senses double‐stranded DNA (dsDNA) in the cytosol and leads to the activation of an inflammatory response. Indeed, inactivation of TREX1, an exonuclease, leads to cytosolic DNA accumulation, TFEB nuclear localization and increased lysosome biogenesis (Hasan et al, 2013). In addition, TFEB plays a role in activated macrophages. Conditional KO mice lacking TFEB in macrophages showed decreased expression of genes encoding pro‐inflammatory cytokines and chemokines. Furthermore, macrophages lacking TFEB displayed impaired autophagy and lysosomal biogenesis (Pastore et al, 2016). Such lysosomal defects arising from TFEB depletion in macrophages have furthermore been shown to impair their ability to upregulate anti‐bacterial activities in response to bacterial exposure (Gray et al, 2016). Further evidence for the role of TFEB in inflammation and immunity came from the study of conditional KO mice in which TFEB was specifically deleted in the intestinal epithelium, which showed increased susceptibility to epithelial cell injury and subsequent colitis (Murano et al, 2017). Recently, by using endothelial cell (EC)‐specific TFEB transgenic and KO mice, TFEB was also shown to positively regulate angiogenesis via activation of AMPK and autophagy (Fan et al, 2018).
Remarkably, TFE3 full KO mice appeared superficially healthy, but in‐depth analysis revealed cellular and metabolic phenotypes that are very similar to TFEB liver‐, muscle‐, and macrophage‐specific conditional KO mice, and such effects are significantly enhanced by the loss of both TFEB/TFE3 in these tissues (Pastore et al, 2016, 2017). These data suggest that these two transcription factors regulate very similar sets of genes in multiple tissues and play a cooperative, rather than redundant role. Alternatively, the lack of redundancy may reflect a high degree of sensitivity to their dosage. Consistent with the idea of dosage, TFEB overexpression via viral‐mediated gene transfer was able to rescue the phenotype of TFE3 KO mice and vice versa (Pastore et al, 2017).
Recently, TFEB and TFE3 were also shown to regulate the induction of protein synthesis via mechanistic target of rapamycin complex 1 (mTORC1) upon amino acid feeding after starvation or physical exercise. This effect is mediated by the transcriptional regulation of RagD GTPase and is important for an efficient mTORC1 recruitment to the lysosomal surface (Di Malta et al, 2017).
Role of MiT‐TFE transcription factors in cancer
Transcription factor EB and other members of the MiT‐TFE family of transcription factors have been known for quite some time for their oncogenic features (Kuiper et al, 2003; Haq & Fisher, 2011; Kauffman et al, 2014). Chromosomal translocations involving the TFEB and TFE3 genes cause a particular type of renal cancer, referred to as translocation carcinoma (Linehan et al, 2010; Malouf et al, 2014). These fusions consistently preserve the TFEB/TFE3 open reading frame and always include the DNA‐binding domains (Kuiper et al, 2003). While a variety of genes can be fused to TFEB or TFE3 a major consequence of the translocation with respect to oncogenic activity is a massive increase in the expression levels of a TFEB or TFE3 protein. This conclusion is supported by translocations that increase TFEB expression without changing the coding sequence (Kuiper et al, 2003). Interestingly, MITF may also be translocated in tRCC (Durinck et al, 2015). tRCCs account for approximately 1–5% of all RCCs, but are particularly prevalent in children and adolescents. TFE3, which is the most common member translocated in tRCC, is also translocated in a particular type of sarcoma (alveolar soft part sarcoma) and in perivascular epithelioid tumors (PEComas).
The ASPSCR1‐TFE3 translocation, which is characteristic of alveolar soft part sarcoma and also frequently observed in tRCC, was modeled in mice (Goodwin et al, 2014). A human cDNA fusion was targeted to the Rosa26 locus, and its expression was activated using both a tamoxifen‐inducible Cre‐recombinase (CreER) also from the Rosa26 locus and a Prx1‐CreERT2. Cre expression in both models removed a stop signal leading to expression of the ASPSCR1‐TFE3 fusion protein. Prx1 is expressed in osteochondral progenitors, neural stem cells, and in some intramuscular pericytes. In the CreER‐driven mice, stochastic, low‐level expression (in the absence of tamoxifen) resulted in tumors in the brain, choroid plexus, and orbit between 3 and 6 months. Prx1‐CreERT2 mice developed tumors in the brain and intracranial periosteum following tamoxifen administration. No skeletal muscle (or kidney) tumors were reported in either strain. Overall, while the anatomic sites markedly differed between human and murine tumors, the murine tumors expressed the fusion cDNA and resembled alveolar soft part sarcomas both histologically and by gene expression. The authors speculate that the sites of tumor development in the mice are the result of a microenvironment rich in lactate, which is used by the tumors as a source of energy.
In addition to translocation, TFEB may also induce renal cancers through amplification (Durinck et al, 2015). Interestingly, the TFEB gene resides in the proximity of VEGFA and both genes may be amplified together (Gupta et al, 2017). Recently, a kidney‐specific TFEB‐overexpressing mouse line was generated using a Cadherin16 Cre (or CreERT2), which recapitulates some features of human TFEB/TFE3‐associated RCC (Calcagni et al, 2016). These mice developed renal cysts and papillary carcinomas, followed by liver metastases. This mouse model shows TFEB‐/TFE3‐dependent induction of both the Wnt‐beta‐catenin and mTORC1 pathways (Calcagni et al, 2016; Di Malta et al, 2017).
MITF, TFEB, and TFE3 overexpression was also observed in pancreatic adenoductal carcinoma, where they appear to support tumor growth via the induction of autophagy (Perera et al, 2015). MITF is an established oncogene and can be found amplified in a subset of melanoma patients (Tsao et al, 2012). Interestingly, tumors in which MiT‐TFE genes are amplified or overexpressed show an induction of RagD, a direct transcriptional target of TFEB, that resulted in mTORC1 hyperactivation (Di Malta et al, 2017). The RagD GTPase is important for an efficient mTORC1 recruitment to the lysosomal surface. Silencing of MiT‐TFE genes in primary cell cultures obtained from tumors resulted in a significant reduction in the hyperproliferative phenotype. Furthermore, xenotransplantation experiments performed using a melanoma cell line showed that silencing of RagD significantly reduced tumor growth (Di Malta et al, 2017). In addition to promoting various cancers, TFEB was also recently identified as being highly expressed in non‐Hodgkins lymphoma where it may render these cancer cells sensitive to apilimod‐mediated inhibition of the PIKfyve lipid kinase (Gayle et al, 2017).
Transcription factor EB subcellular localization
Conditions that promote TFEB nuclear translocation
Initial evidence for the shuttling of TFEB between the cytoplasm and the nucleus was obtained in cells treated with sucrose, which is endocytosed and accumulated in the lysosomes due to their lack of invertase enzymes, and thus provides a cellular model of lysosome storage (Sardiello et al, 2009). While TFEB was primarily localized in the cytoplasm in untreated cells, sucrose treatment was followed by relocation of TFEB from the cytoplasm to the nucleus. Other pharmacological treatments that result in lysosome stress such as chloroquine (a weak base that neutralizes lysosome pH), bafilomycin or trehalose, also cause TFEB nuclear translocation (Roczniak‐Ferguson et al, 2012; Settembre et al, 2012; Palmieri et al, 2017). Both TFE3 and MITF are also subject to nucleo‐cytoplasmic shuttling and behave similarly to TFEB (Bronisz et al, 2006; Roczniak‐Ferguson et al, 2012; Martina & Puertollano, 2013; Martina et al, 2014).
A variety of stimuli have been associated with TFEB nuclear translocation, the best studied of which is nutrient deprivation. When cells are grown in starvation medium, TFEB relocates to the nucleus as early as 1 h after exposure to the nutrient‐deprived (amino acid and serum) medium (Settembre et al, 2011, 2012; Martina et al, 2012; Roczniak‐Ferguson et al, 2012). Figure 1A shows a cytoplasmic localization of TFEB in HeLa cells grown in normal medium and a nuclear localization in cells grown in nutrient‐deprived medium. A nuclear localization of TFEB is also evident in Fig 1B, which shows a super‐resolution image of a HeLa cell stably transfected with a TFEB‐GFP plasmid and grown with a nutrient‐deprived medium. This observation appears to be consistent in all cell types studied. In vivo studies also demonstrated that starvation (i.e., elimination of the food) for approximately 16 h in mice results in a nuclear localization of TFEB in all tissues studied (Settembre et al, 2011; Chen et al, 2017). In cell culture, re‐feeding after starvation results in nucleus‐to‐cytoplasm re‐localization of TFEB within minutes (Settembre et al, 2011, 2012; Martina et al, 2012; Roczniak‐Ferguson et al, 2012). A similar acute behavior was observed for TFE3 (Martina et al, 2014).
Figure 1. TFEB subcellular localization is both nutrient‐ and phosphorylation‐dependent.
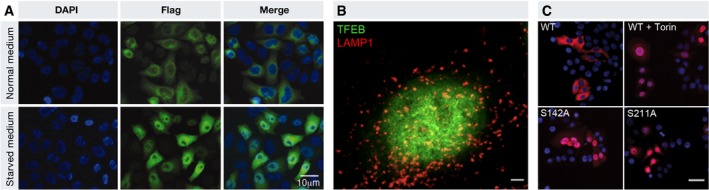
(A) HeLa cells cultured in either normal or starved medium were stained for Flag‐TFEB (green; Settembre et al, 2011). (B) 3D reconstruction of Airyscan super‐resolution imaging of HeLa cells stained for TFEB (green) and LAMP1 (red; Image courtesy of Jlenia Monfregola and A.B.). (C) TFEB phosphorylation of specific serine residues controls its subcellular localization. Flag immunostaining (red) of HeLa cells transfected with WT TFEB and treated with Torin 1 and of HeLa cells transfected with serine‐to‐alanine mutated versions of 3xFlag‐TFEB and grown in normal medium (Settembre et al, 2012).
This acute sensitivity of MiT‐TFE proteins to nutrient availability reflects their dual dependence on the lysosome‐localized and amino acid‐regulated Rag GTPases: first, as TFEB‐Rag interactions underlie the recruitment the transcription factor to the surface of the lysosomes (Martina & Puertollano, 2013), and second, as Rags are also responsible for communicating amino acid availability to mTORC1 (Sancak et al, 2010, 2008). Impaired control of the nuclear versus cytoplasmic localization of TFEB by Rag GTPases may also contribute to human disease. For example, loss‐of‐function mutations in folliculin (FLCN), a GTPase‐activating protein (GAP) for RagC/D, result in constitutively nuclear localization of TFEB and TFE3 and give rise to Birt‐Hogg‐Dube' syndrome, a disease characterized by benign, hair follicle tumors (fibrofolliculomas), risk of pneumothorax, and frequent occurrence of bilateral, multifocal renal carcinoma (Hong et al, 2010; Petit et al, 2013; Tsun et al, 2013; Schmidt & Linehan, 2015).
In addition to lysosomal storage and starvation, other conditions were found to promote TFEB nuclear localization. These include infection (Visvikis et al, 2014; Campbell et al, 2015; Pastore et al, 2016), bacterial phagocytosis (Gray et al, 2016), inflammation (i.e., LPS treatment; Pastore et al, 2016), physical exercise (in muscle; Mansueto et al, 2017), mitochondrial damage (Nezich et al, 2015), PIKfyve inhibition (Gayle et al, 2017), and ER stress (Martina et al, 2016). In addition, inactivation of the cytosolic exonuclease, TREX1, also leads to TFEB nuclear localization and lysosome expansion (Hasan et al, 2013). It is becoming evident that multiple types of cellular stress ultimately induce TFEB nuclear localization, suggesting that TFEB subcellular localization is controlled by a stress response mechanism (Raben & Puertollano, 2016). It remains to be determined, however, to what extent these various stressors impinge on MiT‐TFE proteins via mTOR‐dependent versus mTOR‐independent mechanisms (see below).
It is important that appropriate, and ideally multiple, methods are used to test the effects of a specific condition on TFEB subcellular localization. The most commonly used methods are immuno‐fluorescence and nucleo‐cytoplasmic fractionation. These methods can be used on both endogenous and ectopically overexpressed TFEB (Settembre & Medina, 2015). The low abundance of TFEB, particularly in some cell types, has hampered the analysis of the endogenous protein. Therefore, it is particularly important to use high‐quality antibodies. Due to the difficulties to analyze endogenous TFEB, many studies have relied on cells stably transfected with tagged versions of TFEB. However, when TFEB is overexpressed at high levels, a significant portion of the protein can be found in the nucleus even in normally fed cells.
When using immuno‐fluorescence, it is particularly important to analyze many cells in order to get robust and statistically significant results. This can be achieved by using high‐content analysis with an automated confocal microscope (Settembre & Medina, 2015). This system allows for a completely unbiased analysis of hundreds of cells. It may also be helpful to score not only cells with nuclear or cytoplasmic TFEB, but also ratios of nuclear to cytoplasmic localization in cells with a mixed pattern where TFEB is found in both compartments (Petit et al, 2013). It is also worth considering that while changes in the subcellular distribution suggest alterations in TFEB shuttling from one compartment to the other, compartment‐selective degradation has not been excluded in most contexts and may give the same results. Another aspect to consider when seeking to understand the regulation of endogenous MiT‐TFE family members in diverse cell types is the variability in their relative expression levels such that different family members may predominate in a given cell type.
mTOR‐mediated TFEB phosphorylation
The best‐studied mechanism that regulates TFEB subcellular localization involves the phosphorylation of specific serine residues in the TFEB protein. Other, phosphorylation‐independent, mechanisms have been proposed but never formally demonstrated. Table 2 lists all TFEB and TFE3 phosphorylation sites that have been directly evaluated to date. The mTOR kinase was shown to phosphorylate specific serine residues in TFEB and to play a major role in the regulation of TFEB subcellular localization. The nutrient dependence of mTOR‐mediated TFEB phosphorylation indicated that mTORC1 was the complex involved (Settembre et al, 2011, 2012; Martina et al, 2012; Roczniak‐Ferguson et al, 2012; Vega‐Rubin‐de‐Celis et al, 2017). At least three serines, S122, S142, and S211, in the TFEB protein are phosphorylated by mTORC1 (Settembre et al, 2011, 2012; Martina et al, 2012; Roczniak‐Ferguson et al, 2012; Vega‐Rubin‐de‐Celis et al, 2017). Mutations of either S142 or S211 into alanines (S142A, S211A) result in a constitutively nuclear TFEB, similar to cells treated with the mTOR inhibitor Torin 1, as shown in Fig 1C.
Table 2.
TFEB and TFE3 phosphorylation sites that have been directly evaluated to date
| Site | Kinase | Method used to characterize site | Treatments | Effects of site phosphorylation on TFEB | References |
|---|---|---|---|---|---|
| TFEB | |||||
| S122 | mTOR | In vitro kinase assay, phospho‐antibody, mutation of modification site, Western blotting, immunoprecipitation | Amino acid starvation, glucose starvation, serum starvation, Torin‐1 | Cytoplasmic retention | Vega‐Rubin de Celis et al (2017) |
| S134/S138 | GSK3B | In vitro kinase assay, mutation of modification site | GSK3 inhibitors, PMA, angiotensinII, LPS | Cytoplasmic retention | Li et al (2016) |
| S142 | Erk 1/2, mTOR | Phospho‐antibody, in vitro kinase assay, mutation of modification site, Western blotting | Amino acid starvation, serum starvation, Torin‐1, antimycinA/oligomycin | Cytoplasmic retention | Settembre et al (2011), Settembre et al (2012), Nezich et al (2015) |
| S211 | mTOR | In vitro kinase assay, mass‐spectometry, phospho‐antibody, mutation of the modification site, Western blotting, immunoprecipitation | Amino acid starvation, serum starvation, lysosomal stress, Torin‐1/PP42 | Cytoplasmic retention, 14‐3‐3 binding | Martina et al (2012), Roczniak‐Ferguson et al (2012), Settembre et al (2012), Vega‐Rubin de Celis et al (2017) |
| S462/S463/S467/S469 | PKCB | Mutation of modification site, Western blotting | RANKL | Protein stabilization | Ferron et al (2013) |
| S462/S463/S466/S467/S469 | Hyperactive mTOR (TSC2−/− cells) | Mutation of modification site | Rapamycin, serum starvation | Nuclear translocation | Pena‐Llopis et al (2011) |
| S467 | AKT | In vitro kinase assay, mutation of modification site | Trehalose | Cytoplasmic retention | Palmieri et al (2017) |
| TFE3 | |||||
| S321 | mTOR | In vitro kinase assay, phospho‐antibody, mutation of modification site, Western blotting, immunoprecipitation | Amino acid starvation, serum starvation, Torin‐1, tunicamycin, LPS | Cytoplasmic retention, 14‐3‐3 binding | Martina et al (2014), Martina et al (2016), Pastore et al (2016), Wada et al (2016) |
Phosphorylation of S211 determines TFEB binding with 14‐3‐3 protein. It has been hypothesized that this binding masks an NLS, thus inhibiting TFEB nuclear translocation (Martina et al, 2012; Roczniak‐Ferguson et al, 2012). A recent study also showed that the phosphorylation of serines S142 and S211 also mediates the targeting of TFEB to the ubiquitin proteasome system via the binding to the E3 ubiquitin ligase STUB1, suggesting that phosphorylation may regulate TFEB function not only by determining its subcellular localization but also by modulating its stability (Sha et al, 2017).
In vitro kinase assays showed that S122 is directly phosphorylated by mTORC1 (Vega‐Rubin‐de‐Celis et al, 2017). Mutation of S122 to alanine (S122A) does not, by itself, affect TFEB subcellular localization but appears to enhance the effects of the S211A mutation (Vega‐Rubin‐de‐Celis et al, 2017). However, the phosphomimetic mutation of S122 to aspartic acid (S122D) blocks the effects of the S211A mutation on TFEB nuclear translocation. Nevertheless, whether the mechanism whereby aspartate substitution affects TFEB localization is by mimicking phosphorylation remains to be determined.
The mechanisms by which the phosphorylation of S142 and S122 affect TFEB subcellular localization are still unclear. TFE3 subcellular localization is also regulated by mTORC1‐mediated phosphorylation and involves serine residues that are conserved between TFEB and TFE3 (Martina et al, 2014, 2016; Wada et al, 2016). Data obtained from patient‐derived pancreatic cancer cell lines revealed that TFEB and TFE3 nuclear translocation is mediated by specific importins; however, it is still unclear whether this is a general mechanism relevant in all cell types (Perera et al, 2015).
It is particularly important to test the phosphorylation of TFEB by immunoblotting using standard antibodies directed to TFEB (i.e., looking at the TFEB electrophoretic mobility on SDS–PAGE gels, which is influenced by phosphorylation) and phosphospecific antibodies that recognize the phosphorylation state of specific amino acid residues. Given how widely phosphorylation occurs and the difficulties related to generating highly quality phosphospecific antibodies, their validation using phosphosite mutants (i.e., alanine substitution of the particular site) is advisable. With respect to TFEB, phosphospecific antibodies that detect phosphorylation on S142 and S211 have been reported (Settembre et al, 2011; Petit et al, 2013). An alternative method to measure phosphorylation on S211 that confers binding capabilities to 14‐3‐3 proteins when in the phosphorylated state is the use of an anti‐14‐3‐3‐binding motif antibody on TFEB immunoprecipitates (Martina et al, 2012; Roczniak‐Ferguson et al, 2012). Phosphoproteomic studies have revealed that TFEB is phosphorylated at multiple sites (more than 20; Dephoure et al, 2008; Chen et al, 2009; Mayya et al, 2009; Huttlin et al, 2010; Olsen et al, 2010), many of which are phosphorylated by mTOR (Yu et al, 2011). Figure 2 shows all the serine residues in the TFEB protein that were shown to be subject to phosphorylation by functional studies. It is worth noting that key residues and regulatory mechanisms are conserved between TFEB, TFE3, and MITF (Martina et al, 2016, 2014; Wada et al, 2016; Fig 3).
Figure 2. Relevant TFEB phosphorylation sites and their regulatory role.
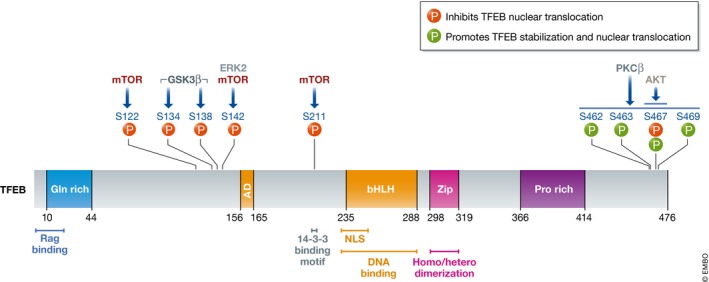
Figure 3. Sequence conservation of TFEB, TFE3, MITF and TFEC phosphorylation sites.
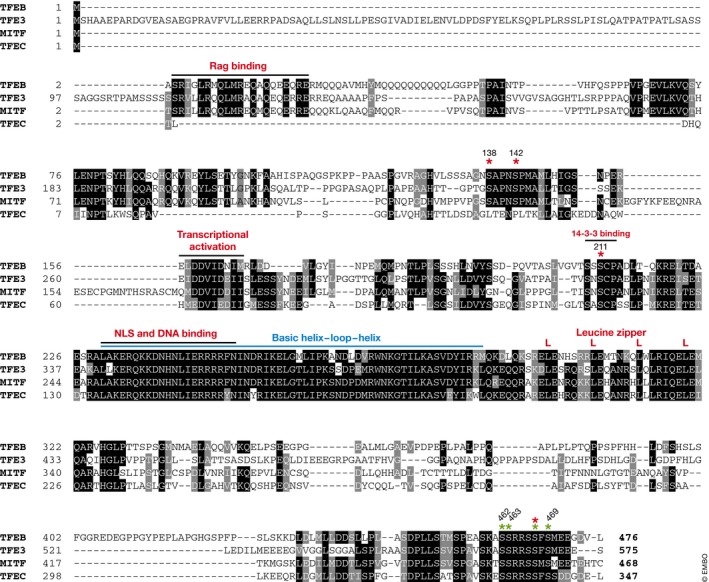
Multiple sequence alignment highlights the conservation of critical domains and phosphorylation sites between human TFEB (Uniprot P19484), TFE3 (Uniprot 19532), MITF (O75030, “D” splice variant shown here), and TFEC (Uniprot O14948) proteins. Red asterisks highlight phosphorylation sites that inhibit nuclear translocation of TFEB, while those shown in green have been found to promote TFEB nuclear localization.
Atypical regulation of TFEB by mTORC1 in TSC1/TSC2‐deficient cells
While mTOR inhibition leads to TFEB nuclear localization and induction of lysosome biogenesis quite broadly across cell types, in some cellular contexts, this is not the case. In an unbiased screen to identify mTORC1‐regulated genes, the expression of genes encoding v‐ATPase subunits was upregulated in TSC1/TSC2‐deficient cells, which retain mTORC1 activity under serum‐starved conditions, but not in wild‐type cells, in which mTORC1 was inhibited. Interestingly, v‐ATPase expression in TSC1/TSC2‐deficient cells could be downregulated by treatment with the mTORC1 inhibitor rapamycin (Pena‐Llopis et al, 2011). v‐ATPase expression in TSC1/TSC2‐deficient cells was associated with nuclear TFEB and was dependent on it. Specifically, TFEB depletion using shRNA led to the downregulation of v‐ATPases. These data suggest that mTORC1 promotes TFEB nuclear localization and expression of target genes such as v‐ATPases in TSC1/TSC2‐deficient cells. Consistent with this notion, siRNA‐mediated silencing of the mTORC1 essential subunit Raptor in these cells excluded TFEB from the nucleus and led to a reduction in v‐ATPase expression. Notably, when a C‐terminal serine‐rich motif (S462/463/466/467/469) was mutated to aspartate, TFEB remained constitutively nuclear. Furthermore, TFEB5xSD localization was unaltered by rapamycin. Why mTORC1 promotes rather than inhibits TFEB in TSC1/TSC2‐deficient cells remains unclear. The data show, however, that TFEB regulation by mTORC1 is plastic and cell context dependent.
Intriguingly, the subcellular localization of TFE3, whose regulation by mTOR is similar to TFEB, appears to be independent from both Rheb and TSC2 activities in adipocytes (Wada et al, 2016). In addition, while the mTORC1/mTORC2 inhibitor Torin 1 inhibits TFEB phosphorylation at low doses, the specific mTORC1 inhibitor rapamycin did not affect TFEB phosphorylation and subcellular localization even at high doses (Settembre et al, 2012; Kang et al, 2013). These data suggest that TFEB may be an atypical mTORC1 substrate and that mTORC1 regulation of TFEB is more complex than generally appreciated and can be affected by cellular context.
Other kinases
In addition to mTORC1, other kinases were found to phosphorylate TFEB. In osteoclasts, PKCβ phosphorylates multiple serine residues (i.e., S462, S463, S467, and S469) located in the C‐terminus of human TFEB. Phosphorylation of these serines by PKCβ is important for TFEB protein stability but does not affect TFEB subcellular localization (Ferron et al, 2013). ERK2 was also found to phosphorylate TFEB. Phosphorylation of TFEB S142 by ERK2 promotes TFEB cytoplasmic retention. Accordingly, treatment with ERK inhibitors and silencing of ERK2 resulted in TFEB nuclear translocation (Settembre et al, 2011). The relationship between mTORC1‐mediated and ERK2‐mediated phosphorylation of TFEB S142 is still unclear. However, ERK‐mediated phosphorylation of the homologous S73 site in MITF was reported to control transcriptional activity and protein stability (Wu et al, 2000).
Transcription factor EB can be phosphorylated by GSK3 at residues S134 and S138 leading to cytoplasmic retention, whereas GSK3 inhibition led to TFEB nuclear translocation (Li et al, 2016). The phosphorylation of these serines by GSK3 mediates the recruitment of TFEB to the lysosome by an unknown mechanism. S134A and S138A mutations impair lysosomal recruitment of TFEB, thus indirectly impairing mTORC1‐mediated phosphorylation. Based on these observations, the effects of GSK3 on TFEB may not be truly independent from mTORC1. Similar GSK3‐dependent phosphorylation on evolutionarily conserved sites in MITF resulted in stabilization and enhanced function (Ploper et al, 2015).
A recent study showed that TFEB is phosphorylated by AKT at serine residue S467 and that treating cells with an AKT inhibitor promotes TFEB nuclear translocation (Palmieri et al, 2017). In addition, trehalose, a known autophagy activator, was shown to inhibit Akt activity, thus promoting TFEB nuclear translocation. A mutant form of TFEB carrying an S467A mutation shows an increased nuclear localization in normally fed cells compared to wild‐type TFEB. This effect appears to be independent from mTOR‐mediated TFEB phosphorylation. However, the effects of AKT inhibition on mTORC1 activity were not tested, nor was the effect of the S467A mutation on TFEB serine S122, S211, or S142 phosphorylation. Thus, also in this case, a dependence on the mTOR pathway cannot be formally excluded (Palmieri et al, 2017).
Finally, another recent study showed that the curcumin analogue C1 promotes TFEB nuclear translocation in a phosphorylation‐independent way. In this case, C1 binds directly to TFEB, manner that interferes with binding of TFEB to 14‐3‐3 proteins (Song et al, 2016).
De‐phosphorylation of TFEB by calcineurin
As discussed above, to be active, TFEB needs to be de‐phosphorylated, at least partially, at some key residues. Therefore, understanding how the process of TFEB de‐phosphorylation occurs, and the phosphatases involved, is of critical importance to understand TFEB regulation. The search for the phosphatase responsible for TFEB de‐phosphorylation was performed by siRNA‐based high‐content screening using an assay based on TFEB subcellular localization. Briefly, TFEB‐GFP‐expressing cells were starved in the presence of siRNAs directed against all the 231 known human phosphatases to identify the phosphatase(s) whose inhibition would prevent TFEB nuclear translocation.
This screening led to the identification of calcineurin as the phosphatase that plays a major role in TFEB de‐phosphorylation (Medina et al, 2015). Interestingly, calcineurin was known to de‐phosphorylate NFAT proteins, another family of transcription factors (Macian, 2005), and to promote their nuclear translocation. Inhibition of both calcineurin and mTORC1 activities results in TFEB cytoplasmic localization, indicating that the effects of calcineurin inhibition override the effects of mTORC1 inhibition. These data suggest that calcineurin acts downstream of mTORC1 in the regulation of TFEB (Medina et al, 2015) or through a parallel pathway. Depletion or inhibition of calcineurin also causes a significant reduction in TFE3 activation in response to ER stress, revealing additional parallels in TFEB and TFE3 regulation (Martina et al, 2016).
Calcineurin is composed of a catalytic and a regulatory, calcium‐dependent, subunit (Hogan & Li, 2005). This suggested that TFEB subcellular localization might be influenced by changes in intracellular calcium levels. Indeed, calcium chelators block starvation‐induced TFEB nuclear translocation, while calcium ionophores promote TFEB nuclear translocation in a calcineurin‐dependent manner (Medina et al, 2015). The lysosomal calcium channel mucolipin 1 (MCOLN1), also known as TRPML1, plays an important role in the activation of calcineurin and consequent TFEB de‐phosphorylation (Medina et al, 2015; Zhang et al, 2016). Lysosomal calcium release via TRPML1 may increase calcium concentration near the lysosomal surface, and this may lead to local calcineurin activation. Thus, the lysosome acts as a calcium signaling hub by regulating TRPML1‐Calcineurin‐TFEB signaling. Interestingly, TRPML1 is also a direct transcriptional target of TFEB (Palmieri et al, 2011), suggesting the possibility of a positive feedback loop that involves TFEB and TRPML1.
Conclusions
Transcription factor EB and other members of the MiT‐TFE family of transcription factors have emerged as important regulators of cellular energy metabolism. These transcription factors also appear to mediate communication between the lysosome and the nucleus in the adaptive response to environmental cues such as nutrient availability. Despite considerable progress made toward our understanding of how signaling pathways regulate TFEB subcellular localization and function via the phosphorylation of specific serine residues, several critical questions still remain unanswered. TFEB has been found to be phosphorylated by several kinases. However, the relationship and interdependence of these phosphorylation events are still unclear. It also remains to be established whether TFEB subcellular localization, in a physiological context, may be modulated by phosphorylation‐independent mechanisms. Another important point to be addressed is how, once in the nucleus, TFEB is exported to the cytoplasm and whether TFEB nuclear export mechanisms add another layer of TFEB function regulation. Finding the answers to these and other critical questions will be of fundamental importance to our understanding of the transcriptional mechanisms that regulate cell metabolism in response to the environment and may lead to the development of powerful tools to modulate these pathways in human diseases.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
We are grateful to Drs. Graciana Diez‐Roux, Gennaro Napolitano, and Carmine Settembre for helpful suggestions. We gratefully acknowledge the support of the Italian Telethon Foundation (Grant TGM11CB6 to A.B.), the European Research Council (Advanced Investigator Grant LYSOSOMICS to A.B.), the US National Institutes of Health (NIH grants R01‐NS078072 to A.B., GM105718 and AG047270 to S.M.F., R01CA175754 and P50 CA196516 to J.B., Intramural Research Program of the National Institutes of Health, National Heart, Lung, and Blood Institute (NHLBI) to R.P.), the Associazione Italiana per la Ricerca sul Cancro (A.I.R.C.) (Grant IG 2015 Id 17639 to A.B), and the Cancer Prevention and Research Institute of Texas (RP16044, RP180191) to J.B.
The EMBO Journal (2018) 37: e98804
Contributor Information
Rosa Puertollano, Email: puertolr@nhlbi.nih.gov.
Shawn M Ferguson, Email: shawn.ferguson@yale.edu.
James Brugarolas, Email: James.Brugarolas@utsouthwestern.edu.
Andrea Ballabio, Email: ballabio@tigem.it.
References
- Beg AA, Ruben SM, Scheinman RI, Haskill S, Rosen CA, Baldwin AS Jr (1992) I kappa B interacts with the nuclear localization sequences of the subunits of NF‐kappa B: a mechanism for cytoplasmic retention. Genes Dev 6: 1899–1913 [DOI] [PubMed] [Google Scholar]
- Bouche V, Espinosa AP, Leone L, Sardiello M, Ballabio A, Botas J (2016) Drosophila Mitf regulates the V‐ATPase and the lysosomal‐autophagic pathway. Autophagy 12: 484–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronisz A, Sharma SM, Hu R, Godlewski J, Tzivion G, Mansky KC, Ostrowski MC (2006) Microphthalmia‐associated transcription factor interactions with 14‐3‐3 modulate differentiation of committed myeloid precursors. Mol Biol Cell 17: 3897–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcagni A, Kors L, Verschuren E, De Cegli R, Zampelli N, Nusco E, Confalonieri S, Bertalot G, Pece S, Settembre C, Malouf GG, Leemans JC, de Heer E, Salvatore M, Peters DJ, Di Fiore PP, Ballabio A (2016) Modelling TFE renal cell carcinoma in mice reveals a critical role of WNT signaling. Elife 5: e17047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell GR, Rawat P, Bruckman RS, Spector SA (2015) Human immunodeficiency virus type 1 Nef inhibits autophagy through transcription factor EB sequestration. PLoS Pathog 11: e1005018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S, Ahmed Z, Bradfute SB, Arko‐Mensah J, Mandell MA, Won Choi S, Kimura T, Blanchet F, Waller A, Mudd MH, Jiang S, Sklar L, Timmins GS, Maphis N, Bhaskar K, Piguet V, Deretic V (2015) Pharmaceutical screen identifies novel target processes for activation of autophagy with a broad translational potential. Nat Commun 6: 8620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RQ, Yang QK, Lu BW, Yi W, Cantin G, Chen YL, Fearns C, Yates JR III, Lee JD (2009) CDC25B mediates rapamycin‐induced oncogenic responses in cancer cells. Cancer Res 69: 2663–2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wang K, Long A, Jia L, Zhang Y, Deng H, Li Y, Han J, Wang Y (2017) Fasting‐induced hormonal regulation of lysosomal function. Cell Res 27: 748–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decressac M, Mattsson B, Weikop P, Lundblad M, Jakobsson J, Bjorklund A (2013) TFEB‐mediated autophagy rescues midbrain dopamine neurons from alpha‐synuclein toxicity. Proc Natl Acad Sci USA 110: E1817–E1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay B, Bove J, Rodriguez‐Muela N, Perier C, Recasens A, Boya P, Vila M (2010) Pathogenic lysosomal depletion in Parkinson's disease. J Neurosci 30: 12535–12544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP (2008) A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA 105: 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Malta C, Siciliano D, Calcagni A, Monfregola J, Punzi S, Pastore N, Eastes AN, Davis O, De Cegli R, Zampelli A, Di Giovannantonio LG, Nusco E, Platt N, Guida A, Ogmundsdottir MH, Lanfrancone L, Perera RM, Zoncu R, Pelicci PG, Settembre C et al (2017) Transcriptional activation of RagD GTPase controls mTORC1 and promotes cancer growth. Science 356: 1188–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, Stawiski EW, Pavia‐Jimenez A, Modrusan Z, Kapur P, Jaiswal BS, Zhang N, Toffessi‐Tcheuyap V, Nguyen TT, Pahuja KB, Chen YJ, Saleem S, Chaudhuri S, Heldens S, Jackson M, Pena‐Llopis S, Guillory J, Toy K, Ha C, Harris CJ et al (2015) Spectrum of diverse genomic alterations define non‐clear cell renal carcinoma subtypes. Nat Genet 47: 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Lu H, Liang W, Garcia‐Barrio MT, Guo Y, Zhang J, Zhu T, Hao Y, Zhang J, Chen YE (2018) Endothelial TFEB (Transcription Factor EB) positively regulates postischemic angiogenesis. Circ Res 122: 945–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron M, Settembre C, Shimazu J, Lacombe J, Kato S, Rawlings DJ, Ballabio A, Karsenty G (2013) A RANKL‐PKCbeta‐TFEB signaling cascade is necessary for lysosomal biogenesis in osteoclasts. Genes Dev 27: 955–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher DE, Carr CS, Parent LA, Sharp PA (1991) TFEB has DNA‐binding and oligomerization properties of a unique helix‐loop‐helix/leucine‐zipper family. Genes Dev 5: 2342–2352 [DOI] [PubMed] [Google Scholar]
- Gayle S, Landrette S, Beeharry N, Conrad C, Hernandez M, Beckett P, Ferguson SM, Mandelkern T, Zheng M, Xu T, Rothberg J, Lichenstein H (2017) Identification of apilimod as a first‐in‐class PIKfyve kinase inhibitor for treatment of B‐cell non‐Hodgkin lymphoma. Blood 129: 1768–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin ML, Jin H, Straessler K, Smith‐Fry K, Zhu JF, Monument MJ, Grossmann A, Randall RL, Capecchi MR, Jones KB (2014) Modeling alveolar soft part sarcomagenesis in the mouse: a role for lactate in the tumor microenvironment. Cancer Cell 26: 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D, Kutay U (1999) Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 15: 607–660 [DOI] [PubMed] [Google Scholar]
- Gray MA, Choy CH, Dayam RM, Ospina‐Escobar E, Somerville A, Xiao X, Ferguson SM, Botelho RJ (2016) Phagocytosis enhances lysosomal and bactericidal properties by activating the transcription factor TFEB. Curr Biol 26: 1955–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Johnson SH, Vasmatzis G, Porath B, Rustin JG, Rao P, Costello BA, Leibovich BC, Thompson RH, Cheville JC, Sukov WR (2017) TFEB‐VEGFA (6p21.1) co‐amplified renal cell carcinoma: a distinct entity with potential implications for clinical management. Mod Pathol 30: 998–1012 [DOI] [PubMed] [Google Scholar]
- Hallsson JH, Haflidadottir BS, Stivers C, Odenwald W, Arnheiter H, Pignoni F, Steingrimsson E (2004) The basic helix‐loop‐helix leucine zipper transcription factor Mitf is conserved in Drosophila and functions in eye development. Genetics 167: 233–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq R, Fisher DE (2011) Biology and clinical relevance of the micropthalmia family of transcription factors in human cancer. J Clin Oncol 29: 3474–3482 [DOI] [PubMed] [Google Scholar]
- Hasan M, Koch J, Rakheja D, Pattnaik AK, Brugarolas J, Dozmorov I, Levine B, Wakeland EK, Lee‐Kirsch MA, Yan N (2013) Trex1 regulates lysosomal biogenesis and interferon‐independent activation of antiviral genes. Nat Immunol 14: 61–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemesath TJ, Steingrimsson E, McGill G, Hansen MJ, Vaught J, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA, Fisher DE (1994) Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev 8: 2770–2780 [DOI] [PubMed] [Google Scholar]
- Henkel T, Zabel U, van Zee K, Muller JM, Fanning E, Baeuerle PA (1992) Intramolecular masking of the nuclear location signal and dimerization domain in the precursor for the p50 NF‐kappa B subunit. Cell 68: 1121–1133 [DOI] [PubMed] [Google Scholar]
- Hill CS (2009) Nucleocytoplasmic shuttling of Smad proteins. Cell Res 19: 36–46 [DOI] [PubMed] [Google Scholar]
- Hodgkinson CA, Moore KJ, Nakayama A, Steingrimsson E, Copeland NG, Jenkins NA, Arnheiter H (1993) Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic‐helix‐loop‐helix‐zipper protein. Cell 74: 395–404 [DOI] [PubMed] [Google Scholar]
- Hogan PG, Li H (2005) Calcineurin. Curr Biol 15: R442–R443 [DOI] [PubMed] [Google Scholar]
- Hong SB, Oh H, Valera VA, Baba M, Schmidt LS, Linehan WM (2010) Inactivation of the FLCN tumor suppressor gene induces TFE3 transcriptional activity by increasing its nuclear localization. PLoS One 5: e15793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP (2010) A tissue‐specific atlas of mouse protein phosphorylation and expression. Cell 143: 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SA, Pacold ME, Cervantes CL, Lim D, Lou HJ, Ottina K, Gray NS, Turk BE, Yaffe MB, Sabatini DM (2013) mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science 341: 1236566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman EC, Ricketts CJ, Rais‐Bahrami S, Yang Y, Merino MJ, Bottaro DP, Srinivasan R, Linehan WM (2014) Molecular genetics and cellular features of TFE3 and TFEB fusion kidney cancers. Nat Rev Urol 11: 465–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick K, Zeng Y, Hancock T, Segatori L (2015) Genetic and chemical activation of TFEB mediates clearance of aggregated alpha‐synuclein. PLoS One 10: e0120819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper RP, Schepens M, Thijssen J, van Asseldonk M, van den Berg E, Bridge J, Schuuring E, Schoenmakers EF, van Kessel AG (2003) Upregulation of the transcription factor TFEB in t(6;11)(p21;q13)‐positive renal cell carcinomas due to promoter substitution. Hum Mol Genet 12: 1661–1669 [DOI] [PubMed] [Google Scholar]
- Lapierre LR, De Magalhaes Filho CD, McQuary PR, Chu CC, Visvikis O, Chang JT, Gelino S, Ong B, Davis AE, Irazoqui JE, Dillin A, Hansen M (2013) The TFEB orthologue HLH‐30 regulates autophagy and modulates longevity in Caenorhabditis elegans . Nat Commun 4: 2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xu M, Ding X, Yan C, Song Z, Chen L, Huang X, Wang X, Jian Y, Tang G, Tang C, Di Y, Mu S, Liu X, Liu K, Li T, Wang Y, Miao L, Guo W, Hao X et al (2016) Protein kinase C controls lysosome biogenesis independently of mTORC1. Nat Cell Biol 18: 1065–1077 [DOI] [PubMed] [Google Scholar]
- Linehan WM, Bratslavsky G, Pinto PA, Schmidt LS, Neckers L, Bottaro DP, Srinivasan R (2010) Molecular diagnosis and therapy of kidney cancer. Annu Rev Med 61: 329–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macian F (2005) NFAT proteins: key regulators of T‐cell development and function. Nat Rev Immunol 5: 472–484 [DOI] [PubMed] [Google Scholar]
- Malouf GG, Su X, Yao H, Gao J, Xiong L, He Q, Comperat E, Couturier J, Molinie V, Escudier B, Camparo P, Doss DJ, Thompson EJ, Khayat D, Wood CG, Yu W, Teh BT, Weinstein J, Tannir NM (2014) Next‐generation sequencing of translocation renal cell carcinoma reveals novel RNA splicing partners and frequent mutations of chromatin‐remodeling genes. Clin Cancer Res 20: 4129–4140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansueto G, Armani A, Viscomi C, D'Orsi L, De Cegli R, Polishchuk EV, Lamperti C, Di Meo I, Romanello V, Marchet S, Saha PK, Zong H, Blaauw B, Solagna F, Tezze C, Grumati P, Bonaldo P, Pessin JE, Zeviani M, Sandri M et al (2017) Transcription factor EB controls metabolic flexibility during exercise. Cell Metab 25: 182–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Chen Y, Gucek M, Puertollano R (2012) MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8: 903–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Puertollano R (2013) Rag GTPases mediate amino acid‐dependent recruitment of TFEB and MITF to lysosomes. J Cell Biol 200: 475–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Diab HI, Lishu L, Jeong AL, Patange S, Raben N, Puertollano R (2014) The nutrient‐responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal 7: ra9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Diab HI, Brady OA, Puertollano R (2016) TFEB and TFE3 are novel components of the integrated stress response. EMBO J 35: 479–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayya V, Lundgren DH, Hwang SI, Rezaul K, Wu L, Eng JK, Rodionov V, Han DK (2009) Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system‐wide modulation of protein‐protein interactions. Sci Signal 2: ra46 [DOI] [PubMed] [Google Scholar]
- Medina DL, Fraldi A, Bouche V, Annunziata F, Mansueto G, Spampanato C, Puri C, Pignata A, Martina JA, Sardiello M, Palmieri M, Polishchuk R, Puertollano R, Ballabio A (2011) Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev Cell 21: 421–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto‐Rosato A, Prezioso C, Forrester A, Settembre C, Wang W, Gao Q, Xu H, Sandri M, Rizzuto R, De Matteis MA, Ballabio A (2015) Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol 17: 288–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murano T, Najibi M, Paulus GLC, Adiliaghdam F, Valencia‐Guerrero A, Selig M, Wang X, Jeffrey K, Xavier RJ, Lassen KG, Irazoqui JE (2017) Transcription factor TFEB cell‐autonomously modulates susceptibility to intestinal epithelial cell injury in vivo . Sci Rep 7: 13938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardozzi JD, Lott K, Cingolani G (2010) Phosphorylation meets nuclear import: a review. Cell Commun Signal 8: 32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezich CL, Wang C, Fogel AI, Youle RJ (2015) MiT/TFE transcription factors are activated during mitophagy downstream of Parkin and Atg5. J Cell Biol 210: 435–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, Mann M (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal 3: ra3 [DOI] [PubMed] [Google Scholar]
- Opdecamp K, Nakayama A, Nguyen MT, Hodgkinson CA, Pavan WJ, Arnheiter H (1997) Melanocyte development in vivo and in neural crest cell cultures: crucial dependence on the Mitf basic‐helix‐loop‐helix‐zipper transcription factor. Development 124: 2377–2386 [DOI] [PubMed] [Google Scholar]
- O'Rourke EJ, Ruvkun G (2013) MXL‐3 and HLH‐30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat Cell Biol 15: 668–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, Ballabio A (2011) Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet 20: 3852–3866 [DOI] [PubMed] [Google Scholar]
- Palmieri M, Pal R, Nelvagal HR, Lotfi P, Stinnett GR, Seymour ML, Chaudhury A, Bajaj L, Bondar VV, Bremner L, Saleem U, Tse DY, Sanagasetti D, Wu SM, Neilson JR, Pereira FA, Pautler RG, Rodney GG, Cooper JD, Sardiello M (2017) mTORC1‐independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat Commun 8: 14338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore N, Brady OA, Diab HI, Martina JA, Sun L, Huynh T, Lim JA, Zare H, Raben N, Ballabio A, Puertollano R (2016) TFEB and TFE3 cooperate in the regulation of the innate immune response in activated macrophages. Autophagy 12: 1240–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore N, Vainshtein A, Klisch TJ, Armani A, Huynh T, Herz NJ, Polishchuk EV, Sandri M, Ballabio A (2017) TFE3 regulates whole‐body energy metabolism in cooperation with TFEB. EMBO Mol Med 9: 605–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena‐Llopis S, Vega‐Rubin‐de‐Celis S, Schwartz JC, Wolff NC, Tran TA, Zou L, Xie XJ, Corey DR, Brugarolas J (2011) Regulation of TFEB and V‐ATPases by mTORC1. EMBO J 30: 3242–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera RM, Stoykova S, Nicolay BN, Ross KN, Fitamant J, Boukhali M, Lengrand J, Deshpande V, Selig MK, Ferrone CR, Settleman J, Stephanopoulos G, Dyson NJ, Zoncu R, Ramaswamy S, Haas W, Bardeesy N (2015) Transcriptional control of autophagy‐lysosome function drives pancreatic cancer metabolism. Nature 524: 361–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit CS, Roczniak‐Ferguson A, Ferguson SM (2013) Recruitment of folliculin to lysosomes supports the amino acid‐dependent activation of Rag GTPases. J Cell Biol 202: 1107–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploper D, Taelman VF, Robert L, Perez BS, Titz B, Chen HW, Graeber TG, von Euw E, Ribas A, De Robertis EM (2015) MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc Natl Acad Sci USA 112: E420–E429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogenberg V, Ogmundsdottir MH, Bergsteinsdottir K, Schepsky A, Phung B, Deineko V, Milewski M, Steingrimsson E, Wilmanns M (2012) Restricted leucine zipper dimerization and specificity of DNA recognition of the melanocyte master regulator MITF. Genes Dev 26: 2647–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polito VA, Li H, Martini‐Stoica H, Wang B, Yang L, Xu Y, Swartzlander DB, Palmieri M, di Ronza A, Lee VM, Sardiello M, Ballabio A, Zheng H (2014) Selective clearance of aberrant tau proteins and rescue of neurotoxicity by transcription factor EB. EMBO Mol Med 6: 1142–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raben N, Puertollano R (2016) TFEB and TFE3: linking lysosomes to cellular adaptation to stress. Annu Rev Cell Dev Biol 32: 255–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rega LR, Polishchuk E, Montefusco S, Napolitano G, Tozzi G, Zhang J, Bellomo F, Taranta A, Pastore A, Polishchuk R, Piemonte F, Medina DL, Catz SD, Ballabio A, Emma F (2016) Activation of the transcription factor EB rescues lysosomal abnormalities in cystinotic kidney cells. Kidney Int 89: 862–873 [DOI] [PubMed] [Google Scholar]
- Rehli M, Den Elzen N, Cassady AI, Ostrowski MC, Hume DA (1999) Cloning and characterization of the murine genes for bHLH‐ZIP transcription factors TFEC and TFEB reveal a common gene organization for all MiT subfamily members. Genomics 56: 111–120 [DOI] [PubMed] [Google Scholar]
- Roczniak‐Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM (2012) The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal 5: 42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar‐Peled L, Sabatini DM (2008) The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320: 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar‐Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM (2010) Ragulator‐Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141: 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, Banfi S, Parenti G, Cattaneo E, Ballabio A (2009) A gene network regulating lysosomal biogenesis and function. Science 325: 473–477 [DOI] [PubMed] [Google Scholar]
- Schmidt LS, Linehan WM (2015) Molecular genetics and clinical features of Birt‐Hogg‐Dube syndrome. Nat Rev Urol 12: 558–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergin I, Evans TD, Zhang X, Bhattacharya S, Stokes CJ, Song E, Ali S, Dehestani B, Holloway KB, Micevych PS, Javaheri A, Crowley JR, Ballabio A, Schilling JD, Epelman S, Weihl CC, Diwan A, Fan D, Zayed MA, Razani B (2016) Exploiting macrophage autophagy‐lysosomal biogenesis as a therapy for atherosclerosis. Nat Commun 8: 1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A (2011) TFEB links autophagy to lysosomal biogenesis. Science 332: 1429–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, Sabatini DM, Ballabio A (2012) A lysosome‐to‐nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 31: 1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Klisch TJ, Wollenberg AC, Di Bernardo D, Chan L, Irazoqui JE, Ballabio A (2013a) TFEB controls cellular lipid metabolism through a starvation‐induced autoregulatory loop. Nat Cell Biol 15: 647–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Fraldi A, Medina DL, Ballabio A (2013b) Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol 14: 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Medina DL (2015) TFEB and the CLEAR network. Methods Cell Biol 126: 45–62 [DOI] [PubMed] [Google Scholar]
- Sha Y, Rao L, Settembre C, Ballabio A, Eissa NT (2017) STUB1 regulates TFEB‐induced autophagy‐lysosome pathway. EMBO J 36: 2544–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Wang F, Savini M, Ake A, di Ronza A, Sardiello M, Segatori L (2013) TFEB regulates lysosomal proteostasis. Hum Mol Genet 22: 1994–2009 [DOI] [PubMed] [Google Scholar]
- Song JX, Sun YR, Peluso I, Zeng Y, Yu X, Lu JH, Xu Z, Wang MZ, Liu LF, Huang YY, Chen LL, Durairajan SS, Zhang HJ, Zhou B, Zhang HQ, Lu A, Ballabio A, Medina DL, Guo Z, Li M (2016) A novel curcumin analog binds to and activates TFEB in vitro and in vivo independent of MTOR inhibition. Autophagy 12: 1372–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spampanato C, Feeney E, Li L, Cardone M, Lim JA, Annunziata F, Zare H, Polishchuk R, Puertollano R, Parenti G, Ballabio A, Raben N (2013) Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol Med 5: 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrimsson E, Moore KJ, Lamoreux ML, Ferre‐D'Amare AR, Burley SK, Zimring DC, Skow LC, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA (1994) Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nat Genet 8: 256–263 [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Tessarollo L, Reid SW, Jenkins NA, Copeland NG (1998) The bHLH‐Zip transcription factor Tfeb is essential for placental vascularization. Development 125: 4607–4616 [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Tessarollo L, Pathak B, Hou L, Arnheiter H, Copeland NG, Jenkins NA (2002) Mitf and Tfe3, two members of the Mitf‐Tfe family of bHLH‐Zip transcription factors, have important but functionally redundant roles in osteoclast development. Proc Natl Acad Sci USA 99: 4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrimsson E, Copeland NG, Jenkins NA (2004) Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet 38: 365–411 [DOI] [PubMed] [Google Scholar]
- Tognon E, Kobia F, Busi I, Fumagalli A, De Masi F, Vaccari T (2016) Control of lysosomal biogenesis and Notch‐dependent tissue patterning by components of the TFEB‐V‐ATPase axis in Drosophila melanogaster . Autophagy 12: 499–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao H, Chin L, Garraway LA, Fisher DE (2012) Melanoma: from mutations to medicine. Genes Dev 26: 1131–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsun ZY, Bar‐Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini DM (2013) The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell 52: 495–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega‐Rubin‐de‐Celis S, Pena‐Llopis S, Konda M, Brugarolas J (2017) Multistep regulation of TFEB by MTORC1. Autophagy 13: 464–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvikis O, Ihuegbu N, Labed SA, Luhachack LG, Alves AF, Wollenberg AC, Stuart LM, Stormo GD, Irazoqui JE (2014) Innate host defense requires TFEB‐mediated transcription of cytoprotective and antimicrobial genes. Immunity 40: 896–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S, Neinast M, Jang C, Ibrahim YH, Lee G, Babu A, Li J, Hoshino A, Rowe GC, Rhee J, Martina JA, Puertollano R, Blenis J, Morley M, Baur JA, Seale P, Arany Z (2016) The tumor suppressor FLCN mediates an alternate mTOR pathway to regulate browning of adipose tissue. Genes Dev 30: 2551–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang R, Carrera I, Xu S, Lakshmana MK (2016) TFEB overexpression in the P301S model of tauopathy mitigates increased PHF1 levels and lipofuscin puncta and rescues memory deficits. eNeuro 3 https://doi.org/10.1523/ENEURO.0042-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Hemesath TJ, Takemoto CM, Horstmann MA, Wells AG, Price ER, Fisher DZ, Fisher DE (2000) c‐Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes Dev 14: 301–312 [PMC free article] [PubMed] [Google Scholar]
- Xiao Q, Yan P, Ma X, Liu H, Perez R, Zhu A, Gonzales E, Burchett JM, Schuler DR, Cirrito JR, Diwan A, Lee JM (2014) Enhancing astrocytic lysosome biogenesis facilitates Abeta clearance and attenuates amyloid plaque pathogenesis. J Neurosci 34: 9607–9620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Massague J (2004) Nucleocytoplasmic shuttling of signal transducers. Nat Rev Mol Cell Biol 5: 209–219 [DOI] [PubMed] [Google Scholar]
- Yasumoto K, Amae S, Udono T, Fuse N, Takeda K, Shibahara S (1998) A big gene linked to small eyes encodes multiple Mitf isoforms: many promoters make light work. Pigment Cell Res 11: 329–336 [DOI] [PubMed] [Google Scholar]
- Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, Blenis J (2011) Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 332: 1322–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Zhou Q, Ogmundsdottir MH, Moller K, Siddaway R, Larue L, Hsing M, Kong SW, Goding CR, Palsson A, Steingrimsson E, Pignoni F (2015) Mitf is a master regulator of the v‐ATPase, forming a control module for cellular homeostasis with v‐ATPase and TORC1. J Cell Sci 128: 2938–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Cheng X, Yu L, Yang J, Calvo R, Patnaik S, Hu X, Gao Q, Yang M, Lawas M, Delling M, Marugan J, Ferrer M, Xu H (2016) MCOLN1 is a ROS sensor in lysosomes that regulates autophagy. Nat Commun 7: 12109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Shibasaki F, Price R, Guillemot JC, Yano T, Dotsch V, Wagner G, Ferrara P, McKeon F (1998) Intramolecular masking of nuclear import signal on NF‐AT4 by casein kinase I and MEKK1. Cell 93: 851–861 [DOI] [PubMed] [Google Scholar]


