Abstract
Ribosome biogenesis is a complex process involving multiple factors. Here, we show that the widely conserved RNA chaperone Hfq, which can regulate sRNA‐mRNA basepairing, plays a critical role in rRNA processing and ribosome assembly in Escherichia coli. Hfq binds the 17S rRNA precursor and facilitates its correct processing and folding to mature 16S rRNA. Hfq assists ribosome assembly and associates with pre‐30S particles but not with mature 30S subunits. Inactivation of Hfq strikingly decreases the pool of mature 70S ribosomes. The reduction in ribosome levels depends on residues located in the distal face of Hfq but not on residues found in the proximal and rim surfaces which govern interactions with the sRNAs. Our results indicate that Hfq‐mediated regulation of ribosomes is independent of its function as sRNA‐regulator. Furthermore, we observed that inactivation of Hfq compromises translation efficiency and fidelity, both features of aberrantly assembled ribosomes. Our work expands the functions of the Sm‐like protein Hfq beyond its function in small RNA‐mediated regulation and unveils a novel role of Hfq as crucial in ribosome biogenesis and translation.
Keywords: Hfq, ribosome biogenesis, rRNA, translation
Subject Categories: Microbiology, Virology & Host Pathogen Interaction; Protein Biosynthesis & Quality Control; RNA Biology
Introduction
Ribosomal RNA (rRNA) represents more than 80% of total RNA in the cell and along with a plethora of ribosomal proteins (r‐proteins) constitutes the ribosome—the biosynthetic machinery of the cell. Ribosome biogenesis is a multi‐step hierarchically ordered process in which processing of rRNA precursor (pre‐rRNA) is a critical step. Emerging evidence suggests that pre‐rRNA maturation serves as a quality control to guarantee the integrity of the functional ribosome. In Escherichia coli, RNase III is responsible for the initial cleavages that separate individual rRNA precursors, followed by subsequent 5′ and 3′ processing by multiple ribonucleases to generate the 16S, 23S, and 5S rRNAs necessary to assemble the mature ribosomal subunits (Deutscher, 2009). Alterations in pre‐rRNA processing cause conformational changes in the final rRNA and lead to aberrantly assembled immature ribosomal particles with largely compromised translational accuracy (Liiv & Remme, 2004; Roy‐Chaudhuri et al, 2010; Yang et al, 2014). A parallel can be drawn to eukaryotes in which rRNA maturation errors lead to the production of defective ribosomal subunits (Cole et al, 2009; Fujii et al, 2012; Karbstein, 2013).
In prokaryotes, the small 30S ribosomal subunit contains 16S rRNA whereas 23S and 5S rRNA are the major components of the large 50S ribosomal subunit. The two asymmetric subunits include numerous r‐proteins and associate to form the functionally active 70S ribosome (Shajani et al, 2011). Many auxiliary ribosome biogenesis factors, including GTPases, rRNA modification enzymes, helicases, and other maturation factors, assist rRNA folding and r‐protein assembly pathway (Davis & Williamson, 2017). Strikingly, mutations affecting many of these accessory proteins cause dysfunctional ribosomes. In humans, such mutations were shown to lead to severe diseases, collectively referred to as ribosomopathies (Narla & Ebert, 2010).
The bacterial RNA‐binding protein Hfq is a member of the Sm/Lsm superfamily of proteins with homologues in all domains of life (Wilusz & Wilusz, 2013). Hfq is an RNA chaperone which facilitates basepairing between small regulatory RNAs (sRNAs) and their mRNA targets. Consequently, Hfq controls the expression of many mRNAs either positively or negatively (Vogel & Luisi, 2011; Hajnsdorf & Boni, 2012; Updegrove et al, 2016). Importantly, in many bacteria, Hfq is not required for the sRNA‐dependent pathways (Christiansen et al, 2006; Rochat et al, 2015), suggesting other yet undefined function(s) of Hfq beyond regulation of sRNA activity.
Hfq interacts in vitro with the 16S rRNA (de Haseth & Uhlenbeck, 1980) although the functional role of this interaction has remained elusive. Furthermore, rRNA molecules are commonly found in Hfq‐enriched co‐immunoprecipitations, what is usually regarded as a background noise in transcriptomic studies (Zhang et al, 2003; Sittka et al, 2008; Bilusic et al, 2014). A cross‐linking‐based study in E. coli suggests interactions of Hfq with rRNA in vivo (Tree et al, 2014). An interaction between Hfq and S12 protein of the 30S ribosomal subunit has been previously reported, yet lacking mechanistic details on its role (Strader et al, 2013). Clearly, Hfq interacts with rRNA but is this a functional or redundant interaction?
Here, we identify a novel role of Hfq in ribosome biogenesis. Inactivation of Hfq leads to accumulation of 17S rRNA and reduced levels of 70S ribosomes in E. coli. Using in vivo and in vitro approaches, including ribosome profiling, we demonstrate that Hfq deletion affects the ribosome pool with direct effects on translation efficiency and fidelity. Our data propose Hfq as a novel auxiliary ribosome biogenesis factor. This expands the functional spectrum of this RNA chaperone beyond the sRNA‐biology with impact on rRNA processing, ribosome biogenesis, and translation fidelity.
Results
Hfq is required for 16S rRNA maturation
The 16S rRNA arises from processing of the 17S rRNA precursor which harbors additional nucleotides (nts) at both extremities (Fig 1A). Hfq is a key regulator of cell physiology affecting gene expression in both exponential and stationary phases (Tsui et al, 1994; Muffler et al, 1997; De Lay et al, 2013). Thus, we compared the total RNA from wild‐type and Δhfq cells extracted from exponential and stationary phase cells by Northern blotting using specific probes complementary to 5′‐ or 3′‐ends of the 17S rRNA. In addition, we used probes corresponding to the internal regions of 16S rRNA or 23S rRNA (Fig 1B). Both 17S‐specific probes hybridized only to 17S rRNA, whereas the 16S‐probe identified both 16S and 17S rRNA. Notably, inactivation of Hfq in both growth phases resulted in higher levels of 17S rRNA with misprocessed extremities (28 and 148% increase in exponential and stationary phases, respectively), suggesting a role for Hfq in 16S rRNA maturation. On the other hand, Hfq did not significantly change the levels of 23S rRNA. The accumulation of 17S in the ∆hfq mutant compared to the wild‐type strain is observed over time in different points of the growth curve (Fig EV1A).
Figure 1. Hfq is required for correct processing and folding of 16S rRNA .
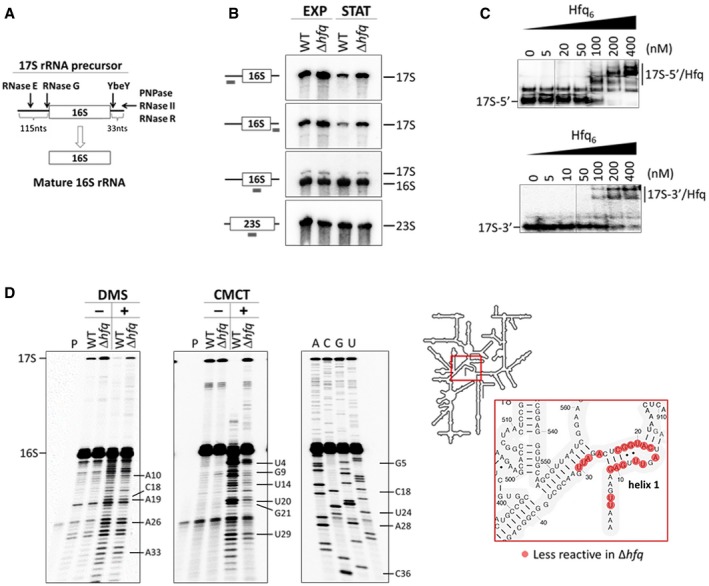
- Schematic representation of the RNase‐mediated processing of the 17S rRNA precursor into mature 16S rRNA.
- Northern blot analysis of total RNA extracted from cells in exponential (EXP) or stationary (STAT) growth phase. Samples were fractionated on a 4% polyacrylamide/7 M urea gel. A scheme of the probes binding to the rRNA sequence is displayed on the side.
- Electrophoretic mobility shift assays of Hfq binding to the 5′ and 3′ extremities of the 17S rRNA. Increasing amounts of Hfq hexamer were mixed with a constant amount of the specific 17S‐flanking sequences and resolved on a 6% (top panel) or 8% (bottom panel) native polyacrylamide gel.
- DMS and CMCT accessibility probing of the 16S rRNA. Reverse‐transcribed cDNA was fractionated on an 10% polyacrylamide/7 M urea gel. Residues with altered reactivities in the Δhfq mutant are indicated. The inset depicts the analyzed region of the 16S rRNA.
Source data are available online for this figure.
Figure EV1. Hfq regulates 17S rRNA levels.
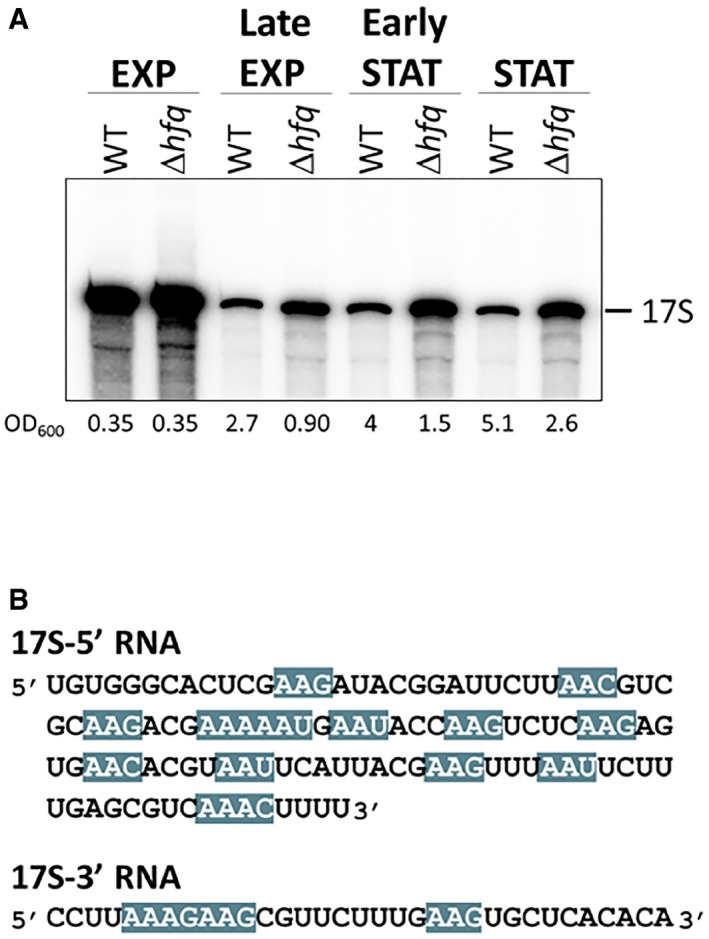
- 17S rRNA accumulates over time in the Δhfq strain. Northern blot analysis of total RNA isolated at different timepoints following the growth curve of wild‐type and Δhfq cells. Samples were separated on a 4% polyacrylamide/7 M urea gel, and a probe specific for the 17S 5′‐end was used.
- Predicted Hfq‐binding motifs within the 17S rRNA flanking sequences. The Hfq‐binding motif (ARN)x is highlighted in blue. (R—purine; N—any nucleotide).
Hfq preferentially binds to the (ARN)x motif in RNAs (Mikulecky et al, 2004; Link et al, 2009; Peng et al, 2014). Strikingly, both 5′ and 3′ extremities of the 17S rRNA carry several of these predicted Hfq‐binding sequences (Fig EV1B). To further assess the Hfq binding to these regions, we performed gel mobility shift experiments with constant amounts of the 5′‐end and 3′‐end sequences of 17S RNA and increasing levels of the Hfq protein (Fig 1C). Indeed, Hfq complexed with both 17S‐end sequences corroborating the idea that Hfq interacts in vitro with 17S rRNA specific sequences.
The additional nucleotides from the 17S rRNA could interact with helix 1 and helix 2 of the mature 16S rRNA inducing alternative structures which would affect the folding of the central pseudoknot (Lodmell & Dahlberg, 1997; Roy‐Chaudhuri et al, 2010). To evaluate whether the deletion of Hfq leads to an altered conformation of the 16S rRNA, we performed RNA mapping using two chemical probes, dimethyl sulfate (DMS) and N‐cyclohexyl‐N′‐(2‐morpholinoethyl)carbodiimide (CMCT), in separate experiments (Fig 1D). A specific antisense primer to the 5′‐end of the 16S rRNA was used in the primer extension reactions which allowed good resolution of the 16S central pseudoknot that consists of helix 1 (nucleotides 9–13/21–25) and helix 2 (nucleotides 17–19/916–918; Brink et al, 1993). Several nucleotides accessible to DMS (adenosines and cytidines) or CMCT (uridines and guanosines) modification in the wild type were less reactive to these probes in the absence of Hfq (Fig 1D). Our data imply that the folding of the 16S rRNA is altered as consequence of Hfq inactivation, resulting in the structural occlusion of those residues. Altogether, our observations indicate that Hfq interacts with 17S rRNA and is necessary for the correct processing and folding of the mature 16S rRNA, and affects the formation of the central pseudoknot.
Inactivation of Hfq results in altered profiles of ribosome sedimentation
Given that misprocessing of rRNA can be consequence of defects in ribosome assembly (Liiv & Remme, 2004; Roy‐Chaudhuri et al, 2010; Shajani et al, 2011), we next examined whether the defects in the maturation of 16S rRNA found in the Δhfq strain had consequences to the total amount of ribosomes. We profiled the ribosomes from exponential and stationary phase cultures of wild‐type and mutant Δhfq strains by sucrose gradient ultracentrifugation (Fig 2A and B). The ribosome identity in the different peaks was further confirmed by analyzing their rRNAs (Appendix Fig S1). In the wild‐type strain, under conditions that favor ribosome association (10 mM Mg2+), the peak corresponding to the small 30S subunits was nearly absent, while the amount of the 70S ribosomes was comparable between exponential and stationary phases (Fig 2A and B, and Appendix Fig S1). In clear contrast, the levels of the mature 70S ribosomes were reduced in the ∆hfq mutant as compared to the wild type, an effect particularly severe in the stationary phase. Additionally, free 30S accumulated in the ∆hfq, which again was more evident in stationary phase (Fig 2A and B). The complementation of the ∆hfq deletion in trans with a plasmid expressing Hfq (pHfq; Andrade et al, 2012) raised the amount of mature ribosomes to levels comparable to that of the wild‐type strain (Fig 2A and B, Appendix Fig S1). Strikingly, the plasmid expressing Hfq rescued the defects in the ribosomal amounts isolated from the Hfq deletion strain. Note that the Δhfq strain transformed with the empty vector was essentially identical to the Δhfq strain suggesting no effects of the transformation itself.
Figure 2. Defective ribosome biogenesis in the Δhfq strain.
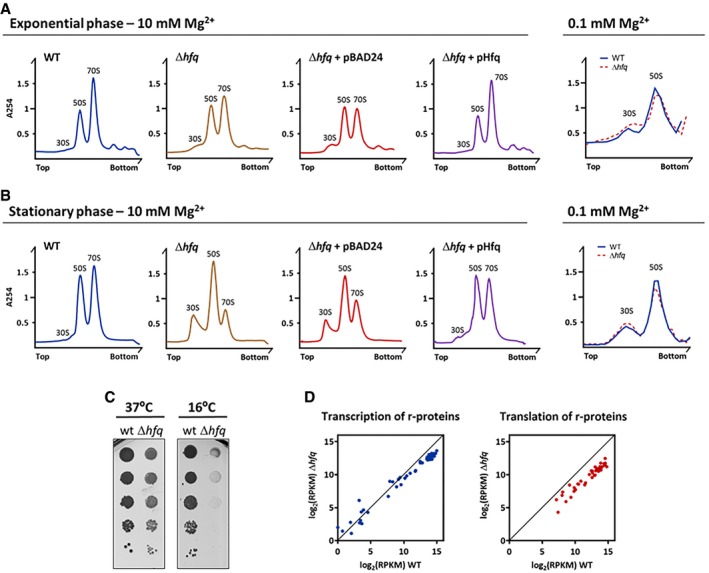
-
A, B(Left panels) Ribosomes from cells in the exponential or stationary phase were fractionated on sucrose density gradients in 10 mM Mg2+ to stabilize 70S particles with and without trans‐complementation of hfq gene using pBAD24 plasmid. Ribosome species are identified over each peak; top and bottom denote the lowest (15%) and highest (45%) sucrose concentration in the gradient, respectively. (Right panels) Ribosomes purified from cells in exponential and stationary phases fractionated on sucrose density gradients at low 0.1 mM Mg2+ concentration to promote 70S dissociation into free 30S and 50S subunits. Top and bottom denote the lowest (10%) and highest (30%) sucrose concentration in the gradient, respectively.
-
CSerial dilutions (with 1:10 steps) of wild‐type and Δhfq strains grown on LB‐agar plates at 37 or 16°C.
-
DComparison of mRNA expression (left) and protein production (right) of ribosomal proteins between wild‐type and Δhfq strains analyzed by RNA‐Seq (left) and ribosome profiling (right), respectively.
Source data are available online for this figure.
Our data clearly demonstrate that the inactivation of Hfq leads to a reduction in the pool of 70S ribosomes in the cell. This could either result from imbalanced production of subunits or the occurrence of major defects in the assembly of the 70S particle upon inactivation of Hfq. To distinguish between these two possibilities, we further analyzed ribosomes under dissociative conditions (0.1 mM Mg2+) to guarantee that all ribosomal subunits would be in their free state. As observed in Fig 2A and B (right panels), both strains displayed comparable contents of 30S and 50S subunits irrespective of the growth phase. Hence, the lower levels of 70S ribosomes in the absence of Hfq (Fig 2A and B, left panels) are a consequence of defects in the 70S assembly.
A well‐known hallmark of ribosome biogenesis defects in bacteria is the cold‐sensitive phenotype (Connolly & Culver, 2009). We next compared the growth of the wild‐type and Δhfq strains at 37 and 16°C (Fig 2C). Clearly, the Δhfq mutant exhibited the cold‐sensitive phenotype, with severe growth defects under cold shock but not at 37°C which correlated with the altered ribosome profile found in the absence of Hfq. This effect is reminiscent of the cold‐sensitive phenotype observed with different ribosome biogenesis factors like RbfA, KsgA, RimM, and RimO (Bylund et al, 1998; Connolly et al, 2008; Leong et al, 2013).
rRNA synthesis feedforwards the synthesis of ribosomal proteins (Scott et al, 2014). Thus, to assess the expression of the r‐proteins, we used ribosome profiling which captures the positions of actively translating ribosomes and the ribosome‐protected fragments (RPFs) reporting on differences in gene expression at the level of translation (Ingolia et al, 2009; Li et al, 2014). This analysis was combined with RNA‐Seq to determine the mRNA expression levels and the regulation of gene expression at the level of transcription. Strikingly, all ribosomal proteins were significantly translationally downregulated in the Δhfq mutant strain while the levels of their transcripts remained unchanged or decreased to much lower extent (Figs 2D and EV2A, and Dataset EV1). Notably, among the significantly enriched gene ontology (GO) terms are genes participating in ribosome assembly (Dataset EV1). Furthermore, within the polycistronic mRNAs, the translation yields of the encoded r‐proteins differed (Fig EV2B and Dataset EV1) implying an independent translation initiation of the r‐proteins (Li et al, 2014). This expression pattern corroborates earlier observations for translational coupling of the expression of the ribosomal proteins and rRNA synthesis (Jinks‐Robertson & Nomura, 1981; Nomura, 1999). Cumulative profiles of all expressed genes do not differ between wild‐type and Δhfq strains, arguing against an effect of Hfq depletion on translation initiation (Fig EV3A).
Figure EV2. Hfq affects the translation of ribosomal proteins.
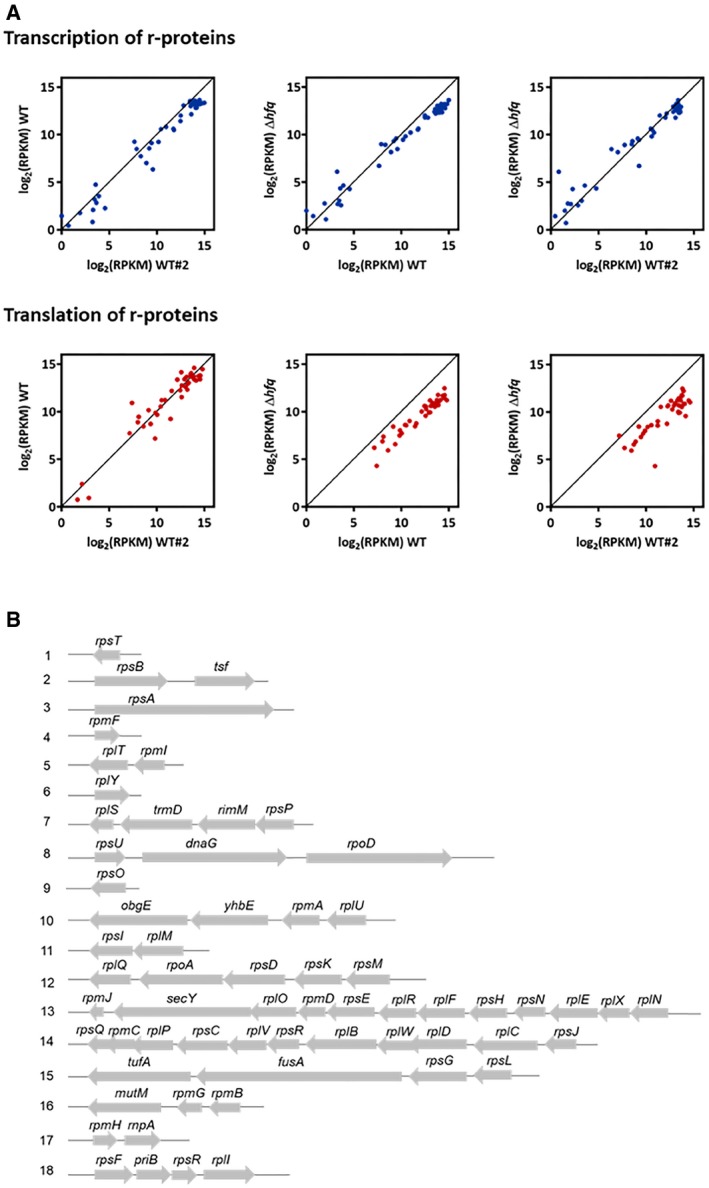
- Translational downregulation of ribosomal protein genes in the Δhfq strain. Comparison of mRNA expression (top panels) and protein production (bottom panels) of ribosomal proteins between the wild‐type and Δhfq strains used in this study and an additional wild‐type data set (WT#2; Hwang & Buskirk, 2017).
- Schematic organization of the ribosomal protein genes their respective operons. The operons were arbitrarily numbered 1–18.
Figure EV3. Cumulative metagene profile and coverage profiles of selected downregulated genes in the wild‐type and Δhfq strain obtained by ribosome profiling.
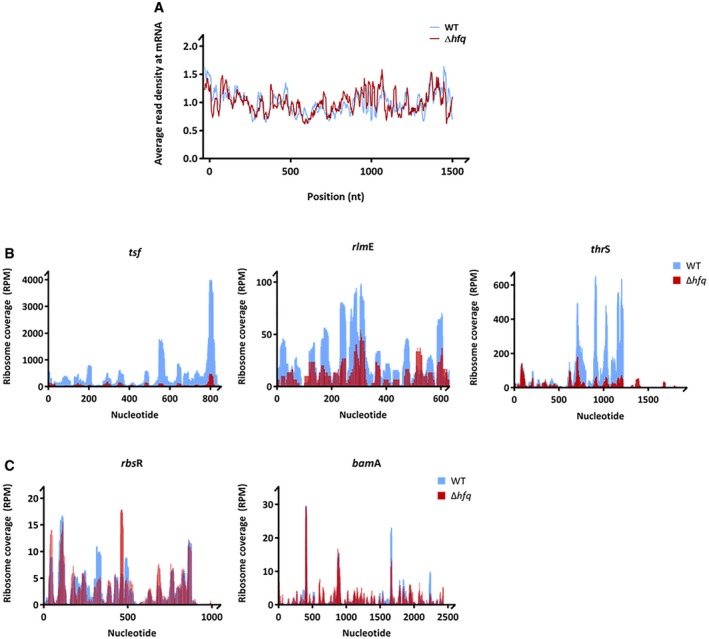
- Cumulative metagene profile of the read density as a function of position for RPFs. The expressed genes were individually normalized, aligned at the start codon, and averaged with equal weight. 1,075 and 1,231 genes from wild‐type and Δhfq strains, respectively, were considered in the analysis.
- Coverage profiles of selected downregulated genes representative of gene categories affected by Hfq deletion (Fig 4D). Elongation factor‐Ts (tsf), 23S rRNA 2′‐O‐ribose U2552 methyltransferase (rlmE) are included in the GO term “translation and ribosome” and threonine‐tRNA ligase (thrS) in the GO term “amino acid biosynthesis”.
- Coverage profiles of exemplified genes (bamA and rbsR) whose expression remained unchanged upon hfq deletion. BamA is an outer membrane protein assembly factor, and RbsR is a transcriptional factor of the operon involved in ribose catabolism and transport.
Overall, our results show that the Hfq depletion leads to defects in ribosome biogenesis with consequences for the pool of mature 70S ribosomes and propose Hfq as an auxiliary factor which regulates ribosome biogenesis.
Hfq copurifies with immature 30S subunits
We hypothesized that Hfq would preferably bind to immature 30S subunits as these can be enriched in 17S RNA. To test this, we purified immature 30S subunits from the knockout mutant of RbfA, a late assembly factor that accumulates pre‐30S particles enriched in 17S rRNA (Jones & Inouye, 1996; Bylund et al, 1998; Thurlow et al, 2016). Compared to the wild‐type, the ΔrbfA mutant showed a similar ribosome profile to the Δhfq mutant, with increasing levels 30S particles and lower levels 70S ribosomes (Fig 3A). The peak corresponding to the 30S fraction was recovered from the sucrose gradients of the ΔrbfA mutant, and the 30S subunits were purified in low salt conditions. In parallel, mature 30S subunits were obtained from dissociation of 70S ribosomes isolated from the wild type, also in low salt conditions. Purified 30S samples were then analyzed by mass spectrometry that identified proteins associated with 30S subunits. Most of the proteins identified corresponded to r‐proteins or known factors associated with ribosomes (Dataset EV2). Strikingly, Hfq was found to copurify only with immature 30S isolated from the ΔrbfA but not with the mature 30S isolated from the wild type (Fig 3B). The same 30S samples were analyzed by Western blotting using an anti‐Hfq antibody. Cell lysates of wild‐type and Δhfq strains and purified His‐tagged Hfq were used as controls. Western blot confirmed the presence of Hfq in the 30S purified from the ΔrbfA but not from the wild type, in total agreement with mass spectrometry data (Fig 3C). Overall, these results show that Hfq is copurifying with precursor 30S ribosomes in vivo and corroborates that Hfq is a novel factor that assists ribosome assembly.
Figure 3. Hfq copurifies with immature 30S subunits.
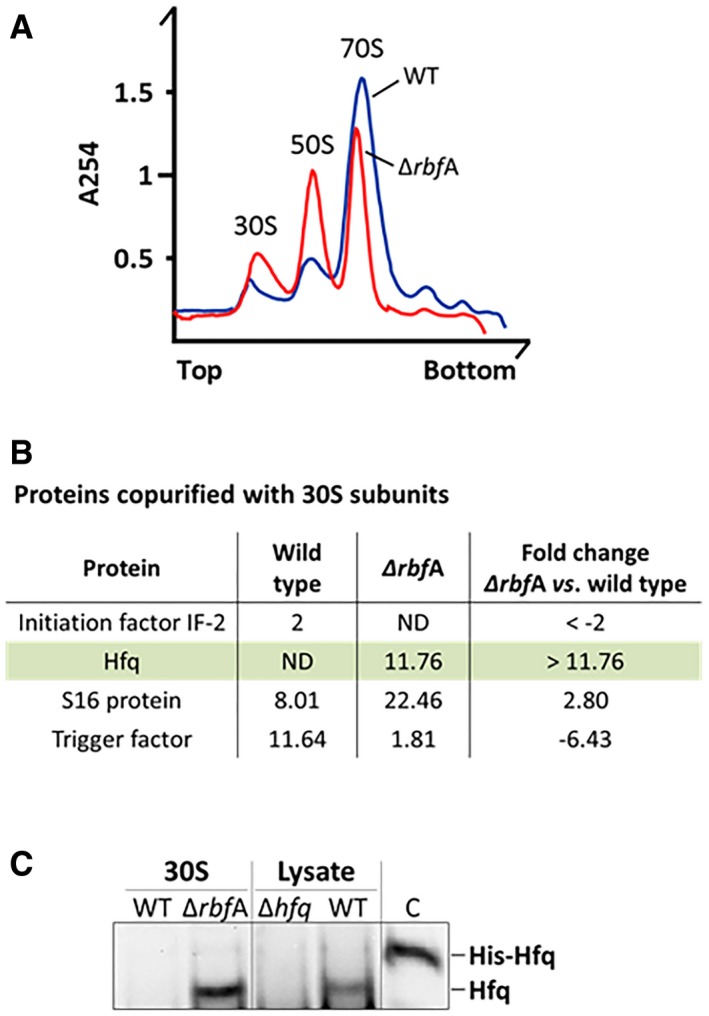
- Ribosomes from wild‐type and ΔrbfA exponential growing cells were analyzed on sucrose density gradients. The ΔrbfA mutant displays an altered ribosome profile with an increase in 30S and 50S subunits and a reduction of 70S ribosomes compared to the wild type.
- Representative proteins identified by mass spectrometry of purified 30S subunits from the wild‐type and ΔrbfA mutant. The measurement of all the peptides identified for each protein is shown as total ProtScore values calculated with the Pro Group™ Algorithm (Sciex), with a 95% confidence. The ratio between the ΔrbfA mutant and wild type are shown as normalized fold changes that are represented by positive or negative values corresponding to an increase or decrease, respectively, of the number of peptides found in the ΔrbfA mutant compared to the wild type. (ND, not detected).
- Western blot analysis of purified 30S subunits using an anti‐Hfq antibody. WT and Δhfq cell lysates as well as purified His‐Hfq protein were loaded as controls.
Source data are available online for this figure.
Hfq affects translation efficiency
Altered ribosome biogenesis can lead to major defects in translation, and thus, we next assessed the translational status in the Δhfq mutant. Firstly, the Δhfq strain showed a reduced polysome fraction compared to that of the wild‐type strain (Fig 4A). Secondly, a global measurement of protein synthesis by pulse metabolic labeling confirmed a significant reduction of translation in Hfq‐depleted background (Fig 4B). Thirdly, the global translation efficiency, which was determined by the density of ribosomes from the ribosome profiling per mRNA from the RNA‐Seq dataset, was significantly reduced (Mann–Whitney U‐test or Wilcoxon rank‐sum test, P = 0.0001996; Fig 4C). Hence, the defects in rRNA precursor processing and ribosome biogenesis in the Δhfq mutant decreased translation volume and efficiency as compared to the parental strain. The well‐known importance of Hfq for stress response in E. coli could arise from this effect on translational capacity, and not only from Hfq's role in sRNA‐dependent regulation.
Figure 4. The Δhfq strain displays reduced translation levels.
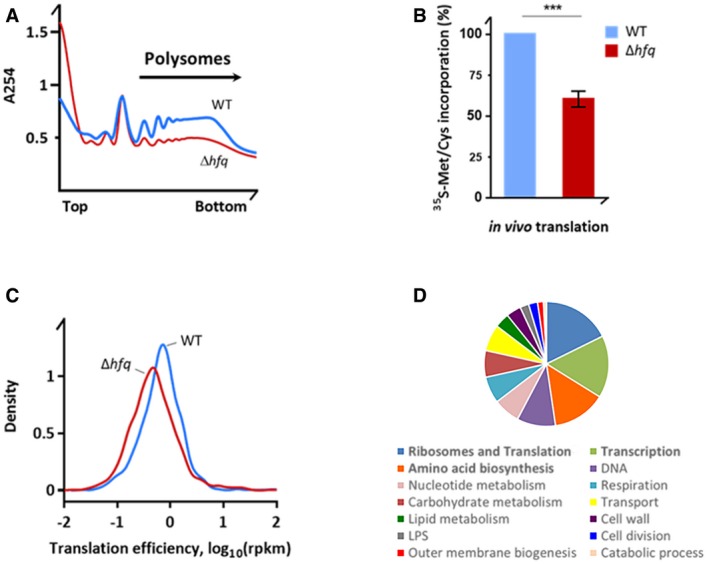
- Polysomal fraction is reduced in Δhfq cells. Polysome profiles of the wild‐type and Δhfq strains were resolved on sucrose density gradient. Top and bottom denote the lowest (15%) and highest (50%) sucrose concentration in the gradient, respectively.
- In vivo incorporation of 35S‐methionine/cysteine translation assay in M9 medium. Data are normalized to the wild‐type strain and are means ± SEM (n = 3). ***P = 0.0004 (paired t‐test).
- Translation efficiency of wild‐type and Hfq‐depleted cells obtained by ribosome profiling.
- GO term analysis of translationally downregulated genes in the Δhfq. The top three affected categories are in bold. Full GO term analysis is included in Dataset EV1.
We next asked whether these changes in translation efficiency are global or a fraction of genes escapes this trend. We performed a fold‐change analysis and ranked the genes according to the fold‐change in translation (i.e., only translationally up‐ or downregulated in the ribosome profiling set) but with unchanged mRNA expression from the RNA‐Seq experiment. Genes with changes in their RPF coverage higher than twofold were considered. The gene ontology (GO) analysis of the downregulated genes in Hfq‐depleted background showed several pathways being affected but with a significant GO term enrichment in genes participating in ribosome biogenesis, translation, and amino acid metabolism (Fig 4D and Dataset EV1). The complete list of genes with the GO categories is summarized in Dataset EV1. For comparison, density plots of representative examples downregulated in the Δhfq mutant (Fig EV3B) or with unaltered translation (Fig EV3C) are included. Notably, inactivation of Hfq augmented the mRNA levels of genes known to be regulated by Hfq‐dependent sRNAs, while their translation was only slightly affected (Dataset EV3).
Hfq affects translation fidelity
The ribosomal tRNA accommodation site (A‐site) is formed by helix 44 of the 16S rRNA of the 30S subunit. Three aminoglycoside antibiotics, neomycin, paromomycin, and kanamycin, interact with the 16S rRNA near the A‐site and induce translational misreading (i.e., shift of the reading frame, stop‐codon readthrough; Foster & Champney, 2008). In the presence of sub‐lethal concentrations of neomycin, paromomycin, or kanamycin, the Δhfq mutant strain showed exacerbated growth defects relative to untreated Δhfq or wild‐type strains, suggesting that Hfq affects translation fidelity (Fig 5A). Additional aminoglycosides were further tested showing similar effect (Fig EV4). As control, the Δhfq strain did not show increased sensitivity to other classes of antibiotics, like colistin, which targets cell membrane (Figs 5A and EV4). We also investigated the misreading using a collection of widely used plasmids bearing lacZ as reporter (O'Connor et al, 1997). When compared with the isogenic parent, the Δhfq mutant showed a substantial increase in frameshifting, aberrant initiation from alternative start codon(s), and stop‐codon readthrough (Fig 5B), indicating that the accuracy of translation in Hfq‐depleted background is severely compromised. In sum, these data suggest that inactivation of Hfq decreases translation efficiency and enhances misreading of mRNA, implying a functional link between Hfq‐dependent alterations in rRNA processing, ribosome biogenesis, and translation fidelity.
Figure 5. Hfq‐depleted cells exhibit increased codon misreading.
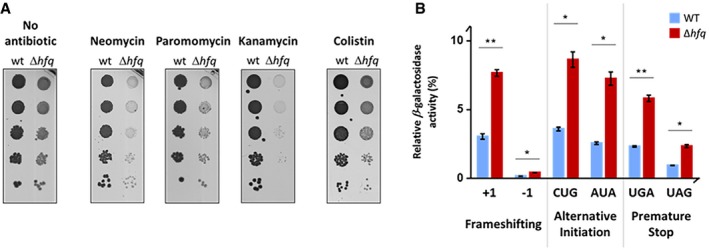
- Serial dilutions (1:10) of wild‐type and Δhfq strains grown on LB‐agar plates at 37°C with and without sub‐lethal concentrations of neomycin (1 μg/ml), paromomycin (1 μg/ml), kanamycin (1 μg/ml), or colistin (0.1 μg/ml).
- Wild‐type and Δhfq strains expressing mutated lacZ gene (pSG plasmids) were tested for a frameshift mutation (+1 or −1), alternative initiation codons (CUG or AUA), or a non‐sense stop‐codon mutation (UGA or UAG). For each strain, the β‐galactosidase activity (in Miller units) was normalized to that of strain expressing the wild‐type lacZ. Data are means ± SEM (n = 3). **P < 0.01; *P < 0.02 (paired t‐test).
Source data are available online for this figure.
Figure EV4. Additional Δhfq antibiotic sensitivity tested by serial dilution platting.
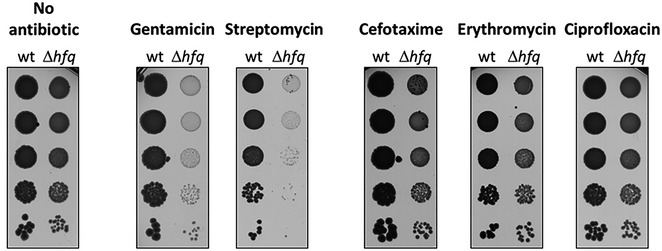
Serial dilutions (1:10) of wild‐type and Δhfq strains grown on LB‐agar plates at 37°C with and without sub‐lethal concentrations of gentamicin (0.1 μg/ml), streptomycin (1 μg/ml), cefotaxime (0.01 μg/ml), erythromycin (2 μg/ml), or ciprofloxacin (0.002 μg/ml).
The distal face is critical for the Hfq‐dependent regulation of ribosome biogenesis
The Sm‐like Hfq assembles into a hexamer with a ring‐like shape that displays at least three RNA‐binding surfaces which confer Hfq the ability to bind simultaneously different RNA substrates. The proximal face and a charge patch in the outer rim of the hexamer bind preferably U‐rich sRNAs while the distal face binds to A‐rich sequences in target mRNAs (Mikulecky et al, 2004; Link et al, 2009; Otaka et al, 2011; Sauer & Weichenrieder, 2011; Sauer et al, 2012; Panja et al, 2013; Zhang et al, 2013).
To evaluate which binding surface would be responsible for the newly identified Hfq‐dependent regulation of ribosome biogenesis, representative Hfq variants with mutations in the different surfaces (Zhang et al, 2013) were tested. Ribosome sedimentation profiles of proximal (Q8A and F39A), rim (R16A), and distal (Y25D and K31A) mutants isolated from exponential cultures were compared to that of the wild‐type strain (Figs 6A and EV5). In addition, rRNAs from each fraction were isolated to confirm the ribosome identity in each peak (Appendix Fig S2). Strikingly, only mutations in the distal face caused reduction in the 70S ribosome levels, which were similar to those we observed for the Hfq deletion mutant (Fig 2A and B). The ribosome profiles of mutants in the proximal or rim surface were similar to that of the wild type, and these surfaces were shown to govern interactions with sRNAs (Sauer & Weichenrieder, 2011; Sauer et al, 2012; Panja et al, 2013; Zhang et al, 2013). Altogether, from these data, we conclude that the distal face of Hfq is critical for the regulation of the rRNA maturation and ribosome biogenesis and propose that the novel function of Hfq in the ribosome biogenesis might be independent of sRNA binding.
Figure 6. The distal face of Hfq is required for correct ribosome biogenesis and translation fidelity.
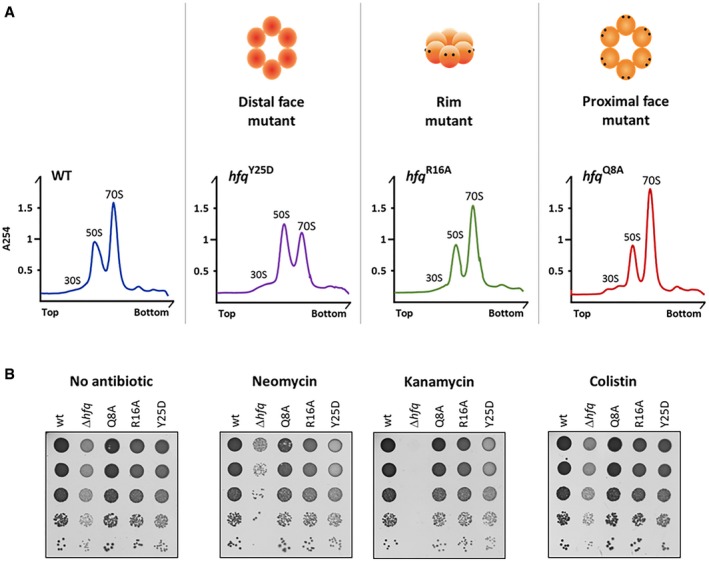
- Ribosomes purified from strains with specific point mutations in the hfq gene were fractionated on sucrose density gradients and compared to the wild‐type strain. The binding surface affected by each mutation is schematically depicted on the top.
- Serial dilutions (1:10) of wild‐type, Δhfq, and Hfq variants grown on LB‐agar plates at 37°C with and without sub‐lethal concentrations of neomycin (1 μg/ml), kanamycin (1 μg/ml), or colistin (0.1 μg/ml).
Source data are available online for this figure.
Figure EV5. Sucrose density gradients of the K31A and F39A Hfq mutants.
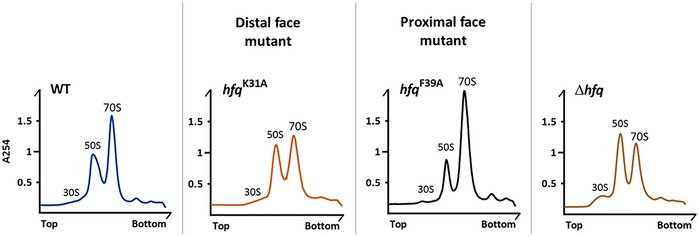
Ribosomes purified from strains with specific point mutations in the hfq gene (alleles K31A and F39A) were resolved in 15–45% sucrose density gradients. Gradients from wild‐type and Δhfq strains are included for comparison. The binding surface affected by each mutation is indicated.
The sensitivity of the Hfq variant strains against different antibiotics was also tested (Fig 6B). The proximal and rim mutants (Q8A and R16A) did not show significant growth difference to the wild type. Only the distal Hfq‐Y25D variant showed increased susceptibility to aminoglycosides, like neomycin or kanamycin, suggesting that Hfq‐Y25D is impaired in translation efficiency. However, the growth defect of Hfq‐Y25D strain is not as severe as the one found in the knockout Δhfq mutant, which suggest that Y25D is an important residue but is not the sole responsible for the increased susceptibility to aminoglycosides. This phenotype was not observed when other classes of antibiotics were tested, such as polypeptide antimicrobials like colistin. These results from antibiotic sensitivity further support the importance of the distal face of Hfq in translation.
Discussion
Here, we present results that support a novel role of Hfq in bacterial ribosome biogenesis with important consequences for translation (Fig 7). Hfq is a widely conserved RNA‐binding protein of the Sm/Lsm family of proteins (Wilusz & Wilusz, 2013) that it is mostly known for promoting sRNA basepairing with target mRNAs (Updegrove et al, 2016). However, a role of Hfq in regulating rRNA processing and folding has not been proposed. Our work unveils previously undescribed roles of Hfq in ribosome biogenesis and expands the repertoire of Hfq functions in the cell.
Figure 7. Model for the Hfq regulation of ribosome biogenesis.
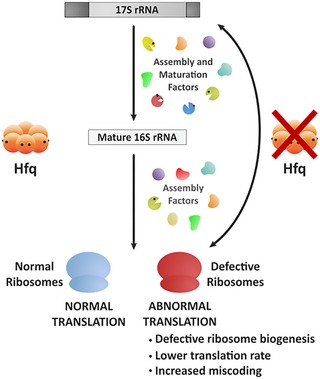
- Hfq assists ribosome biogenesis together with other ribosome assembly and maturation factors. Hfq depletion exhibits critical consequences for ribosome biogenesis and cellular translation. The Δhfq mutant affects the correct maturation of 30S subunits and accumulates unprocessed 17S rRNA precursor leading to a general translation deficiency.
We show that Hfq is a new regulator of rRNA maturation. Hfq depletion results in loss of normal processing of rRNA, leading to the accumulation of unprocessed 17S rRNA precursor. Earlier cross‐linking studies in E. coli identified interactions between Hfq and rRNA (Tree et al, 2014), but did not analyze it further. We find that Hfq directly interacts with the 17S rRNA and Hfq inactivation results in the misprocessing of both 17S extremities. Our data align well with observations made for Lsm proteins, the evolutionarily conserved eukaryotic counterparts of the bacterial Hfq; depletion of Lsm proteins causes defects in the processing of pre‐rRNAs (Kufel et al, 2003; Beggs, 2005) supporting the notion for an evolutionary conserved function of the members of the Sm/Lsm protein family in rRNA processing.
Accumulation of the 17S rRNA precursor is usually linked to problems in formation of mature 30S subunit, and most likely because maturation of 16S rRNA is a final step in ribosome biogenesis (Srivastava & Schlessinger, 1988; Shetty & Varshney, 2016). rRNA synthesis and maturation are tightly intertwined with the r‐protein biosynthesis (Jinks‐Robertson & Nomura, 1981; Nomura, 1999; Scott et al, 2014). In the Δhfq background, accumulation of unprocessed 17S rRNA is accompanied with significant reduction of r‐proteins synthesis, substantial reduction in the levels of 70S ribosomes, and concomitant accumulation of immature ribosomal subunits. The initiation of translation is the most sensitive node in translation regulation and defects during this process could lead to a similar phenotype (Laursen et al, 2005). However, translation initiation remains unaffected upon Hfq inactivation (Fig EV3A). Along with the fact that in trans‐complementation with Hfq rescues the defective ribosome assembly, we show that Hfq is needed for proper ribosome biogenesis but unessential for proper initiation. Hfq‐depleted cells show phenotypes typically found in mutants of ribosome biogenesis factors, namely defects in rRNA maturation, accumulation of rRNAs precursors, and cold sensitivity (Kaczanowska & Rydén‐Aulin, 2007; Shajani et al, 2011). rRNA precursors compete with mature rRNAs for binding to r‐proteins, although pre‐rRNA containing ribosomes are conformationally defective (Liiv & Remme, 2004; Yang et al, 2014).
Several auxiliary factors associate with the ribosome during the intricate process of ribosome assembly assisting in r‐protein binding and rRNA folding steps (Kaczanowska & Rydén‐Aulin, 2007). Hfq is a novel assembly factor that preferentially binds immature 30S subunits, like other chaperones such as RimM or RbfA. Like these factors, Hfq probably acts to facilitate or proofread folding of the pre‐rRNA and pre‐30S assembly. Hfq is a well‐known RNA chaperone able to remodel RNA secondary structures (Moll et al, 2003; Wroblewska & Olejniczak, 2016); hence, it is conceivable that it might be essential for the correct processing and folding of 16S rRNA into 30S subunits. In fact, cells lacking Hfq present an altered rRNA folding as suggested by our RNA‐structure mapping. This is likely a consequence of the additional nucleotides from the 17S rRNA precursor which perturb the formation of helixes 1 and 2 of the 16S rRNA (Lodmell & Dahlberg, 1997; Roy‐Chaudhuri et al, 2010). This in turn affects the folding of the central pseudoknot, a universally conserved structural element that establish long‐range interactions within the 16S rRNA and that is critical for the overall folding of the small subunit (Brink et al, 1993). Moreover, alterations in the secondary structure of the pseudoknot result in error‐prone ribosomes (Lodmell & Dahlberg, 1997) as we observed in the Δhfq mutant. Alternatively, Hfq may promote RNA‐protein interactions that are important for the correct rRNA processing. Notably, Hfq was previously shown to bind to the S12 protein of the 30S small subunit in E. coli (Strader et al, 2013). The S12 protein is a key mediator of translation fidelity in both prokaryotes and eukaryotes and is positioned in helix 44 of the 16S rRNA that is known to form extensive contacts with the large subunit (Yusupov et al, 2001; Cukras et al, 2003). Association of Hfq with S12 is suggested to be important for the correct folding of 16S rRNA and formation of interface between ribosomal subunits and consequently the assembly of 70S ribosomes. Moreover, the sensitivity of the Δhfq strain to aminoglycosides and the cumulative translation errors induced by hfq deletion corroborate the observation that the conserved helix 44 of 16S rRNA maintains translation fidelity and serve as aminoglycoside target (Davis, 1987).
The role of Hfq in promoting the basepairing between regulatory small RNAs and their target mRNAs constitutes the most well‐known function of this RNA‐binding protein. An interesting feature of Hfq is that it is possible to uncouple its multiple functions by introducing point mutations in each of its RNA‐binding surfaces: the distal face of Hfq recognizes and binds to trinucleotide ARN repeats in mRNA, while the proximal and rim faces bind preferably to U‐rich sequences in small RNAs (Link et al, 2009; Otaka et al, 2011; Sauer & Weichenrieder, 2011; Sauer et al, 2012; Panja et al, 2013). Strikingly, we found that the reduced levels of the 70S ribosomes in Hfq‐depleted cells is dependent on residues located at the distal face of Hfq but not on those in the proximal and rim RNA‐binding faces suggesting that rRNA regulation is independent of sRNA binding of Hfq. Despite Hfq being widely conserved, Hfq‐dependent regulation of sRNAs is not a common feature; for example, this function is missing in many bacteria like Bacillus subtilis and Listeria monocytogenes (Christiansen et al, 2006; Rochat et al, 2015). Hfq is known to act independently of an sRNA as partner in a variety of cellular functions. Namely, Hfq stimulates the addition of poly(A) tails to the 3′ end of mRNAs containing Rho‐independent transcription terminators, promoting their degradation in E. coli (Le Derout et al, 2003; Mohanty et al, 2004; Folichon et al, 2005; Régnier & Hajnsdorf, 2013). Also, Hfq inhibits translation by binding directly to mRNAs, independent of a sRNA partner (Salvail et al, 2013; Ellis et al, 2015).
In summary, we have demonstrated that Hfq is a new ribosome assembly factor. Cells lacking Hfq exhibit diverse hallmarks of ribosome biogenesis defects, namely (i) misprocessing of rRNA and accumulation of 17S rRNA precursor; (ii) reduced pool of 70S ribosomes; (iii) an abnormal translation and compromised translation fidelity; and (iv) cold‐sensitive phenotype, typically associated with ribosome biogenesis factor mutants. This work expands the functions of Hfq beyond the regulation of small non‐coding RNA biology and unveils unprecedented roles in ribosome biogenesis and translation.
Materials and Methods
Bacterial strains, plasmids, and oligonucleotides
All bacterial strains, plasmids, and oligonucleotides are listed in Appendix Tables S1–S3, respectively. All E. coli K‐12 strains used in this study are derivatives of strains MG1693 or MC1061. Deletion of hfq was obtained using the λ‐Red recombination (Datsenko & Wanner, 2000). The hfq point mutant alleles (Zhang et al, 2013) were P1‐transduced to our parental strain, following selection on glucose minimal plates and screening for sensitivity to chloramphenicol. The ΔrbfA mutant was obtained from the Keio collection (Baba et al, 2006). All mutations were confirmed by PCR and sequencing.
Bacterial growth
Strains were grown in LB medium (Difco) supplemented with thymine (50 μg/ml) at 37°C, unless otherwise stated. Overnight cultures of single freshly grown colonies were diluted to an initial OD600 ~ 0.03. Cultures were collected either at exponential phase (OD600 ~ 0.5) or stationary phase (after ~ 14 h growth). Antibiotics were present at the following concentrations when needed: 25 μg/ml chloramphenicol, 50 μg/ml kanamycin, 10 μg/ml tetracycline, and 100 μg/ml ampicillin. For the dilution plating assays, serial dilutions were made in 10‐fold increments and immediately spotted onto LB‐agar plates. Sub‐lethal concentrations of antibiotics were added when relevant: 1 μg/ml neomycin, 1 μg/ml paromomycin, 1 μg/ml kanamycin, 0.1 μg/ml colistin, 0.1 μg/ml gentamicin, 1 μg/ml streptomycin, 0.01 μg/ml cefotaxime, 2 μg/ml erythromycin, and 0.002 μg/ml ciprofloxacin.
RNA analysis
For Northern blots, total RNA was extracted as previously described (Andrade et al, 2012). One microgram of total RNA was resolved on 4% polyacrylamide/7 M urea gels in TBE 1× buffer, transferred to a nylon membrane (GE Healthcare) and UV cross‐linked. Membranes were hybridized with PerfectHyb Plus (Sigma‐Aldrich) and probed with 32P‐5′‐end‐labeled DNA oligonucleotides. Blots were analyzed on the Fuji TLA‐5100 imaging system (GE Healthcare). RNAs collected from ribosome sedimentation fractions were extracted using TRI Reagent (Sigma‐Aldrich) and resolved on agarose gels stained with ethidium bromide.
Electrophoretic mobility shift assays
Binding assays were performed essentially as previously described (Andrade et al, 2013). The 17S rRNA extremities were generated by in vitro transcription with T7 RNAP (Promega) and [α‐32P]‐UTP (Perkin Elmer). EMSA samples were electrophoresed on native 6% or 8% polyacrylamide gels in TBE 1× buffer in a cold room. Gels were exposed to a PhosphorImager screen (GE Healthcare).
RNA mapping
Chemical modification reactions were carried out with DMS (diluted 1:6 in ethanol) or CMCT (1 mg/ml) following protocols described in Andrade et al, 2013 and Caprara, 2011; respectively. Total RNA (10 μg) extracted from exponential phase cultures (OD600 ~ 0.35–0.40) of wild‐type and ∆hfq strains was used. Primer extension reactions were carried out using the 32P‐5′‐end‐labeled primer 46 (Clatterbuck Soper et al, 2013) and 100 U of reverse transcriptase SuperScript III or IV (Thermo Fisher Scientific). Samples were analyzed on 10% polyacrylamide/7 M urea gels run in TBE 1× buffer.
Ribosome extraction and sucrose sedimentation
Ribosome isolation was adapted from (Powers & Noller, 1991). Cell pellets were resuspended in ice‐cold buffer A (50 mM Tris–Cl at pH 7.5, 10 mM MgCl2, 100 mM NH4Cl, 0.5 mM EDTA, and 6 mM 2‐mercaptoethanol) with the addition of Complete Mini Protease Inhibitor cocktail EDTA‐free (Roche) and lysed by French press. After TurboDNase (Ambion) digestion, the clarified lysate was layered over a 36% sucrose cushion composed of buffer B (50 mM Tris–Cl at pH 7.5, 10 mM MgCl2, 500 mM NH4Cl, 0.5 mM EDTA, and 6 mM 2‐mercaptoethanol) and spun at 120,000 g for 16 h in a Beckman ultracentrifuge 90Ti rotor at 4°C. The ribosome pellets were washed once with buffer C (50 mM Tris–Cl at pH 7.5, 10 mM MgCl2, 100 mM NH4Cl, and 6 mM 2‐mercaptoethanol) and then resuspended in the same buffer by gentle rocking at 4°C. Purified ribosomes were analyzed in 15–50% (w/v) sucrose gradients prepared in buffer C with 10 mM MgCl2 (associative conditions) or in 10–30% (w/v) sucrose gradients prepared in buffer C with 0.1 mM MgCl2 (dissociative conditions). Associative samples were centrifuged in a Beckman ultracentrifuge SW41 rotor for 16 h at 71,000 g at 4°C and analyzed by UV using the AKTA system (GE Healthcare). Dissociative samples were centrifuged in a Beckman ultracentrifuge SW28 rotor for 16 h at 76,000 g at 4°C and fractions collected from the top were quantified on Nanodrop.
Ribosome profiling, RNA‐Seq, and data analysis
Ribosome‐protected fragments and randomly fragmented mRNA for ribosome profiling and RNA‐Seq, respectively, were isolated as described previously (Del Campo et al, 2015). Briefly, cells cultured to the exponential phase (OD600 0.35–0.40) in LB medium were split into two aliquots. From one aliquot, total RNA was extracted using TRI Reagent (Sigma‐Aldrich), enriched by depleting small RNAs with GeneJET Purification Kit (Fermentas) and rRNA with MICROBExpress Bacterial mRNA Enrichment Kit (Ambion), and fragmented in alkaline solution (2 mM EDTA and 100 mM Na2CO3 pH 9.2 for 40 min at 95°C) to fragments with size of 24–35 nts. The second aliquot was used to isolate mRNA‐bound ribosome complexes. Cells were collected by filtration and flash‐frozen without preincubation with antibiotics. Cells were lysed by freeze‐rupturing (Retch Mill), and 100 A260 units of ribosome‐bound mRNA fraction were directly used for polysomal analysis or subjected to nucleolytic digestion with 10 units/μl micrococcal nuclease (Fermentas) for 10 min at room temperature in buffer with pH 9.2 (10 mM Tris pH 11 containing 50 mM NH4Cl, 10 mM MgCl2, 0.2% Triton X‐100, 100 μg/ml chloramphenicol, and 20 mM CaCl2) to obtain the monosomal fraction. Separation was obtained by sucrose density gradient (15–50% w/v). Subsequently, 20–35‐nt RNA fragments from the monosomal fraction were size selected on a denaturing 15% polyacrylamide gel. For both ribosome‐protected fragments and mRNA fragments, the libraries were prepared by direct ligation of the adaptors (Del Campo et al, 2015) and sequenced on the Illumina GAIIx platform. Sequenced reads were quality trimmed using fastx‐toolkit (0.0.13.2; quality threshold: 20), and sequencing adapters were cut using cutadapt (1.8.3); minimal overlap: 1 nt) and mapped to the E. coli genome (strain MG1655, version U00096.3, NCBI) using Bowtie (1.1.2) allowing a maximum of two mismatches. The number of raw reads was used to generate gene read counts for each ORF, by counting the number of reads whose middle nucleotide (for even read length the nucleotide 5′ of the mid‐position) fell in the CDS. Gene read counts were normalized by the length of the unique CDS per kilobase (RPKM) and the total mapped reads per million (RPM; Mortazavi et al, 2008). Spike‐ins (ERCC, Thermo, Germany) were added to the RNA‐Seq data set upon rRNA depletion with MICROBExpress kit and used to set the detection threshold in each sequencing set. The same detection threshold was used for the corresponding ribosome profiling experiment. Furthermore, to determine the reproducibility of our sequencing data sets, we used published data set serving as a truly independent biological replicate in which bacteria were grown under identical conditions (GEO accession number, GSE85540; Hwang & Buskirk, 2017). The reproducibility is very high, R 2 = 0.865 and R 2 = 0.816 (Spearman correlation coefficient) for the RNA‐Seq and ribosome profiling data sets, respectively. The correlation is even higher for the r‐proteins only. For fold‐change analysis, we used a threshold of 2. Cumulative profiles of read density for RPFs have been computed as described (Ingolia et al, 2009). The overlapping genes were excluded from this analysis as initiation of the downstream gene is within the open‐reading frame of the upstream gene and the RPFs in this region cannot be unambiguously assigned to either gene. Gene ontology enrichment including statistical analysis was performed using the tools and gene lists from Gene Ontology Consortium (http://geneontology.org/).
Analysis of purified 30S‐associated proteins
For 30S purification, cells were grown in 1 l of LB medium at 37°C and 160 rpm agitation to an OD600 ~ 0.6 for the wild‐type strain and OD600 ~ 0.2 in the case of the ΔrbfA strain, as previously described with minor modifications (Thurlow et al, 2016). Ribosomes were isolated in a similar manner as detailed above. However, “low salt conditions” were used to allow mass spectrometry analysis, meaning that all buffers contained only 60 mM of NH4Cl. Isolated ribosomes were then quantified and separated on 15–45% (w/v) sucrose gradients under dissociative (0.1 mM MgCl2) and associative conditions (10 mM MgCl2) for the wild‐type and ΔrbfA strain, respectively. Gradients were centrifuged in a Beckman ultracentrifuge SW41 rotor at 71,000 g and 4°C for 16 h and analyzed by UV using the AKTA system (GE Healthcare). Fractions corresponding to 30S peak were collected and spun in a Beckman 90Ti rotor at 120,000 g and 4°C for 16 h to remove the sucrose buffer from the 30S particles. The pellet was then resuspended in buffer D (10 mM Tris–Cl at pH 7.5, 10 mM MgCl2, 60 mM NH4Cl, and 3 mM 2‐mercaptoethanol) and stored at −80°C. Mature 30S subunits isolated from the wild‐type strain and immature 30S particles isolated from the ΔrbfA strain were quantified on Nanodrop. Mass spectrometry data were obtained by the UniMS service (Mass Spectrometry Unit, ITQB/iBET, Oeiras, Portugal). Peptides were analyzed using the Pro Group™ Algorithm (Sciex), and for each protein, two types of scores were obtained: unused and total ProtScore. While the latter is a sum of the ion scores of all identified peptide evidence for a protein, the unused ProtScore reflects the amount of total unique peptide evidence related to the same protein. The confidence threshold was set at unused score of 2 and 1.3 with 99 and 95% confidence, respectively. A ratio from the ΔrbfA strain over the wild‐type control was used to identify fold‐change variation of proteins. Positive or negative fold‐change values correspond to an increase or decrease, respectively, of the number of peptides found in the ΔrbfA mutant compared to the wild type. Hfq presence in purified 30S samples (2.5 μg) was further analyzed by Western blot using an anti‐Hfq antibody (Ziolkowska et al, 2006).
Pulse‐labeling assay
Bacteria were grown in M9 medium supplemented with 0.02% casaminoacids (Difco) in an orbital shaker at 37°C. Exponential phase cells were centrifuged, resuspended in M9 medium supplemented with 0.15 mM amino acid mix without methionine and cysteine (Promega), and incubated for 60 min in a water bath at 37°C. Labeling with 35S‐radiolabeled L‐Met/L‐Cys mix (Perkin Elmer) proceeded for 30 s at 37°C. Reaction was stopped with addition of TCA to a final concentration of 5%, and samples were spotted onto GF/C glass microfibers filters (Millipore). Filters were washed four times with TCA 5%, once with ethanol, and then dried under vacuum. 35S signal on filters was quantified by scintillation counting using the Ready Safe Liquid Scintillation cocktail (Beckman Coulter).
β‐Galactosidase assay
Translation fidelity was analyzed by measurement of the β‐galactosidase activity using the pSG plasmid series (O'Connor et al, 1997). Cells were grown to log phase (OD600 ~ 0.35–0.40) in LB medium at 37°C. β‐galactosidase activity from the plasmid encoding WT lacZ was used for normalization in the respective set of MC1061 strains or MC1061 Δhfq mutant strains. Paired t‐test statistical analysis performed using GraphPad Prism 6 software.
Statistical analysis and data deposition
The sequencing data were also submitted to GEO under the accession number GSE100373.
Author contributions
JMA conceived the project; JMA, ZI, and CMA designed experiments; JMA, RFdS, and IC performed experiments; all authors analyzed data; JMA, ZI, and CMA supervised the work; JMA and RFdS prepared the figures; JMA wrote the article, and all authors edited the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 5
Source Data for Figure 6
Acknowledgements
We thank Susan Gottesman (National Cancer Institute, USA) and Eric Brown (McMaster University, Canada) for kindly providing strains. We are grateful to Sarah Woodson (Johns Hopkins University, USA) and Murray Deutscher (University of Miami, USA) for insightful comments. We thank Ricardo Gomes (UniMS, ITQB NOVA/iBET, Portugal) for performing mass spectrometry services. Cristian del Campo is acknowledged for his help with the ribosome profiling protocol. This work was financially supported by Project LISBOA‐01‐0145‐FEDER‐007660 (Microbiologia Molecular, Estrutural e Celular) funded by FEDER through COMPETE2020—Programa Operacional Competitividade e Internacionalização (POCI) and by FCT—Fundação para a Ciência e a Tecnologia (Portugal), including project PTDC/BIA‐MIC/1399/2014 to C.M.A., project PTDC/IMI‐MIC/4463/2014 and FCT Investigator Programme (IF/00961/2014) to J.M.A.; European Union Horizon 2020 Research and Innovation Programme (grant agreement no. 635536) to C.M.A. and by the Deutsche Forschungsgemeinschaft (FOR1805) and European Union (grant NICHE ITN) to Z.I. R.F.dS. is recipient of a FCT Doctoral fellowship (PD/BD/105733/2014) in frame of the ITQB PhD Program in Molecular Biosciences.
The EMBO Journal (2018) 37: e97631
See also: https://doi.org/10.15252/embj.201899616 (June 2018)
Contributor Information
Zoya Ignatova, Email: Zoya.Ignatova@chemie.uni-hamburg.de.
Cecília M Arraiano, Email: cecilia@itqb.unl.pt.
References
- Andrade JM, Pobre V, Matos AM, Arraiano CM (2012) The crucial role of PNPase in the degradation of small RNAs that are not associated with Hfq. RNA 18: 844–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade JM, Pobre V, Arraiano CM (2013) Small RNA modules confer different stabilities and interact differently with multiple targets. PLoS One 8: e52866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K‐12 in‐frame, single‐gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs JD (2005) Lsm proteins and RNA processing. Biochem Soc Trans 33: 433–438 [DOI] [PubMed] [Google Scholar]
- Bilusic I, Popitsch N, Rescheneder P, Schroeder R, Lybecker M (2014) Revisiting the coding potential of the E. coli genome through Hfq co‐immunoprecipitation. RNA Biol 11: 641–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink MF, Verbeet MP, de Boer HA (1993) Formation of the central pseudoknot in 16S rRNA is essential for initiation of translation. EMBO J 12: 3987–3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylund GO, Wipemo LC, Lundberg LA, Wikström PM (1998) RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli . J Bacteriol 180: 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprara M (2011) RNA structure determination using chemical and nuclease digestion methods In RNA: a laboratory manual, Rio D, Hannon G, Ares M, Nilsen T. (eds), pp 269–275. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Christiansen JK, Nielsen JS, Ebersbach T, Valentin‐Hansen P, Søgaard‐Andersen L, Kallipolitis BH (2006) Identification of small Hfq‐binding RNAs in Listeria monocytogenes . RNA 12: 1383–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clatterbuck Soper SF, Dator RP, Limbach PA, Woodson SA (2013) In vivo X‐ray footprinting of pre‐30S ribosomes reveals chaperone‐dependent remodeling of late assembly intermediates. Mol Cell 52: 506–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SE, LaRiviere FJ, Merrikh CN, Moore MJ (2009) A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol Cell 34: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly K, Rife JP, Culver G (2008) Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA. Mol Microbiol 70: 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly K, Culver G (2009) Deconstructing ribosome construction. Trends Biochem Sci 34: 256–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukras AR, Southworth DR, Brunelle JL, Culver GM, Green R (2003) Ribosomal proteins S12 and S13 function as control elements for translocation of the mRNA:tRNA complex. Mol Cell 12: 321–328 [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One‐step inactivation of chromosomal genes in Escherichia coli K‐12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BD (1987) Mechanism of bactericidal action of aminoglycosides. Microbiol Rev 51: 341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JH, Williamson JR (2017) Structure and dynamics of bacterial ribosome biogenesis. Philos Trans R Soc Lond B Biol Sci 372: 20160181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lay N, Schu DJ, Gottesman S (2013) Bacterial small RNA‐based negative regulation: Hfq and its accomplices. J Biol Chem 288: 7996–8003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Campo C, Bartholomäus A, Fedyunin I, Ignatova Z (2015) Secondary structure across the bacterial transcriptome reveals versatile roles in mRNA regulation and function. PLoS Genet 11: e1005613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP (2009) Maturation and degradation of ribosomal RNA in bacteria. Prog Mol Biol Transl Sci 85: 369–391 [DOI] [PubMed] [Google Scholar]
- Ellis MJ, Trussler RS, Haniford DB (2015) Hfq binds directly to the ribosome‐binding site of IS 10 transposase mRNA to inhibit translation. Mol Microbiol 96: 633–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folichon M, Allemand F, Régnier P, Hajnsdorf E (2005) Stimulation of poly(A) synthesis by Escherichia coli poly(A)polymerase I is correlated with Hfq binding to poly(A) tails. FEBS J 272: 454–463 [DOI] [PubMed] [Google Scholar]
- Foster C, Champney WS (2008) Characterization of a 30S ribosomal subunit assembly intermediate found in Escherichia coli cells growing with neomycin or paromomycin. Arch Microbiol 189: 441–449 [DOI] [PubMed] [Google Scholar]
- Fujii K, Kitabatake M, Sakata T, Ohno M (2012) 40S subunit dissociation and proteasome‐dependent RNA degradation in nonfunctional 25S rRNA decay. EMBO J 31: 2579–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnsdorf E, Boni IV (2012) Multiple activities of RNA‐binding proteins S1 and Hfq. Biochimie 94: 1544–1553 [DOI] [PubMed] [Google Scholar]
- de Haseth PL, Uhlenbeck OC (1980) Interaction of Escherichia coli host factor protein with Q beta ribonucleic acid. Biochemistry 19: 6146–6151 [DOI] [PubMed] [Google Scholar]
- Hwang J‐Y, Buskirk AR (2017) A ribosome profiling study of mRNA cleavage by the endonuclease RelE. Nucleic Acids Res 45: 327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS (2009) Genome‐wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinks‐Robertson S, Nomura M (1981) Regulation of ribosomal protein synthesis in an Escherichia coli mutant missing ribosomal protein L1. J Bacteriol 145: 1445–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PG, Inouye M (1996) RbfA, a 30S ribosomal binding factor, is a cold‐shock protein whose absence triggers the cold‐shock response. Mol Microbiol 21: 1207–1218 [DOI] [PubMed] [Google Scholar]
- Kaczanowska M, Rydén‐Aulin M (2007) Ribosome biogenesis and the translation process in Escherichia coli . Microbiol Mol Biol Rev 71: 477–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbstein K (2013) Quality control mechanisms during ribosome maturation. Trends Cell Biol 23: 242–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel J, Allmang C, Petfalski E, Beggs J, Tollervey D (2003) Lsm Proteins are required for normal processing and stability of ribosomal RNAs. J Biol Chem 278: 2147–2156 [DOI] [PubMed] [Google Scholar]
- Laursen BS, Sørensen HP, Mortensen KK, Sperling‐Petersen HU (2005) Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev 69: 101–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Derout J, Folichon M, Briani F, Dehò G, Régnier P, Hajnsdorf E (2003) Hfq affects the length and the frequency of short oligo(A) tails at the 3′ end of Escherichia coli rpsO mRNAs. Nucleic Acids Res 31: 4017–4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong V, Kent M, Jomaa A, Ortega J (2013) Escherichia coli rimM and yjeQ null strains accumulate immature 30S subunits of similar structure and protein complement. RNA 19: 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G‐W, Burkhardt D, Gross C, Weissman JS (2014) Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157: 624–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liiv A, Remme J (2004) Importance of transient structures during post‐transcriptional refolding of the pre‐23S rRNA and ribosomal large subunit assembly. J Mol Biol 342: 725–741 [DOI] [PubMed] [Google Scholar]
- Link TM, Valentin‐Hansen P, Brennan RG (2009) Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci USA 106: 19292–19297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodmell JS, Dahlberg AE (1997) A conformational switch in Escherichia coli 16S ribosomal RNA during decoding of messenger RNA. Science 277: 1262–1267 [DOI] [PubMed] [Google Scholar]
- Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL (2004) Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat Struct Mol Biol 11: 1206–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty BK, Maples VF, Kushner SR (2004) The Sm‐like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli . Mol Microbiol 54: 905–920 [DOI] [PubMed] [Google Scholar]
- Moll I, Leitsch D, Steinhauser T, Bläsi U (2003) RNA chaperone activity of the Sm‐like Hfq protein. EMBO Rep 4: 284–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA‐Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Muffler A, Traulsen DD, Fischer D, Lange R, Hengge‐Aronis R (1997) The RNA‐binding protein HF‐I plays a global regulatory role which is largely, but not exclusively, due to its role in expression of the sigmaS subunit of RNA polymerase in Escherichia coli . J Bacteriol 179: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narla A, Ebert BL (2010) Ribosomopathies: human disorders of ribosome dysfunction. Blood 115: 3196–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M (1999) Regulation of ribosome biosynthesis in Escherichia coli and Saccharomyces cerevisiae: diversity and common principles. J Bacteriol 181: 6857–6864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor M, Thomas CL, Zimmermann RA, Dahlberg AE (1997) Decoding fidelity at the ribosomal A and P sites: influence of mutations in three different regions of the decoding domain in 16S rRNA. Nucleic Acids Res 25: 1185–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaka H, Ishikawa H, Morita T, Aiba H (2011) PolyU tail of rho‐independent terminator of bacterial small RNAs is essential for Hfq action. Proc Natl Acad Sci USA 108: 13059–13064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panja S, Schu DJ, Woodson SA (2013) Conserved arginines on the rim of Hfq catalyze base pair formation and exchange. Nucleic Acids Res 41: 7536–7546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Soper TJ, Woodson SA (2014) Positional effects of AAN motifs in rpoS regulation by sRNAs and Hfq. J Mol Biol 426: 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T, Noller HF (1991) A functional pseudoknot in 16S ribosomal RNA. EMBO J 10: 2203–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Régnier P, Hajnsdorf E (2013) The interplay of Hfq, poly(A) polymerase I and exoribonucleases at the 3′ ends of RNAs resulting from Rho‐independent termination: a tentative model. RNA Biol 10: 602–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochat T, Delumeau O, Figueroa‐Bossi N, Noirot P, Bossi L, Dervyn E, Bouloc P (2015) Tracking the elusive function of Bacillus subtilis Hfq. PLoS One 10: e0124977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy‐Chaudhuri B, Kirthi N, Culver GM (2010) Appropriate maturation and folding of 16S rRNA during 30S subunit biogenesis are critical for translational fidelity. Proc Natl Acad Sci USA 107: 4567–4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvail H, Caron M‐P, Bélanger J, Massé E (2013) Antagonistic functions between the RNA chaperone Hfq and an sRNA regulate sensitivity to the antibiotic colicin. EMBO J 32: 2764–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer E, Weichenrieder O (2011) Structural basis for RNA 3′‐end recognition by Hfq. Proc Natl Acad Sci USA 108: 13065–13070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer E, Schmidt S, Weichenrieder O (2012) Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc Natl Acad Sci USA 109: 9396–9401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M, Klumpp S, Mateescu EM, Hwa T (2014) Emergence of robust growth laws from optimal regulation of ribosome synthesis. Mol Syst Biol 10: 747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajani Z, Sykes MT, Williamson JR (2011) Assembly of bacterial ribosomes. Annu Rev Biochem 80: 501–526 [DOI] [PubMed] [Google Scholar]
- Shetty S, Varshney U (2016) An evolutionarily conserved element in initiator tRNAs prompts ultimate steps in ribosome maturation. Proc Natl Acad Sci USA 113: E6126–E6134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JCD, Vogel J (2008) Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post‐transcriptional regulator, Hfq. PLoS Genet 4: e1000163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AK, Schlessinger D (1988) Coregulation of processing and translation: mature 5′ termini of Escherichia coli 23S ribosomal RNA form in polysomes. Proc Natl Acad Sci USA 85: 7144–7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader MB, Hervey Iv WJ, Costantino N, Fujgaki S, Chen CY, Akal‐Strader A, Ihunnah CA, Makusky AJ, Court D, Markey SP, Kowalak JA (2013) A coordinated proteomic approach for identifying proteins that interact with the E. coli ribosomal protein S12. J Proteome Res 12: 1289–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurlow B, Davis JH, Leong VF, Moraes T, Williamson JR, Ortega J (2016) Binding properties of YjeQ (RsgA), RbfA, RimM and Era to assembly intermediates of the 30S subunit. Nucleic Acids Res 44: 9918–9932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tree JJ, Granneman S, McAteer SP, Tollervey D, Gally DL (2014) Identification of bacteriophage‐encoded anti‐sRNAs in pathogenic Escherichia coli . Mol Cell 55: 199–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui HC, Leung HC, Winkler ME (1994) Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K‐12. Mol Microbiol 13: 35–49 [DOI] [PubMed] [Google Scholar]
- Updegrove TB, Zhang A, Storz G (2016) Hfq: the flexible RNA matchmaker. Curr Opin Microbiol 30: 133–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Luisi BF (2011) Hfq and its constellation of RNA. Nat Rev Microbiol 9: 578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz CJ, Wilusz J (2013) Lsm proteins and Hfq: life at the 3′ end. RNA Biol 10: 592–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewska Z, Olejniczak M (2016) Hfq assists small RNAs in binding to the coding sequence of ompD mRNA and in rearranging its structure. RNA 22: 979–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Guo Q, Goto S, Chen Y, Li N, Yan K, Zhang Y, Muto A, Deng H, Himeno H, Lei J, Gao N (2014) Structural insights into the assembly of the 30S ribosomal subunit in vivo: functional role of S5 and location of the 17S rRNA precursor sequence. Protein Cell 5: 394–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF (2001) Crystal structure of the ribosome at 5.5 A resolution. Science 292: 883–896 [DOI] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S (2003) Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol 50: 1111–1124 [DOI] [PubMed] [Google Scholar]
- Zhang A, Schu DJ, Tjaden BC, Storz G, Gottesman S (2013) Mutations in interaction surfaces differentially impact E. coli Hfq association with small RNAs and their mRNA targets. J Mol Biol 425: 3678–3697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziolkowska K, Derreumaux P, Folichon M, Pellegrini O, Régnier P, Boni IV, Hajnsdorf E (2006) Hfq variant with altered RNA binding functions. Nucleic Acids Res 34: 709–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 5
Source Data for Figure 6


