Abstract
Skeletal muscle hypertrophy is a widely sought exercise adaptation to counteract the muscle atrophy of aging and disease, or to improve athletic performance. While this desired muscle enlargement is a well-known adaptation to resistance exercise training (RT), the mechanistic underpinnings are not fully understood. The purpose of this review is thus to provide the reader with a summary of recent advances in molecular mechanisms—based on the most current literature—that are thought to promote RT-induced muscle hypertrophy. We have therefore focused this discussion on the following areas of fertile investigation: ribosomal function and biogenesis, muscle stem (satellite) cell activity, transcriptional regulation, mechanotransduction, and myokine signaling.
Whether to counteract disease-related muscle atrophy or improve athletic performance, skeletal muscle hypertrophy is among the primary goals and a well-known adaptation to resistance exercise training (RT). The purpose of this review is to provide the reader with a summary of molecular mechanisms—based on the most current literature—that are thought to drive resistance training–induced muscle hypertrophy. Because muscle cells—known as myofibers—are terminally differentiated and thus cannot reenter the cell cycle, RT-induced muscle enlargement is dependent on the cellular hypertrophy of existing myofibers. Herein we therefore focus on mechanisms considered central to the hypertrophy of individual myofibers. We published an extensive review of this topic in 2012 (Adams and Bamman 2012); thus, our emphasis in this article is on scientific advances over the past 4 years.
The skeletal myofiber is a complex but well-characterized, highly-organized structure packed with myofibrils, each consisting of sarcomeres (i.e., contractile units) in a series that define a registered ultrastructure of contractile, anchoring, and tethering proteins. The myofiber plasma membrane (i.e., sarcolemma) is home to a dynamic network of transmembrane receptors and transport proteins that respond to stimuli and communicate signals to intracellular organelles and to the extensive syncytium of peripherally located nuclei dispersed along the full length of the myofiber. Mechanically loaded contractions during resistance exercise result in a unique cascade of signaling events that promote the net accumulation of intracellular proteins and thus myofiber enlargement along with remodeling of the extracellular matrix. While the entirety of the molecular responses driving myofiber hypertrophy are not fully understood, there have been important advances in recent years—particularly in the areas of ribosomal function and biogenesis, muscle stem (satellite) cell activity, transcriptional regulation, mechanotransduction, and myokine signaling—that are the focuses of this work. It should be noted that this discussion is centered on available findings in humans adapting to RT (which involves preferential hypertrophy of type II myofibers), supplemented by complementary results in animal models as warranted.
We wish to emphasize that the intent of this relatively brief article is to highlight recent advances; thus, the reader should not consider this an exhaustive review. For example, a rich literature has accumulated that establishes a putative central role for cellular hypertrophy–promoting processes regulated by the master kinase, mammalian target of rapamycin (mTOR) or more specifically its complex 1 (mTORC1). It is beyond the scope of the present work to review the evidence base of mTOR in sufficient detail. As such, the reader is referred to thorough reviews (Hornberger 2011; Adams and Bamman 2012; Jacobs et al. 2014). Discussion of mTOR and related cell signaling—including phosphorylation/activation of its major downstream readout p70S6K—herein is limited to recent advances within the focus areas of this article.
LEVERAGING HUMAN INTERINDIVIDUAL RESPONSE HETEROGENEITY
Interrogating potential regulatory processes of RT-induced hypertrophy directly in humans is of great value but certainly not without limitations. For obvious reasons, definitive cause–effect determinations cannot be made in the absence of the ability to genetically or pharmacologically manipulate a target gene or signaling pathway. On the other hand, the human relevance of even correlational/associative findings is equally obvious and can be leveraged for mechanistic “reverse translation” studies in lower animals. One extremely valuable strategy representing a significant advance over the past few years has been the utilization of human responder phenotyping and subsequent molecular interrogation. In other words, although we cannot manipulate the human phenotype, we can leverage similarities and differences in exercise responsiveness among human subjects (i.e., interindividual response heterogeneity) to then identify common and differential patterns ranging from molecular to behavioral phenotyping.
To better understand potential processes modulating RT-induced myofiber hypertrophy, a decade ago we began using cluster analysis approaches to classify human subjects as nonresponders, moderate responders, and extreme responders based on each subject’s degree of myofiber hypertrophy following standardized and well-controlled RT interventions (Bamman et al. 2007). This strategy has yielded valuable insight into potential cellular and molecular regulators of human RT-induced myofiber hypertrophy (Bamman et al. 2007; Kim et al. 2007; Petrella et al. 2008; Mayhew et al. 2011; Thalacker-Mercer et al. 2013). We have since validated the utility of the model (Stec et al. 2016) and today similar strategies used by a number of research teams continue to bear fruit (Davidsen et al. 2011; Prestes et al. 2015; Ahtiainen et al. 2016; Ogasawara et al. 2016). Of course all participants in RT trials benefit to varying degrees in other ways (Bamman et al. 2007; Bickel et al. 2011; Churchward-Venne et al. 2015) (e.g., strength gains, functional mobility improvements, etc.) but clustering specifically on myofiber hypertrophy reveals clear differential adaptations (including magnitude of functional gains) (Bamman et al. 2007) that enable investigations of potential mechanistic underpinnings (molecular and otherwise) to help advance the field. It should be noted that, to date, no follow-up studies have been completed to determine whether high and low responders—based on a single defined outcome (e.g., myofiber hypertrophy)—consistently maintain their responder status. There are numerous approaches to manipulating the stimulus (i.e., RT prescription) that may influence an individual’s response, which opens the door to the attractive application of a precision medicine strategy.
RIBOSOMAL FUNCTION AND BIOGENESIS
Central to myofiber hypertrophy is the accretion of cellular component proteins in various pools and compartments (e.g., myofibrillar/contractile, mitochondrial, sarcoplasmic reticulum, cytoskeletal, etc.). This accretion requires an overall net positive protein balance, with anabolism exceeding catabolism over an extended time period. The short-term induction of mixed muscle and myofibrillar protein synthesis after a bout of resistance exercise has been documented numerous times, with elevations seen within an hour postexercise and persisting for as many as 48 h (Phillips et al. 1997; Wolfe 2006; Mayhew et al. 2009; Burd et al. 2011; Schoenfeld et al. 2013). This has led to a flurry of research focused on translational control, with particular emphasis on the molecular regulation of translation initiation. However, recent results question the degree to which protein synthesis and/or translation initiation signaling responses to an initial exercise exposure are prognostic of eventual RT-induced myofiber hypertrophy as an adaptation to several weeks of training. For example, Mitchell et al. reported no correlation between the myofibrillar protein synthesis (MyoPS) response 1–6 h after an initial resistance exercise bout and the magnitude of long-term, RT-induced muscle hypertrophy (Mitchell et al. 2014). In an interesting subsequent study, Damas et al. (2016) confirmed no relationship between the 24–48 h MyoPS response and eventual myofiber hypertrophy when studying MyoPS during the first week of training; however, strong correlations were noted when MyoPS was quantified in response to exercise during weeks 3 and 10 of a 10-week RT program. The investigators’ assessments of muscle damage suggest that the week-1 MyoPS response was driven largely by damage/repair, whereas the later responses after a few weeks of training were more reflective of anabolism supporting myofiber hypertrophy (Damas et al. 2016). Thus, much like the magnitude of myofiber hypertrophy (Bamman et al. 2007), the acute muscle protein synthesis response to RT is variable between individuals and the relationship between the two appears to strengthen as training progresses.
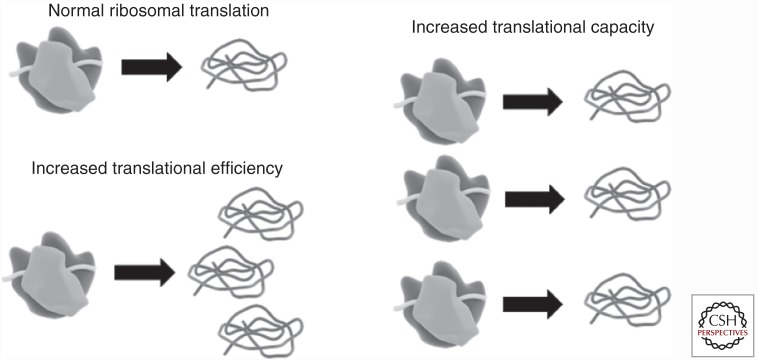
Hypertrophy driven by net protein synthesis requires increased ribosomal function. To that end, enhanced protein translation rates can be achieved by increased ribosomal efficiency (i.e., greater translation of mRNA per ribosome) and/or elevated ribosomal capacity via ribosome biogenesis (Fig. 1). Both processes are regulated at least in part by mTORC1 activity. Results of a recent study of compensatory overload hypertrophy in a murine model suggest enhanced mTORC1 activity (by knocking out a major repressor called regulated in development and DNA damage 1 [REDD1]) has a greater impact on translational efficiency than capacity during the first few days following loading (Gordon et al. 2016); however, mTORC1 also plays a central role in ribosome biogenesis by regulating the synthesis of both ribosomal proteins and ribosomal RNAs (rRNAs) (reviewed in Mayer and Grummt 2006). In humans, acute responses to resistance exercise and long-term RT have focused largely on ribosomal efficiency until recently. Several lines of recent evidence suggest ribosomal biogenesis may be a key rate-limiting factor in the regulation of RT-induced myofiber hypertrophy (discussed below).
Figure 1.
Ribosomal function. Skeletal myofiber hypertrophy as an adaptation to resistance exercise training is accomplished by cellular protein accretion over time, which requires increased ribosomal function. Enhanced protein translation rates can be achieved by increased ribosomal efficiency (i.e., more translation per ribosome) and/or elevated ribosomal capacity via ribosome biogenesis (i.e., de novo synthesis of ribosomes). Current evidence indicates that both of these routes to enhanced translation are involved in resistance training hypertrophy, and support for the latter has been building rapidly in recent years.
Ribosome biogenesis is a complicated and tightly regulated process. As summarized elsewhere (Wen et al. 2016), synthesis of a single functional ribosome requires transcription of four ribosomal RNAs (rRNAs 28S, 18S, 5S, and 5.8S) and translation of approximately 80 ribosomal proteins. The process is highly regulated, energetically expensive, and supported by hundreds of proteins involved in the production, assembly, and transport of the ribosome. Transcription of ribosomal DNA (rDNA) to the rRNAs by polymerase I is considered a major rate-limiting step in ribosome biogenesis (Mayer and Grummt 2006).
Using total muscle RNA as a surrogate of rRNA (80%–85% of total RNA), the magnitude of change in total muscle RNA over the course of RT appears linked to the magnitude of myofiber (Stec et al. 2016) and muscle (Figueiredo et al. 2015) hypertrophy. Via K-means cluster analysis to phenotype human subjects based on the degree of type II myofiber hypertrophy, Stec et al. found that only extreme responders successfully up-regulated muscle rRNA expression (indicative of ribosomal biogenesis), and the biopsied muscles of both moderate and extreme responders increased c-Myc protein content (a major upstream regulator of ribosome biogenesis). Additionally, follow-up studies in vitro showed that growth factor–induced human myotube hypertrophy was prevented when de novo ribosome biogenesis was blocked by a specific inhibitor of rRNA transcription (Stec et al. 2016), a finding recently advanced and linked to mTOR regulation during hypertrophy in C2C12 myotubes (von Walden et al. 2016).
Transcriptome profiling of pretraining skeletal muscle among three RT hypertrophy responder clusters revealed higher pretraining expression of several ribosomal protein mRNAs and higher expression of major upstream regulators of ribosome biogenesis (c-Myc and n-Myc) in moderate and extreme responders compared to nonresponders (Thalacker-Mercer et al. 2013). It is also noteworthy that only the extreme responders showed sustained phosphorylation 24 h after their first full exercise bout of the autoinhibitory domain of p70 S6 kinase, a key signaling intermediate known to modulate both translation initiation and ribosome biogenesis (Mayhew et al. 2011). The human findings are corroborated by dose-dependent induction of muscle hypertrophy and ribosome biogenesis across four doses of mechanical overload via graded synergist ablation (i.e., graded overload of plantaris by ablation of various amounts of gastrocnemius and/or soleus) in rats (Nakada et al. 2016).
Additional support for ribosome biogenesis as a major regulator of myofiber hypertrophy is provided by studies of aging. The well-known blunted capacity for RT-induced myofiber hypertrophy with advancing age is associated with blunted induction of key regulatory processes: (1) In response to a single bout of resistance exercise, expression of 45S pre-rRNA was up-regulated in adults 39 years of age but not in 76-year-olds (Stec et al. 2015); (2) during 6 weeks of RT, surrogate indices of ribosome biogenesis (RNA:DNA-ratio and c-Myc induction) were impaired in conjunction with blunted muscle hypertrophy in old versus young adults (Brook et al. 2016); and (3) using the synergist ablation model, the blunted hypertrophy noted in aged mice was linked to blunted ribosome biogenesis as indicated by impaired induction of both pre-rRNA and rRNA (Kirby et al. 2015). Further, it was recently noted that muscle expression levels of 45S pre-rRNA and c-myc increase substantially after an acute resistance exercise bout in untrained subjects (Nader et al. 2014), although these responses are blunted in the trained state. Overall, the recent findings in humans discussed throughout this section, supported by both animal models and in vitro studies, strongly implicate ribosomal biogenesis as a central regulator of RT-induced myofiber hypertrophy.
MUSCLE STEM (SATELLITE) CELL ACTIVITY
Located within the basal lamina and just exterior to the myofiber sarcolemma are resident multipotent muscle stem cells known as satellite cells. While not fully differentiated, satellite cells are largely committed to the myogenic lineage, and their essential role during muscle regeneration following damage is well established (reviewed in Dumont et al. 2015). As a brief summary, on significant injury/damage, quiescent satellite cells enter the cell cycle and some of the cell progeny differentiate into fusion-competent myoblasts and form new myofibers or fuse into existing damaged fibers to promote repair. Regarding this role there is no debate.
On the other hand, one of the most controversial topics in the field of muscle biology centers on a potential role of satellite cells in the process of RT-induced myofiber hypertrophy. As we reviewed in detail previously (Adams and Bamman 2012), findings supporting a key role of satellite cells are based on the tenet that each nucleus in the multinucleated myofiber is limited in its capacity to transcriptionally regulate a finite volume of cytoplasm termed the myonuclear domain. Thus, significant myofiber hypertrophy beyond the domain ceiling of each myonucleus requires the addition of nuclei, which are thought to result from the differentiation of satellite cells into fusion-competent myoblasts that fuse into the growing myofiber as nuclear donors. Indeed, a strong linear correlation between the human myofiber cross-sectional area and the number of myonuclei per fiber was recently noted (Karlsen et al. 2015). The concept of a ceiling on the nuclear DNA/cytoplasm volume relationship is not unique to skeletal muscle and is actually a primary driver of cell division in mitotically active mononucleated cells. In fact, some current strategies in cancer-targeting therapeutics are focused on disrupting the protein synthetic machinery in cancer cells to limit cellular hypertrophy, thereby mitigating proliferation (Devlin et al. 2016; Rebello et al. 2016).
There are numerous lines of evidence indicating resistance exercise, and long-term RT increase the prevalence of muscle satellite cells in humans (Dreyer et al. 2006; Mackey and Robinovitch 2006; Petrella et al. 2008; Verdijk et al. 2009; Snijders et al. 2014, 2016; Nederveen et al. 2015). Correlative evidence supporting satellite cell–mediated myonuclear addition during human RT-induced myofiber hypertrophy gained traction in the mid-2000s (e.g., Olsen et al. 2006; Petrella et al. 2006, 2008) and studies reporting modest hypertrophy with no appreciable myonuclear addition (Petrella et al. 2006, 2008; Verdijk et al. 2009) fueled the concept of reaching a domain ceiling that led to a “demand” for nuclear addition. This concept was reaffirmed recently as moderate myofiber hypertrophy was accomplished in older adults in the absence of myonuclear addition, whereas nuclear addition accompanied greater hypertrophy (Stec et al. 2016). New research leveraging the Pax7-DTA satellite cell depletion mouse and myonuclear transcription rates lends additional support for myonuclear reserve capacity (i.e., expansion of the domains before demanding additional nuclei) (Kirby et al. 2016). In the absence of satellite cell–mediated myonuclear accretion, the number of transcriptionally active myonuclei and the amount of RNA generated by each myonucleus increased during hypertrophy (Kirby et al. 2016). When the average myonuclear domain is already near the ceiling at the outset of a hypertrophic stimulus, there appears to be a need for myonuclear addition early during the intervention. For example, in studying young adults with myofibers near the theoretical domain ceiling in the pretraining state, Snijders et al. (2016) reported myonuclear addition in conjunction with RT-induced myofiber hypertrophy but no significant expansion of the nuclear domain, suggesting that the myofibers needed nuclear addition to enlarge. Very recently, myonuclear addition in conjunction with extreme RT-induced human myofiber hypertrophy was hypothesized to be a means of meeting the demand for more rDNA templates toward facilitating ribosome biogenesis (Stec et al. 2016).
Interestingly, results in humans not experiencing appreciable myofiber hypertrophy suggest myonuclear addition also occurs in response to stimuli and/or demand for additional nuclear control (e.g., high metabolic demand) other than reaching the theoretical domain ceiling to support hypertrophy. Interesting recent work by Frese et al. showed marked myonuclear addition during the competition training phase among young highly competitive endurance cyclists with no change in myofiber size, resulting in a more than 50% contraction of the myonuclear domain (Frese et al. 2015). The finding is complemented by a thorough investigation of fusion-promoting processes, implicating the fusogenic genes Syncytin-1 and Syncytin-3 and corresponding protein signaling cascades in the accumulation of myonuclei during the metabolically demanding exercise training period. Further, in a nonexercise study of monozygotic twins, older women on hormone replacement therapy (HRT) showed smaller myonuclear domains in type I myofibers compared to twins not on HRT, while fiber size was not different (Qaisar et al. 2013).
The findings in humans notwithstanding, innovative work in 2011 from McCarthy et al. (2011) called into question the need for satellite cell–mediated myonuclear addition during the early phase of myofiber hypertrophy. After developing a novel tamoxifen-inducible mouse strain (Pax7-DTA) enabling conditional ablation of >90% of muscle satellite cells in adult mice, the team found no impact of satellite cell depletion on mechanical overload-induced muscle hypertrophy. A similar magnitude of muscle hypertrophy (via synergist ablation) was noted in tamoxifen-treated (i.e., satellite cell depleted) versus vehicle-treated control mice. There was, however, a profound impact of satellite cell depletion on muscle regeneration following myotoxin injury. The hypertrophy results of McCarthy et al. in the Pax7-DTA mouse were recently challenged (Egner et al. 2016). In this follow-up comparison, Egner et al. reported a stronger dependence on satellite cells to achieve myofiber hypertrophy consequent to 2 weeks of compensatory overload; however, a number of methodological differences between the two studies make direct comparisons difficult. Extending the duration of overload from 2 weeks (McCarthy et al. 2011) to 8 weeks (Fry et al. 2014), the team who developed the Pax7-DTA mouse noted successful but blunted myofiber hypertrophy, coupled with lesser myonuclear addition, reduced muscle specific force, and increased fibrosis (i.e., extracellular matrix deposition). It is noteworthy that functional muscle quality in mice induced to hypertrophy appears partially dependent on successful myonuclear addition even in models not driven by mechanical overload. For example, Qaisar et al. (2013) recently reported similar magnitudes of myofiber hypertrophy in the extensor digitorum longus muscle of myostatin knockout (−/−) mice and muscle-specific insulin-like growth factor I (IGF-I) overexpressing (+/+) mice; yet myonuclear domain and specific force were maintained only in IGF-I+/+, suggesting functional muscle quality suffered in myostatin−/− at least in part caused by failed myonuclear addition.
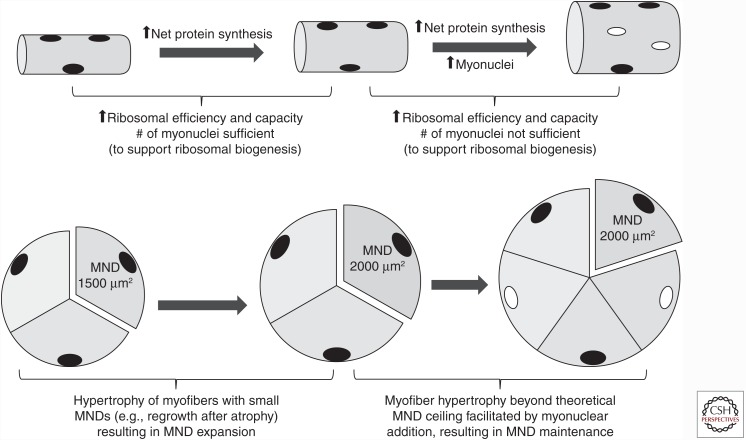
Overall, we interpret the findings in both humans and rodents as generally in agreement. In fact, the latest research findings seem to largely reaffirm the model summarized in our 2012 review (Fig. 2) (Adams and Bamman 2012). (1) The initial phase of mechanical overload-induced myofiber hypertrophy is not necessarily satellite cell–dependent—particularly if the myonuclear domains have room to expand before reaching the postulated ceiling—thus, in this early phase, myofiber enlargement can occur primarily by protein accretion alone; and (2) with more extended exposure to the mechanical overload stimulus—as myofiber enlargement progresses and myonuclear domain size expands—satellite cell–mediated myonuclear addition may be essential for continued hypertrophy.
Figure 2.
Distinguishing satellite cell–independent and –dependent myofiber hypertrophy. In smaller myofibers with small myonuclear domains (MNDs), resistance exercise training (RT)-induced cellular hypertrophy is not necessarily satellite cell–dependent and can be accomplished by protein accretion alone (via increased ribosomal efficiency and capacity), as each myonucleus has “room to expand” both its transcriptional activity and cytoplasmic domain. In contrast, in larger myofibers with MNDs at or approaching a ceiling size, additional nuclei are needed to support enhanced ribosomal function and biogenesis en route to protein accretion; thus, for successful RT-induced hypertrophy of fibers with larger MNDs, satellite cell–mediated myonuclear addition may be essential and rate-limiting. (From Adams and Bamman 2012; adapted, with permission.)
TRANSCRIPTIONAL REGULATION
Based on the evidence available, we approached this article from the position that the effectiveness of RT-induced myofiber hypertrophy largely rests with processes regulating ribosomal capacity, which at some stage appears facilitated by satellite cell–mediated myonuclear addition. The full gamut of molecular cues regulating these processes in response to RT is, however, not entirely clear. Novel findings over the past few years suggest that control of the muscle transcriptome may play a central role. Indeed, an elegant recent study by the McCarthy laboratory showed a robust up-regulation of myonuclear transcriptional activity during the early stages of myofiber hypertrophy (Kirby et al. 2016), which led to altered expression of genes known to modulate myofiber size.
Transcriptome Profiling
In 2012, the Trappe group published a thorough investigation of muscle transcriptome profiling focused on the impacts of RT and acute resistance exercise in young and older adults (Raue et al. 2012). A remarkable 661 genes were identified for which (1) transcription was altered by acute resistance exercise in both the pre-RT and post-RT states; and (2) the changes in expression correlated with individual magnitudes of RT-induced muscle hypertrophy. Follow-up fiber-type-specific microarrays indicated that the transcriptome profile of type IIa myofibers was impacted by resistance exercise far greater than that of type I myofibers (463 vs. 26 differentially expressed genes in type IIa vs. type I myofibers in young). Further, based on transcriptome alterations, the muscles of young were much more responsive than those of old. Because type II myofibers hypertrophy substantially more than type I myofibers during RT, and young tend to hypertrophy more than old, these findings strongly suggest that transcriptional regulation plays a key role in determining RT hypertrophy potential. Using cluster analysis of myofiber hypertrophy, we found remarkably different basal transcriptome profiles in non- versus extreme RT responders (Thalacker-Mercer et al. 2013). In addition, follow-up functional annotation revealed networks favoring growth, ribosomal activity, and stem cell activity in extreme responders versus proinflammatory processes in nonresponders, which suggests that the pretraining muscle transcriptome profile is highly influential in the RT myofiber hypertrophy adaptation (Thalacker-Mercer et al. 2013).
MicroRNAs
Small noncoding RNA molecules (i.e., microRNAs [miRNAs]), modulate gene expression via RNA silencing/posttranscriptional regulation. The first evidence that miRNAs may modulate skeletal muscle hypertrophy appeared in 2007, when McCarthy and Esser (2007) noted a substantial drop in miR-1 expression concomitant with increased IGF-I expression during hypertrophy in mice, suggesting miR-1 may repress IGF-I, with derepression fueled by the loading-mediated drop in miR-1. Differential muscle expression of four other miRNAs (miR-378, miR-29a, miR-26a, miR-451) was first noted in human high versus low responders based on RT-induced muscle hypertrophy in 2011 (Davidsen et al. 2011). Subsequent gene ontology analysis suggested that the miRNA changes seen in low responders may limit the ability to express genes that promote hypertrophy and muscle remodeling and, in support of this, targeted polymerase chain reaction (PCR) showed that only high responders increased IGF-I mRNA expression (Davidsen et al. 2011). In a recent small study of high versus low RT responders, a screen of 800 miRNAs revealed that 26 were differentially expressed by muscle hypertrophy responder classification (Ogasawara et al. 2016). These studies have revealed novel targets for future studies deemed necessary to fully appreciate the role of miRNA biology in muscle hypertrophy.
Transcriptional Coregulators
Because of the growing evidence suggesting that muscle transcriptome influences RT-induced myofiber hypertrophy and is itself substantially altered by single exposures to resistance exercise, recent attention has been given to transcriptional coregulators (i.e., proteins that interact with transcription factors to either activate or repress the transcription of particular genes). For example, a splice variant of the transcriptional coactivator, peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) known as PGC-1α4, was found to influence RT-induced muscle hypertrophy, potentially by up-regulating muscle IGF-I expression (Ruas et al. 2012). Resistance exercise–induced up-regulation of PGC-1α4 was indeed affirmed recently (Samuelsson et al. 2016); interestingly, the effect appeared uncoupled from nutritionally enhanced mTORC1 signaling. Recent evidence suggests the muscle hypertrophy-promoting effects of PGC-1α4 may be through the actions of one of its transcriptional targets, G-protein-coupled receptor 56 (GPR56), which is induced by mechanical overload and up-regulates IGF-I (White et al. 2014). With only one identified thus far, this series of studies should fuel further interrogation into the potential role of transcriptional coregulators in the muscle hypertrophy adaptation.
Overall, these recent advances support a role for the muscle transcriptome and its molecular regulation in the RT hypertrophy adaptation. Much more work is clearly needed to elucidate the mechanisms of transcriptional control, which in our view presents an opportunity for fertile research.
MECHANOTRANSDUCTION
There is a general consensus that the efficacy of RT-induced myofiber hypertrophy is load-dependent. In other words, the common view is that a minimum threshold of mechanical loading must be met to result in the adaptive process of hypertrophy. We previously covered the concept of RT dosing in detail (Adams and Bamman 2012), and the idea of load-dependent dosing to maximize muscle growth was reaffirmed in a recent animal model (Eftestol et al. 2016). Using an in vivo electrical activity model, Eftestol et al. found approximately twofold greater hypertrophy and substantially more myonuclear addition in response to maximal isometric contractions (vs. the same number of submaximal contractions). Remaining elusive for decades, however, are the precise mechanisms by which increased mechanical loading transduces biochemical responses that ultimately lead to the biological adaptation of myofiber hypertrophy: the quintessential “black box.” Since our previous review of mechanotransduction and potential mechanosensing processes (Adams and Bamman 2012), a bit of progress has led to the recognition of a few molecules and communication networks in the extracellular matrix (ECM), sarcolemma, sarcoplasm, and sarcomere that appear to be involved in transduction of mechanosensing to hypertrophy signaling (Fig. 3).
Figure 3.
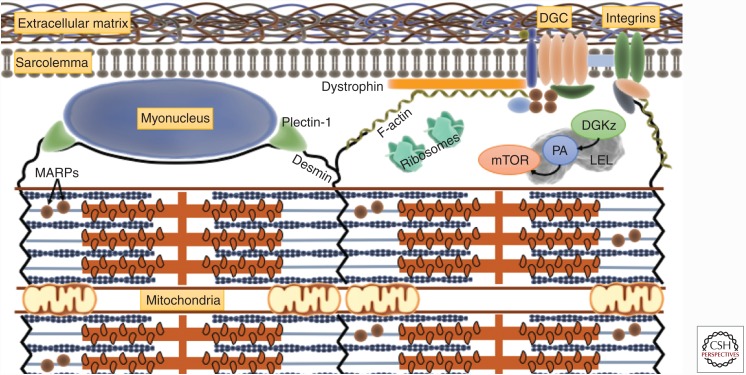
Schematic of mechanosensing elements supported by recent evidence. The latest studies of molecules and communication networks localized to the extracellular matrix (ECM), sarcolemma, sarcoplasm, and sarcomere suggest the transduction of mechanosensing to hypertrophy signaling involves (1) increased phosphatidic acid (PA) via the mechanosensitive diacylglycerol kinase ζ (DGKz) and PA-induced mammalian target of rapamycin (mTOR) signaling, localized to late endosome–lysosomal (LEL) hybrid organelles; (2) signaling initiated by the mechanosensitive costamere complexes dystrophin-associated glycoprotein complex (DGC) and focal adhesion vinculin–talin–integrin complex; (3) signaling and nuclear transcriptional activity initiated by intermediate filaments (e.g., desmin, plectin-1); and (4) contraction-induced mobilization of muscle ankryn repeat proteins (MARPs) from titin filaments.
Phosphatidic Acid
Just as mTORC1 is considered a master regulator of muscle protein synthesis and myofiber hypertrophy, data have emerged—largely from the Hornberger laboratory—implicating the lipid second messenger phosphatidic acid (PA) as a mechanically sensitive signal that directly activates mTORC1 in response to mechanical load (Hornberger 2011; Jacobs et al. 2014). In an elegant series of ex vivo studies exposing isolated muscles to mechanical stretch, supplemented with in vitro myoblast studies, the Hornberger group showed quite convincingly that mechanical stimulation up-regulates PA, and that PA activates mTORC1 signaling and protein synthesis independent of intermediate signaling (e.g., extracellular regulated kinase [ERK]) (You et al. 2012). The team subsequently determined the mechanically sensitive up-regulation of PA results from its synthesis by the ζ isoform of diacylglycerol kinase ζ (DGKz) (which synthesizes PA via the phosphorylation of diacylglycerol [DAG]), suggesting that DGKz is a mechanosensitive enzyme that, on mechanical load, increases PA and ultimately mTOR signaling (You et al. 2014). Further interrogation suggests that PA-mTORC1 signaling axis may be localized to hybrid organelles consisting of late endosomes coupled to lysozomes (LELs) (Jacobs et al. 2014). PA treatment indeed increases muscle protein synthesis and mTOR signaling in vivo (Mobley et al. 2015). Translating these findings to human RT, supplementation with PA was recently found to drive greater RT-induced muscle hypertrophy than placebo (Joy et al. 2014). The remaining knowledge gap is the precise mechanism(s) by which DGKz-synthesized PA is activated by mechanical load.
Costameres
Costameres are highly ordered macromolecular protein complexes that connect the z-discs of sarcomeres to the sarcolemma and ultimately the ECM, and are therefore central to lateral force transmission and overall myofiber integrity. As the structural anchor for the myofibrils, costameres are certainly mechanosensory and some protein components are known to function in signaling (i.e., mechanotransduction). Two major protein complexes make up the costameres in skeletal muscle: dystrophin-associated glycoprotein complex (DGC) and the focal adhesion vinculin–talin–integrin complex, which is tethered to desmin intermediate filaments at the z-disc. Protein turnover in these complexes is regulated by integrin-associated focal adhesion kinase (FAK). Indeed, one group identified FAK as an upstream regulator of the mechanosensory mTOR pathway (Klossner et al. 2009). The DGC, a complex of numerous structural and signaling proteins linking the contractile apparatus to the myofiber membrane (sarcolemma), is critical for maintaining myofiber integrity. Indeed, lack of the primary DGC protein dystrophin, as seen in Duchenne muscular dystrophy, causes irreparable myofiber damage, and declining levels of dystrophin in aging muscle increase susceptibility to damage (Hughes et al. 2015). Interestingly, dystrophin itself is capable of mediating signals relevant to muscle hypertrophy (Acharyya et al. 2005). The α7β1 integrin complex also seems to play a role in muscle hypertrophy and has been shown to modulate myoblast adhesion, migration, proliferation, and differentiation (Boppart et al. 2006). Among humans, changes in β1 integrin protein levels during RT correlate with changes in muscle size (Li et al. 2013). While both of these costameric protein complexes (DGC and integrins) are attractive candidate mechanotransducers involved in hypertrophy signaling, their interrogation has not advanced much since our last review (Adams and Bamman 2012).
Intermediate Filaments
Among the intermediate filaments, desmin is emerging as a player of multiple roles beyond maintenance of myofibrillar integrity and sarcomeric structure at the z-disc. Novel findings in 2015 indicate that desmin is also important for myonuclear positioning, deformation during mechanical load, and JNK signaling (Palmisano et al. 2015). Additional recent findings point to plectin-1, which is linked to desmin and may be a key signal transducer (Staszewska et al. 2015). Plectin-1 appears to be largely responsible for maintaining the architecture and docking of the subsarcolemmal myonuclei, and is linked to sarcomeres via desmin. The novel findings of Staszewska et al. indicate quite convincingly that plectin-1 plays a central role in mechanosensitive signaling induction, as knockout models markedly reduce the nuclear transcription of known mechanosensitive components. To date, plectin biology has not been studied in the context of RT and, more broadly, a search for “plectin and muscle hypertrophy” returned zero publications. In our view, plectin–desmin interactions and signal transduction are ripe for investigation.
Muscle Ankyrin Repeat Proteins
As reviewed elsewhere recently (Lun et al. 2014), muscle ankyrin repeat proteins (MARPs) are linked to titin in the sarcomere and perform a variety of functions, including modulating the activity of transcription factors. Using the mouse compensatory overload (synergist ablation) model and gene expression profiling, Chaillou et al. identified a functional group of up-regulated genes that included three MARPs thought to be involved in mechanotransduction: ankyrin repeat domain 1 (Ankrd1), Ankrd2, and cysteine and glycine-rich protein 3 (Csrp3). During compensatory overload, Ankrd1 mRNA is increased over 50-fold at day 1 and remains elevated ∼20-fold at day 3, whereas Ankrd2 and Csrp3 are also increased (Chaillou et al. 2013, 2015). When muscle is loaded, MARPs migrate from the sarcomere into the nucleus to regulate numerous transcription factors, some of which are involved in myofiber hypertrophy (Kojic et al. 2010; Belgrano et al. 2011). The majority of published literature on MARPs in muscle hypertrophy is focused on cardiac muscle. However, it is noteworthy that polymorphic variants of Ankrd6 in humans are associated with skeletal muscle size and the magnitude of RT-induced muscle hypertrophy (Van Deveire et al. 2012). Cumulatively, the findings suggest that these sarcomere-related proteins may play a role in RT hypertrophy signaling, although more work is needed.
MYOKINE SIGNALING
Myokines are cytokines, growth factors, and other proteins secreted by skeletal muscle. They can act in an autocrine, paracrine, or endocrine manner to affect the muscle itself, surrounding cells, or cells at distal locations in nonmuscle tissues. Myokines are now recognized as potentially important for cross talk between skeletal muscle and the liver, adipose tissue, bone, and the brain (Pedersen and Hojman 2012; Kaji 2016). For several myokines, contractile activity is a key regulator of gene expression, translation, and secretion, although in many cases their physiologic function(s) are not fully understood. In our 2012 review (Adams and Bamman 2012), we highlighted what was known about a few myokines and associated signaling cascades in the context of exercise-induced myofiber hypertrophy (e.g., fibroblast growth factors [FGFs], interleukin [IL]-4, IL-6, IL-8, IL-15, IGF-I, hepatocyte growth factor [HGF], leukemia inhibitory factor [LIF], and myostatin). To our knowledge, there have been no major advances since then. However, it is noteworthy that recent studies—leveraging “omics” approaches—have led to the identification of over 500 myokines induced by muscle contraction (Norheim et al. 2011; Raschke et al. 2013; Catoire et al. 2014; Hartwig et al. 2014). These recent discoveries open the door to promising research in molecular muscle biology to functionally characterize the role(s) specific myokines may play in driving the molecular mechanisms of RT myofiber hypertrophy.
CONCLUSIONS AND FUTURE DIRECTIONS
In recent years, the black box representing the regulation of loading-induced muscle hypertrophy has begun to be populated with an array of putative mechanisms. In the 4 years since we last reviewed this topic, there have been a number of refinements to proposed mechanisms and the identification of previously unsuspected contributors.
One of the most powerful approaches used to investigate RT-induced compensatory hypertrophy has been the application of statistical methods in larger human studies that allow the identification of distinct subpopulations with regard to the hypertrophic training response. Use of naturally occurring experimental groups (i.e., nonresponders vs. responders) identified by these means has provided many new insights as well as confirming previous hypotheses.
Recent important observations have affirmed of the role of ribosomal biogenesis as a significant contributor to sustained functional hypertrophic responses. Many of the regulatory elements that have been identified converge on some aspect of translational capacity and specifically ribosomal capacity and /or activity.
High-quality human RT studies have also contributed significantly to the maturation of our understanding of the role of muscle satellite cell activity in the hypertrophy process. The nature of the contribution of myonuclear domain size in human skeletal myofibers has been clarified and strongly supported by recent work.
Moving forward, the research topics that stood out to us as ripe for further investigation include the relatively recently elucidated agents of transcriptional regulation/modulation such as miRNAs and coregulators exemplified by the PGC-1α family. A second direction of advance leads through one of the most fundamental processes—mechanotransduction. Recent identification of messenger molecules such as PA, as well as the elucidation of key structurally linked components, suggests that this component of the regulatory scheme may finally yield its secrets.
Footnotes
Editors: Juleen R. Zierath, Michael J. Joyner, and John A. Hawley
Additional Perspectives on The Biology of Exercise available at www.perspectivesinmedicine.org
REFERENCES
- Acharyya S, Butchbach ME, Sahenk Z, Wang H, Saji M, Carathers M, Ringel MD, Skipworth RJ, Fearon KC, Hollingsworth MA, et al. 2005. Dystrophin glycoprotein complex dysfunction: A regulatory link between muscular dystrophy and cancer cachexia. Cancer Cell 8: 421–432. [DOI] [PubMed] [Google Scholar]
- Adams GR, Bamman MM. 2012. Characterization and regulation of mechanical loading-induced compensatory muscle hypertrophy. Compr Physiol 2: 2829–2870. [DOI] [PubMed] [Google Scholar]
- Ahtiainen JP, Walker S, Peltonen H, Holviala J, Sillanpaa E, Karavirta L, Sallinen J, Mikkola J, Valkeinen H, Mero A, et al. 2016. Heterogeneity in resistance training-induced muscle strength and mass responses in men and women of different ages. Age 38: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamman MM, Petrella JK, Kim JS, Mayhew DL, Cross JM. 2007. Cluster analysis tests the importance of myogenic gene expression during myofiber hypertrophy in humans. J Appl Physiol (1985) 102: 2232–2239. [DOI] [PubMed] [Google Scholar]
- Belgrano A, Rakicevic L, Mittempergher L, Campanaro S, Martinelli VC, Mouly V, Valle G, Kojic S, Faulkner G. 2011. Multi-tasking role of the mechanosensing protein Ankrd2 in the signaling network of striated muscle. PLoS ONE 6: e25519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel CS, Cross JM, Bamman MM. 2011. Exercise dosing to retain resistance training adaptations in young and older adults. Med Sci Sports Exerc 43: 1177–1187. [DOI] [PubMed] [Google Scholar]
- Boppart MD, Burkin DJ, Kaufman SJ. 2006. α7β1-integrin regulates mechanotransduction and prevents skeletal muscle injury. Am J Physiol Cell Physiol 290: C1660–C1665. [DOI] [PubMed] [Google Scholar]
- Brook MS, Wilkinson DJ, Mitchell WK, Lund JN, Phillips BE, Szewczyk NJ, Greenhaff PL, Smith K, Atherton PJ. 2016. Synchronous deficits in cumulative muscle protein synthesis and ribosomal biogenesis underlie age-related anabolic resistance to exercise in humans. J Physiol 594: 7399–7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd NA, West DW, Moore DR, Atherton PJ, Staples AW, Prior T, Tang JE, Rennie MJ, Baker SK, Phillips SM. 2011. Enhanced amino acid sensitivity of myofibrillar protein synthesis persists for up to 24 h after resistance exercise in young men. J Nutr 141: 568–573. [DOI] [PubMed] [Google Scholar]
- Catoire M, Mensink M, Kalkhoven E, Schrauwen P, Kersten S. 2014. Identification of human exercise-induced myokines using secretome analysis. Physiol Genomics 46: 256–267. [DOI] [PubMed] [Google Scholar]
- Chaillou T, Lee JD, England JH, Esser KA, McCarthy JJ. 2013. Time course of gene expression during mouse skeletal muscle hypertrophy. J Appl Physiol (1985) 115: 1065–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaillou T, Jackson JR, England JH, Kirby TJ, Richards-White J, Esser KA, Dupont-Versteegden EE, McCarthy JJ. 2015. Identification of a conserved set of upregulated genes in mouse skeletal muscle hypertrophy and regrowth. J Appl Physiol (1985) 118: 86–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchward-Venne TA, Tieland M, Verdijk LB, Leenders M, Dirks ML, de Groot LC, van Loon LJ. 2015. There are no nonresponders to resistance-type exercise training in older men and women. J Am Med Dir Assoc 16: 400–411. [DOI] [PubMed] [Google Scholar]
- Damas F, Phillips SM, Libardi CA, Vechin FC, Lixandrao ME, Jannig PR, Costa LA, Bacurau AV, Snijders T, Parise G, et al. 2016. Resistance training-induced changes in integrated myofibrillar protein synthesis are related to hypertrophy only after attenuation of muscle damage. J Physiol 594: 5209–5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidsen PK, Gallagher IJ, Hartman JW, Tarnopolsky MA, Dela F, Helge JW, Timmons JA, Phillips SM. 2011. High responders to resistance exercise training demonstrate differential regulation of skeletal muscle microRNA expression. J Appl Physiol (1985) 110: 309–317. [DOI] [PubMed] [Google Scholar]
- Devlin JR, Hannan KM, Hein N, Cullinane C, Kusnadi E, Ng PY, George AJ, Shortt J, Bywater MJ, Poortinga G, et al. 2016. Combination therapy targeting ribosome biogenesis and mRNA translation synergistically extends survival in MYC-driven lymphoma. Cancer Discov 6: 59–70. [DOI] [PubMed] [Google Scholar]
- Dreyer HC, Blanco CE, Sattler FR, Schroeder ET, Wiswell RA. 2006. Satellite cell numbers in young and older men 24 hours after eccentric exercise. Muscle Nerve 33: 242–253. [DOI] [PubMed] [Google Scholar]
- Dumont NA, Bentzinger CF, Sincennes MC, Rudnicki MA. 2015. Satellite cells and skeletal muscle regeneration. Comp Physiol 5: 1027–1059. [DOI] [PubMed] [Google Scholar]
- Eftestol E, Egner IM, Lunde IG, Ellefsen S, Andersen T, Sjaland C, Gundersen K, Bruusgaard JC. 2016. Increased hypertrophic response with increased mechanical load in skeletal muscles receiving identical activity patterns. Am J Physiol Cell Physiol 311: C616–C629. [DOI] [PubMed] [Google Scholar]
- Egner IM, Bruusgaard JC, Gundersen K. 2016. Satellite cell depletion prevents fiber hypertrophy in skeletal muscle. Development 143: 2898–2906. [DOI] [PubMed] [Google Scholar]
- Figueiredo VC, Caldow MK, Massie V, Markworth JF, Cameron-Smith D, Blazevich AJ. 2015. Ribosome biogenesis adaptation in resistance training-induced human skeletal muscle hypertrophy. Am J Physiol Endocrinol Metab 309: E72–E83. [DOI] [PubMed] [Google Scholar]
- Frese S, Ruebner M, Suhr F, Konou TM, Tappe KA, Toigo M, Jung HH, Henke C, Steigleder R, Strissel PL, et al. 2015. Long-term endurance exercise in humans stimulates cell fusion of myoblasts along with fusogenic endogenous retroviral genes in vivo. PLoS ONE 10: e0132099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry CS, Lee JD, Jackson JR, Kirby TJ, Stasko SA, Liu H, Dupont-Versteegden EE, McCarthy JJ, Peterson CA. 2014. Regulation of the muscle fiber microenvironment by activated satellite cells during hypertrophy. FASEB J 28: 1654–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon BS, Liu C, Steiner JL, Nader GA, Jefferson LS, Kimball SR. 2016. Loss of REDD1 augments the rate of the overload-induced increase in muscle mass. Am J Physiol Regul Integr Comp Physiol 311: R545–R557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig S, Raschke S, Knebel B, Scheler M, Irmler M, Passlack W, Muller S, Hanisch FG, Franz T, Li X, et al. 2014. Secretome profiling of primary human skeletal muscle cells. Biochim Biophys Acta 1844: 1011–1017. [DOI] [PubMed] [Google Scholar]
- Hornberger TA. 2011. Mechanotransduction and the regulation of mTORC1 signaling in skeletal muscle. Int J Biochem Cell Biol 43: 1267–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes DC, Wallace MA, Baar K. 2015. Effects of aging, exercise, and disease on force transfer in skeletal muscle. Am J Physiol Endocrinol Metab 309: E1–E10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs BL, Goodman CA, Hornberger TA. 2014. The mechanical activation of mTOR signaling: An emerging role for late endosome/lysosomal targeting. J Muscle Res Cell Motil 35: 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joy JM, Gundermann DM, Lowery RP, Jager R, McCleary SA, Purpura M, Roberts MD, Wilson SM, Hornberger TA, Wilson JM. 2014. Phosphatidic acid enhances mTOR signaling and resistance exercise induced hypertrophy. Nutr Metab (Lond) 11: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji H. 2016. Effects of myokines on bone. Bonekey Rep 5: 826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsen A, Couppe C, Andersen JL, Mikkelsen UR, Nielsen RH, Magnusson SP, Kjaer M, Mackey AL. 2015. Matters of fiber size and myonuclear domain: Does size matter more than age? Muscle Nerve 52: 1040–1046. [DOI] [PubMed] [Google Scholar]
- Kim JS, Petrella JK, Cross JM, Bamman MM. 2007. Load-mediated downregulation of myostatin mRNA is not sufficient to promote myofiber hypertrophy in humans: A cluster analysis. J Appl Physiol (1985) 103: 1488–1495. [DOI] [PubMed] [Google Scholar]
- Kirby TJ, Lee JD, England JH, Chaillou T, Esser KA, McCarthy JJ. 2015. Blunted hypertrophic response in aged skeletal muscle is associated with decreased ribosome biogenesis. J Appl Physiol (1985) 119: 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby TJ, Patel RM, McClintock TS, Dupont-Versteegden EE, Peterson CA, McCarthy JJ. 2016. Myonuclear transcription is responsive to mechanical load and DNA content but uncoupled from cell size during hypertrophy. Mol Biol Cell 27: 788–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klossner S, Durieux AC, Freyssenet D, Flueck M. 2009. Mechano-transduction to muscle protein synthesis is modulated by FAK. Eur J Appl Physiol 106: 389–398. [DOI] [PubMed] [Google Scholar]
- Kojic S, Nestorovic A, Rakicevic L, Belgrano A, Stankovic M, Divac A, Faulkner G. 2010. A novel role for cardiac ankyrin repeat protein Ankrd1/CARP as a co-activator of the p53 tumor suppressor protein. Arch Biochem Biophys 502: 60–67. [DOI] [PubMed] [Google Scholar]
- Li R, Narici MV, Erskine RM, Seynnes OR, Rittweger J, Pisot R, Simunic B, Fluck M. 2013. Costamere remodeling with muscle loading and unloading in healthy young men. J Anat 223: 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun AS, Chen J, Lange S. 2014. Probing muscle ankyrin-repeat protein (MARP) structure and function. Anat Rec (Hoboken) 297: 1615–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey DC, Robinovitch SN. 2006. Mechanisms underlying age-related differences in ability to recover balance with the ankle strategy. Gait Posture 23: 59–68. [DOI] [PubMed] [Google Scholar]
- Mayer C, Grummt I. 2006. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 25: 6384–6391. [DOI] [PubMed] [Google Scholar]
- Mayhew DL, Kim JS, Cross JM, Ferrando AA, Bamman MM. 2009. Translational signaling responses preceding resistance training-mediated myofiber hypertrophy in young and old humans. J Appl Physiol (1985) 107: 1655–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayhew DL, Hornberger TA, Lincoln HC, Bamman MM. 2011. Eukaryotic initiation factor 2Bɛ induces cap-dependent translation and skeletal muscle hypertrophy. J Physiol 589: 3023–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy JJ, Esser KA. 2007. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol (1985) 102: 306–313. [DOI] [PubMed] [Google Scholar]
- McCarthy JJ, Mula J, Miyazaki M, Erfani R, Garrison K, Farooqui AB, Srikuea R, Lawson BA, Grimes B, Keller C, et al. 2011. Effective fiber hypertrophy in satellite cell-depleted skeletal muscle. Development 138: 3657–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell CJ, Churchward-Venne TA, Parise G, Bellamy L, Baker SK, Smith K, Atherton PJ, Phillips SM. 2014. Acute post-exercise myofibrillar protein synthesis is not correlated with resistance training-induced muscle hypertrophy in young men. PLoS ONE 9: e89431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley CB, Hornberger TA, Fox CD, Healy JC, Ferguson BS, Lowery RP, McNally RM, Lockwood CM, Stout JR, Kavazis AN, et al. 2015. Effects of oral phosphatidic acid feeding with or without whey protein on muscle protein synthesis and anabolic signaling in rodent skeletal muscle. J Int Soc Sports Nutr 12: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nader GA, von Walden F, Liu C, Lindvall J, Gutmann L, Pistilli EE, Gordon PM. 2014. Resistance exercise training modulates acute gene expression during human skeletal muscle hypertrophy. J Appl Physiol (1985) 116: 693–702. [DOI] [PubMed] [Google Scholar]
- Nakada S, Ogasawara R, Kawada S, Maekawa T, Ishii N. 2016. Correlation between ribosome biogenesis and the magnitude of hypertrophy in overloaded skeletal muscle. PLoS ONE 11: e0147284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nederveen JP, Joanisse S, Seguin CM, Bell KE, Baker SK, Phillips SM, Parise G. 2015. The effect of exercise mode on the acute response of satellite cells in old men. Acta Physiol (Oxf) 215: 177–190. [DOI] [PubMed] [Google Scholar]
- Norheim F, Raastad T, Thiede B, Rustan AC, Drevon CA, Haugen F. 2011. Proteomic identification of secreted proteins from human skeletal muscle cells and expression in response to strength training. Am J Physiol Endocrinol Metab 301: E1013–E1021. [DOI] [PubMed] [Google Scholar]
- Ogasawara R, Akimoto T, Umeno T, Sawada S, Hamaoka T, Fujita S. 2016. MicroRNA expression profiling in skeletal muscle reveals different regulatory patterns in high and low responders to resistance training. Physiol Genomics 48: 320–324. [DOI] [PubMed] [Google Scholar]
- Olsen S, Aagaard P, Kadi F, Tufekovic G, Verney J, Olesen JL, Suetta C, Kjaer M. 2006. Creatine supplementation augments the increase in satellite cell and myonuclei number in human skeletal muscle induced by strength training. J Physiol 573: 525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmisano MG, Bremner SN, Hornberger TA, Meyer GA, Domenighetti AA, Shah SB, Kiss B, Kellermayer M, Ryan AF, Lieber RL. 2015. Skeletal muscle intermediate filaments form a stress-transmitting and stress-signaling network. J Cell Sci 128: 219–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen L, Hojman P. 2012. Muscle-to-organ cross talk mediated by myokines. Adipocyte 1: 164–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella JK, Kim JS, Cross JM, Kosek DJ, Bamman MM. 2006. Efficacy of myonuclear addition may explain differential myofiber growth among resistance-trained young and older men and women. Am J Physiol Endocrinol Metab 291: E937–E946. [DOI] [PubMed] [Google Scholar]
- Petrella JK, Kim JS, Mayhew DL, Cross JM, Bamman MM. 2008. Potent myofiber hypertrophy during resistance training in humans is associated with satellite cell-mediated myonuclear addition: A cluster analysis. J Appl Physiol (1985) 104: 1736–1742. [DOI] [PubMed] [Google Scholar]
- Phillips SM, Tipton KD, Aarsland A, Wolf SE, Wolfe RR. 1997. Mixed muscle protein synthesis and breakdown after resistance exercise in humans. Am J Physiol 273: E99–E107. [DOI] [PubMed] [Google Scholar]
- Prestes J, da Cunha Nascimento D, Tibana RA, Teixeira TG, Vieira DC, Tajra V, de Farias DL, Silva AO, Funghetto SS, de Souza VC, et al. 2015. Understanding the individual responsiveness to resistance training periodization. Age 37: 9793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qaisar R, Renaud G, Hedstrom Y, Pollanen E, Ronkainen P, Kaprio J, Alen M, Sipila S, Artemenko K, Bergquist J, et al. 2013. Hormone replacement therapy improves contractile function and myonuclear organization of single muscle fibres from postmenopausal monozygotic female twin pairs. J Physiol 591: 2333–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschke S, Eckardt K, Bjorklund Holven K, Jensen J, Eckel J. 2013. Identification and validation of novel contraction-regulated myokines released from primary human skeletal muscle cells. PLoS ONE 8: e62008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raue U, Trappe TA, Estrem ST, Qian HR, Helvering LM, Smith RC, Trappe S. 2012. Transcriptome signature of resistance exercise adaptations: Mixed muscle and fiber type specific profiles in young and old adults. J Appl Physiol (1985) 112: 1625–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebello RJ, Kusnadi E, Cameron DP, Pearson HB, Lesmana A, Devlin JR, Drygin D, Clark AK, Porter L, Pedersen J, et al. 2016. The dual inhibition of RNA Pol I transcription and PIM kinase as a new therapeutic approach to treat advanced prostate cancer. Clin Cancer Res 22: 5539–5552. [DOI] [PubMed] [Google Scholar]
- Ruas JL, White JP, Rao RR, Kleiner S, Brannan KT, Harrison BC, Greene NP, Wu J, Estall JL, Irving BA, et al. 2012. A PGC-1 α isoform induced by resistance training regulates skeletal muscle hypertrophy. Cell 151: 1319–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelsson H, Moberg M, Apro W, Ekblom B, Blomstrand E. 2016. Intake of branched-chain or essential amino acids attenuates the elevation in muscle levels of PGC-1α4 mRNA caused by resistance exercise. Am J Physiol Endocrinol Metab 311: E246–E251. [DOI] [PubMed] [Google Scholar]
- Schoenfeld BJ, Aragon AA, Krieger JW. 2013. The effect of protein timing on muscle strength and hypertrophy: A meta-analysis. J Int Soc Sports Nutr 10: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders T, Verdijk LB, Smeets JS, McKay BR, Senden JM, Hartgens F, Parise G, Greenhaff P, van Loon LJ. 2014. The skeletal muscle satellite cell response to a single bout of resistance-type exercise is delayed with aging in men. Age 36: 9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders T, Smeets JS, van Kranenburg J, Kies AK, van Loon LJ, Verdijk LB. 2016. Changes in myonuclear domain size do not precede muscle hypertrophy during prolonged resistance-type exercise training. Acta Physiol (Oxf) 216: 231–239. [DOI] [PubMed] [Google Scholar]
- Staszewska I, Fischer I, Wiche G. 2015. Plectin isoform 1-dependent nuclear docking of desmin networks affects myonuclear architecture and expression of mechanotransducers. Hum Mol Genet 24: 7373–7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stec MJ, Mayhew DL, Bamman MM. 2015. The effects of age and resistance loading on skeletal muscle ribosome biogenesis. J Appl Physiol (1985) 119: 851–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stec MJ, Kelly NA, Many GM, Windham ST, Tuggle SC, Bamman MM. 2016. Ribosome biogenesis may augment resistance training-induced myofiber hypertrophy and is required for myotube growth in vitro. Am J Physiol Endocrinol Metab 310: E652–E661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalacker-Mercer A, Stec M, Cui X, Cross J, Windham S, Bamman M. 2013. Cluster analysis reveals differential transcript profiles associated with resistance training-induced human skeletal muscle hypertrophy. Physiol Genomics 45: 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deveire KN, Scranton SK, Kostek MA, Angelopoulos TJ, Clarkson PM, Gordon PM, Moyna NM, Visich PS, Zoeller RF, Thompson PD, et al. 2012. Variants of the ankyrin repeat domain 6 gene (ANKRD6) and muscle and physical activity phenotypes among European-derived American adults. J Strength Cond Res 26: 1740–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdijk LB, Gleeson BG, Jonkers RA, Meijer K, Savelberg HH, Dendale P, van Loon LJ. 2009. Skeletal muscle hypertrophy following resistance training is accompanied by a fiber type-specific increase in satellite cell content in elderly men. J Gerontol A Biol Sci Med Sci 64: 332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Walden F, Liu C, Aurigemma N, Nader GA. 2016. mTOR signaling regulates myotube hypertrophy by modulating protein synthesis, rDNA transcription and chromatin remodeling. Am J Physiol Cell Physiol 311: C663–C672. [DOI] [PubMed] [Google Scholar]
- Wen Y, Alimov AP, McCarthy JJ. 2016. Ribosome biogenesis is necessary for skeletal muscle hypertrophy. Exerc Sport Sci Rev 44: 110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JP, Wrann CD, Rao RR, Nair SK, Jedrychowski MP, You JS, Martinez-Redondo V, Gygi SP, Ruas JL, Hornberger TA, et al. 2014. G protein-coupled receptor 56 regulates mechanical overload-induced muscle hypertrophy. Proc Natl Acad Sci 111: 15756–15761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe RR. 2006. The underappreciated role of muscle in health and disease. Am J Clin Nutr 84: 475–482. [DOI] [PubMed] [Google Scholar]
- You JS, Frey JW, Hornberger TA. 2012. Mechanical stimulation induces mTOR signaling via an ERK-independent mechanism: Implications for a direct activation of mTOR by phosphatidic acid. PLoS ONE 7: e47258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You JS, Lincoln HC, Kim CR, Frey JW, Goodman CA, Zhong XP, Hornberger TA. 2014. The role of diacylglycerol kinase ζ and phosphatidic acid in the mechanical activation of mammalian target of rapamycin (mTOR) signaling and skeletal muscle hypertrophy. J Biol Chem 289: 1551–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]