Abstract
RAS proteins are key signaling switches essential for control of proliferation, differentiation, and survival of eukaryotic cells. RAS proteins are mutated in 30% of human cancers. In addition, mutations in upstream or downstream signaling components also contribute to oncogenic activation of the pathway. RAS proteins exert their functions through activation of several signaling pathways and dissecting the contributions of these effectors in normal cells and in cancer is an ongoing challenge. In this review, we summarize our current knowledge about how RAS regulates type I phosphatidylinositol 3-kinase (PI3K), one of the main RAS effectors. RAS signaling through PI3K is necessary for normal lymphatic vasculature development and for RAS-induced transformation in vitro and in vivo, especially in lung cancer, where it is essential for tumor initiation and necessary for tumor maintenance.
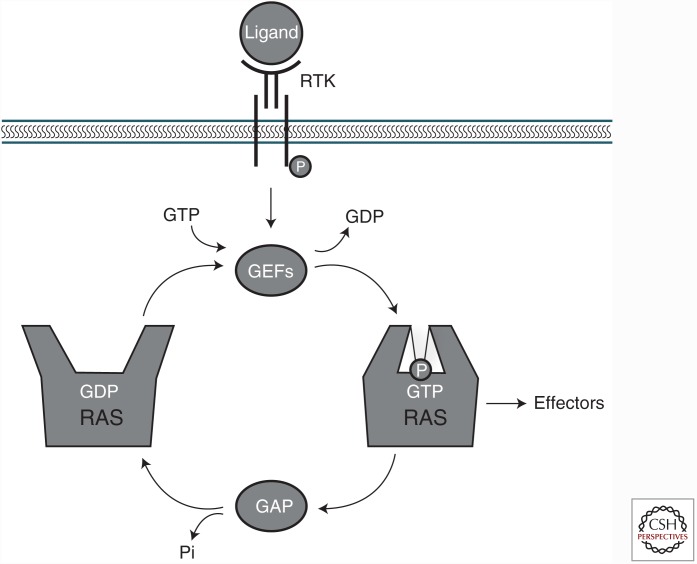
The superfamily of small guanosine triphosphate (GTP)-binding proteins plays a pivotal role in cellular signal transduction. Within this superfamily, RAS proteins (HRAS, NRAS, and Kirsten rat sarcoma [KRAS]) are the key regulators of a large number of processes such as cell-cycle progression, cell division, and apoptosis. RAS cycles between an active, GTP-bound state (RAS-GTP), and an inactive, guanosine diphosphate (GDP)-bound state (RAS-GDP) (Buday and Downward 2008; Vigil et al. 2010; Rojas et al. 2011). Guanine nucleotide exchange factors (GEFs) catalyze the exchange of GDP for GTP, whereas GTPase-activating proteins (GAPs) increase the rate of GTP hydrolysis to GDP (Fig. 1) (Quilliam et al. 1995).
Figure 1.
RAS activation/deactivation cycle by guanine nucleotide exchange factors (GEFs) and guanosine triphosphate (GTP)ase-activating proteins (GAPs). RAS proteins cycle between an inactive (guanosine diphosphate [GDP]-bound) and an active (GTP-bound) state. After receptor tyrosine kinase (RTK) engagement, RAS are activated by GEFs, which stimulate the exchange of GDP for GTP, and inactivated by GAPs that bind to RAS and stimulate its intrinsic GTPase activity.
RAS proteins have >80% homology sequence and differ mostly at the carboxyl terminus hypervariable region (HVR), a stretch of 25 amino acids known as the CAAX box (C, cysteine; A, aliphatic amino-acid; X, any amino acid) (Parker and Mattos 2015). To become fully active, newly synthetized RAS proteins undergo a number of posttranslational modifications at the CAAX box, including the addition of a farnesyl group to the cysteine residue of the CAAX box, the cleavage of the three terminal amino residues, and, finally, the carboxymethylation of the new carboxy-terminal cysteine residue (Tamanoi et al. 1988). HRAS and NRAS undergo an additional palmitoylation modification (Dudler and Gelb 1996). All these modifications are required for binding to the lipid membrane and dictate membrane localization of RAS proteins to specific microdomains. Each RAS isoform localizes to a distinct, nonoverlapping plasma membrane microdomain, where it contacts a unique pool of effector and regulatory proteins (Parker and Mattos 2015). The hypothesis that each isoform recruits effector proteins to the membrane via lipid and structural reorganization (Li and Gorfe 2012; Zhou et al. 2014) has emerged as an organizational scheme to produce overlapping but distinct signaling outputs from RAS nanoclusters. However, isoform-specific protein–lipid interaction and its connection to RAS-mediated signaling cascades have not yet been explored.
Wild-type RAS proteins are crucial for several physiological processes, but it is their involvement in cancer that has attracted the most attention (Land et al. 1983a,b; Ruley 1983; Trahey and McCormick 1987; Hirakawa and Ruley 1988). RAS proteins are mutated in ∼30% of human cancers. Point mutations in certain amino acid residues, most commonly at positions G12, G13, and Q61, lead to the expression of constitutively active proteins (Bos 1989; Prior et al. 2012) that signal to downstream effectors even in the absence of extracellular stimuli (Barbacid 1987; Riely et al. 2009). Remarkably, these mutations predominantly affect the KRAS locus, with oncogenic KRAS mutations being detected in 25%–30% of all screened tumor samples (Forbes et al. 2011). RAS oncoproteins have also been implicated in the development of cancer by signaling downstream with increased intensity or duration (Downward 2003). The importance of RAS signaling in cancer is accentuated by the incidence of mutations in many of its regulators or effector pathways.
Aberrant RAS signaling is also implicated in several developmental disorders known as cardio-facio-cutaneous diseases (i.e., neurofibromatosis-type 1, Costello syndrome, and Noonan syndrome) (Cox and Der 2010; Fernandez-Medarde and Santos 2011).
RAS EFFECTOR PATHWAYS
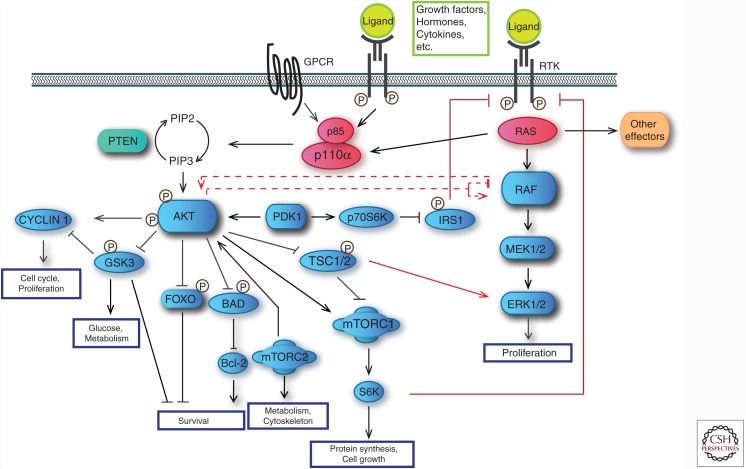
RAS proteins play a key part in multiple signaling networks, linking upstream signals to a broad set of downstream pathways that control cell-cycle progression (Coleman et al. 2004), growth, migration, apoptosis, and more (Fig. 2) (Fernandez-Medarde and Santos 2011). The cross talk and relative balance between these pathways determines cellular responses (Fig. 2) (Rajalingam et al. 2007; Stites and Ravichandran 2009).
Figure 2.
RAS effector pathways. Once activated, RAS proteins signal through multiple effector pathways, thus activating many different signal transduction pathways, such as mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3K). The RAS signaling network gets more complex because PI3K is also directly activated by receptor tyrosine kinases (RTKs) or G protein–coupled receptors (GPCRs). Besides, there are several points in which the MAPK and PI3K pathways cross talk, leading to either activation or inhibition at different levels. Raf can be inhibited by PI3K in different situations. PI3K pathway can also be activated or inhibited by Raf under certain conditions. Another level of connection of these two pathways involves target of rapamycin complex 1 (mTORC1) and insulin receptor substrate 1 (IRS1). When these proteins get inhibited, a feedback signal through RTKs is initiated that activates MAPK in a RAS-dependent manner, thus provoking extracellular signal-regulated kinase (ERK) and AKT phosphorylation and activation of both pathways. PTEN, Phosphatase and tensin homolog; PDK1, PI3K-dependent kinase 1; MEK1/2, mitogen-activated protein kinases 1 and 2.
RAS effectors share little sequence homology but all contain a RAS-binding domain (RBD) that shows a ubiquitin fold (Herrmann 2003). Recent nuclear magnetic resonance (NMR) studies provide evidence that RAS prefers certain effectors more than others in the presence of multiple binding partners. This preference changes with oncogenic mutations such as G12V, explaining how small changes in RAS affinity could substantially modify the outcome of integrated RAS signaling networks (Marshall et al. 2012a; Smith and Ikura 2014). The enhancement of specific effector pathways plays a critical role in maintaining an appropriate biological response (Morrison and Davis 2003). The specificity in RAS-induced signaling is primarily determined by the balance between RAS affinity for each of its effectors and the local concentrations of those effectors (Rodriguez-Viciana et al. 2004). Scaffold proteins also guide activation of specific effector pathway(s) (Li et al. 2000; Elion 2001).
In humans, there are ∼100 genes containing putative RAS-association (RA) or RBDs, although some of these proteins might not actually bind to RAS with physiologically relevant affinities. Actual interactions of RAS with about 20 effectors have been confirmed by direct affinity measurements (Kiel et al. 2005; Wohlgemuth et al. 2005).
One of the first identified mammalian effectors of RAS was Raf (Fig. 2) (Moodie et al. 1993; Warne et al. 1993). There are three closely related RAF proteins, C-Raf1, B-Raf, and A-Raf, which are known to be activated by RAS-GTP (Roskoski 2010). Activated Raf phosphorylates and activates downstream targets such as mitogen-activated protein kinases 1 and 2 (MEK1 and MEK2). MEK1 and MEK2 then phosphorylate and activate other mitogen-activated protein kinases (MAPKs), such as extracellular signal-regulated kinases 1 and 2 (ERK1 and ERK2) (Alessi et al. 1994). The substrates for ERK1/2 include both nuclear and cytosolic proteins, of which transcription factors, such as ELK1 and c-Jun, have been widely studied (Murphy and Blenis 2006; Yoon and Seger 2006). Activation of all of these transcription factors promotes cell-cycle progression and proliferation of the cells (Pruitt and Der 2001).
In addition to the RAF/MAPK effector pathway, RAS can also interact with phosphatidyl inositol 3-kinases (PI3Ks) (Rodriguez-Viciana et al. 1994; Pacold et al. 2000). PI3K activity is a necessity for the activation of Akt family Ser/Thr kinases, which, in turn, are necessary in the inhibition of apoptosis and the promotion of cell survival (Khwaja et al. 1997; Datta et al. 1999). Other PI3K targets include the Akt-activating kinase PDK1 (PI3K-dependent kinase 1) and GEFs that target members of the Rho family of RAS superfamily GTPases (Rodriguez-Viciana et al. 1996a; Downward 1997, 2004; Hamad et al. 2002; Karnoub and Weinberg 2008; Yuan and Cantley 2008; Zhao and Vogt 2008). The functional importance of the RAS/PI3K signaling pathway will be more extensively discussed in the next sections.
The third class of well-known effector of RAS includes three exchange factors for RAS-related RAL proteins: RAL guanine nucleotide dissociation stimulator (RALGDS), RGL2/RLF, and RALGDS-like protein (RGL/RSB2) (Hofer et al. 1994; Feig et al. 1996; Takaya et al. 2007). RAL proteins regulate vesicular trafficking within the cell and nuclear factor (NF)-κB activation.
Other proteins known to directly interact with RAS-GTP are PLC, AF6, TIAM1, RIN, NORE1, among others (Castellano and Downward 2010; Stephen et al. 2014). The functional significance of many of these interactions has not been properly determined yet.
PI3K SIGNALING PATHWAY
PI3Ks are members of a conserved family of lipid kinases that are split in three classes according to their substrate preference and sequence homology (Vanhaesebroeck et al. 2010b). The class I PI3Ks (the most studied in cancer) comprises p110α (encoded by PIK3CA), p110β (PIK3CB), p110γ (PIK3CG), and p110δ (PIK3CD). p110 subunits were originally divided into a class IA group (p110α, p110β, and p110δ), which bind the p85 type of regulatory subunit, and a class IB group (p110γ), which binds to either p101 or p87. Class II PI3Ks includes PI3K-C2α, PI3K-C2β, and PI3K-C2γ. “Signals feeding into vacuolar protein sorting 34 (VPS34), the only class III PI3K, are becoming apparent, but its physiological importance is unclear” (Vanhaesebroeck et al. 2010a).
Dissecting the role of RAS proteins in the activation of PI3K has been particularly complicated because these kinases can be activated by multiple upstream signals, both dependent and independent of RAS proteins: G protein–coupled receptors (GPCRs) and receptor tyrosine kinases (RTKs) can activate PI3K independently of RAS by its binding to p85. However, RTKs can also bind to RAS, which in turn bind to p110α subunit and activate PI3K signaling (Fig. 2) (Hemmings and Restuccia 2015). In addition, although H-, N-, and KRAS activate p110α and p110γ subunits of PI3K (Rodriguez-Viciana et al. 2004), the closely related R-RAS proteins activate the p110α, p110γ, and p110δ subunits (Marte et al. 1997), and p110β is regulated by the related proteins Rac1 and Cdc42 (Yang et al. 2012; Fritsch et al. 2013).
RAS-dependent activation of PI3K requires a cascade of events that start with the dimerization of RTKs in response to growth factor stimulation. This leads to autophosphorylation of the RTKs at tyrosine residues and activates Src homology 2 (SH2) domain-containing molecules, like GRB2. The signaling cascade then proceeds through Son of Sevenless (SOS) onto RAS, which then interacts directly with the RBD in the p110 subunit of PI3K and activates it. This process does not depend on the p85 regulatory subunit of PI3K, although GRB2 can be found in a complex with a scaffolding protein (e.g., GAB), which interacts with p85, enhancing the chance of p110 activation (Ong et al. 2001; Castellano and Downward 2010).
When active, PI3Ks phosphorylate phosphatidylinositol (4,5)-bisphosphate (PI-4,5-P2) and phosphatidylinositol (4)-phosphate (PI(4)P) on the inositol 3′-hydroxyl group, thereby generating phosphatidylinositol (3,4,5)-trisphosphate (PIP3) and phosphatidylinositol (3,4)-bisphosphate (PI(3,4)P2), respectively (Fig. 2). An important second messenger is PIP3, which binds to pleckstrin homology (PH) domains of effector proteins, causing their activation and/or translocation (Zhao et al. 2006; Fritsch and Downward 2013; Cescon et al. 2015). These lipids act on different pathways often through activation of either the PDK1, the Ser/Thr kinase Akt, or the mammalian target of rapamycin complex 1 (mTORC1) (Martini et al. 2014; Gyori et al. 2017). Akt regulates prosurvival signaling pathways by mediating various cellular processes such as epithelial to mesenchymal transition, glycogen metabolism, autophagy, and glucose uptake, whereas mTORC1 stimulates protein synthesis by phosphorylating the eukaryotic initiation factor 4E and the ribosomal S6 protein, among others. Although mTORC1 relays signals following PI3K/Akt activation, a second mTOR complex, mTORC2, contributes to complete Akt activation by phosphorylating Akt on serine 473. Activation of the mTORC1 target S6 kinase negatively feeds back to decrease PI3K activation (Polivka and Janku 2014; Mayer and Arteaga 2016).
Negative regulation of PI3K is accomplished primarily via action of the phosphatase and tensin homolog (PTEN) deleted on chromosome ten. PTEN encodes a lipid and protein phosphatase whose primary lipid substrate is PIP3. Mutations within the phosphatase domain of PTEN are frequent in many human cancers and impair its function resulting in elevated Akt activity and abnormal cell proliferation (Vanhaesebroeck et al. 2012; Martini et al. 2014).
A common feature of cellular signaling systems is feedback control, and MEK and PI3K signaling networks provides many examples. PI3K/Akt and RAS/MAPK pathways influence each other, both negatively and positively, at different stages of signal propagation resulting in dynamic and complex cross talk whose coordinated action determines the cell fate (Fig. 2). Such cross talk is often revealed when one pathway is chemically blocked, thereby releasing the cross-inhibition and effectively activating the other pathway (Castellano and Downward 2010; Mendoza et al. 2011; Aksamitiene et al. 2012; Fruman and Rommel 2014).
Oncogenic compensation can severely limit the anticancer efficacy of PI3K/Akt/mTOR inhibitors. Conversely, active PI3K signaling is a central mechanism of resistance to various targeted therapies (Tan and Yu 2013; Wood 2015). An important consequence of this feedback is that inhibitors of different components of the PI3K signaling pathway can cause elevated expression and activity of growth factor receptors, which leads to increased PI3K activity and RAS signaling, in addition to alternative survival pathways in cancer cells (Chandarlapaty et al. 2011; Rodrik-Outmezguine et al. 2011). There are multiple potential strategies to overcome the “rebound” signaling that occurs in response to PI3K/Akt/mTOR inhibitors. These include vertical inhibition of several signaling nodes and combination approaches.
RAS–PI3K INTERACTION: STRUCTURE AND BINDING INTERFACES
RAS-GTP binds to class I PI3K through its RBD, present in the amino terminal; p110α, p110δ, and p110γ are each stimulus-dependent RAS effectors (Vanhaesebroeck et al. 2010a). In contrast, the p110β RBD interacts with Rab5 GTPase (Christoforidis et al. 1999) and Rho GTPase family members (Fritsch et al. 2013) and, specifically, Rac potentiates p110β GPCR responses (Guillermet-Guibert et al. 2008; Dbouk et al. 2012; Fritsch et al. 2013). Class II PI3Ks also harbor an RBD, although these signaling inputs are not well characterized; the RBD domain is not present in class III PI3K (Vanhaesebroeck et al. 2010a; Thorpe et al. 2015).
RAS isoforms HRAS, NRAS, and KRAS share 100% sequence identity in the effector lobe and interact with effectors and regulators via conserved switch I and switch II regions. The switch I and II regions change conformation on GTP binding and serve as binding sites for a diverse array of downstream effectors.
PI3K’s RBD is a small globular domain of ∼100 residues in length that consists of a five-stranded mixed β sheet (Rb1-Rb5) flanked by two α helices (Rα1 and Rα2) (Walker et al. 1999; Pacold et al. 2000). RAS complexes with RBDs that come from different effectors form a similar model in which β sheets from RAS and the RBD align to form a single β-sheet connecting both proteins. Contacts between the switch I region of RAS and the RBD stabilize the interaction and ensure its dependence on RAS-GTP. Despite the commonalities of different RAS–RBD interactions, RAS discriminates among its effectors by rotating with respect to the RBD of a given effector. In the case of PI3Kγ, the length of the Rα1 helix and the size of the subsequent loop cause a significant rotation of RAS relative to the PI3Kγ RBD and results in switch II interactions for which there are no equivalents in the other RAS effector RBDs (Pacold et al. 2000; Djordjevic and Driscoll 2002). In addition to the allosteric effect, RAS orients the PI3K molecule in a way that allows it to more efficiently access its lipid substrate inserted in the membrane (Herrmann 2003).
Until recently, little was known about the considerably weaker interactions between RAS-GDP and RAS effectors. A double mutant of c-Raf N71R/A85K designed to bind RAS-GDP with a 100-fold increased affinity made it possible to solve for the first time the structure of a RAS-GDP/effector complex (Filchtinski et al. 2010). Surprisingly, this revealed that when bound to the N71R/A85K Raf mutant the switch I region of RAS-GDP is found in a conformation similar to that of RAS-GTP, and not RAS-GDP. Moreover, the structure indicates an increased mobility of the switch I region as shown by increased B factors. This increased flexibility compared with that of the same loop in RAS-GTP likely explains the low affinity of RAS effectors toward RAS-GDP.
RAS/PI3K SIGNALING IN HEALTH
After nearly 20 years of research, we are now getting a clearer picture of how and when RAS proteins mediate PI3K activation in normal and cancer cells. The use of RAS mutants that are defective in PI3K binding, but retain other effector functions, has significantly added to our knowledge of the specific functional role of RAS/PI3K signaling. Suire et al. (2006) generated a mouse model with a mutated p110γ RBD in which neutrophils presented decreased production of PI(3,4)P2 and PIP3, decreased activation of Akt, and diminished chemotaxis compared with wild-type cells. Additionally, the production of reactive oxygen species in response to agonist stimulation was decreased in p110γ RBD mutant mice. This revealed a significant role for the interaction in the normal function of neutrophils (Suire et al. 2006).
Further evidence of the importance of the RAS–PI3K interaction in vivo has been provided by genetic mouse models in which the RBD of p110α has two point mutations (T208D and K227A) that prevent its interaction with RAS without affecting its enzymatic activity (referred to herein as RBD-mutant model) (Gupta et al. 2007). Homozygous RBD-mutant animals often died shortly after birth owing to defects in the branching and development of the lymphatic vasculature system; however, surviving pups reached adulthood with no further obvious defects (Gupta et al. 2007). Similar observations were seen in mice expressing a kinase-dead version of p110α, a modification that is embryonic lethal because of impaired angiogenic remodeling defects (Graupera et al. 2008). Furthermore, deletion of Pik3ca alleles exclusively in endothelial cells resulted in a similar outcome. Intact RAS/PI3K signaling was also found to be essential for proper angiogenesis in zebrafish (Liu et al. 2008).
The role of RAS/PI3K/Akt signaling in cell survival and prevention of apoptosis is well known and was long established (Cox and Der 2003; Downward 2004; Zhang et al. 2011). Activation of Akt leads to both inhibition of proapoptotic proteins and activation or antiapoptotic proteins. Thus, by increasing the ability for growth and decreasing the capacity for apoptosis, RAS/PI3K signaling supports tumorigenesis.
Autophagy is a process by which cytoplasmic components (macromolecules and organelles) are degraded by the lysosome. Autophagy is essential in physiological processes that range from adaptation to starvation, cell differentiation and development, tumor suppression, innate and adaptive immunity, life span extension, and cell death (Mizushima 2007; Mizushima and Levine 2010; Wang et al. 2011; Boya et al. 2013; Nezis et al. 2014). The cross talk between RAS and autophagy is highly complex. RAS has been shown to be both a positive and negative regulator of autophagy, depending on the cell type, cellular context, and effector activation (Schmukler et al. 2014). However, it is clear that RAS inhibition of autophagy is controlled through PI3K activation and subsequent inhibition of the ULK1/Atg13/FIP200 complex (Furuta et al. 2004; Berry and Baehrecke 2007; Shao et al. 2016).
RAS signaling through PI3K is essential for hematopoiesis. The first evidence that RAS regulates eritropoiesis came from studies using primary fetal liver erythroid progenitors, in which knockout of KRAS impaired Akt activation and delayed erythroid differentiation (Zhang and Lodish 2005). Similar observations in zebrafish confirmed that KRAS knockout resulted in hematopoietic and angiogenic defects, including impaired expression of the erythroid-specific genes gata1 and βe3-hemoglobin, reduced blood circulation and disorganized blood vessels (Liu et al. 2008). Recently, the role of RAS–PI3K interaction in hematopoiesis has been shown in a mouse model of myeloid leukemia in which RAS/PI3K signaling is necessary for maturation and maintenance of erythroblasts (Gritsman et al. 2014).
RAS/PI3K SIGNALING IN CANCER
Activating point mutations in the genes encoding RAS proteins contribute to the formation of a large proportion of human tumors. The repertoire of their downstream effectors, upstream activators, and molecular players regulating their signaling pathway are genetically modified in an even larger proportion of human tumors.
The most frequently mutated kinase in human cancer is P110α with particularly high incidences in cancers of the breast, the colon, or the endometrium; alterations are also found in tumors of the brain, liver, stomach, lung, and ovary (Ligresti et al. 2009). There are three hot-spot mutations (E542K, E545K, and H1047R) that comprise 80% of all defects found in the PIk3ca gene. The E542 and E545K mutations map to the helical domain of p110α, whereas H1047R is located in the kinase domain. These mutations have been shown to increase proliferation and invasion of cells in vitro and in vivo by hyperactivating the downstream target Akt (Kang et al. 2005; Samuels et al. 2005; Zhao and Vogt 2010). Helical and kinase domain mutations induce gains-of-function by different molecular mechanisms. Activity of kinase domain mutants is RAS-independent, but requires p85 binding, whereas the helical domain mutants are p85-independent but their full activation requires RAS (Zhao and Vogt 2010). Pang et al. (2009) showed that expression of p110α E545K in a breast cancer cell line leads to a more severe metastatic phenotype than that induced by expressing p110α H1047R. On the other hand, H1047R, but not E545K, enhances HER2-mediated transformation of immortalized mammary epithelial cells (Chakrabarty et al. 2010). Recently, it was shown that p110α helical domain mutants bind to insulin receptor substrate 1 (IRS1) in the membrane, but the mutant proteins also require RAS-binding to fully exert their oncogenic function (Hao et al. 2013).
RTKs and RAS have been shown to cooperate in activating PI3K in KRAS mutant human colorectal (Ebi et al. 2011) and lung cancer cell lines (Molina-Arcas et al. 2013). Insulin-like growth factor 1 receptor (IGF-1R) was predominantly responsible for PI3K activation through binding to IRS adaptor proteins, with KRAS making a relatively minor contribution. It is likely that both RAS and receptor-mediated p110 activation is required to achieve PI3K activation levels high enough for tumor growth, which can be further amplified by loss of PTEN. The fact that alternative means of PI3K activation persist in the context of oncogenic KRAS may be caused by cancer cells losing their addiction to KRAS.
A useful insight about the importance mutant RAS interacting with endogenous p110α during tumor development has come from the use of KRAS mutants that do not bind PI3K but retain other effector functions. Shannon and colleagues showed that these mutants can still drive T-lineage acute lymphoblastic leukemias, with loss of PTEN compensating for the lack of RAS-p110α binding (Shieh et al. 2013).
Functional studies show that PI3K activation is vital for tumor initiation. Expression of a dominant-negative p85α lacking the p110-binding domain inhibited RAS-mediated transformation (Rodriguez-Viciana et al. 1997; Katso et al. 2001). Additionally, RBD-mutant mice are highly resistant to formation of lung and skin tumors induced by mutant KRAS (Gupta et al. 2007). Similarly, deletion of Pik3r1 and Pik3r2 abrogated KRASG12D-induced lung tumorigenesis (Engelman et al. 2008). Although PI3K activation may be necessary for KRAS-induced tumorigenesis, different studies have shown that inhibition of PI3K by itself is not sufficient to shrink established tumors in vivo or effectively treat KRAS mutated cancer cells in vitro (Engelman et al. 2008; Ihle et al. 2009). Experiments that use an inducible version of the RBD-mutant mice showed that disruption of RAS/PI3K in established lung tumors causes partial regression of tumors, followed by long-term stasis, but it is insufficient to cause complete regression of established tumors in the autochthonous setting, underscoring the differences between blocking tumorigenesis and eliminating established tumors.
Tumors harboring mutations in p110α also garner mutations in components of the PI3K pathway, like RAS or loss of PTEN (Cescon et al. 2015). Mutations of p110α and RAS are mutually exclusive in breast cancer, but often coexist in colorectal, endometrial, and lung cancers (Parsons et al. 2005; Oda et al. 2008; Janku et al. 2011; Chaft et al. 2012; Wang et al. 2014; Zhang et al. 2015). It is plausible that additional mutations enhance oncogenic transformation by increasing PI3K pathway activity levels beyond those caused by oncogenic RAS, thus activating additional downstream pathways. Recently, it was described that inhibition of p110α in colorectal cancer cell (CRC) lines harboring KRAS mutations or concomitant KRAS/PIK3CA mutations does not have the same effect. Functional responses to PI3K p110α silencing were distinct depending on the genetic background for KRAS and PIK3CA. PIK3CA silencing in CRC cells harboring KRAS/PIK3CA mutations induced apoptosis; in contrast, cell-cycle arrest at G2/M and a concomitant reduction in the number of cells at G1 phase was observed in CRC cells harboring KRAS mutations (Fernandes et al. 2016).
RAS/PI3K Signaling in the Tumor Stroma
RAS has been the prime example of a potent cell-autonomous oncogene for decades, but it is now evident that its effects stretch further to include non-cell-autonomous changes in the cellular microenvironment that have essential roles in both tumor initiation and progression.
The mechanisms by which RAS activation initiates and sustains proangiogenic processes are complex and rely on the modulation of endothelial growth factors and also on the increase of local inflammation and stromal remodeling (Pylayeva-Gupta et al. 2011). Blocking RAS signaling through PI3K in the tumor stroma impairs growth of tumors in which RAS/PI3K signaling is intact, as a consequence of inefficient angiogenesis (Murillo et al. 2014). Similar tumor growth impairment was observed in a p110α kinase-dead mouse model (Soler et al. 2013). Accordingly, PI3K inhibitors block tumor growth not only by targeting tumor cells but also by impairing tumor vasculature formation (Garlich et al. 2008; Schnell et al. 2008; Yuan et al. 2008; Lane et al. 2009; Soler et al. 2015).
Data obtained both in human cancer and transgenic mouse models indicate that RAS-driven tumors are able to overcome host-protecting adaptive immune responses (Clark et al. 2009). One mechanism by which oncogenic RAS may impair antitumor immune response is through recruitment of immunosuppressive regulatory T cells (Tregs) and myeloid-derived suppressor cells (MDSCs) (Clark et al. 2007; Tran Thang et al. 2010). Recent evidence suggests that PI3Ks play an important role in regulating the immune-suppressive, tumor-associated myeloid cell and Tregs, respectively (Gyori et al. 2017). Interestingly, RAS/PI3K signaling, either in the tumor (Castellano et al. 2013) or stromal compartment (Murillo et al. 2014), is required for macrophage recruitment, which may explain at least in part the decrease in tumor growth and angiogenesis observed on blocking the interaction between RAS and PI3K.
Remodeling of the extracellular matrix (ECM) is a key feature of cancer regulated by cancer-associated fibroblasts (CAFs), which secrete ECM components and ECM-modifying enzymes such as matrix metalloproteinases (MMPs). The different RAS isoforms control ECM synthesis through activation of Akt and transforming growth factor β (TGF-β) expression in a model of renal fibrosis (Fuentes-Calvo et al. 2012, 2013). PI3Ks also control the interaction between cancer cells and the tumor microenvironment (TME), namely, the production of MMP in normal and cancer cells (Chen et al. 2009; Awad et al. 2010; Kim et al. 2010; Hutti et al. 2012). In particular, MMP-14/CD44 binding results in activation of epidermal growth factor receptor (EGFR) and downstream engagement of PI3K signaling pathway in cancer cells, thus promoting tumor cell migration (Zarrabi et al. 2011).
Thus, RAS and PI3K are important players in the formation of a functional stroma that supports tumor growth and dissemination through a complex signaling network. A better understanding of the many ways by which PI3K contributes to the stromal remodeling activity of oncogenic RAS may uncover new avenues for therapeutic interventions targeting tumor stromal components.
RAS/PI3K Signaling in Migration and Metastasis Promotion
The most life-threatening consequence of an evolving tumor is the acquisition of metastatic properties. Many metastatic tumors (lung, pancreas, and colon) contain RAS mutations, which also confer metastatic properties to mouse cells in culture. The PI3K pathway has also been shown to exert a fundamental role regulating some of the steps of this process.
One of the first requirements for metastasis formation is the acquisition of a migratory phenotype accompanied by extensive remodeling of the actin cytoskeleton. The involvement of oncogenic RAS and PI3K in migration and remodeling of actin cytoskeleton has been well established through their interaction with Rac and Rho proteins both in mammalian cells and in the slime mold Dictyostelium discoideum (Campbell and Der 2004; Giehl 2005; Pollock et al. 2005; Sasaki and Firtel 2006; Castellano and Downward 2011; Devreotes and Horwitz 2015; Di Blasio et al. 2017). However, whether all these mechanisms are directly related to RAS signaling through PI3K is not clear.
RAS activation of Rac requires PI3K, and actin rearrangement correlates with the ability of RAS mutants to activate PI3K. Inhibition of PI3K activity blocks RAS induction of membrane ruffling, whereas activated PI3K is sufficient to induce membrane ruffling, acting through Rac (Han et al. 1998; Nimnual et al. 1998). Migration of endothelial cells is necessary for the creation of new vessels and repair of lesions, a process that is dependent on nitric oxide (NO). Signal transduction mediated by NO can trigger the process of cell adhesion and migration, releasing traction forces exerted by the endothelial cells on a surface and resulting in their spreading (Chatterjee et al. 2008). A recent report suggests that NO activates RAS, localizing PI3K to the plasma membrane and facilitating the interaction between PI3K and Rac, which is essential for the migration of endothelial cells (Eller-Borges et al. 2015).
RAS, through its interaction with PI3K, regulates migration of fibroblasts in response to several growth factors by regulating Rac activation and inhibiting Reelin expression (Castellano et al. 2016). When Reelin is expressed it prevents cells from migrating and up-regulates of E-cadherin, thereby impacting on cell–cell interaction and migration. Loss of E-cadherin is associated with increased cell migration and invasion (Kardash et al. 2010; Canel et al. 2013), a key event in the acquisition of invasive capacity, because reexpression of E-cadherin suppresses the invasion of tumor cells in vitro (Kardash et al. 2010; Canel et al. 2013). Mouse lung tumors in which the RAS–PI3K interaction is blocked display an increase in Reelin and E-cadherin expression and expression analysis in human samples showed that both lung adenocarcinoma and breast carcinoma patients whose tumors express high levels of Reelin have a better prognosis. These results suggest that inhibition of Reelin expression by RAS/PI3K signaling may be part of a mechanism by which cancer cells acquire the enhanced motility necessary to metastasize.
Another component of these mechanisms may be Sur8, a RAS-Raf scaffolding protein that also binds to p110α; this interaction is important for Sur8-mediated cell migration and invasion, along with tumor metastasis. Sur8 regulates migration and invasion of cells in response to RAS/PI3K signaling through activation of Rac and MMPs (Kaduwal et al. 2015; Lee et al. 2016).
Formation of metastasis also requires tumor cells to avoid anoikis or apoptosis caused by matrix deprivation (Paoli et al. 2013). Oncogenic RAS and PI3K can promote the loss of anchorage-dependent growth. For example, in Madin–Darby canine kidney (MDCK) epithelial cells, PI3K, but not RAF, is both necessary and sufficient for the protection provided by RAS from anoikis (Frisch et al. 1996; Khwaja et al. 1997). In tumors, PI3K signaling downstream from EGFR assists cancer cells in overcoming detachment-induced metabolic stress and, in turn, prevents programmed cell death (Schafer et al. 2009) and tumor cells growing in anchorage-independent conditions are addicted to PI3K signaling, and thus more sensitive to PI3K/mTOR inhibitors than matrix adherent cells (Muranen et al. 2012).
TARGETING THE RAS/PI3K PATHWAY IN CANCER THERAPY
For many years, oncogenic RAS proteins have been considered a key but “undruggable” cancer target because of the abundance of its substrate GTP, the high-affinity with which RAS binds it (Takashima and Faller 2013), and the apparent lack of suitable surfaces in critical regions necessary for small-molecule binding.
However, recently several groups have published on small molecule inhibitors of KRAS, which has restored hope that direct targeting of this protein can be achieved. Genentech identified a compound called DCAI that binds to a pocket located between the α2 helix and the β-sheet of KRAS4B, which is the binding site for the RAS guanidine exchange factor SOS1. In live-cell experiments, DCAI prevents membrane recruitment of the RBD of c-Raf kinase (Maurer et al. 2012). Independently, a group from Vanderbilt University discovered several molecules that weakly bind to GDP-loaded KRAS (Sun et al. 2012) into the same pocket as DCAI. Shokat and colleagues (Ostrem et al. 2013) screened a library of small molecules with GDP-bound KRASG12C, followed by the design and synthesis of analogs. The compounds discovered were found to bind to a pocket between the α2 and α3 helices, on the other side of the SII region from the Genentech and Vanderbilt compounds. As predicted, the compounds blocked the SOS1-mediated nucleotide exchange and decreased the binding of RAS to both BRAF and CRAF. It also seemed to selectively kill cancer cells harboring the G12C mutation (Ostrem et al. 2013). However, none of these compounds are very potent.
An effective RAS inhibitor must prevent the activation by RAS of its downstream binding partners, which could be achieved through direct inhibition of RAS GTPase activity, impairment of RAS localization or trafficking (Gysin et al. 2011; Wang et al. 2012; Bhattacharya et al. 2015; Singh et al. 2015; Ostrem and Shokat 2016; Asati et al. 2017). Recently, immunological approaches to target mutant RAS have also been investigated (Meyer et al. 2007; Rahma et al. 2014; Singh et al. 2015; Gil-Bazo et al. 2016).
An alternate approach to inhibit oncogenic RAS is to directly inhibit effector pathways downstream from RAS (Athuluri-Divakar et al. 2016; Keeton et al. 2017). However, the critical downstream effector for a given tumor may vary with the tissue type and even the specific mutational variant of RAS (Ihle et al. 2012; Alagesan et al. 2015).
Although several studies showed a role for PI3K signaling in RAS-mediated tumorigenesis (Rodriguez-Viciana et al. 1996b, 1997; Sheng et al. 2001; Li et al. 2004; Gupta et al. 2007; Castellano et al. 2013), PI3K pathway inhibitors used as single agents are not effective against RAS-driven cell lines and xenotransplantation models. Resistance to these inhibitors has been ascribed to negative feedback mechanisms and reflexive activation of other downstream signaling partners of RAS (Engelman et al. 2008; Ihle et al. 2009; Sos et al. 2009; Dan et al. 2010). Conversely, treating RAS mutant cancer cells with MEK inhibitors results in increased phosphorylation of the PI3K pathway effector Akt (Hoeflich et al. 2009; Mirzoeva et al. 2009; Sos et al. 2009). Thus, dual inhibition of both pathways may be required to block the growth of RAS-driven tumors. In support of this hypothesis, several studies showed that combined inhibition of MEK and PI3K pathway in RAS mutant background is superior to single-agent inhibition several in vitro and in vivo models, resulting in a synergistic inhibition of tumor growth, cell viability, and increased apoptosis (Engelman et al. 2008; Hoeflich et al. 2009; Mirzoeva et al. 2009; Sos et al. 2009; Castellano et al. 2013; Qu et al. 2014; Jifu et al. 2015; Schrauwen et al. 2015; Talbert et al. 2017). However, in a KRAS-driven pancreatic model combined therapy showed only modest activity (Alagesan et al. 2015), suggesting that there may be tissue and/or tumor specificity of the various downstream effector pathways of RAS and stressing the obvious need to better understand signaling downstream from oncogenic RAS in different tumor types.
Data from human cancers and transgenic mouse models indicate that RAS-driven cancers have the ability to overcome host-protecting adaptive immune responses (Clark et al. 2009). There is increasing appreciation for how small molecules targeting the cancer cell affect the immune context of the tumor (Vanneman and Dranoff 2012; Zitvogel et al. 2013). Extensive studies of the PI3K/Akt network in immune cells have shown that PI3K activation is not a simple on/off switch (Fruman and Bismuth 2009; Okkenhaug 2013; Okkenhaug et al. 2016). Inhibiting the pathway can either suppress immune responses or them through effects on diverse subsets of innate and adaptive immune cells and therefore in KRAS mutant cancers it may be possible to implement treatment regimens that increase immune rejection of tumors with concomitant direct antitumor effect. Proper assessment of candidate inhibitor compounds therefore requires testing in genetically engineered mouse models, with extensive monitoring of tumor infiltration and activity of diverse immune subsets including macrophages, T cells, and natural killer cells.
Tumor-immune infiltration studies have shown that macrophage recruitment to tumors is severely impaired in animals in which RAS cannot bind and activate PI3K (Castellano et al. 2013; Murillo et al. 2014). It will be crucial to determine which agents targeting PI3K/Akt enhance or suppress the efficacy of emerging cancer immunotherapies and vaccines. In mouse models, PI3K inhibitors can enhance the efficacy of immune-directed therapies (Marshall et al. 2012b). It is relevant to consider that isoform-selective agents minimize immune suppressive effects on lymphocytes compared with pan-class I inhibitors (So et al. 2013). Thus, pan-PI3K inhibitors are more likely to enforce or accelerate the immune exhaustion state, whereas isoform-selective PI3K are likely to be better suited for use in combination with cancer immunotherapy. Matching patients to the right combinations will require knowledge of the genomic driver and the immune fingerprint of the tumor.
Interruption of RAS-effector binding is also a promising strategy for drug design against RAS-driven cancers and other diseases associated with RAS. Strong support for the concept of disrupting activated RAS/effector protein complexes as a therapeutic strategy came from studies using a membrane-directed, single immunoglobulin antibody domain (iDab#6-memb). This antibody fragment binds the switch I and II domains of active HRAS predicted to be mutually exclusive of the amino-terminal RAS-binding domain of RAF1 (RAF-RBD), RALGDS, and PI3K. In a transgenic mouse model of KRAS-driven lung cancer, tumor initiation was dramatically reduced by expressing the antibody fragment (Tanaka and Rabbitts 2010). Although RAS-effector domain antibodies provide convincing proof-of-concept, isolated antibody fragments are not currently a viable clinical approach for disrupting intracellular signaling.
However, interfering with protein–protein interactions is, by its nature, difficult. Specifically, the binding of small molecules to protein–protein contact surfaces remains a major challenge (Scott et al. 2016). In the case of RAS and other GTPases, allosteric inhibition of protein–protein interactions may be a more plausible approach. Multiple lines of evidence point to this being the case for RAS. First, switch I acts as the main protein–protein interface, and it is known to be highly dynamic, undergoing substantial conformational rearrangements between nucleotide states. Second, mutations at sites outside the interface have been shown to affect the conformation in ways that impair effector binding. Finally, structural and NMR studies have enabled the identification of multiple inactive conformations. Compounds that bind to and stabilize these inactive states have the potential to potently and effectively inhibit RAS. One example of this kind of blockade comes from small molecules that bind to the pseudokinase “kinase suppressor of RAS” (KSR) (Brennan et al. 2011). The kinase inhibitor ASC24 binds to an inactive conformation of KSR and blocks the allosteric transition that is induced on heterodimerization of KSR with BRAF.
The switch region of RAS interacts with a great number of effector proteins that contain a common RBD. Because RBD-mediated interactions are necessary for RAS signaling, blocking RBD association with small molecules makes an attractive therapeutic course. Mutational studies of the RBD in the p110α catalytic subunit of PI3K have shown the importance of its RAS-binding domain in the establishment and maintenance of KRAS-driven lung tumors, validating this protein–protein interaction as a therapeutic target for RAS-driven cancers (Gupta et al. 2007; Castellano et al. 2013). Additionally, a recent report described an unexpected interaction between the Polo-like kinase inhibitor, Rigosertib, with several RAS effector proteins. Rigosertib acts as a RAS mimetic that binds to the RBDs of the RAS effectors and interferes with their ability to bind to RAS, resulting in the inhibition of RAS-RAF/MEK and PI3K/Akt signaling pathways. The compound is able to inhibit the interactions of both mutant and wild-type isoforms of K- or NRAS isoforms (Athuluri-Divakar et al. 2016). The real test for this drug will most likely be whether or not it has efficacy in patients with KRAS mutant tumors. This question will be solved relatively soon because multiple clinical trials (some of them in phase III) for hematologic and solid malignancies are ongoing at this time.
CONCLUDING REMARKS
RAS proteins activate multiple effector pathways. Dissecting the contributions of these interactions in normal cells and in cancer is an ongoing challenge. The role of RAS proteins in regulating PI3K has been particularly complicated, because these kinases can be activated by multiple upstream signals, some of which depend on cooperation with RAS proteins, whereas others do not. However, research over the last decade has shown, in a variety of systems, that this signaling pathway plays essential roles in both normal and oncogenic signaling. Recent data has broadened our knowledge on a RAS/PI3K signaling pathway, suggesting that this pathway is equally active in cancer cells and in the major components of the TME, controlling not only tumor growth but also tumor-associated immune responses, tumor angiogenesis, tumor-induced ECM reorganization, and tumor dissemination. The ability of RAS/PI3K signaling to promote tumorigenesis by acting both in “seed” and “soil”-dependent mechanisms indicates that therapeutic PI3K inhibition can be seen as a plausible opportunity to attack RAS-driven tumors on multiple fronts. More research is needed in this front to understand the specific molecular mechanisms by which RAS/PI3K signaling exerts control over the host responses. Targeting pathways that depend on RAS/PI3K signaling, instead of direct components of the pathway, will avoid feedback loops and could be a way forward to treat RAS-driven cancers.
ACKNOWLEDGMENTS
We thank Claudio Santos and Fran Balkwill for assistance and feedback on this review. This work is funded by Queen Mary University of London. The authors have no conflicts of interest to declare.
Footnotes
Editors: Linda VanAelst, Julian Downward, and Frank McCormick
Additional Perspectives on Ras and Cancer in the 21st Century available at www.perspectivesinmedicine.org
REFERENCES
- Aksamitiene E, Kiyatkin A, Kholodenko BN. 2012. Cross-talk between mitogenic Ras/MAPK and survival PI3K/Akt pathways: A fine balance. Biochem Soc Trans 40: 139–146. [DOI] [PubMed] [Google Scholar]
- Alagesan B, Contino G, Guimaraes AR, Corcoran RB, Deshpande V, Wojtkiewicz GR, Hezel AF, Wong KK, Loda M, Weissleder R, et al. 2015. Combined MEK and PI3K inhibition in a mouse model of pancreatic cancer. Clin Cancer Res 21: 396–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alessi DR, Saito Y, Campbell DG, Cohen P, Sithanandam G, Rapp U, Ashworth A, Marshall CJ, Cowley S. 1994. Identification of the sites in MAP kinase kinase-1 phosphorylated by p74raf-1. EMBO J 13: 1610–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asati V, Mahapatra DK, Bharti SK. 2017. K-Ras and its inhibitors towards personalized cancer treatment: Pharmacological and structural perspectives. Eur J Med Chem 125: 299–314. [DOI] [PubMed] [Google Scholar]
- Athuluri-Divakar SK, Vasquez-Del Carpio R, Dutta K, Baker SJ, Cosenza SC, Basu I, Gupta YK, Reddy MV, Ueno L, Hart JR, et al. 2016. A small molecule RAS-mimetic disrupts RAS association with effector proteins to block signaling. Cell 165: 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad AE, Kandalam V, Chakrabarti S, Wang X, Penninger JM, Davidge ST, Oudit GY, Kassiri Z. 2010. Tumor necrosis factor induces matrix metalloproteinases in cardiomyocytes and cardiofibroblasts differentially via superoxide production in a PI3Kγ-dependent manner. Am J Physiol Cell Physiol 298: C679–C692. [DOI] [PubMed] [Google Scholar]
- Barbacid M. 1987. ras genes. Annu Rev Biochem 56: 779–827. [DOI] [PubMed] [Google Scholar]
- Berry DL, Baehrecke EH. 2007. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell 131: 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Socinski MA, Burns TF. 2015. KRAS mutant lung cancer: Progress thus far on an elusive therapeutic target. Clin Transl Med 4: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL. 1989. ras oncogenes in human cancer: A review. Cancer Res 49: 4682–4689. [PubMed] [Google Scholar]
- Boya P, Reggiori F, Codogno P. 2013. Emerging regulation and functions of autophagy. Nat Cell Biol 15: 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan DF, Dar AC, Hertz NT, Chao WC, Burlingame AL, Shokat KM, Barford D. 2011. A Raf-induced allosteric transition of KSR stimulates phosphorylation of MEK. Nature 472: 366–369. [DOI] [PubMed] [Google Scholar]
- Buday L, Downward J. 2008. Many faces of Ras activation. Biochim Biophys Acta 1786: 178–187. [DOI] [PubMed] [Google Scholar]
- Campbell PM, Der CJ. 2004. Oncogenic Ras and its role in tumor cell invasion and metastasis. Semin Cancer Biol 14: 105–114. [DOI] [PubMed] [Google Scholar]
- Canel M, Serrels A, Frame MC, Brunton VG. 2013. E-cadherin-integrin crosstalk in cancer invasion and metastasis. J Cell Sci 126: 393–401. [DOI] [PubMed] [Google Scholar]
- Castellano E, Downward J. 2010. Role of RAS in the regulation of PI 3-kinase. Curr Top Microbiol Immunol 346: 143–169. [DOI] [PubMed] [Google Scholar]
- Castellano E, Downward J. 2011. RAS interaction with PI3K: More than just another effector pathway. Genes Cancer 2: 261–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano E, Sheridan C, Thin MZ, Nye E, Spencer-Dene B, Diefenbacher ME, Moore C, Kumar MS, Murillo MM, Gronroos E, et al. 2013. Requirement for interaction of PI3-kinase p110α with RAS in lung tumor maintenance. Cancer Cell 24: 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano E, Molina-Arcas M, Krygowska AA, East P, Warne P, Nicol A, Downward J. 2016. RAS signalling through PI3-kinase controls cell migration via modulation of reelin expression. Nat Commun 7: 11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cescon DW, Gorrini C, Mak TW. 2015. Breaking up is hard to do: PI3K isoforms on the rebound. Cancer Cell 27: 5–7. [DOI] [PubMed] [Google Scholar]
- Chaft JE, Arcila ME, Paik PK, Lau C, Riely GJ, Pietanza MC, Zakowski MF, Rusch V, Sima CS, Ladanyi M, et al. 2012. Coexistence of PIK3CA and other oncogene mutations in lung adenocarcinoma—Rationale for comprehensive mutation profiling. Mol Cancer Ther 11: 485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty A, Rexer BN, Wang SE, Cook RS, Engelman JA, Arteaga CL. 2010. H1047R phosphatidylinositol 3-kinase mutant enhances HER2-mediated transformation by heregulin production and activation of HER3. Oncogene 29: 5193–5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, Majumder PK, Baselga J, Rosen N. 2011. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell 19: 58–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Black SM, Catravas JD. 2008. Endothelial nitric oxide (NO) and its pathophysiologic regulation. Vascul Pharmacol 49: 134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JS, Wang Q, Fu XH, Huang XH, Chen XL, Cao LQ, Chen LZ, Tan HX, Li W, Bi J, et al. 2009. Involvement of PI3K/PTEN/AKT/mTOR pathway in invasion and metastasis in hepatocellular carcinoma: Association with MMP-9. Hepatol Res 39: 177–186. [DOI] [PubMed] [Google Scholar]
- Christoforidis S, Miaczynska M, Ashman K, Wilm M, Zhao L, Yip SC, Waterfield MD, Backer JM, Zerial M. 1999. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat Cell Biol 1: 249–252. [DOI] [PubMed] [Google Scholar]
- Clark CE, Hingorani SR, Mick R, Combs C, Tuveson DA, Vonderheide RH. 2007. Dynamics of the immune reaction to pancreatic cancer from inception to invasion. Cancer Res 67: 9518–9527. [DOI] [PubMed] [Google Scholar]
- Clark CE, Beatty GL, Vonderheide RH. 2009. Immunosurveillance of pancreatic adenocarcinoma: Insights from genetically engineered mouse models of cancer. Cancer Lett 279: 1–7. [DOI] [PubMed] [Google Scholar]
- Coleman ML, Marshall CJ, Olson MF. 2004. RAS and RHO GTPases in G1-phase cell-cycle regulation. Nat Rev Mol Cell Biol 5: 355–366. [DOI] [PubMed] [Google Scholar]
- Cox AD, Der CJ. 2003. The dark side of Ras: Regulation of apoptosis. Oncogene 22: 8999–9006. [DOI] [PubMed] [Google Scholar]
- Cox AD, Der CJ. 2010. Ras history: The saga continues. Small GTPases 1: 2–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan S, Okamura M, Seki M, Yamazaki K, Sugita H, Okui M, Mukai Y, Nishimura H, Asaka R, Nomura K, et al. 2010. Correlating phosphatidylinositol 3-kinase inhibitor efficacy with signaling pathway status: In silico and biological evaluations. Cancer Res 70: 4982–4994. [DOI] [PubMed] [Google Scholar]
- Datta SR, Brunet A, Greenberg ME. 1999. Cellular survival: A play in three Akts. Genes Dev 13: 2905–2927. [DOI] [PubMed] [Google Scholar]
- Dbouk HA, Vadas O, Shymanets A, Burke JE, Salamon RS, Khalil BD, Barrett MO, Waldo GL, Surve C, Hsueh C, et al. 2012. G protein–coupled receptor-mediated activation of p110β by Gβγ is required for cellular transformation and invasiveness. Sci Signal 5: ra89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devreotes P, Horwitz AR. 2015. Signaling networks that regulate cell migration. Cold Spring Harb Perspect Biol 7: a005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Blasio L, Gagliardi PA, Puliafito A, Primo L. 2017. Serine/threonine kinase 3-phosphoinositide-dependent protein kinase 1 (PDK1) as a key regulator of cell migration and cancer dissemination. Cancers (Basel) 9: E25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic S, Driscoll PC. 2002. Structural insight into substrate specificity and regulatory mechanisms of phosphoinositide 3-kinases. Trends Biochem Sci 27: 426–432. [DOI] [PubMed] [Google Scholar]
- Downward J. 1997. Role of phosphoinositide-3-OH kinase in Ras signaling. Adv Second Messenger Phosphoprotein Res 31: 1–10. [DOI] [PubMed] [Google Scholar]
- Downward J. 2003. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer 3: 11–22. [DOI] [PubMed] [Google Scholar]
- Downward J. 2004. PI 3-kinase, Akt and cell survival. Semin Cell Dev Biol 15: 177–182. [DOI] [PubMed] [Google Scholar]
- Dudler T, Gelb MH. 1996. Palmitoylation of Ha-Ras facilitates membrane binding, activation of downstream effectors, and meiotic maturation in Xenopus oocytes. J Biol Chem 271: 11541–11547. [DOI] [PubMed] [Google Scholar]
- Ebi H, Corcoran RB, Singh A, Chen Z, Song Y, Lifshits E, Ryan DP, Meyerhardt JA, Benes C, Settleman J, et al. 2011. Receptor tyrosine kinases exert dominant control over PI3K signaling in human KRAS mutant colorectal cancers. J Clin Invest 121: 4311–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion EA. 2001. The Ste5p scaffold. J Cell Sci 114: 3967–3978. [DOI] [PubMed] [Google Scholar]
- Eller-Borges R, Batista WL, da Costa PE, Tokikawa R, Curcio MF, Strumillo ST, Sartori A, Moraes MS, de Oliveira GA, Taha MO, et al. 2015. Ras, Rac1, and phosphatidylinositol-3-kinase (PI3K) signaling in nitric oxide induced endothelial cell migration. Nitric Oxide 47: 40–51. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M, McNamara K, Perera SA, Song Y, et al. 2008. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med 14: 1351–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig LA, Urano T, Cantor S. 1996. Evidence for a Ras/Ral signaling cascade. Trends Biochem Sci 21: 438–441. [DOI] [PubMed] [Google Scholar]
- Fernandes MS, Melo S, Velho S, Carneiro P, Carneiro F, Seruca R. 2016. Specific inhibition of p110α subunit of PI3K: Putative therapeutic strategy for KRAS mutant colorectal cancers. Oncotarget 7: 68546–68558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Medarde A, Santos E. 2011. Ras in cancer and developmental diseases. Genes Cancer 2: 344–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filchtinski D, Sharabi O, Ruppel A, Vetter IR, Herrmann C, Shifman JM. 2010. What makes Ras an efficient molecular switch: A computational, biophysical, and structural study of Ras-GDP interactions with mutants of Raf. J Mol Biol 399: 422–435. [DOI] [PubMed] [Google Scholar]
- Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al. 2011. COSMIC: Mining complete cancer genomes in the catalogue of somatic mutations in cancer. Nucleic Acids Res 39: D945–D950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch SM, Vuori K, Ruoslahti E, Chan-Hui PY. 1996. Control of adhesion-dependent cell survival by focal adhesion kinase. J Cell Biol 134: 793–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch R, Downward J. 2013. SnapShot: Class I PI3K isoform signaling. Cell 154: 940–940.e1. [DOI] [PubMed] [Google Scholar]
- Fritsch R, de Krijger I, Fritsch K, George R, Reason B, Kumar MS, Diefenbacher M, Stamp G, Downward J. 2013. RAS and RHO families of GTPases directly regulate distinct phosphoinositide 3-kinase isoforms. Cell 153: 1050–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruman DA, Bismuth G. 2009. Fine tuning the immune response with PI3K. Immunol Rev 228: 253–272. [DOI] [PubMed] [Google Scholar]
- Fruman DA, Rommel C. 2014. PI3K and cancer: Lessons, challenges and opportunities. Nat Rev Drug Discov 13: 140–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes-Calvo I, Blazquez-Medela AM, Eleno N, Santos E, Lopez-Novoa JM, Martinez-Salgado C. 2012. H-Ras isoform modulates extracellular matrix synthesis, proliferation, and migration in fibroblasts. Am J Physiol Cell Physiol 302: C686–C697. [DOI] [PubMed] [Google Scholar]
- Fuentes-Calvo I, Crespo P, Santos E, Lopez-Novoa JM, Martinez-Salgado C. 2013. The small GTPase N-Ras regulates extracellular matrix synthesis, proliferation and migration in fibroblasts. Biochim Biophys Acta 1833: 2734–2744. [DOI] [PubMed] [Google Scholar]
- Furuta S, Hidaka E, Ogata A, Yokota S, Kamata T. 2004. Ras is involved in the negative control of autophagy through the class I PI3-kinase. Oncogene 23: 3898–3904. [DOI] [PubMed] [Google Scholar]
- Garlich JR, De P, Dey N, Su JD, Peng X, Miller A, Murali R, Lu Y, Mills GB, Kundra V, et al. 2008. A vascular targeted pan phosphoinositide 3-kinase inhibitor prodrug, SF1126, with antitumor and antiangiogenic activity. Cancer Res 68: 206–215. [DOI] [PubMed] [Google Scholar]
- Giehl K. 2005. Oncogenic Ras in tumour progression and metastasis. Biol Chem 386: 193–205. [DOI] [PubMed] [Google Scholar]
- Gil-Bazo I, Sweet-Cordero A, Vicent S. 2016. The present and the future of the RAS pathway: From function and genomics to inhibition. Cancer Biol Ther 17: 719–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graupera M, Guillermet-Guibert J, Foukas LC, Phng LK, Cain RJ, Salpekar A, Pearce W, Meek S, Millan J, Cutillas PR, et al. 2008. Angiogenesis selectively requires the p110α isoform of PI3K to control endothelial cell migration. Nature 453: 662–666. [DOI] [PubMed] [Google Scholar]
- Gritsman K, Yuzugullu H, Von T, Yan H, Clayton L, Fritsch C, Maira SM, Hollingworth G, Choi C, Khandan T, et al. 2014. Hematopoiesis and RAS-driven myeloid leukemia differentially require PI3K isoform p110α. J Clin Invest 124: 1794–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillermet-Guibert J, Bjorklof K, Salpekar A, Gonella C, Ramadani F, Bilancio A, Meek S, Smith AJ, Okkenhaug K, Vanhaesebroeck B. 2008. The p110β isoform of phosphoinositide 3-kinase signals downstream of G protein-coupled receptors and is functionally redundant with p110γ. Proc Natl Acad Sci 105: 8292–8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, Nicke B, Nye E, Stamp G, Alitalo K, Downward J. 2007. Binding of ras to phosphoinositide 3-kinase p110α is required for Ras-driven tumorigenesis in mice. Cell 129: 957–968. [DOI] [PubMed] [Google Scholar]
- Gyori D, Chessa T, Hawkins PT, Stephens LR. 2017. Class (I) phosphoinositide 3-kinases in the tumor microenvironment. Cancers (Basel) 9: E24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gysin S, Salt M, Young A, McCormick F. 2011. Therapeutic strategies for targeting ras proteins. Genes Cancer 2: 359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamad NM, Elconin JH, Karnoub AE, Bai W, Rich JN, Abraham RT, Der CJ, Counter CM. 2002. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev 16: 2045–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Luby-Phelps K, Das B, Shu X, Xia Y, Mosteller RD, Krishna UM, Falck JR, White MA, Broek D. 1998. Role of substrates and products of PI 3-kinase in regulating activation of Rac-related guanosine triphosphatases by Vav. Science 279: 558–560. [DOI] [PubMed] [Google Scholar]
- Hao Y, Wang C, Cao B, Hirsch BM, Song J, Markowitz SD, Ewing RM, Sedwick D, Liu L, Zheng W, et al. 2013. Gain of interaction with IRS1 by p110α–helical domain mutants is crucial for their oncogenic functions. Cancer Cell 23: 583–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmings BA, Restuccia DF. 2015. The PI3K-PKB/Akt pathway. Cold Spring Harb Perspect Biol 7: a026609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann C. 2003. Ras-effector interactions: After one decade. Curr Opin Struct Biol 13: 122–129. [DOI] [PubMed] [Google Scholar]
- Hirakawa T, Ruley HE. 1988. Rescue of cells from ras oncogene-induced growth arrest by a second, complementing, oncogene. Proc Natl Acad Sci 85: 1519–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeflich KP, O’Brien C, Boyd Z, Cavet G, Guerrero S, Jung K, Januario T, Savage H, Punnoose E, Truong T, et al. 2009. In vivo antitumor activity of MEK and phosphatidylinositol 3-kinase inhibitors in basal-like breast cancer models. Clin Cancer Res 15: 4649–4664. [DOI] [PubMed] [Google Scholar]
- Hofer F, Fields S, Schneider C, Martin GS. 1994. Activated Ras interacts with the Ral guanine nucleotide dissociation stimulator. Proc Natl Acad Sci 91: 11089–11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutti JE, Pfefferle AD, Russell SC, Sircar M, Perou CM, Baldwin AS. 2012. Oncogenic PI3K mutations lead to NF-κB-dependent cytokine expression following growth factor deprivation. Cancer Res 72: 3260–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihle NT, Lemos R Jr., Wipf P, Yacoub A, Mitchell C, Siwak D, Mills GB, Dent P, Kirkpatrick DL, Powis G. 2009. Mutations in the phosphatidylinositol-3-kinase pathway predict for antitumor activity of the inhibitor PX-866 whereas oncogenic Ras is a dominant predictor for resistance. Cancer Res 69: 143–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihle NT, Byers LA, Kim ES, Saintigny P, Lee JJ, Blumenschein GR, Tsao A, Liu S, Larsen JE, Wang J, et al. 2012. Effect of KRAS oncogene substitutions on protein behavior: Implications for signaling and clinical outcome. J Natl Cancer Inst 104: 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janku F, Lee JJ, Tsimberidou AM, Hong DS, Naing A, Falchook GS, Fu S, Luthra R, Garrido-Laguna I, Kurzrock R. 2011. PIK3CA mutations frequently coexist with RAS and BRAF mutations in patients with advanced cancers. PLoS ONE 6: e22769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jifu E, Xing J, Gong H, He J, Zhang W. 2015. Combine MEK inhibition with PI3K/mTOR inhibition exert inhibitory tumor growth effect on KRAS and PIK3CA mutation CRC xenografts due to reduced expression of VEGF and matrix metallopeptidase-9. Tumour Biol 36: 1091–1097. [DOI] [PubMed] [Google Scholar]
- Kaduwal S, Jeong WJ, Park JC, Lee KH, Lee YM, Jeon SH, Lim YB, Min do S, Choi KY. 2015. Sur8/Shoc2 promotes cell motility and metastasis through activation of Ras-PI3K signaling. Oncotarget 6: 33091–33105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Bader AG, Vogt PK. 2005. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci 102: 802–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardash E, Reichman-Fried M, Maitre JL, Boldajipour B, Papusheva E, Messerschmidt EM, Heisenberg CP, Raz E. 2010. A role for Rho GTPases and cell–cell adhesion in single-cell motility in vivo. Nat Cell Biol 12: 47–53. [DOI] [PubMed] [Google Scholar]
- Karnoub AE, Weinberg RA. 2008. Ras oncogenes: Split personalities. Nat Rev Mol Cell Biol 9: 517–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katso R, Okkenhaug K, Ahmadi K, White S, Timms J, Waterfield MD. 2001. Cellular function of phosphoinositide 3-kinases: Implications for development, homeostasis, and cancer. Annu Rev Cell Dev Biol 17: 615–675. [DOI] [PubMed] [Google Scholar]
- Keeton AB, Salter EA, Piazza GA. 2017. The RAS–effector interaction as a drug target. Cancer Res 77: 221–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khwaja A, Rodriguez-Viciana P, Wennstrom S, Warne PH, Downward J. 1997. Matrix adhesion and Ras transformation both activate a phosphoinositide 3-OH kinase and protein kinase B/Akt cellular survival pathway. EMBO J 16: 2783–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel C, Wohlgemuth S, Rousseau F, Schymkowitz J, Ferkinghoff-Borg J, Wittinghofer F, Serrano L. 2005. Recognizing and defining true Ras binding domains II: In silico prediction based on homology modelling and energy calculations. J Mol Biol 348: 759–775. [DOI] [PubMed] [Google Scholar]
- Kim S, Huang W, Mottillo EP, Sohail A, Ham YA, Conley-Lacomb MK, Kim CJ, Tzivion G, Kim HR, Wang S, et al. 2010. Posttranslational regulation of membrane type 1-matrix metalloproteinase (MT1-MMP) in mouse PTEN null prostate cancer cells: Enhanced surface expression and differential O-glycosylation of MT1-MMP. Biochim Biophys Acta 1803: 1287–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land H, Parada LF, Weinberg RA. 1983a. Cellular oncogenes and multistep carcinogenesis. Science 222: 771–778. [DOI] [PubMed] [Google Scholar]
- Land H, Parada LF, Weinberg RA. 1983b. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature 304: 596–602. [DOI] [PubMed] [Google Scholar]
- Lane HA, Wood JM, McSheehy PM, Allegrini PR, Boulay A, Brueggen J, Littlewood-Evans A, Maira SM, Martiny-Baron G, Schnell CR, et al. 2009. mTOR inhibitor RAD001 (everolimus) has antiangiogenic/vascular properties distinct from a VEGFR tyrosine kinase inhibitor. Clin Cancer Res 15: 1612–1622. [DOI] [PubMed] [Google Scholar]
- Lee YM, Kaduwal S, Lee KH, Park JC, Jeong WJ, Choi KY. 2016. Sur8 mediates tumorigenesis and metastasis in colorectal cancer. Exp Mol Med 48: e249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Gorfe AA. 2012. What drives the clustering of membrane-bound Ras? Small GTPases 3: 244–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Han M, Guan KL. 2000. The leucine-rich repeat protein SUR-8 enhances MAP kinase activation and forms a complex with Ras and Raf. Genes Dev 14: 895–900. [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhu T, Guan KL. 2004. Transformation potential of Ras isoforms correlates with activation of phosphatidylinositol 3-kinase but not ERK. J Biol Chem 279: 37398–37406. [DOI] [PubMed] [Google Scholar]
- Ligresti G, Militello L, Steelman LS, Cavallaro A, Basile F, Nicoletti F, Stivala F, McCubrey JA, Libra M. 2009. PIK3CA mutations in human solid tumors: Role in sensitivity to various therapeutic approaches. Cell Cycle 8: 1352–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhu S, Gong Z, Low BC. 2008. K-ras/PI3K-Akt signaling is essential for zebrafish hematopoiesis and angiogenesis. PLoS ONE 3: e2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CB, Meiri D, Smith MJ, Mazhab-Jafari MT, Gasmi-Seabrook GM, Rottapel R, Stambolic V, Ikura M. 2012a. Probing the GTPase cycle with real-time NMR: GAP and GEF activities in cell extracts. Methods 57: 473–485. [DOI] [PubMed] [Google Scholar]
- Marshall NA, Galvin KC, Corcoran AM, Boon L, Higgs R, Mills KH. 2012b. Immunotherapy with PI3K inhibitor and Toll-like receptor agonist induces IFN-γ+IL-17+ polyfunctional T cells that mediate rejection of murine tumors. Cancer Res 72: 581–591. [DOI] [PubMed] [Google Scholar]
- Marte BM, Rodriguez-Viciana P, Wennstrom S, Warne PH, Downward J. 1997. R-Ras can activate the phosphoinositide 3-kinase but not the MAP kinase arm of the Ras effector pathways. Curr Biol 7: 63–70. [DOI] [PubMed] [Google Scholar]
- Martini M, De Santis MC, Braccini L, Gulluni F, Hirsch E. 2014. PI3K/AKT signaling pathway and cancer: An updated review. Ann Med 46: 372–383. [DOI] [PubMed] [Google Scholar]
- Maurer T, Garrenton LS, Oh A, Pitts K, Anderson DJ, Skelton NJ, Fauber BP, Pan B, Malek S, Stokoe D, et al. 2012. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc Natl Acad Sci 109: 5299–5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer IA, Arteaga CL. 2016. The PI3K/AKT pathway as a target for cancer treatment. Annu Rev Med 67: 11–28. [DOI] [PubMed] [Google Scholar]
- Mendoza MC, Er EE, Blenis J. 2011. The Ras-ERK and PI3K-mTOR pathways: Cross-talk and compensation. Trends Biochem Sci 36: 320–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RG, Korn S, Micke P, Becker K, Huber C, Wolfel T, Buhl R. 2007. An open-label, prospective phase I/II study evaluating the immunogenicity and safety of a ras peptide vaccine plus GM-CSF in patients with non-small cell lung cancer. Lung Cancer 58: 88–94. [DOI] [PubMed] [Google Scholar]
- Mirzoeva OK, Das D, Heiser LM, Bhattacharya S, Siwak D, Gendelman R, Bayani N, Wang NJ, Neve RM, Guan Y, et al. 2009. Basal subtype and MAPK/ERK kinase (MEK)-phosphoinositide 3-kinase feedback signaling determine susceptibility of breast cancer cells to MEK inhibition. Cancer Res 69: 565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. 2007. Autophagy: Process and function. Genes Dev 21: 2861–2873. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B. 2010. Autophagy in mammalian development and differentiation. Nat Cell Biol 12: 823–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Arcas M, Hancock DC, Sheridan C, Kumar MS, Downward J. 2013. Coordinate direct input of both KRAS and IGF1 receptor to activation of PI3 kinase in KRAS-mutant lung cancer. Cancer Discov 3: 548–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moodie SA, Willumsen BM, Weber MJ, Wolfman A. 1993. Complexes of Ras·GTP with Raf-1 and mitogen-activated protein kinase kinase. Science 260: 1658–1661. [DOI] [PubMed] [Google Scholar]
- Morrison DK, Davis RJ. 2003. Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu Rev Cell Dev Biol 19: 91–118. [DOI] [PubMed] [Google Scholar]
- Muranen T, Selfors LM, Worster DT, Iwanicki MP, Song L, Morales FC, Gao S, Mills GB, Brugge JS. 2012. Inhibition of PI3K/mTOR leads to adaptive resistance in matrix-attached cancer cells. Cancer Cell 21: 227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murillo MM, Zelenay S, Nye E, Castellano E, Lassailly F, Stamp G, Downward J. 2014. RAS interaction with PI3K p110α is required for tumor-induced angiogenesis. J Clin Invest 124: 3601–3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy LO, Blenis J. 2006. MAPK signal specificity: The right place at the right time. Trends Biochem Sci 31: 268–275. [DOI] [PubMed] [Google Scholar]
- Nezis IP, Vaccaro MI, Devenish RJ, Juhasz G. 2014. Autophagy in development, cell differentiation, and homeodynamics: From molecular mechanisms to diseases and pathophysiology. Biomed Res Int 2014: 349623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimnual AS, Yatsula BA, Bar-Sagi D. 1998. Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science 279: 560–563. [DOI] [PubMed] [Google Scholar]
- Oda K, Okada J, Timmerman L, Rodriguez-Viciana P, Stokoe D, Shoji K, Taketani Y, Kuramoto H, Knight ZA, Shokat KM, et al. 2008. PIK3CA cooperates with other phosphatidylinositol 3′-kinase pathway mutations to effect oncogenic transformation. Cancer Res 68: 8127–8136. [DOI] [PubMed] [Google Scholar]
- Okkenhaug K. 2013. Signaling by the phosphoinositide 3-kinase family in immune cells. Annu Rev Immunol 31: 675–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkenhaug K, Graupera M, Vanhaesebroeck B. 2016. Targeting PI3K in cancer: Impact on tumor cells, their protective stroma, angiogenesis, and immunotherapy. Cancer Discov 6: 1090–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SH, Hadari YR, Gotoh N, Guy GR, Schlessinger J, Lax I. 2001. Stimulation of phosphatidylinositol 3-kinase by fibroblast growth factor receptors is mediated by coordinated recruitment of multiple docking proteins. Proc Natl Acad Sci 98: 6074–6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrem JM, Shokat KM. 2016. Direct small-molecule inhibitors of KRAS: From structural insights to mechanism-based design. Nat Rev Drug Discov 15: 771–785. [DOI] [PubMed] [Google Scholar]
- Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. 2013. K-RasG12C inhibitors allosterically control GTP affinity and effector interactions. Nature 503: 548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacold ME, Suire S, Perisic O, Lara-Gonzalez S, Davis CT, Walker EH, Hawkins PT, Stephens L, Eccleston JF, Williams RL. 2000. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase γ. Cell 103: 931–943. [DOI] [PubMed] [Google Scholar]
- Pang H, Flinn R, Patsialou A, Wyckoff J, Roussos ET, Wu H, Pozzuto M, Goswami S, Condeelis JS, Bresnick AR, et al. 2009. Differential enhancement of breast cancer cell motility and metastasis by helical and kinase domain mutations of class IA phosphoinositide 3-kinase. Cancer Res 69: 8868–8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoli P, Giannoni E, Chiarugi P. 2013. Anoikis molecular pathways and its role in cancer progression. Biochem Biophys Acta 1833: 3481–3498. [DOI] [PubMed] [Google Scholar]
- Parker JA, Mattos C. 2015. The Ras-membrane interface: Isoform-specific differences in the catalytic domain. Mol Cancer Res 13: 595–603. [DOI] [PubMed] [Google Scholar]
- Parsons DW, Wang TL, Samuels Y, Bardelli A, Cummins JM, DeLong L, Silliman N, Ptak J, Szabo S, Willson JK, et al. 2005. Colorectal cancer: Mutations in a signalling pathway. Nature 436: 792. [DOI] [PubMed] [Google Scholar]
- Polivka J Jr, Janku F. 2014. Molecular targets for cancer therapy in the PI3K/AKT/mTOR pathway. Pharmacol Ther 142: 164–175. [DOI] [PubMed] [Google Scholar]
- Pollock CB, Shirasawa S, Sasazuki T, Kolch W, Dhillon AS. 2005. Oncogenic K-RAS is required to maintain changes in cytoskeletal organization, adhesion, and motility in colon cancer cells. Cancer Res 65: 1244–1250. [DOI] [PubMed] [Google Scholar]
- Prior IA, Lewis PD, Mattos C. 2012. A comprehensive survey of Ras mutations in cancer. Cancer Res 72: 2457–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K, Der CJ. 2001. Ras and Rho regulation of the cell cycle and oncogenesis. Cancer Lett 171: 1–10. [DOI] [PubMed] [Google Scholar]
- Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. 2011. RAS oncogenes: Weaving a tumorigenic web. Nat Rev Cancer 11: 761–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Wu X, Yin Y, Yang Y, Ma D, Li H. 2014. Antitumor activity of selective MEK1/2 inhibitor AZD6244 in combination with PI3K/mTOR inhibitor BEZ235 in gefitinib-resistant NSCLC xenograft models. J Exp Clin Cancer Res 33: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilliam LA, Khosravi-Far R, Huff SY, Der CJ. 1995. Guanine nucleotide exchange factors: Activators of the Ras superfamily of proteins. BioEssays 17: 395–404. [DOI] [PubMed] [Google Scholar]
- Rahma OE, Hamilton JM, Wojtowicz M, Dakheel O, Bernstein S, Liewehr DJ, Steinberg SM, Khleif SN. 2014. The immunological and clinical effects of mutated ras peptide vaccine in combination with IL-2, GM-CSF, or both in patients with solid tumors. J Transl Med 12: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajalingam K, Schreck R, Rapp UR, Albert S. 2007. Ras oncogenes and their downstream targets. Biochem Biophys Res Commun 1773: 1177–1195. [DOI] [PubMed] [Google Scholar]
- Riely GJ, Marks J, Pao W. 2009. KRAS mutations in non-small cell lung cancer. Proc Am Thoracic Soc 6: 201–205. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Warne PH, Dhand R, Vanhaesebroeck B, Gout I, Fry MJ, Waterfield MD, Downward J. 1994. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature 370: 527–532. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Marte BM, Warne PH, Downward J. 1996a. Phosphatidylinositol 3′ kinase: One of the effectors of Ras. Phil Trans R Soc Lond B 351: 225–231; discussion 231–222. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Warne PH, Vanhaesebroeck B, Waterfield MD, Downward J. 1996b. Activation of phosphoinositide 3-kinase by interaction with Ras and by point mutation. EMBO J 15: 2442–2451. [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Warne PH, Khwaja A, Marte BM, Pappin D, Das P, Waterfield MD, Ridley A, Downward J. 1997. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell 89: 457–467. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Sabatier C, McCormick F. 2004. Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors they regulate. Mol Cell Biol 24: 4943–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrik-Outmezguine VS, Chandarlapaty S, Pagano NC, Poulikakos PI, Scaltriti M, Moskatel E, Baselga J, Guichard S, Rosen N. 2011. mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer Discov 1: 248–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas JM, Oliva JL, Santos E. 2011. Mammalian son of sevenless guanine nucleotide exchange factors: Old concepts and new perspectives. Genes Cancer 2: 298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskoski R Jr. 2010. RAF protein-serine/threonine kinases: Structure and regulation. Biochem Biophys Res Commun 399: 313–317. [DOI] [PubMed] [Google Scholar]
- Ruley HE. 1983. Adenovirus early region 1A enables viral and cellular transforming genes to transform primary cells in culture. Nature 304: 602–606. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Diaz LA Jr., Schmidt-Kittler O, Cummins JM, Delong L, Cheong I, Rago C, Huso DL, Lengauer C, Kinzler KW, et al. 2005. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell 7: 561–573. [DOI] [PubMed] [Google Scholar]
- Sasaki AT, Firtel RA. 2006. Regulation of chemotaxis by the orchestrated activation of Ras, PI3K, and TOR. Eur J Cell Biol 85: 873–895. [DOI] [PubMed] [Google Scholar]
- Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, Gao S, Puigserver P, Brugge JS. 2009. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature 461: 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmukler E, Kloog Y, Pinkas-Kramarski R. 2014. Ras and autophagy in cancer development and therapy. Oncotarget 5: 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell CR, Stauffer F, Allegrini PR, O’Reilly T, McSheehy PM, Dartois C, Stumm M, Cozens R, Littlewood-Evans A, Garcia-Echeverria C, et al. 2008. Effects of the dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor NVP-BEZ235 on the tumor vasculature: Implications for clinical imaging. Cancer Res 68: 6598–6607. [DOI] [PubMed] [Google Scholar]
- Schrauwen S, Depreeuw J, Coenegrachts L, Hermans E, Lambrechts D, Amant F. 2015. Dual blockade of PI3K/AKT/mTOR (NVP-BEZ235) and Ras/Raf/MEK (AZD6244) pathways synergistically inhibit growth of primary endometrioid endometrial carcinoma cultures, whereas NVP-BEZ235 reduces tumor growth in the corresponding xenograft models. Gynecol Oncol 138: 165–173. [DOI] [PubMed] [Google Scholar]
- Scott DE, Bayly AR, Abell C, Skidmore J. 2016. Small molecules, big targets: Drug discovery faces the protein-protein interaction challenge. Nat Rev Drug Discov 15: 533–550. [DOI] [PubMed] [Google Scholar]
- Shao J, Yang X, Liu T, Zhang T, Xie QR, Xia W. 2016. Autophagy induction by SIRT6 is involved in oxidative stress-induced neuronal damage. Protein Cell 7: 281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng H, Shao J, DuBois RN. 2001. Akt/PKB activity is required for Ha-Ras-mediated transformation of intestinal epithelial cells. J Biol Chem 276: 14498–14504. [DOI] [PubMed] [Google Scholar]
- Shieh A, Ward AF, Donlan KL, Harding-Theobald ER, Xu J, Mullighan CG, Zhang C, Chen SC, Su X, Downing JR, et al. 2013. Defective K-Ras oncoproteins overcome impaired effector activation to initiate leukemia in vivo. Blood 121: 4884–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H, Longo DL, Chabner BA. 2015. Improving prospects for targeting RAS. J Clin Oncol 33: 3650–3659. [DOI] [PubMed] [Google Scholar]
- Smith MJ, Ikura M. 2014. Integrated RAS signaling defined by parallel NMR detection of effectors and regulators. Nat Chem Biol 10: 223–230. [DOI] [PubMed] [Google Scholar]
- So L, Yea SS, Oak JS, Lu M, Manmadhan A, Ke QH, Janes MR, Kessler LV, Kucharski JM, Li LS, et al. 2013. Selective inhibition of phosphoinositide 3-kinase p110α preserves lymphocyte function. J Biol Chem 288: 5718–5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler A, Serra H, Pearce W, Angulo A, Guillermet-Guibert J, Friedman LS, Vinals F, Gerhardt H, Casanovas O, Graupera M, et al. 2013. Inhibition of the p110α isoform of PI 3-kinase stimulates nonfunctional tumor angiogenesis. J Exp Med 210: 1937–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler A, Angulo-Urarte A, Graupera M. 2015. PI3K at the crossroads of tumor angiogenesis signaling pathways. Mol Cell Oncol 2: e975624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sos ML, Fischer S, Ullrich R, Peifer M, Heuckmann JM, Koker M, Heynck S, Stuckrath I, Weiss J, Fischer F, et al. 2009. Identifying genotype-dependent efficacy of single and combined PI3K- and MAPK-pathway inhibition in cancer. Proc Natl Acad Sci 106: 18351–18356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen AG, Esposito D, Bagni RK, McCormick F. 2014. Dragging Ras back in the ring. Cancer Cell 25: 272–281. [DOI] [PubMed] [Google Scholar]
- Stites EC, Ravichandran KS. 2009. A systems perspective of Ras signaling in cancer. Clin Cancer Res 15: 1510–1513. [DOI] [PubMed] [Google Scholar]
- Suire S, Condliffe AM, Ferguson GJ, Ellson CD, Guillou H, Davidson K, Welch H, Coadwell J, Turner M, Chilvers ER, et al. 2006. Gβγs and the Ras binding domain of p110γ are both important regulators of PI3Kγ signalling in neutrophils. Nat Cell Biol 8: 1303–1309. [DOI] [PubMed] [Google Scholar]
- Sun Q, Burke JP, Phan J, Burns MC, Olejniczak ET, Waterson AG, Lee T, Rossanese OW, Fesik SW. 2012. Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew Chem Int Ed Engl 51: 6140–6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima A, Faller DV. 2013. Targeting the RAS oncogene. Expert Opin Ther Targets 17: 507–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaya A, Kamio T, Masuda M, Mochizuki N, Sawa H, Sato M, Nagashima K, Mizutani A, Matsuno A, Kiyokawa E, et al. 2007. R-Ras regulates exocytosis by Rgl2/Rlf-mediated activation of RalA on endosomes. Mol Biol Cell 18: 1850–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert EE, Yang J, Mace TA, Farren MR, Farris AB, Young GS, Elnaggar O, Che Z, Timmers CD, Rajasekera P, et al. 2017. Dual inhibition of MEK and PI3K/Akt rescues cancer cachexia through both tumor-extrinsic and -intrinsic activities. Mol Cancer Ther 16: 344–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamanoi F, Hsueh EC, Goodman LE, Cobitz AR, Detrick RJ, Brown WR, Fujiyama A. 1988. Posttranslational modification of ras proteins: Detection of a modification prior to fatty acid acylation and cloning of a gene responsible for the modification. J Cell Biochem 36: 261–273. [DOI] [PubMed] [Google Scholar]
- Tan J, Yu Q. 2013. Molecular mechanisms of tumor resistance to PI3K-mTOR-targeted therapy. Chin J Cancer 32: 376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Rabbitts TH. 2010. Interfering with RAS-effector protein interactions prevent RAS-dependent tumour initiation and causes stop-start control of cancer growth. Oncogene 29: 6064–6070. [DOI] [PubMed] [Google Scholar]
- Thorpe LM, Yuzugullu H, Zhao JJ. 2015. PI3K in cancer: Divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer 15: 7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trahey M, McCormick F. 1987. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science 238: 542–545. [DOI] [PubMed] [Google Scholar]
- Tran Thang NN, Derouazi M, Philippin G, Arcidiaco S, Di Berardino-Besson W, Masson F, Hoepner S, Riccadonna C, Burkhardt K, Guha A, et al. 2010. Immune infiltration of spontaneous mouse astrocytomas is dominated by immunosuppressive cells from early stages of tumor development. Cancer Res 70: 4829–4839. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. 2010a. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol 11: 329–341. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Vogt PK, Rommel C. 2010b. PI3K: From the bench to the clinic and back. Curr Top Microbiol Immunol 347: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Stephens L, Hawkins P. 2012. PI3K signalling: The path to discovery and understanding. Nat Rev Mol Cell Biol 13: 195–203. [DOI] [PubMed] [Google Scholar]
- Vanneman M, Dranoff G. 2012. Combining immunotherapy and targeted therapies in cancer treatment. Nat Rev Cancer 12: 237–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigil D, Cherfils J, Rossman KL, Der CJ. 2010. Ras superfamily GEFs and GAPs: Validated and tractable targets for cancer therapy? Nat Rev Cancer 10: 842–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker EH, Perisic O, Ried C, Stephens L, Williams RL. 1999. Structural insights into phosphoinositide 3-kinase catalysis and signalling. Nature 402: 313–320. [DOI] [PubMed] [Google Scholar]
- Wang C, Wang Y, McNutt MA, Zhu WG. 2011. Autophagy process is associated with anti-neoplastic function. Acta Biochim Biophys Sin (Shanghai) 43: 425–432. [DOI] [PubMed] [Google Scholar]
- Wang W, Fang G, Rudolph J. 2012. Ras inhibition via direct Ras binding—Is there a path forward? Bioorg Med Chem Lett 22: 5766–5776. [DOI] [PubMed] [Google Scholar]
- Wang L, Hu H, Pan Y, Wang R, Li Y, Shen L, Yu Y, Li H, Cai D, Sun Y, et al. 2014. PIK3CA mutations frequently coexist with EGFR/KRAS mutations in non-small cell lung cancer and suggest poor prognosis in EGFR/KRAS wildtype subgroup. PLoS ONE 9: e88291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warne PH, Viciana PR, Downward J. 1993. Direct interaction of Ras and the amino-terminal region of Raf-1 in vitro. Nature 364: 352–355. [DOI] [PubMed] [Google Scholar]
- Wohlgemuth S, Kiel C, Kramer A, Serrano L, Wittinghofer F, Herrmann C. 2005. Recognizing and defining true Ras binding domains. I: Biochemical analysis. J Mol Biol 348: 741–758. [DOI] [PubMed] [Google Scholar]
- Wood KC. 2015. Mapping the pathways of resistance to targeted therapies. Cancer Res 75: 4247–4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HW, Shin MG, Lee S, Kim JR, Park WS, Cho KH, Meyer T, Heo WD. 2012. Cooperative activation of PI3K by Ras and Rho family small GTPases. Mol Cell 47: 281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S, Seger R. 2006. The extracellular signal-regulated kinase: Multiple substrates regulate diverse cellular functions. Growth Factors 24: 21–44. [DOI] [PubMed] [Google Scholar]
- Yuan TL, Cantley LC. 2008. PI3K pathway alterations in cancer: Variations on a theme. Oncogene 27: 5497–5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan TL, Choi HS, Matsui A, Benes C, Lifshits E, Luo J, Frangioni JV, Cantley LC. 2008. Class 1A PI3K regulates vessel integrity during development and tumorigenesis. Proc Natl Acad Sci 105: 9739–9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrabi K, Dufour A, Li J, Kuscu C, Pulkoski-Gross A, Zhi J, Hu Y, Sampson NS, Zucker S, Cao J. 2011. Inhibition of matrix metalloproteinase 14 (MMP-14)-mediated cancer cell migration. J Biol Chem 286: 33167–33177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Lodish HF. 2005. Identification of K-ras as the major regulator for cytokine-dependent Akt activation in erythroid progenitors in vivo. Proc Natl Acad Sci 102: 14605–14610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Tang N, Hadden TJ, Rishi AK. 2011. Akt, FoxO and regulation of apoptosis. Biochim Biophys Acta 1813: 1978–1986. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zheng J, Yang Y, Lu J, Gao J, Lu T, Sun J, Jiang H, Zhu Y, Zheng Y, et al. 2015. Molecular spectrum of KRAS, NRAS, BRAF and PIK3CA mutations in Chinese colorectal cancer patients: Analysis of 1,110 cases. Sci Rep 5: 18678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Vogt PK. 2008. Class I PI3K in oncogenic cellular transformation. Oncogene 27: 5486–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Vogt PK. 2010. Hot-spot mutations in p110α of phosphatidylinositol 3-kinase (pI3K): Differential interactions with the regulatory subunit p85 and with RAS. Cell Cycle 9: 596–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JJ, Cheng H, Jia S, Wang L, Gjoerup OV, Mikami A, Roberts TM. 2006. The p110α isoform of PI3K is essential for proper growth factor signaling and oncogenic transformation. Proc Natl Acad Sci 103: 16296–16300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liang H, Rodkey T, Ariotti N, Parton RG, Hancock JF. 2014. Signal integration by lipid-mediated spatial cross talk between Ras nanoclusters. Mol Cell Biol 34: 862–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitvogel L, Galluzzi L, Smyth MJ, Kroemer G. 2013. Mechanism of action of conventional and targeted anticancer therapies: Reinstating immunosurveillance. Immunity 39: 74–88. [DOI] [PubMed] [Google Scholar]