Abstract
Two sisters (ages 16 yr and 15 yr) have been followed by our clinical genetics team for several years. Both girls have severe intellectual disability, hypotonia, seizures, and distinctive craniofacial features. The parents are healthy and have no other children. Oligo array, fragile X testing, and numerous single-gene tests were negative. All four family members underwent research exome sequencing, which revealed a heterozygous nonsense mutation in ASXL3 (p.R1036X) that segregated with disease. Exome data and independent Sanger sequencing confirmed that the variant is de novo, suggesting possible germline mosaicism in one parent. The p.R1036X variant has never been observed in healthy human populations and has been previously reported as a pathogenic mutation. Truncating de novo mutations in ASXL3 cause Bainbridge–Ropers syndrome (BRPS), a developmental disorder with similarities to Bohring–Opitz syndrome. Fewer than 30 BRPS patients have been described in the literature; to our knowledge, this is the first report of the disorder in two related individuals. Our findings lend further support to intellectual disability, absent speech, autistic traits, hypotonia, and distinctive facial appearance as common emerging features of Bainbridge–Ropers syndrome.
Keywords: absent speech; aplasia/hypoplasia of the corpus callosum; autism; broad nasal tip; cleft of chin; clinodactyly of the 5th finger; downslanted palpebral fissures; generalized hirsutism; high forehead; high, narrow palate; hypertelorism; incisor macrodontia; intellectual disability, severe; prominent nasal bridge; recurrent hand flapping; severe global developmental delay; severe muscular hypotonia; short stature; thick eyebrow
CASE PRESENTATION
Patient 1 is a 16-yr-old Caucasian female born at term after an uncomplicated pregnancy. Her 15-yr-old sister was born at 36 weeks’ gestation by emergency cesarean section due to fetal distress. Both sisters have severe intellectual disability, no language, autistic features, hypotonia, and a thin habitus. They share distinctive craniofacial features including a broad/prominent forehead, hypertelorism, downslanting palpebral fissures, prominent nasal root, thick eyebrows, hirsutism, cleft chin, and strikingly prominent upper central incisors (Table 1). They have a history of feeding difficulties, seizures, and developmental delay. The parents were unaffected, and there was no relevant family history.
Table 1.
Clinical features of proband (16-yr-old female) and affected sister (15 yr old)
| Phenotypic feature | Patient 1 (proband) | Patient 2 (sibling) |
|---|---|---|
| Intellectual disability | Yes | Yes |
| Seizures | Yes | Yes |
| Autistic features | Yes | Yes |
| Global developmental delay | Yes | Yes |
| Language impairment | No speech | No speech |
| Recurrent hand flapping | Yes | Yes |
| Hypoplasia of corpus callosum | Yes | No |
| Ventriculomegaly | Yes | No |
| Feeding difficulties | Yes | Yes |
| Short stature | Yes | Yes |
| Face | Long | Rectangular |
| Forehead | Prominent | Broad |
| Downslanted palpebral fissures | Yes | Yes |
| Abnormality of the pinna | Yes | Yes |
| Prominent nasal bridge | Yes | Yes |
| Underdeveloped nasal alae | Yes | Yes |
| Broad nasal tip | Yes | Yes |
| Macrodontia of permanent maxillary central incisor | Yes | Yes |
| High, narrow palate | Yes | Yes |
| Cleft of chin | Yes | Yes |
| Micrognathia | Yes | Yes |
| Clinodactyly | Fifth finger | Fifth finger |
| Hirsutism | Yes | Yes |
The proband underwent a series of genetic tests including oligo array, fragile X, MECP2, CDKL5, SLC9A6, and COH1, all of which were negative. The sibling also underwent genetic testing (ZEB2 and MECP2). Both sisters had metabolic studies. After these tests failed to provide a molecular diagnosis, the family underwent research exome sequencing in 2014.
TECHNICAL ANALYSIS AND METHODS
The proband, her affected sibling, and both parents underwent exome sequencing as follows. Exome capture was performed using Agilent SureSelect v5 reagents according to manufacturer protocols. Exome libraries underwent paired-end sequencing (2 × 100 bp) on an Illumina HiSeq 2500 instrument. We generated ∼6.2 Gbp of uniquely mapped reads per sample (Table 2). Reads were mapped to the GRCh37 reference sequence and secondary data analysis was performed using Churchill (Kelly et al. 2015), which implements the GATK “best practices” workflow for alignment, variant discovery and genotyping. Sequencing metrics are provided in Supplemental Table 1. Variants were called in all four samples simultaneously, yielding 612,356 variants of which 574,390 (506,121 single nucleotide variants and 68,269 indels) passed minimum quality filters (QUAL > 100). Family relatedness was confirmed using the KING algorithm (v2.0; see Supplemental Table 2; Manichaikul et al. 2010).
Table 2.
Exome sequencing metrics for the proband, sibling, and parents described in this study
| Sample | Reads | Mapped | Duplicate | Map rate (%) | Dup. rate | Avg. depth |
|---|---|---|---|---|---|---|
| Proband | 56,019,520 | 55,815,413 | 1,577,081 | 99.64 | 2.83% | 55.0 |
| Mother | 66,027,995 | 65,781,485 | 3,173,546 | 99.63 | 4.82% | 64.3 |
| Father | 69,163,438 | 68,854,038 | 3,044,286 | 99.55 | 4.42% | 65.4 |
| Sibling | 63,049,513 | 62,811,744 | 2,033,149 | 99.62 | 3.24% | 62.3 |
SNPeff, ANNOVAR, and custom in-house scripts were used to annotate SNPs/indels with gene, transcript, function class, damaging scores, and population allele frequencies. Some 32,095 variants mapped to the exons or splice regions of known protein-coding genes. After removing common variants (MAF > 0.01 in the ExAC, ESP, or 1000 Genomes databases), we selected for further analysis all splice site, frameshift, and nonsense variants, as well as missense variants predicted to be damaging by SIFT (score < 0.05), PolyPhen (score > 0.453), GERP (score > 2.0), or CADD (Phred score > 15).
We searched for variants consistent with recessive inheritance that were present in both patients, finding no compound-heterozygous variants but two homozygous recessive variants of uncertain significance (described in the Supplemental Text and Supplemental Table 3). We also searched for candidate de novo mutations and identified a candidate nonsense mutation at the same position in both patients.
VARIANT INTERPRETATION
A heterozygous nonsense variant in ASXL3 (Table 3) was present in both sisters but absent from the parents. Manual review of the exome data verified the variant in both patients but showed no evidence of the alternate allele in either parent (Supplemental Fig. 1). The de novo status was independently confirmed by Sanger sequencing of all four individuals (Supplemental Fig. 2).
Table 3.
Genomic findings and variant interpretation
| Gene | Genomic location | HGVS cDNA | HGVS protein | Zygosity (pro/sib) | Parent of origin | Interpretation |
|---|---|---|---|---|---|---|
| ASXL3 | Chr 18:31322918 C>T (GRCh37) | NM_030632.1: c.3106C>T | p.R1036X | Het/Het | De novo | Pathogenic (PVS1, PS2, PM2, PP1, PP5) |
Criteria: PVS1, null variant; PS2, de novo in a patient with disease and no family history; PM2, absent from population controls; PP1, cosegregation with disease in a gene definitively known to cause the disease; PP5, reputable source recently reports the variant as pathogenic, but evidence was not available for us to perform an independent evaluation.
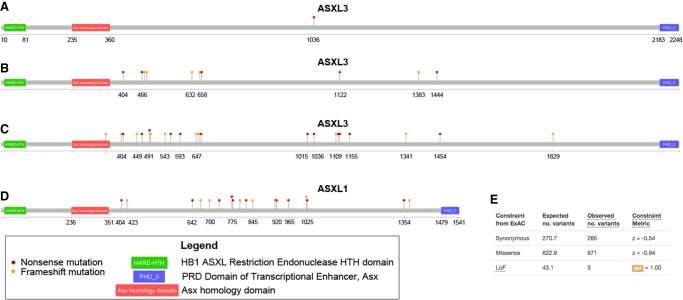
The variant is predicted to cause a stop-gain at amino acid 1036 (NM_030632.1 c.3106C>T, p.R1036X), at ∼46% of its wild-type length (Fig. 1A). It has never been observed in 122,882 individuals in the gnomAD database, making it extremely rare in human populations. However, truncating mutations in ASXL3 were recently reported as the cause of Bainbridge–Ropers syndrome (BRPS, OMIM #615485), a disorder similar to Bohring–Opitz syndrome, which is caused by truncating mutations in ASXL1 (Bainbridge et al. 2013).
Figure 1.
Graphical view of disease-causing mutations. (A) The de novo nonsense mutation in ASXL3 detected in the proband. (B) Pathogenic mutations in ASXL3 reported to the HGMD Pro database. (C) Pathogenic and Likely Pathogenic nonsense/frameshift variants in ASXL3 reported in ClinVar as of November 2017. (D) Pathogenic mutations in ASXL1 in Bohring–Opitz syndrome patients as reported to the HGMD Pro database. (E) Constraint metrics for ASXL3 from the ExAC database.
The Human Gene Mutation Database (HGMD) contains 10 nonsense/frameshift ASXL3 variants (five nonsense variants and five frameshift indels) reported in patients with Bainbridge–Ropers syndrome or Bohring–Opitz-like syndrome (Fig. 1B). The ClinVar database contains 22 nonsense/frameshift variants (12 nonsense variants including p.R1036X, and 10 frameshift indels), all of which are reported as Pathogenic or Likely Pathogenic (Fig. 1C). Of note, many of the pathogenic mutations reported in ASXL1 in Bohring–Opitz syndrome also truncate ∼50% of the encoded protein (Fig. 1D). Constraint metrics from the ExAC database (Lek et al. 2016) likewise indicate that ASXL3 is extremely intolerant of loss-of-function (LoF) mutations (43.1 variants expected, 3 observed, pLI = 1.00; Fig. 1E).
In summary, this de novo nonsense mutation is present in both affected individuals, absent from databases of population controls, and consistent with previously reported pathogenic mutations in ASXL3. We therefore conclude that it is pathogenic, and we provide a molecular diagnosis of Bainbridge–Ropers syndrome to the proband and her affected sibling.
SUMMARY
Bainbridge–Ropers syndrome (BRPS) was first described in 2013, when Bainbridge et al. reported de novo truncating mutations in four unrelated probands with feeding difficulties, failure to thrive, neurological abnormalities, and significant developmental delay. To date, fewer than 30 cases of BRPS have been described in the literature (Bainbridge et al. 2013; Dinwiddie et al. 2013; Srivastava et al. 2016; Balasubramanian et al. 2017; Kuechler et al. 2017). The report by Kuechler et al. includes a 4-yr old female with the p.R1036X mutation. She was not reported to have seizures (Supplemental Table 4), but she and both of our patients share the six most emerging hallmarks of BRPS: severe intellectual disability, poor/absent speech, autistic traits, distinct face, hypotonia, and significant feeding difficulties. We assess our patients for the clinical features highlighted by Balasubramanian et al. (2017) in Supplemental Table 5.
We recently diagnosed a third (unrelated) child at our institution, suggesting that the prevalence of BRPS is likely to be higher than currently reported in the medical literature, and the diagnosis will be made more frequently as WES is incorporated more routinely into clinical practice. Indeed, the membership of a support group for BRPS families suggests that as many as 200 patients may have been diagnosed to date (M Bainbridge, pers. comm.).
To our knowledge, this is the first report of BRPS caused by the same de novo mutation in two related individuals (siblings). Although it is theoretically possible that the same mutation arose independently in two different embryos, germline mosaicism in one of the parents seems a more likely explanation. The family declined further testing to confirm the origin of the mutation.
ADDITIONAL INFORMATION
Data Deposition and Access
The ASXL3 variant and information about its interpretation were submitted to the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/) on September 29, 2017 (SCV000605939) and accepted/released on October 9, 2017. Unfortunately, the informed consent does not allow for deposition of the raw sequencing data into public databases.
Ethics Statement
Written consent was obtained prior to enrolling subjects into a research protocol approved by the Institutional Review Board at Nationwide Children's Hospital (IRB11-00215 Study: Using Genome Sequencing to Identify Causes of Rare Birth Defects and Rare Disorders).
Acknowledgments
We thank the patients and their family for participation in research. We also thank Matthew Bainbridge for discussions about the BRPS patient population.
Author Contributions
D.C.K. contributed to genomic analysis, variant interpretation, and manuscript preparation. T.M.M. contributed to patient enrollment and phenotyping. B.J.K. contributed to genomic analysis and variant interpretation. P.W. and R.K.W. contributed to project supervision and manuscript preparation. K.M., E.S., D.B., and S.E.H. contributed to patient phenotyping and variant interpretation. All authors contributed to final manuscript review.
Funding
This work was supported by The Research Institute at Nationwide Children's Hospital.
Competing Interest Statement
The authors have declared no competing interest.
Referees
Darrell L. Dinwiddie
Anshika Srivastava
Anonymous
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Bainbridge MN, Hu H, Muzny DM, Musante L, Lupski JR, Graham BH, Chen W, Gripp KW, Jenny K, Wienker TF, et al. 2013. De novo truncating mutations in ASXL3 are associated with a novel clinical phenotype with similarities to Bohring–Opitz syndrome. Genome Med 5: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian M, Willoughby J, Fry AE, Weber A, Firth HV, Deshpande C, Berg JN, Chandler K, Metcalfe KA, Lam W, et al. 2017. Delineating the phenotypic spectrum of Bainbridge–Ropers syndrome: 12 new patients with de novo, heterozygous, loss-of-function mutations in ASXL3 and review of published literature. J Med Genet 54: 537–543. [DOI] [PubMed] [Google Scholar]
- Dinwiddie DL, Soden SE, Saunders CJ, Miller NA, Farrow EG, Smith LD, Kingsmore SF. 2013. De novo frameshift mutation in ASXL3 in a patient with global developmental delay, microcephaly, and craniofacial anomalies. BMC Med Genomics 6: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly BJ, Fitch JR, Hu Y, Corsmeier DJ, Zhong H, Wetzel AN, Nordquist RD, Newsom DL, White P. 2015. Churchill: an ultra-fast, deterministic, highly scalable and balanced parallelization strategy for the discovery of human genetic variation in clinical and population-scale genomics. Genome Biol 16: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuechler A, Czeschik JC, Graf E, Grasshoff U, Hüffmeier U, Busa T, Beck-Woedl S, Faivre L, Rivière JB, Bader I, et al. 2017. Bainbridge–Ropers syndrome caused by loss-of-function variants in ASXL3: a recognizable condition. Eur J Hum Genet 25: 183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. 2010. Robust relationship inference in genome-wide association studies. Bioinformatics 26: 2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A, Ritesh KC, Tsan YC, Liao R, Su F, Cao X, Hannibal MC, Keegan CE, Chinnaiyan AM, Martin DM, et al. 2016. De novo dominant ASXL3 mutations alter H2A deubiquitination and transcription in Bainbridge–Ropers syndrome. Hum Mol Genet 25: 597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.