Abstract
Mutations that activate the protease calpain-5 (CAPN5) cause a nonsyndromic adult-onset autoinflammatory eye disease characterized by uveitis, altered synaptic signaling, retinal degeneration, neovascularization, and intraocular fibrosis. We describe a pediatric patient with severe inflammatory vitreoretinopathy accompanied by hearing loss and developmental delay associated with a novel, de novo CAPN5 missense mutation (c.865C>T, p.Arg289Trp) that shows greater hyperactivation of the calpain protease, indicating a genotype–phenotype correlation that links mutation severity to proteolytic activity and the possibility of earlier onset syndromic disease with auditory and neurological abnormalities.
Keywords: degenerative vitreoretinopathy, delayed social development, mid-frequency hearing loss, neovascularization of peripheral and posterior retina, retrobulbar optic neuritis, uveitis
INTRODUCTION
The calpain family of cysteine proteases mediates intracellular regulatory proteolysis in response to calcium signaling (Campbell and Davies 2012). Increased calpain protease activity is associated with numerous pathologies, such as myocardial infarction, stroke, diabetes, thrombosis, neuronal injury, and neurodegeneration (Artal-Sanz and Tavernarakis 2005; Huang et al. 2010; Randriamboavonjy and Fleming 2010; Ono et al. 2016). The only inherited calpain disease characterized by protease hyperactivity is autosomal dominant neovascular inflammatory vitreoretinopathy (ADNIV; OMIM #193235), which is due to mutations in CAPN5. Despite the widespread expression of CAPN5 (Dear et al. 1997; Dear and Boehm 1999; Singh et al. 2014; Schaefer et al. 2016), to date only ophthalmological manifestations have been reported in ADNIV. In their second or third decade, ADNIV patients develop intraocular inflammation (uveitis) and retinal signaling defects (Mahajan et al. 2012). The disease continues to progress sequentially through a series of pathological stages, each sharing characteristics with common eye diseases (e.g., retinitis pigmentosa, proliferative diabetic retinopathy, and proliferative vitreoretinopathy), ultimately culminating in blindness in the fifth decade. Here we report a child with early-onset, severe inflammatory vitreoretinopathy, profound hearing loss, and developmental delay associated with a novel de novo CAPN5 mutation.
RESULTS
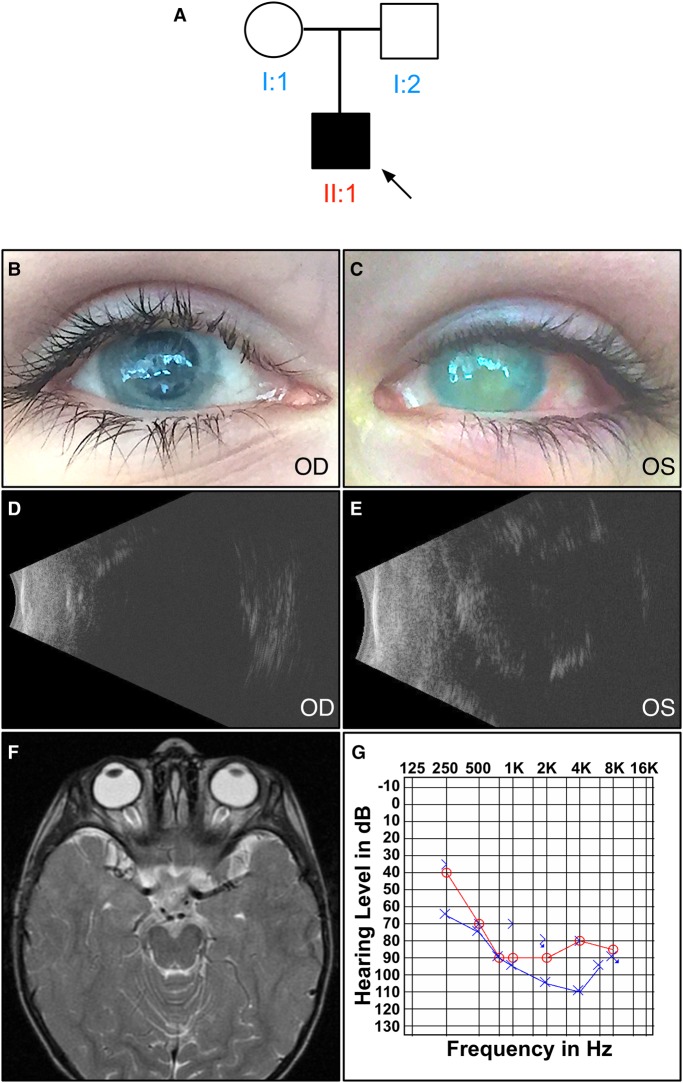
The proband (Fig. 1A) presented to us at the age of 10 yr with end-stage chronic uveitis and progressive sensorineural hearing loss. He had no light perception in both eyes. There was bilateral band keratopathy (Fig. 1B,C) without any view to the posterior pole. Ultrasound B-scans showed a silicon-filled right eye and a phthisical left eye (Fig. 1D,E). His visual issues were first noted at age 2, where he showed vertical nystagmus, bilateral pupillary membranes, anterior and posterior inflammatory cells, retinal membranes, and optic disc edema. His magnetic resonance imaging (MRI), which revealed thickening of the optic chiasm and enhancement affecting the prechiasmatic optic nerves, was consistent with optic neuritis (Fig. 1F). Over the next several years, he underwent multiple procedures to treat recurrent pupillary membranes, cataracts, glaucoma, and retinal detachment. His uveitis was treated with ocular prednisolone, atropine, and a short trial of oral methotrexate. By age 5, he had lost all vision and his eyes were prephthisical. Follow-up MRIs revealed resolution of optic neuritis but mild chronic ocular inflammation (Supplemental Fig. 1).
Figure 1.
Clinical examination of a patient with inflammatory vitreoretinopathy, hearing loss, and developmental delay. (A) Pedigree of the affected proband. Black symbols represent clinically affected subjects. Open symbols represent unaffected subjects. Both parents had normal eye and hearing exams. There were no extended family members with similar vision loss, sensorineural hearing loss, neurologic disorder, or developmental delay. (B,C) External photo showing bilateral band keratopathy. B-scan ultrasonography showed a (D) silicon-filled right eye and (E) phthisical left eye. (F) T2-weighted MRI imaging of the proband at age 2 (no fat suppression) reveals that optic chiasm thickening and enhancement affecting the prechiasmatic optic nerves was consistent with optic neuritis. (G) Audiogram at age 10 reveals significant sensorineural hearing loss.
The patient met his developmental milestones until the age of 2 yr, but has he had developmental delays with regard to language, socialization, and quantitative reasoning. He had displayed slow progression in his learning, which appeared to plateau in the most recent years. There was less delay in the patient's gross motor development, although he was noted to have mild truncal hypotonia and displayed repetitive hand motions. He had unexplained behavioral outbursts, therefore an electroencephalography (EEG) was performed. EEG showed nonspecific temporal lobe abnormalities as well as generalized low-amplitude β frequency activity. Hearing difficulties began at age 6, and audiograms revealed mild high- to mid-range sensorineural hearing loss. There was progressive decline in his hearing from mild, to moderate, to severe, requiring hearing aids (Fig. 1G). He had lifelong gastrointestinal difficulties, including episodes of chronic diarrhea and constipation, without a clear etiology. A brain MRI showed normal inner ear structures and seventh and eighth cranial nerves, bilateral phthisis bulbi, and no optic nerve enhancement (Supplemental Fig. 2).
We performed whole-exome sequencing (WES) on this patient and the unaffected parents. Samples were sequenced with a mean depth of coverage of 104× (Supplemental Table 1). Bidirectional sequences were assembled, aligned to reference gene sequences based on GRCh37/UCSC hg19, and analyzed for sequence variants using XomeDx software (GeneDx). We identified 140,798 coding variants in the proband (Supplemental Table 2). Variants were then filtered based on predicted loss-of-function or missense mutation, the presence of gene or variant in the Human Gene Mutation Database, and minor allele frequency (MAF) of <0.01 (Stenson et al. 2014). MAFs were obtained from the 1000 Genomes Project using the Ensembl Variant Effect Predictor (VEP) tool (McLaren et al. 2016).
The proband harbored a de novo, heterozygous, missense variant in the CAPN5 gene (NM_004055.4), c.865C>T, p.Arg289Trp (Table 1). For this gene, 98.9% of the coding region was covered at a minimum of 10×. Other mutations in CAPN5 are associated with autoinflammatory disease restricted to the eye (Mahajan et al. 2012; Bassuk et al. 2015; Randazzo et al. 2017). In view of the patient's extraocular phenotype, additional review of 133 genes associated with sensorineural hearing loss, global developmental delay, and Usher syndrome was performed (Supplemental Table 3; Shearer et al. 1993; Miclea et al. 2015). Identified variants in these genes were reviewed for inheritance pattern, phenotypic correlation, and predicted loss of function. These remaining candidate variants were either predicted to be tolerated by SIFT and PolyPhen-2 analysis or did not match the mode of inheritance for their respective phenotype (Supplemental Table 4; Sim et al. 2012; Adzhubei et al. 2013)
Table 1.
CAPN5 variant information
| Gene | Chr: position GRCh37 (hg19) | HGVS DNA reference | HGVS protein reference | Gene coverage | Variant read depth | Variant type | Genotype | ClinVar accession | Inheritance |
|---|---|---|---|---|---|---|---|---|---|
| CAPN5 | Chr 11: 76826606 | c.865C>T | p.Arg289Trp | 98.9% | 10× | Missense | Heterozygous | SCV000329671.5 | De novo |
The patient's mutation and constellation of symptoms suggested an underlying autoinflammatory disease and prompted further rheumatologic and neurophysiologic evaluation. With the recent auditory and behavioral decline, the patient was started on monthly intravenous immunoglobulin (IVIG), high-dose steroids followed by 3 mo of oral prednisolone, which appeared to stabilize his hearing loss. Eight months later, mycophenolate mofetil was added. Despite 15 mo of treatment, his hearing continued to decline, and he became profoundly deaf in both ears.
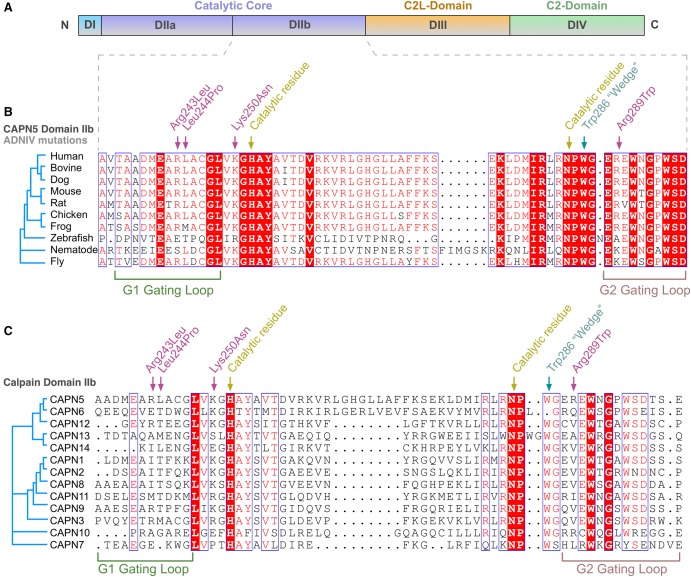
CAPN5 encodes the protein calpain-5 (CAPN5), an intracellular calcium-activated cysteine protease with evolutionarily conserved domains and residues required for its activity. CAPN5's catalytic core is encoded by exon 6, where we previously identified three disease-causing mutations (Fig. 2A). The new c.865C>T variant mutates Arg289, which is five residues away from a catalytic residue (Asn284) and only three residues away from a highly conserved residue (Trp286). Arg289 is conserved in 70% of CAPN5 orthologs but poorly conserved among human paralogs (Fig. 2B).
Figure 2.
The CAPN5 gene harbors a novel mutation in domain II. (A) CAPN5 is composed of four domains, and the novel mutation was located in catalytic domain II (violet). (B) Primary protein sequence alignment of calpain-5 orthologs shows 70% evolutionary conservation of the new mutated p.Arg289Trp residue, along with the three prior ADNIV mutations, p.Arg243Leu, p.Leu244Pro, and p.Lys250Asn (magenta arrow), in catalytic domain IIb. (C) Alignment of 13 human calpain family paralogs show the p.Arg289Trp is poorly conserved (15%).
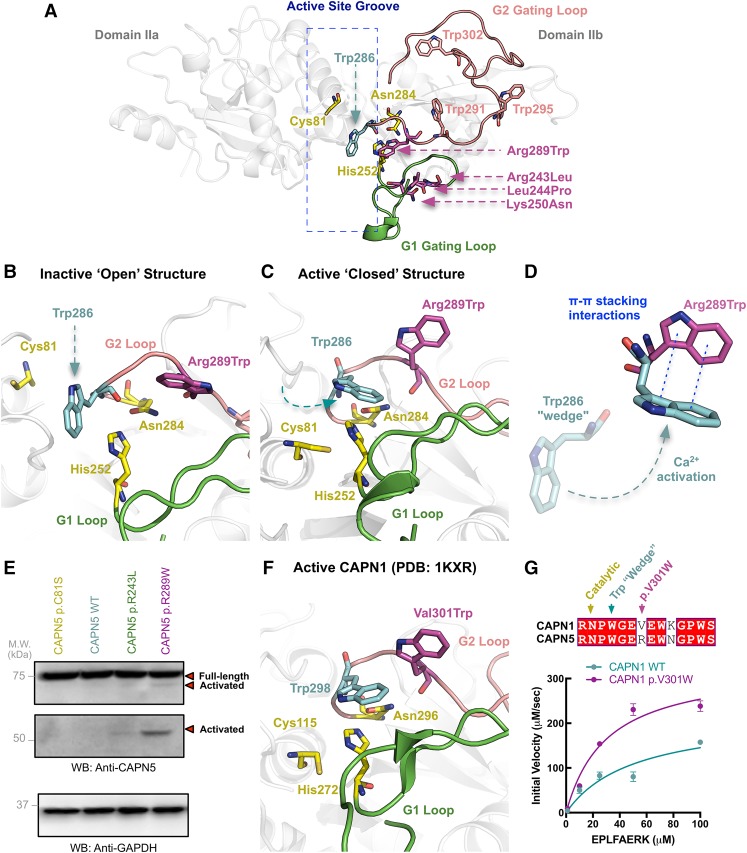
We generated a structural model for CAPN5 (Gakhar et al. 2016). Three ADNIV mutations are located on a flexible loop (G1 loop) (Fig. 3A) that undergoes calcium-induced conformation changes during enzyme activation (Moldoveanu et al. 2002; Campbell and Davies 2012). This loop is located on the amino-terminal side of the catalytic groove and acts as a gatekeeper to control substrate access (Fig. 3A). The first three CAPN5 mutants significantly alter the G1 loop structure (Mahajan et al. 2012; Bassuk et al. 2015). The new CAPN5 mutation (p.Arg289Trp) is located on a complementary flexible loop (G2 loop) on the carboxy-terminal side of the catalytic groove. Introduction of the mutant tryptophan places it near three other tryptophans, which can engage in noncovalent π–π stacking interactions. One of these tryptophans (Trp286) is the critical “wedge” tryptophan that inhibits enzymatic activity by inserting itself in the catalytic groove to prevent bonding between the catalytic triad (Fig. 3B; Moldoveanu et al. 2002). Normally, only calcium-induced conformational changes remove this inhibitory wedge from the active site (Fig. 3C). Tryptophan has strong stacking interactions because of the large surface area and dipole moment of its side chain (Qian et al. 2008). This property has been exploited in the design of α-ketoamide inhibitors for CAPN1 and CAPN2. These compounds (e.g., ZLAK-3001 and ZLAK-3002) contain adenine and piperazyl side chains that stabilize the inhibitory tryptophan wedge in the inactive state (Qian et al. 2008). Our model indicated introduction of an additional tryptophan on the G2 loop could cause stacking interactions that stabilize the Trp286 “wedge” in calpain's active conformation (Fig. 3D). Disrupting this regulatory mechanism could lead to reduced calcium requirements for activation.
Figure 3.
The p.Arg289Trp mutation causes calpain hyperactive proteolysis. (A) Structural model of the inactive CAPN5 catalytic core generated using closely related CAPN9 (PDB ID: 1ZIV) as a template. The catalytic core is comprised of two globular subdomains (DIIa and DIIb) separated by a flexible linker. Movement of DIIa and DIIb with respect to the linker region regulates formation of the active site. The catalytic residues (yellow) are in the active site groove. The G1 gating loop (green) contains previously identified ADNIV mutations (p.Arg243Leu, p.Leu244Pro, p.Lys250Asn; magenta). The G2 gating loop (pink) contains the novel p.Arg289Trp mutation along with the Trp286 “wedge” residue (blue) and three additional tryptophans. (B) Close-up view of the CAPN5 catalytic groove in its inactive state. In the absence of activating calcium, the Trp286 residue acts as a wedge and prevents the formation of the catalytic triad. (C) Close-up view of the CAPN5 catalytic groove in its active state modeled off CAPN9 (PDB ID: 2P0R). Calcium binding causes a structural rearrangement that removes the Trp286 “wedge” away from the active site and facilitates the formation of the catalytic triad. (D) Tryptophans engage in π–π stacking interactions. Substitution of Arg289 with Trp on the G2 gating loop may cause stacking interactions that stabilize the Trp286 “wedge” in its active conformation. Disruption of this inhibitory mechanism may reduce the calcium requirement for CAPN5 activation and lead to hyperactive proteolysis of substrates in affected tissues. (E) Whereas wild-type and inactive versions (p.Cys81Ser) of CAPN5 show no autoproteolytic activity at normal cellular calcium levels, the syndromic CAPN5 p.Arg289Trp mutant displays autoproteolytic activity. This proteolytic activity is higher than that observed for the nonsyndromic CAPN5 p.Arg243Leu mutant. (F) Close-up view of the crystal structure of active rat CAPN1 (PDB ID: 1KXR) shows similarities to our CAPN5 homology model. (G) A homologous mutation (p.Val301Trp) to p.Arg289Trp was introduced to purified recombinant rat CAPN1, which resulted in increased enzymatic activity compared to wild type in a fluorescence-based enzymatic assay. Results are reported as initial velocity versus substrate (EPLFAERK) concentration.
We used CAPN5 autoproteolysis to measure its proteolytic activity, because CAPN5 does not proteolyze known calpain substrates (Wert et al. 2015). Increased intracellular calcium triggers calpain activation and subsequent autoproteolysis and release of calpain's catalytic core (Campbell and Davies 2012). We expressed CAPN5 mutants in cells to determine their activity (Fig. 3E). At normal intracellular calcium, both a catalytically inactive mutant (p.Cys81Ser) and the wild-type (WT) CAPN5 were inactive. The p.Arg243Leu mutant showed slight activation, whereas the p.Arg289Trp disease variant showed strong activation. Thus, CAPN5 p.Arg289Trp was hyperactive, autoproteolyzed even in the absence of calcium levels required to activate WT CAPN5, and displayed more proteolytic fragments than p.Arg243Leu (Fig. 3E; Supplemental Fig. 3), consistent with the more severe phenotype seen in our proband. At elevated intracellular calcium, the WT CAPN5 became activated as expected, and the p.Arg289Trp maintained high hyperactivity (data not shown). Interestingly, introduction of the homologous mutation (p.Val301Trp) (Fig. 3F) (Moldoveanu et al. 2002) into purified CAPN1 catalytic core also led to increased enzymatic activity, further confirming that disruption of the Trp “wedge” by this mutation is a critical, conserved regulatory mechanism across calpains (Fig. 3G; Supplemental Fig. 4).
DISCUSSION
In this report, we present a 10-yr-old boy with a novel CAPN5 mutation (p.Arg289Trp) that is associated with early-onset severe inflammatory vitreoretinopathy, optic neuritis, sensorineural hearing loss, developmental delay, mild nonspecific EEG abnormalities, and GI disturbances. The clinical phenotype in this patient is distinct from previous ADNIV kindreds (Mahajan et al. 2012; Bassuk et al. 2015; Randazzo et al. 2017). In these families, disease onset did not begin until the second decade of life, there was no optic neuritis, and blindness typically occurred after the fifth decade. These ADNIV pedigrees did not manifest disease outside the eye, despite CAPN5 expression during embryogenesis, in the colon, kidney, liver, trachea, uterus, thymus, and subsets of CNS neurons (Dear et al. 1997; Dear and Boehm 1999; Singh et al. 2014; Schaefer et al. 2016).
The proteotype–phenotype difference between previous ADNIV kindreds and the proband is noteworthy. Nonsyndromic ADNIV patients affected by G1 loop mutations have a nearly identical phenotype. The earlier onset and more severe phenotype associated with p.Arg289Trp is in a different region of the protein (G2 loop). The p.Arg289Trp falls into a peptide loop that changes conformation upon calcium binding, similar to prior ADNIV mutations (Qian et al. 2008; Mahajan et al. 2012; Bassuk et al. 2015). This finding indicates a functional proteotype–phenotype correlation for CAPN5 mutations in which the disease severity follows the level of protease hyperactivity. In fact, the structural mechanisms seem to be conserved across calpains and can be used to infer functional consequences on other isoforms. Interestingly, variants in the CAPN5 G2 loop have been previously reported in the GnoMAD browser, suggesting that some mutations in this region of the protein may be tolerated (Supplemental Table 5; Lek et al. 2016). However, none of these variants add an additional tryptophan residue that would similarly disrupt the regulatory mechanism of the G2 loop.
The presence of sensorineural hearing loss in our patient is interesting, as CAPN5 has not been found to be expressed in cochlear hair cells (Thul and Lindskog 2018). The auditory and CNS phenotype may be due to synaptic signaling defects, because CAPN5 has been found to be expressed in neuronal synapses and mitochondria. These compartments are rich in calcium and may be susceptible to damage by a hyperactive CAPN5 (Vosler et al. 2008). The combination of hearing loss and blindness recalls Usher syndrome. However, identified variants in genes associated with Usher syndrome, sensorineural hearing loss, and developmental delay in our patient were predicted to be nonpathogenic (Supplemental Table 4; Petit 2001). Dysfunction in these systems may be due to dysregulation of other calpain effector pathways, such as synaptic signaling, or late clinical examination may have missed these signs. Maintenance of gut pacemaker activity is heavily dependent on intracellular calcium signaling and dysregulation is associated with gut motility dysfunction (Takaki 2003).
In summary, our findings suggest that the CAPN5 mutation in this patient may be biologically more pathogenic than those identified in previous ADNIV kindreds. This also suggests that hyperactive CAPN5 can cause disease in other tissues and that it is involved in numerous pathological pathways. There is some precedent for this, as increased calpain activity has been implicated in other autoinflammatory diseases. The presence of a single patient with the described mutation limits the conclusions of the current study. Thus, identification of additional patients harboring the p.Arg289Trp mutation will add confidence to our findings and further expand the ADNIV phenotype. Although unlikely, we cannot exclude the possibility that the patient's sensorineural hearing loss is due to an additional identified variant (Supplemental Table 4). Nevertheless, these mutations would not account for all the patient's extraocular phenotypes, such as developmental delay and GI disturbance. Developmental delay is multifactorial and may be explained by an additional unknown genetic variant or environmental factor (Smith et al. 2017). Further, we did not account for potential copy-number variants (CNVs) or gene deletions, which are known to contribute to a large percentage of hearing loss and developmental delay cases (Cooper et al. 2011; Perry et al. 2014; Shearer et al. 2014; Hladilkova et al. 2015). Additionally, although neither parent was found to carry the p.Arg289Trp likely pathogenic variant, the possibility of germline mosaicism cannot be excluded in this case. Finally, although 95.3% of the exomes were covered with at least 10 sequence reads, we cannot rule out the possibility that a small portion of the target region was not covered with sufficient depth or quality to confidently call some variant positions. Therefore, we anticipate that further mechanistic studies on CAPN5 function in different tissues may validate this novel variant and offer therapeutic insight into various diseases.
METHODS
Study Approval
The study was approved by the University of Iowa's Institutional Review Board and adheres to the tenets set forth in the Declaration of Helsinki. Informed consent was obtained from study participants. The subject underwent eye exams that included slit-lamp examination, dilated retinal biomicroscopy, indirect ophthalmoscopy, and B-scan ultrasonography. MRI examination of the brain was performed utilizing axial T2 weighted images obtained prior to and following intravenous Gadolinium DTPA administration. In addition, axial and coronal T2 weighed, axial diffusion weighted and axial FLAIR images of the posterior periventricular white matter were obtained. This was supplemented with sagittal and coronal T1 weighted Gadolinium enhanced images.
Whole-Exome Sequencing and Analysis
Genomic DNA from the proband and parents were processed using the Agilent Clinical Research Exome kit. The targeted regions (exonic regions and flanking splicing junctions) were sequenced simultaneously by massively parallel (NextGen) sequencing on an Illumina HiSeq sequencing system with 100-bp paired-end reads with a mean depth of coverage of 104× (quality threshold 95.3%) (Supplemental Table 1). Bidirectional sequences were assembled, aligned to reference gene sequences based on GRCh37/UCSC hg19, and analyzed for sequence variants using XomeDx software (GeneDx). Variants were filtered based on predicted loss-of-function or missense mutation, presence of gene or variant in the Human Gene Mutation Database, and MAF of <0.01 (Stenson et al. 2014). MAFs were obtained from the 1000 Genomes Project using the Ensembl Variant Effect Predictor (VEP) tool. The remaining variants were analyzed by SIFT and PolyPhen-2 (Supplemental Table 4; Sim et al. 2012; Adzhubei et al. 2013). Capillary sequencing was used to confirm all potentially pathogenic variants identified in the proband and parent samples. The identified variant was submitted to ClinVar (ID 279987) (Landrum et al. 2014).
Structural Modeling of CAPN5
Primary and secondary structure protein alignments and trees were created with Geneious R10 and visualized using ESPript 3.0. Homology models were generated for the human calpain-5 catalytic core using MODELLER 9.14, as previously described (Bassuk et al. 2015; Gakhar et al. 2016). The structures of human calpain-9 (42% sequence identity) were used as templates (PDB ID 2P0R; calcium and leupeptin bound closed and PDB ID 1ZIV; calcium and β-mercaptoethanol bound open) (Davis et al. 2007). Ten models were generated off each template. Because there was little variation around the ADNIV mutation sites, we chose the top models from each template generated by MODELLER for figures. To better visualize the G2 gating loop, five different conformations of the loop (residues 286–320) were generated using the refinement function of the MODELLER graphical user interface in UCSF Chimera (Pettersen et al. 2004). PyMOL was used to generate all structure figures.
Cell Transfections
Of note, 6 × 105 HEK-293T cells (ATCC) were seeded into each well of six-well plates and grown overnight in Dulbecco's modified Eagle medium (DMEM, GIBCO) supplemented with 10% fetal bovine serum (GIBCO), 1% penicillin–streptomycin (10,000 U/ml; GIBCO). After 24 h, vectors were transfected into cells using PolyFect (QIAGEN) as per the manufacturer's protocol. Cells were incubated for 24 h before being harvested by lysis in 200 µl of TNE buffer (50 mM Tris, 100 mM NaCl, 5 mM EDTA).
Western Blots
Protein levels were detected by western blotting. Total cellular protein was measured using NanoDrop 2000c (ThermoScientific). Reactions were carried out with 4× LDS-sample buffer, 10× reducing agent and equal amounts of protein. Reactions were denatured for 5 min at 90°C. Samples were then separated electrophoretically on a 15-well 4%–12% Bis–Tris gel (Life Sciences) and transferred onto a nitrocellulose membrane using an iBlot dry blotting system (ThermoScientific). Proteins were immunostained with anti-CAPN5 (GTX103264, GeneTex; 1: 1000 dilution) and anti-GAPDH (sc-25778, Santa Cruz Biotechnology; 1: 1000 dilution) and goat anti-rabbit secondary antibody (sc-2004, Santa Cruz Biotechnology; 1: 1000 dilution). The immunoreactive bands were detected with Supersignal West Dura Extended Duration Substrate (ThermoScientific). Visualization was performed with MYECL Imager (ThermoScientific).
Cloning
The rat CAPN1 WT and p.V301W catalytic core sequences were cloned into a pUC57 vector with an XhoI restriction site, with an amino-terminal thrombin cleavage site and a carboxy-terminal TEV protease cleavage site followed by a 6xHis tag, as previously described (Moldoveanu et al. 2002; Gakhar et al. 2016). Calpain constructs were transferred into the pMAL-C5X vector to obtain an amino-terminal maltose-binding protein (MBP) as a fusion partner. Sequence of the calpain-flanking regions was confirmed by sequencing of constructs. Plasmids were amplified and isolated from DH5α cells and then were transformed into Escherichia coli BL21 (DE3; New England Biolabs).
Protein Purification
Escherichia coli BL21 (DE3; New England Biolabs) cells expressing rat CAPN1 WT and p.V301W catalytic core sequences were grown in 1 l shake-flask cultures, at 250 rpm in Terrific Broth (Invitrogen), at 37°C until an OD600 of 2.0 and then induced with 0.2 mM IPTG (Sigma-Aldrich). Cells were grown for 19 h at 16°C , harvested, and centrifuged at 4000g for 20 min and pellet frozen at −20°C. Cells were resuspended in 35 ml of lysis buffer (20 mM Tris, 300 mM NaCl, 1 mM DTT, pH 7.5) and one tablet of EDTA-free protease inhibitor (Roche) and lysed using sonicator (Fisher Scientific Model 705) and centrifuged for 1 h at 18,000 RPM. Cell debris were discarded and the supernatant (30 ml) loaded onto a column packed with 5 ml (1CV) amylose resin (New England Biolabs). The column was washed with 12 CV of wash buffer (20 mM Tris, 300 mM NaCl, pH 7.5) and eluted with 6 CV of elution buffer (20 mM Tris, 300 mM NaCl, 10 mM maltose, pH 7.5). Eluted fractions were pooled and concentrated to 2 ml at 10 mg/ml (30 kDa NMWL spin concentrator; Millipore) and passed over a Superdex 200 (GE) size-exclusion column to remove residual impurities. The column was equilibrated with 20 mM Tris, 300 mM NaCl, 2 mM DTT, pH of 7.5. The final sample was concentrated with a 30 kDa NMWL spin concentrator and stored in 50% glycerol at −80°C before use.
Proteolysis Assay
Purified calpain activity was measured by cleavage of a FRET-tagged calpain peptide substrate (EPLFAERK; Biopeptek) as previously described (Wert et al. 2015). Briefly, 5 µg of purified calpain was added to a reaction buffer containing 20 mM Tris, 300 mM NaCl, 2 mM DTT, and 1 mM CaCl2 (pH 7.5). Different concentrations of substrate (0–100 µM) were added and the reaction was incubated for 15 min at 37°C. Proteolysis was quantified as raw fluorescence units on a fluorimetric plate reader (Tecan Spark).
ADDITIONAL INFORMATION
Data Deposition and Access
The CAPN5 variant has been registered in ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) under accession number SCV000329671.5. Patient consent could not be obtained for deposition of raw sequencing data.
Ethics Statement
The study was approved by the University of Iowa's Institutional Review Board and adheres to the tenets set forth in the Declaration of Helsinki. Informed consent was obtained from study participants. Both the patient and his parents have consented to publication of this case history with patient pictures.
Acknowledgments
We thank Jing Yang and Carin Yates for technical assistance.
Author Contributions
V.B.M. had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design were contributed by V.B.M., S.H.T., P.J.F., and A.G.B. Data were acquired by G.V., A.G.B., L.G., M.M., P.J.F., K.A.S., and V.B.M. Analysis and interpretation of data was performed by G.V., A.G.B., K.A.S., L.G., M.M., P.J.F., and V.B.M. The manuscript was drafted by G.V., A.G.B., L.G., M.M., P.J.F., and V.B.M. S.H.T., P.J.F., L.G., B.B., P.K., A.G.B., and V.B.M. critically revised the manuscript for important intellectual content. Bioinformatics analysis was performed by G.V., L.G., and V.B.M. V.B.M. obtained funding. Administrative, technical, and material support were provided by and the study was supervised by V.B.M.
Funding
V.B.M. and A.G.B. are supported by National Institutes of Health (NIH) grants R01EY026682, R01EY024665, R01EY025225, R01EY024698, R21AG050437, and P30EY026877, The Doris Duke Charitable Foundation Grant #2013103, and Research to Prevent Blindness (RPB), New York, New York. G.V. is supported by NIH grants F30EYE027986 and T32GM007337. The Barbara & Donald Jonas Laboratory of Regenerative Medicine and Bernard & Shirlee Brown Glaucoma Laboratory are supported by NIH grants 5P30EY019007, R01EY018213, R01EY024698, and R21AG050437, National Cancer Institute Core grant 5P30CA013696, the Research to Prevent Blindness (RPB) Physician-Scientist Award, unrestricted funds from RPB, New York, New York. S.H.T. is a member of the RD-CURE Consortium and is supported by the Tistou and Charlotte Kerstan Foundation, the Schneeweiss Stem Cell Fund, New York State (C029572), the Joel Hoffman Fund, the Professor Gertrude Rothschild Stem Cell Foundation, and the Gebroe Family Foundation. P.J.F. is supported by NIH grant R01AR059703.
Role of the Sponsor
The funding organizations had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Competing Interest Statement
The authors have declared no competing interest.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Adzhubei I, Jordan DM, Sunyaev SR. 2013. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet Chapter 7: Unit7 20 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artal-Sanz M, Tavernarakis N. 2005. Proteolytic mechanisms in necrotic cell death and neurodegeneration. FEBS Lett 579: 3287–3296. [DOI] [PubMed] [Google Scholar]
- Bassuk AG, Yeh S, Wu S, Martin DF, Tsang SH, Gakhar L, Mahajan VB. 2015. Structural modeling of a novel CAPN5 mutation that causes uveitis and neovascular retinal detachment. PLoS One 10: e0122352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RL, Davies PL. 2012. Structure–function relationships in calpains. Biochem J 447: 335–351. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V, et al. 2011. A copy number variation morbidity map of developmental delay. Nat Genet 43: 838–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TL, Walker JR, Finerty PJ Jr, Mackenzie F, Newman EM, Dhe-Paganon S. 2007. The crystal structures of human calpains 1 and 9 imply diverse mechanisms of action and auto-inhibition. J Mol Biol 366: 216–229. [DOI] [PubMed] [Google Scholar]
- Dear TN, Boehm T. 1999. Diverse mRNA expression patterns of the mouse calpain genes Capn5, Capn6 and Capn11 during development. Mech Dev 89: 201–209. [DOI] [PubMed] [Google Scholar]
- Dear N, Matena K, Vingron M, Boehm T. 1997. A new subfamily of vertebrate calpains lacking a calmodulin-like domain: implications for calpain regulation and evolution. Genomics 45: 175–184. [DOI] [PubMed] [Google Scholar]
- Gakhar L, Bassuk AG, Velez G, Khan S, Yang J, Tsang SH, Mahajan VB. 2016. Small-angle X-ray scattering of calpain-5 reveals a highly open conformation among calpains. J Struct Biol 196: 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hladilkova E, Baroy T, Fannemel M, Vallova V, Misceo D, Bryn V, Slamova I, Prasilova S, Kuglik P, Frengen E. 2015. A recurrent deletion on chromosome 2q13 is associated with developmental delay and mild facial dysmorphisms. Mol Cytogenet 8: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CJ, Gurlo T, Haataja L, Costes S, Daval M, Ryazantsev S, Wu X, Butler AE, Butler PC. 2010. Calcium-activated calpain-2 is a mediator of β cell dysfunction and apoptosis in type 2 diabetes. J Biol Chem 285: 339–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR. 2014. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 42(Database issue): D980–D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan VB, Skeie JM, Bassuk AG, Fingert JH, Braun TA, Daggett HT, Folk JC, Sheffield VC, Stone EM. 2012. Calpain-5 mutations cause autoimmune uveitis, retinal neovascularization, and photoreceptor degeneration. PLoS Genet 8: e1003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F. 2016. The ensembl variant effect predictor. Genome Biol 17: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miclea D, Peca L, Cuzmici Z, Pop IV. 2015. Genetic testing in patients with global developmental delay/intellectual disabilities. A review. Clujul Med 88: 288–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldoveanu T, Hosfield CM, Lim D, Elce JS, Jia Z, Davies PL. 2002. A Ca2+ switch aligns the active site of calpain. Cell 108: 649–660. [DOI] [PubMed] [Google Scholar]
- Ono Y, Saido TC, Sorimachi H. 2016. Calpain research for drug discovery: challenges and potential. Nat Rev Drug Discov 15: 854–876. [DOI] [PubMed] [Google Scholar]
- Perry BP, Sebold C, Hasi M, Heard P, Carter E, Hill A, Gelfond J, Hale DE, Cody JD. 2014. Sensorineural hearing loss in people with deletions of 18q: hearing in 18q. Otol Neurotol 35: 782–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit C. 2001. Usher syndrome: from genetics to pathogenesis. Annu Rev Genomics Hum Genet 2: 271–297. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612. [DOI] [PubMed] [Google Scholar]
- Qian J, Cuerrier D, Davies PL, Li Z, Powers JC, Campbell RL. 2008. Cocrystal structures of primed side-extending α-ketoamide inhibitors reveal novel calpain-inhibitor aromatic interactions. J Med Chem 51: 5264–5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo NM, Shanks ME, Clouston P, MacLaren RE. 2017. Two novel CAPN5 variants associated with mild and severe autosomal dominant neovascular inflammatory vitreoretinopathy phenotypes. Ocul Immunol Inflamm 1–6. 10.1080/09273948.2017.1370651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randriamboavonjy V, Fleming I. 2010. The role of calpain in diabetes-associated platelet hyperactivation. Adv Pharmacol 59: 235–257. [DOI] [PubMed] [Google Scholar]
- Schaefer KA, Toral MA, Velez G, Cox AJ, Baker SA, Borcherding NC, Colgan DF, Bondada V, Mashburn CB, Yu CG, et al. 2016. Calpain-5 expression in the retina localizes to photoreceptor synapses. Invest Ophthalmol Vis Sci 57: 2509–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer AE, Hildebrand MS, Smith RJH. 1993. Hereditary hearing loss and deafness overview. In GeneReviews (ed. Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A). University of Washington, Seattle. [PubMed] [Google Scholar]
- Shearer AE, Kolbe DL, Azaiez H, Sloan CM, Frees KL, Weaver AE, Clark ET, Nishimura CJ, Black-Ziegelbein EA, Smith RJ. 2014. Copy number variants are a common cause of non-syndromic hearing loss. Genome Med 6: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. 2012. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res 40(Web Server issue): W452–W457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Brewer MK, Mashburn CB, Lou D, Bondada V, Graham B, Geddes JW. 2014. Calpain 5 is highly expressed in the central nervous system (CNS), carries dual nuclear localization signals, and is associated with nuclear promyelocytic leukemia protein bodies. J Biol Chem 289: 19383–19394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L, van Jaarsveld CHM, Llewellyn CH, Fildes A, López Sánchez GF, Wardle J, Fisher A. 2017. Genetic and environmental influences on developmental milestones and movement: results from the Gemini Cohort Study. Res Q Exerc Sport 88: 401–407. [DOI] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN. 2014. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet 133: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaki M. 2003. Gut pacemaker cells: the interstitial cells of Cajal (ICC). J Smooth Muscle Res 39: 137–161. [DOI] [PubMed] [Google Scholar]
- Thul PJ, Lindskog C. 2018. The human protein atlas: a spatial map of the human proteome. Protein Sci 27: 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosler PS, Brennan CS, Chen J. 2008. Calpain-mediated signaling mechanisms in neuronal injury and neurodegeneration. Mol Neurobiol 38: 78–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wert KJ, Bassuk AG, Wu WH, Gakhar L, Coglan D, Mahajan M, Wu S, Yang J, Lin CS, Tsang SH, et al. 2015. CAPN5 mutation in hereditary uveitis: the R243L mutation increases calpain catalytic activity and triggers intraocular inflammation in a mouse model. Hum Mol Genet 24: 4584–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.