Abstract
Ribosome biogenesis is a complex process that is facilitated by a large number of assembly factors. In this issue, Andrade et al (2018) provide evidence that a widely conserved RNA chaperone, Hfq, acts as a ribosomal assembly factor in bacteria. Hfq is known to support regulation of stress response genes by small RNAs. Andrade et al (2018) show that the absence of Hfq results in higher levels of immature 30S ribosomes and error‐prone translation, suggesting that Hfq globally affects the quality of protein synthesis when bacteria are under stress.
Subject Categories: Microbiology, Virology & Host Pathogen Interaction; Protein Biosynthesis & Quality Control; RNA Biology
Ribosomes are macromolecular machines composed of RNA and proteins that translate messenger RNA (mRNA) into proteins in all living cells. Bacterial ribosomes are made up of two asymmetrical subunits. The small subunit (30S) is made up of a 1,542 nucleotide (nt) 16S ribosomal RNA (rRNA) and 21 ribosomal proteins (r‐proteins), whereas the large subunit (50S) is made up of the 2,904 nt 23S rRNA, 120 nt 5S rRNA, and 34 proteins. Owing to this complexity, the production of new ribosomal subunits is assisted by a number of assembly factors and tightly regulated with respect to cell growth. In favorable growth conditions, the rRNA is highly transcribed, and assembly of the 30S and 50S subunits begins while the rRNA is synthesized. During assembly, the r‐proteins bind the 16S or 23S rRNA in a hierarchically ordered fashion that is coupled to folding of the rRNA (Shajani et al, 2011). In unfavorable growth conditions, rRNA transcription is repressed through the stringent response and by reduced pools of initiating nucleotides (Paul et al, 2004).
Although the RNA and protein components of bacterial ribosomes can reassemble in the test tube, cellular factors and modification enzymes facilitate maturation of the ribosome by lowering the activation barriers for rRNA folding. Proper rRNA folding and assembly are coupled to 5′ and 3′ processing of the pre‐rRNA by ribonucleases (Fig 1). These factors bind to the immature rRNA–protein complexes and dissociate when they are no longer needed, leaving behind a functional ribosomal subunit. Bacteria depleted of these factors exhibit phenotypic hallmarks of ribosome assembly defects, such as cold sensitivity and accumulation of unprocessed pre‐16S or pre‐23S rRNA (Shajani et al, 2011).
Figure 1. Hypothesized interactions of Hfq with pre‐30S ribosomes.
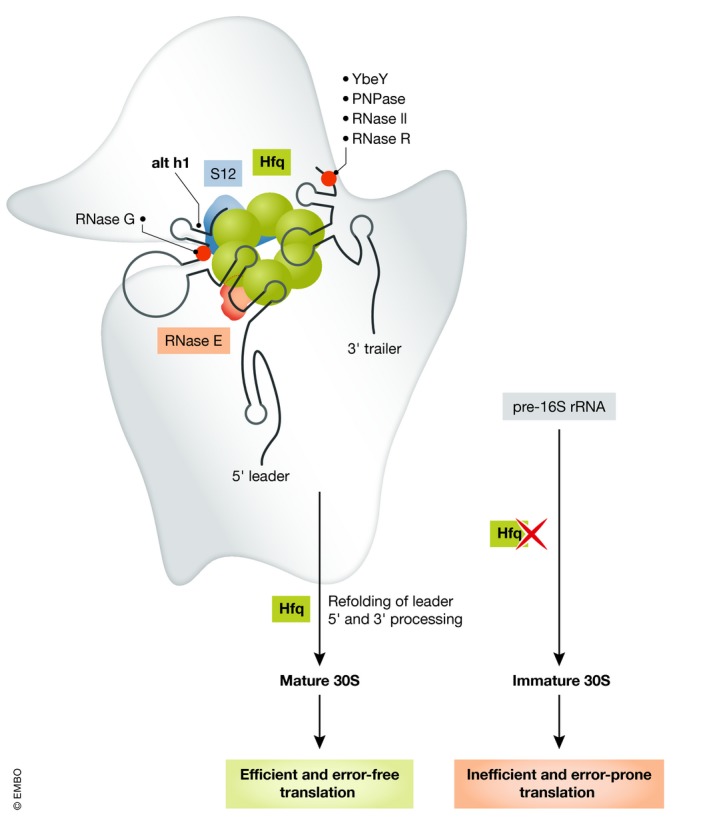
Hfq may facilitate maturation of 30S ribosomes by refolding the leader and trailer segments of pre‐16S rRNA, allowing access for RNase E and conversion of alternative helix 1 (alt h1) to h1. Proper maturation of the 30S subunit is essential for efficient and correct translation. In the absence of Hfq and in stationary phase, unfolded leader and trailer intermediates cannot be processed and therefore block ribosome maturation. When such immature 30S particles enter the translation cycle, they reduce the efficiency and fidelity of protein synthesis.
Hfq is a conserved RNA chaperone that was first identified as a Host Factor for replication of bacteriophage Qβ in Escherichia coli. Hfq assembles into a hexameric ring that binds RNA on both sides and that is homologous to eukaryotic Sm/Lsm proteins (Vogel & Luisi, 2011; Santiago‐Frangos & Woodson, 2018). Hfq has been widely studied for its role in facilitating post‐transcriptional gene regulation by small non‐coding RNA (sRNA). In addition to facilitating sRNA regulation, Hfq can directly inhibit translation of certain mRNAs by blocking ribosome binding and promoting turnover of mRNAs terminated by Rho ATPase (Vogel & Luisi, 2011). Because Hfq participates in post‐transcriptional regulation of many stress response genes, hfq mutations render bacteria sensitive to many types of environmental stress. Moreover, Hfq is important for the virulence of bacterial pathogens such as Vibrio cholera, Salmonella typhimurium, and Listeria monocytogenes.
Although Hfq is well known to interact with sRNA and mRNA, early reports of biochemical interactions between Hfq and 16S rRNA and r‐protein S12 hinted that Hfq might also interact with ribosomes (de Haseth & Uhlenbeck, 1980; Strader et al, 2013). The relevance of such interactions, however, was unclear. In this issue, Andrade et al (2018) bridge this gap by providing evidence that Hfq is directly involved in the biogenesis and maturation of 30S ribosomes in E. coli. This connection between Hfq and the translational machinery provides an additional explanation for the pleiotropic effects of hfq deletion on bacterial growth and stress response (Vogel & Luisi, 2011). It also raises many intriguing questions of how bacterial cells maintain their capacity for protein synthesis when nutrients are limited.
Andrade et al (2018) show that E. coli cells lacking hfq grow poorly at 16°C and contain higher levels of unprocessed pre‐16S rRNA, especially during stationary phase. The amount of 23S rRNA is unaffected, suggesting that Hfq specifically acts on maturation of 30S ribosomal subunits. These phenotypes are similar to those produced by deletions of 30S assembly factors, indicating a link between Hfq and ribosome assembly. Andrade et al (2018) also found that an amino acid substitution that weakens RNA binding to the distal side of the Hfq ring also inhibited 30S maturation. By contrast, no ribosomal defects arose from amino acid substitutions on the rim or the proximal face of Hfq that inhibit sRNA binding. These results ruled out an indirect effect of Hfq on ribosome biogenesis through sRNA regulation and instead suggested that the distal face of Hfq interacts directly with the rRNA. This possibility was supported by the observation that Hfq co‐purifies with immature 30S subunits, but not with 70S ribosomes or polysomes.
How might Hfq increase the numbers of mature 30S ribosomes, particularly during stationary phase? First, E. coli Hfq can accelerate RNA base pairing and exchange of RNA helices in vitro (Santiago‐Frangos & Woodson, 2018). One possibility is that Hfq helps refold the pre‐16S rRNA into a form that can be processed by RNase E and RNase G. Dammel and Noller (1993) proposed that competition between an alternative helix in the 5′ leader upstream of the 16S coding sequence and helix 1 in the 16S rRNA impairs 30S maturation at cold temperatures. Hfq may facilitate refolding of the pre‐16S rRNA, thereby ensuring that the immature rRNA is processed.
Second, Hfq may recruit the processing enzyme RNase E to the pre‐30S complex. Hfq is known to stimulate mRNA turnover by RNase E, either alone or in tandem with a complementary sRNA (Vogel & Luisi, 2011). A candidate binding site for the distal face of Hfq lies in the region of the RNase E cleavage site in the pre‐16S rRNA, in a configuration very like that observed in mRNA targets of Hfq and sRNA regulation.
Third, Hfq may act more generally to block translation initiation by immature 30S ribosomes, as proposed for several well‐studied assembly factors. Immature 30S complexes can participate in translation in certain strains of E. coli (Clatterbuck Soper et al, 2013), but are thought to have lower activity and translational fidelity (Roy‐Chaudhuri et al, 2010). Interestingly, Andrade et al (2018) also observed that global synthesis of highly expressed r‐proteins was diminished in hfq − bacteria. Moreover, hfq − strains were more sensitive to antibiotics targeting the 30S ribosome and had increased levels of frameshifting, alternative initiation, and premature stops.
This study places Hfq in a growing list of ribosome biogenesis factors in bacteria, yet raises many more questions to be addressed, such as the timing of Hfq binding with pre‐30S complexes during 30S biogenesis, and how Hfq interacts with the RNA and protein components of the ribosome. Nevertheless, the importance of Hfq for translation during stationary phase observed by Andrade et al (2018) points to a wider role for Hfq during bacterial stress response than previously suspected.
The EMBO Journal (2018) 37: e99616
See also: https://doi.org/10.15252/embj.201797631 (June 2018)
References
- Andrade JM, dos Santos RF, Chelysheva I, Ignatova Z, Arraiano CM (2018) The RNA‐binding protein Hfq is important for ribosome biogenesis and affects translation fidelity. EMBO J 37: e97631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clatterbuck Soper SF, Dator RP, Limbach PA, Woodson SA (2013) In vivo X‐ray footprinting of Pre‐30S ribosomes reveals chaperone‐dependent remodeling of late assembly intermediates. Mol Cell 52: 506–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammel CS, Noller HF (1993) A cold‐sensitive mutation in 16S rRNA provides evidence for helical switching in ribosome assembly. Genes Dev 7: 660–670 [DOI] [PubMed] [Google Scholar]
- de Haseth PL, Uhlenbeck OC (1980) Interaction of Escherichia coli host factor protein with Q beta ribonucleic acid. Biochemistry 19: 6146–6151 [DOI] [PubMed] [Google Scholar]
- Paul BJ, Ross W, Gaal T, Gourse RL (2004) rRNA transcription in Escherichia coli . Annu Rev Genet 38: 749–770 [DOI] [PubMed] [Google Scholar]
- Roy‐Chaudhuri B, Kirthi N, Culver GM (2010) Appropriate maturation and folding of 16S rRNA during 30S subunit biogenesis are critical for translational fidelity. Proc Natl Acad Sci USA 107: 4567–4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago‐Frangos A, Woodson SA (2018) Hfq chaperone brings speed dating to bacterial sRNA. Wiley Interdiscip Rev RNA e1475 https://doi.org/10.1002/wrna.1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajani Z, Sykes MT, Williamson JR (2011) Assembly of bacterial ribosomes. Annu Rev Biochem 80: 501–526 [DOI] [PubMed] [Google Scholar]
- Strader MB, Hervey WJ IV, Costantino N, Fujigaki S, Chen CY, Akal‐Strader A, Ihunnah CA, Makusky AJ, Court DL, Markey SP, Kowalak JA (2013) A coordinated proteomic approach for identifying proteins that interact with the E. coli ribosomal protein S12. J Proteome Res 12: 1289–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Luisi BF (2011) Hfq and its constellation of RNA. Nat Rev Microbiol 9: 578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]


