SUMMARY
Microtubules act as “railways” for motor-driven intracellular transport, interact with accessory proteins to assemble into larger structures such as the mitotic spindle, and provide an organizational framework to the rest of the cell. Key to these functions is the fact that microtubules are “dynamic.” As with actin, the polymer dynamics are driven by nucleotide hydrolysis and influenced by a host of specialized regulatory proteins, including microtubule-associated proteins. However, microtubule turnover involves a surprising behavior—termed dynamic instability—in which individual polymers switch stochastically between growth and depolymerization. Dynamic instability allows microtubules to explore intracellular space and remodel in response to intracellular and extracellular cues. Here, we review how such instability is central to the assembly of many microtubule-based structures and to the robust functioning of the microtubule cytoskeleton.
1. INTRODUCTION
Along with actin (Pollard 2016) and intermediate filaments (Herrmann and Aebi 2016; Hol and Capetanaki 2016; Jacob et al. 2016; Jones et al. 2016; Yuan et al. 2016), microtubules (Figs. 1 and 2) constitute one of the three main classes of cytoskeletal filaments in eukaryotic cells. Microtubules are found in all characterized eukaryotic organisms. Thus, the last common ancestor of eukaryotes had microtubules; this ancestor also had the dynein and kinesin motors that operate on the microtubule cytoskeleton (Sweeney and Holzbaur 2016). Many prokaryotes have at least one gene homologous to that encoding tubulin, the most common of which encodes FtsZ, a protein that forms polymers involved in cytokinesis. These observations suggest that the tubulin gene family appeared very early, perhaps in the last common ancestor of all forms of life on Earth (Pollard and Goldman 2016). Even without knowing anything else about microtubules, the maintenance of these structures and their constituent proteins across such a large span of time and in highly divergent organisms indicates that they have a fundamental role in eukaryotic cell biology.
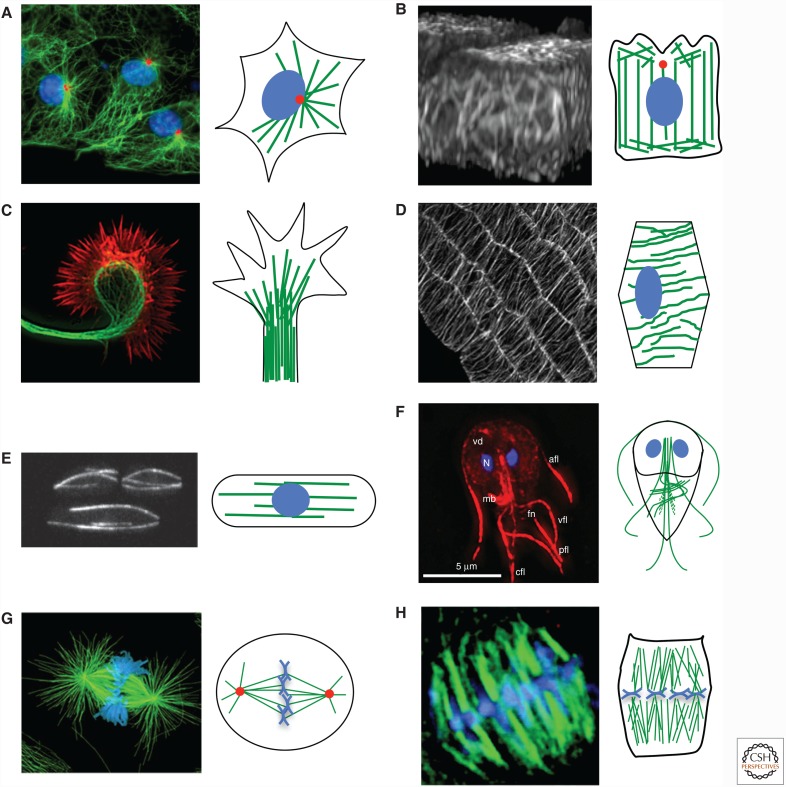
Figure 1.
The microtubule cytoskeleton in various cell types. Each pair of panels contains a fluorescence microscopy image of a specific cell/group of cells (left) with a cartoon depicting the generalized microtubule organization in that cell type (right). The color schemes for the microscope images are described below. In the cartoons, microtubules are shown in green, the DNA in blue, and centrosomes in red. Noncentrosomal microtubule nucleation machinery exists in many cell types (see text) but is not depicted. (A) Radial microtubule array in interphase cells. Microtubules (green), DNA (blue), microtubule-organizing center (MTOC; red). (B) Columnar microtubule array in polarized epithelial cells (green-fluorescent protein [GFP]–tubulin expressed in Madin–Darby canine kidney [MDCK] cells). (C) Microtubules in a neuronal growth cone. Microtubules (green) and actin filaments (red). (D) Cortical microtubule array in plant cells (GFP–tubulin expressed in Arabidopsis cells). (E) Fission yeast interphase microtubules (GFP–tubulin). (F) Microtubule cytoskeleton in Giardia. Microtubules (red), DNA (blue). (G) Animal cell mitotic spindle. Microtubules (green) and DNA (blue). (H) Metaphase plant mitotic spindle. Microtubules (green) and DNA (blue). (A, Reproduced from Gundersen laboratory website [http://www.columbia.edu/~wc2383/pictures.html]; B, reprinted from Reilein et al. 2005; C, reprinted from Kalil et al. 2011; D, reprinted from Ehrhardt and Shaw 2006, with permission of Annual Review of Plant Biology; E, reprinted, with permission, from Chang and Martin 2009, © Cold Spring Harbor Laboratory Press; F, reprinted, with permission, from Dawson 2010, © John Wiley & Sons Inc.; G, left image, reprinted from O’Connell and Khodjakov 2007, with permission from Elsevier, originally from Cell Motility and Cytoskeleton [1999] V.43[3] [cover], with permission from Wiley; H, reproduced from Yu et al. 1999.)
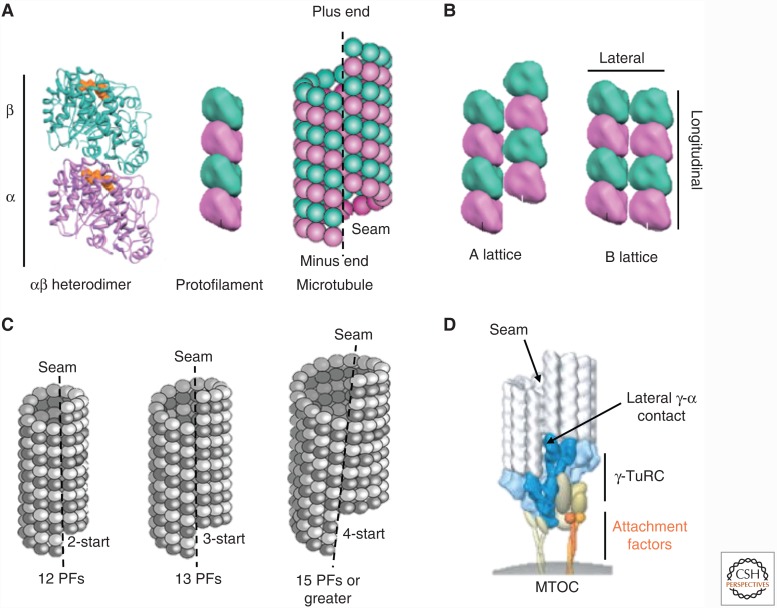
Figure 2.
Microtubule structure. (A,B) Key aspects of microtubule structure, as indicated. (C) Diagram of the relationship between protofilament number and microtubule structure. (D) Model of the γ-tubulin ring complex (γ-TuRC) associated with the minus end of a microtubule (gray). MTOC, microtubule-organizing center. (A,B, Modified from Kollman et al. 2011, with permission from Macmillan Publishers; C, modified, with permission, from Amos 2004, with permission of The Royal Society of Chemistry, http://dx.doi.org/10.1039/B403634D.)
Indeed, microtubules and their accessory proteins form the mitotic spindle—the dynamic self-organized machine that separates the chromosomes during mitosis, arguably the most important of all eukaryotic cell processes (McIntosh 2016). In addition, complexes of microtubules and motors form the core of cilia and flagella (Viswanadha et al. 2016), making microtubules essential for the motility of many organisms, including numerous protists and most metazoan sperm. Microtubules also provide tracks for motors that catalyze the movement of organelles, transport vesicles, and other structures. This microtubule-based intracellular transport contributes to the efficient function of many organisms and cell types, but it is crucial for the dramatically elongated neurons of animals (reviewed by Barlan and Gelfand 2016). Microtubules also play fundamental roles in cell organization by localizing organelles and establishing the polarity of a wide variety of cells in both animals and plants.
How do microtubules contribute to these diverse cellular activities? As reviewed here and elsewhere, there are many answers to this question, only some of which are well understood. One central theme is that the dynamic behavior of microtubule polymers is essential to many microtubule-based processes. Briefly, structures assembled from microtubules and actin filaments usually have longer lifetimes than the individual polymers from which they are assembled. In both cases, hydrolysis of nucleotides bound to the polymer subunits drives their turnover. However, the patterns of turnover differ. Actin filaments in vivo typically grow at their barbed end and disassemble at their pointed end as a result of multiple reactions, including severing by accessory proteins.1 In contrast, microtubule polymers usually (but not exclusively) display a surprising behavior termed “dynamic instability,” in which individual polymers switch stochastically (i.e., randomly) between growth and shortening. As explained below, this dynamic instability behavior is fundamental to many of the functions and properties of the microtubule cytoskeleton.
Because microtubule function and behavior derive ultimately from the structure and biochemistry of the microtubule filaments, this review starts with a description of their structure and biochemistry. It then discusses the subcellular structures that form from microtubules and returns to examine in more depth dynamic instability and its mechanism. Finally, we shall discuss the proteins that regulate microtubule dynamics and interact with microtubules to interface with the rest of the cell.
2. PHYSICAL ATTRIBUTES OF MICROTUBLES AND TUBULIN
2.1. Microtubule Structure
Microtubules are assembled from heterodimers of α- and β-tubulin into long hollow polymers that are ∼25 nm wide and range in length from <1 μm to >100 µm (Fig. 2). These heterodimeric tubulin subunits are referred to as αβ-tubulin, tubulin dimers, or simply “tubulin.”
Microtubule structure is most straightforwardly described as comprising approximately 13 linear protofilaments (PFs) that are associated laterally and closed into a hollow tube. The resulting polymer is polar, with a fast-growing plus end that has exposed β-tubulin and a slow-growing minus end with exposed α-tubulin (Fig. 2A).
In typical 13-PF microtubules, the boundary where the tube closes, unlike the other interfaces between PFs, creates a “seam” where the lateral interactions among PFs differ from those elsewhere in the microtubule (Fig. 2A). This seam is generally believed to be a weak point of the microtubule structure, although there is some evidence to the contrary (Sui and Downing 2010). Although most microtubules in vivo have 13 PFs, there are some exceptions, and microtubules assembled in vitro can have a wide range of PF numbers. One striking difference between the typical 13-PF microtubules and those with 15 or 16 PFs is that the 15- and 16-PF microtubules do not have a recognizable seam (Fig. 2C).
The number of PFs and presence/absence of a seam might seem like arcane details, but the idea that the PF number can have physiological relevance is supported by the observation that microtubules with consistently different numbers of PFs do appear in nature (reviewed by Sui and Downing 2010). In addition, the issue of PF number has practical significance for researchers because the presence of a seam interferes with structure determination by methods based on helical reconstruction. In contrast, the seamless body of 15- and 16-PF microtubules is fully symmetric (Fig. 2C). Thus, in vitro methods to produce (limited numbers of) 15- and 16-PF microtubules can be quite useful for determination of the structure of microtubules and their binding proteins, such as motors (e.g., see Arnal et al. 1996).
Although PF-based descriptions of microtubule structure are easiest to visualize, microtubules are sometimes described in terms of their lattice structure and/or helical structure (Box 1).
BOX 1.
DESCRIPTION OF MICROTUBULE STRUCTURE IN TERMS OF LATTICES AND HELICES
The main body of a typical 13-PF microtubule can be described as being composed of a so-called B lattice, in which α-subunits are next to α-subunits (α–α) and β next to β (β–β). However, at the seam, the α-subunits associate laterally with β-subunits in the adjacent PFs (α–β) in an A lattice (Fig. 2A,B). The lattice structure changes at the seam because each PF in the main body of a microtubule is shifted slightly relative to its neighbor, resulting in an offset at the seam of 1.5 dimers for a 13-PF microtubule (Fig. 2C). Changing the number of PFs changes the offset, so that the offset for microtubules with 15 or 16 PFs is two full subunits, resulting in a situation in which there is no discernable seam (these microtubules comprise entirely B lattice: α–α and β–β).
In addition, microtubules are sometimes described as helices, although the presence of a seam means that a 13-PF microtubule is not a true helix; it just appears to be one in low-resolution electron microcopy images in which α- and β-tubulin are indistinguishable. Given this ambiguity, a 13-PF microtubule can be described as a left-handed three-start helix because each of the three monomers in the 1.5 tubulin dimer offset at the seam can be viewed as starting a new helix (Fig. 2C). As noted above, changing the number of PFs changes the offset, so that the offset for microtubules with 15 or 16 PFs is two full subunits. This situation results in a four-start helix, in which, as noted above, B lattice is found uniformly through the microtubule (Fig. 2C; see also Amos 2004).
Although considering microtubules as (pseudo)helices can be useful in structural studies, it is usually more informative to view microtubules as PF-based structures because a consensus is building that microtubules grow by adding subunits to these linear PFs, not by extending the helices. This idea is based, in part, on evidence that the longitudinal interactions between subunits within a PF are stronger and more extensive than the lateral bonds between subunits in different PFs (Sept et al. 2003; Zhang et al. 2015). The predominance of these longitudinal bonds means that interactions within a given PF are more significant than those within a given helix.
Several additional aspects of microtubule structure are functionally significant. First, the surface of microtubules is negatively charged, because the carboxy-terminal tails of α- and β-tubulins contain several acidic residues and are located on the outer surface. These carboxy-terminal tails (also called E-hooks because they are glutamate-rich) are key sites of interaction for many microtubule-binding proteins (MTBPs) (reviewed by Roll-Mecak 2015). Second, although the walls of microtubules are often represented as being solid, high-resolution structural analysis shows that microtubule walls contain holes large enough to allow diffusion of water and small molecules such as taxol (Li et al. 2002). The inside of a microtubule is an intriguing space that is, as yet, relatively unexplored in terms of its significance and potential interactions. Some electron microscopy images of microtubule cross sections have visualized densities of unknown composition inside microtubules (Garvalov et al. 2006); one explanation for these lumenal structures is that they are enzyme complexes involved in posttranslational modification of tubulin subunits such as acetylation (e.g., see Soppina et al. 2012).
2.2. Microtubules Are Structurally Rigid
Microtubules and actin filaments both have physical properties similar to the stiff plastic Plexiglas, but microtubules are much more rigid owing to their larger diameter and tubular construction. To make this comparison more quantitative, the persistence length of microtubules is ∼5000 µm compared with ∼20 µm for actin filaments (Gittes et al. 1993; Hawkins et al. 2010). The rigid nature of microtubules is hard to reconcile with the observation that microtubules are often highly curved in vivo. One explanation is that the curvature results from forces exerted by motors and connections to other cytoskeletal elements. Alternative explanations include the idea that curvature is induced by lattice defects (i.e., missing and/or misincorporated subunits) and/or binding of MTBPs. The mechanical aspects of microtubules are discussed in more depth elsewhere (Hawkins et al. 2010).
2.3. Isoforms of Tubulin Are Specialized for Specific Functions
Microtubules comprise subunits consisting of heterodimers of αβ-tubulin, an arrangement that appears to be ubiquitous in eukaryotes (see Pollard and Goldman 2016 for a discussion of microtubule-related proteins in prokaryotes). However, most eukaryotic cells also contain multiple tubulin isoforms, many of which are both ancient in origin and fundamental to aspects of microtubule function, as explained briefly below. For more detailed discussion of the tubulin superfamily, see McKean et al. (2001) and Findeisen et al. (2014).
α- and β-tubulin assemble into obligate heterodimers that form the body of cytoplasmic microtubules (Fig. 2A) in all eukaryotic organisms characterized thus far. Both bind the nucleotide GTP, but only the β-subunit hydrolyzes its GTP in the course of normal microtubule polymerization. Folding of α- and β-tubulin requires the assistance of a set of dedicated chaperones (Tian and Cowan 2013).
γ-tubulin is a key part of the machinery that nucleates the growth of new microtubule structures (Fig. 2D; see below for details). Like α- and β-tubulin, γ-tubulin has been found in all free-living (i.e., nonparasitic) eukaryotic organisms examined, and the combination of these three proteins appears to form the minimal set of eukaryotic tubulins (Findeisen et al. 2014; Gull 2001).
δ-, ɛ-, and ζ-tubulin isoforms are found in cilia, flagella, and/or basal bodies, and in general they are specific to organisms with these structures. These ancient proteins have been identified in some of the most divergent known eukaryotic organisms, but they have also been lost from many lineages (McKean et al. 2001; Findeisen et al. 2014).
Other tubulin isoforms (e.g., η-tubulin) have been described, but appear to be limited to specific lineages (Findeisen et al. 2014).
2.4. New Microtubules Appear through the Process of Nucleation
Given the complex structure of a microtubule, one might imagine that generation of new microtubules is a difficult process. Indeed, spontaneous appearance of new microtubules is rare in solutions of pure αβ-tubulin heterodimers unless tubulin concentrations are high. Nucleation of pure tubulin is an unfavorable and highly cooperative process, with the nucleation rate depending on a large power (6–12) of the tubulin concentration (Caudron et al. 2002). This naturally suppresses spontaneous nucleation and allows cells to regulate when and where new microtubules will be assembled. Careful control of the localization and activity of microtubule nucleators plays an important role in generating different types of microtubule arrays (e.g., Fig. 1).
Generation of new microtubules in vivo involves the action of specialized nucleation machinery, generally associated with γ-tubulin. The most well-known and intuitively understandable nucleator is the γ-tubulin ring complex (γ-TuRC), a lock-washer-shaped structure that appears to act as both a template for microtubule assembly and a cap for the minus end (Fig. 2D). γ-TuRC is well-characterized biochemically in vertebrates, but components are found in many organisms, including fungi and plants, and so involvement of γ-TuRC in nucleation appears to be ancient and widespread (Kollman et al. 2011).
Although most well-characterized examples of microtubule nucleation in vivo involve γ-tubulin, additional mechanisms for increasing microtubule numbers exist. Many microtubule-stabilizing proteins have nucleation activity in vitro. However, it is not clear whether these proteins nucleate microtubules in vivo or simply stabilize new microtubules nucleated by other mechanisms. More significantly, augmin can promote nucleation of new microtubules from the sides of preexisting microtubules (Petry et al. 2013). The significance of this side nucleation is still being debated, but it has been implicated in the functioning of the mitotic spindle and beyond (Sanchez-Huertas and Luders 2015). Finally, microtubule-severing proteins, such as katanin, can potentially increase microtubule numbers by breaking existing microtubules into multiple pieces (discussed in Ehrhardt and Shaw 2006). The contributions of these mechanisms to the function of the microtubule cytoskeleton are currently being elucidated.
2.5. Microtubules Undergo Posttranslational Modifications
One curious and as yet poorly understood aspect of microtubule structure is that tubulin subunits undergo a series of posttranslational modifications. These include common modifications such as phosphorylation, acetylation, and sumoylation, as well as more unusual and/or tubulin-specific modifications such as detyrosination (removal of the carboxy-terminal tyrosine) and polyglutamylation (addition of free glutamates to the side-chain of a glutamate in the polypeptide; reviewed by Janke and Bulinski 2011; Garnham and Roll-Mecak 2012). It is interesting to note that most of these modifications occur on the residues of the carboxy-terminal E-hook; an exception is acetylation, which occurs in the microtubule lumen (Soppina et al. 2012). Experiments in a wide range of organisms and cell types have shown that, in many cases, the modifications are more common in older (stabilized) polymers. It is tempting to think on the basis of this observation that the modification(s) cause the stabilization. However, tests of this hypothesis have generally been inconclusive, and so at present it is safest to conclude only that many modifications correlate with polymer stabilization.
In contrast to the above, it is clear that the modifications can alter the affinity of binding proteins for microtubules, and hence it is possible that the partial correlation between modification and stability is mediated by MTBPs. For example, detyrosination inhibits binding of both the plus end–tracking protein EB1 and depolymerizing motor MCAK while increasing binding of the transport motor kinesin-1 (reviewed by Garnham and Roll-Mecak 2012). A number of studies have suggested that other posttranslational modifications influence the affinities of motors for microtubules, although interpreting this work is complicated (reviewed by Janke and Bulinski 2011). Although much work remains to be performed, an emerging notion is that posttranslational modifications create chemical marks along stabilized microtubules that specialize them for specific functions. Clearly, determining the functional significance and spatiotemporal regulation of microtubule modifications will be an important focus for future study.
3. STRUCTURES FORMED FROM MICROTUBULES
3.1. The Interphase Microtubule Array
The interphase array of microtubules (usually dynamic, sometimes stabilized) helps to determine cell shape and organization and acts as a substrate for motor-driven intracellular transport. The organization of the interphase microtubule array varies by cell type and organism (Fig. 1). In many cell types, ranging from vertebrate fibroblasts to Dictyostelium amoebas, the interphase microtubule cytoskeleton is a radially organized structure that emanates from a centrally located microtubule-organizing center (MTOC; see below) (Fig. 1A). In radially organized cells, the plus ends of the microtubules are oriented toward the cell boundary, with the minus ends embedded in the MTOC.
Although radial organization of the microtubule cytoskeleton is common and is sometimes presented as canonical, it is by no means universal. As one example of a different arrangement, the microtubule array in vertebrate polarized epithelial cells has a more parallel organization, with the microtubule minus ends located toward the apical membrane and the plus ends located toward the base of the cell (Fig. 1B) (Bartolini and Gundersen 2006). In higher-plant cells (e.g., Arabidopsis), microtubules are found in cortically associated parallel arrays oriented transverse to the axis of cell elongation (Fig. 1D) (Ehrhardt and Shaw 2006).
The organization of the microtubule array is important in most cells because it plays a central role in determining the organization of the rest of the cell. For example, when microtubules are oriented radially, as in fibroblasts, the membranes of the Golgi apparatus are generally located near the MTOC, with most other membranes distributed more peripherally. In contrast, the parallel microtubule arrays endow polarized epithelial cells with a more linear internal organization, with the Golgi membranes at the apical face, and other membranes located more basolaterally (Bartolini and Gundersen 2006). In these cells and others, loss of microtubules leads to loss of normal internal organization, as well as other aspects of cell polarity (de Forges et al. 2012). For example, many animal cells can move without microtubules, but lose directionality (Ganguly et al. 2012), and plant cells with depolymerized microtubules grow aberrantly (Ehrhardt and Shaw 2006).
How is microtubule organization established and maintained? These processes are not yet well understood, but the microtubule array, as observed in a particular cell, emerges from interactions between dynamic microtubules, their nucleators, their regulators, microtubule motors, and the cell boundary (Box 2).
BOX 2.
HOW IS THE ORGANIZATION OF THE MICROTUBULE NETWORK ESTABLISHED AND MAINTAINED?
The obvious answer is that microtubule organization depends on localization of the microtubule organizing center, but functional bipolar spindles can form in cells even after the centrosomes have been removed by microsurgery (Khodjakov and Rieder 2001), and a number of cell types (most obviously plant cells) lack centrosomes (Ehrhardt and Shaw 2006). Moreover, spindle-like bipolar structures can form around artificial chromosomes in vitro in the absence of centrosomes (Heald et al. 1996). Examination of the localization of γ-tubulin in these and other noncentrosomal systems leads to a related proposal: Perhaps microtubule organization depends on the localization of the microtubule nucleators. Centralized nucleation machinery correlates with radial organization, and distributed nucleation machinery correlates with distributed networks (Fig. 1). Although compelling, this explanation raises the next question: What determines the localization of the nucleators?
A hint to resolving this conundrum is provided by the observation that mixtures of stabilized microtubules and purified active motors can spontaneously self-organize into a range of different structures; the details of these structures depend on the specific activities and ratios of the proteins involved (Surrey et al. 2001). Motor-driven microtubule organization is also involved in the formation of dynamic radial microtubule arrays in melanophore cell fragments (Vorobjev et al. 2001).
However, it is important to remember that other aspects of the cellular environment can also influence the microtubule cytoskeleton. For instance, the pigment granules in melanophores appear to participate in microtubule organization (Vorobjev et al. 2001), and the Golgi apparatus can nucleate microtubules and/or act as a MTOC (de Forges et al. 2012). Moreover, the signal transduction pathways that were originally hypothesized by Mitchison and Kirschner to allow selective stabilization of microtubules near particular regions of the cell boundary (Kirschner and Mitchison 1986) are becoming elucidated (Akhmanova et al. 2009). Finally, it is important to understand that the physical barrier presented by the cell boundary can itself influence microtubule organization and dynamics (Maly and Borisy 2002; Gregoretti et al. 2006; Dogterom and Surrey 2013).
In summary, it is becoming apparent that the interphase microtubule array emerges from dynamic interactions between microtubules, motors, their regulators, and their physical environment. Gaining a deeper understanding of how particular large-scale structures emerge from local interactions will likely require computational modeling and other approaches used to study complex systems.
3.2. Other Subcellular Microtubule-Based Structures
3.2.1. Mitotic Spindle
The mitotic spindle (Fig. 1G,H) is the complex and beautiful self-assembled machine that separates the chromosomes in all eukaryotic cells. The mitotic spindle comprises dynamic microtubules, a wide array of motors, and a series of other microtubule-associated proteins (MAPs). The mitotic spindle forms when the interphase microtubule array undergoes a dramatic reorganization upon entry into mitosis. The assembly, activities, and properties of the mitotic spindle have been, and continue to be, the subjects of intense study, and they are discussed in more detail elsewhere (McIntosh 2016).
3.2.2. Microtubule-Organizing Centers, Centrosomes, and Spindle Pole Bodies
MTOCs, centrosomes, and spindle pole bodies (SPBs) are various names given to localized foci of microtubule-nucleating machinery (see Vertii et al. 2016). “Microtubule-organizing center” is a term that applies to all of these structures, whereas “centrosome” usually applies more specifically to the perinuclear MTOC of radially organized cells, and “spindle pole body” applies to the nuclear-membrane-embedded MTOC of fungi such as the budding yeast Saccharomyces cerevisiae and the fission yeast Schizosaccharomyces pombe. Although centrosomes and SPBs have similar functions in terms of microtubule nucleation and contain many similar proteins, their ultrastructure is quite different (Ada and Kilmartin 2000). MTOCs contain γ-tubulin and the γ-TURC (Fig. 2D), but can contain a complex array of other proteins, such as motors and +TIPs (microtubule plus end–tracking proteins; see Sec. 5 below), and can include centrioles (see below). In the past, centrioles were thought to be fundamental to the function of MTOCs, but many organisms (e.g., most higher plants) lack centrioles, and fly mutants lacking centrioles develop in a largely normal way (Basto et al. 2006). One explanation for the frequent association between centrioles and centrosomes is that the colocalization of these structures keeps the microtubule nucleating activities organized in a single focus.
3.2.3. Flagella and Cilia
Flagella and cilia (reviewed by Viswanadha et al. 2016) are complex organelles that comprise highly organized arrangements of microtubules, motors, and other proteins. Structurally they are similar, but they can differ in terms of attributes such as their function, motion, length, and details of their protein composition. Flagella and cilia are highly conserved and ancient organelles, existing in similar form in organisms ranging from human to some of the most divergent protists (Carvalho-Santos et al. 2010).
3.2.4. Centrioles and Basal Bodies
Centrioles and basal bodies are complex structures that typically comprise nine sets of triplet microtubules and a set of well-conserved associated proteins (Vertii et al. 2016), although with some variation (Carvalho-Santos et al. 2011). They are found at the base of flagella and cilia (where they are called basal bodies) and in centrosomes (where they are called centrioles). Centrioles and basal bodies can interconvert as cells pass through the cell cycle. Centrioles undergo a mysterious form of replication in which new centrioles appear at 90° angles from the parent centrioles, and this replication is typically tightly coordinated with cell replication and assembly of the mitotic spindle (Nigg and Stearns 2011).
3.2.5. The Midbody
The midbody is an enigmatic structure comprising bundled microtubules and associated proteins derived from the mitotic spindle. The midbody forms during cytokinesis at the point of abscission (separation) of the two daughter cells. Midbodies have often been viewed as waste depots for the cell-division process, but increasing evidence suggests that these structures are transient organelles with still-mysterious functions of their own (Chen et al. 2013).
3.2.6. Organism-Specific Structures
Protists contain a wide variety of complex microtubule-based structures that are important for their viability and/or pathogenicity. Striking examples include the Toxoplasma conoid (Morrissette 2015), the Giardia ventral disk (i.e., the suction plate) (Fig. 1F) (Schwartz et al. 2012), and the cilia array of ciliates (Winey et al. 2012). The diversity of cellular architecture in protists is remarkable, and the mechanisms leading to generation of these structures are only beginning to be defined (Slabodnick and Marshall 2014).
4. MICROTUBULE ASSEMBLY AND DYNAMICS
4.1. Introduction to Microtubule Dynamics
The term “cytoskeleton” brings to mind a static structure, an idea that is reinforced by immunofluorescence images such as those in Figure 1. Nothing could be farther from the truth: As with actin (reviewed by Pollard 2016), the microtubule cytoskeleton in eukaryotic cells is constantly turning over in a process that is driven by nucleotide hydrolysis (GTP hydrolysis in the case of microtubules). However, the energy of the nucleotides is not used to build actin filaments and microtubules but rather to destroy them. This concept can be shown by replacing the GTP in the tubulin subunits with the slowly hydrolyzable GTP analog GMPCPP (guanylyl 5′-α,β-methylenediphosphonate): The microtubules grow, but fail to disassemble normally (Hyman et al. 1992). Where does the energy for building the filament come from? The answer is that assembly of GTP–tubulin into microtubules is a spontaneous process that is driven primarily by the hydrophobic effect (Vulevic and Correia 1997), like many other forms of macromolecular assembly.
Microtubules in most cell types display a behavior known as dynamic instability, in which the ends of individual polymers transition randomly between periods of growth and shortening (Fig. 3). In many animal cells, the minus ends of most microtubules are embedded in the MTOC (see discussion above), and so dynamic instability occurs primarily at the plus ends, but uncapped minus ends can also show dynamic instability, at least in vitro (Fig. 3A). Some microtubules can also treadmill, a behavior that is particularly important in the cortical microtubule array of plants (Ehrhardt and Shaw 2006) but is also seen in more-limited cases in animal cells (e.g., Rodionov and Borisy 1997; Vorobjev et al. 2001).
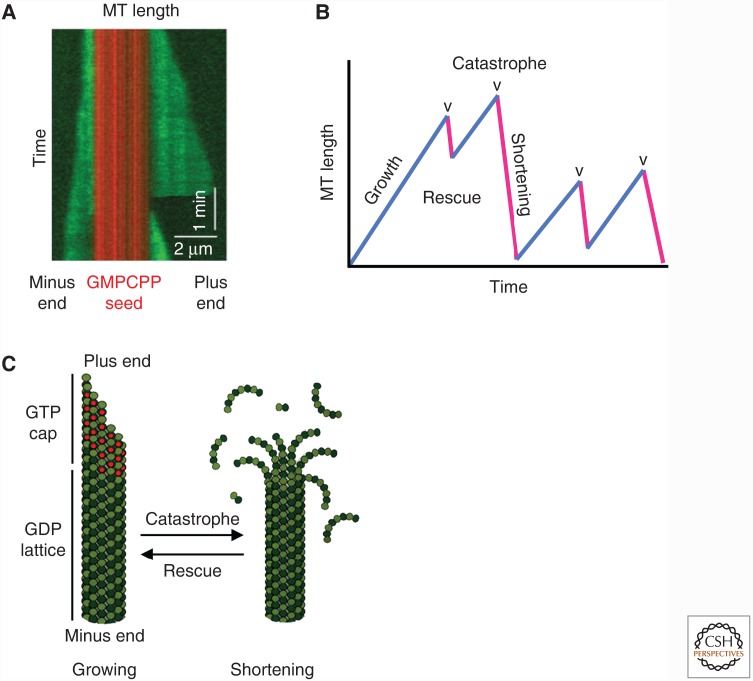
Figure 3.
Microtubule (MT) dynamics and assembly. (A) Kymograph (length/time plot derived from a movie) of a microtubule undergoing dynamic instability in vitro, with dynamics at both the minus (left) and plus (right) ends. Green represents Alexa488-labeled tubulin, and red represents tetra-rhodamine-labeled tubulin GMPCPP (guanylyl 5′-α,β-methylenediphosphonate)-stabilized microtubule seeds. (B) Cartoon of a length-history plot (also called a life-history plot) of a microtubule undergoing dynamic instability. The key processes of microtubule dynamics are indicated. (C) Standard model of dynamic instability. As long as the microtubule has a GTP cap, it can grow, but it transitions to rapid depolymerization (catastrophe) on loss of the GTP cap. (A, Adapted, with permission, from Zanic et al. 2013.)
The constant turnover of cytoskeletal structures might seem wasteful—just consider the amount of GTP that is “burned” by a single microtubule undergoing dynamic instability. Why would cells expend so much energy constantly destroying structures that they have just built? It turns out that microtubule turnover is necessary for many aspects of cell physiology and is an essential aspect of the microtubule cytoskeleton. Some examples include the following.
The dynamic nature of the microtubule cytoskeleton allows cells to adapt to changes in cell shape and environment.
The random probing generated by dynamic instability allows individual microtubules to explore cellular space and bring the microtubule “train tracks” into contact with cargo such as vesicles, organelles, and chromosomes, which are too large to diffuse effectively in the highly crowded environment of the cytoplasm.
Spatially localized (“selective”) stabilization of dynamic microtubules provides a mechanism for generating morphological change in response to internal or external signals (Kirschner and Mitchison 1986).
The combination of spatial exploration and selective stabilization (sometimes summarized by the phrase “search–capture”) plays a fundamental role in the self-assembly of structures such as the mitotic spindle (reviewed by Mogilner et al. 2006).
The process of building and maintaining such a dynamic microtubule cytoskeleton might be energy-intensive, but it is profoundly robust—in other words, it is unlikely to fail even if significant perturbations (e.g., changes in cell shape, number of microtubules) occur to the system. This realization is important for understanding how the cytoskeleton works, but it also has implications for understanding the process by which the cytoskeleton evolved (Kirschner and Gerhart 2005).
4.2. Microtubule Assembly Can Be Altered by Changes to the Environment and by Drugs
Microtubules assembled from pure GTP–tubulin are remarkably unstable polymers—reduce the concentration of GTP–tubulin subunits for a few seconds and they disappear. Rapid depolymerization can also be induced by reducing the temperature or shifting other aspects of the environment (e.g., an increase in Ca2+). The transient and unstable nature of microtubules is all the more striking when considered alongside the ability of these structures to withstand physical perturbations (bending, tension), as outlined above.
Researchers can manipulate the assembly state of microtubules through the use of small molecules. The natural product taxol and its relatives induce microtubule assembly and can stabilize microtubules against dilution-induced depolymerization and (to a lesser degree) cold temperatures, whereas molecules such as nocodazole, colchicine, vinblastine, and vincristine destabilize microtubules. The mechanism of taxol-mediated stabilization has been unclear (Amos 2011), but recent high-resolution structural data might be resolving this issue (Alushin et al. 2014).
Microtubule-altering drugs are very important in agriculture and medicine, especially cancer chemotherapy, at least partly because of the role microtubules play in spindle assembly. Microtubule-directed drugs can be somewhat organism specific, and some commercially significant compounds target microtubules of fungi or plants (e.g., Benomyl). Hence, work continues on developing new compounds that target microtubules (Amos 2011). Researchers are also developing drugs that target MTBPs—for example, motors (as possible cancer-fighting agents) and tau (as possible treatments for neurodegenerative disease) (Rath and Kozielski 2012).
4.3. Mechanism of Microtubule Dynamic Instability
As discussed above, the apparently random2 transitions between growth and depolymerization that characterize microtubule dynamic instability are functionally significant, but they are also intriguing. Transitions from growth to depolymerization are termed “catastrophes,” whereas those from depolymerization to growth are called “rescues” (Fig. 3B). What could cause such abrupt switching? In other words, what is the “mechanism” of dynamic instability? The answers to these questions have become clearer in the 30 years that have passed since dynamic instability was first recognized (Mitchison and Kirschner 1984), but some important questions still remain.
Some generally accepted experimental observations relevant to the mechanism of dynamic instability include the following.
As discussed above, soluble tubulin binds to GTP. The α-subunit binds GTP without hydrolyzing or exchanging. The exchangeable GTP on the β-subunit hydrolyzes and releases its phosphate quickly after polymerization (with rates on the order of 0.5/sec) but slowly in the absence of polymer (Melki et al. 1998; see also Margolin et al. 2012 and Seetapun et al. 2012).
Assembly promotes hydrolysis of the GTP bound to the subunits. This occurs because the incoming subunit functions as a GAP (GTPase-activating protein) for the subunit by completing its nucleotide-active site (Nogales et al. 1998).
GTP–tubulin will assemble into microtubules if the free-tubulin concentration is sufficiently high (i.e., above the critical concentration). GDP–tubulin will not assemble into more than oligomers (its critical concentration is impractically high; Howard 2001).
Growing microtubules have slightly curved PFs and/or sheet-like extensions at their tips (Chretien et al. 1995), but depolymerizing microtubules have at their tips tightly curled “ram’s horns” consisting of curved PFs that appear to be peeling off the microtubules. Isolated GDP–tubulin can form rings similar to these rams’ horns.
These observations led early on to the idea that GTP–tubulin and GDP–tubulin have different preferred conformations. Initially, it was proposed that GTP–tubulin is straight and GDP–tubulin is curved. Later structural data suggested that both are curved, but that GTP–tubulin can become straight in the context of a microtubule. The bending appears to occur both within and between subunits (reviewed by Brouhard and Rice 2014).
Microtubules assembled from the slowly hydrolyzing GTP-like analog GMPCPP polymerize as normal but depolymerize much more slowly (Hyman et al. 1992). This is a key point: It means that the energy of GTP is used to destroy the microtubule not to build it.
Microtubule growth and dynamic instability can occur at either end, although the kinetics of the two ends differ (e.g., the plus end grows faster than the minus end).
For further information and citations not given above, see Desai and Mitchison 1997, Howard 2001, and Howard and Hyman 2003.
These and other experiments have led to the standard textbook model of dynamic instability, which typically includes the following ideas.
After a new subunit adds to a growing microtubule tip, a short delay in hydrolysis and phosphate release result in a GTP-rich region (the GTP cap3) at the growing tip, whereas older parts of the microtubule primarily comprise GDP–tubulin (this region is sometimes referred to as the “GDP lattice”) (Fig. 3C).
GTP hydrolysis promotes depolymerization in at least two ways: First, the GDP–tubulin in the microtubule lattice is under strain because it is forced to be in an unfavorable straight conformation. This strain effectively weakens the already weak lateral bonds between PFs (Zhang et al. 2015). In addition, conformational changes associated with hydrolysis and phosphate loss weaken the longitudinal bond (Rice et al. 2008; Zhang et al. 2015). (Note: It used to be thought that the lateral bonds of the GTP and GDP lattices are significantly different, but, as has been seen from recent cryo-electron microscopy [EM] work, the lateral contacts of the GTP and GDP lattices are similar; GTP hydrolysis further weakens the lateral bonds by increasing the strain on these bonds [Zhang et al. 2015].)
When the GTP cap is present (i.e., as long as new GTP–tubulin is added to the tip faster than it is lost through dissociation and/or hydrolysis), its relatively strong lateral bonds maintain the tubular structure and allow continued polymerization. However, if the cap is lost, a so-called catastrophe occurs: The GDP–tubulin is exposed, the PFs splay apart as the GDP subunits adopt their preferred bent conformation, and the microtubule rapidly depolymerizes.
According to this model, the GDP lattice below the GTP cap stores the energy of GTP hydrolysis, allowing the depolymerizing microtubule to do work (Grishchuk et al. 2005; Mogilner and Oster 2003).
Although this conceptual model is attractive, it leaves many questions unresolved. For example, what are the size and shape of the GTP cap? Are GTP and GDP the only important conformations, or do intermediates (i.e., GDP-Pi) play an important role? Why do the minus and plus ends show quantitatively different dynamic instability behaviors?4 What are the detailed molecular mechanisms of catastrophe and rescue? One can imagine fluctuations in tubulin addition and first-order GTP hydrolysis leading to catastrophe, but what series of events could cause a rapidly depolymerizing microtubule to start polymerizing again?
The sudden and apparently unpredictable nature of catastrophe and rescue have been so puzzling that some researchers have proposed that they result entirely from “outside” influences, such as lattice defects or thermal fluctuations, and/or require the action of MTBPs. Alternatively, it has been proposed that microtubules normally grow as open sheets, and that catastrophe might be caused by tube closure, which could be related in an unspecified way to GTP hydrolysis (Chretien et al. 1995; Wieczorek et al. 2015). It is important to note that some of these ideas are not mutually exclusive with the standard model presented above and that multiple mechanisms are likely at work (e.g., catastrophe can be both a spontaneous event and one caused by MTBPs).
4.4. Current Research into the Mechanism of Dynamic Instability
Recently an array of new experiments, such as nanoscale assembly measurements, superresolution microscopy, and localization of conformation-specific binding proteins, has provided new insight into the microtubule assembly processes (e.g., Dimitrov et al. 2008; Gardner et al. 2011; Seetapun et al. 2012; Coombes et al. 2013). These approaches have been complemented by determination of microtubule structure at higher resolution (Alushin et al. 2014; Zhang et al. 2015) and improved computer simulations (e.g., VanBuren et al. 2005; Margolin et al. 2012; Bowne-Anderson et al. 2013). The picture that is emerging is broadly consistent with the classical model of dynamic instability outlined above, but it contains additional molecular detail, and it is beginning to address some of the open questions.
More specifically, the model that emerges from multiple experimental and theoretical studies is one in which rapidly exchanging GTP subunits add to PF tips (with only a small fraction being incorporated), and the nonterminal subunits undergo first-order (not vectorial5) GTP hydrolysis. The resulting microtubule structure has an unstable GDP lattice capped by a stabilizing region rich in GTP (or GDP-Pi). This GTP cap has a size that depends on the elongation rate, as well as an approximate exponential shape with a poorly defined lower boundary, and it can extend approximately 10 or more subunits into the microtubule (Margolin et al. 2012; Seetapun et al. 2012; Coombes et al. 2013; Bowne-Anderson et al. 2015). In these models, open sheets are not observed, but instead flat extensions protruding from a closed tube are seen; these sheets might explain the age-dependent catastrophe that has been observed experimentally (Coombes et al. 2013).
Dimer-based computational simulations founded on this conceptual model recapitulate many aspects of experimentally observed dynamic instability, supporting its broad structure, but they also make new predictions. For example, one of these models suggests that the rapid subunit exchange mentioned above occurs because cracks (laterally unbonded regions) exist between adjacent PFs in the region close to the tip, allowing subunits to exchange until they form lateral bonds and become incorporated into the lattice (Margolin et al. 2012; Li et al. 2014). One appealing aspect of this and related ideas is that the healing of these cracks by MTBPs and the resulting suppression of loss of recently added subunits could potentially account for the otherwise surprising ability of some MTBPs to increase the rate of microtubule growth (Howard and Hyman 2009; Gardner et al. 2011).
These computer simulations are also providing specific hypotheses about the molecular-scale mechanisms of catastrophe and rescue. One idea is that catastrophe results from stochastic fluctuations in the extent to which interprotofilament cracks extend into the GDP-rich region (Margolin et al. 2012; Li et al. 2014) and/or the lengths of the PF extensions (Coombes et al. 2013). Rescue (a rare event in the absence of MTBPs) might be promoted by stochastic blunting of the microtubule tip, followed by reestablishment of regions of laterally bonded GTP–tubulin (Margolin et al. 2012; Li et al. 2014). Regulatory proteins could potentially promote or suppress transitions by altering the stability of lateral bonding between subunits or the tip extensions (e.g., Gupta et al. 2013) or by altering delivery of tubulin subunits to the tip (Ayaz et al. 2014). In addition, the ability of computer simulations to follow a system simultaneously at different scales means that these models have the potential to provide insight into long-standing questions about how the properties of the tubulin dimers relate to the observed dynamic instability parameters and bulk-scale properties such as the critical concentration.
5. MICROTUBULE-BINDING PROTEINS
5.1. Introduction to MTBPs
Naked microtubules assembled from pure tubulin are unstable structures that are poised, quite literally, at the edge of catastrophe. This situation enables microtubule assembly to respond quickly to changes in the environment and the influence of regulatory proteins. Some microtubule regulatory proteins are widespread in eukaryotes, but others vary significantly by organism and cell type. Even among the organism-specific proteins there are some common motifs and themes, as discussed below.
The term MTBP applies broadly to any protein that can be shown experimentally to bind to microtubules. Another term, MAP, is often used to describe the subset of MTBPs that cosediment with microtubules through multiple rounds of polymerization and depolymerization, a group that includes proteins such as MAP2 and tau.
Broadly speaking, the MTBPs that regulate assembly can be categorized functionally as stabilizers, destabilizers (including severing proteins), capping proteins, and bundlers/cross-linkers (Fig. 4). Other MTBPs include motors that use microtubules as tracks for intracellular transport (reviewed by Sweeney and Holzbaur 2016) and cytoplasmic linker proteins (CLIPs), which anchor organelles to microtubules to promote cell organization. Some MTBPs are cytoskeletal integrators (i.e., proteins that connect to other components of the cytoskeleton). In addition, some proteins involved in signal transduction, translation, and metabolism bind microtubules or other components of the cytoskeleton. Many MTBPs have multiple activities. Determining the functions of MTBPs can be challenging because the activity observed can depend on the details of the assay or on other aspects of the experimental conditions.
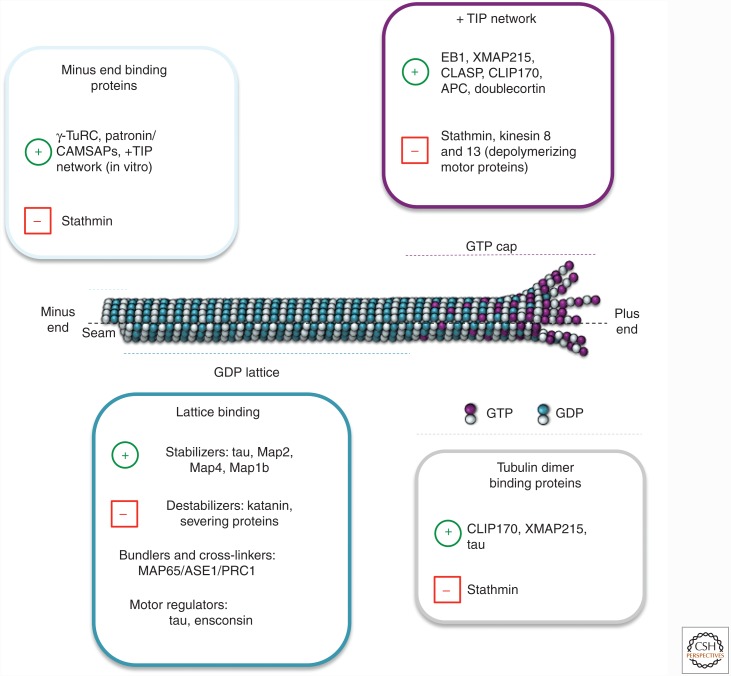
Figure 4.
Microtubule-binding proteins. Model summarizing some of the major microtubule-binding proteins according to their localization on the microtubule and their activities. The green plus symbol (+) means positive regulation and the red minus symbol (–) means negative regulation. At the plus end (fast growing), the members of the +TIP network (EB1, XMAP215, CLASP, CLIP170, doublecortin, and others not shown) associate with the stabilizing (GTP or GDP-Pi) cap of the growing microtubule and stabilize this dynamic structure to promote growth. Conversely, proteins such as the depolymerizing kinesins and stathmin facilitate microtubule disassembly. At the minus end (slow growing), proteins such as γ-TURC and Patronin/CAMSAP associate with the α-tubulin subunit to cap the end of the filament to prevent depolymerization, which is promoted by stathmin. In the central part of the microtubule, the GDP microtubule lattice can be stabilized by the activities of classical MAPs (tau, Map2, Map4, stop proteins) or destabilized by severing proteins (e.g., katanin). Microtubule-binding proteins that regulate the activity of microtubule motors also bind along the GDP lattice. Microtubules can form large networks through the activities of bundlers/cross-linkers, such as MAP65/ASE1/PRC1. Tubulin dimer binding proteins include stathmin (which promotes depolymerization by sequestering tubulin), as well as CLIP-170, tau, and XMAP-215 (which promote polymerization). Further detail and discussion are available in the main text and references.
Microtubule regulatory proteins can also be categorized as to where they localize on dynamic microtubules (Fig. 4). Lattice-binding proteins associate with microtubules along their length, whereas end-binding proteins localize more specifically to one or both of the microtubule extremities. Microtubule +TIPs are a subset of end-binding proteins that dynamically track growing microtubule ends, which in vivo are typically the plus ends (Akhmanova and Steinmetz 2015). The recently recognized microtubule minus end–targeting proteins are specifically recruited to minus ends (Akhmanova and Hoogenraad 2015).
5.2. Specific Classes of MTBPs
For the purposes of the discussion that follows, major classes of MTBPs are listed in the headings, subclasses are in italics, and individual proteins or protein families are underlined. Figure 4 lists some of the major MTBPs along with their predominant activities (plus and minus signs indicate positive and negative regulation, respectively).
5.2.1. MTBPs Categorized by Activity
5.2.1.1. Stabilizers
Stabilizers are proteins that promote polymerization and/or slow depolymerization. Although these two activities are similar, they are not necessarily identical: A protein could potentially stabilize a microtubule by inducing pause (inhibiting shortening but also inhibiting growth) without promoting polymerization. In practice, it can be difficult to distinguish between these two activities.
Many microtubule stabilizers can be categorized into one or another broad group based on shared sequences or behaviors. Microtubule stabilizers are often less conserved than analogous actin stabilizers, but relatedness can still be recognized across organisms by the presence of conserved domains, which are frequently found repeated. For example, multi-TOG-domain proteins, such as XMAP215/DIS1 and CLASP, are found in a wide range of organisms (Al-Bassam and Chang 2011), as are calponin-homology (CH)-domain-containing proteins such as the +TIP EB1 (Komarova et al. 2009) and the kinetochore–microtubule linker NDC80 (Varma and Salmon 2012). CAP-GLY-containing proteins are found in most if not all eukaryotes, although the CAP-GLY proteins with stabilizing activity (e.g., the +TIP CLIP-170) might be restricted to the animal–fungi lineage (Steinmetz and Akhmanova 2008; HV Goodson, unpubl.).
Animal cells also contain a host of more lineage-restricted protein families with stabilizing activities. This group includes the classical MAPs (tau, MAP2, and MAP4), which are poorly conserved but do contain common repeat structures (Dehmelt and Halpain 2005); the STOP proteins, which are particularly effective in stabilizing microtubules against cold (Bosc et al. 2003); and the doublecortin and EMAP families, which have roles in neuronal development (Fourniol et al. 2013) and cancer (Bayliss et al. 2016), respectively.
Exactly how most stabilizers work has not been determined, but the common presence of multiple microtubule-binding domains suggests that they work, at least in part, by cross-linking PFs laterally or longitudinally. Such cross-linking could stabilize the structure of the microtubule and thus prevent catastrophe, promote rescue, or both. Some proteins might stabilize microtubules by suppressing GTP hydrolysis, but none with this activity has (as yet) been clearly identified. Some proteins (e.g., XMAP-215) promote polymerization in part by increasing the microtubule growth rate, but the mechanism is still debated. One possibility is that these proteins bind free tubulin dimers and help deposit them at the tip (Ayaz et al. 2014). Alternatively, a tip-localized subunit cross-linker could potentially increase the growth rate by increasing the fraction of incoming tubulin subunits that incorporate into the lattice (Gardner et al. 2011). Stabilizers might work synergistically to promote microtubule polymerization (Zanic et al. 2013; Gupta et al. 2014). Such cooperation is just starting to be investigated. This issue is discussed more in the section on +TIPs below (see Sec. 5.2.2.2).
5.2.1.2. Destabilizers
Destabilizers shift a pool of dynamic microtubules toward free subunits by one or more mechanisms.
Sequestering proteins depolymerize microtubules indirectly by binding free tubulin subunits and preventing them from polymerizing. The best-characterized sequestering protein is the animal protein stathmin, which works in part by binding two dimers in a curved conformation that cannot incorporate into a microtubule (Cassimeris 2002).
Tip destabilizers act by directly attacking the sensitive microtubule tip. The best-characterized examples of these are the depolymerizing kinesins (e.g., kinesin-13), which use cycles of ATP hydrolysis to actively remove subunits. This activity causes net depolymerization, in part, by promoting catastrophe, but these proteins can even depolymerize taxol- and GMPCPP-stabilized microtubules (Walczak et al. 2013). Stathmin can also destabilize microtubule tips, acting, at least in part, by interfering with lateral bonding between subunits (Gupta et al. 2013).
Microtubule severing proteins use the energy of ATP to cut microtubules into pieces. Katanin, spastin, fidgetin, and related proteins are AAA ATPases that sever microtubules and are found in a wide range of organisms (reviewed by Roll-Mecak and McNally 2010). Katanin and at least some members of this family appear to work by using ATP hydrolysis to extract tubulin dimers from the lattice and destabilize the polymer. The new ends created by severing lack GTP caps, so they typically depolymerize rapidly (Sharp and Ross 2012).
Other possible mechanisms for promoting depolymerization include increasing the tubulin GTPase (proposed for stathmin; Cassimeris 2002) and capping PF ends (see below).
5.2.1.3. Capping proteins
Capping proteins adhere to the microtubule plus end or minus end and thus have the potential to stop both dimer association and dissociation. Although actin filament capping proteins are well characterized (see Pollard 2016), less is known about microtubule capping proteins, perhaps because of the greater size and complexity of the microtubule tip. For example, the only known minus-end capping proteins are large complexes rather than individual proteins. The best-characterized examples are the related complexes γ-TuRC and γ-TuSC, which not only cap minus ends but also nucleate microtubules (Kollman et al. 2011). Some evidence suggests that stathmin can cap PFs and suppress subunit addition without stabilizing the polymer (Gupta et al. 2013). Patronin and other proteins known as CAMSAPs are sometimes considered capping proteins, although they appear to associate laterally with the minus end (see also minus end–targeting proteins below).
5.2.1.4. Bundlers and cross-linkers
Bundlers and cross-linkers associate microtubules laterally. Proteins variously called MAP65/Ase1/PRC1 preferentially bundle antiparallel microtubules (Walczak and Shaw 2010), an activity important in the mitotic spindle. Most stabilizers have some bundling activity, but whether this activity is physiologically relevant is unclear. Simply coating negatively charged microtubules with tau peptides can result in bundling (Melki et al. 1991), as can addition of crowding agents such as polyethylene glycol (Sanchez et al. 2012).
5.2.1.5. Cytoskeletal integrators
Cytoskeletal integrators bind to and/or modulate microtubules and at least one other cytoskeletal element. This diverse category contains large scaffolding molecules such as the cancer-associated protein APC and the “plakin” family (Suozzi et al. 2012). It also includes the actin-nucleating formin family (Bartolini et al. 2008; Gaillard et al. 2011), myosin 10 (Weber et al. 2004), and even classic MAPs such as tau (Gallo 2007). Communication between the actin and microtubule cytoskeletons is essential for proper functioning of processes such as cytokinesis and generation and maintenance of cell polarity, and these proteins play fundamental, yet still poorly understood, roles in these processes (Rodriguez et al. 2003).
5.2.1.6. Other microtubule-associated activities
Additional microtubule-associated activities include the following.
Microtubule motors kinesin, dynein, and their accessories such as dynactin complex (reviewed by Barlan and Gelfand 2016 and Sweeney and Holzbaur 2016).
Motor modulators are proteins such as tau (Dixit et al. 2008) and ensconcin (Barlan et al. 2013) that bind the microtubule lattice and alter the behavior of motors acting on those microtubules.
Membrane–microtubule linkers such as CLIMP63 (Vedrenne et al. 2005) provide organelles with alternative (nonmotor) connections to microtubules (Gurel et al. 2014).
Metabolic proteins, including most enzymes of the glycolytic pathway, often bind to microtubules. One possible physiological function of this interaction is to increase the local concentration of proteins in the same biochemical pathway, but some of these proteins are capable of altering microtubule assembly (at least in vitro or on overexpression). These and related observations suggest that the metabolic state of the cell can influence microtubules and vice versa (reviewed by Cassimeris et al. 2012).
5.2.2. MTBPs as Categorized by Localization
5.2.2.1. Lattice-binding proteins
Lattice-binding proteins bind along the body of the microtubule and include all proteins that do not target either the plus or minus ends. As a result, this category includes proteins with a range of activities. Well-recognized examples of lattice-binding proteins include the classical MAPs tau, Map2, and Map4 (Dehmelt and Halpain 2005). Tau and Map2 are neuronal microtubule stabilizers that are localized to axons and dendrites, respectively, whereas Map4 is expressed in most tissues. Tau is the focus of much research because of its involvement in Alzheimer’s disease (Iqbual et al. 2016). Tau and MAP2 also seem to play a role in the spacing of microtubules in tightly packed neuronal extensions, as well as in regulating motor activity (Dixit et al. 2008).
5.2.2.2. Microtubule plus end–trafficking proteins
Microtubule +TIPs dynamically track growing microtubule ends. As the configuration of the microtubule tip determines whether a microtubule grows or shrinks, +TIP behavior allows a polymerization regulator to be dynamically localized where it needs to act. Although the abbreviation “+TIP” is attractive, it can be misleading: The canonical +TIP EB1 is specific not to plus ends but to growing ends, as it will track growing minus ends if presented with the opportunity, at least in vitro (Akhmanova and Steinmetz 2008). The set of +TIPs includes some of the most conserved and significant polymerization-promoting MTBPs known, including EB1, XMAP-215, CLASP, CLIP-170, and their relatives. It is important to note that not all +TIPs promote polymerization—some +TIPs, including kinesin-13, depolymerize microtubules (for an overview of +TIPs, see Akhmanova and Steinmetz 2008, 2015).
A small number of +TIPs (most notably EB1) track growing ends by binding preferentially to a short-lived tip-specific conformation (Maurer et al. 2014). Most other +TIPs localize to ends by binding to EB1, so EB1 is often considered to be “the master +TIP” (reviewed by Akhmanova and Steinmetz 2015). Consistent with the idea that EB1 has a central and ancient role in microtubule dynamics, EB1 is found across the spectrum of eukaryotic organisms, including Giardia (Dawson 2010). In this wide diversity of organisms, EB1 has a canonical homodimeric structure consisting of a CH domain followed by an apparently unstructured region, a short and highly conserved coiled-coil by which the protein dimerizes, and a tail that folds back to make a four-helix bundle. This four-helix bundle contains the hydrophobic pocket into which inserts the SxIP motifs of most EB1-binding +TIPs. In many organisms, EB1 terminates with an EEY motif that mimics the tubulin carboxy-terminal tail; this appears to be involved in autoinhibition (for reviews of EB1 structure and function, see Akhmanova and Steinmetz 2008, 2015).
Most if not all +TIPs bind other +TIPs, and so they are said to form the “+TIP network”—a loose web of interacting proteins that work together to regulate microtubule dynamics and integrate microtubules with the rest of the cell (Akhmanova and Steinmetz 2015). These interactions localize many of the proteins indirectly to growing microtubule tips and often release autoinhibition, allowing the +TIP network to integrate signals from many pathways (Akhmanova and Steinmetz 2015). The +TIP network might also act as a superstructure that promotes microtubule growth by stabilizing the structure of the microtubule tip and thus helping to promote microtubule polymerization (Gupta et al. 2014).
5.2.2.3. Minus end–targeting proteins
Two types of proteins localize to microtubule minus ends—the γ-TuRC (Kollman et al. 2011) and a more recently discovered set of proteins known collectively as CAMSAPs (which includes the Drosophila protein Patronin) (Akhmanova and Hoogenraad 2015). γ-TuRC blocks both subunit association and dissociation by interacting with all of the PFs (Fig. 2D). CAMSAPs bind at the minus end and effectively cap it, stabilizing it against depolymerization and stopping or slowing tubulin addition; some CAMSAPs can even track growing minus ends (Akhmanova and Hoogenraad 2015). CAMSAPs consist of an amino-terminal CH domain, some regions of coiled-coil, and a conserved carboxy-terminal CKK motif, and they seem to accomplish their stabilization of minus ends by assembling laterally at the minus end instead of by creating a classical cap. CAMSAPs are found in most metazoans and appear to be absent from plants and fungi, but they might be more widely distributed because some sequence motifs are present in the genomes of organisms such as ciliates (Akhmanova and Hoogenraad 2015; HV Goodson, unpubl.). As another example of a minus end binding protein, some evidence suggests that stathmin binding might be biased to the minus end (cited by Gupta et al. 2013).
6. CONCLUSION
The microtubule cytoskeleton is one of the most remarkable components of eukaryotic cells: During interphase, the microtubule array explores the cytoplasm, finds cargo, provides a substrate for transport, adjusts to internal and external signals, and directs the organization of the rest of the cell. Then, upon transitioning into mitosis, it disassembles and reassembles into the strikingly different mitotic spindle, a structure with the profoundly important purpose of precisely and consistently segregating to each daughter cell the correct set of chromosomes. Central to all of these processes are microtubule dynamics.
In reflecting on the role of dynamics in the function of the microtubules, it is interesting to realize that the combination of random probing and selective stabilization, as seen in the microtubule cytoskeleton, is a theme that occurs throughout biology: Random exploration of space followed by reinforcement/selection of the optimized variants is found in processes as diverse as acquired immunity, insect forging behavior, and even Darwinian evolution itself. This strategy produces highly robust systems (i.e., systems that are adaptable and hard to break), and its recurring appearance in biology provides an explanation for the robustness of life itself (Kirschner and Gerhart 2005; Karsenti 2008).
Much has been learned in the 30 years or so since dynamic instability was discovered, but a great deal remains to be understood about microtubules, how their assembly is regulated, how they are dynamically organized, and how their organization drives the organization of the rest of the cell. At the molecular scale, key problems include establishing the mechanisms of the catastrophe and rescue transitions, determining how MTBPs alter these transitions, and understanding how groups of MTBPs work together to create particular behaviors. At the cell scale, major challenges include understanding the assembly and dynamics of large-scale structures such as the mitotic spindle and flagellum and elucidating the organizational cross talk between microtubules and the rest of the cell. In both cases, an important goal will be to obtain a quantitative and predictive understanding of these processes. Gaining this knowledge will be important for cell biology, but the resulting information about self-assembling systems also has the potential to impact fields as distant as nanotechnology and synthetic biology.
To make these advances will require a multidisciplinary strategy that is founded on the classical approaches of biochemistry, cell biology, and genetics, but includes new types of thinking and approaches imported from the study of physics, chemistry, and complex systems. Already, input from these fields has made significant contributions and promises to make many more (e.g., Karsenti 2008; Sanchez et al. 2011). Now that efforts over the past half-century have identified most of the cellular components and made progress in determining their activities, it is time to start understanding how these components work together to create a dynamic and functional cell. The microtubule cytoskeleton will be a central focus for this endeavor.
ACKNOWLEDGMENTS
We apologize to those investigators whose work we were not able to cite for reasons of space. We thank members of the Goodson laboratory for critically reading the manuscript. This work was supported by a grant from the National Science Foundation (NSF) (MCB-1244593) to H.V.G.
In vitro, purified actin can by itself slowly treadmill (undergo assembly at one end and disassembly at the other). In vivo treadmilling of the actin network is driven by proteins that regulate assembly, capping, severing, and depolymerization (reviewed by Pollard 2016).
The transitions are often described as random, but this is not completely accurate: Microtubules that have been growing longer are more likely to undergo catastrophe (Odde et al. 1995; Coombes et al. 2013).
It is not clear whether it is actually the GTP form, the GDP-Pi form, or both that have the stabilizing activity. For simplicity, most publications refer to this stabilizing GTP- and/or GDP-Pi-rich structure as the “GTP cap.”
One reasonable (but not necessarily complete) explanation for the asymmetry is that a subunit attaching at the plus end does not hydrolyze its GTP until after a new subunit attaches to it, whereas one attaching at the minus end will start the process of first-order hydrolysis as soon as it attaches.
Early conceptual models of microtubule dynamic instability assumed that the GTP hydrolysis occurred vectorially—that is, in a wave that travels up from the base toward the tip. Later mathematical modeling (Flyvbjerg et al. 1994) showed that this mechanism was not consistent with the data from sudden dilution experiments (Walker et al. 1991). See Bowne-Anderson et al. 2015 for more discussion.
Editors: Thomas D. Pollard and Robert D. Goldman
Additional Perspectives on The Cytoskeleton available at www.cshperspectives.org
REFERENCES
*Reference is in this collection.
- Ada R, Kilmartin JV. 2000. Spindle pole body duplication: A model for centrosome duplication? Trends Cell Biol 10: 329–335. [DOI] [PubMed] [Google Scholar]
- Akhmanova A, Hoogenraad CC. 2015. Microtubule minus-end-targeting proteins. Curr Biol 25: R162–R171. [DOI] [PubMed] [Google Scholar]
- Akhmanova A, Steinmetz MO. 2008. Tracking the ends: A dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol 9: 309–322. [DOI] [PubMed] [Google Scholar]
- Akhmanova A, Steinmetz MO. 2015. Control of microtubule organization and dynamics: Two ends in the limelight. Nat Rev Mol Cell Biol 16: 711–726. [DOI] [PubMed] [Google Scholar]
- Akhmanova A, Stehbens SJ, Yap AS. 2009. Touch, grasp, deliver and control: Functional cross-talk between microtubules and cell adhesions. Traffic 10: 268–274. [DOI] [PubMed] [Google Scholar]
- Al-Bassam J, Chang F. 2011. Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol 21: 604–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin GM, Lander GC, Kellogg EH, Zhang R, Baker D, Nogales E. 2014. High-resolution microtubule structures reveal the structural transitions in αβ-tubulin upon GTP hydrolysis. Cell 157: 1117–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos LA. 2004. Microtubule structure and its stabilisation. Org Biomol Chem 2: 2153–2160. [DOI] [PubMed] [Google Scholar]
- Amos LA. 2011. What tubulin drugs tell us about microtubule structure and dynamics. Semin Cell Dev Biol 22: 916–926. [DOI] [PubMed] [Google Scholar]
- Arnal I, Metoz F, DeBonis S, Wade RH. 1996. Three-dimensional structure of functional motor proteins on microtubules. Curr Biol 6: 1265–1270. [DOI] [PubMed] [Google Scholar]
- Ayaz P, Munyoki S, Geyer EA, Piedra FA, Vu ES, Bromberg R, Otwinowski Z, Grishin NV, Brautigam CA, Rice LM. 2014. A tethered delivery mechanism explains the catalytic action of a microtubule polymerase. Elife 3: e03069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Barlan K, Gelfand VI. 2016. Microtubule-based transport and the distribution, tethering, and organization of organelles. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a021998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlan K, Lu W, Gelfand VI. 2013. The microtubule-binding protein ensconsin is an essential cofactor of kinesin-1. Curr Biol 23: 317–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini F, Gundersen GG. 2006. Generation of noncentrosomal microtubule arrays. J Cell Sci 119: 4155–4163. [DOI] [PubMed] [Google Scholar]
- Bartolini F, Moseley JB, Schmoranzer J, Cassimeris L, Goode BL, Gunderson GG. 2008. The formin mDia2 stabilizes microtubules independently of its actin nucleation activity. J Cell Biol 181: 523–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. 2006. Flies without centrioles. Cell 125: 1375–1386. [DOI] [PubMed] [Google Scholar]
- Bayliss R, Choi J, Fennell DA, Fry AM, Richards MW. 2016. Molecular mechanisms that underpin EML4-ALK driven cancers and their response to targeted drugs. Cell Mol Life Sci 73: 1209–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosc C, Andrieux A, Job D. 2003. STOP proteins. Biochemistry 42: 12125–12132. [DOI] [PubMed] [Google Scholar]
- Bowne-Anderson H, Zanic M, Kauer M, Howard J. 2013. Microtubule dynamic instability: A new model with coupled GTP hydrolysis and multistep catastrophe. BioEssays 35: 452–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowne-Anderson H, Anneke H, Howard J. 2015. Regulation of microtubule growth and catastrophe: Unifying theory and experiment. Trends Cell Biol 25: 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard GJ, Rice LM. 2014. The contribution of αβ-tubulin curvature to microtubule dynamics. J Cell Biol 207: 323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Machado P, Branco P, Tavares-Cadete F, Rodrigues-Martins A, Pereira-Leal JB, Bettencourt-Dias M. 2010. Stepwise evolution of the centriole-assembly pathway. J Cell Sci 123: 1414–1426. [DOI] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, Bettencourt-Dias M. 2011. Evolution: Tracing the origins of centrioles, cilia, and flagella. J Cell Biol 194: 165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris L. 2002. The oncoprotein 18/stathmin family of microtubule destabilizers. Curr Opin Cell Biol 14: 18–24. [DOI] [PubMed] [Google Scholar]
- Cassimeris L, Silva VC, Miller E, Ton Q, Molnar C, Fong J. 2012. Fueled by microtubules: Does tubulin dimer/polymer partitioning regulate intracellular metabolism? Cytoskeleton 69: 133–143. [DOI] [PubMed] [Google Scholar]
- Caudron N, Arnal I, Buhler E, Job D, Valiron O. 2002. Microtubule nucleation from stable tubulin oligomers. J Biol Chem 277: 50973–50979. [DOI] [PubMed] [Google Scholar]
- Chang F, Martin SG. 2009. Shaping fission yeast with microtubules. Cold Spring Harb Perspect Biol 1: a001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CT, Ettinger AW, Huttner WB, Doxsey SJ. 2013. Resurrecting remnants: The lives of post-mitotic midbodies. Trends Cell Biol 23: 118–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone MA, DuPage AG, Goode BL. 2010. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol 11: 62–74. [DOI] [PubMed] [Google Scholar]
- Chretien D, Fuller SD, Karsenti E. 1995. Structure of growing microtubule ends: Two-dimensional sheets close into tubes at variable rates. J Cell Biol 129: 1311–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes CE, Yamamoto A, Kenzie MR, Odde DJ, Gardner MK. 2013. Evolving tip structures can explain age-dependent microtubule catastrophe. Curr Biol 23: 1342–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson SC. 2010. An insider’s guide to the microtubule cytoskeleton of Giardia. Cell Microbiol 12: 588–598. [DOI] [PubMed] [Google Scholar]
- de Forges H, Bouissou A, Perez F. 2012. Interplay between microtubule dynamics and intracellular organization. Int J Biochem Cell Biol 44: 266–274. [DOI] [PubMed] [Google Scholar]
- Dehmelt L, Halpain S. 2005. The MAP2/Tau family of microtubule-associated proteins. Genome Biol 6: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ. 1997. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol 13: 83–117. [DOI] [PubMed] [Google Scholar]
- Dimitrov A, Quesnoit M, Moutel S, Cantaloube I, Pous C, Perez F. 2008. Detection of GTP-tubulin conformation in vivo reveals a role for GTP remnants in microtubule rescues. Science 322: 1353–1356. [DOI] [PubMed] [Google Scholar]
- Dixit R, Ross JL, Goldman YE, Holzbaur EL. 2008. Differential regulation of dynein and kinesin motor proteins by tau. Science 319: 1086–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogterom M, Surrey T. 2013. Microtubule organization in vitro. Curr Opin Cell Biol 25: 23–29. [DOI] [PubMed] [Google Scholar]
- Ehrhardt DW, Shaw SL. 2006. Microtubule dynamics and organization in the plant cortical array. Annu Rev Plant Biol 57: 859–875. [DOI] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S, Cobb G, Compton JT, Li XH, Rhoades E. 2014. Tau mutants bind tubulin heterodimers with enhanced affinity. Proc Natl Acad Sci 111: 6311–6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findeisen P, Muhlhausen S, Dempewolf S, Hertzog J, Zietlow A, Carlomagno T, Kollmar M. 2014. Six subgroups and extensive recent duplications characterize the evolution of the eukaryotic tubulin protein family. Genome Biol Evol 6: 2274–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyvbjerg H, Holy TE, Leibler S. 1994. Stochastic dynamics of microtubules: A model for caps and catastrophes. Phys Rev Lett 73: 2372–2375. [DOI] [PubMed] [Google Scholar]
- Folker ES, Baker BM, Goodson HV. 2005. Interactions between CLIP-170, tubulin, and microtubules: Implications for the mechanism of Clip-170 plus-end tracking behavior. Mol Biol Cell 16: 5373–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourniol F, Perderiset M, Houdusse A, Moores C. 2013. Structural studies of the doublecortin family of MAPs. Methods Cell Biol 115: 27–48. [DOI] [PubMed] [Google Scholar]
- Gaillard J, Ramabhadran V, Neumanne E, Gurel P, Blanchoin L, Vantard M, Higgs HN. 2011. Differential interactions of the formins INF2, mDia1, and mDia2 with microtubules. Mol Biol Cell 22: 4575–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo G. 2007. Tau is actin up in Alzheimer’s disease. Nat Cell Biol 9: 133–134. [DOI] [PubMed] [Google Scholar]
- Ganguly A, Yang H, Sharma R, Patel KD, Cabral F. 2012. The role of microtubules and their dynamics in cell migration. J Biol Chem 287: 43359–43369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, Charlebois BD, Janosi IM, Howard J, Hunt AJ, Odde DJ. 2011. Rapid microtubule self-assembly kinetics. Cell 146: 582–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnham CP, Roll-Mecak A. 2012. The chemical complexity of cellular microtubules: Tubulin post-translational modification enzymes and their roles in tuning microtubule functions. Cytoskeleton 69: 442–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvalov BK, Zuber B, Bouchet-Marquis C, Kudryashev M, Gruska M, Beck M, Leis A, Frischknecht F, Bradke F, Baumeister W, et al. 2006. Luminal particles within cellular microtubules. J Cell Biol 174: 759–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittes F, Mickey B, Nettleton J, Howard J. 1993. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J Cell Biol 120: 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoretti IV, Margolin G, Alber MS, Goodson HV. 2006. Insights into cytoskeletal behavior from computational modeling of dynamic microtubules in a cell-like environment. J Cell Sci 119: 4781–4788. [DOI] [PubMed] [Google Scholar]
- Grishchuk EL, Molodtsov MI, Ataullakhanov FI, McIntosh JR. 2005. Force production by disassembling microtubules. Nature 438: 384–388. [DOI] [PubMed] [Google Scholar]
- Gull K. 2001. Protist tubulins: New arrivals, evolutionary relationships and insights to cytoskeletal function. Curr Opin Microbiol 4: 427–432. [DOI] [PubMed] [Google Scholar]
- Gupta KK, Li C, Duan A, Alberico EO, Kim OV, Alber MS, Goodson HV. 2013. Mechanism for the catastrophe-promoting activity of the microtubule destabilizer Op18/stathmin. Proc Natl Acad Sci 110: 20449–20454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta KK, Alberico EO, Nathke IS, Goodson HV. 2014. Promoting microtubule assembly: A hypothesis for the functional significance of the +TIP network. BioEssays 36: 818–826. [DOI] [PubMed] [Google Scholar]
- Gurel PS, Hatch AL, Higgs HN. 2014. Connecting the cytoskeleton to the endoplasmic reticulum and Golgi. Curr Biol 24: R660–R672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins T, Mirigian M, Selcuk Yasar M, Ross JL. 2010. Mechanics of microtubules. J Biomech 43: 23–30. [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. 1996. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature 382: 420–425. [DOI] [PubMed] [Google Scholar]
- *.Herrmann H, Aebi U. 2016. Intermediate filaments: Structure and assembly. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TL. 1987. Linear aggregation theory in cell biology. Springer, New York. [Google Scholar]
- *.Hol EM, Capetanaki Y. 2016. Type III intermediate filaments desmin, glial fibrillary acidic protein (GFAP), vimentin, and peripherin. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a021642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J. 2001. Mechanics of motor proteins and the cytoskeleton. Sinauer Associates, Inc, Sunderland, MA. [Google Scholar]
- Howard J, Hyman AA. 2003. Dynamics and mechanics of the microtubule plus end. Nature 422: 753–758. [DOI] [PubMed] [Google Scholar]
- Howard J, Hyman AA. 2009. Growth, fluctuation and switching at microtubule plus ends. Nat Rev Mol Cell Biol 10: 569–574. [DOI] [PubMed] [Google Scholar]
- Hyman AA, Salser S, Drechsel DN, Unwin N, Mitchison TJ. 1992. Role of GTP hydrolysis in microtubule dynamics: Information from a slowly hydrolyzable analogue, GMPCPP. Mol Biol Cell 3: 1155–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbual K, Liu F, Gong CX. 2016. Tau and neurodegenerative disease: The story so far. Nat Rev Neurol 12: 15–27. [DOI] [PubMed] [Google Scholar]
- *.Jacob JT, Coulomb PA, Kwan R, Omary MB. 2016. Types I and II keratins intermediate filaments. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Bulinski JC. 2011. Post-translational regulation of the microtubule cytoskeleton: Mechanisms and functions. Nat Rev Mol Cell Biol 12: 773–786. [DOI] [PubMed] [Google Scholar]
- Johnson KA, Borisy GG. 1975. The equilibrium assembly of microtubules in vitro. Soc Gen Physiol Ser 30: 119–141. [PubMed] [Google Scholar]
- *.Jones JCR, Kam CY, Harmon RM, Woychek AV, Hopkinson SB, Green KJ. 2016. Intermediate filaments and the plasma membrane. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a025866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalil K, Li L, Hutchins BI. 2011. Signaling mechanisms in cortical axon growth, guidance, and branching. Front Neuroanat 5: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenti E. 2008. Self-organization in cell biology: A brief history. Nat Rev Mol Cell Biol 9: 255–262. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL. 2001. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J Cell Biol 153: 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner MW, Gerhart JC. 2005. The plausibility of life: Resolving Darwin’s dilemma. Yale University Press, New Haven. [Google Scholar]
- Kirschner M, Mitchison T. 1986. Beyond self-assembly: From microtubules to morphogenesis. Cell 45: 329–342. [DOI] [PubMed] [Google Scholar]
- Kollman JM, Merdes A, Mourey L, Agard DA. 2011. Microtubule nucleation by γ-tubulin complexes. Nat Rev Mol Cell Biol 12: 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova Y, De Groot CO, Grigoriev I, Gouveia SM, Munteanu EL, Schober JM, Honnappa S, Buey RM, Hoogenraad CC, Dogterom M, et al. 2009. Mammalian end binding proteins control persistent microtubule growth. J Cell Biol 184: 691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, DeRosier DJ, Nicholson WV, Nogales E, KH Downing. 2002. Microtubule structure at 8 Å resolution. Structure 10: 1317–1328. [DOI] [PubMed] [Google Scholar]
- Li C, Li J, Goodson HV, Alber MS. 2014. Microtubule dynamic instability: The role of cracks between protofilaments. Soft Matter 10: 2069–2080. [DOI] [PubMed] [Google Scholar]
- Maly IV, Borisy GG. 2002. Self-organization of treadmilling microtubules into a polar array. Trends Cell Biol 12: 462–465. [DOI] [PubMed] [Google Scholar]
- Margolin G, Gregoretti IV, Cickovski TM, Li C, Shi W, Alber MS, Goodson HV. 2012. The mechanisms of microtubule catastrophe and rescue: Implications from analysis of a dimer-scale computational model. Mol Biol Cell 23: 642–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer SP, Cade NI, Bohner G, Gustafsson N, Boutant E, Surrey T. 2014. EB1 accelerates two conformational transitions important for microtubule maturation and dynamics. Curr Biol 24: 372–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.McIntosh JR. 2016. Mitosis. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a023218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKean PG, Vaughan S, Gull K. 2001. The extended tubulin superfamily. J Cell Sci 114: 2723–2733. [DOI] [PubMed] [Google Scholar]
- Melki R, Kerjan P, Waller JP, Carlier MF, Pantaloni D. 1991. Interaction of microtubule-associated proteins with microtubules: Yeast lysyl- and valyl-tRNA synthetases and tau 218-235 synthetic peptide as model systems. Biochemistry 30: 11536–11545. [DOI] [PubMed] [Google Scholar]
- Melki R, Fievez S, Carlier MF. 1998. Continuous monitoring of Pi release following nucleotide hydrolysis in actin or tubulin assembly using 2-amino-6-mercapto-7-methylpurine ribonucleoside and purine-nucleoside phosphorylase as an enzyme-linked assay. Biochemistry 35: 12038–12045. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. 1984. Dynamic instability of microtubule growth. Nature 312: 237–242. [DOI] [PubMed] [Google Scholar]
- Mogilner A, Oster G. 2003. Polymer motors: Pushing out the front and pulling up the back. Curr Biol 13: R721–R733. [DOI] [PubMed] [Google Scholar]
- Mogilner A, Wollman R, Civelekoglu-Scholey G, Scholey J. 2006. Modeling mitosis. Trends Cell Biol 16: 88–96. [DOI] [PubMed] [Google Scholar]
- Morrissette N. 2015. Targeting Toxoplasma tubules: Tubulin, microtubules, and associated proteins in a human pathogen. Eukaryot Cell 14: 2–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA, Stearns T. 2011. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol 13: 1154–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Downing KH, Amos LA, Lowe J. 1998. Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol 5: 451–458. [DOI] [PubMed] [Google Scholar]
- O’Connell CB, Khodjakov AL. 2007. Cooperative mechanisms of mitotic spindle formation. J Cell Sci 120: 1717–1722. [DOI] [PubMed] [Google Scholar]
- Odde DJ, Cassimeris L, Buettner HM. 1995. Kinetics of microtubule catastrophe assessed by probabilistic analysis. Biophys J 69: 796–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odde DJ, Cassimeris L, Buettner HM. 2005. Kinetics of microtubule catastrophe assessed by probabilistic analysis. Biophys J 69: 796–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosawa F, Kasai M. 1962. A theory of linear and helical aggregations of macromolecules. J Mol Biol 4: 10–21. [DOI] [PubMed] [Google Scholar]
- Petry S, Groen AC, Ishihara K, Mitchison TJ, Vale RD. 2013. Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell 152: 768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Pollard TD. 2016. Actin and actin-binding proteins. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Pollard TD, Goldman RD. 2016. Overview of the cytoskeleton from an evolutionary perspective. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a030288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath O, Kozielski F. 2012. Kinesins and cancer. Nat Rev Cancer 12: 527–539. [DOI] [PubMed] [Google Scholar]
- Reilein A, Yamada S, Nelson WJ. 2005. Self-organization of an acentrosomal microtubule network at the basal cortex of polarized epithelial cells. J Cell Biol 171: 845–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice LM, Montabana EA, Agard DA. 2008. The lattice as allosteric effector: Structural studies of αβ- and γ-tubulin clarify the role of GTP in microtubule assembly. Proc Natl Acad Sci 105: 5378–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov VI, Borisy GG. 1997. Microtubule treadmilling in vivo. Science 275: 215–218. [DOI] [PubMed] [Google Scholar]
- Rodriguez OC, Schaefer AW, Mandato CA, Forscher P, Bement WM, Waterman-Storer CM. 2003. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat Cell Biol 5: 599–609. [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A. 2015. Intrinsically disordered tubulin tails: Complex tuners of microtubule functions? Semin Cell Dev Biol 37C: 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A, McNally FJ. 2010. Microtubule-severing enzymes. Curr Opin Cell Biol 22: 96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez T, Welch D, Nicastro D, Dogic Z. 2011. Cilia-like beating of active microtubule bundles. Science 333: 456–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez T, Chen DT, DeCamp SJ, Heymann M, Dogic Z. 2012. Spontaneous motion in hierarchically assembled active matter. Nature 491: 431–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Huertas C, Luders J. 2015. The augmin connection in the geometry of microtubule networks. Curr Biol 25: R294–R299. [DOI] [PubMed] [Google Scholar]
- Schwartz CL, Heumann JM, Dawson SC, Hoenger A. 2012. A detailed, hierarchical study of Giardia lamblia’s ventral disc reveals novel microtubule-associated protein complexes. PLoS ONE 7: e43783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seetapun D, Castle BT, McIntyre AJ, PT Tran, Odde DJ. 2012. Estimating the microtubule GTP cap size in vivo. Curr Biol 22: 1681–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sept D, Baker NA, McCammon JA. 2003. The physical basis of microtubule structure and stability. Protein Sci 12: 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, Ross JL. 2012. Microtubule-severing enzymes at the cutting edge. J Cell Sci 125: 2561–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabodnick MM, Marshall WF. 2014. Stentor coeruleus. Curr Biol 24: R783–R784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM, Hedman AC, Sacks DB. 2015. IQGAPs choreograph cellular signaling from the membrane to the nucleus. Trends Cell Biol 25: 171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppina V, Herbstman JF, Skiniotis G, Verhey KJ. 2012. Luminal localization of α-tubulin K40 acetylation by cryo-EM analysis of fab-labeled microtubules. PloS ONE 7: e48204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz MO, Akhmanova A. 2008. Capturing protein tails by CAP-Gly domains. Trends Biochem Sci 33: 535–545. [DOI] [PubMed] [Google Scholar]
- Sui H, Downing KH. 2010. Structural basis of interprotofilament interaction and lateral deformation of microtubules. Structure 18: 1022–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suozzi KC, Wu X, Fuchs E. 2012. Spectraplakins: Master orchestrators of cytoskeletal dynamics. J Cell Biol 197: 465–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surrey T, Nedelec F, Leibler S, Karsenti E. 2001. Physical properties determining self- organization of motors and microtubules. Science 292: 1167–1171. [DOI] [PubMed] [Google Scholar]
- *.Sweeney HL, Holzbaur ELF. 2016. Motor proteins. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Cowan NJ. 2013. Tubulin-specific chaperones: Components of a molecular machine that assembles the α/β heterodimer. Methods Cell Biol 115: 155–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Titus MA. 2016. Myosin-driven intracellular transport. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a021972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBuren V, Cassimeris L, Odde DJ. 2005. Mechanochemical model of microtubule structure and self-assembly kinetics. Biophys J 89: 2911–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D, Salmon ED. 2012. The KMN protein network—Chief conductors of the kinetochore orchestra. J Cell Sci 125: 5927–5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedrenne C, Klopfenstein DR, Hauri HP. 2005. Phosphorylation controls CLIMP-63- mediated anchoring of the endoplasmic reticulum to microtubules. Mol Biol Cell 16: 1928–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Vertii A, Hehnly H, Doxsey S. 2016. The centrosome, a multitalented renaissance organelle. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a025049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Viswanadha R, Sale WS, Porter ME. 2016. Ciliary motility: Regulation of axonemal dynein motors. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev I, Malikov V, Rodionov V. 2001. Self-organization of a radial microtubule array by dynein-dependent nucleation of microtubules. Proc Natl Acad Sci 98: 10160–10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulevic B, Correia JJ. 1997. Thermodynamic and structural analysis of microtubule assembly: The role of GTP hydrolysis. Biophys J 72: 1357–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CE, Shaw SL. 2010. A MAP for bundling microtubules. Cell 142: 364–367. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Gayek S, Ohi R. 2013. Microtubule-depolymerizing kinesins. Annu Rev Cell Dev Biol 29: 417–441. [DOI] [PubMed] [Google Scholar]
- Walker RA, O’Brien ET, Pryer NK, Soboeiro MF, Voter WA, Erickson HP, Salmon ED. 1988. Dynamic instability of individual microtubules analyzed by video light microscopy: Rate constants and transition frequencies. J Cell Biol 107: 1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker RA, Pryer NK, Salmon ED. 1991. Dilution of individual microtubules observed in real time in vitro: Evidence that cap size is small and independent of elongation rate. J Cell Biol 114: 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber KL, Sokac AM, Berg JS, Cheney RE, Bement WM. 2004. A microtubule-binding myosin required for nuclear anchoring and spindle assembly. Nature 431: 325–329. [DOI] [PubMed] [Google Scholar]
- Wieczorek M, Bechstedt S, Chaaban S, Brouhard GJ. 2015. Microtubule-associated proteins control the kinetics of microtubule nucleation. Nat Cell Biol 17: 907–916. [DOI] [PubMed] [Google Scholar]
- Winey M, Stemm-Wolf AJ, Giddings THJ, Pearson CG. 2012. Cytological analysis of Tetrahymena thermophila. Methods Cell Biol 109: 357–378. [DOI] [PubMed] [Google Scholar]
- Woolner S, Bement WM. 2009. Unconventional myosins acting unconventionally. Trends Cell Biol 19: 245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HG, Muszynski MG, Dawe RK. 1999. The maize homologue of the cell cycle checkpoint protein MAD2 reveals kinetochore substrate and contrasting mitotic and meiotic localization patterns. J Cell Biol 145: 425–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Yuan A, Rao MV, Veeranna, Nixon RA. 2016. Neurofilaments and neurofilament proteins in health and disease. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanic M, Widlund PO, Hyman AA, Howard J. 2013. Synergy between XMAP215 and EB1 increases microtubule growth rates to physiological levels. Nat Cell Biol 15: 688–693. [DOI] [PubMed] [Google Scholar]
- Zhang R, Alushin GM, Brown A, Nogales E. 2015. Mechanistic origin of microtubule dynamic instability and its modulations by EB proteins. Cell 162: 849–859. [DOI] [PMC free article] [PubMed] [Google Scholar]