Abstract
Cytokines and long noncoding RNAs (lncRNAs) are intertwined in the regulatory circuit controlling immunity. lncRNA expression levels are altered following cytokine stimulation in a cell-type-specific fashion. lncRNAs, in turn, regulate the inducible expression of cytokines following immune activation. These studies position lncRNAs as important regulators of gene expression within the complex pathways of the immune system. Our understanding of the functions of lncRNAs is just beginning. Current methodologies for functionally understanding how these transcripts mediate their effects are unable to keep up with the speed of genomic outputs cataloging thousands of these novel genes. In this review, we highlight the interplay between cytokines and lncRNAs and speculate on the future utility of these genes as potential biomarkers and therapeutic targets for the treatment of inflammatory disorders.
Cytokines are a large group of small proteins (5 to 20 kda) that are critical in controlling a plethora of biological and physiological processes, including metabolism, inflammation, and blood pressure, to name just a few. They provide the necessary balance to maintain homeostasis at the cellular, tissue, and organismal level. Alterations in the balance between pro- and anti-inflammatory cytokines can have devastating consequences, leading to a myriad of inflammatory diseases (Vilcek and Feldmann 2004; Lin and Karin 2007). Cytokine gene expression is very tightly regulated at both the transcriptional and posttranscriptional level. Although the role of noncoding RNAs such as microRNAs (miRNAs) as regulators of cytokine gene expression is well appreciated, much less is known regarding the contribution of other classes of noncoding RNAs to the control of cytokines. Regulation of long noncoding RNAs (lncRNAs) and cytokine expression is emerging as a reciprocal feedforward/feedback relationship, which will be detailed in this review.
RNA sequencing (RNA-seq) technology has revolutionized the field of genomics and provides a high-throughput approach to define and measure all transcripts expressed in a given cell type. RNA-seq allows for the characterization of annotated transcripts in addition to the identification of novel transcripts. lncRNAs are transcripts that exceed 200 nucleotides in length, a size cutoff that distinguishes these transcripts from small RNAs such as transfer RNA (tRNA), small nucleolar (snoRNA), and miRNAs. To date, there are close to 16,000 lncRNAs that have been cataloged in the human genome (Gencode Version 24). This number is likely to increase as lncRNAs are found to be highly restricted in their expression to specific cell types, and therefore the more sequencing that is performed the more lncRNAs will likely be identified. A recent study from Iyer et al. (2015) identified >58,000 lncRNAs from >7000 human RNA-seq data sets. Based on these data, it appears that lncRNAs greatly outnumber protein-coding genes in the human genome. lncRNAs can arise from regions between protein-coding genes, and these transcripts are referred to as “intergenic” lncRNAs. There are also a large number of antisense transcripts, with recent studies showing that the majority of protein-coding genes contain an antisense transcript owing to the bidirectionality of RNA-polymerase II (Seila et al. 2008; Flynn et al. 2011). lncRNAs can also arise from intronic regions or enhancer regions of the genome (eRNAs) (Lam et al. 2014; Kim et al. 2015). lncRNAs are often named based on their proximity to the nearest protein-coding gene; however, this naming scheme only provides information on the location within the genome where the lncRNA resides, and does not necessarily provide information on the function of a given lncRNA. Until such time as a consensus on nomenclature is reached, there are no formal guidelines on naming lncRNAs. Owing to the large number of lncRNAs present in the genome, it is unlikely that they will follow the naming scheme of miRNAs. However, the current trend of naming based on position has its drawbacks as it suggests their functions are based on or connected to the neighboring gene, which is not the case for trans-acting lncRNAs. lncRNAs were initially thought to represent transcriptional noise arising from imprecise transcriptional initiation; however, there is more and more evidence that these transcripts can have important regulatory functions in a wide variety of biological processes.
One of the most intriguing and also daunting aspects of lncRNAs is their wide variety of potential functions. lncRNAs can function through interactions with proteins, DNA, or RNA (Rinn and Chang 2012). Although lncRNAs are still being described and characterized, some transcripts have been studied for decades. For example, X-inactive specific transcript (Xist) is a large 20-kb transcript that has been studied intensively for the past 20 years. Xist is involved in dosage compensation and X chromosome inactivation in females (Lessing et al. 2013). We continue to learn interesting biology about this critical lncRNA. McHugh et al. (2015) elegantly showed that Xist can directly bind to the protein SHARP (SMRT and HDAC Associated Repressor Protein, also called SPEN) to silence the X chromosome. This was the first demonstration of a protein that binds to Xist required for silencing. Although Xist was previously shown to bind to the polycomb repressor complex 2 (PRC2), knockdown studies of PRC2 complex components failed to interfere with Xist-mediated silencing. Instead, SHARP binds to the SMRT corepressor, which activates HDAC3 mediating silencing and exclusion of RNA pol II from the X chromosome (McHugh et al. 2015). There is an exciting emerging role for Xist in T and B cells. Wang et al. 2016b showed that many immune-related genes are bi-allelically expressed in female lymphocytes, and it appeared that Xist RNA was down-regulated, thus enabling increased expression of these genes. They conclude that in female lymphocytes the X inactivation center is predisposed to become partially reactivated, which could account for why females display enhanced immunity over men and also why females are more prone to autoimmunity (Wang et al. 2016b).
lncRNAs also function as posttranscriptional regulators of gene expression involved in messenger RNA (mRNA) decay (1/2-sbsRNAs), mRNA stabilization (BACE-AS), as well as splicing regulation (MALAT1) (Rashid et al. 2016). A number of lncRNAs have been shown to act as miRNA sponges, and there is a database (miRSponge), which is a manually curated database for all miRNA sponges and competitive endogenous RNAs (ceRNAs) that have experimental data to support their function (Wang et al. 2015a).
A significant challenge in studying lncRNAs is to prove that the transcript is indeed noncoding. A number of bioinformatics tools have been used to determine the possibility of a transcript containing an open reading frame that will produce a protein or peptide. Codon substitution frequency and its most recent version (PhyloCSF) is a comparative genomics method that can analyze nucleotide sequence alignment across multiple species to determine whether a transcript is coding or noncoding (Lin et al. 2011). The development of ribosomal footprinting has increased our ability to experimentally validate whether a transcript is indeed noncoding (Ingolia et al. 2009, 2011, 2012). Ribosome profiling involves the mapping of translating ribosomes across any transcript by deep sequencing of the mRNA footprints that are occupied by ribosomes and thus protected from nuclease digestion (Ingolia et al. 2009). Analysis of the ribosome-protected RNA fragments allows for a map of translation to be identified at a single nucleotide level. Ingolia et al. (2014) have shown that many noncoding regions, including 5′ UTRs and lncRNAs, are found to produce footprints, raising the possibility that some of these regions could be translated producing proteins or small peptides. Guttman et al. (2013) showed that ribosome occupancy of lncRNAs and 5′ UTRs does not necessarily lead to the designation of these transcripts as coding. They developed the ribosome release score (RRS) that allows for lncRNAs and 5′ UTRs to be classified into defined categories that are unique to coding genes (Guttman et al. 2013). The release score indicates the time a ribosome is released once it hits a stop codon. These data indicate that the majority of noncoding transcripts do not function through the production of a protein. A number of additional studies have examined lncRNAs binding to ribosomes in human cells and zebrafish. Although many lncRNAs associate with ribosomes, it is hypothesized that the vast majority of these transcripts fail to be translated, or if they generate small peptides, these are rapidly degraded possibly through the nonsense-mediated decay (NMD) pathway (Bánfai et al. 2012; Chew et al. 2013; Carlevaro-Fita et al. 2016).
Recent studies from the Olsen laboratory at the University of Texas Southwestern have shown that at least two lncRNAs can produce micropeptides in the mammalian system (Anderson et al. 2015; Nelson et al. 2016). Myoregulin (MLN) is a peptide encoded by a skeletal muscle–specific lncRNA. Genetic deletion of MLN-enhanced calcium handling in skeletal muscle in mice and improved exercise performance (Anderson et al. 2015). Dwarf open reading frame (DWORF) is a muscle-specific 34-amino-acid micropeptide encoded within an lncRNA. DWORF functions to regulate SERCA activity and enhances muscle contractility (Nelson et al. 2016). Here, we provide an overview of the cross talk between cytokines and lncRNAs and how these RNAs and the cytokines they regulate can impact the development and activation status of immune cells. We will also discuss the emerging interest in lncRNAs in disease and their potential as therapeutic targets.
lncRNAs, CYTOKINES, AND IMMUNE CELL DIFFERENTIATION
Cell differentiation is a tightly controlled process regulated both transcriptionally and posttranscriptionally. Cell-type specificity is governed through tight control of transcription factors and cytokine production. Recent studies have highlighted the importance of lncRNAs in controlling hematopoiesis through their ability to bind other RNAs, DNA, or proteins and modulate the chromatin state, mRNA stability, protein expression, and signaling. Interestingly, lncRNAs appear to be expressed in a much more cell-type-specific manner than transcription factors and other protein-coding genes, making them attractive targets to understand various stages of cell differentiation (Guttman et al. 2010; Cabili et al. 2011; Washietl et al. 2014).
Hematopoiesis involves the formation of all blood cell components. Hematopoietic stem cells (HSCs) are self-renewing cells that reside within the medulla of the bone marrow. HSCs differentiate in specific restricted stages to give rise to three lineages, erythroid cells (red blood cells and platelets), lymphocytes (T, B, and natural killer [NK] cells), and myeloid cells (monocytes, neutrophils, basophils, and eosinophils (Orkin and Zon 2008). The role of transcription factors in determining cell fate during hematopoiesis has been studied intensely for a number of years. Mouse knockouts of transcription factors confirm their importance in lineage specification. In addition, mutations within some transcription factors are associated with malignancies such as leukemia (Orkin and Zon 2008). Protein-based assays, including flow cytometry and, more recently, CyTOF, are revealing new markers of each cell lineage (Bendall et al. 2011). The contribution of noncoding RNA to these processes is also now emerging.
Cytokines are essential regulators of hematopoiesis. Many cytokines can be used in culture to differentiate HSCs into specific cell types. Cytokines function by binding to specific receptors present on HSCs, thus controlling self-renewal, differentiation, mobility, and death. In this section, we cover the emerging roles for lncRNAs within hematopoiesis and place them in the context of cytokine signaling and cell-type specificity for immune-cell subtypes.
ERYTHROCYTES
Recent studies have provided a comprehensive overview of the erythrocyte transcriptome (Alvarez-Dominguez et al. 2014; Doss et al. 2015). To differentiate cells from peripheral blood into erythrocytes, cells must be treated with interleukin (IL)-3, human stem cell factor, and erythropoietin. Close to 100 lncRNAs were shown to be differentially expressed during erythrocyte development (Alvarez-Dominguez et al. 2014). The top candidate lncRNAs from this study, based on their expression pattern and the transcription factors that regulate them (Gata1, Tal1, and KLF1), were all shown to play a role in controlling erythrocyte development. One of these RNAs (artificial long noncoding RNA [alncRNA]-EC7), an enhancer RNA neighboring the protein-coding gene BAND3, was found to function in cis to control BAND3 expression, which is critical for erythrocyte differentiation. lncRNA-erythroid prosurvival (EPS) was also identified as a highly induced lncRNA during the terminal differentiation of erythrocytes. Knockdown of lncRNA-EPS led to apoptosis, whereas overexpression of the transcript promoted erythrocyte survival. lncRNA-EPS expression repressed expression of proapoptotic genes (Hu et al. 2011). A long intervening noncoding RNA (lincRNA)-EPS-deficient mouse was recently generated by removal of the 4-kb region harboring the lincRNA-EPS locus and replacing it with a neomycin resistance cassette. Surprisingly, this mouse did not manifest any developmental defect, and the process of erythropoiesis was intact (Atianand et al. 2016). This observation indicates potential redundancy regarding the contribution of lincRNA-EPS to erythropoiesis, suggesting that more than one lincRNA may be involved. An intriguing role for lincRNA-EPS in controlling immune signaling has been identified in this animal model (Atianand et al. 2016), and will be discussed in detail later in this review.
SHORT-LIVED MYELOID CELLS
In the peripheral blood, 70% of the leukocytes include neutrophils, eosinophils, and classical monocytes that represent the short-lived myeloid cells. Kotzin et al. (2016) have identified an lncRNA they named “Morrbid” (myeloid RNA regulator of Bim-induced death) that functions to control the short-lived myeloid cell life span. Morrbid is highly conserved across species. The authors generated a Morrbid knockout mouse and observed a dramatic decrease in the short-lived myeloid cell population circulating in the blood. They showed that Morrbid is specifically expressed in these myeloid cells where its expression levels are dependent on the common cytokine receptor β-chain cytokines (IL-3, IL-5, and granulocyte macrophage colony-stimulating factor [GMCSF]). Morrbid localized to the nucleus and bound to chromatin. Mechanistically, Morrbid functions in cis to suppress the expression of its neighboring proapoptotic gene Bcl2l11 (Bim) by recruiting the PRC2 complex to the promoter of Bcl2l11 in short-lived myeloid cells.
MACROPHAGES
Monocytes circulate in the periphery and, when activated by specific cytokines, differentiate into monocyte-derived macrophages. In vitro, human monocytes can be differentiated into classically activated macrophages or M1 cells using GMCSF or stimulation with lipopolysaccharide/interferon γ (LPS/IFN-γ). M2 cells (or alternatively activated cells) are obtained following macrophage colony-stimulating factor (MCSF) stimulation or activation with IL-4. The history behind the classification of macrophages into M1 and M2 is somewhat controversial and is reviewed extensively by Martinez and Gordon (2014) and Murray et al. (2014). PU.1 is the master transcription factor associated with macrophage identity. Polarization of M1 and M2 cells relies on distinct additional transcription factors. The M1 cell phenotype requires IRF5 and signal transducers and activators of transcription (STAT)1, whereas M2 cells use STAT6, IRF4, and PPARG. Activated M1 cells, which produce tumor necrosis factor (TNF), IL-6, and IL-12, display increased antimicrobial activity and antigen presentation, whereas M2 cells are associated with the production of IL-10 and IL-4, T helper (Th) 2 cell activation, and defense against parasitic infections (Lawrence and Natoli 2011).
Long noncoding monocyte (lnc-MC) is the first lncRNA associated with macrophage differentiation. lnc-MC is transcriptionally regulated by PU.1. It appears to be involved in differentiation in THP1 and HL-60 cells. lnc-MC functions as a competitive endogenous RNA (ceRNA) that sponges up miR-199a-5p, which in turn targets activin A receptor type 1B (ACVR1B). This protein is a known regulator of monocyte to macrophage differentiation; therefore, the sponging effect of lnc-MC on miR199-5p ensures its expression and allows for the differentiation program to occur (Chen et al. 2015).
Huang et al. (2016) differentiated macrophages into M1 (IFN-γ+LPS) and M2 (IL-4) and profiled the expressed lncRNAs using an lncRNA-specific microarray. They identified 9343 lncRNAs with twofold differential expression in M1 macrophages and 4592 in M2 macrophages when compared to monocyte-derived macrophages (Huang et al. 2016). They focused on one lncRNA (TCONS_00019715) that was higher in M1 compared to M2 macrophages. Knocking down this lncRNA in human THP1 cells resulted in an increase in M2 markers compared to M1 markers. Whether this lncRNA is directly involved in M1 polarization remains to be determined.
DENDRITIC CELLS
Dendritic cells (DCs) are the professional antigen-presenting cells of the immune system. There are a large number of DC cell subtypes, including conventional DCs, plasmacytoid DCs, and monocyte-derived DCs (Shortman and Liu 2002). In mice, DCs can be cultured from bone marrow using GMCSF. Although, in humans, monocytes from the periphery are differentiated into DCs using GMCSF and IL-4 to produce monocyte-derived dendritic cells (MoDCs) (Shortman and Liu 2002).
Long noncoding dendritic cell (lnc-DC) was identified in human conventional DCs among a collection of lncRNAs whose expression increased during the process of DC differentiation. Knocking down lnc-DC in human monocytes inhibited their ability to differentiate into MoDCs. A similar result was observed when lnc-DC was knocked down in murine bone marrow–derived DCs, which resulted in an impairment in their ability to activate T cells. lnc-DC was shown to interact with STAT3 in the cytoplasm preventing SHP1 from dephosphorylating STAT3, suggesting that the function of lnc-DC is to enhance the activation of STAT3 in the cytoplasm, enabling the STAT3 transcriptional program to occur (Wang et al. 2014). There have been some questions raised concerning the identification and characterization of the murine ortholog of lnc-DC. The gene at this locus was previously identified as Wdnm1-like and possesses protein-coding capacity and is expressed in adipocytes (Wu and Smas 2008). These data suggest that a functional lncRNA is present only in humans, whereas other mammals encode the protein-coding transcript Wdnm1-like (Dijkstra and Ballingall 2014).
T CELLS
Naïve Th cells differentiate into effector cells through the coordinated expression of specific transcription factors and cytokines. The transcription factors orchestrate complex epigenetic alterations of gene loci to specifically activate or repress genes required for lineage specification. Naïve CD4+ T cells differentiate into effector T subsets, including Th1, Th2, Th17, and regulatory T cells (Tregs), which express the cytokines IFN-γ (Th1); IL-4, IL-5, and IL-13 (Th2); IL-17 (Th17); or transforming growth factor β (TGF-β) and IL-10 (Treg cells). In contrast, naïve CD8+ T cells differentiate into cytotoxic CD8 T cells following IL-2 stimulation, and they produce IFN-γ and TNF. This differentiation step is essential for the adaptive immune system to provide protection against infection. Table 1 outlines the cytokines involved in T-cell differentiation, cytokines produced by each T-cell subset, and the lncRNAs identified to function in each subset (also depicted in Fig. 1).
Table 1.
Cytokines and long noncoding RNAs (lncRNAs) involved in immune cell development and function
| Cell type | Transcription factors | Cytokines required for differentiation | Cytokines produced | lncRNAs | Cell functions | References |
|---|---|---|---|---|---|---|
| Macrophage (M1) | PU1 | GMCSF, LPS+IFN-γ | TNF, iNOS | lnc-MC | Proinflammatory cytokine production | Chen et al. 2015 |
| Macrophage (M2) | PU1 | MCSF, IL-4 | IL-4, IL-13, IL-10 | None identified to date | Resolution of inflammation | |
| Dendritic cells | PU1 | GMCSF, IL-4 | lnc-DC | Professional antigen-presenting cells | Wang et al. 2014 | |
| CD8+ T Cells | T-bet, Eomes | IL-2 | IFN-γ, TNF, IL-2 | lncRNA-CD244, | Antiviral activity | Wang et al. 2015b |
| Th1 cells | STAT1, 4, T-bet | IFN-γ, IL-12 | IFN-γ, IL-4, IL-5, IL-13 | NeST | Target intracellular pathogens | Collier et al. 2012; Gomez et al. 2013 |
| Th2 cells | GATA-3, STAT6 | IL-4 | IL-4, IL-13, IL-5 | lincR-Ccr2-5′AS, GATA3-AS, Th2-LCR | Fight parasitic infections | Hu et al. 2013; Spurlock et al. 2015 |
| Th17 cells | Rorγt, BATF | IL-17 | IL-17, IL-22 | RMRP | Target extracelluar bacteria and fungi | Huang et al. 2015 |
| Tregs | FoxP3 | TGF-β, IL-2 | TGF-β, IL-10 | DQ786243 | Maintain self-tolerance and homeostasis | Qiao et al. 2013 |
TGF-β, Transforming growth factor β; GMCSF, granulocyte macrophage colony-stimulating factor; LPS, lipopolysaccharide; IFN-γ, interferon γ; TNF, tumor necrosis factor; iNOS, inducible nitric oxide synthase; lnc-MC, Long noncoding monocyte; MCSF, macrophage colony-stimulating factor; IL, interleukin; lnc-DC, long noncoding dendritic cell; lncRNA; long noncoding RNA; STAT1, signal transducers and activators of transcription 1; LCR, locus control region; BATF, basic leucine zipper ATF-like; Treg, regulatory T cell.
Figure 1.
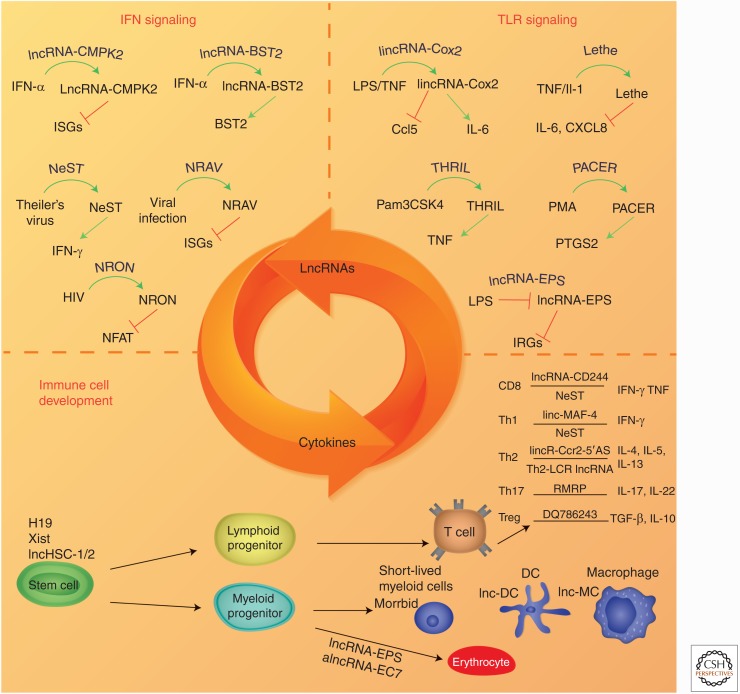
Cytokine/long noncoding RNA (lncRNA) interplay in controlling immune cell development and function. This figure shows lncRNAs that are controlling cytokine production and cytokines controlling lncRNA expression. (A) Interferon (IFN)/lncRNA regulatory loop, (B) lncRNAs induced by Toll-like receptor (TLR) signaling pathway to control cytokine production, and (C) those lncRNAs critical for immune cell development. TNF, Tumor necrosis factor; ISG, interferon-stimulated gene; IL, interleukin; NeST, nettoie Salmonella pas Theiler’s; NRAV, negative regulator of antiviral response; THRIL, TNF-α and heterogeneous nuclear ribonucleoprotein L (hnRNPL)-related immunoregulatory lincRNA; PACER, p50-associated COX-2 extragenic RNA; NRON, noncoding repressor or nuclear factor of activated T (NFAT) cells; IRG, interferon regulated gene; Th, T helper cell; LCR, locus control region; EPS, erythroid prosurvival; HSC, hematopoietic stem cells; Xist, X-inactive specific transcript; alnc, artificial long noncoding; DC, dendritic cell; MC, monocyte.
CD8 T Cells
RNA-seq analysis of various T-cell subsets have identified a large collection of lncRNAs that are both specific for each subset and also play critical roles in T-cell development and differentiation. The first comprehensive study on lncRNAs in CD8+ T cells identified >1000 lncRNAs in human and murine CD8+ T cells that displayed cell-type-specific expression, with many close to protein-coding genes with known functions in T-cell biology (Pang et al. 2009). Patients suffering from severe asthma have increased activation of circulating CD8+ T cells compared to CD4+ T cells. 167 lncRNAs show differential expression in activated CD8+ T cells; however, their roles in regulating these responses were not investigated (Tsitsiou et al. 2012).
lncRNA-CD244 (lncRNA-AS-GSTT1(1-72)) was found to be expressed at high levels in CD8+ T cells that are infected with Mycobacterium tuberculosis, where it functions to impair T-cell antigen receptor (TCR) signaling. Knockdown of lncRNA-CD244 resulted in elevated expression of IFN-γ and TNF-α. Expression of this lncRNA was induced following activation with CD244 (a known TCR inhibitor that is induced following tuberculosis [TB] infection) and the lncRNA, in turn, represses expression of IFN-γ and TNF-α through its interactions with enhancer of zeste homolog 2 (EZH2), a major component of the PRC2 complex (Wang et al. 2015b).
Th1 Cells
Th1 cells require the transcription factor T-bet for their differentiation and production of IFN-γ. A number of studies have cataloged levels of lncRNAs in different T-cell subsets. Xia et al. (2014) studied lncRNA expressed in CD4−CD8− (DN) cells compared to CD4+CD8+ (DP), and CD4+CD8− and activated CD4+CD8− T cells. They identified a total of 788 lncRNAs between all of these cell types and 746 mRNAs (Xia et al. 2014). Hu et al. (2013) systemically studied lncRNA expression in a number of T-cell subsets. They identified 354 lncRNAs that were expressed in Th1 cells. When they compared lncRNAs expressed in Th1, Th2, naïve CD4+, Th17, or inducible Treg (iTreg) cells, they found only 100 lncRNAs that were common to all cell types. Nearly 50% of lncRNAs were unique to a given T-cell subset. Comparing this to >78% of mRNAs that are commonly expressed across all T-cell subtypes confirmed that lncRNAs are expressed in a more cell-type-specific manner than protein-coding genes (Hu et al. 2013). More than 56% of lncRNAs expressed in Th1 cells are controlled by the transcription factor STAT4, whereas STAT6 controls Th2-expressing lncRNAs.
NeST
NeST (nettoie Salmonella pas Theiler’s [cleanup Salmonella not Theiler’s], also called lincR-ifng-3′AS), was originally described as TMEVG, identified in Th1 cells, and expressed in a manner dependent on T-bet (Hu et al. 2013). In more recent studies, NeST was identified as a gene associated with persistence of Theiler’s virus in mice. In these studies, NeST was found to associate with the WDR subunit of mixed-lineage leukemia histone H3 Lys 4 (H3K4) methyltransferase to increase H3K4 methylation at the IFN-γ locus in CD8 T cells. Mice lacking NeST are capable of being persistently infected with Theiler’s virus, while being resistant to lethal Salmonella enterica infection. These data were the first to identify roles for noncoding RNAs in host defense to infectious pathogens (Collier et al. 2012; Gomez et al. 2013). Recently, Li et al. (2016) have shown that expression of NeST is altered in patients with immune thrombocytopenia (IT). Peripheral blood mononuclear cells (PBMCs) from these patients showed decreased NeST expression, whereas IFN-γ was elevated. This was not the case for healthy controls in which NeST expression and IFN-γ expression are positively associated. They conclude that NeST and IFN-γ are in a regulatory loop in which NeST is initially needed for IFN-γ expression, and this increased expression subsequently inhibits NeST and IFN-γ expression levels in patients with IT (Li et al. 2016). NeST also shows elevated expression levels in patients suffering from Sjorgren’s syndrome and ulcerative colitis (Padua et al. 2016; Wang et al. 2016a). The exact mechanism or role of NeST in the pathogenesis of these diseases remains to be determined.
Th2 Cells
The transcription factors STAT6 and GATA3 are critical for Th2 cell differentiation via their regulating the expression of many Th2-specific genes. LincR-Ccr2-5′AS is regulated by GATA3, with reduced expression in a GATA3 knockout Th2 cell. In a more global analysis, GATA3 regulated ∼28% of Th2-specific lncRNAs (Hu et al. 2013). LincR-Ccr2-5′AS can regulate migration of Th2 cells to the lung. Knocking down this lncRNA using short-hairpin (shRNA) did not inhibit expression of the Th2 cytokine IL-4 but did result in decreased expression of its neighboring protein-coding genes, including Ccr1-Ccr5. There is an antisense lncRNA neighboring GATA3 (GATA3-AS) that is also Th2-cell restricted. The lncRNA Th2 locus control region (LCR) is also Th2-specific in expression and is localized at the 3′ end of the Rad50 gene (Spurlock et al. 2015). This lncRNA is coexpressed with IL-4, IL-5, and IL-13, and knockdown of this lncRNA greatly impacted expression levels of these cytokines. The current mechanism for how lncRNA Th2-LCR controls these genes is through its recruitment of the WDR5-containing complexes that regulate H3K4me3 patterns at the affected target gene loci.
Th17
Th17 cells are specified by RORC (retinoic acid receptor-related orphan nuclear receptor γt) and BATF (basic leucine zipper ATF-like) transcription factors and they produce IL-17 and IL-22. Spurlock et al. (2015) identified 56 lncRNAs that are expressed in primary Th17 cells and 61 that are expressed in Th17 effector cells. These two cell states share a total of 31 lncRNAs. They found that lncRNAs are expressed in a highly lineage-specific manner with many Th17-specific lncRNAs encoded on chromosome 2, 16, and 17 (Spurlock et al. 2015).
Huang et al. (2015) have identified a role for lncRNA RMRP in controlling gene expression programs in Th17 cells. They identified DEAD-box helicase DD5 as the binding partner of the transcription factor RORγt that controls transcription of target genes in Th17 cells. The interaction between these two proteins is mediated by the lncRNA Rmrp. Rmrp is highly conserved, and mutations in Rmrp are linked to cartilage hair hyperplasia (CHH), a genetic disorder of bone growth that leads to short stature and other skeletal abnormalities, fine hair, and immune deficiencies. This is the first genetic disorder in which the causative mutation lies within an lncRNA. Mice carrying the point mutation present in CHH had reduced binding to DDX5, resulting in a decrease in RORC target genes (Huang et al. 2015). This discovery provides new insights in the regulation and complexity of Th17 cell functions.
Tregs
T regulatory cells act as suppressors of immune responses in which they function to down-regulate the actions of effector T cells. Natural Tregs originate in the thymus, whereas iTregs arise from naïve T cells in the periphery. Tregs require the transcription factor Foxp3 and the cytokine TGF-β for their differentiation. Tregs can produce TGF-β, IL-10, and IL-35, which all aid in suppressing immune responses. Hu et al. (2013) cataloged a total of 278 lncRNAs in iTregs; however, the functions for these genes in iTreg development or effector functions were not examined. DQ786243 is a lncRNA that was shown to be significantly overexpressed in patients with clinically activated Crohn’s disease (CD). High expression levels of DQ786243 appear to affect expression of CREB and Foxp3 therefore impacting Treg functions. The results here are correlative, and the mechanism by which this lncRNA might impact gene expression within CD remains to be determined (Qiao et al. 2013).
CYTOKINE REGULATION OF lncRNAS
TNF-α Stimulation
Activation of mouse embryonic fibroblasts with TNF-α regulates the expression of 3596 protein-coding genes, 112 lncRNAs, and 54 pseudogene lncRNAs. Rapicavoli et al. (2013) identified Lethe (after the river of forgetfulness in Greek mythology) as a TNF-α-inducible and IL-1B-inducible lncRNA. Lethe is nuclear localized and binds to chromatin. Lethe acts as an inhibitor of nuclear factor κB (NF-κB)-mediated transcriptional activation by sequestering RelA (p65) and preventing its translocation into the nucleus (Rapicavoli et al. 2013). Increased NF-κB activity is associated with aging, and the authors showed that Lethe expression levels decrease dramatically with age because of increased NF-κB activity.
The Interferon/lncRNA Regulatory Loop
Activation of the interferon response (types I, II, and III) is critical for protection against infection by viruses and other pathogens. The type I IFNs bind to their receptor, IFN-α/BR, and trigger activation of the Janus kinase/signal transducers and activators of transcription (JAK-STAT) signaling pathway, leading to the production of interferon-stimulated genes (ISGs). IFN signaling can lead to the activation of a number of lncRNAs, which, in turn, act to control these signaling pathways. There were a number of high-throughput RNA-sequencing studies performed using a variety of viral infections, showing that 1000s of lncRNAs were regulated in virus-infected cells. Profiling of lncRNAs has been performed at a vast rate, yet the mechanistic understanding of these lncRNAs and their functions has lagged far behind. Most of these studies provide information on the differential regulation of lncRNAs, with no insight into the roles these lncRNAs might play in host defense. The database MONOCLdb contains annotated lncRNAs that are induced by IFN-α stimulation (Josset et al. 2014). The numbers of lncRNAs identified are outlined in Table 2. In this section, we will focus on IFN-regulated lncRNAs, with experimental data supporting biological activity in these pathways.
Table 2.
RNA-seq profiling of long noncoding RNAs (lncRNAs) following immune activation
| Stimulant | Species | Cell/tissue type | lncRNAs (up-regulated) | lncRNAs (down-regulated) | References |
|---|---|---|---|---|---|
| TNF-α | Mouse | MEFs | 166 | Rapicavoli et al. 2013 | |
| Pam3CSK4 | Mouse | BMDMs | 62 | 62 | Carpenter et al. 2013 |
| Pam3CSK4 | Human | THP1s (monocytic cell line) | 156 | Li et al. 2014 | |
| IFN-α | Human | Hepatocytes | ∼120 | ∼100, exact number not reported | Kambara et al. 2014 |
| IAV | Human | Alveolar cells | 494 | 413 | Ouyang et al. 2014 |
| EV71 | Human | Rhabdomyosarcoma cells | 2990 | 1876 | Yin et al. 2013 |
| SARS-CoV | Mouse | Lung | 1500 | Peng et al. 2010 |
TNF-α, Tumor necrosis factor α; MEFs, murine embryo fibroblasts; BMDMs, bone-marrow-derived macrophages; IFN-α, interferon α; IAV, influenza A virus; SARS-CoV, severe acute respiratory syndrome coronavirus.
lncRNA-CMPK2
lncRNA-CMPK2 is a polyadenylated, spliced nuclear lncRNA that is induced by IFN-α in a number of cell lines in both humans and mice. lncRNA-CMPK2 has been shown to be elevated in the livers of patients with hepatitis C virus (HCV) infection (Kambara et al. 2014). lncRNA-CMPK2 can inhibit the IFN response as knockdown of this gene in hepatocytes resulted in a reduction in HCV replication and an up-regulation of ISGs. The mechanism by which this lncRNA mediates its inhibitory effects remains unclear.
lncRNA-BST2
Two groups identified the same IFN-α inducible lncRNA known as BST2 IFN-stimulated positive regulator (BISPR) (Barriocanal et al. 2015; Kambara et al. 2015). A bidirectional promoter of the protein-coding gene BST2 transcribes BISPR that then acts as a positive regulator for BST2 expression. Small interfering RNA (siRNA)-mediated knockdown of BISPR had a dramatic impact on BST2 protein expression levels.
NeST
As mentioned earlier, the lncRNA NeST (also known as TMEVPG1 and LincR-Ifng-3′AS′) is expressed in the Th1 subset of helper T cells and is critical for controlling infection with Theiler’s virus. NeST controls the expression of IFN-γ through WDR5-dependent histone methylation at the Ifng locus in CD8+ T cells.
NRAV
Negative regulator of antiviral response (NRAV) is a lncRNA that is dramatically down-regulated in cells following viral infection. It functions to suppress ISG expression through epigenetic regulation of these genes. Overexpressing human NRAV in mice results in enhanced influenza A virus (IAV) virulence (Ouyang et al. 2014).
NRON
The expression of lncRNA NRON (noncoding repressor or nuclear factor of activated T [NFAT] cells) is controlled by HIV. NRON is localized to the cytosol where it can bind to the transcription factor NFAT and repress its activity. Knockdown of NRON using siRNA resulted in enhanced HIV replication because of increased activity of NFAT (Imam et al. 2015).
lncRNA-CD244
lncRNA-CD244 (lncRNA-AS-GSTT1[1-72)] is found at high levels in CD8+ T cells that are infected with M. tuberculosis where it inhibits T-cell signaling. Knocking down lncRNA-CD244 results in increased expression of IFN-γ and TNF-α. The authors find that this lncRNA expression is driven by CD244 signaling and functions to repress IFN-γ and TNF-α through its interactions with EZH2, a component of the PRC2 complex (Wang et al. 2015b).
lncRNAs THAT REGULATE CYTOKINE PRODUCTION
Innate immune signaling pathways are complex, consisting of feedforward and feedback mechanisms, which function in a temporally regulated manner to elicit effective host defenses and ensure that immune homeostasis is maintained. lncRNAs are now emerging as crucial players in controlling these inducible transcriptional networks. In this section, we will outline those lncRNAs with validated experimental data supporting their roles in controlling cytokine expression downstream from innate immune receptor activation.
lincRNA-Cox2
Studies in our laboratory identified lincRNA-Cox2 as a dynamically regulated lincRNA in murine macrophages stimulated with multiple Toll-like receptor (TLR) ligands, including lipopolysaccharide, which activates TLR4/MD2 and Pam3CSK4, a synthetic bacterial lipopeptide that acts as TLR1/2 ligand (Carpenter et al. 2013). Prior studies had identified this lincRNA in DCs exposed to lipopolysaccharide, which engages TLR4/MD2 (Guttman et al. 2009). lincRNA-Cox2 acts as both a repressor and an activator of distinct clusters of innate immune genes. lincRNA-Cox2 can form a complex with hnRNP-A/B and A2/B1 to mediate its repressive effects on interferon-stimulated genes. Knockdown of lincRNA-Cox2 also severely impaired the production of additional immune genes, including the proinflammatory cytokine IL-6; however, the precise molecular mechanism by which lincRNA-Cox2 mediates these latter effects in macrophages is still unclear.
Two recent studies have shed new light on the function of lincRNA-Cox2. Chen and colleagues have shown that TNF-α induced lincRNA-Cox2 functions to control IL-12B levels in intestinal epithelial cells. In a series of elegant biochemical studies, these authors showed that lincRNA-Cox2 facilitates the recruitment of Mi-2/nucleosome remodeling and deacetylate (Mi2/NuRD) complex to the IL-12B promoter region, resulting in inhibition of IL-12B expression (Tong et al. 2016). They also showed that lincRNA-Cox2 can promote immune gene activation through binding to the SWI/SNF chromatin remodeling complex. Both of these studies classify lincRNA-Cox2 as a primary response gene because they observed early induction (2 h) of this transcript in intestinal epithelial cells or RAWs (murine macrophage cell line). In our own studies, in macrophages, we found that lincRNA-Cox2 reached maximal expression following 5 h of stimulation with TLR2 ligands in primary murine macrophages. Indeed, we observe superinduction of lincRNA-Cox2 in murine macrophages treated with TLR2 ligands in the presence of cycloheximide (S Carpenter and KA Fitzgerald, unpubl.), indicating this gene may be a secondary response gene in murine macrophages. It is possible that lincRNA-Cox2 has varying expression profiles in different cell types. The exact mediators that collaborate with lincRNA-Cox2 to exert these regulatory functions remain to be further elucidated.
THRIL
TNF-α and heterogeneous nuclear ribonucleoprotein L (hnRNPL)-related immunoregulatory lincRNA (THRIL) was identified in the human monocyte cell line THP1 (Li et al. 2014). A total of 156 lncRNAs were found to be differentially regulated in THP1 cells following stimulation with the TLR1/2 ligand Pam3CSK4. THRIL is an antisense transcript that partially overlaps the 3′ UTR of the gene encoding BRI3-binding protein (Bri3bp). THRIL functions to regulate TNF-α expression through interactions with hnRNPL. RNA-seq analysis on THRIL knockdown cells showed that this lncRNA has a broad impact on expression of a large number of immune genes (454). Interestingly 98% of these genes were down-regulated when THRIL was knocked down, strongly suggesting that this lncRNA is critical for controlling basal expression of these genes (Li et al. 2014). The exact mechanism of action of this lincRNA and whether all of these effects are direct or indirectly regulated by THRIL remains to be clarified.
PACER
The p50-associated COX-2 extragenic RNA (PACER) is transcribed in the antisense direction directly upstream of the PTGS2 (Cox2) transcription start site (Krawczyk and Emerson 2014). To date, there is no evidence that PACER is conserved across species. PACER functions to sequester p50, the repressive subunit of the NF-κB complex, preventing p50 from binding to the PTGS2 promoter. The authors show that the presence of CTCF at the PTGS2 promoter facilitates chromatin accessibility, resulting in transcription of PACER. PACER then functions to recruit p300 and RNA Pol II complexes, leading to the induction of Cox2. PACER’s interaction with p50 permits access to the activating complex of NF-κB (p50-p65) at the PTGS2 promoter following inflammatory stimulation (Krawczyk and Emerson 2014).
NEAT1
NEAT1 is a 4-kb unspliced nonpolyadenylated transcript critical for the formation of nuclear paraspeckles (Clemson et al. 2009). NEAT1 is evolutionarily conserved and the murine homolog displays two small regions of high sequence conservation (Clemson et al. 2009). NEAT1 is induced by influenza virus and herpes simplex virus in addition to PolyI:C (synthetic RNA ligand that activates TLR3). NEAT1 is critical for the expression of IL-8 through its interactions with the splicing protein splicing factor proline/glutamine-rich (SFPQ) (Imamura et al. 2014). Chromatin immunoprecipitation (ChIP) assays confirmed that at basal levels SFPQ binds to the promoter of the gene encoding IL-8 and inhibits transcription. NEAT1 can interact with SFPQ, resulting in the protein relocating to paraspeckles releasing its repression of IL-8.
Recent evidence suggests that NEAT1 expression is elevated in patients suffering from systemic lupus erythematosus (SLE), and the expression is predominantly present in monocytes. Knockdown of NEAT1 resulted in decreased expression of proinflammatory genes, including IL6 and CXCL10, and the authors propose a mechanism by which NEAT1 affects the late-stage mitogen-activated protein kinases (MAPK) pathway following LPS stimulation. They conclude that NEAT1 could act as a regulatory contributor to the increased cytokine signature associated with the pathogenesis of SLE (Zhang et al. 2016).
lnc-IL-7R
lnc-IL-7R is an LPS-inducible transcript that overlaps the 3′ UTR of IL-7R, with both transcripts sharing the same polyA tail. Knockdown of lnc-IL-7R led to an increase in expression of a number of genes, including IL-7R, IL-8, VCAM-1, and E-selectin. This knockdown was accompanied by a decrease in the transcriptional histone repressor mark H3K27 trimethylation at the promoters of affected genes (Cui et al. 2014). The exact mechanism of how lnc-IL-7R regulates H3K27 at its target genes still requires mechanistic understanding.
IL-1β-RBT46
IL-1β-RBT46 originates from a region of bidirectional transcription upstream of the transcription start site of IL-1B. It is inducible following LPS stimulation or infection with Listeria monocytogenes. Knocking down this transcript with shRNA resulted in a reduction in IL-1B and CXCL8 expression at both the RNA and protein levels (Ilott et al. 2014).
NKILA
NF-κB-interacting lncRNA (NKILA) acts as a direct inhibitor of NF-κB signaling. NKILA binds to the NF-κB/IκB complex inhibiting phosphorylation and subsequent degradation of IκB, resulting in the p65 subunit of NF-κB remaining in the cytoplasm and therefore inhibiting signaling (Liu et al. 2015). A recent article has questioned some of these findings by Liu et al. and provided some additional information on this locus, showing that NKILA is transcribed in the opposite orientation to the protein-coding gene PMEPA1 (Dijkstra and Alexander 2015).
AS-IL-1α
Studies in our own laboratory identified a natural antisense transcript that is transcribed at the IL-1A locus. The lncRNA contains partial sequence complementarity to IL-1A. AS-IL-1α is a nuclear localized transcript that is highly inducible following TLR stimulation or following L. monocytogenes infection in vivo (Chan et al. 2015). Knockdown studies have shown that AS-IL-1α is required for transcription of the IL-1A protein-coding gene.
lincRNA-EPS
The vast majority of studies of lncRNAs in immunity have focused on inducible lncRNAs and their role in controlling immune gene expression. However, it is clear from all of the RNA-seq data generated that there are just as many lncRNAs that are down-regulated following inflammatory stimulation of innate immune cells. One such example is lincRNA-EPS. As mentioned earlier, this lncRNA was first identified as an erythrocyte-expressed lncRNA crucial for during red blood cell development. A recent study expanded on this and showed that lincRNA-EPS has additional functions beyond those proposed for erythrocytes (Atianand et al. 2016). lincRNA-EPS is expressed in macrophages and DCs and is rapidly down-regulated following inflammatory stimulation. lincRNA-EPS-deficient mice display enhanced inflammation and succumb to endotoxin-mediated lethality faster than their corresponding littermate controls. lincRNA-EPS-deficient macrophages have elevated expression of ∼200 immune response genes. lincRNA-EPS associates with chromatin at regulatory regions of these genes where it controls nucleosome positioning to restrain transcription of these genes. lincRNA-EPS mediates some of these functions through its interaction with heterogeneous nuclear ribonucleoprotein L, which occurs via a CANACA motif, known to be enriched in RNAs that bind hnRNPL located in its 3′ end (Atianand et al. 2016). The identification of lincRNA-EPS as a repressor of inflammatory responses further highlights the importance of lincRNAs in the immune system.
EMERGING ROLES FOR lncRNAs IN DISEASE
Remarkably, 93% of disease-associated SNPs lie in noncoding regions of the genome, some of which impact the expression of lncRNAs (Pennisi 2007; Kumar et al. 2013). Approximately 10% of autoimmune and immune-related disorder (AID) SNPs are present in lncRNA genes (Ricaño-Ponce and Wijmenga 2013). Gaining a better understanding of the molecular mechanisms of lncRNA could help define their roles in inflammatory and autoimmune disease pathologies. Many of the studies to date are descriptive and correlate expression of a particular lncRNA associated with specific diseases. Table 3 outlines lncRNAs that have been functionally associated with autoimmune disorders. Only one lncRNA with a known SNP has been shown to be the direct cause of a genetic disorder and that is lncRNA RMRP in CHH as mentioned previously. The majority of autoimmune conditions are not monogenic disorders, and therefore it is possible that many lncRNAs could impact disease pathogenesis in these conditions. Just this year, Lnc13 was identified and associated with susceptibility to celiac disease (Castellanos-Rubio et al. 2016). Similar to what has been reported for lincRNA-EPS, Lnc13 is down-regulated in myeloid cells exposed to inflammatory stimuli. This down-regulation, in turn, results in elevated expression levels of immune genes. Lnc13 partly overlaps with the protein-coding gene sIl18rap. The 5′ end of Lnc13 overlaps the 3′ end of IL-18rap. Like other lincRNAs, Lnc13 interacts with heterogeneous nuclear ribonucleoprotein D (hnRNPD), and this complex is involved in suppressing immune gene expression. Lnc13 is encoded within a celiac disease–associated haplotype. In patients suffering from celiac disease, the levels of Lnc13 are decreased in the small intestine, which could account for the increase in expression of inflammatory genes in these patients. In addition, the Lnc13 variant present in celiac disease patients interacts less efficiently with hnRNPD than its wild-type counterpart, which may account for its inability to mediate its repressor functions.
Table 3.
Disease-associated long coding RNAs (lncRNAs) and their functions
| Disease condition | lncRNA | Function | References |
|---|---|---|---|
| Cartilage hair hypoplasia | RMRP | Controlling gene expression in Th17 cells | Huang et al. 2015 |
| Celiac disease | Lnc13 | Mediates repression of a subset of celiac-associated genes through its interaction with hnRNPD | Castellanos-Rubio et al. 2016 |
| Kawasaki disease | THRIL | Regulates TNF-α through its interactions with hnRNPL in monocytes | Li et al. 2014 |
| Psoriasis | PRINS | Down-regulates the antiapoptotic effects of G1P3 in keratinocytes | Sonkoly et al. 2005; Szegedi et al. 2010 |
| Angelman syndrome | Ube3a-ATS | Silences paternal Ube3a through imprinting | Meng et al. 2015 |
| Systemic lupus erythematosus | NEAT1 | Regulates MAPK signaling | Zhang et al. 2016 |
TNF, Tumor necrosis factor; THRIL, TNF-α and heterogeneous nuclear ribonucleoprotein L (hnRNPL)-related immunoregulatory lincRNA; hnRNPL, heterogeneous nuclear ribonucleoprotein L; MAPK, mitogen-activated protein kinase.
THERAPEUTIC TARGETING OF CYTOKINES AND lncRNAs
Cytokines are a major target for therapeutic intervention in a number of inflammatory conditions with targeted therapeutics for TNF-α, IFN, and IL-17 already in clinical use (Moreland et al. 1997; Lipsky et al. 2000; Plosker 2011). There is increasing interest in therapeutic targeting of noncoding RNAs. A number of companies are currently developing lncRNA therapeutic targeting methods, including RaNA Therapeutics, OPKU-CURNA, and IONIS Pharmaceuticals. All companies are taking similar approaches and targeting lncRNAs that typically mediate repression of genes through their interaction with repressor complexes such as the PRC2 complex to down-regulate expression of specific protein-coding genes. By interfering with the expression or function of these lncRNAs, the goal is to redirect expression of target protein-coding genes. RaNA Therapeutics just received a patent for Polycomb-associated noncoding RNAs, which function through the PRC2 complex (Lee et al. 2016).
All companies are making use of antisense technology to repress or interfere with lncRNAs and their target repressor complexes using antisense oligonucleotides. Specifically, they are developing antagoNATs or gapmers, which are single-stranded DNA oligos with locked nucleic acids (LNAs) that can act either as steric blockers (if LNAs are distributed throughout the oligo) or directly bind to lncRNAs and mediate their degradation through RNase H cleavage (if LNAs are placed at either end of the oligo). The LNAs help to protect the oligo against exonuclease cleavage.
Last year, IONIS Pharmaceuticals in collaboration with a laboratory at Baylor College of Medicine published a study describing the possible uses of lncRNA targeting in Angelman syndrome, a syndrome that is a monogenic disorder caused by maternal deficiency of the imprinted gene UBE3A, which is an E3 ubiquitin ligase. Patients suffering from Angelman syndrome possess one paternal copy of UBE3A; however, this is silenced by the lncRNA UBE3A-ATS (Meng et al. 2015). This disorder results in developmental delay, seizures, and ataxia and there are currently very few treatment options available. In this study, they made use of antisense oligonucleotides (ASOs) designed to target the lncRNA Ube3a-ATS in a mouse model of Angelman syndrome. Even partial restoration of the UBE3A protein resulted in an improvement in the cognitive defects of this condition. Like other oligo-based therapeutics (e.g., siRNA), one of the major obstacles is delivery and specificity of targeting the gene of interest in a specific location. The initial focus of many lncRNA therapeutics is on the central nervous system (CNS) diseases for which there are currently no treatment options. In the case of the Angelman syndrome study, they are delivering the oligo directly to the CNS with some success. Much work is needed to increase the efficiency of the delivery mechanisms and this will open a huge opportunity for RNA-based therapeutics.
CONCLUSIONS AND FUTURE PERSPECTIVES
lncRNAs are quickly emerging as key regulators of a wide variety of biological processes (Rinn and Chang 2012). Because lncRNAs display more cell-type specificity in expression patterns compared to protein-coding genes, it is estimated that the number of annotated lncRNAs will continue to expand as more sequencing is performed. As exciting as this field is, it is also a daunting challenge to begin to understand the biological importance of thousands of novel genes. Some of the challenges include understanding the exact annotations of full-length lncRNA transcripts. Chip-seq, Gro-seq, and RNA-seq technologies have greatly helped in the initial annotations. However, because lncRNAs are cell-type specific in expression patterns, we often need to perform these expensive and sometimes laborious experiments on each cell type of interest to fully understand the start sites of lncRNAs. This information becomes even more pertinent when using Cas9/CRISPR-based techniques such as CRISPRi and CRISPRa, which heavily rely on knowledge of the transcriptional start sites of target genes. Annotating lncRNAs are also made difficult owing to the levels of repeat elements present in their sequences (Kim et al. 2016). Novel long read technologies such as Pac-Bio and Oxford Nanopore sequencing will enable better definition of the full-length sequences and expressed isoforms of lncRNAs in our cells of interest. The more we understand about the primary sequences of lncRNAs, the easier it will be to determine their exact biological functions. Cas9/CRISPR techniques will allow for rapid high-throughput studies of lncRNAs in a variety of biological contexts.
An exciting avenue of lncRNA research relates to disease and possible use as therapeutic targets in the future. Because the majority of disease-associated SNPs lie within noncoding regions, it is critical to attempt to understand what role lncRNAs, enhancers, and enhancer RNAs play in disease pathogenesis. We outlined the current efforts to use lncRNAs for therapeutic manipulation; however, there is also growing interest in studying lncRNAs as biomarkers of disease. It is possible that lncRNA expression will be disease specific. LncRNAs have also been shown to be highly stable in bodily fluids and therefore offer an easy PCR-based target as a biomarker. To date, one lncRNA has been approved by the Food and Drug Administration (FDA) for use as a biomarker in prostate cancer, and this is the lncRNA PCA3 (de Kok et al. 2002). We believe this is just the beginning in terms of using these genes as biomarkers and possible therapeutic targets in the near future.
Once the genomic profiling phase of discovery is complete, we need to understand the specific functions for all these genes. Extensive functional studies are required to better understand the roles of lncRNA in controlling biological processes in addition to understanding their functions in disease settings. Future discoveries of lncRNAs will provide us with a greater understanding of the molecular mechanisms that govern the transcriptional regulation of genes in inflammatory and infectious diseases, thus providing a better platform from which to develop novel diagnostics and therapeutics.
Footnotes
Editors: Warren J. Leonard and Robert D. Schreiber
Additional Perspectives on Cytokines available at www.cshperspectives.org
REFERENCES
- Alvarez-Dominguez JR, Hu W, Yuan B, Shi J, Park SS, Gromatzky AA, van Oudenaarden A, Lodish HF. 2014. Global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood 123: 570–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel-Duby R, et al. 2015. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 160: 595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atianand MK, Hu W, Satpathy AT, Shen Y, Ricci EP, Alvarez-Dominguez JR, Bhatta A, Schattgen SA, McGowan JD, Blin J, et al. 2016. A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell 165: 1672–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bánfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE, Kundaje A, Gunawardena HP, Yu Y, Xie L, et al. 2012. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res 22: 1646–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriocanal M, Carnero E, Segura V. 2015. Long non-coding RNA BST2/BISPR is induced by IFN and regulates the expression of the antiviral factor tetherin. Front Immunol 5: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall SC, Simonds EF, Qiu P, Amir E-AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, et al. 2011. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332: 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. 2011. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlevaro-Fita J, Rahim A, Guigó R, Vardy LA, Johnson R. 2016. Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA 22: 867–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, et al. 2013. A long noncoding RNA mediates both activation and repression of immune response genes. Science 341: 789–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellanos-Rubio A, Fernandez-Jimenez N, Kratchmarov R, Luo X, Bhagat G, Green PHR, Schneider R, Kiledjian M, Bilbao JR, Ghosh S. 2016. A long noncoding RNA associated with susceptibility to celiac disease. Science 352: 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J, Atianand M, Jiang Z, Carpenter S, Aiello D, Elling R, Fitzgerald KA, Caffrey DR. 2015. Cutting edge: A natural antisense transcript, AS-IL1α, controls inducible transcription of the proinflammatory cytokine IL-1α. J Immunol 195: 1359–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MT, Lin HS, Shen C, Ma YN, Wang F, Zhao HL, Yu J, Zhang JW. 2015. The PU.1-regulated long noncoding RNA lnc-MC controls human monocyte/macrophage differentiation through interaction with microRNA-199a-5p. Mol Cell Biol 35: 3212–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew GL, Pauli A, Rinn JL, Regev A, Schier AF, Valen E. 2013. Ribosome profiling reveals resemblance between long non-coding RNAs and 5′ leaders of coding RNAs. Development 140: 2828–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. 2009. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33: 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM. 2012. Cutting edge: Influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J Immunol 189: 2084–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Xie N, Tan Z, Banerjee S, Thannickal VJ, Abraham E, Liu G. 2014. The human long noncoding RNA lnc-IL7R regulates the inflammatory response. Eur J Immunol 44: 2085–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kok JB, Verhaegh GW, Roelofs RW, Hessels D, Kiemeney LA, Aalders TW, Swinkels DW, Schalken JA. 2002. DD3PCA3, a very sensitive and specific marker to detect prostate tumors. Cancer Res 62: 2695–2698. [PubMed] [Google Scholar]
- Dijkstra JM, Alexander DB. 2015. The “NF-ĸB interacting long noncoding RNA” (NKILA) transcript is antisense to cancer-associated gene PMEPA1. F1000Res 4: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra JM, Ballingall KT. 2014. Non-human lnc-DC orthologs encode Wdnm1-like protein. F1000Res 3: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doss JF, Corcoran DL, Jima DD, Telen MJ, Dave SS, Chi JT. 2015. A comprehensive joint analysis of the long and short RNA transcriptomes of human erythrocytes. BMC Genomics 16: 952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RA, Almada AE, Zamudio JR, Sharp PA. 2011. Antisense RNA polymerase II divergent transcripts are P-TEFb dependent and substrates for the RNA exosome. Proc Natl Acad Sci 108: 10460–10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. 2013. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell 152: 743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. 2010. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 28: 503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. 2013. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 154: 240–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Yuan B, Flygare J, Lodish HF. 2011. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev 25: 2573–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Tang Q, Sharma S, Yu F, Escobar TM, Muljo SA, Zhu J, Zhao K. 2013. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol 14: 1190–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Thomas B, Flynn RA, Gavzy SJ, Wu L, Kim SV, Hall JA, Miraldi ER, Ng CP, Rigo FW, et al. 2015. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature 528: 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Huang Z, Luo Q, Yao F, Qing C, Ye J, Deng Y, Li J. 2016. Identification of differentially expressed long non-coding RNAs in polarized macrophages. Sci Rep 6: 19705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilott NE, Heward JA, Roux B, Tsitsiou E, Fenwick PS, Lenzi L, Goodhead I, Hertz-Fowler C, Heger A, Hall N, et al. 2014. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat Commun 5: 3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam H, Shahr Bano A, Patel P, Holla P, Jameel S. 2015. The lncRNA NRON modulates HIV-1 replication in a NFAT-dependent manner and is differentially regulated by early and late viral proteins. Sci Rep 5: 8639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, Kato A, Kawaguchi Y, Sato H, Yoneda M, et al. 2014. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell 53: 393–406. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. 2012. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc 7: 1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJS, Jackson SE, Wills MR, Weissman JS. 2014. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep 8: 1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et al. 2015. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 47: 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josset L, Tchitchek N, Gralinski LE, Ferris MT, Eisfeld AJ, Green RR, Thomas MJ, Tisoncik-Go J, Schroth GP, Kawaoka Y, et al. 2014. Annotation of long non-coding RNAs expressed in collaborative cross founder mice in response to respiratory virus infection reveals a new class of interferon-stimulated transcripts. RNA Biol 11: 875–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambara H, Niazi F, Kostadinova L, Moonka DK, Siegel CT, Post AB, Carnero E, Barriocanal M, Anthony DD, Valadkhan S. 2014. Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucleic Acids Res 42: 10668–10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambara H, Gunawardane L, Zebrowshi E, Valadkhan S. 2015. Regulation of interferon-stimulated gene BST2 by a lncRNA transcribed from a shared bidirectional promoter. Front Immunol 5: 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM. 2015. Enhancer RNAs: A class of long noncoding RNAs synthesized at enhancers. Cold Spring Harb Perspect Biol 7: a018622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EZ, Wespiser AR, Caffrey DR. 2016. The domain structure and distribution of Alu elements in long noncoding RNAs and mRNAs. RNA 22: 254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzin JJ, Spencer SP, McCright SJ, Kumar DBU, Collet MA, Mowel WK, Elliott EN, Uyar A, Makiya MA, Dunagin MC, et al. 2016. The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature 537: 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk M, Emerson BM. 2014. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-κB complexes. eLife 3: e01776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Westra H-J, Karjalainen J, Zhernakova DV, Esko T, Hrdlickova B, Almeida R, Zhernakova A, Reinmaa E, Võsa U, et al. 2013. Human disease-associated genetic variation impacts large intergenic non-coding RNA expression. PLoS Genet 9: e1003201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MTY, Li W, Rosenfeld MG, Glass CK. 2014. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci 39: 170–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence T, Natoli G. 2011. Transcriptional regulation of macrophage polarization: Enabling diversity with identity. Nat Rev Immunol 11: 750–761. [DOI] [PubMed] [Google Scholar]
- Lee JT, Zhao J, Sarma K, Borowsky M, Ohsumi TK. 2016. Polycomb-associated non-coding RNAs. U.S. Patent 9,328,346 filed November 12, 2011, and issued May 3, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessing D, Anguera MC, Lee JT. 2013. X chromosome inactivation and epigenetic responses to cellular reprogramming. Annu Rev Genomics Hum Genet 14: 85–110. [DOI] [PubMed] [Google Scholar]
- Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, Head SR, Burns JC, Rana TM. 2014. The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc Natl Acad Sci 111: 1002–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Hao Y, Zhang D, Fu R, Liu W, Zhang X, Xue F, Yang R. 2016. Aberrant expression of long noncoding RNA TMEVPG1 in patients with primary immune thrombocytopenia. Autoimmunity 49: 496–502. [DOI] [PubMed] [Google Scholar]
- Lin W-W, Karin M. 2007. A cytokine-mediated link between innate immunity, inflammation, and cancer. J Clin Invest 117: 1175–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, Kellis M. 2011. PhyloCSF: A comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27: i275–i282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsky PE, van der Heijde DMFM, St Claire W, Furst DE, Breedveld FC, Kalden JR, Smolen JS, Weisman M, Emery P, Feldmann M, et al. 2000. Infliximab and methotrexate in the treatment of rheumatoid arthritis. Anti-tumor necrosis factor trial in rheumatoid arthritis with concomitant therapy study group. N Engl J Med 343: 1594–1602. [DOI] [PubMed] [Google Scholar]
- Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L, Yao H, Su F, Li D, et al. 2015. A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 27: 370–381. [DOI] [PubMed] [Google Scholar]
- Martinez FO, Gordon S. 2014. The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000Prime Rep 6: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, et al. 2015. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521: 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Ward AJ, Chun S, Bennett CF, Beaudet AL, Rigo F. 2015. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature 518: 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreland LW, Baumgartner SW, Schiff MH, Tindall EA, Fleischmann RM, Weaver AL, Ettlinger RE, Cohen S, Koopman WJ, Mohler K, et al. 1997. Treatment of rheumatoid arthritis with a recombinant human tumor necrosis factor receptor (p75)-Fc fusion protein. N Engl J Med 337: 141–147. [DOI] [PubMed] [Google Scholar]
- Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, et al. 2014. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 41: 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BR, Makarewich CA, Anderson DM, Winders BR, Troupes CD, Wu F, Reese AL, McAnally JR, Chen X, Kavalali ET, et al. 2016. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351: 271–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. 2008. Hematopoiesis: An evolving paradigm for stem cell biology. Cell 132: 631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang J, Zhu X, Chen Y, Wei H, Chen Q, Chi X, Qi B, Zhang L, Zhao Y, Gao GF, et al. 2014. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe 16: 616–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padua DM, Mahurkar-Joshi S, Law IKM, Polytarchou C, Vu JP, Pisegna JR, Shih DQ, Iliopoulos D, Pothoulakis C. 2016. A long noncoding RNA signature for ulcerative colitis identifies IFNG-AS1 as an enhancer of inflammation. Am J Physiol Gastrointest Liver Physiol 311: G446–G457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang KC, Dinger ME, Mercer TR, Malquori L, Grimmond SM, Chen W, Mattick JS. 2009. Genome-wide identification of long noncoding RNAs in CD8+ T cells. J Immunol 182: 7738–7748. [DOI] [PubMed] [Google Scholar]
- Peng X, Gralinski L, Armour CD, Ferris MT, Thomas MJ, Proll S, Bradel-Tretheway BG, Korth MJ, Castle JC, Biery MC, et al. 2010. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. MBio 1: e00206–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E. 2007. Breakthrough of the year. Human genetic variation. Science 318: 1842–1843. [DOI] [PubMed] [Google Scholar]
- Plosker GL. 2011. Interferon-β-1b: A review of its use in multiple sclerosis. CNS Drugs 25: 67–88. [DOI] [PubMed] [Google Scholar]
- Qiao YQ, Huang ML, Xu AT, Zhao D, Ran ZH, Shen J. 2013. LncRNA DQ786243 affects Treg related CREB and Foxp3 expression in Crohn’s disease. J Biomed Sci 20: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge R-M, Chang HY. 2013. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. eLife 2: e00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid F, Shah A, Shan G. 2016. Long non-coding RNAs in the cytoplasm. Genomics Proteomics Bioinformatics 14: 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricaño-Ponce I, Wijmenga C. 2013. Mapping of immune-mediated disease genes. Annu Rev Genomics Hum Genet 14: 325–353. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. 2012. Genome regulation by long noncoding RNAs. Annu Rev Biochem 81: 145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. 2008. Divergent transcription from active promoters. Science 322: 1849–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortman K, Liu YJ. 2002. Mouse and human dendritic cell subtypes. Nat Rev Immunol 2: 151–161. [DOI] [PubMed] [Google Scholar]
- Sonkoly E, Bata-Csorgo Z, Pivarcsi A, Polyanka H, Kenderessy-Szabo A, Molnar G, Szentpali K, Bari L, Megyeri K, Mandi Y, et al. 2005. Identification and characterization of a novel, psoriasis susceptibility-related noncoding RNA gene, PRINS. J Biol Chem 280: 24159–24167. [DOI] [PubMed] [Google Scholar]
- Spurlock CF, Tossberg JT, Guo Y, Collier SP, Crooke PS, Aune TM. 2015. Expression and functions of long noncoding RNAs during human T helper cell differentiation. Nat Commun 6: 6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szegedi K, Sonkoly E, Nagy N, Németh IB, Bata-Csorgo Z, Kemeny L, Dobozy A, Szell M. 2010. The anti-apoptotic protein G1P3 is overexpressed in psoriasis and regulated by the non-coding RNA, PRINS. Exp Dermatol 19: 269–278. [DOI] [PubMed] [Google Scholar]
- Tong Q, Gong AY, Zhang XT, Lin C, Ma S, Chen J, Hu G, Chen XM. 2016. LincRNA-Cox2 modulates TNF-α-induced transcription of Il12b gene in intestinal epithelial cells through regulation of Mi-2/NuRD-mediated epigenetic histone modifications. FASEB J 30: 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitsiou E, Williams AE, Moschos SA, Patel K, Rossios C, Jiang X, Adams OD, Macedo P, Booton R, Gibeon D, et al. 2012. Transcriptome analysis shows activation of circulating CD8+ T cells in patients with severe asthma. J Allergy Clin Immunol 129: 95–103. [DOI] [PubMed] [Google Scholar]
- Vilcek J, Feldmann M. 2004. Historical review: Cytokines as therapeutics and targets of therapeutics. Trends Pharmacol Sci 25: 201–209. [DOI] [PubMed] [Google Scholar]
- Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X. 2014. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 344: 310–313. [DOI] [PubMed] [Google Scholar]
- Wang P, Zhi H, Zhang Y, Liu Y, Zhang J, Gao Y, Guo M, Ning S, Li X. 2015a. miRSponge: A manually curated database for experimentally supported miRNA sponges and ceRNAs. Database (Oxford) 2015: bav098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhong H, Xie X, Chen CY, Huang D, Shen L, Zhang H, Chen ZW, Zeng G. 2015b. Long noncoding RNA derived from CD244 signaling epigenetically controls CD8+ T-cell immune responses in tuberculosis infection. Proc Natl Acad Sci 112: E3883–E3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Peng H, Tian J, Ma J, Tang X, Rui K, Tian X, Wang Y, Chen J, Lu L, et al. 2016a. Upregulation of long noncoding RNA TMEVPG1 enhances T helper type 1 cell response in patients with Sjögren syndrome. Immunol Res 64: 489–496. [DOI] [PubMed] [Google Scholar]
- Wang J, Syrett CM, Kramer MC, Basu A, Atchison ML, Anguera MC. 2016b. Unusual maintenance of X chromosome inactivation predisposes female lymphocytes for increased expression from the inactive X. Proc Natl Acad Sci 113: E2029–E2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S, Kellis M, Garber M. 2014. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res 24: 616–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Smas CM. 2008. Wdnm1-like, a new adipokine with a role in MMP-2 activation. Am J Physiol Endocrinol Metab 295: E205–E215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia F, Dong F, Yang Y, Huang A, Chen S, Sun D, Xiong S, Zhang J. 2014. Dynamic transcription of long non-coding RNA genes during CD4+ T cell development and activation. PLoS ONE 9: e101588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Guan D, Fan Q, Su J, Zheng W, Ma W, Ke C. 2013. lncRNA expression signatures in response to enterovirus 71 infection. Biochem Biophys Res Commun 430: 629–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wu L, Qian J, Qu B, Xia S, La T, Wu Y, Ma J, Zeng J, Guo Q, et al. 2016. Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. J Autoimmun 75: 96–104. [DOI] [PubMed] [Google Scholar]