Abstract
Staphylococcus aureus, a major cause of bovine mastitis, produces a wide range of immune-evasion molecules. The bi-component leukocidin LukMF’ is a potent killer of bovine neutrophils in vitro. Since the role of LukMF’ in development of bovine mastitis has not been studied in natural infections, we aimed to clarify whether presence of the lukM-lukF’ genes and production levels of LukMF’ are associated with clinical severity of the disease. Staphylococcus aureus was isolated from mastitis milk samples (38 clinical and 17 subclinical cases) from 33 different farms. The lukM-lukF’ genes were present in 96% of the isolates. Remarkably, 22% of the lukM-lukF’-positive S. aureus isolates displayed a 10-fold higher in vitro LukMF’ production than the average of the lower-producing ones. These high producing isolates were cultured significantly more frequently from clinical than subclinical mastitis cases. Also, the detection of LukM protein in milk samples was significantly associated with clinical mastitis and high production in vitro. The high producing LukMF’ strains all belonged to the same genetic lineage, spa-type t543. Analysis of their global toxin gene regulators revealed a point mutation in the Repressor of toxins (rot) gene which results in a non-functional start codon, preventing translation of rot. This mutation was only identified in high LukMF’ producing isolates and not in low LukMF’ producing isolates. Since rot suppresses the expression of various toxins including leukocidins, this mutation is a possible explanation for increased LukMF’ production. Identification of high LukMF’ producing strains is of clinical relevance and can potentially be used as a prognostic marker for severity of mastitis.
Keywords: Staphylococcus aureus, bovine mastitis, clinical severity, LukMF’, repressor of toxins, phage encoded leukocidin
1. Introduction
Mastitis—inflammation of the mammary gland—is a major cause of economic losses in the dairy industry associated with costs of treatment, reduced milk yield, discarded milk and premature culling of animals [1,2]. In addition, mastitis has severe impact on animal welfare [3]. Staphylococcus aureus is one of the major causes of mastitis in cows [4]. These infections are mostly subclinical and often chronic, but may also result in clinical mastitis [5]. Staphylococcus aureus possesses many virulence factors, some of which enable it to manipulate the innate and adaptive immune responses of the host [6]. These virulence factors vary widely between lineages [7] and include immunomodulatory proteins [8], proteases [9], factors that impede phagocytosis [10] and cytotoxins [11]. Some virulence factors are phage-encoded which allows horizontal transfer of virulence genes between bacteria, resulting in genetic diversity between S. aureus lineages [12]. An important group of S. aureus immune evasion molecules is that of the leukocidins: pore-forming, bi-component toxins that specifically target immune cells [13]. So far, seven different leukocidins have been described [13,14], of these, the phage-encoded leukocidins Panton-Valentine leukocidin (PVL), LukPQ, and LukMF’ most strongly affect immune cells from a limited host species range [6,13,14]. LukMF’ is almost exclusively present on prophages carried by S. aureus strains of ruminant origin [15,16,17] and it is a potent killer of bovine neutrophils, macrophages and monocytes, but not of human neutrophils [18,19]. LukM binds to the CCR1 receptor, which is highly present on bovine neutrophils, but absent on human neutrophils [19]. Since neutrophils are key players in initial immune responsiveness during inflammation of the mammary gland [20], it is expected that potent killing of neutrophils by LukMF’ reduces its effectiveness and, therefore, influences the clinical outcome of infection. Indeed, experimental intramammary challenge with high LukMF’ producing S. aureus strains resulted in more severe mastitis compared to challenge with intermediate producing S. aureus strains [21]. Also, high LukM levels in milk have been associated with clinical mastitis [21]. It is, however, unclear whether differences in LukMF’ production are of clinical significance in natural infections.
In this study, we investigated the impact of LukMF’ on clinical severity of bovine mastitis under field conditions. We identified a genetic lineage of S. aureus with increased LukMF’ production that is associated with clinical rather than subclinical mastitis. In this lineage, we found a nonsense mutation in the start codon of the global expression regulator Repressor of toxins (rot). This mutation is the likely cause of increased toxin production associated with severity of clinical signs of mastitis.
2. Results
2.1. Prevalence of lukM-lukF’ Genes among Bovine Mastitis Isolates
Fifty-five S. aureus isolates were collected at 33 Dutch dairy farms by bacteriological culture of milk samples from clinical (n = 37) and subclinical (n = 18) cases of bovine mastitis and the presence of the femA, lukM, and lukF’ genes was detected using PCR. All isolates were positive for the S. aureus-specific gene femA, confirming that the selected bacteria were S. aureus, and 53/55 (96%) of isolates were positive for lukM and lukF’. The proportion of lukM-lukF’ positive S. aureus was similar for clinical (37/38, 97%) and subclinical (16/17, 94%) mastitis cases.
2.2. Production of LukM In Vitro and In Vivo
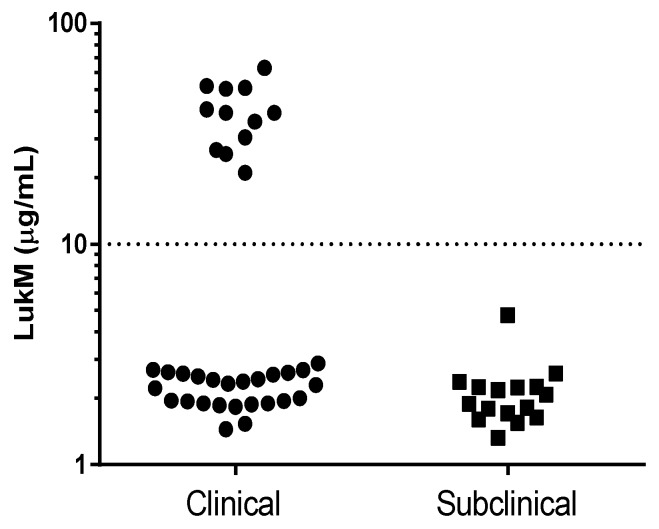
The in vitro LukMF’ production potential was investigated by growing S. aureus isolates for eight hours under controlled conditions and measuring the LukM concentration in supernatant using ELISA. All lukM-lukF’ positive S. aureus isolates produced LukM in vitro, with levels ranging from 1.5 to 60 µg/mL. The production levels had a clear bimodal distribution (Figure 1). Based on this distribution, isolates were categorized into two groups: LukMF’ high producers (>10 µg/mL) and LukMF’ low producers (<10 µg/mL). Most S. aureus (n = 41) were LukMF’ low producers (mean = 2.2 µg/mL LukM, SD = 0.6). The remaining isolates (n = 12) were LukMF’ high producers (mean = 39.6 µg/mL LukM, SD = 12.6) and were significantly more often isolated from cases of clinical mastitis (Fisher’s exact test, p = 0.011). Although some of the 55 S. aureus isolates originated from the same farm, the 12 high producing isolates were cultured from cows on 12 different farms. To compensate for a possible farm effect, we randomly selected one isolate per farm (n = 33) and found the same association between the LukMF’ high producing isolates and clinical mastitis (Fisher’s exact test, p = 0.033).
Figure 1.
LukM levels in vitro after eight hours of culture of 53 lukM-lukF’ positive S. aureus isolates from milk of cows with clinical or subclinical mastitis, measured by ELISA.
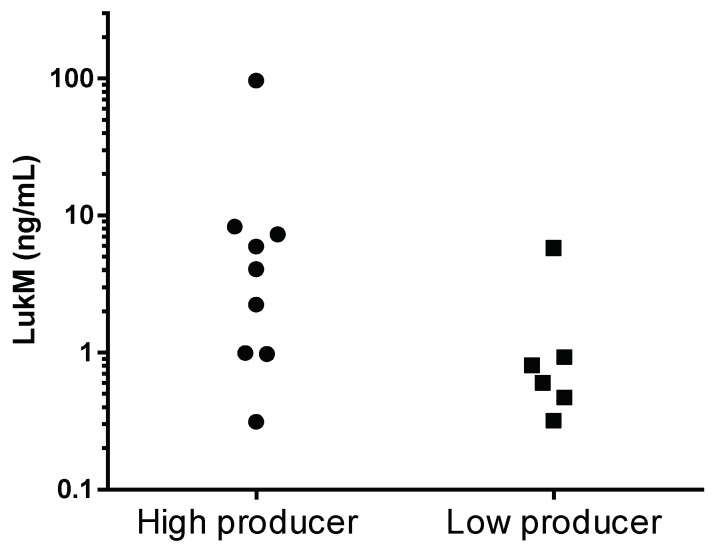
Next, in vivo LukMF’ production during mastitis was assessed by measuring LukM in the milk samples corresponding to each of the S. aureus isolates. Eight milk samples (seven clinical and one subclinical) could not be tested for LukM because the milk was clotted and therefore not fit for use in the ELISA, or because insufficient milk was available. LukM was detected in 15/45 milk samples, with concentrations ranging from 0.31 ng/mL to 96 ng/mL, significantly more often in milk from clinical mastitis cases (13/29 = 45%) than from subclinical cases (2/16 = 12%, Fisher’s exact test, p = 0.046) and in samples that yielded LukMF’ high producing isolates (9/11 = 82%) compared to samples that yielded LukMF’ low producing isolates (6/34 = 18%, Fisher’s exact test, p < 0.001). After again selecting one isolate per farm, the association between high in vitro production and LukM in milk, ex vivo, remained (Fisher’s exact test, p = 0.007), but the association between LukM in milk and clinical severity was no longer statistically significant (Fisher’s exact test, p = 0.22). The average LukM concentration in milk of cows that hosted LukMF’ high producing isolates (mean = 14.2 ng/mL LukM, SD = 31.1) was higher than of those carrying LukMF’ low producing isolates (mean = 1.49 ng/mL LukM, SD = 2.12) (Mann–Whitney test, p = 0.049) (Figure 2).
Figure 2.
LukM concentration in milk samples of mastitis cases (in vivo) caused by LukMF’ high (>10 µg/mL production in vitro after eight hours of culture) and low (<10 µg/mL production in vitro after eight hours of culture) producing S. aureus (p = 0.049, Mann–Whitney test).
2.3. Genotyping of S. aureus Isolates
All isolates were spa-typed to investigate whether differences in LukMF’ production associated with certain genetic lineages. Table 1 gives the spa-types identified in our isolate collection, and shows that all high producing S. aureus isolates belonged to spa-type t543. Whole genome sequencing was performed on a subset of seven isolates, selected based on differences in LukM production levels, spa-type and farm. Sequencing results showed that all three spa-type t543 strains belonged to multilocus sequence type (ST) 479, which belongs to clonal complex (CC) 479. The three spa-type t529 isolates all belonged to CC151, with two isolates identified as ST151 and one as ST504, a double-locus variant (DLV) of ST151. The single spa-type t1403 strain belonged to ST133 (CC133).
Table 1.
Spa-types and in vitro LukMF’ production of field isolates from bovine clinical and subclinical mastitis cases.
| Spa-Type | Isolates N | Clinical Mastitis N (%) | lukM-lukF’ Postive Isolates N (%) 1 | High LukMF’ Production Isolates N (%) 2 | Clonal Complex 3 |
|---|---|---|---|---|---|
| t529 | 40 | 23 (58) | 40 (100) | 0 (0) | CC151 |
| t543 | 12 | 12 (100) | 12 (100) | 12 (100) | CC479 |
| t524 | 1 | 1 (100) | 0 (0) | 0 (0) | ND 4 |
| t1403 | 1 | 1 (100) | 1 (100) | 0 (0) | CC133 |
| t015 | 1 | 0 (0) | 0 (0) | 0 (0) | ND 4 |
| Total | 55 | 37 (67) | 53 (96) | 12 (22) |
1 Number of lukM-lukF’ positive isolates. Genes detected by PCR. 2 Number of samples with >10 µg/mL LukMF’ production after eight hours culture, measured in supernatant by ELISA. 3 Clonal complex determined by MLST based on whole genome sequences of subset of isolates from this spa-type. 4 Not determined.
2.4. Analysis of the lukM-lukF’ Operon and saeS, saeR, rot Genes
To identify genetic factors that were associated with the LukMF’ production phenotype, we compared whole genome sequences of three high producing and four low producing S. aureus isolates. First, the prophages containing lukM-lukF’ were identified using PHAST [22]. Prophages encoding lukM-lukF’ were indeed present in all sequenced isolates, and were identified by PHAST to be most similar to reference phage phiPV83 (Genbank accession NC_002486.1). Prophages from CC479 and CC133 isolates were very similar to each other in size and gene content, but both differed from the smaller CC151 prophage. Variations were observed in lukM (four synonymous and five non-synonymous single nucleotide polymorphisms (SNPs)) and lukF’ (three synonymous and six non-synonymous SNPs) among the different CCs (Supplementary Figure S1a,b). The putative promotor region of the lukM-lukF’ operon (up to 200 bp upstream from the start codon) contained five SNPs, but none were exclusively present in high producing CC479 strains (Supplementary Figure S1c).
Next, we examined genes putatively involved in the regulation of lukM expression. Genes involved in LukMF’ expression are unknown, but analogous to the regulation of the expression of PVL, we investigated Repressor of toxins (rot) and the exoprotein two-component system SaeRS (saeS and saeR) [23,24]. These genes were present in all sequenced strains and no variation was found in the saeR gene, whereas a single non-synonymous SNP (not associated with CC479) was found in the saeS gene (data not shown). Two non-synonymous SNPs (position 2 and 452) were found in the rot gene, exclusively present in the CC479 isolates. The SNP at position 2 renders the rot gene non-functional due to the loss of a start codon. To corroborate these findings, the region surrounding the start codon of rot was identified in five additional S. aureus isolates (three LukMF’ high producing isolates and two LukMF’ low producing isolates) using PCR and sequencing. In these additional isolates, the mutation in the rot start codon was also only present in the high producing S. aureus isolates (Supplementary Figure S1d).
3. Discussion
We observed a high lukM-lukF’ carriage (96%) among S. aureus field isolates cultured both from cases of clinical and subclinical mastitis. A subpopulation of S. aureus, spa-type t543-ST479, with a very high in vitro LukMF’ production was significantly associated with clinical rather than subclinical mastitis. A mutation in the rot gene, leading to loss of the primary start codon, found exclusively in spa-type t543-ST479 isolates, may be functionally linked to the increased LukMF’ production.
All sequence types and spa-types identified in this study have previously been associated with bovine mastitis [25,26,27,28,29]. Several authors have reported about sequence types and lukM-lukF’ carriage of S. aureus, and this reveals that it is strongly associated with specific ruminant-associated clonal complexes, namely CC151, CC133, CC705, and CC479 [16,30,31,32,33,34] (see supplementary Table S1 for a detailed overview). Within the bovine associated CC97, only one specific lineage (ST352-CC97) showed a high prevalence of lukM-lukF’ [30,31]. This demonstrates that the carriage of the lukM-lukF’ harboring (pro)phage [17] is lineage specific, since the genes are only present in certain sequence types and absent in others within the same CC.
LukMF’ is a potent killer of bovine neutrophils, which play a critical role in the innate immune defense against S. aureus [5,19]. We hypothesize that killing of neutrophils by LukMF’ reduces the overall phagocytic activity in the mammary gland, resulting in survival of S. aureus, and hence pro-inflammatory responsiveness increasing the clinical severity of mastitis. In our study, LukMF’ high production in vitro by ST479 S. aureus is indeed associated with clinical rather than subclinical mastitis. High production was also strongly correlated with substantial levels of LukM present in milk in vivo. Still, also the presence of LukMF’ low producing S. aureus in milk may lead to detectable levels. Although differences in bacterial load of LukMF’ producing bacteria in the mammary gland could also explain differences in milk LukM levels, we assume that ST479 are also LukMF’ high producers in vivo during the course of infection. In a recent study, cattle were challenged intramammary with LukMF’ high and low producing S. aureus strains [21]. Quarters challenged with the high producer developed more severe clinical symptoms and higher bacterial loads compared to the other, low producing, strains [21].
Although, to our knowledge, the regulation of LukMF’ expression has not been studied, it is plausible to assume that the regulation system of LukMF’ expression is similar to that of other leukocidins, which are controlled by global gene regulators Agr, Rot and SaeRS [13]. We observed a mutation in the rot start codon that was strongly associated with high LukMF’ production. Rot is a global regulator of S. aureus virulence gene expression and can directly bind to the promotor region of various toxin genes, such as hla, hlgC-hlgB, lukE-lukD, and lukA-lukB [23,35]. Mutations in rot can both activate or repress gene expression, depending on the site of the mutation and the target gene [35]. The expression of leukocidins (LukAB, LukED, PVL) by S. aureus increases when the rot gene is made inoperative [23,36]. The hypervirulence of the community-associated methicillin-resistant S. aureus (CA-MRSA) clone USA500 is believed to be a result of increased leukocidin (LukAB, LukED, hlgCB) production compared to other CA-MRSA, induced by an insertion in the promotor region of rot that prevents expression of this gene [36]. The LukMF’ high producing strain used in a previous study [21] also contained the same mutation in rot as the one identified in our work. This demonstrates that the absence of, or impaired or altered function of rot likely explains the increased LukMF’ production of ST479 strains observed in our study. To further substantiate this, ST479 could be complemented with a functional rot copy which should lead to decreased LukMF’ production compared to the WT ST479. Likewise, rot knockout strains of ST151 or ST133 are expected to produce higher amounts of LukMF’ compared to the WT. These strains could also be used to identify how this mutation in rot affects the regulation of other leukocidins. Because of the limited geographic range from which our samples originated, it is unclear how prevalent the mutation in rot is in CC479 isolates cultured from cattle in other countries. Still, the strain S1444 [21], a high producing CC479 isolate was originally cultured from a German sample, suggesting that the mutation is not restricted to the region of this study.
Mastitis milk samples used in our study as source of S. aureus isolates were submitted by farmers, sometimes in consultation with their veterinarian, hence not randomly collected and not likely to be representative for the population in the field. Due to that approach, under- or overestimation of the actual proportions of LukMF’ high producing isolates in the population as well as of isolates belonging to the various spa-types cannot be excluded. The association between high LukMF’ production and clinical versus subclinical mastitisis, however, not likely to be affected by this sampling bias, but the clinical importance of this association in terms of the population attributable fraction depends on the prevalence of LukMF’ high producing isolates in the field and cannot be calculated from our data.
Since the rot gene is part of the core genome of S. aureus, transfer of the ability of ST479 to produce high levels of LukMF’ seems unlikely. It is unclear if increased production of LukMF’ by ST479 also increases the transmission of this lineage in the population, as a severe clinical infection is expected to result in a quicker death or culling from the herd of the host or quicker treatment with antibiotics, reducing the chances to further spread in the population. In previous studies, ST479 made up 26% of Dutch [28] and 17% of German S. aureus mastitis isolates [16], suggesting that this lineage can persist within a population despite its association with clinical mastitis.
Screening tools, like loop-mediated isothermal amplification (LAMP) [37], currently exist that allow for quick identification of mastitis pathogens [38]. However, these tools mostly identify the pathogen on a species level. Since our research shows that the sequence type of S. aureus is strongly associated to the type of mastitis, sequence typing of mastitis isolates would enable farmers to implement specific interventions in case of infection with the high LukMF’ producing lineages.
4. Materials and Methods
4.1. Collection of S. aureus Bovine Mastitis Isolates
Quarter milk samples from cows with subclinical or clinical mastitis were aseptically collected by farmers belonging to the University Farm Animal Practice (Harmelen, the Netherlands) and sent in for bacteriological culture and species identification, which were performed according to National Mastitis Council protocols [39]. As participating farms (n = 33) belonged to the same veterinary practice, the farms were geographically clustered around the Utrecht region in the Netherlands. Samples were collected between April 2014 and December 2015. Clinical or subclinical mastitis was diagnosed by the farmer, with clinical mastitis defined as visibly abnormal appearance of the udder, the milk or both. Subclinical mastitis was characterized by absence of clinical signs and generally were animals with a high somatic cell count. A total of 55 S. aureus positive milk samples from cases of bovine mastitis (38 clinical and 17 subclinical cases) were used for this study and stored at −18 °C before further use. After thawing of the milk samples, 200 µL aliquots of the S. aureus positive milk samples were plated on sheep blood agar plates and cultured overnight at 37 °C to isolate fresh bacterial colonies. Of milk samples showing signs of a clinical mastitis (clots, flakes, discolored milk), a smaller volume of 50 µL was used to prevent bacterial overgrowth of the plate. Single colonies were picked and added to 2 mL T1438 Todd Hewitt Broth (THB) (Sigma, St. Louis, MO, USA) and incubated overnight at 37 °C with agitation. Bacterial glycerol stocks (25% glycerol) were made by adding 0.5 mL of bacterial broth to 0.5 mL 50% glycerol solution in distilled water. Stocks were stored at −80 °C before use in further experiments.
4.2. DNA Extraction and Amplification of lukM, lukF’, femA and rot Genes
Bacterial isolates were plated from glycerol stocks on blood agar plates and cultured overnight at 37 °C. Single colonies were picked for DNA extraction and washed in 1 mL distilled water. After centrifugation (17,000× g for 1 min), bacteria were resuspended in 200 µL distilled water and heated at 100 °C for 10 min, centrifuged at 17,000× g for 1 min, and diluted 1:10 in distilled water and stored at −20 °C.
Primers to amplify femA, lukM, lukF’ and rot were designed or taken from literature (Table 2). The primers for femA and lukF’ were used together in a duplex PCR, and the other primers (rot, lukM) in separate, single PCR. The reaction was performed in a total volume of 25 µL containing 10 µL 1:10 diluted boiled DNA sample, 5 µL GoTaQ Green buffer 5× (Promega, Madison, WI, USA), 1.5 mM MgCl2 (Promega), 0.2 mM dNTPs (Promega, Madison, WI, USA), 0.4 µM of the lukF’ primer pair and 0.6 µM of the other primer pairs (Invitrogen, Carlsbad, CA, USA) and 0.625 U of GoTaQ DNA polymerase (Promega, Madison, WI, USA). After an initial denaturation step at 95 °C for 2 min, 35 cycles (30 s at 95 °C, 35 s at various annealing temperatures (Table 2, column 4) and 35–60 s at 72 °C) were performed in a T100 Thermal Cycler (Bio-Rad, Hercules, CA, USA). Electrophoresis on 1.5% agarose gel was used to visualize PCR products.
Table 2.
Primers and annealing temperature used in this study.
| Gene | Sequence | Product Size (bp) | Annealing Temperature (°C) | Reference |
|---|---|---|---|---|
| femA | f: 5′-tgcctttacagatagcatgcca-3′ | 142 | 59.5 | [40] |
| r: 5′-agtaagtaagcaagctgcaatgacc-3′ | ||||
| lukM | f: 5′-aaacgcgcagttaataaaaag-3′ | 975 | 55 | This study |
| r: 5′-agcattaggtcctcttgtcg-3′ | ||||
| lukF’ | f: 5′-actcaggctatacccaaccca-3′ | 472 | 59.5 | This study |
| r: 5′-cgagctactctgtctgccac-3′ | ||||
| rot | f: 5′-accaatttagcctcattcggtttg-3′ | 705 | 55 | This study |
| r: 5′-catcgtcaacaggacgctct-3′ |
4.3. In Vitro and In Vivo LukM Production
Bacteria were cultured from glycerol stock on blood agar plates overnight at 37 °C. Single colonies were picked and added to 1.5 mL THB. Bacteria were incubated with agitation for 30 min at 37 °C. Optical density (OD) at 600 nm was measured and samples were diluted to an OD of 0.01 in 2.5 mL of THB. Next, bacteria were incubated with agitation for 8 h at 37 °C. After incubation, samples were centrifuged (4000× g for 10 min) and supernatant was collected. The supernatant was sterilized using a microfilter (0.20 µm; Corning Incorporated, Corning, NY, USA) and stored at −20 °C before use in further experiments. Bacterial supernatants were produced in triplicate using separate, single colonies from the same cultured plate.
LukM in supernatant and bovine mastitis milk samples was measured by ELISA, according to the method described by Vrieling et al. [21]. In short, LukM is captured using LukM specific polyclonal bovine IgG isolated from the colostrum of a cow with high LukM antibody titers, and captured LukM is detected using the LukM specific monoclonal antibody LM43.F8 [21]. Milk samples were heated to 95 °C for 10 min to prevent interference from antibodies in the milk.
4.4. Genotyping of Mastitis Strains and Genomic Analyses
The polymorphic X-region of the Staphylococcal Protein A (spa) gene of all S. aureus isolates was amplified according to the Ridom StaphType standard protocol (www.ridom.org). PCR amplicons were purified using ExoSAP-IT PCR Cleanup Reagent (Affymetrix, Santa Clara, CA, USA) according to manufacturer’s instructions and sequenced using Sanger sequencing (Baseclear, Leiden, The Netherlands). BioNumerics v7.5 software (Applied Maths, Sint-Martens-Latem, Belgium) was used to analyze sequence data and to assign spa-types.
Whole genome sequencing was performed on seven isolates, selected from LukMF’ high and low producers of different spa-type, and each selected isolate originated from a different farm. DNA was isolated with the Ultra Clean Microbial DNA isolation kit (Mo-Bio, Carlsbad, CA, USA). MiSeq sequencing (Illumina, San Diego, CA, USA) was performed at the Utrecht Sequencing Facility (UMC Utrecht, the Hubrecht institute and Utrecht University, the Netherlands), using 300 bp paired end reads. Reads were assembled into a scaffold genome using SPAdes v3.1.1. [41]. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession QFCT00000000-QFCZ00000000. The version described in this paper is the first version. MLST was determined using the MLST tool of the Center of Genomic Epidemiology (accessible on https://cge.cbs.dtu.dk/services/MLST/) [42]. Prophages containing the LukM-lukF’ genes were identified using PHAST (accessible on http://phast.wishartlab.com/index.html) [22]. The lukM-lukF’ encoding gene sequences and putative promotor region (200 bp upstream of start LukM gene) were identified by BLASTN using reference sequences for lukM (GenBank accession: 1262967) and lukF’ (GenBank accession: 1262954). Sequences for candidate LukMF’ regulator genes were extracted from available genomes using reference gene sequences of rot (Genbank accession: AF189239.2) and saeRS locus (Genbank accession: AF129010.1). Nucleic acid sequences were translated to their corresponding protein sequences using EMBOSS Transeg software (accessible on http://www.ebi.ac.uk/Tools/st/emboss_transeq/). Gene and protein sequences were aligned using MegALIGN Pro software (DNAstar Incorporate, Madison, WI, USA).
4.5. Statistical Analysis
Statistical analysis was performed using GraphPad Prism 7 software (GraphPad Software, La Jolla, CA, USA). In vitro differences in LukM levels between clinical/subclinical isolates and in vivo LukM levels between LukMF’ high/low producers were compared using the Mann–Whitney U test. The associations between in vitro LukMF’ production levels, presence of detectable LukM in milk and mastitis type were tested using Fisher’s exact test. A subset of the dataset with a single sample per farm was assembled using the random number generator function in Microsoft Excel (Microsoft Corporation, Redmond, WA, USA).
Supplementary Materials
The following are available online at http://www.mdpi.com/2072-6651/10/5/200/s1, Figure S1: (a) Alignment of lukM of seven S. aureus isolates obtained from cases of bovine mastitis; (b) Alignment of lukF’ of seven S. aureus isolates obtained from cases of bovine mastitis; (c) Alignment of the putative promotor region of lukM-lukF’ operon of seven S. aureus isolates obtained from cases of bovine mastitis; (d) Alignment of the first 20 nucleotides of rot in 12 S. aureus isolates obtained from cases of bovine mastitis. Table S1: Number of S. aureus of different lineages found among bovine isolates, with percentage of lukM-lukF’ positive S. aureus among these lineages.
Author Contributions
Conceptualization, L.B., G.K., J.H. and T.v.W.; Methodology, J.H., G.K., L.B., V.R. and B.D.; Validation, J.H. and L.B.; Formal Analysis, J.H. and G.K.; Investigation, J.H., M.S., T.v.W. and L.S.; Resources, T.v.W. and B.D.; Data Curation, J.H. and B.D.; Writing-Original Draft Preparation, J.H.; Writing-Review & Editing, G.K., L.B., V.R. and B.D.; Visualization, J.H.; Supervision, G.K. and L.B.; Project Administration, L.B. and G.K.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Key Contribution
A lineage of Staphylococcus aureus cultured from intramammary infections in cows was found to produce the bovine-associated leukocidin LukMF’ at substantially higher levels than other lineages, possibly explained by a nonsense mutation in one of the toxin-regulation genes. This high LukMF’ producing lineage was significantly more often cultured from clinical than from subclinical mastitis cases in the field.
References
- 1.Hogeveen H., Huijps K., Lam T.J.G.M. Economic aspects of mastitis: New developments. N. Z. Vet. J. 2011;59:16–23. doi: 10.1080/00480169.2011.547165. [DOI] [PubMed] [Google Scholar]
- 2.Halasa T., Huijps K., Østerås O., Hogeveen H. Economic effects of bovine mastitis and mastitis management: A review. Vet. Q. 2007;29:18–31. doi: 10.1080/01652176.2007.9695224. [DOI] [PubMed] [Google Scholar]
- 3.Fogsgaard K.K., Bennedsgaard T.W., Herskin M.S. Behavioral changes in freestall-housed dairy cows with naturally occurring clinical mastitis. J. Dairy Sci. 2015;98:1730–1738. doi: 10.3168/jds.2014-8347. [DOI] [PubMed] [Google Scholar]
- 4.Peton V., Le Loir Y. Staphylococcus aureus in veterinary medicine. Infect. Genet. Evol. 2014;21:602–615. doi: 10.1016/j.meegid.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 5.Wellnitz O., Bruckmaier R.M. The innate immune response of the bovine mammary gland to bacterial infection. Vet. J. 2012;192:148–152. doi: 10.1016/j.tvjl.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 6.Koymans K.J., Vrieling M., Gorham R.D., Jr., van Strijp J.A. Staphylococcal Immune Evasion Proteins: Structure, Function, and Host Adaptation. Curr. Top. Microbiol. Immunol. 2017;409:441–489. doi: 10.1007/82_2015_5017. [DOI] [PubMed] [Google Scholar]
- 7.Thammavongsa V., Kim H.K., Missiakas D., Schneewind O. Staphylococcal manipulation of host immune responses. Nat. Rev. Microbiol. 2015;13:529–543. doi: 10.1038/nrmicro3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spaulding A.R., Salgado-Pabón W., Kohler P.L., Horswill A.R., Leung D.Y.M., Schlievert P.M. Staphylococcal and streptococcal superantigen exotoxins. Clin. Microbiol. Rev. 2013;26:422–447. doi: 10.1128/CMR.00104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bukowski M., Wladyka B., Dubin G. Exfoliative toxins of Staphylococcus aureus. Toxins. 2010;2:1148–1165. doi: 10.3390/toxins2051148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Riordan K., Lee J.C. Staphylococcus aureus Capsular Polysaccharides. Clin. Microbiol. Rev. 2004;17:218–234. doi: 10.1128/CMR.17.1.218-234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DuMont A.L., Torres V.J. Cell targeting by the Staphylococcus aureus pore-forming toxins: It’s not just about lipids. Trends Microbiol. 2014;22:21–27. doi: 10.1016/j.tim.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deghorain M., Van Melderen L. The staphylococci phages family: An overview. Viruses. 2012;4:3316–3335. doi: 10.3390/v4123316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alonzo F., 3rd, Torres V.J. The bicomponent pore-forming leucocidins of Staphylococcus aureus. Microbiol. Mol. Biol. Rev. 2014;78:199–230. doi: 10.1128/MMBR.00055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koop G., Vrieling M., Storisteanu D.M.L., Lok L.S.C., Monie T., Van Wigcheren G., Raisen C., Ba X., Gleadall N., Hadjirin N., et al. Identification of LukPQ, a novel, equid-adapted leukocidin of Staphylococcus aureus. Sci. Rep. 2017;7:40660. doi: 10.1038/srep40660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bar-Gal G.K., Blum S.E., Hadas L., Ehricht R., Monecke S., Leitner G. Host-specificity of Staphylococcus aureus causing intramammary infections in dairy animals assessed by genotyping and virulence genes. Vet. Microbiol. 2015;176:143–154. doi: 10.1016/j.vetmic.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Schlotter K., Ehricht R., Hotzel H., Monecke S., Pfeffer M., Donat K. Leukocidin genes lukF-P83 and lukM are associated with Staphylococcus aureus clonal complexes 151, 479 and 133 isolated from bovine udder infections in Thuringia, Germany. Vet. Res. 2012;43 doi: 10.1186/1297-9716-43-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamada T., Tochimaru N., Nakasuji S., Hata E., Kobayashi H., Eguchi M., Kaneko J., Kamio Y., Kaidoh T., Takeuchi S. Leukotoxin family genes in Staphylococcus aureus isolated from domestic animals and prevalence of lukM-lukF-PV genes by bacteriophages in bovine isolates. Vet. Microbiol. 2005;110:97–103. doi: 10.1016/j.vetmic.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 18.Fromageau A., Gilbert F.B., Prévost G., Rainard P. Binding of the Staphylococcus aureus leucotoxin LukM to its leucocyte targets. Microb. Pathog. 2010;49:354–362. doi: 10.1016/j.micpath.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Vrieling M., Koymans K.J., Heesterbeek D.A., Aerts P.C., Rutten V.P., de Haas C.J., van Kessel K.P.M., Koets A.P., Nijland R., van Strijp J.A.G. Bovine Staphylococcus aureus Secretes the Leukocidin LukMF' To Kill Migrating Neutrophils through CCR1. MBio. 2015;6:e00335-15. doi: 10.1128/mBio.00335-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rainard P., Riollet C. Innate immunity of the bovine mammary gland. Vet. Res. 2006;37:369–400. doi: 10.1051/vetres:2006007. [DOI] [PubMed] [Google Scholar]
- 21.Vrieling M., Boerhout E.M., van Wigcheren G.F., Koymans K.J., Mols-Vorstermans T.G., de Haas C.J., Aerts P.C., Daemen I.J.J.M., van Kessel K.P.M., Koets A.P., et al. LukMF’ is the major secreted leukocidin of bovine Staphylococcus aureus and is produced in vivo during bovine mastitis. Sci. Rep. 2016;25:37759. doi: 10.1038/srep37759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Y., Liang Y., Lynch K.H., Dennis J.J., Wishart D.S. PHAST: A Fast Phage Search Tool. Nucleic Acids Res. 2011;39(Suppl. 2):W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mootz J.M., Benson M.A., Heim C.E., Crosby H.A., Kavanaugh J.S., Dunman P.M., Kielian T., Torres V.J., Horswill A.R. Rot is a key regulator of Staphylococcus aureus biofilm formation. Mol. Microbiol. 2015;96:388–404. doi: 10.1111/mmi.12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montgomery C.P., Boyle-Vavra S., Daum R.S. Importance of the global regulators agr and SaeRS in the pathogenesis of CA-MRSA USA300 infection. PLoS ONE. 2010;5:e15177. doi: 10.1371/journal.pone.0015177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haran K.P., Godden S.M., Boxrud D., Jawahir S., Bender J.B., Sreevatsan S. Prevalence and characterization of Staphylococcus aureus, including methicillin-resistant Staphylococcus aureus, isolated from bulk tank milk from Minnesota dairy farms. J. Clin. Microbiol. 2012;50:688–695. doi: 10.1128/JCM.05214-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veh K.A., Klein R.C., Ster C., Keefe G., Lacasse P., Scholl D., Roy J.P., Haine D., Dufour S., Talbot B.G., et al. Genotypic and phenotypic characterization of Staphylococcus aureus causing persistent and nonpersistent subclinical bovine intramammary infections during lactation or the dry period. J. Dairy Sci. 2015;98:155–168. doi: 10.3168/jds.2014-8044. [DOI] [PubMed] [Google Scholar]
- 27.Johler S., Layer F., Stephan R. Comparison of virulence and antibiotic resistance genes of food poisoning outbreak isolates of Staphylococcus aureus with isolates obtained from bovine mastitis milk and pig carcasses. J. Food Prot. 2011;74:1852–1859. doi: 10.4315/0362-028X.JFP-11-192. [DOI] [PubMed] [Google Scholar]
- 28.Ikawaty R., Brouwer E.C., Jansen M.D., van Duijkeren E., Mevius D., Verhoef J., Fluit A.C. Characterization of Dutch Staphylococcus aureus from bovine mastitis using a Multiple Locus Variable Number Tandem Repeat Analysis. Vet. Microbiol. 2009;136:277–284. doi: 10.1016/j.vetmic.2008.10.034. [DOI] [PubMed] [Google Scholar]
- 29.Merz A., Stephan R., Johler S. Staphylococcus aureus isolates from goat and sheep milk seem to be closely related and differ from isolates detected from bovine milk. Front. Microbiol. 2016;7:319. doi: 10.3389/fmicb.2016.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt T., Kock M.M., Ehlers M.M. Molecular characterization of Staphylococcus aureus isolated from bovine mastitis and close human contacts in South African dairy herds: Genetic diversity and inter-species host transmission. Front. Microbiol. 2017;8:511. doi: 10.3389/fmicb.2017.00511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hata E., Katsuda K., Kobayashi H., Uchida I., Tanaka K., Eguchi M. Genetic variation among Staphylococcus aureus strains from bovine milk and their relevance to methicillin-resistant isolates from humans. J. Clin. Microbiol. 2010;48:2130–2139. doi: 10.1128/JCM.01940-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Artursson K., Söderlund R., Liu L., Monecke S., Schelin J. Genotyping of Staphylococcus aureus in bovine mastitis and correlation to phenotypic characteristics. Vet. Microbiol. 2016;193:156–161. doi: 10.1016/j.vetmic.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 33.Monecke S., Kuhnert P., Hotzel H., Slickers P., Ehricht R. Microarray based study on virulence-associated genes and resistance determinants of Staphylococcus aureus isolates from cattle. Vet. Microbiol. 2007;125:128–140. doi: 10.1016/j.vetmic.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 34.Conceição T., De Lencastre H., Aires-De-Sousa M. Healthy Bovines as Reservoirs of Major Pathogenic Lineages of Staphylococcus aureus in Portugal. Microb. Drug Resist. 2017;23:845–851. doi: 10.1089/mdr.2017.0074. [DOI] [PubMed] [Google Scholar]
- 35.Killikelly A., Benson M.A., Ohneck E.A., Sampson J.M., Jakoncic J., Spurrier B., Torres V.J., Kong X.-P. Structure-based functional characterization of repressor of toxin (Rot), a central regulator of Staphylococcus aureus virulence. J. Bacteriol. 2015;197:188–200. doi: 10.1128/JB.02317-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benson M.A., Ohneck E.A., Ryan C., Alonzo F., Smith H., Narechania A., Kolokotronis S.O., Satola S.W., Uhlemann A.C., Sebra R. Evolution of hypervirulence by a MRSA clone through acquisition of a transposable element. Mol. Microbiol. 2014;93:664–681. doi: 10.1111/mmi.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang T., Wang C., Wei X., Zhao X., Zhong X. Loop-mediated isothermal amplification for detection of Staphylococcus aureus in dairy cow suffering from mastitis. J. Biomed. Biotechnol. 2012;2012:435982. doi: 10.1155/2012/435982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Britten A.M. The role of diagnostic microbiology in mastitis control programs. Vet. Clin. N. Am. Food Anim. Pract. 2012;28:187–202. doi: 10.1016/j.cvfa.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 39.Hogan J.S., National Mastitis Council (U.S.) Laboratory Handbook on Bovine Mastitis. National Mastitis Council; Madison, WI, USA: 1999. [Google Scholar]
- 40.Francois P., Pittet D., Bento M., Pepey B., Vaudaux P., Lew D., Schrenzel J. Rapid detection of methicillin-resistant Staphylococcus aureus directly from sterile or nonsterile clinical samples by a new molecular assay. J. Clin. Microbiol. 2003;41:254–260. doi: 10.1128/JCM.41.1.254-260.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larsen M.V., Cosentino S., Rasmussen S., Friis C., Hasman H., Marvig R.L., Jelsbakc L., Sicheritz-Ponténa T., Usserya D.W., Aarestrup F.M., et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.