Abstract
Background
Norwalk virus causes outbreaks of acute non-bacterial gastroenteritis in humans. The virus capsid is composed of a single 60 kDa protein. In a previous study, the capsid protein of recombinant Norwalk virus genogroup II was expressed in an E. coli system and monoclonal antibodies were generated against it. The analysis of the reactivity of those monoclonal antibodies suggested that the N-terminal domain might contain more antigenic epitopes than the C-terminal domain. In the same study, two broadly reactive monoclonal antibodies were observed to react with genogroup I recombinant protein.
Results
In the present study, we used the recombinant capsid protein of genogroup I and characterized the obtained 17 monoclonal antibodies by using 19 overlapping fragments. Sixteen monoclonal antibodies recognized sequential epitopes on three antigenic regions, and the only exceptional monoclonal antibody recognized a conformational epitope. As for the two broadly reactive monoclonal antibodies generated against genogroup II, we indicated that they recognized fragment 2 of genogroup I. Furthermore, genogroup I antigen from a patient's stool was detected by sandwich enzyme-linked immunosorbent assay using genogroup I specific monoclonal antibody and biotinated broadly reactive monoclonal antibody.
Conclusion
The reactivity analysis of above monoclonal antibodies suggests that the N-terminal domain may contain more antigenic epitopes than the C-terminal domain as suggested in our previous study. The detection of genogroup I antigen from a patient's stool by our system suggested that the monoclonal antibodies generated against E. coli expressed capsid protein can be used to detect genogroup I antigens in clinical material.
Background
Norwalk viruses (NVs) are a small positive-stranded RNA virus within the family Caliciviridae and consist of genogroups I and II (GI and GII). An enzyme-linked immunosorbent assay (ELISA) system to detect all types of NVs has not yet been established due to various clusters of NVs in two genogroups. The lack of an in vitro culture system might have hindered establishing a good detection system.
The genome of NV is approximately 7.6 kb in size, and consists of three open reading frames (ORFs). The first ORF encodes non-structural proteins, a capsid protein is encoded by ORF2 and a function of the protein encoded by ORF3 has been unknown [1,2]. However, recently it was suggested that the ORF3 protein is a minor structural protein of the virion [3].
Among the expression of cDNAs that encode NV capsid proteins, the baculovirus recombinant system is most common. Since all expressed recombinant proteins spontaneously assembled into empty virus-like particles (VLPs) in this system [4-9], immunological reaction of generating antibodies using these VLPs is considered to be similar to that when using native NV particles. The availability of large amounts of recombinant NV (rNV) VLPs has facilitated the production of polyclonal antibodies as well as monoclonal antibodies (MAbs) useful to develop immunological detection systems and characterize antigenic sites of the virus. However, the antisera generated against VLPs showed the specific reactivity. These sera only reacted towards the closely related strains of the same clusters [10-12].
In contrast to the specific reactivity of antibodies generated against VLPs, we showed broad reactivity of rabbit IgG as well as two MAbs (generated against the E. coli-expressed rNV GII capsid protein resulting in soluble proteins without assembly into VLPs) in a previous study [13,14]. We indicated the primary antigenic map of the rNV36 (GII) capsid protein using MAbs generated against it [14]. In the present study, 17 MAbs were generated against rNV96-908 (GI) capsid protein and 19 truncated rNV fragments in addition to the whole capsid protein were expressed in E. coli as antigens. The GI antigenic map of rNV capsid protein using these Mabs was constructed and the results compared with the former GII antigenic map. We also clearly showed that two MAbs generated against GII rNV36 reacted with fragment 2 of rNV96-908 (GI). We obtained good results of detecting NV in clinical material by sandwich ELISA using anti-G I MAb as the captured antibody and biotinated-broadly reactive MAb generated against GII, 1F6 (B-1F6), as the detecting antibody. These results encouraged us to establish a good immunological detection system.
Materials and Methods
Viruses, plasmid and E. coli
NV strains 36, 21, 114, 96–908, expression vector pTrxFus*, E. coli (GI 724) strain and anti-NV36 MAbs used in this study were prepared as described previously [13,14]. Other NV strains 99–8, and 00–001 were detected from patients in our institute and strain Gifu'96 was isolated and submitted to the Gifu Prefectural Institute for Bio-industrial Technology.
Immunization of mice
MAbs specific to rNV96-908 were prepared from female BALB/c mice immunized with recombinant thioredoxin (TRX)-fused NV96-908 capsid protein (rTRX-NV96-908) that had been partially purified as described previously [13]. Female BALB/c mice were immunized as described previously [14].
Preparation of hybridoma
Cell fusion between lymphocytes and mouse myeloma cells was performed as described previously [14].
Cloning of the rNV96-908 fragment gene in pTrx-Fus* expression vectors
The expression vector, pTrxFus* was used to construct fusion proteins between TRX (thioredoxin, approximately 12 kDa) and NV96-908 fragment proteins. A total of 19 overlapping fragments of the protein as well as the whole NV96-908 capsid protein were expressed in E. coli (Fig. 1, though two large domains are not shown).
Figure 1.
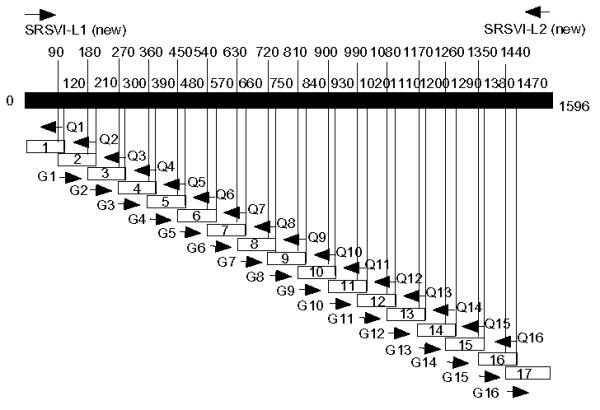
Schematic diagram of rNV96-908 capsid fragments expressed in E. coli. The solid bar represents the full length of the rNV96-908 capsid protein. Numbers besides the bar show nucleotide numbers of NV96-908 capsid. Numbers above the bar indicate the planned position of the fragment edges not the actual positions. Small open boxes with numbers indicate each fragment. The N-terminal fragment consists of fragments 1–7 and the C-terminal fragment consists of fragments 8–17. These fragments were constructed using restriction enzyme sites in primers for ligation convenience (Table 2).
These 19 overlapping fragments consisted of two large fragments corresponding to the N-terminal domain composed of residues 1–226 and the C-terminal domain composed of residues 208–531 and 17 small fragments of about 40 a.a. residues. All of the NV96-908 fragments were generated using PCR with LA Taq DNA polymerase (Takara Biomedicals, Japan). Most of the PCR experiments consisted of 35 cycles of denaturation (94°C for 30 sec), primer annealing (55°C for 30 sec) and extension (72°C for 1 min). For the primers G-l, -2, -3, -4, -5, -11, -14 and Q-2, -3, -4, -5, -6, -12 and -15, PCR conditions were as given above, except for primer annealing at 50°C. The primers used in this experiment are shown in Table 1. All PCR products were digested with SmaI and XbaI followed by purification on agarose gel, and were ligated with SmaI and XbaI-digested pTrxFus* vectors. The ligation mixtures were used to transform GI724 competent cells.
Table 1.
Primers used in this experiment
| LI: | TCC CCCGGG ATG ATG ATG GCG TCT AAG GAC | L2: | GC TCTAGA TTA TWW WCG GCG CAV WCC AAG CC |
| G1: | TCC CCCGGG CTT GCA ATG GAT CCT GTG GC | Ql: | GC TCTAGA ACT GCT GTT GAA GAA CCC GC |
| G2: | TCC CCCGGG CAA GGT GAA TTT ACT ATT TC | Q2: | GC TCTAGA AAT AGT AAA TTC ACC TTG GG |
| G3: | TCC CCCGGG CTT AAT CCC TTC TTG TTA C | Q3: | GC TCTAGA TGC CAA CCC AGC CAT TAT AC |
| G4: | TCC CCCGGG GGT AAA ATT ATA GTT TCT TGC | Q4: | GC TCTAGA AAT CAC ATG CGG GAA GAG AG |
| G5: | TCC CCCGGG GAT GTA AGG AAT GTT CTC TTT C | Q5: | GC TCTAGA CTT ACA TCT ACT AAG GGT AC |
| G6: | TCC CCCGGG CAA ACA ATG CGC CTT GTG TGC | Q6: | GC TCTAGA GGA GGG GGG TGT AAA GCA TG |
| G7: | TCC CCCGGG ACT TGT CCT AGC CCC GAT TTC | Q7: | GC TCTAGA GTT CCA CTG TGG GAG GAA CC |
| G8: | TCC CCCGGG TTG TCA AAT TCA CGT GCC CC | Q8: | GC TCTAGA AAT TGG AAG AGG GGC ACG TG |
| G9: | TCC CCCGGG TGT ACC TTA GAT GGA CGA C | Q9: | GC TCTAGA GGT GGT ACC AAC AAG TCG TC |
| G10: | TCC CCCGGG GGT ACT TCT AAT GGT ACT GTG | Q10: | GC TCTAGA GGG GCA GGG CCT TCA AAA GG |
| G11: | TCC CCCGGG GAT TGG CAT ATT AAC ATG AC | Q11: | GC TCTAGA TGA GTC TGG CTA GAA TGT CC |
| G12: | TCC CCCGGG GGT TCA ATC CAG GCG AAT GG | Q12: | GC TCTAGA TAA GAA CAC CAA TGT AGT TG |
| G13: | TCC CCCGGG AAG ATC CCC AAC TAT GGG TC | Q13: | GC TCTAGA CTG TGA TGC TAG ACC CAT AG |
| G14: | TCC CCCGGG TTC ATG TCA AAG ATA CCA GG | Q14: | GC TCTAGA AAG CAC CAG GGC CTG GTA TC |
| G15: | TCC CCCGGG CCC ACT GTT GGT GAA GCC GC | Q15: | GC TCTAGA TAG TGG AGT AGG GCG GCT TC |
| G16: | TCC CCCGGG GAT GGT TTC CTA ACC TGT GTC | Q16: | GC TCTAGA GCT GGC CCC GTT AGG GAC AC |
| Trx-up: TTC CTC GAC GCT AAC CTG GC | Trx-DW: GTA AA CGA CGG CCA GTG CC | ||
G, L and Q primers were used for amplifying NV96-908 fragments, and Trx-up and Trx-dw primers for generating the insert and junction sequence of recombinant plasmids. These Trx primers have also been used for sequencing. Underlined sequences of G primers and LI primer shows the SmaI site, and that of bo th L2 primer and Q primers the XbaI site.
Induction and analysis of recombinants
The resulting colonies carrying the expression constructs were incubated, induced and analyzed as described previously [13]. The expression products were analyzed on SDS-PAGE (Fig. 2). Both strands of the insert and junction of recombinant plasmids were sequenced using the PRISM™ Dye-Deoxy Terminator Cycle Sequencing Kit (Perkin Elmer ABI, USA) with the Trx-up and Trx-dw and analyzed using the 310 Genetic analyzer (Perkin Elmer ABI) as described previously [15].
Figure 2.
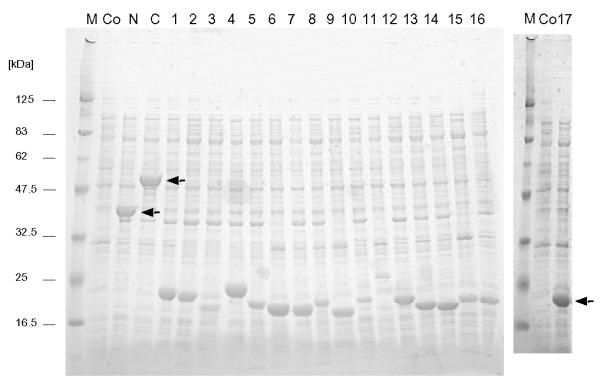
Expression of the rNV96-908 fragments in E. coli. Production of TRX-fused capsid protein fragments in E. coli was analyzed by staining two SDS-PAGE gels. From the left, lane 1 and lane 21 : marker [kDa], lane 2 and lane 2: control recombinant E. coli lysate (before induction). From lane 3 to 23 except 21 and 22 are TRX-NV96-908 N-terminal domain, C-terminal domain, fragment 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, and 17, respectively. The positions of the recombinant NV capsid fragments are indicated by arrows (the small fragments show similar molecular weights).
Production and partial purification of recombinant proteins
Recombinant proteins were produced from a 100 ml tryptophan-induced culture of transformed E. coli. Purification of TRX-fusion proteins was performed by sonication and centrifugation once, as described previously [13]. Partial purification of the recombinant proteins was performed as described before [14].
SDS-PAGE and immunoblotting analysis
Epitope mapping analysis
Purity of the fusion protein and protein concentrations was estimated using Coomasie brilliant blue-stained SDS-PAGE gels. WB assays were carried out according to the methods described previously [13]. Briefly, the proteins were transferred onto a NitranR membrane (Schleicher & Schuell, Germany) using a Western transfer buffer with semi-dry equipment, BP-312 (Bio-Craft, Japan). After blocking with Block Ace (Snow Brand, Japan), the membranes were incubated for 1 hr at 37°C with MAbs diluted at various ratios in working buffer. The dilution ratios used were: 1:100,000 to 1:250,000 for cloned MAbs (3E7, 4E6, 4F7, 7D2 and 13F9), 1:100 to 1:1,000 for culture sup. (1G9, 3D10, 4B10, 5B6, 6C4, 8C7, 8D2, 10F8, 10G10, 12D4 and 15G5). The membranes were washed and incubated with horseradish peroxidase (HRP)-conjugated anti-mouse-IgG Fc specific (Sigma) diluted at 1:1,000 for 1 hr at room temperature (RT). Bound antibodies were detected using 3, 3', 5, 5'-tetramethylbenzidine (TMB) and membrane peroxidase substrate system (Kirkegaard Perry Laboratories, USA). The MAb, 7E9 was not tested, since it did not react in immunoblotting.
ELISA
A direct ELISA was carried out to determine the reactivity of the mouse MAbs with TRX-NV96-908 and TRX-NV96-908 fragments. All of the reactions in ELISA were performed in triplicate according to a methodology similar to that described previously, except for adsorption with 10 μg/ml TRX [13]. Briefly, 96-well microplates were coated with 1 μg of the different antigens, TRX-NV96-908, TRX-NV96-908 fragments and TRX, overnight at 4°C. After washing 3 times, microplates were incubated with anti-rNV96-908 mouse MAbs at various concentrations for 1 hr at 37°C (Fig. 3a and 3b). HRP-labeled mouse IgG diluted at 1:5,000 was used as the second antibody. After incubation for 1 hr at 37°C, bound antibodies were detected by adding TMB as a substrate followed by incubation for 30 min at RT. The enzyme reaction was then stopped with 1 M phosphoric acid and the absorbance measured at 450 nm in an ELISA microplate reader. Model 550 (Bio-Rad, USA). The dilution ratios of the MAbs were adjusted so that the OD value showed less than 5 at 450 nm as indicated in Fig. 3. A double antibody sandwich-ELISA (DAS-ELISA) was used to determine the existence inhibition between the reaction of rGI specific MAbs and the reaction of the MAb 1F6 using rNV96-908. The detection sensitivity to different GI rNV strains was also examined in the same DAS-ELISA. The MAb 1F6 was biotinated using EZ-Link NHS-Biotin (PIERCE, USA) according to the manufacture's protocol. Then, 96-well microplates were coated with 1 μg of two MAbs (3E7 and 4E6). After washing, various concentrations (0, 3.125, 6.25, 12.5, 25, 50, 100 and 200 ng/ml) of rNVs (TRX-NV36, 96–908, and 00–002) and TRX were added to each well and incubated for 1 hr at 37°C. After washing, the microplates were incubated for 1 hr at 37°C with the biotinated (B)-antibody, B-1F6. The Vectastain ABC Kit PK-6100 (Vector Laboratories, USA) was used as a conjugate. After incubation with Vectastain ABC for 30 min at 37°C, bound antibodies were detected by adding TMB, as in the direct ELISA.
Figure 3.
Antigenic sites on rNV96-908 defined by MAbs using the N-terminal (a) and the C-terminal fragments (b) as antigens. Reactivity of MAbs with rNV96-908 derived fragments was measured by ELISA. Names of the individual MAbs and their dilutions used are shown at the bottom of the figures.
Detection of Norwalk virus from a patient's stool sample by sandwich ELISA
A DAS-ELISA was performed to detect NVs in a patient's stool sample. The entire procedure in DAS-ELISA was similar to other DAS-ELISA except for using clinical materials as antigens (100 μl/well).
Pre-treatment of clinical material (patient's stool)
After making 10% homogenized solution using saline, centrifuging at 15,000 rpm. for 20 min at 4°C, the supernatant was put into new tubes containing Block Ace (final concentration 20%), and 2-Mercaptoethanol (2ME, Wako Pure Chemical Industries, LTD, Japan) to reach a final concentration of 2.5%. Four types of treatments were carried out. 1) Heat treatment: incubate sample for 5 min at 100°C. 2) SDS/heat treatment: Add SDS into sample to reach a final concentration of 1%, incubate for 5 min at 100°C. 3) Alkaline treatment: add 1 M NaOH into the sample to reach a final concentration of 0.18 M (approximately pH 10), incubate for 10 min at RT. Then, add 1.5 fold vol. of 1 M Tris-HCl (pH 6.8) into the sample to make pH approximately 8.4. 4) Freeze and thaw treatment: samples were frozen and thawed twice after alkaline and neutralization treatment (freeze at -80°C until the samples become frozen, then thaw at 37°C).
Results and Discussion
Recombinant Proteins
Nineteen trumcated pTrx-Fus recombinant plasmids derived from pTrx-Fus-NV96-908 were constructed containing overlapping fragments of NV96-908 at various lengths, as shown in Fig. 1. Production of rNV96-908 fragments in E. coli were verified by Coomassie brilliant blue staining of SDS-PAGE gels (Fig. 2). Expression levels varied widely, with the amount of purified fragment ranging from 0.1 to 8 mg/ml (yield 0.05 mg to 4 mg). The concentration of purified fragments was estimated by visual comparison with known quantities of bovine serum albumin (BSA) in SDS-PAGE gels.
Production and preliminary characterization of rNV-specific MAbs
A total of 17 anti-rNV96-908 MAbs have been obtained. Their specificity in preliminary immuno-reactions to whole NV capsid protein, as well as to the N-terminal domain (NV96-908 residues 1–226) and C-terminal domain (NV96-908 residues 208–531), using ELISA and immunoblot analysis is shown in Table 2. In ELISA, 14 MAbs reacted with the N-terminal domain and 3 MAbs reacted with the C-terminal domain. The findings of the immunoblotting corresponded well with those of ELISA except one MAb, 7E9. The MAb, 7E9 might recognize the conformational epitope, since it did not react in immunoblotting.
Table 2.
Characterization and reactivity of MAbs to whole rNV96-908, and its N-terminal and C-terminal fragments(rNV-N, rNV-C).
| Hybridoma | Isotype | Reactivity in ELISA | Reactivity in WB | ||||
| (type of light chains) | whole rNV96-908 | rNV-N | rNV-C | whole rNV96-908 | rNV-N | rNV-C | |
| 3E7 | IgG1 (k) | ++ | ++ | - | ++ | ++ | - |
| 4E6 | lgG2b (k) | ++ | ++ | - | ++ | ++ | - |
| 4F7 | lgG2a (k) | ++ | ++ | - | ++ | ++ | - |
| 7D2 | lgG2b (k) | ++ | ++ | - | ++ | + | - |
| 7E9 | lgG2b (k) | ++ | ++ | - | - | - | - |
| 13F9 | lgG2b (k) | ++ | ++ | - | ++ | ++ | - |
| 1G9 | Not cloned | ++ | ++ | - | ++ | ++ | - |
| 3D10 | Not cloned | ++ | ++ | - | ++ | ++ | - |
| 4B10 | Not cloned | ++ | ++ | - | ++ | ++ | - |
| 5B6 | Not cloned | ++ | - | ++ | ++ | - | ++ |
| 6C4 | Not cloned | ++ | ++ | - | ++ | ++ | - |
| 8C7 | Not cloned | ++ | - | ++ | ++ | - | ++ |
| 8D2 | Not cloned | ++ | ++ | - | ++ | ++ | - |
| 10F8 | Not cloned | ++ | ++ | - | ++ | ++ | - |
| 10G10 | Not cloned | ++ | - | ++ | ++ | - | ++ |
| 12D4 | Not cloned | ++ | ++ | - | ++ | ++ | - |
| 15G5 | Not cloned | ++ | ++ | - | ++ | ++ | - |
- Negative reaction; + positive reaction; ++ strong positive reaction
Reactivity of rNV96-908-specific MAbs with the recombinant fragments determined with ELISA
To complete the epitope mapping, the reactivity of MAbs to different areas of the N-terminal domain (fragments 1–7) and C-terminal domain (fragments 8–17) was determined using ELISA (Fig. 3a and 3b). Thirteen of 14 MAbs (that reacted with the N-terminal domain) reacted with fragment 1. One MAb (7E9) was suggested to recognize the conformational epitope, since it reacted with the N-terminal domain, but reacted with none of the fragments (fragments 1–7) as shown in Fig. 3a. These results explained why 7E9 did not react in the immunoblotting test (Table 2).
Among the C-terminal domain recognition MAbs, two MAbs, 10G10 and 5B6 showed a clear reactivity with fragment 11, and one MAb, 8C7, reacted with fragment 16.
Characterization of rNV96-908 epitopes by immuoblotting
The findings of immunoblotting corresponded well to those of ELISA with only one exception, MAb 7E9. Since all MAbs except 7E9 recognized the sequential epitopes, similar findings between imrnunoblotting and ELISA should be as no surprise.
Antigenic domains of rNV96-908
The antigenic structure of the rNV96-908 capsid protein appears to be characterized by the presence of three immunodominant regions based on the immuno-reactivity of mouse MAbs towards the capsid protein (rNV96-908) fragments (Fig. 4). Thirteen of 17 MAbs recognized fragment 1, composed of 43 a.a. residues. Two of the 17 MAbs recognized fragment 11, consisting of 51 a.a. residues, and one MAb recognized fragment 16, with 39 a.a. residues. Comparisons of a.a. sequences of fragments 1,11 and 16 were carried out among various rNVs and reference NV strains (Fig. 5). Fragment 1 was suggested to contain an antigenically dominant region that probably includes several epitopes, since the MAbs reacted differently towards a different type of GI rNV002 (Chiba type). Although data is not shown, three patterns of reaction were observed; those that reacted with similar sensitivity as rNV96-908, approximately 50% sensitivity, and those that did not react. There might not be common epitopes between GI and GII in fragment 1, since only one MAb, 3E7 has weak cross-reactivity with one of the rNVs, GII rNV36 (Mexico virus type). It is unclear whether those MAbs that recognized fragment 1 on rNV96-908 had a broad recognition range of GI rNV, since the cross-reactivity with other GI rNV had not been examined except for the Chiba type. Fine epitope mapping of cloned MAbs that recognized fragment 1 of NV96-908 should be helpful to elucidate the possibility of MAbs application to detect a broad range of GI NVs.
Figure 4.
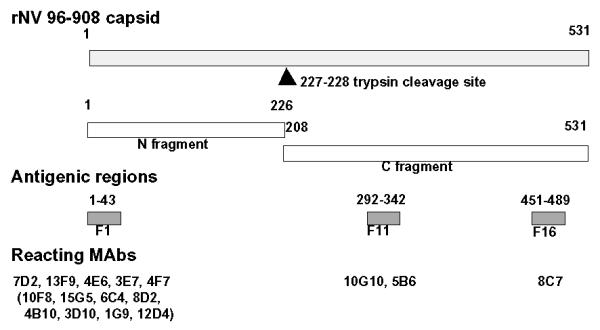
Diagram of rNV antigenic domains. The gray bar at the top represents the full length of the rNV96-908 capsid protein. The arrowhead shows the putative position of the trypsin cleavage site. The N-terminal and C-terminal parts are shown as open bars. The antigenic regions of fragments 1, 11 and 16 are indicated as dark gray color boxes. Reacting MAbs are shown below the corresponding fragments. Numbers on the top of the rNV capsid and the antigenic fragments indicate the first and last residues.
Figure 5.

Comparison of amino acid sequences of fragments 1 (a), 11 (b) and 16 (c) in two rNVs of GI strains and the related reference NV strains. The dotted lines show deletions and under lined a.a. sequences are unique parts of the individual fragments. Other parts are overlapping with proximal fragments. Accession numbers for NV96-908, NV002, KY89 and Chiba virus are AB028247, AB063325, L23828 and AB022679, respectively.
The MAbs, 10G10 and 5B6 recognized fragment 11 of NV96-908, but did not react with GI rNV002 (Chiba type). This finding suggested that fragment 11 of NV96-908 probably did not contain common epitopes among GI NVs. The MAb (8C7) that recognized fragment 16 reacted well with GI rNV002 (Chiba type). It might recognize common epitopes among GI NVs.
Reactivity of broadly reactive MAbs generated against rNV36 (GII) with the recombinant fragments of NV96-908 (GI) determined by ELISA
Since the cross-reactive MAbs generated against rNV36 (1B4 and 1F6) showed broad reactivity towards different types of rNV capsid proteins by direct ELISA as indicated in Fig. 6, ELISA was performed to determine epitopes of these MAbs on rNV96-908 fragments. As expected, both of the MAbs reacted with the N-terminal domain, but not with the C-terminal domain, moreover, both reacted with fragment 2 specifically among fragments 1 to 7 of rNV96-908 as shown in Fig. 7. Comparison of amino acid sequences of fragment 2 of rNV96-908 (GI) and that of rNV36 (GII) with other rNVs, and their related reference strains are shown in Fig. 8. It might be difficult to determine the fine epitopes of these two MAbs (1B4 and 1F6) with the present findings. However, it appears to be important to elucidate fine epitopes of these MAbs for further study.
Figure 6.
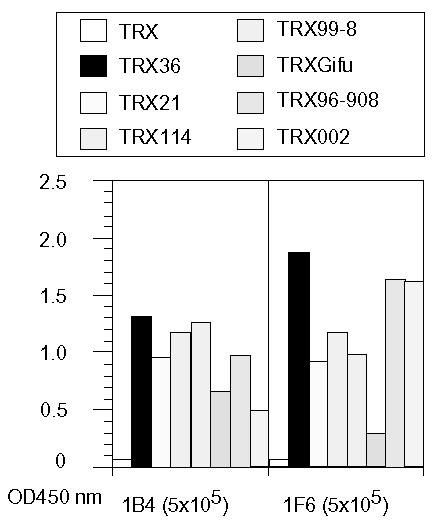
Reactivity comparison of the broadly reactive MAbs generated against GII rNV36 towards various rNVs. Reactivity of the MAbs (1B4 and 1F6) was measured by direct ELISA. Among rNVs, strains 002 and 96–908 are GI and the others are GII. Names of the individual MAbs and their dilutions (in the parentheses) are indicated at the bottom of the figure.
Figure 7.
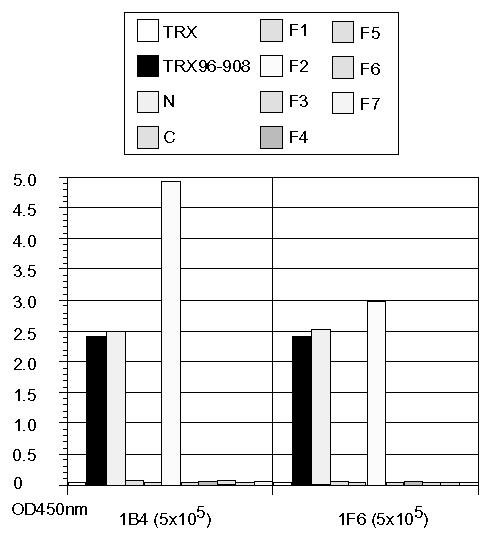
Antigenic sites on rNV96-908 were defined by the broadly reactive MAbs using the N- and C-terminal fragments as well as fragments 1 to 7 as antigens. Reactivity of the MAbs towards rNV96-908 derived fragments was measured by ELISA. Names of the MAbs and their dilutions used are shown at the bottom of the figure.
Figure 8.
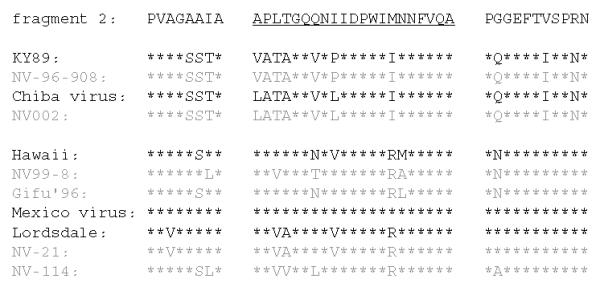
Comparison of amino acid fragment 2 of NV36 with other rNVs and the reference NV strains. The underlined a.a. sequences are unique parts of the individual fragments. Other parts are overlapping with proximal fragments. Accession numbers for NV21, NV114, Gifu'96, Mexico virus and Lordsdale virus are AB028245, AB028246, AB045603, U22498 and X86557, respectively.
Double antibody sandwich ELISA (DAS-ELISA)
Since one of the broadly reactive MAbs generated against GII, 1F6, reacted with GI rNVs better than 1B4 as shown in Fig. 6, 1F6 was used for a DAS-ELISA. MAbs, 3E7 and 4E6 were chosen from rNV96-908 specific MAbs since these two MAbs reacted well with rNV002 (Chiba type). Due to the close location between the epitopes 3E7 or 4E6 (fragment 1 of rNV96-908) and 1F6 (fragment 2 of rNV96-908), a DAS-ELISA was carried out to examine whether MAbs (3E7 and 4E6) inhibit the reaction of the Biotinated-lF6 (B-1F6). Two rGI NV specific MAbs were used as captured Abs, and B-1F6 was used as detecting Ab. The results indicated that these two anti-GI NV MAbs did not inhibit the reaction of B-lF6 as indicated in Fig. 9a and 9b. Recombinant NV002 can be detected as sensitive as NV96-908 in an appropriate detection area in these systems. Since one of the anti-GI NV MAbs, 3E7 showed weak cross-reactivity towards GII rNV36, rNV 36 could be detected as shown in Fig. 9(b) but not in 9(a).
Figure 9.
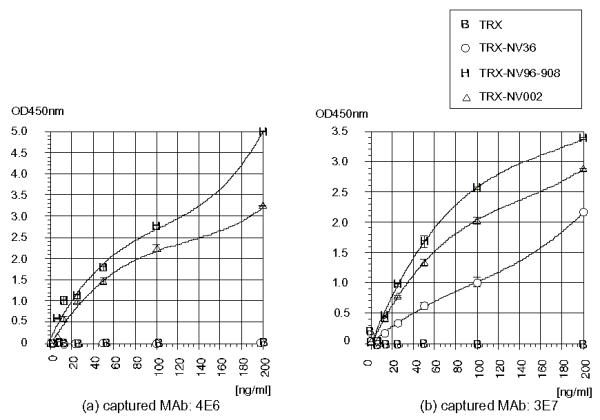
A double antibody sandwich ELISA for detecting various rNV antigens. The captured antibody in (a): 4E6 (GI specific MAb) and (b): 3E7 (GI specific MAb), B-1F6 was used as the detecting antibody in both of the reaction.
Detection of Norwalk virus from a patient's stool by sandwich ELISA
A sandwich ELISA was performed to detect NV from a patient's stool using the rGI NV specific monoclonal antibody (3E7) as the captured antibody, biotinated 1F6 (generated against GII) as the detecting antibody. The nucleotide sequence (nt) of equivalent parts in NV strains from patients were obtained and a. a. residues were compared with that of fragment 1 in rNV96-908 and that of fragment 2 in rNV36 prior to use. Amino acid residues of the selected strain, 00–001 was similar to fragments 1 and 2 of rNV96-908 (and suggested to be recognized by both MAbs in this ELISA). As shown as Fig. 10(a), the alkaline treatment appeared best for specific detection of the GI NV antigen among four pre-treatments of the clinical material. On the other hand, freezing and thawing after alkaline and neutralization treatment appeared to change appropriate denaturation of the NV antigen in the materials. Heat treatment with or without SDS did not work at all. The existence of 2-Mercaptoethanol (2ME) during these treatments was significantly important, since none of the treatments without 2ME could detect the GI NV antigen from clinical material. Various concentrations of rNV protein were used to obtain a calibration curve shown in Fig. 10(b). Only some of the NV antigens in 00–001 sample were considered to be detected by this system, because among NV particles and capsid proteins in 00–001, some of the particles and capsid proteins were denatured in alkaline buffer and partly maintained an adequate structure to be detected by MAbs after neutralization. Although it is impossible to estimate the precise amount of NV capsid protein in 00–001 from the calibration curve in (b), it was estimated at approximately 22 ng/ml. Application of this system should be examined using more clinical materials.
Figure 10.
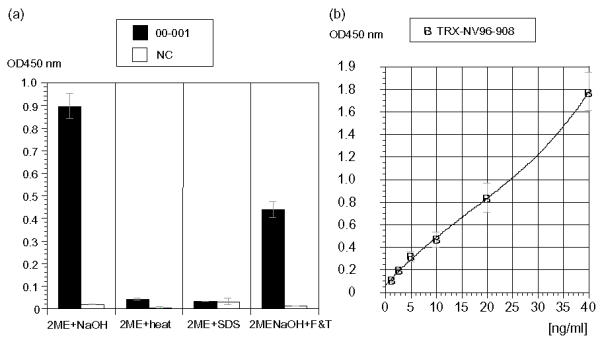
A double antibody sandwich ELISA for detecting the clinical material (NV from a patient's stool) in (a). GI specific MAb, 3E7 was used as the captured antibody, and B-1F6 was used as the detecting antibody, (a) Comparison of NV GI positive and NV negative clinical materials in 4 types of pre-treatments. (b) A calibration curve to detect rNV96-908 by the same system as (a).
Conclusions
Fourteen of the 17 MAbs generated against the E. coli expressed GI NV96-908 capsid protein recognized the N-terminal domain (equivalent residues 1–226). Thirteen of 14 MAbs recognized fragment 1 (residues 1–43), and only one exceptional MAb recognized the conformational epitope. The remaining 3 MAbs recognized two different fragments within the C-terminal domain (residues 208–531). These results suggest that the N-terminal domain may contain more antigenic epitopes than the C-terminal domain. This suggestion appears different from MAbs generated against rNV VLPs [16]. It might be better using baculovirus expressed rNV VLPs for generating Abs, since the immunological reaction appears similar to that when using native NV particles. In contrast to the highly specific reactivity of Abs generated against baculovirus expressed rNV VLPs [10-12], Abs generated against E. coli expressed GII rNV showed broad reactivity. Broad and strong cross-reactivity of these MAbs generated against GII NV towards GI NV was clearly elucidated by showing their specific reaction to fragment 2 on rNV96-908 in this study.
Instead of indicating NV Ag detection by WB analysis after purification [14], we indicated NV Ag detection by ELISA using a simple pre-treated patient's stool sample in this study. This is the first study that reported the application of GI NV antigen detection from clinical material using the MAbs generated against E. coli expressed NV capsid protein. Furthermore complicated pre-treatment of stool samples was not necessary and there appears to be the possibility to improve the NV Ag detection system. Due to the lack of a good immunological detection system, RT-PCR has been most common NV Ag detection test. However, it is time consuming with a complicated procedure. It appears to be significantly important to indicate the application possibility of establishing a good immunological system using MAbs generated against E. coli expressed rNV.
List of abbreviations
2ME, 2-Mercaptoethanol; a.a., amino acid; B, biotinated; BSA, bovine serum albumin; E. coli, Escherichia coli; DAS-ELISA, a double antibody sandwich-ELISA; ELISA, enzyme-linked immunosorbent assay;
GI, genogroup I; GII, genogroup II; HRP, horseradish peroxidase; Igs, immunoglobulins; MAb, monoclonal antibody; NV, Norwalk virus; PCR, polymerase chain reaction; rNV, recombinant NV; RT, room temperature; rTRX, recombinant TRX; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; Sup., supernatant; TMB, 3,3', 5, 5'-tetramethylbenzidine; TRX, thioredoxin; VLP, virus-like particle; WB, Western blot;
Acknowledgments
Acknowledgments
We thank Dr. M. Oseto, Ehime Prefectural Institute of Public Health and Environmental Science, for kindly providing us with specimen NV96-908 (GI type). Our thanks also go to Mr. A. Shimada, K. Hishida, and Ms. N. Goto at JBS, for their devoted support for this study.
Contributor Information
Tomoko Yoda, Email: yoda@iph.pref.osaka.jp.
Yoshitake Terano, Email: terano@wombat.or.jp.
Yasuhiko Suzuki, Email: suzuki@iph.pref.osaka.jp.
Kenji Yamazaki, Email: knyamaza@iph.pref.osaka.jp.
Isao Oishi, Email: ooisi@iph.pref.osaka.jp.
Tsuyoshi Kuzuguchi, Email: kuzuguti@bio.rd.pref.gifu.jp.
Hiroyoshi Kawamoto, Email: kawamoto@bio.rd.pref.gifu.jp.
Etsuko Utagawa, Email: etu@nih.go.jp.
Koichi Takino, Email: jbsbio@po.mirai.ne.jp.
Hajime Oda, Email: hjoda@iph.pref.osaka.jp.
Tadayoshi Shibata, Email: sibata@iph.pref.osaka.jp.
References
- Lambden PR, Caul EO, Ashley CR, Clarke IN. Sequence and genome organization of a human small round-structured (Norwalk-like) virus. Science. 1993;259:516–519. doi: 10.1126/science.8380940. [DOI] [PubMed] [Google Scholar]
- Jiang X, Wang M, Wang K, Estes MK. Sequence and genomic organization of Norwalk Virus. Virology. 1993;195:51–61. doi: 10.1006/viro.1993.1345. [DOI] [PubMed] [Google Scholar]
- Glass PJ, White LJ, Ball JM, Leparc-Goffart I, Hardy ME, Estes MK. Norwalk Virus Open Reading Frame 3 Encodes a Minor Structural Protein. J Virol. 2000;74:6581–6591. doi: 10.1128/jvi.74.14.6581-6591.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale AD, Crawford SE, Ciarlet M, Green J, Gallimore C, Brown DWG, Jiang X, Estes MK. Expression and self-assembly of Grimsby Virus: Antigenic distinction from Norwalk and Mexico viruses. Clin Diagn Lab Immunol. 1999;6:142–145. doi: 10.1128/cdli.6.1.142-145.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite JPG, Ando T, Noel JS, Jiang B, Humphrey CD, Lew JF, Green KY, Glass RI, Monroe SS. Characterization of Toronto virus capsid protein expressed in baculovirus. Arch Virol. 1996;141:865–875. doi: 10.1007/BF01718161. [DOI] [PubMed] [Google Scholar]
- Dingle KE, Lambden PR, Caul EO, Clarke IN. Human enteric Caliciviridae: the complete genome sequence and expression of virus-like particles from a genetic group II small round structured virus. J Gen Virol. 1995;76:2349–2355. doi: 10.1099/0022-1317-76-9-2349. [DOI] [PubMed] [Google Scholar]
- Green KY, Kapikian AZ, Valdesuso J, Sonsnovtsev S, Treanor JJ, Lew JF. Expression and self-assembly of recombinant capsid protein from the antigenically distinct Hawaii human calicivirus. J Clin Microbiol. 1997;35:1909–1914. doi: 10.1128/jcm.35.7.1909-1914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Matson DO, Ruiz-Palacios GM, Hu J, Treanor J, Pickering LK. Expression, self-assembly and antigenicity of a Snow Mountain Agent-like calicivirus capsid protein. J Clin Microbiol. 1995;33:1452–1455. doi: 10.1128/jcm.33.6.1452-1455.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Wang M, Graham DY, Estes MK. Expression, self-assembly and antigenicity of the Norwalk Virus capsid protein. J Virol. 1992;66:6527–6532. doi: 10.1128/jvi.66.11.6527-6532.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Cubitt D, Hu J, Dai X, Treanor J, Matson DO, Pickering LK. Development of an ELISA to detect MX Virus, a human calicivirus in the Snow Mountain Agent genogroup. J Gen Virol. 1995;76:2739–2747. doi: 10.1099/0022-1317-76-11-2739. [DOI] [PubMed] [Google Scholar]
- Jiang X, Matson DO, Cubitt WD, Estes MK. Genetic and antigenic diversity of human caliciviruses (HuCVs) using RT-PCR and new ElAs. Arch Virology [supplement] 1996;12:251–262. doi: 10.1007/978-3-7091-6553-9_27. [DOI] [PubMed] [Google Scholar]
- Jiang X, Wang J, Estes MK. Characterization of SRSVs using RT-PCR and a new antigen ELISA. Arch Virol. 1995;140:363–374. doi: 10.1007/BF01309870. [DOI] [PubMed] [Google Scholar]
- Yoda T, Terano Y, Shimada A, Suzuki Y, Yamazaki K, Sakon N, Oishi I, Utagawa ET, Okuno Y, Shibata T. Expression of recombinant Norwalk-like virus capsid proteins using a bacterial system and the development of its immunologic detection. J Med Virol. 2000;60:475–481. doi: 10.1002/(sici)1096-9071(200004)60:4<475::aid-jmv17>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Yoda T, Terano Y, Suzuki Y, Yamazaki K, Oishi I, Utagawa E, Shimada A, Matsuura S, Nakajima M, Shibata T. Characterization of Monoclonal Antibodies Generated against Norwalk virus GII Capsid Protein Expressed in Escherichia coli. Microbiol Immunol. 2000;44:905–914. doi: 10.1111/j.1348-0421.2000.tb02582.x. [DOI] [PubMed] [Google Scholar]
- Cauchi MR, Doultree JC, Marshall JA, Wright PJ. Molecular characterization of Camberwell virus and sequence variation in ORF3 of small round-structured (Norwalk-like) viruses. J Med Virol. 1996;49:70–76. doi: 10.1002/(SICI)1096-9071(199605)49:1<70::AID-JMV12>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Hardy M, Tanaka TN, Kitamoto N, White LJ, Ball JM, Jiang X, Estes MK. Antigenic mapping of the recombinant Norwalk Virus capsid protein using monoclonal antibodies. Virology. 1996;217:252–261. doi: 10.1006/viro.1996.0112. [DOI] [PubMed] [Google Scholar]


