Abstract
Adult vascular smooth muscle cells (VSMCs) dedifferentiate in response to extracellular cues such as vascular damage and inflammation. Dedifferentiated VSMCs are proliferative, migratory, less contractile, and can contribute to vascular repair as well as to cardiovascular pathologies such as intimal hyperplasia/restenosis in coronary artery and arterial aneurysm. We here demonstrate the role of ubiquitin-like containing PHD and RING finger domains 1 (UHRF1) as an epigenetic master regulator of VSMC plasticity. UHRF1 expression correlated with the development of vascular pathologies associated with modulation of noncoding RNAs, such as microRNAs. miR-145 — pivotal in regulating VSMC plasticity, which is reduced in vascular diseases — was found to control Uhrf1 mRNA translation. In turn, UHRF1 triggered VSMC proliferation, directly repressing promoters of cell-cycle inhibitor genes (including p21 and p27) and key prodifferentiation genes via the methylation of DNA and histones. Local vascular viral delivery of Uhrf1 shRNAs or Uhrf1 VSMC-specific deletion prevented intimal hyperplasia in mouse carotid artery and decreased vessel damage in a mouse model of aortic aneurysm. Our study demonstrates the fundamental role of Uhrf1 in regulating VSMC phenotype by promoting proliferation and dedifferentiation. UHRF1 targeting may hold therapeutic potential in vascular pathologies.
Keywords: Cell Biology, Vascular Biology
Keywords: Cardiovascular disease, Epigenetics, Mouse models
Introduction
Cardiovascular diseases (CVDs) are the major cause of morbidity and mortality in the Western world. Among the different cell types involved in such pathologies, vascular smooth muscle cells (VSMCs) present a unique characteristic: they show a remarkable plasticity that enables them to switch between a differentiated and a dedifferentiated status in response to different stimuli (1). In a normal, healthy vessel, VSMCs have low levels of proliferation and apoptosis and are able to contract and maintain the vascular tone. However, in pathological conditions such as atherosclerosis and restenosis following surgery, VSMCs become increasingly proliferative and, as a consequence, also less contractile. Epigenetic modulation of gene expression is centrally involved in this process. Among the known epigenetic mechanisms taking place in vascular diseases, histone acetylation has been studied in depth (2, 3), whereas only a few studies have focused on histone and DNA methylation (4). In order to further investigate the importance of epigenetics in regulating VSMC phenotypic switch, we screened for the expression of key epigenetic-related genes during this process. Ubiquitin-like containing PHD and RING finger domains 1 (Uhrf1) came out as the most-modulated gene induced by PDGF-BB–mediated (PDGF-cytokine composed by two B subunits) dedifferentiation.
UHRF1, also known as NP95, does not have intrinsic enzymatic activity but is able to repress expression by directly interacting with DNA methyltransferase 1 (DNMT1) and histone deacetylase 1 (HDAC1) (5, 6), both markers of gene repression. Through its set and ring associated (SRA) domain, UHRF1 specifically binds to hemimethylated DNA and has an essential role in the maintenance of DNA methylation by recruiting DNMT1 to these DNA sites (7). Besides coupling with enzymes for DNA methylation, UHRF1 might also interact directly with modified histones (e.g., H3K9) to promote its dimethylation (8).
Another class of VSMC phenotype regulator recently identified is microRNAs (9), which mainly inhibit the expression of genes involved in cardiovascular pathologies (10, 11). Among various microRNAs, miR-145 was demonstrated to play a pivotal role in VSMC phenotypic switch and, consequently, in mouse and human vascular diseases (12–16). In the present study, we demonstrate that miR-145 regulates the epigenetic modulator Uhrf1, adding an additional layer of complexity to VSMC phenotype regulation. We further show the importance of Uhrf1 in modulating disease conditions, such as vascular stenosis, through in vivo knockdown and VSMC-specific KO mouse models.
Results
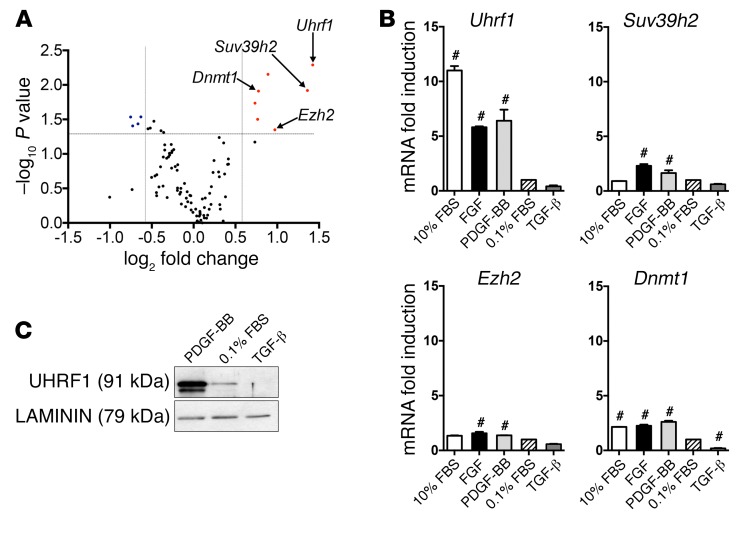
Uhrf1 is the most upregulated gene during VSMC dedifferentiation.
In order to identify genes involved in the regulation of VSMC phenotypic switching, we used quantitative real-time PCR (qRT-PCR) to profile the expression of 96 epigenetic genes, comparing quiescent cells (cultured in 0.1% FBS) with proliferating cells (cultured in 25 ng/ml PDGF-BB for 24 hours). Results show that the expression level of most epigenetic genes was not modified by PDGF-BB, whereas Uhrf1 was the most upregulated gene (Figure 1A). In order to validate this result, we compared the effect of prodifferentiation agents (TGF-β and starvation with 0.1% FBS), which trigger a contractile phenotype, and of PDGF-BB, FGF, and 10% FBS, which activate VSMC proliferation and migration, on the 7 most upregulated genes. Again, Uhrf1 was the gene most transcriptionally upregulated by all antidifferentiation agents (Figure 1B and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI96121DS1). Interestingly, rapamycin, a cell-cycle inhibitor and prodifferentiation agent used in clinical practice (17), is able to decrease Uhrf1 expression in VSMCs cultured under proliferating conditions (10% FBS) (Supplemental Figure 1B). Moreover, the UHRF1 protein level was elevated in proliferating VSMCs, but repressed after treatment with prodifferentiation agents (Figure 1C and Supplemental Figure 1C).
Figure 1. Regulation of Uhrf1 expression in response to mediators contributing to VSMC phenotypic switching.
(A) Analysis of the expression of 96 epigenetic genes in PDGF-BB–treated primary murine VSMCs with microfluidic cards. After global normalization, the statistical analysis was performed on 2 experimental duplicates compared with control cells (grown in a serum-deprived condition) and plotted versus the values of fold changes. A gene was arbitrarily considered modulated if fold induction was <0.6 to >2 compared with control and if the P value of such a difference was less than 0.05. (B) RT-qPCR gene expression analysis of VSMCs treated with different stimuli. The control sample (0.1% FBS) was set as 1. Results are the average of at least 3 independent experiments, and error bars indicate SD. #P < 0.05. (C) Representative Western blot showing UHRF1 levels in VSMCs treated with PDGF-BB (25 ng/ml) or TGF-β (10 ng/ml). Unpaired 2-tailed Student’s t test was used to compare means for A, and 1-way ANOVA with Tukey’s multiple comparisons test was used for B. #Adjusted P < 0.05.
Time- and dose-dependent experiments were performed using PDGF-BB (Supplemental Figure 1, D–G) and TGF-β (Supplemental Figure 2, A and B). Interestingly, Uhrf1 expression inversely correlated temporally and in a dose-dependent manner with the VSMC differentiation markers Myh11 and Acta2. Furthermore, using a public RNA sequencing data set (18), UHRF1 upregulation was also confirmed in PDGF-BB–treated human VSMCs (Supplemental Figure 2C). Taken together, these data demonstrate the direct association between Uhrf1 expression and VSMC differentiation status.
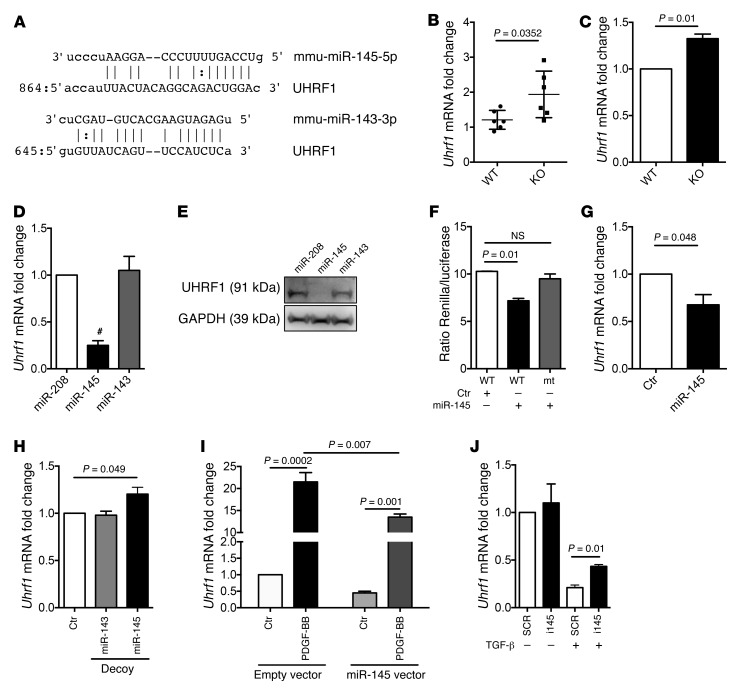
miR-145 contributes to Uhrf1 expression.
miR-143 and miR-145 are master regulators of VSMC biology (12–16). Interestingly, an in silico analysis predicted Uhrf1 as a putative miR-143 and miR-145 target gene (Figure 2A). In order to validate this, we first measured Uhrf1 RNA levels in vessels of miR-143– and miR-145–KO mice (19) as well as in miR-143– and miR-145–KO VSMCs in vitro. We found an increased expression of this gene in both cases (Figure 2, B and C). Next, miR-143– and miR-145–KO VSMCs (12) were transduced with adenoviruses expressing miR-208, miR-143, or miR-145. Decreased UHRF1 RNA and protein were observed after miR-145 but not miR-143 or miR-208 overexpression (Figure 2, D and E). The direct binding of miR-145 on Uhrf1 mRNA was then validated through a luciferase assay in which its 3′ UTR sequence was cloned downstream to the reporter Renilla gene (Figure 2F). miR-145 inhibited luciferase activity when the reporter construct included the WT but not the mutated (mt) putative microRNA seed sequence. Gain- and loss-of-function experiments using a lentivirus expressing miR-145, an inhibitor sponge sequence (19), or an anti-miR oligonucleotide confirmed that Uhrf1 expression is inversely correlated to the level of miR-145 (Figure 2, G and H and Supplemental Figure 2D). Finally, since miR-145 expression is downregulated upon PDGF-BB stimulation (16) and upregulated upon TGF-β treatment (19), we evaluated whether Uhrf1 modulation during PDGF-BB (upregulation) or TGF-β (downregulation) treatments might be at least partially mediated by modulation of miR-145 expression. To this aim, miR-145 gain- and loss-of-function experiments were performed on VSMCs incubated with PDGF-BB or TGF-β for 18 hours. As expected, PDGF-BB treatment induced Uhrf1 statistically less in VSMCs overexpressing miR-145, whereas the ability of TGF-β to decrease Uhrf1 expression was statistically less efficient in VSMCs treated with a miR-145 inhibitor when compared with VSMCs transduced with a control lentivirus or transfected with a scrambled (SCR) oligonucleotide (Figure 2, I and J).
Figure 2. miR-145–mediated regulation of Uhrf1.
(A) Putative binding sites for miR-143 and miR-145 on the 3′ UTR of Uhrf1. (B) Uhrf1 expression in aortas of miR-143– and miR-145–KO mice, measured by RT-qPCR. (C) Uhrf1 expression measured by RT-qPCR in 3 different preparations of VSMCs isolated from miR-143– and miR-145–KO mice, cultured in medium with 10% FBS. (D and E) Representative RT-qPCR RNA analysis and immunoblot of the target gene UHRF1 in miR-143– and miR-145–KO VSMCs transduced with adenovirus expressing miR-208, miR-145, or miR-143 and grown in medium with 10% FBS. (F) Luciferase reporter assay on A7r5 cells stably expressing miR-145 (Ctr, cells transduced with the empty vector) with a Renilla reporter gene linked to the WT or mutated (mt) Uhrf1 3′ UTR. (G) Uhrf1 expression in WT VSMCs transduced with a lentivirus expressing miR-145 (Ctr, cells transduced with the empty vector and cells cultured in medium with 10% FBS). (H) Uhrf1 expression in WT VSMCs transduced with lentivirus expressing sponge sequences (Decoys) targeting miR-143 or miR-145 and empty vector, as measured by RT-qPCR (cells cultured in medium with 10% FBS). (I) Uhrf1 expression in WT VSMCs transduced with a lentivirus expressing miR-145 and treated with PDGF-BB (25 ng/ml) (Ctr, cells treated with vehicle). (J) Uhrf1 expression in WT VSMCs transfected with an anti–miR-145 LNA (i145) oligo and treated with TGF-β (10 ng/ml) (SCR, cells treated with scrambled oligo). All results are the average of at least 3 independent experiments and error bars indicate SD. To compare means, an unpaired 2-tailed Student’s t test was used for B, C, D, G, and J, whereas 1-way ANOVA with Tukey’s multiple comparisons t test was used for F, H, and I. #P < 0.05. Adjusted P value is shown in F, H, and I.
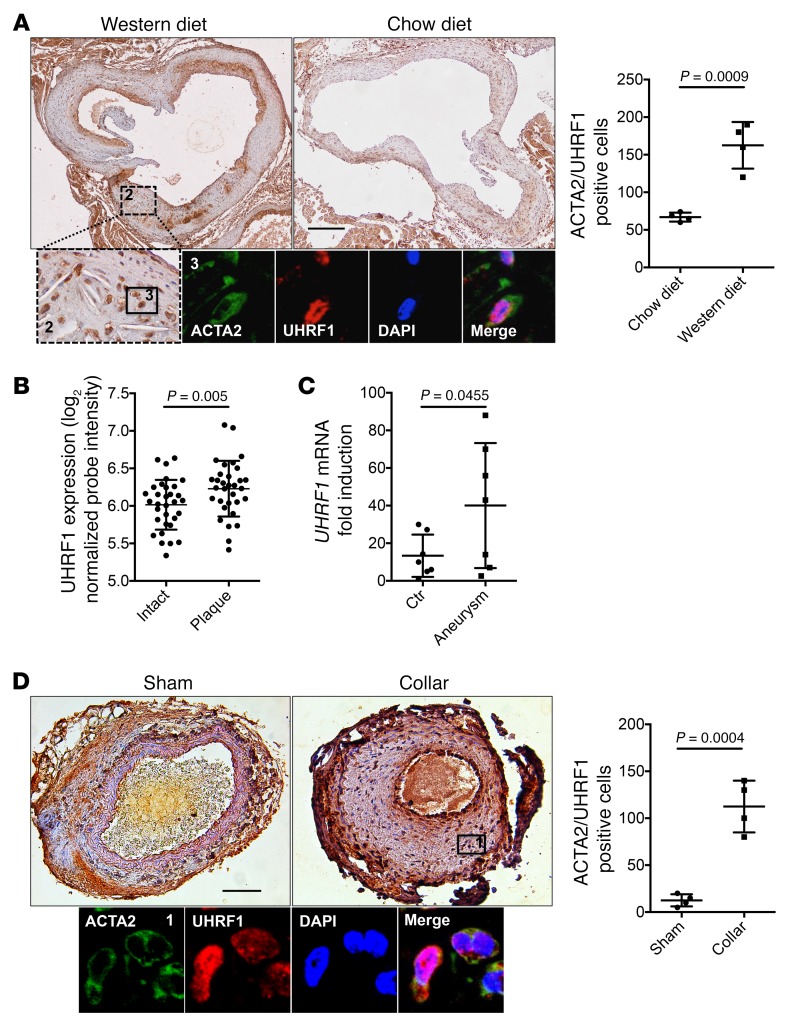
UHRF1 is upregulated in diseased vascular tissues.
To determine whether VSMCs express UHRF1 in vivo, we performed cell tracing using a VSMC-specific reporter mouse model. Healthy adult VSMCs express very little UHRF1, so we triggered its expression by performing transverse aortic constriction (TAC) in mice expressing the tomato-red (TR) reporter gene under the SM-MYH11 promoter. This confirmed that VSMCs express UHRF1 (Supplemental Figure 3, A and B).
To determine whether UHRF1 is regulated in vascular diseases, we investigated its expression in normal and pathological vascular samples. First, we sought to assess whether UHRF1 is deregulated in atherosclerotic lesions. Cross sections of aorta were prepared from ApoE–/– mice fed on a normal chow diet (CD) or a hypercholesterolemic Western diet (WD). As expected, CD-fed mice showed no or small atherosclerotic lesions, whereas WD-fed mice exhibited lesions that progressed in size. Immunohistochemistry revealed a strong increase in the number of cells expressing UHFR1 in the plaques of WD-fed mice (Figure 3A). Colocalization experiments of UHRF1 with ACTA2 showed that UHRF1 is principally, but not exclusively, expressed in ACTA2-positive cells in atherosclerotic plaques (Figure 3A and Supplemental Figure 3, B and C). Taking advantage of the SM-MYH11 TR mouse model, we confirmed that ACTA2-positive cells are indeed VSMCs (TR positive) (Supplemental Figure 3D).
Figure 3. UHRF1 is upregulated during atherosclerosis development and vascular injury.
(A) Representative immunostaining and quantification for UHRF1 on aortic cross sections of ApoE–/– mice fed with chow and Western diets (colorimetric images showing UHRF1 in brown, immunofluorescence showing UHRF1 in green and ACTA2 in red). Scale bar: 1 mm. Labels 2 and 3 indicate the specific areas from where the insets have been obtained. (B and C) UHRF1 expression analysis on atheroma plaque versus paired macroscopically intact tissue of 32 patients (GSE43292). The y axis indicates normalized UHRF1 probe intensity (ID 8024900) measured by Affymetrix Human Gene 1.0 ST Array (B) or abdominal aortic aneurytic vessels measured by RT-qPCR (C). (D) Representative immunostaining and quantification for UHRF1 on carotid sections of ApoE–/– mice undergoing collar placement (Collar) or a sham procedure (Sham) (colorimetric images showing UHRF1 in brown, immunofluorescence showing UHRF1 in green and ACTA2 in red). Scale bar: 100 μm. Error bars indicate SD. To compare means, we used the Wilcoxon matched paired test in B and the unpaired 2-tailed Student’s t test in A, C, and D.
To corroborate these results, we looked for UHRF1 levels in a public data set of plaques from atherosclerotic patients (20). We found that there was a statistically significant increase in UHRF1 gene expression in the human setting (Figure 3B). Furthermore, we compared UHRF1 levels in aortic samples taken from patients undergoing open abdominal aortic aneurysm (AAA) repair with those in explant samples from organ donors (12). We found a marked and significant increase in UHRF1 mRNA levels in pathological vessels (Figure 3C). These results were confirmed by systematically reviewing a public data set of patients with brain aneurysm (21) (Supplemental Figure 3E) and a published data set of a murine model of aneurysm (22) (Supplemental Figure 3F).
We then analyzed the expression levels of DNMT1 and HDAC1, 2 fundamental UHRF1 interactors, documented in the above-mentioned databases. We found a statistical increase of DNMT1 in all cases (Supplemental Figure 4, A–C). Interestingly, a linear correlation between DNMT1 and UHRF1 was observed in human atherosclerotic plaques and aneurysms (Supplemental Figure 4, D and E), but not in the mouse model of aneurysm (Supplemental Figure 4F). HDAC1 expression was not statistically increased in these pathological specimens, except for atherosclerotic plaques (Supplemental Figure 4, G–I).
We next aimed to assess whether Uhrf1 plays a role in a murine model of vascular injury. Experimental arterial intimal hyperplasia was induced by placement of a silicone collar around the carotid artery (23), and samples were taken 30 days after surgery. A significant difference in UHRF1 expression was observed in the stented arteries when compared with the nonstented carotid segments (Figure 3D). Moreover, colocalization experiments of UHRF1 with ACTA2 further demonstrated that UHRF1 was expressed by VSMCs in stenotic vessels and sham controls (Figure 3D and Supplemental Figure 3G).
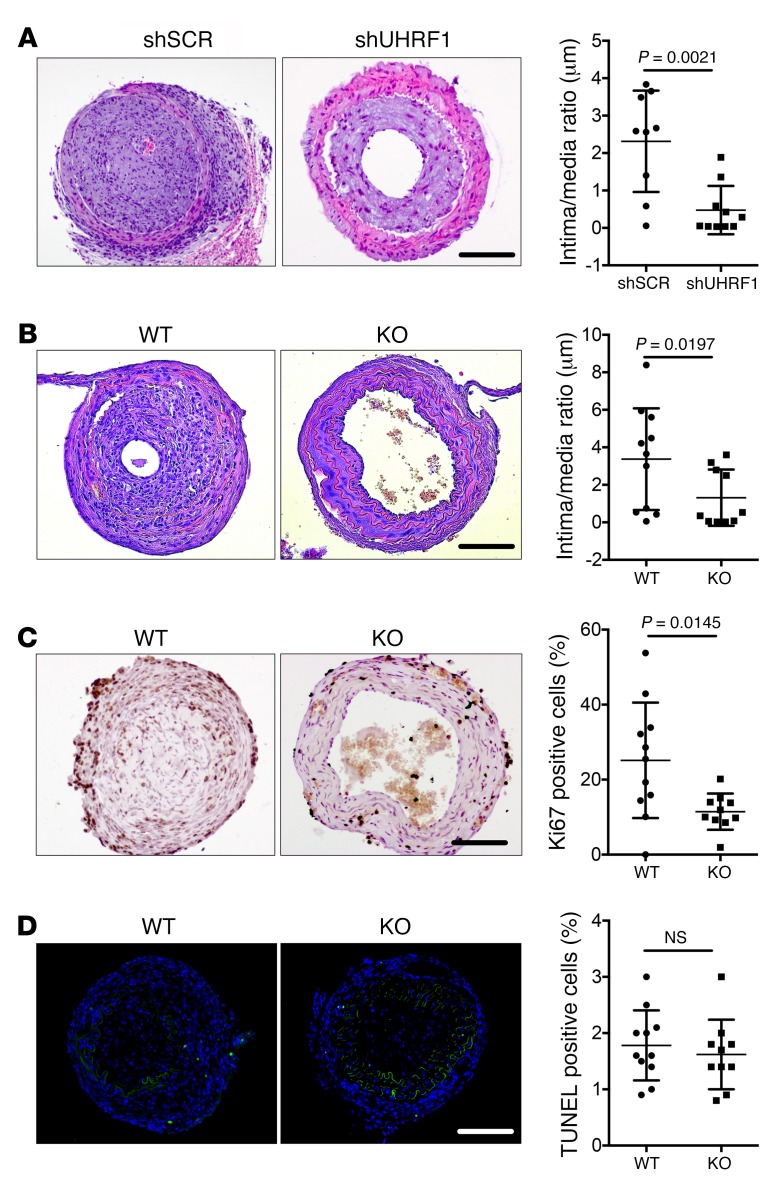
Localized reduction of Uhrf1 improves stenosis development.
To test the hypothesis that Uhrf1 plays an important role in VSMC plasticity in vivo, we examined the antiproliferative effect of specifically knocking down Uhrf1 in a model of stenosis induced by perivascular carotid collar placement in ApoE−/− mice (23). We performed local delivery of scrambled shRNA (shSCR) or Uhrf1-shRNA (shUHRF1) viruses with 2 different types of shRNA sequences and determined the effects on the vessels 4 weeks after collar placement. Stented carotids treated with a control virus showed strong intimal hyperplasia, whereas delivery of the Uhrf1 shRNA virus markedly decreased Uhrf1 expression and, consequently, the neointimal area (Figure 4A and Supplemental Figure 5A). To further corroborate this result, we generated a conditional KO mouse model in an ApoE–/– background in which Uhrf1 could be specifically deleted in VSMCs, obtaining the strains ApoE–/–UHRF1fl/flSM-MYH11-cre/ERT2 (KO) and ApoE–/–UHRFwt/wtSM-MYH11-cre/ERT2 (WT) (Supplemental Figure 5, B–D). When stenosis formation was induced in these models, Uhrf1-KO mice exhibited a decreased intimal-to-medial ratio when compared with control mice (WT) 4 weeks after vascular injury (Figure 4B and Supplemental Figure 5, E–I). In addition, Uhrf1-KO mice displayed decreased vascular cell proliferation in response to injury (Figure 4C), whereas apoptotic rate was unchanged (Figure 4D and Supplemental Figure 5J), as measured by Ki67 and TUNEL staining.
Figure 4. Effect of Uhrf1 modulation in an in vivo model of restenosis.
(A) Representative H&E staining and quantification of carotid sections of ApoE–/– mice infused with shSCR or shUHRF1 viruses undergoing collar placement. (B) Representative H&E staining and experimental quantification of carotid sections of Uhrf1 WT (ApoE–/–UHRF1wt/wtcre/ERT2) and KO (ApoE–/–UHRF1fl/flcre/ERT2) mice undergoing collar placement. (C) Representative immunostaining and quantification for Ki67 on carotid sections of Uhrf1 WT and KO mice undergoing collar placement (Ki67 in brown). (D) Representative TUNEL staining and quantification of carotid sections from Uhrf1 WT and KO mice undergoing collar placement (TUNEL+ cells in green). Error bars indicate SD. To compare means, we used the Mann Whitney U test in A and B and the unpaired 2-tailed Student’s t test in C and D. Scale bars: 100 μm.
Uhrf1 regulates proliferation and migration in cultured VSMCs.
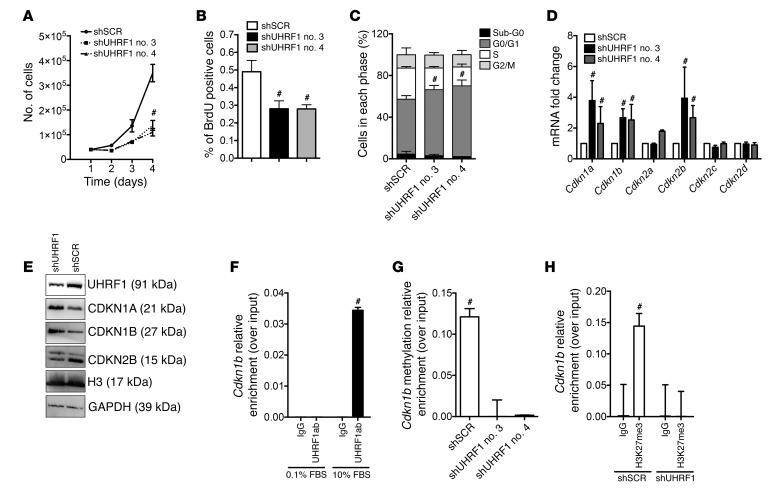
In order to dissect the mechanism by which Uhrf1 affects VSMC phenotype, we performed Uhrf1 gain- and loss-of-function experiments in primary VSMCs and studied the effects on proliferation and migration. Uhrf1-silenced VSMCs showed a reduced proliferation rate, as determined by growth curve and BrdU incorporation assay (cells cultured in 10% FBS) (Figure 5, A and B), whereas UHRF1 upregulation determined an increase of proliferation in WT and Uhrf1-silenced cells (Supplemental Figure 6, A and B). Moreover, Uhrf1 downregulation was able to inhibit the effect of PDGF-BB on cell proliferation, as measured by a BrdU incorporation assay (Supplemental Figure 6C). Interestingly, UHRF1 overexpression also increased the proliferation rate of miR-145–overexpressing VSMCs (Supplemental Figure 6D), indicating that part of the phenotypic effect of miR-145 could be mediated by Uhrf1. The cell cycle was impaired in Uhrf1-silenced cells. In particular, a significantly higher percentage of cells were in the G0/G1 phase, with a concomitant reduction of cells in the S phase (Figure 5C). Subsequently, the mRNA levels of several cyclin-dependent kinase inhibitors were measured. We found specific upregulation of Cdkn1a and Cdkn1b, 2 inhibitors of the cell cycle, a result in line with the decrease of proliferation (Figure 5D). UHRF1 upregulation, on the other hand, determined a decrease in Cdkn1a and Cdkn1b mRNA in WT cells as well as in Uhrf1-silenced cells (Supplemental Figure 6, E and F). Western blotting analysis corroborated the results on CDKN1A and CDKN1B (Figure 5E and Supplemental Figure 6G). Since Cdkn1a has been already shown to be directly regulated by UHRF1 (24), we therefore conducted a ChIP analysis on the promoter of Cdkn1b, using an anti–UHRF1 antibody. As expected, there was increased binding of UHRF1 to the Cdkn1b promoter in proliferating VSMCs (cultured in 10% FBS) (Figure 5F). Since UHRF1 binding is associated with increased 5-methylcytosine (5-mC) DNA content (25), we studied the Cdkn1b promoter with a methyl capture technique. Indeed, 5-mC enrichment at the promoter was decreased in Uhrf1-silenced VSMCs (Figure 5G). Methylation of 5-cytosine on promoters is associated with an increase of the histone repressive mark H3K27me3 (26), we sought to determine whether, together with a decreased level of DNA methylation, Uhrf1 knockdown caused a reduction of H3K27me3. We found that, indeed, Uhrf1 knockdown combined with decreased DNA methylation reduced H3K27me3 (Figure 5H). Finally, suppression of Uhrf1 with specific shRNAs was associated with a significant decrease of VSMC migration, as assessed by scratch assay (Supplemental Figure 6H).
Figure 5. Uhrf1 regulates VSMC plasticity in vitro.
(A) Proliferation curve of VSMCs stably expressing shSCR or shUHRF1. To determine growth curves, 2 × 104 cells/ml were plated in 6-well plates and cultured with 10% FBS. The number of viable cells was counted for 3 days. (B) Proliferation measured by BrdU incorporation in shSCR and shUHRF1 SMCs. (C) Cell-cycle analysis of shSCR- and shUHRF1-expressing SMCs. (D and E) RNA and protein analysis of different cyclins involved in VSMC phenotypic switch. (F) ChIP analysis showing UHRF1 enrichment on the Cdkn1b promoter in proliferating cells. Data are presented as mean relative enrichment over input ± SD of 3 biological repeats. (G) Results from methyl-ChIP experiments showing reduction of methylation at the promoter in the absence of Uhrf1. Data are presented as mean relative enrichment over input ± SD of 3 biological repeats. (H) ChIP showing the enrichment of H3K27me3 in control VSMCs compared with UHRF1-silenced cells. Data are presented as mean relative enrichment over input ± SD of 3 biological repeats. If not otherwise stated, the results are the average of at least 3 independent experiments and error bars indicate SD. To compare means, 1-way ANOVA with Tukey’s multiple comparisons test was used. #Adjusted P < 0.05.
Reduction of UHRF1 results in an increased contractile phenotype.
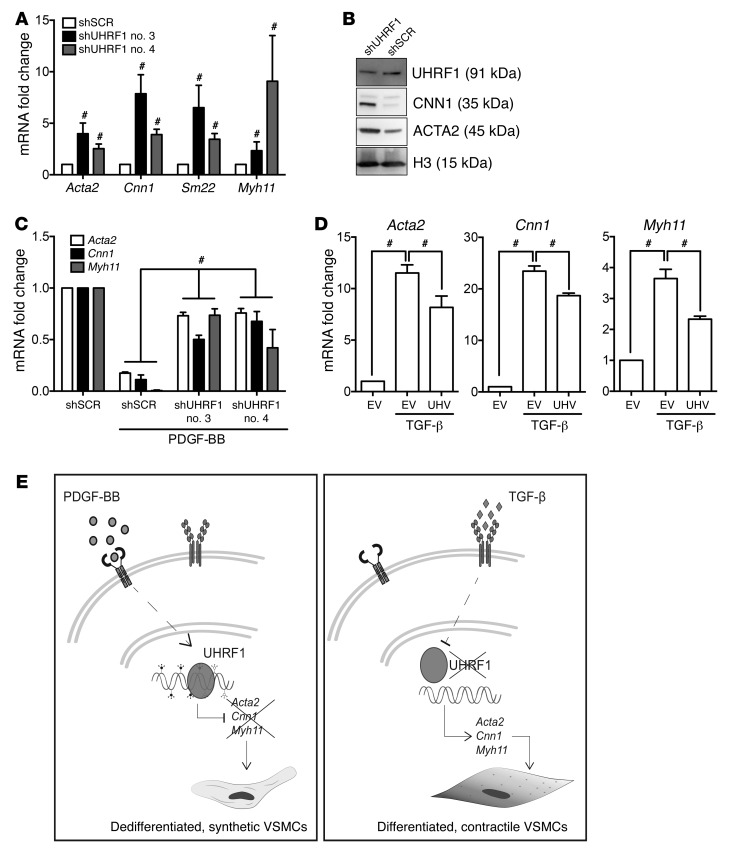
Since Uhrf1 expression inversely correlated with VSMC-specific gene expression (Supplemental Figure 1, D–G), it is conceivable that it could directly repress the expression of these genes. Indeed, a significant increase in expression of key VSMC differentiation genes was observed upon Uhrf1 knockdown at the mRNA level in VSMCs cultured in a proliferating condition (namely, in 10% FBS) (Figure 6A). Western blotting for representative proteins (CNN1, ACTA2, and MYH11) confirmed these results at the protein level (Figure 6B and Supplemental Figure 7A). Moreover, compared with shSCR-transduced cells, Uhrf1 knockdown also decreased the expression of osteopontin (Opn), a marker associated with the synthetic VSMC phenotype (Supplemental Figure 7B), and increased the expression of Srf, Tet2, Myoc, and Mrtf-a (Supplemental Figure 7C), but did not significantly change the expression of other synthetic markers such as Klf4 (data not shown). On the other hand, overexpression of UHRF1 caused decreased expression of key VSMC differentiation genes in serum-deprived VSMC cultures (Supplemental Figure 7D). A similar effect could be detected in VSMCs overexpressing miR-145 (Supplemental Figure 7E), confirming the inverse correlation between miR-145 and Uhrf1 in phenotype regulation. Interestingly, when treated with PDGF-BB for 24 hours, VSMCs in which Uhrf1 expression was crippled by a specific shRNA showed a significant increase in the level of some differentiation markers when compared with scrambled shRNA–expressing (shSCR-expressing) cells (Figure 6C). Conversely, UHRF1-overexpressing VSMCs treated with TGF-β for 24 hours showed a significant decrease in the expression level of VSMC differentiation markers compared with cells expressing an empty vector (Figure 6D). Altogether, these results demonstrate that Uhrf1 plays a crucial role in VSMC phenotype modulation by mediating the dedifferentiation triggered by PDGF-BB as well as by antagonizing the differentiation triggered by TGF-β, as summarized in Figure 6E.
Figure 6. Role of Uhrf1 on VSMC function and differentiation.
(A and B) RT-qPCR and Western blot showed an increase in several differentiation markers in Uhrf1-silenced VSMCs (cells cultured in medium with 10% FBS). (C) VSMC differentiation marker expression in Uhrf1-silenced cells treated with PDGF-BB (cells cultured in medium with 0.1% FBS). (D) VSMC differentiation marker expression in cells overexpressing human UHRF1 treated with TGF-β (cells cultured in medium with 0.1% FBS). EV, empty vector; UHV, UHRF1 vector. (E) Schematic illustration representing data shown in A–D. The results are the average of at least 3 independent experiments. Error bars indicate SD. To compare means, unpaired 2-tailed Student’s t test was used for D and 1-way ANOVA with Tukey’s multiple comparisons t test was used for A and C. #P < 0.05 (P value is adjusted only in A and C).
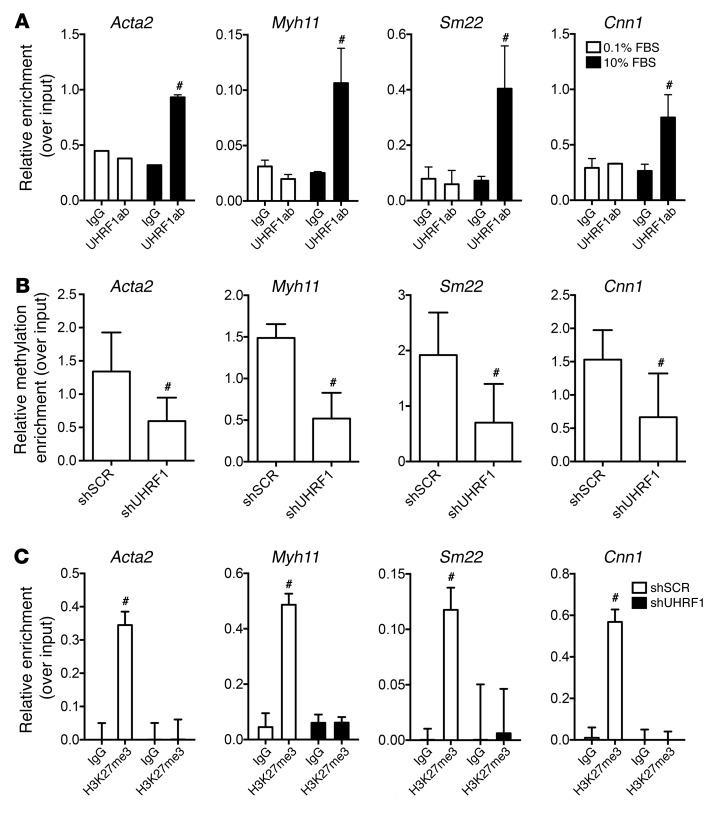
The effects of Uhrf1 on VSMC phenotypic modulation suggested the possibility of a direct interaction between the protein and the promoter of at least some critical VSMC differentiation genes. To test this hypothesis, we performed ChIP experiments using an anti–UHRF1 antibody to pull down protein/DNA complexes of proliferative (10% FBS) or resting (0.1% FBS) VSMCs. Compared with resting cells, an enrichment of the genomic regions bound to UHRF1 was observed for the genes Acta2, Cnn1, Myh11, and Sm22 in proliferative VSMCs (Figure 7A). These data were consistent with a decrease of UHRF1 expression upon serum-deprived VSMC differentiation (Figure 1C and Supplemental Figure 1C). In line with the previous results (Figure 7A), we found that the 5-mC content of the promoters of these genes depended upon the activity of UHRF1 (Figure 7B). In line with the Cdkn1b promoter results, Uhrf1 knockdown caused a decrease in H3K27me3, confirming transcriptional reactivation of VSMC-specific genes (Figure 7C).
Figure 7. UHRF1 binding on VSMC differentiation genes.
(A) ChIP assay showing UHRF1 enrichment at the Myh11, Acta2, Cnn1, and Sm22 promoters during VSMC dedifferentiation. Mouse primary VSMCs were cultured in a serum-deprived condition (0.1% FBS) or with 10% FBS. (B) Methyl ChIP assay showing reduced methylation at the Myh11, Acta2, Cnn1, and Sm22 promoters in the absence of Uhrf1. (C) ChIP assay showing H3K27me3 enrichment at the Myh11, Acta2, Cnn1, and Sm22 promoters in the absence of Uhrf1. Data are presented as mean relative enrichment over input ± SD of 3 biological repeats. To compare means, unpaired 2-tailed Student’s t test was used. #P < 0.05.
RNA interference of Uhrf1 and gene expression profiling reveal direct regulation of the TGF-β pathway by Uhrf1.
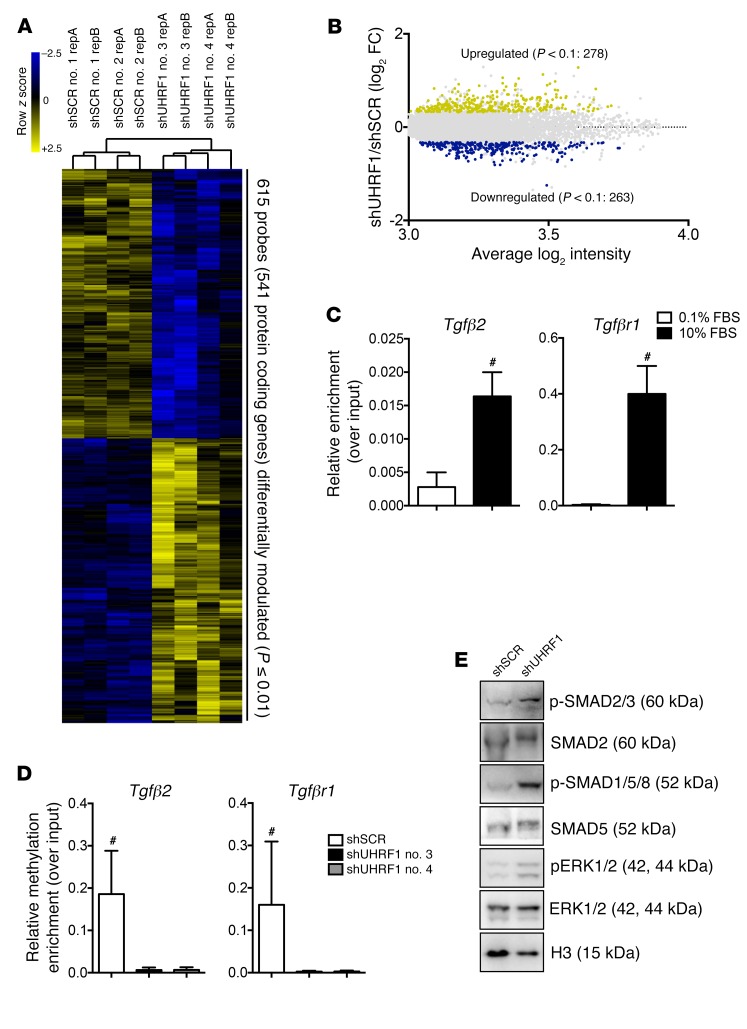
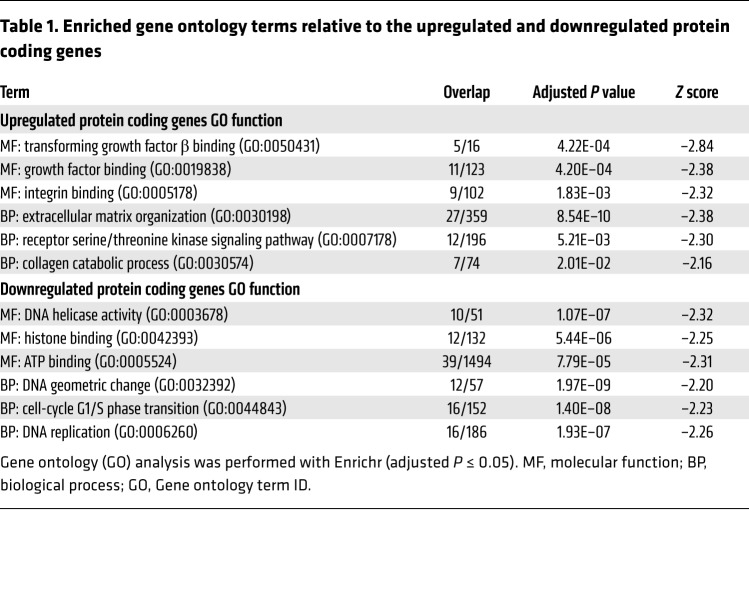
To gain deeper insight into the biological role of Uhrf1 in VSMCs, we suppressed endogenous Uhrf1 expression and carried out global gene expression profiling with an Illumina mouse array. We observed perturbed expression of 541 genes (>1.3-fold change, P < 0.01), of which 278 genes were upregulated and 263 genes were downregulated (Figure 8, A and B). Perturbed genes were subjected to hierarchical cluster analysis, and different sets of gene ontology (GO) terms (i.e., biological processes and molecular functions) were identified (adjusted P ≤ 0.05). As expected, downregulated genes were enriched in GO terms related to cell proliferation and cell-cycle regulation, whereas upregulated genes were enriched in terms involving extracellular matrix organization (Table 1). Moreover, TGF-β signaling pathway was indeed the top enriched molecular function in the upregulated genes (adjusted P = 4.22 × 10–4). Genes encoding proteins involved in the activation of TGF-β pathway signaling, such as Tgfβ2 and Tgfβr1, and genes downstream from the TGF-β signaling pathway, such as Col1a1 and Cdkn1a, were present within the perturbed gene list. For a subset of transcripts included in the most-represented categories, such as extracellular matrix (ECM) remodeling and TGF-β signaling pathway, quantitative RT-qPCR was used to validate microarray data. The results of this validation on 6 biological replicates demonstrated a positive correlation with the microarray data for the analyzed genes (Supplemental Figure 7F and Supplemental Figure 8A). Moreover, ChIP experiments using an anti–UHRF1 antibody on proliferative VSMCs and MeCapture assay on Tgfβ2 and Tgfβr1 promoters suggested there was direct regulation of these genes by UHRF1, which bound to their promoters, modulating their methylation status (Figure 8, C and D). Western blotting analysis showed increased SMAD 2/3 and SMAD 1/5/8 phosphorylation levels, suggesting consequent activation of the TGF-β signaling pathways (Figure 8E and Supplemental Figure 8B). Furthermore, Uhrf1-silenced cells treated for 48 hours with a specific inhibitor of the TGF-β pathway (SB431542) expressed lower levels of VSMC differentiation markers and of representative TGF-β pathway-specific targets (Cola1) compared with vehicle-treated cells (Supplemental Figure 8C). These data confirm the involvement of Uhrf1 in the downstream regulation of the TGF-β pathway.
Figure 8. Effects of Uhrf1 absence in vitro.
(A) Hierarchical clustering heat map of 615 probes differentially expressed between shUHRF1 and shSCR (P ≤ 0.1, –1.3 ≤ fold-change ≥ 1.3). (B) Dot plot, with blue dots representing downregulated protein-coding genes (263) and yellow dots representing upregulated protein-coding genes (278) in shUHRF1 and shSCR cells. (C) ChIP assay showing UHRF1 enrichment at the Tgfβ2 and Tgfβr1 promoters in proliferating VSMCs (cells cultured with 10% FBS). Data are presented as mean relative enrichment over input ± SD of 3 biological repeats. (D) Methyl ChIP assay showing reduced methylation at the Tgfβ2 and Tgfβr1 promoters in the absence of UHRF1 (cells cultured with 10% FBS). Data are presented as mean relative enrichment over input ± SD of 3 biological repeats. (E) Representative Western blots showing activation of the canonical TGF-β pathway in Uhrf1-silenced VSMCs. Error bars indicate SD. To compare means, we used unpaired 2-tailed Student’s t test in C and 1-way ANOVA with Tukey’s multiple comparisons t test in D. #P < 0.05 (P value is only adjusted in D).
Table 1. Enriched gene ontology terms relative to the upregulated and downregulated protein coding genes.
Uhrf1-KO improves aortic wall homeostasis and function.
Since we found UHRF1 upregulated in human and mouse aneurysms (Figure 3, C–E), we evaluated whether there was an inverse correlation between genes modulated by UHRF1 (Figure 8A) and genes deregulated in published aneurysm data sets (Figure 3, D and E). More than 30% of genes affected by Uhrf1 silencing were statistically deregulated in the mouse aneurysm data set (180 of 541, P ≤ 0.1). Spearman’s rank correlation coefficient indicated a weak-to-moderate anticorrelation (r = –0.37). As expected, 119 genes were negatively correlated in the 2 data sets (Supplemental Figure 9).
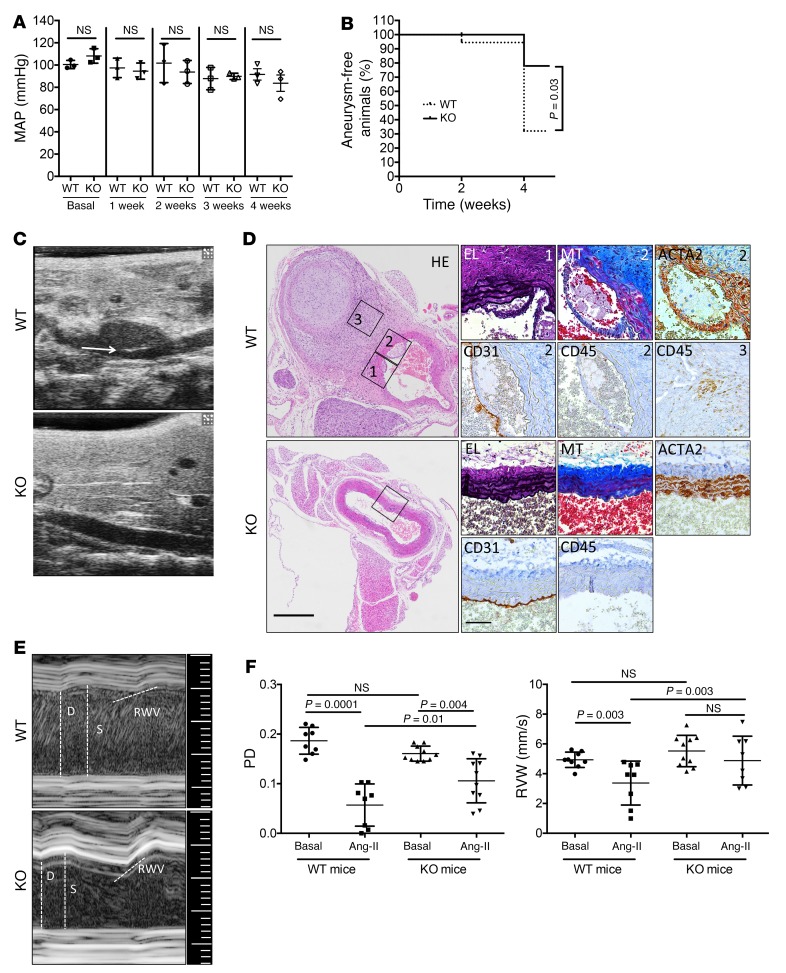
Next, to determine whether Uhrf1-KO mice have any major structural defects of the aorta in basal conditions, gene deletion was induced at different developmental stages. No major structural alteration of the aorta in P7 (1 week), P21 (3 weeks), and adult (8 weeks) mice could be detected (Supplemental Figure 10, A–F and Supplemental Figure 11, A–D). In adult animals, smooth muscle (SM) cell number and size (Supplemental Figure 11, E and F) were normal. Similarly, mean arterial blood (MAP) pressure in adult animals was comparable to that in control WT animals (Figure 9A).
Figure 9. Effects of Uhrf1 absence in vivo.
(A) Mean arterial blood pressure (MAP) of WT and KO mice as measured by telemetry. Measurements started before tamoxifen injection (Basal) and were continued for 4 weeks after induction. Symbols represent individual mice; horizontal bars indicate means (n = 3 KO and 3 WT animals). (B) Survival curve for the appearance of aneurysm formation in WT and KO animals, y axis shows the percentage of mice that did not develop the pathology (n = 11 KO and 10 WT animals). (C) Representative bidimensional echo-Doppler images of abdominal aortas of Ang-II–infused KO (Ang-II) mice compared with controls (WT). The white arrow indicates an example of aortic rupture in WT control. (D) Representative cross sections of aneurysms forming in abdominal aortas of WT and KO mice. HE, H&E; MT, ECM deposition by Masson Trichrome staining; EL, elastin by van Gieson’s staining; ACTA2, immune-staining for smooth muscle actin; CD31, immune-staining for platelet and endothelial cell adhesion molecule 1 (PECAM1); CD45, immune-staining for leukocyte common antigen (LCA). Scale bars 400 μm (big panels) and 50 μm (small panels). Labels 1, 2 and 3 indicate the specific areas from where the insets have been obtained. (E) Representative echo-Doppler images of abdominal aortas of Ang-II–infused KO mice compared with controls (WT), and (F) quantitative analysis of the pulse diameters (PD) and radial wall velocity (RWV). To compare groups, we used the log-rank Mantel-Cox test in B and unpaired 2-way ANOVA in A and F. Adjusted P value shown for A and F. NS indicates not statistically significant.
Next, we tested whether Uhrf1-KO could improve aortic wall homeostasis in vivo, triggering the formation of suprarenal aneurysms in ApoE-deficient mice via angiotensin II (Ang-II) infusion (27). At the 4-week endpoint, WT and KO mice presented with remodeling and dilatations along segments of the aorta. Although the average diameter of the KO vessel was lower compared with controls (25% reduction in diastole), no statistical differences were observed between the 2 groups of mice (Supplemental Figure 12, A and B). One mouse (1/10) in the control group died because of aortic rupture before the end of the experiment (second week post–Ang-II infusion). None (0/11) of the mice in the KO group died before the end of the experiment. The overall analysis of event appearance measured by echo-Doppler analysis showed a marked reduction of aneurysm formation in KO mice compared with WT mice (Figure 9B). Furthermore, at the end of the 4-week experiment, aortic rupture was observed, as expected, in 30% of WT mice (28), whereas there were no ruptures in KO animals (Figure 9C). The data obtained by echo-Doppler analysis was confirmed by evaluating event appearance in aorta cross sections of WT and KO mice infused with Ang-II. WT animals showed rupture of the vascular wall with immune cell infiltration, whereas none of these events was observed in KO mice (Figure 9D). SMAD pathway activation in Ang-II–infused mice was also assessed. Although not statistically significant, a trend toward an increase in phosphorylated SMAD 2/3 level in KO animals compared with WT animals (Supplemental Figure 12C) was observed. We also measured pulse diameter (PD) as a functional analysis for assessing arterial wall weakening and elasticity of the vessel wall (29). Mean PD of aneurysm-prone aortic areas 4 weeks after Ang-II infusion was significantly lower in WT and KO mice when compared with baseline (Figure 9, E and F, left panel, and Supplemental Figure 12D). However, at 4 weeks, mean PD in KO mice was consistently elevated when compared with WT mice (Figure 9, E and F, left panel, and Supplemental Figure 12D), confirming a protective role for the absence of Uhrf1 in this pathological model.
These functional data were then corroborated by measuring radial wall velocity (RWV), a parameter associated with the wall motility speed during the cardiac contraction phase. Reductions of RWV are usually caused by increased infiltration of inflammatory cells accompanied by changes in the viscoelastic characteristics of the arterial wall (30). When compared with the basal condition, WT mice showed a marked reduction in RWV, whereas KO mice did not (Figure 9, E and F, right panel, and Supplemental Figure 12D), thus strengthening the aneurysm penetrance and PD results, which suggested a negative role for Uhrf1 in the vessel response to stress.
Discussion
In the present study, we demonstrate that Uhrf1 plays a fundamental role in the regulation of VSMC plasticity in physiology and in vascular disease. VSMC gene transcription plays an important role in vascular development and pathophysiology and is finely regulated by epigenetic mechanisms (31). The binding of serum response factor (SRF) and myocardin to CArG box DNA sequences in VSMC gene promoters, for instance, is dependent on the structure of the chromatin. In particular, acetylation of lysine 9, methylation of lysine 4, and dimethylation of lysine 79 on histone H3 are positive regulators of SRF binding (2). Conversely, class II HDACs interact with a myocardin domain and suppress VSMC activation (2). HDACs have also been implicated in the proliferation of VSMCs, modulating histone acetylation on genes controlling the cell cycle (32). Furthermore, it has been shown that the 5-cytosine hydroxymethylation (5-hmC) enzyme TET2 induces Myocd and Srf expression together with other genes involved in VSMC contractility, while Klf4, one of the most important dedifferentiation genes, is concomitantly repressed (33). These reports highlight the complexity of the epigenetic landscape regulating VSMC plasticity and show that understanding of the global picture is still limited.
Here, we contribute to the comprehension of the regulation of VSMC gene transcription by identifying the involvement of UHRF1 in the modulation of DNA methylation and histone modifications in VSMCs. We demonstrate that UHRF1 directly binds to DNA, promoting DNA and histone modifications at the promoters of genes that are hallmarks of SMC differentiation as well as at key inhibitors of the cell cycle. UHRF1 binding was strongly increased in dedifferentiated VSMCs, and UHRF1 was found to regulate the expression of the key transcription factors Srf and Myocd. Interestingly, we also show that Uhrf1 repression triggers the upregulation of Tet2, which was shown to positively modulate VSMC contraction, suggesting a collaboration between these 2 genes.
Uhrf1 expression is finely controlled. Many transcription factors have been shown to positively regulate Uhrf1, including specificity protein 1 (SP1) (34), E2F1, and E2F8 (35, 36). Notably, E2F proteins are important regulators of VSMC proliferation in vitro and in vivo (37). Besides transcription factors, several microRNAs have been shown to regulate the expression of Uhrf1 in different types of cancer cells — miR-146a in gastric (38) and renal (39) carcinomas, miR-193a-3p in lung cancer (40), and miR-124 and miR-145 in bladder cancer (41, 42). Our data show that environmental signals modulate Uhrf1 expression in VSMCs. PDGF-BB, a potent VSMC dedifferentiator, is able to upregulate UHRF1 levels. Furthermore, our group and others have shown that miR-145 inhibition promotes VSMC dedifferentiation and is also involved in the development of different vascular pathologies (12, 16). Here, we corroborated this finding by showing that Uhrf1 expression is, at least in part, regulated by miR-145 and that miR-145 overexpression attenuates PDGF-induced Uhrf1 expression, making UHRF1 one of the functional mediators of miR-145 in VSMC biology.
CVDs are a vast health problem, and improving therapeutic and preventive measures for CVDs is necessary. Despite efforts aimed at finding epigenetic therapies for the primary prevention of CVDs, no epigenetically active agents have entered into clinical trials for CVDs thus far. Our work provides strong evidence of a pivotal role for Uhrf1 in the development of vascular pathologies. Environmental stimuli such as PDGF-BB and TGF-β precisely modulate Uhrf1 expression in VSMCs. Whereas Uhrf1 expression is almost undetectable in healthy vessels, it is highly expressed in diseased tissues. The identification of the molecular pathways regulated by UHRF1 expands our understanding of the hierarchal complexity of gene regulation in VSMCs. Interestingly, we found that UHRF1-silenced VSMCs increased expression of TGF-β pathway components, including augmented activation of SMAD signaling in vitro. In an in vivo model of Ang-II–induced aneurysm, the number of P-SMAD–positive cells was slightly increased in UHRF1-deficient mice, but no statistical differences were observed when compared with WT littermates. The role of TGF-β in aneurysm development is quite controversial. Li et al. showed that TGFβR2 disruption in SMCs of weaned mice is responsible for aneurysm formation and that it correlates with a decrease in SMAD signaling (43). On the other hand, Gallo et al. developed a mouse model resembling the Loeys-Dietz syndrome phenotype in which the pathology was associated with an aberrant activation of the SMAD signaling pathway (44). Unfortunately, our in vivo model did not completely clarify the differences observed in the aforementioned studies, perhaps due to poor understanding of the activation kinetics of the SMAD pathway in the Ang-II–induced aneurysm model. Further studies are necessary to clarify the timing of UHRF1-mediated intervention on TGF-β signaling in vivo. New animal models with multiple gene deletions are needed to clarifying the interplay between UHRF1 and the TGF-β signal transduction cascade.
Drugs such as rapamycin and taxol that target the cell cycle are already in use in medicated stents for coronary artery disease (CAD) treatment, so envisioning the targeting of UHRF1 for treating stenosis or other forms of atherosclerosis is not unreasonable.
The link between UHRF1 expression and aortic aneurysm merits further discussion. Aneurysm formation in humans has been found to be associated with mutations on MYH11 and ACTA2 (45). Here, we demonstrate that the expression of these contractile proteins is directly regulated by UHRF1. Moreover, we show that inhibition of VSMC proliferative functions through specific KO of Uhrf1 not only increases the contractile phenotype of VSMCs, but also preserves vessel wall integrity, reducing fatal transmural rupture.
Lifestyle modifications such as nutritional habits and exercise are well-known first protective measures for reducing the risk of CVDs. Interestingly, several recent studies have described the potential of adding natural polyphenols to the diet to markedly downregulate UHRF1 in cancer cells, with subsequent arrest of cell cycle and apoptosis. Luteolin, for instance, is able to trigger apoptosis of colorectal cancer and human cervical cancer by downregulating UHRF1 and DNMT1 (46, 47). It may therefore be possible to target this multifunctional protein with natural dietary polyphenols for the prevention of CVDs.
Taken together, our data not only show that Uhrf1 orchestrates the VSMC phenotype but also suggest that UHRF1 is a potential target for therapeutic intervention, because it may normalize a VSMC phenotypic switch and reduce the vascular complications of atherosclerosis.
Methods
ChIP-qPCR.
ChIP was performed as previously described (48). Briefly, 2.1 × 106 cells were cross-linked and used for each immunoprecipitation. DNA was sheared to 500–1,000 bp by sonication. Protein G (Seracare, catalog 223-51-01) was used to pull down the antibody–antigen complexes immunoprecipitated with antibodies against UHRF1 (49), H3K4me3 (Cell Signaling, catalog 9751, clone C42D8), and H3K27me3 (Cell Signaling, catalog 9733, clone C36B11). H3 (Cell Signaling, catalog 4499, clone D1H2) and IgG (Life Technologies, catalog 026102) were included as positive and negative controls, respectively. Immunoprecipitated DNA was extracted with phenol-chloroform, precipitated with ethanol, and eluted. Recovered DNA was analyzed by qPCR. Primers spanning the promoter regions (within 2,500 bp of transcription start site) of the Myh11, Cdkn1b, Acta2, Sm22, Cnn1, Tgfβ2, and Tgfβr1 genes were used to amplify input and immunoprecipitated DNA. Primers were designed to span a CArG [CC(A/T)6GG] element. Sequences are listed in Supplemental Table 1. All samples were performed at least in triplicate from at least 2 independent experiments, and data were normalized to percentage of input.
Methyl-capture assay.
The MethylCollector Ultra kit (Active Motif, catalog 55005) was used according to the manufacturer’s instructions.
Cell culture.
Primary SMCs were generated as previously described (19). Adventitia and intima layers were surgically separated from the media, which was subsequently digested with collagenase and dispase for 1 hour at 37°C. Digestion was then blocked by adding medium supplemented with 10% FBS, and cells were plated on plastic dishes precoated with gelatin 0.1%. VSMCs were characterized by qPCR and immunofluorescence analysis for the expression of specific markers such as ACTA2 and SM22. A7r5 was purchased from ATCC (catalog CRL-1444). Both cell lines were cultured in DMEM with 10% FBS. For virus production, HEK-293T cells were cultured in DMEM with 10% FBS as above. All cultures were maintained in a humidified 5% CO2 atmosphere at 37°C. For treatments, cells were washed with PBS, serum-starved for 48 hours, and then treated with 10 ng/ml TGF-β (Miltenyi Biotec, catalog 130-095-067), 100 ng/ml rapamycin (Sigma-Aldrich, catalog 553210), or different doses of PDGF-BB (Sigma-Aldrich, catalog P4306). Primary VSMCs were infected with lentivirus expressing UHRF1-specific shRNA or scrambled shRNA, selected with puromycin for 2 days, and RNA was collected after 5 days. Lentiviruses expressing miR-145 or empty control and RNA were collected after 5 days. Primary VSMCs were transfected with Lipofectamine LTX or Lipofectamine RNAiMAX (Life Technologies) following the manufacturer’s protocols. Briefly, the day prior to transfection, cells were detached using trypsin, counted, and plated in 6-well plates at the concentration of 192 × 103 cells per well. The next day, we used Lipofectamine LTX+PLUS reagent (Life Technologies) for plasmid transfection and Lipofectamine RNAiMAX for oligonucleotides. Control, miR-145 mimic, and inhibitor LNA oligonucleotides were purchased from Exiqon (catalog YI00199006, YM00479902, YM00470014, and YI04102426).
Virus production.
shRNA, empty, miR-145, decoy miR-143, and decoy miR-145 lentivirus viruses were generated using standard protocols (19). Plasmids were transfected into HEK-293T cells using the CaCl2 method, and the viral supernatant was collected by ultracentrifugation. Viruses were titrated using a qPCR Lentivirus Titration(Titer) Kit (ABM, catalog LV900) following the manufacturer’s instructions. Adenoviruses were produced by Viraquest (12).
BrdU incorporation assay.
The BrdU incorporation assay was performed as previously described (19).
Bioinformatics.
BeadChips (Illumina) were scanned with the Illumina iScan system. Raw data underwent variance-stabilizing transformation (VST) and were normalized using the quantile normalization method of the lumi R bioconductor package (50). Normalized data were filtered for genes with significant expression levels compared with negative control beads. Differentially expressed genes (P < 0.01) were identified with the Illumina t test error model (limma R bioconductor package) (51). Functional annotation of differentially expressed protein coding genes identified by microarray analysis was performed with the web-accessible tool Enrichr (52).
Immunocytochemistry and immunofluorescence.
Cells and tissues were fixed and stained using a standard protocol. Samples were incubated with antibodies against UHRF1 (1:250) (ref. 49), Ki67 (1:500, Cell Signaling, catalog 12202, clone D3B5), ACTA2 (1:300, Biocare Medical, catalog CME305B, clone E184), CD45 (1:300, R&D Systems, catalog MAB114, clone 30-F11), and CD31 (1:300, R&D Systems, catalog AF3628).
Extracellular matrix deposition and elastin structure were evaluated using the Gomori Trichrome Kit (Histo-Line, catalog GT-100T) and the Weigert-Van Gieson Kit (Histo-Line, catalog WVG-100T), respectively.
Apoptosis was evaluated using the DeadEnd Fluorometric TUNEL System (Promega, catalog G3250), following the manufacturer’s instructions. Images were taken using an IX81-FV1000 Olympus spectral confocal laser scanning microscope.
RNA extraction and reverse transcription–quantitative polymerase chain reaction (RT-qPCR).
Total RNA was extracted from primary VSMCs using NucleoZOL reagent (Macherey-Nagel) and then reverse-transcribed into complementary DNA (Life Technologies, catalog 4368813). The relative expression level of miRNA was normalized to the expression of the internal controls (CHPa or U6) using the 2−ΔΔCt method. Primers sequences are listed in Supplemental Table 2.
Custom microfluidic cards.
A list of included genes was previously described (53). Cards were purchased from Life Technologies.
Cell-cycle analysis.
Cell-cycle analysis was performed as previously described (54).
Animals.
Eight-week-old mice were used for the experiments. Special attention was paid to animal welfare and to minimize the number of animals used and their suffering. For Uhrf1 knockdown on mouse carotids, 1 × 107 viral particles were mixed with 50% Pluronic F127 gel (Sigma-Aldrich, catalog P2443) at 4°C and the solution applied to the mouse vessels (Tygon collar placed 2 weeks after carotid transduction). For Uhrf1 genetic knockout, SM-MYH11-cre/ERT2 mice were donated by Stefan Offermanns (Max-Planck Institute for Heart and Lung, Bad Nauheim, Germany) and crossed with B6.Cg-UHRF1fl/fl (Riken, catalog RBRC06536)(24) and ApoE–/– mice (Charles River Laboratories). ApoE–/–UHRF1fl/flSM-MYH11-cre/ERT2 mice and relative controls were housed under specific pathogen–free conditions. Gene knockdown was induced by intraperitoneal injections of tamoxifen (Sigma-Aldrich, catalog T5648) for 5 consecutive days (1 mg total per mouse). Ang-II infusion (Sigma-Aldrich, catalog A9525; 1,000 ng/kg/min) was performed as previously described (28) using osmotic minipumps (Alzet, model 2004). For TR reporter mice, Cg-Gt(ROSA)26Sortm9(CAG–tdTomato)Hze (The Jackson Laboratory, catalog 007909) were crossed with SM-MYH11-cre/ERT2 and then fluorescent reporter was activated with tamoxifen injection as described above. Transaortic constriction was performed as previously described (55) and aortas were collected 2 weeks after surgery.
Ultrasound imaging, analysis, and blood pressure measurement.
A Vevo 2100 high-resolution in vivo imaging system (Fujifilm VisualSonics) with an MS550S probe high-frame scan head was used for echocardiographic analysis. The abdominal aorta was identified by Doppler signal and longitudinal images were acquired between the renal arteries and the diaphragm. All measurements were performed with VisualSonics proprietary software using multiple frames, and analyses were performed as previously described (28). Briefly, cross-sectional diastolic and systolic diameter, together with the maximal aortic RWV [the first derivative (slope) of the aortic diameter with respect to time (dD/dt)], were determined from M-mode acquisitions and used to assess the consistency and viscoelastic behavior of the arterial wall (30). PD was calculated by subtracting diastolic from systolic aortic diameter and the resulting value was normalized to the maximum systolic diameter to account for vessel size.
For telemetry analysis, an HD-X11 transmitter (DSI Data Sciences International) was positioned subcutaneously and a catheter was implanted in the left carotid. The basal pressure measurements were registered 1 week before tamoxifen injection, and subsequent measurements were started 1 week after gene deletion induction. All measurements were recorded at the same time of the day. MAP was measured using the following formula: MAP = (systolic pressure + [2 × diastolic pressure])/3.
Western blotting.
Cells were lysed in RIPA buffer and sonicated for 5 cycles (30 seconds on, 30 seconds off) using a Biorupture Next Gen (Diagenode). After quantification using DCTM Protein Assay (BioRad), the same quantity of protein was separated in 4–12% NuPAGE Bis-Tris precast gel (Life Technologies) and transferred to a nitrocellulose membrane (BioRad). Afterwards, anti-UHRF1 (1:1,000) (49), anti-CDKN1A (1:1,000, catalog sc-528, clone C-19), anti-CDKN1B (1:1,000, catalog sc-397, clone C-19), anti-CDKN2B (1:1,000, catalog sc-377412, clone C-7), anti-H3 (1:5,000, catalog sc-517385, clone 1B1-B2), anti-CNN1 (1:1,000, catalog sc-136987, clone G-10) (all from Santa Cruz Biotechnologies); anti-ACTA2 (1:1,000, Sigma-Aldrich, catalog A5228, clone 1A4); anti-GAPDH (1:1,000, catalog 2118, clone 14C10), anti-pSMAD1/5/8 (1:1,000, catalog 9511), anti-pSMAD 2/3 (1:1,000, catalog 8828, clone D27F4), anti-SMAD2 (1:1,000, catalog 5339, clone D43B4), anti-SMAD5 (1:1,000, catalog 12534, clone D4G2) (all from Cell Signaling); and anti-MYH11 (1:1,500, Protein Tech, catalog 21404-1-AP) were incubated overnight at 4°C, and secondary peroxidase-conjugated antibodies were incubated for 1 hour at room temperature. Immobilon Western Chemiluminescent HRP (Millipore) was used for protein visualization using a ChemiDoc (BioRad). Bands were quantified using ImageJ software. Results are expressed relative to the control. See complete unedited blots in the supplemental material.
Statistics.
Normality of the data was calculated with the Kolmogorov-Smirnov test. Statistical comparison was performed on at least 3 independent experiments with the Mann-Whitney U test (nonparametric unpaired), Wilcoxon’s test (nonparametric paired), 2-tailed t test (parametric unpaired, 2 groups of analysis), or repeated-measures ANOVA combined with Tukey’s multicomparison t test (comparisons among more than 2 groups, parametric unpaired). Prism 6.0 software (GraphPad) was used to assess the normality of the data and for statistical calculation. P < 0.05 was considered significant. Statistical analysis data are mean ± SD.
Data and materials availability.
The gene expression profile data set is available from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nih.gov/geo/) under the accession number GSE99472. The authors declare that all data supporting the findings of this study are available within the article and its supplementary information files.
Study approval.
Study protocols (15/2014 and 17/2014) were approved by the Italian Ministry of Health. All procedures involving mice were performed according to institutional guidelines in compliance with national and international law and policies and the Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011).
Author contributions
LE performed the experiments, interpreted the data, and wrote the manuscript. PK performed the bioinformatics analysis. FMF, IFH, PC, SM, MV, and CP performed the experiments and interpreted the data. RP interpreted the data. GC contributed to hypothesis formulation, project development, and manuscript writing. MQ conceived the initial idea, designed the research, performed the experiments, interpreted the data, and wrote the manuscript.
Supplementary Material
Acknowledgments
We would like to thank Ian Marc Bonapace (Università degli studi dell’Insubria, Varese, Italy) for the anti–UHRF1 antibody. This work was supported by grants from the Italian Ministry of Health (GR2011_02349075) and the CARIPLO Foundation (2014-1108) (to MQ), and by funding from the European Research Council Advanced Grant (CardioEpigen, 294609), ERC Proof of Concept (713734), the Italian Ministry of Health (PE-2013-02356818), the CARIPLO Foundation (2015-0573), the Italian Ministry of Education, University and Research (2015583WMX), and the Italian Association for Cancer Research (AIRC) (IG-15801) (to GC).
Version 1. 03/20/2018
In-Press Preview
Version 2. 05/07/2018
Electronic publication
Version 3. 06/01/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(6):2473–2486.https://doi.org/10.1172/JCI96121.
Contributor Information
Leonardo Elia, Email: leonardo.elia@unibs.it.
Paolo Kunderfranco, Email: paolo.kunderfranco@humanitasresearch.it.
Pierluigi Carullo, Email: pierluigi.carullo@humanitasresearch.it.
Marco Vacchiano, Email: marco.vacchiano@humanitasresearch.it.
Floriana Maria Farina, Email: Floriana.Farina@humanitasresearch.it.
Ignacio Fernando Hall, Email: Ignacio_Fernando.Hall_Balcazar@humanitasresearch.it.
Stefano Mantero, Email: stefano.mantero@humanitasresearch.it.
Cristina Panico, Email: cristina.panico@humanitas.it.
Roberto Papait, Email: roberto.papait@humanitasresearch.it.
Gianluigi Condorelli, Email: gianluigi.condorelli@hunimed.eu.
Manuela Quintavalle, Email: manuela.quintavalle@humanitasresearch.it.
References
- 1.Owens GK. Regulation of differentiation of vascular smooth muscle cells. Physiol Rev. 1995;75(3):487–517. doi: 10.1152/physrev.1995.75.3.487. [DOI] [PubMed] [Google Scholar]
- 2.McDonald OG, Wamhoff BR, Hoofnagle MH, Owens GK. Control of SRF binding to CArG box chromatin regulates smooth muscle gene expression in vivo. J Clin Invest. 2006;116(1):36–48. doi: 10.1172/JCI26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Findeisen HM, Kahles FK, Bruemmer D. Epigenetic regulation of vascular smooth muscle cell function in atherosclerosis. Curr Atheroscler Rep. 2013;15(4):319. doi: 10.1007/s11883-013-0319-7. [DOI] [PubMed] [Google Scholar]
- 5.Nishiyama A, et al. Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature. 2013;502(7470):249–253. doi: 10.1038/nature12488. [DOI] [PubMed] [Google Scholar]
- 6.Bostick M, Kim JK, Estève PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317(5845):1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 7.Sharif J, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450(7171):908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 8.Rothbart SB, et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol. 2012;19(11):1155–1160. doi: 10.1038/nsmb.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis-Dusenbery BN, Wu C, Hata A. Micromanaging vascular smooth muscle cell differentiation and phenotypic modulation. Arterioscler Thromb Vasc Biol. 2011;31(11):2370–2377. doi: 10.1161/ATVBAHA.111.226670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thum T, Condorelli G. Long noncoding RNAs and microRNAs in cardiovascular pathophysiology. Circ Res. 2015;116(4):751–762. doi: 10.1161/CIRCRESAHA.116.303549. [DOI] [PubMed] [Google Scholar]
- 11.Elia L, Quintavalle M. Epigenetics and vascular diseases: influence of non-coding RNAs and their clinical implications. Front Cardiovasc Med. 2017;4:26. doi: 10.3389/fcvm.2017.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elia L, et al. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 2009;16(12):1590–1598. doi: 10.1038/cdd.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cordes KR, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460(7256):705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xin M, et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23(18):2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boettger T, et al. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J Clin Invest. 2009;119(9):2634–2647. doi: 10.1172/JCI38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quintavalle M, Elia L, Condorelli G, Courtneidge SA. MicroRNA control of podosome formation in vascular smooth muscle cells in vivo and in vitro. J Cell Biol. 2010;189(1):13–22. doi: 10.1083/jcb.200912096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Kim SG, Blenis J. Rapamycin: one drug, many effects. Cell Metab. 2014;19(3):373–379. doi: 10.1016/j.cmet.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ballantyne MD, et al. Smooth muscle enriched long noncoding RNA (SMILR) regulates cell proliferation. Circulation. 2016;133(21):2050–2065. doi: 10.1161/CIRCULATIONAHA.115.021019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Climent M, Quintavalle M, Miragoli M, Chen J, Condorelli G, Elia L. TGFβ triggers miR-143/145 transfer from smooth muscle cells to endothelial cells, thereby modulating vessel stabilization. Circ Res. 2015;116(11):1753–1764. doi: 10.1161/CIRCRESAHA.116.305178. [DOI] [PubMed] [Google Scholar]
- 20.Ayari H, Bricca G. Identification of two genes potentially associated in iron-heme homeostasis in human carotid plaque using microarray analysis. J Biosci. 2013;38(2):311–315. doi: 10.1007/s12038-013-9310-2. [DOI] [PubMed] [Google Scholar]
- 21.Bekelis K, et al. MicroRNA and gene expression changes in unruptured human cerebral aneurysms. J Neurosurg. 2016;125(6):1390–1399. doi: 10.3171/2015.11.JNS151841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maegdefessel L, et al. miR-24 limits aortic vascular inflammation and murine abdominal aneurysm development. Nat Commun. 2014;5:5214. doi: 10.1038/ncomms6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baetta R, et al. Perivascular carotid collar placement induces neointima formation and outward arterial remodeling in mice independent of apolipoprotein E deficiency or Western-type diet feeding. Atherosclerosis. 2007;195(1):e112–e124. doi: 10.1016/j.atherosclerosis.2007.03.035. [DOI] [PubMed] [Google Scholar]
- 24.Obata Y, et al. The epigenetic regulator Uhrf1 facilitates the proliferation and maturation of colonic regulatory T cells. Nat Immunol. 2014;15(6):571–579. doi: 10.1038/ni.2886. [DOI] [PubMed] [Google Scholar]
- 25.Liu X, et al. UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat Commun. 2013;4:1563. doi: 10.1038/ncomms2562. [DOI] [PubMed] [Google Scholar]
- 26.Babbio F, et al. The SRA protein UHRF1 promotes epigenetic crosstalks and is involved in prostate cancer progression. Oncogene. 2012;31(46):4878–4887. doi: 10.1038/onc.2011.641. [DOI] [PubMed] [Google Scholar]
- 27.Daugherty A, Manning MW, Cassis LA. Angiotensin II promotes atherosclerotic lesions and aneurysms in apolipoprotein E-deficient mice. J Clin Invest. 2000;105(11):1605–1612. doi: 10.1172/JCI7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gavish L, et al. Low-level laser irradiation inhibits abdominal aortic aneurysm progression in apolipoprotein E-deficient mice. Cardiovasc Res. 2009;83(4):785–792. doi: 10.1093/cvr/cvp149. [DOI] [PubMed] [Google Scholar]
- 29.Dobrin PB, Mrkvicka R. Failure of elastin or collagen as possible critical connective tissue alterations underlying aneurysmal dilatation. Cardiovasc Surg. 1994;2(4):484–488. [PubMed] [Google Scholar]
- 30.Armentano R, Megnien JL, Simon A, Bellenfant F, Barra J, Levenson J. Effects of hypertension on viscoelasticity of carotid and femoral arteries in humans. Hypertension. 1995;26(1):48–54. doi: 10.1161/01.HYP.26.1.48. [DOI] [PubMed] [Google Scholar]
- 31.McDonald OG, Owens GK. Programming smooth muscle plasticity with chromatin dynamics. Circ Res. 2007;100(10):1428–1441. doi: 10.1161/01.RES.0000266448.30370.a0. [DOI] [PubMed] [Google Scholar]
- 32.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370(Pt 3):737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu R, et al. Ten-eleven translocation-2 (TET2) is a master regulator of smooth muscle cell plasticity. Circulation. 2013;128(18):2047–2057. doi: 10.1161/CIRCULATIONAHA.113.002887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu SM, et al. Negative modulation of the epigenetic regulator, UHRF1, by thyroid hormone receptors suppresses liver cancer cell growth. Int J Cancer. 2015;137(1):37–49. doi: 10.1002/ijc.29368. [DOI] [PubMed] [Google Scholar]
- 35.Unoki M, Nishidate T, Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23(46):7601–7610. doi: 10.1038/sj.onc.1208053. [DOI] [PubMed] [Google Scholar]
- 36.Park SA, Platt J, Lee JW, López-Giráldez F, Herbst RS, Koo JS. E2F8 as a novel therapeutic target for lung cancer. J Natl Cancer Inst. 2015;107(9):djv151. doi: 10.1093/jnci/djv151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giangrande PH, et al. Distinct roles of E2F proteins in vascular smooth muscle cell proliferation and intimal hyperplasia. Proc Natl Acad Sci U S A. 2007;104(32):12988–12993. doi: 10.1073/pnas.0704754104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou L, et al. Regulation of UHRF1 by miR-146a/b modulates gastric cancer invasion and metastasis. FASEB J. 2013;27(12):4929–4939. doi: 10.1096/fj.13-233387. [DOI] [PubMed] [Google Scholar]
- 39.Wotschofsky Z, et al. Integrated microRNA and mRNA signature associated with the transition from the locally confined to the metastasized clear cell renal cell carcinoma exemplified by miR-146-5p. PLoS One. 2016;11(2):e0148746. doi: 10.1371/journal.pone.0148746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng W, et al. Quantitative proteomic analysis of the metastasis-inhibitory mechanism of miR-193a-3p in non-small cell lung cancer. Cell Physiol Biochem. 2015;35(5):1677–1688. doi: 10.1159/000373981. [DOI] [PubMed] [Google Scholar]
- 41.Wang X, et al. MiR-124 exerts tumor suppressive functions on the cell proliferation, motility and angiogenesis of bladder cancer by fine-tuning UHRF1. FEBS J. 2015;282(22):4376–4388. doi: 10.1111/febs.13502. [DOI] [PubMed] [Google Scholar]
- 42.Matsushita R, et al. Regulation of UHRF1 by dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p): inhibition of bladder cancer cell aggressiveness. Oncotarget. 2016;7(19):28460–28487. doi: 10.18632/oncotarget.8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li W, et al. Tgfbr2 disruption in postnatal smooth muscle impairs aortic wall homeostasis. J Clin Invest. 2014;124(2):755–767. doi: 10.1172/JCI69942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gallo EM, et al. Angiotensin II-dependent TGF-β signaling contributes to Loeys-Dietz syndrome vascular pathogenesis. J Clin Invest. 2014;124(1):448–460. doi: 10.1172/JCI69666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Milewicz DM, et al. Genetic basis of thoracic aortic aneurysms and dissections: focus on smooth muscle cell contractile dysfunction. Annu Rev Genomics Hum Genet. 2008;9:283–302. doi: 10.1146/annurev.genom.8.080706.092303. [DOI] [PubMed] [Google Scholar]
- 46.Krifa M, Leloup L, Ghedira K, Mousli M, Chekir-Ghedira L. Luteolin induces apoptosis in BE colorectal cancer cells by downregulating calpain, UHRF1, and DNMT1 expressions. Nutr Cancer. 2014;66(7):1220–1227. doi: 10.1080/01635581.2014.951729. [DOI] [PubMed] [Google Scholar]
- 47.Krifa M, et al. Limoniastrum guyonianum aqueous gall extract induces apoptosis in human cervical cancer cells involving p16 INK4A re-expression related to UHRF1 and DNMT1 down-regulation. J Exp Clin Cancer Res. 2013;32:30. doi: 10.1186/1756-9966-32-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Papait R, et al. Genome-wide analysis of histone marks identifying an epigenetic signature of promoters and enhancers underlying cardiac hypertrophy. Proc Natl Acad Sci U S A. 2013;110(50):20164–20169. doi: 10.1073/pnas.1315155110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papait R, et al. The PHD domain of Np95 (mUHRF1) is involved in large-scale reorganization of pericentromeric heterochromatin. Mol Biol Cell. 2008;19(8):3554–3563. doi: 10.1091/mbc.E07-10-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 51.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21(9):2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 52.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cattaneo P, et al. DOT1L-mediated H3K79me2 modification critically regulates gene expression during cardiomyocyte differentiation. Cell Death Differ. 2016;23(4):555–564. doi: 10.1038/cdd.2014.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Farina FM, Inguscio A, Kunderfranco P, Cortesi A, Elia L, Quintavalle M. MicroRNA-26a/cyclin-dependent kinase 5 axis controls proliferation, apoptosis and in vivo tumor growth of diffuse large B-cell lymphoma cell lines. Cell Death Dis. 2017;8(6):e2890. doi: 10.1038/cddis.2017.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elia L, et al. Reciprocal regulation of microRNA-1 and insulin-like growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation. 2009;120(23):2377–2385. doi: 10.1161/CIRCULATIONAHA.109.879429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.