Abstract
Triple-negative breast cancer (TNBC) is a heterogeneous disease with poor prognosis that lacks targeted therapies, especially in patients with chemotherapy-resistant disease. Since DNA methylation-induced silencing of tumor suppressors is common in cancer, reversal of promoter DNA hypermethylation by 5-aza-2′-deoxycytidine (decitabine), an FDA-approved DNA methyltransferase (DNMT) inhibitor, has proven effective in treating hematological neoplasms. However, its antitumor effect varies in solid tumors, stressing the importance of identifying biomarkers predictive of therapeutic response. Here, we focused on the identification of biomarkers to select decitabine-sensitive TNBC through increasing our understanding of the mechanism of decitabine action. We showed that protein levels of DNMTs correlated with response to decitabine in patient-derived xenograft (PDX) organoids originating from chemotherapy-sensitive and -resistant TNBCs, suggesting DNMT levels as potential biomarkers of response. Furthermore, all 3 methytransferases, DNMT1, DNMT3A, and DNMT3B, were degraded following low-concentration, long-term decitabine treatment both in vitro and in vivo. The DNMT proteins could be ubiquitinated by the E3 ligase, TNF receptor–associated factor 6 (TRAF6), leading to lysosome-dependent protein degradation. Depletion of TRAF6 blocked decitabine-induced DNMT degradation, conferring resistance to decitabine. Our study suggests a potential mechanism of regulating DNMT protein degradation and DNMT levels as response biomarkers for DNMT inhibitors in TNBCs.
Keywords: Oncology, Therapeutics
Keywords: Breast cancer, Drug therapy, Mouse models
Introduction
Breast cancer is the second leading cause of cancer death in women, with 40,610 women predicted to die from breast cancer in 2017 (1). Triple-negative breast cancer (TNBC), characterized by absence or minimal expression of estrogen receptor (ER) and progesterone receptor (PR) and lack of the human epidermal growth factor receptor 2 (her2) amplification, represents 15%–20% of all breast cancers. TNBC is generally more aggressive; although many patients respond well to standard anthracycline- and taxane-based chemotherapy, long-term patient survival is poor due to high rates of relapse and recurrence (2, 3). Furthermore, there are few treatment options for TNBC patients whose tumors are resistant to standard chemotherapy (4). Therefore, there is an urgent need to identify additional drugs and drug targets for TNBC patients, who have a very poor prognosis. DNA methylation, the transfer of a methyl group to a cytosine residue to form 5-methylcytosine (5mC), is a heritable epigenetic process regulating gene function in normal mammalian development. However, hypermethylation of the CpG islands in the promoter regions of critical growth regulators, such as tumor suppressors, is a major event in the origin of many cancers (5). TNBC tumors show extensive promoter hypermethylation of epigenetic biomarker genes compared with other breast cancer subtypes in which promoter hypermethylation events were less frequent (6, 7), suggesting that targeting the epigenetic machinery of TNBCs may have clinical benefits.
5-Aza-2′-deoxycytidine (decitabine) is a demethylating agent that is approved by many international regulatory agencies, including the US FDA and the European Commission (EC), for treating hematological malignancies (8–10) and that has become a standard of care for patients with higher risk myelodysplastic syndrome (MDS) (11, 12). Decitabine suppresses DNA methyltransferases (DNMTs) (13, 14). Three active DNMTs have been identified in humans, DNMT1, DNMT3A, and DNMT3B (15). DNMT1 recognizes hemimethylated DNA generated during DNA replication and then methylates newly synthesized CpG dinucleotides (16). DNMT3A and DNMT3B mainly work as de novo methyltransferases to establish DNA methylation, and they can also methylate hemimethylated DNA (17). Decitabine converts to the active thiphosphorylated nucleotides, incorporates to DNA as a cytosine substitute, irreversibly binds to DNMTs, and traps the enzymes on DNA. This causes subsequent depletion of the DNMT pool and inhibits the DNMT function (18). High micromolar doses of decitabine rapidly induce DNA damage and cytotoxicity (19, 20). This mechanism is different from the mechanisms observed at lower nanomolar concentrations. While low-dose decitabine treatment does not inhibit DNA synthesis, clinical trials in patients with hematological malignancies demonstrated improved safety and efficacy compared with those who received high-dose treatment (21–23), suggesting unexplored mechanisms of action for decitabine at lower doses (24).
In solid cancers, decitabine has been studied mainly using high doses, and these studies have shown substantial toxicity (25), thereby prohibiting the clinical development of this drug in solid tumors. Despite the toxicity, antitumor activity has been observed, with decitabine response rates ranging from 6% in female reproductive cancer (n = 35) to 50% in breast cancer (n = 4) (26). Even though the number of patients in these studies is small, the interindividual variation in response to decitabine emphasizes the clinical importance of identifying effective markers for selecting patients who may benefit from this drug.
Unlike in hematological malignancies, few studies have explored the mechanisms of low-dose decitabine in solid tumors. Tsai et al. have shown that exposure of epithelial tumor cells to decitabine at nanomolar doses produced an antitumor “memory” response by sustained decrease in genome-wide promoter DNA methylation and gene reexpression in key cellular regulatory pathways (27). These data suggest that a better understanding of decitabine mechanism of action is essential for its application in the treatment of solid tumors.
DNMT1 and DNMT3A are reported to be overexpressed in 5% and 3% of breast carcinomas (6), whereas DNMT3B is overexpressed in 30% of breast cancers (28). The relationship between DNMTs and response to decitabine in solid cancer remains controversial. Previous studies suggested that DNMT1 mRNA expression is positively correlated with decitabine sensitivity, while others suggested a negative correlation between DNMT3B mRNA expression and decitabine sensitivity in ovarian (29) and pancreatic cancers (30). In the current study, we sought to systematically explore the relationship of different DNMT isoforms with decitabine-treatment response in TNBCs and to further investigate the mechanism of decitabine action in the treatment of TNBC. We found that DNMT protein levels correlated with decitabine response in a breast cancer organoid model derived from patient-derived xenograft (PDX) tumors. This effect was mediated by decitabine-induced protein degradation of all 3 DNMTs, and this process was dependent on the E3 ligase TNF receptor–associated factor 6 (TRAF6). Collectively, our results point to decitabine as a potential new therapeutic for TNBC.
Results
DNMT expression levels dictate tumor sensitivity to decitabine treatment.
3D organoid cultures recapitulate in vivo tissue structural organization and more closely resemble the actual tumor biology than 2D cultures of primary or immortalized cells (31), thereby providing an attractive platform for testing cancer cell response to drugs (32). As previously described, we generated a panel of PDX models from patients with primary breast cancer recruited in a prospective neoadjuvant study of anthracycline- and taxane-based chemotherapy, with PDX models derived from both baseline percutaneous biopsy prior to chemotherapy and postchemotherapy surgical samples (33, 34). In addition to in vivo PDX studies, we also successfully grew organoids using the PDX tumors to test response to therapies and to identify molecular markers that might be associated with response (Figure 1A).
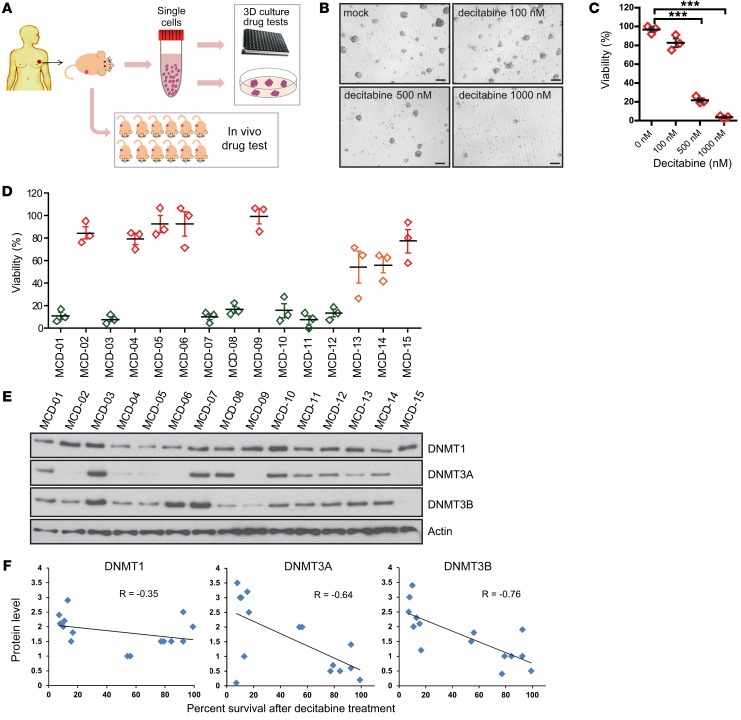
Figure 1. Protein levels of DNMTs correlated with decitabine sensitivity in a collection of breast cancer PDX–derived organoids.
(A) Schematic representation of the work flow for generating organoid culture from TNBC PDX tumors. (B) Representative figures of organoid cultures derived from TNBC PDX tumor (MCD-02, resistant to decitabine treatment). Organoids were treated with 0 nM, 100 nM, 500 nM, or 1000 nM decitabine for 7 days and recovered for 2 days (n = 3 independent experiments). Scale bars: 20 μm. (C) Quantification of tumorsphere in B. Data represent mean ± SEM of control from n = 3 independent experiments. ***P < 0.001, 2-tailed t tests. (D) Organoids from 15 different TNBC PDX tumors were cultured and tested for decitabine sensitivity at a concentration of 100 nM for 7 days, followed by 2 days of recovery. Colors represent the degree of decitabine sensitivity. Red or orange indicates decitabine resistant; green indicates decitabine sensitive. Data represent mean ± SEM of n = 3 independent experiments when compared with control. (E) Protein levels of DNMT1, DNMT3A, and DNMT3B detected in the cell lysates isolated from untreated organoids. (F) Protein level correlation with organoid percentage of survival after decitabine treatment. The y axis represents the relative quantification of protein levels and the x axis represents the percentage of survival after 100 nM decitabine as in D.
Previous studies demonstrated that decitabine exhibited antitumor activity in breast cancer (30, 35). To identify biomarkers for decitabine response, we tested decitabine sensitivity in a panel of 15 organoid cultures developed from 15 distinct TNBC PDX tumors from patients whose disease was either sensitive (n = 9) or resistant (n = 6) to standard anthracycline- and taxane-based neoadjuvant chemotherapy (NAC) (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI97924DS1) (33). As shown in Figure 1, B–D, we observed great variability in decitabine response in these breast cancer organoids. Interestingly, we observed that decitabine response highly correlated with DNMT3A and DNMT3B protein levels and, to a lesser extent, with DNMT1 protein levels (Figure 1, D–F). Tumors with high expression of all 3 DNMTs were more sensitive to decitabine treatment (Figure 1, D and E). These findings suggest that protein levels of these DNMTs might influence breast-tumor response to decitabine treatment.
PDXs are considered to be more representative of human tumors and are increasingly used as in vivo models for preclinical drug testing (36). To further determine whether the decitabine response in the organoids represents the drug sensitivity in the preclinical models, we selected 3 PDX models that were predicted to be “sensitive” (MCD-01, MCD-11, MCD-03) and 3 PDX models predicted to be “resistant” (MCD-05, MCD-02, MCD-04) based on the organoid testing results (Figure 1D) for the in vivo drug tests. Mice were treated with 5 mg/kg decitabine according to the dosing schedule shown in Figure 2A. As shown in Figure 2B and Supplemental Table 1, the response pattern to decitabine in the PDX models was consistent with our observations in the organoid models.
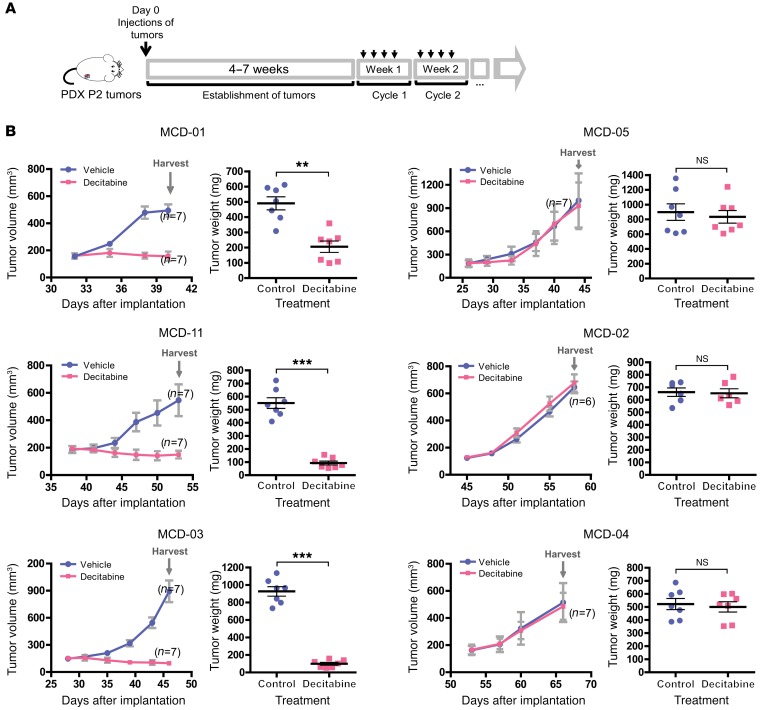
Figure 2. PDXs expressing high levels of DNMTs showed greater sensitivity to decitabine treatment.
(A) Schematic outline of decitabine treatment for the PDX tumors. Small pieces of passage 2 tumors were injected s.c. into the back flanks of SCID mice (n = 6–7 respectively). Drug was administered i.p. and started when the tumors grew to 150–200 mm3. Mice were treated with decitabine for 4 days per cycle, followed by 3 days of recovery. (B) Tumor-response curve for PDX tumors. Mice bearing passage 3 breast cancer PDX tumors were treated with either vehicle (blue) or 5 mg/kg decitabine (pink) according to the time schedule described in A. Tumors were measured twice weekly. Data represent mean tumor volume ± SEM for all tumors at each indicated day (n = 6–7 mice respectively). PDX line ID MCD numbers correspond to MCD-IDs shown in Figure 1D. Tumors were harvested at the time when experiments were terminated, and tumor weight was measured. Data represent mean ± SEM. **P < 0.01; ***P < 0.001, 2-tailed t test.
Furthermore, we tested decitabine sensitivity in PDX organoids and PDX lines derived from both pre- and post-NAC tumor (chemotherapy-resistant tumor) and found that the sensitivity to decitabine is dependent on the DNMT levels regardless of the tumors’ sensitivity to chemotherapy (Supplemental Table 1), suggesting that decitabine might be effective in both settings.
Taken together, the findings that organoids and PDX models expressing elevated levels of DNMT proteins are more sensitive to decitabine than those expressing low levels of DNMTs suggest that DNMT protein levels might serve as biomarkers for decitabine-treatment response in TNBC.
Decitabine treatment induces lysosomal degradation of DNMTs.
Decitabine, when administered at high concentrations, is known to degrade DNMT1 protein, resulting in DNA damage at a high dose and short-term treatment setting (37). However, the effect of decitabine action at low concentrations has not been fully elucidated (22). To explore the mechanism, we first determined the protein levels of 3 DNMTs in both control and decitabine-treated PDX groups. DNMT1 levels decreased upon low-concentration decitabine treatment. Interestingly, both DNMT3A and DNMT3B protein levels were also dramatically reduced (Figure 3A) after decitabine treatment, whereas decitabine treatment did not affect the mRNA levels of all 3 genes (Figure 3B).
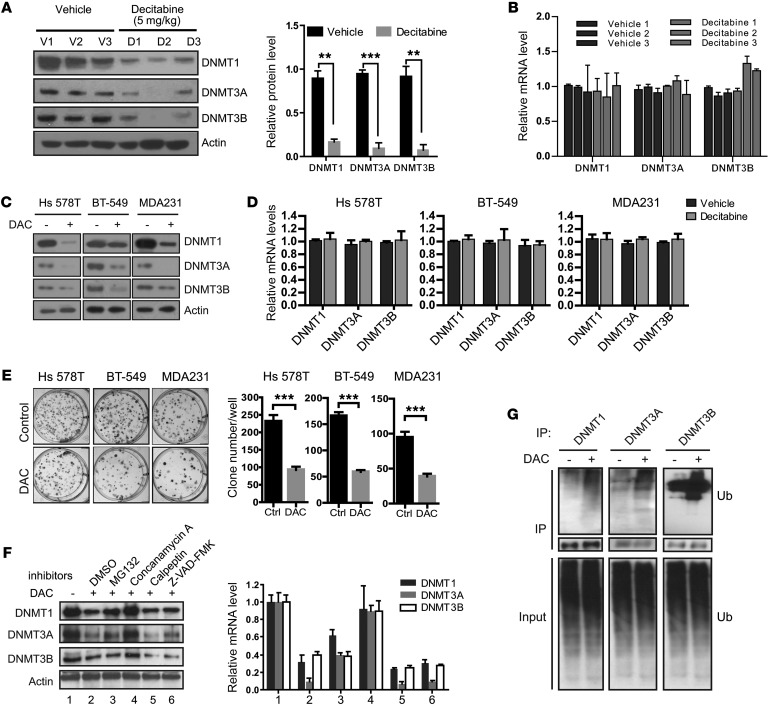
Figure 3. Decitabine treatment induced lysosome-dependent degradation of DNMTs.
(A) DNMT-specific Western blot using whole-tissue lysate from the indicated PDX tumors treated with either vehicle (n = 3 mice) or decitabine (n = 3 mice). Protein was quantified using ImageJ software (NIH), and relative protein levels were corrected for the vehicle group. Data represent mean ± SD from n = 3 different samples. **P < 0.01, ***P < 0.001, 2-tailed t tests. (B) Relative mRNA levels of DNMT1, DNMT13A and DNMT13B genes in tumor samples after decitabine treatment compared with samples treated with vehicle control group (n = 3 independent experiments). (C) Breast cancer cells were treated with 100 nM decitabine (DAC) for 7 days, and Western blot analysis was performed using the indicated antibodies. (D) Breast cancer cells were treated with 100 nM decitabine for 7 days, and mRNA levels were quantified by qRT-PCR (n = 3 independent experiments). (E) Representative well from clonogenic assay of 3 different breast cancer cell lines 14 days after decitabine treatment. Average colony number was quantified for each well, and each experiment was conducted as n = 3 independent experiments in triplicate. ***P < 0.001, 2-tailed t test. (F) MDA-MB-231 cells were treated with decitabine for 7 days and treated with concanamycin A, calpetin, Z-VAD-FAM, and MG132 for an additional 24 hours. The protein levels of DNMT1, DNMT3A, and DNMT3B were analyzed by Western blot and quantified by ImageJ software (n = 3). (G) MDA-MB-231 cells were treated with decitabine for 7 days, and the ubiquitination of DNMTs was determined by pulling down DNMTs with the indicated antibodies followed by Western blot analysis.
To further confirm these results, we treated 3 different TNBC cancer cell lines, Hs 578T, BT-549, and MDA-MB-231 cells, with 100 nM decitabine for 7 days. A reduction of all 3 DNMT proteins, but not the mRNAs, in these cells was observed (Figure 3, C and D). Clonogenic assay also showed that fewer clones formed after decitabine treatment compared with the control group, indicating that decitabine inhibits cell proliferation (Figure 3E). Because protein can be degraded through several proteolytic pathways, we utilized various inhibitors to block protein degradation: MG132 (proteasome), concanamycin A (lysosome), calpeptin (calpain), and Z-VAD-FMK (caspase) to identify the specific pathway. MG132 only partially rescued the DNMT protein levels, while concanamycin A treatment resulted in the most substantial reversal of DNMT protein levels. Moreover, we did not observe any marked effect with calpain or caspase inhibitors (Figure 3F). Proteins are marked for degradation by the attachment of ubiquitin to the side chain of lysine residues called ubiquitination (38). We also detected enhanced ubiquitination of DNMTs in the presence of decitabine (Figure 3G). These results suggest that low-concentration and long-term decitabine treatment induce degradation of all 3 DNMT isoforms predominantly in a lysosome-dependent manner.
The ubiquitin E3 ligase TRAF6 interacts with DNMTs.
The E3 ligases are responsible for recognizing specific substrates and catalyzing their ubiquitination (38). To determine the potential E3 ligase that is responsible for the degradation of DNMTs, we analyzed the protein sequence for all 3 DNMT isoforms and found that all 3 DNMTs harbored a conserved TRAF6-binding motif (PxExx [aromatic (F/W/Y)/acidic residue (D/E)]) (39), as shown in Figure 4A. We thus hypothesized that TRAF6 might be the E3 ligase that mediates DNMTs’ ubiquitination. To test this hypothesis, we first determined whether TRAF6 and DNMTs interact by performing immunoprecipitation assays. Coimmunoprecipitation revealed the interaction of endogenous TRAF6 and DNMTs (Figure 4B). To further confirm these interactions, we carried out in vitro protein-protein interaction assays. As shown in Figure 4C, bacterial expressed and purified GST-DNMT1 could pull down purified Myc-TRAF6, indicating a direct interaction. Similar in vitro results were also observed between TRAF6 and DNMT3B or DNMT3A, respectively (Figure 4, D and E).
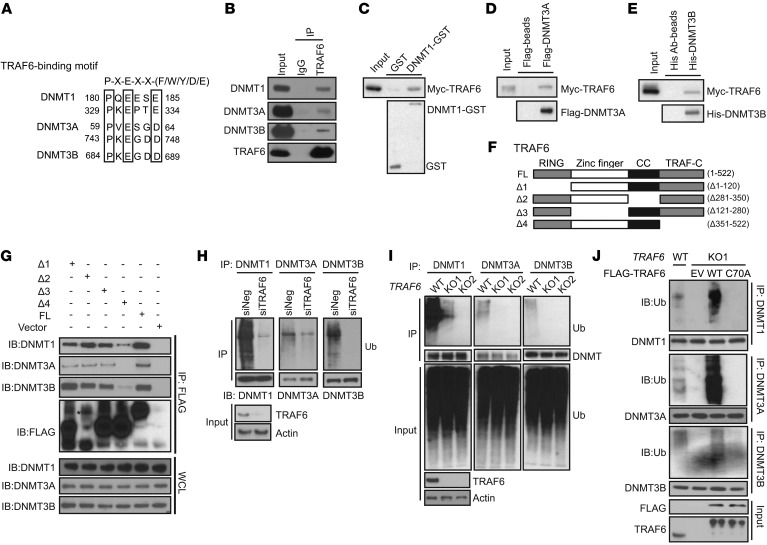
Figure 4. The E3 ubiquitin-protein ligase TRAF6 interacts with and ubiquitinates DNMTs.
(A) DNMT1, DNMT3A, and DNMT3B proteins contain a potential TRAF6-binding site. The consensus TRAF6-binding site motif has been described previously (78). (B) Endogenous interactions of TRAF6 with DNMT1, DNMT3A, and DNMT3B. MDA-MB-231 cells were used to perform endogenous immunoprecipitation with TRAF6 antibody, followed by Western blot with indicated DNMT antibodies. (C–E) In vitro binding assays of TRAF6 and DNMTs. Myc-TRAF6 expressed in E. coli was purified and incubated with GST-DNMT1, His-DNMT3B, and Flag-DNMT3A with agarose beads. The precipitated samples were analyzed by Western blot using indicated antibodies. (F) Schematic representation of WT TRAF6 and its different deletion mutants. (G) FLAG-tagged TRAF6 constructs were transfected into 293T cells, and FLAG beads were used for immunoprecipitation. Precipitates were subjected to Western blot analysis with indicated antibodies. WCL, whole-cell lysates. (H) MDA-MB-231 cells were transiently transfected with negative control or TRAF6-specific siRNAs for 72 hours. DNMT protein from the cell lysates was immunoprecipitated with corresponding anti-DNMT antibody. The ubiquitinated DNMTs were detected by Western blot using anti-ub antibody at high molecular weight ladders. (I) Crispr/Cas9 TRAF6-KO cell line was generated and WT and Crispr/Cas9 TRAF6-KO cell lines were subjected to immunoprecipitation using anti-DNMT antibodies. The ubiquitinated DNMTs were detected by Western blot analysis. (J) MDA-MB-231 TRAF6 Crispr/Cas9-KO cells (KO1) were transfected with FLAG-tagged WT TRAF6 (WT) and catalytic-dead CA mutant (C70A). DNMTs from the cell lysates were immunoprecipitated with anti-DNMT1, anti-DNMT13A, and anti-DNMT13B antibodies. The ubiquitinated DNMTs were detected by Western blot analysis.
To map the region or regions of TRAF6 involved in the interaction with DNMTs, we generated 4 TRAF6 deletion constructs, based on TRAF6 domain architecture: RING domain deletion (Δ1), the internal CC domain deletion (Δ2), Zinc finger deletion (Δ3), and TRAF-C domain deletion (Δ4) (40) (Figure 4F). Deletion of the TRAF-C domain (Δ351–522aa) abrogated the interaction among TRAF6 and DNMT1, DNMT3A and DNMT3B, respectively (Figure 4G). Taken together, these data indicate that TRAF6 directly interacts with DNMTs and that the interaction is dependent on the TRAF6 TRAF-C domain.
TRAF6 ubiquitinates DNMTs and induces protein degradation.
As TRAF6 is a RING-domain E3 ubiquitin ligase, we next tested to determine whether TRAF6 was the E3 ligase for the DNMTs’ ubiquitination. We first tested the effect of knockdown of TRAF6 on DNMT ubiquitination. As shown in Figure 4H, in MDA-MB-231 cells transfected with TRAF6 siRNA, ubiquitinated DNMTs decreased in TRAF6–knocked down cells. Consistently, TRAF6 CRISPR/Cas9-KO cells also showed defects in ubiquitination of DNMTs compared with WT control cells (Figure 4I). To further confirm that TRAF6 E3 ligase activity is essential for this process, we overexpressed WT TRAF6 and the TRAF6 with an E3-inactive RING mutant (TRAF6C70A) in TRAF6-KO cells. We found that overexpression of WT, but not the E3-inactive RING mutant (TRAF6C70A) TRAF6, in TRAF6-KO cells reversed the decrease in DNMT1 ubiquitination caused by TRAF6 depletion (Figure 4J).
Lys-48–linked ubiquitination mainly targets substrates for proteasome-mediated degradation, whereas Lys-63–linked polyubiquitination is primarily associated with nonproteolytic functions, such as regulation of protein activity and lysosomal degradation (41). TRAF6 has been reported as mainly mediating Lys-63–linked ubiquitination (42). Based on the phenotype, it is possible that TRAF6 might facilitate lysosomal degradation of DNMTs. To determine whether TRAF6 regulates protein stability of DNMTs, we depleted TRAF6 in different breast cancer cell lines. Interestingly, knockdown of TRAF6 upregulated the protein levels of all 3 DNMT members (Figure 5A). Similar findings were observed in TRAF6 CRISPR/Cas9-KO MDA-MB-231 breast cancer cells (Figure 5B). To determine whether the changes in 3 DNMT protein levels caused by TRAF6 depletion translated to changes in global DNA methylation, we determined the global methylation in TRAF6-WT and TRAF6-KO cells by dot blot analysis using anti-5mC antibody (Figure 5C). Consistent with the increased DNMT levels in the TRAF6-KO cells, global methylation levels of 5mC were substantially increased in these cells when compared with the WT cells (Figure 5C). To further confirm that TRAF6-dependent degradation of all 3 DNMTs depends on its E3 ligase activity, we transfected WT and E3 ligase catalytic inactive forms of TRAF6 into TRAF6-KO cells and found that only the WT TRAF6 promoted degradation of DNMTs, but not the TRAF6 C70A mutant that lacks E3 ligase activity (Figure 5D). Taken together, these results suggest that TRAF6 is the E3 ubiquitin ligase for all 3 DNMTs and that it regulates their protein stability through a lysosome-mediated degradation pathway.
Figure 5. TRAF6 E3-ligase activity is essential for the ubiquitination.
(A) Breast cancer cell lines were transfected with negative or TRAF6-specific siRNAs for 72 hours. Cell lysates were subjected to Western blot to determine DNMT protein levels with the indicated antibodies. (B) Cell lysates from the WT and Crispr/Cas9 TRAF6-KO (KO1) MDA-MB-231 cell lines were subjected to Western blot analysis as described above. (C) Dot blot for detecting global DNA methylation. Genomic DNA from WT and Crispr/Cas9 TRAF6-KO cells were isolated and dot blotted with specific anti-5mC antibody. Two-fold dilution of 1 μg of genomic DNA isolated from each sample was spotted on the membrane, followed by incubation with the antibody. (D) MDA-MB-231 Crispr/Cas9 TRAF6-KO cells were transfected with FLAG-tagged WT TRAF6 (WT) and catalytic-dead CA mutant (C70A). Cell lysates were subjected to Western blot analysis with the indicated antibodies. (E) MDA-MB-231 TRAF6-WT and Crispr/Cas9 TRAF6-KO cells were treated with decitabine for 6 days with or without concanamycin A for an additional 24 hours. Cell lysates were subjected to Western blot analysis with the indicated antibodies. (F) MDA-MB-231 TRAF6-WT and Crispr/Cas9 TRAF6-KO cells were treated with the indicated doses of decitabine for 7 days, and colony-formation assay was performed. Crispr/Cas9 TRAF6-KO clone 1 (KO1) or clone 2 (KO2) were used. Data represent mean ± SD (n = 3 independent experiments). (G) Crispr/Cas9 TRAF6-KO cells were transfected with TRAF6-WT and CA mutants. Cells were treated with the indicated doses of decitabine for 7 days, and colony-formation assay was performed as above. Data represent mean ± SD (n = 3 independent experiments).
Decitabine induces ubiquitination and degradation of DNMTs through TRAF6.
Our data demonstrate that decitabine induces ubiquitination of all 3 DNMTs (Figure 3G) and mediates their protein degradation in a lysosome-dependent manner (Figure 3F). To determine whether the degradation of DNMTs following decitabine treatment is dependent on TRAF6, we treated control and TRAF6-depleted cells with decitabine (Figure 5E). Depletion of TRAF6 reversed the degradation of DNMTs induced by decitabine (Figure 5E). These results reveal that decitabine-induced DNMT degradation is dependent on TRAF6.
To further determine whether the degradation process might affect decitabine efficacy, we performed colony-formation assays. In the TRAF6-KO group, more colonies formed following decitabine treatment compared with in the control group (Figure 5F), suggesting that the intact TRAF6 gene is essential for decitabine’s inhibition of breast cancer cell growth. Reconstitution of WT TRAF6, but not the TRAF6 C70A mutant, restored the sensitivity to decitabine treatment, further confirming that TRAF6 E3 ligase activity is essential in this process (Figure 5G). These results suggest that TRAF6 determines breast cancer cell sensitivity to decitabine treatment through the regulation of protein stability of DNMT isoforms.
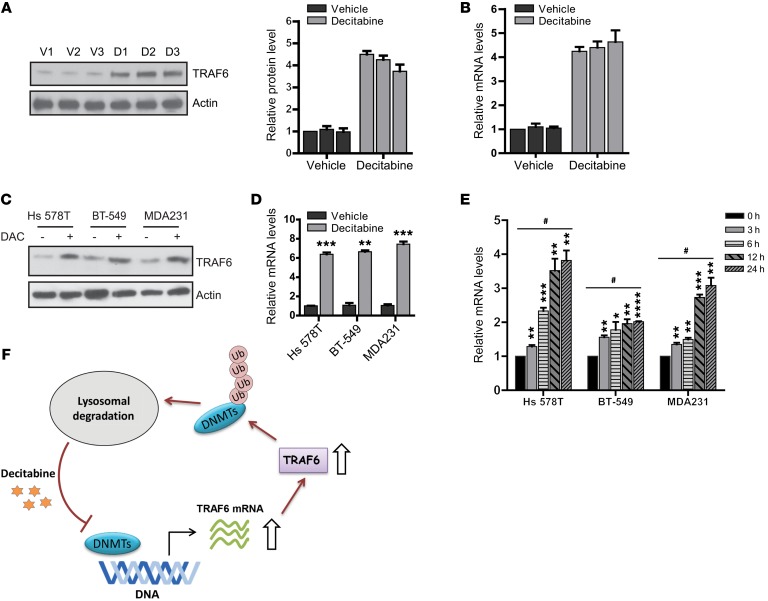
Decitabine treatment induces TRAF6 mRNA expression.
To further explore the mechanism of how decitabine induces TRAF6-mediated degradation of DNMTs, we assessed TRAF6 expression in vivo both before and after decitabine treatment. We observed elevated TRAF6 protein levels in PDX tumors after decitabine treatment, as shown in Figure 6A. We further examined TRAF6 mRNA expression by real-time reverse transcriptase-PCR (qRT-PCR) analysis. Consistent with elevated TRAF6 protein levels, a significant increase in mRNA levels was observed in PDX tumors treated with decitabine compared with vehicle (Figure 6B). These results were confirmed in 3 different TNBC cell lines (Figure 6, C and D). Furthermore, TRAF6 expression was induced by decitabine in a time-dependent manner (Figure 6E). These data show that decitabine treatment induces TRAF6 expression, resulting in increased protein levels.
Figure 6. Decitabine treatment induces TRAF6 expression.
(A) Tumor samples isolated from 3 PDX models treated with vehicle (V) or decitabine (D) were lysed and analyzed with the indicated antibodies. The protein level of TRAF6 was quantified. (B) Quantification of TRAF6 mRNA levels by qRT-PCR. Data represent mean ± SD from 3 independent experiments. (C) Hs 578T, BT-549, and MDA-MB-231 cells were treated with decitabine and the cell lysates were analyzed by the indicated antibodies. (D) Quantification of TRAF6 mRNA levels in Hs 578T, BT-549, and MDA-MB-231 cells by qRT-PCR. Data represent mean ± SD from 3 independent experiments. **P < 0.01; ***P < 0.001, 2-tailed t tests. (E) MDA-MB-231 was treated with decitabine, and the cells were harvested at the indicated time point. Quantification of TRAF6 mRNA levels was performed by qRT-PCR. Data represent mean ± SD from n = 3 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, 2-tailed t tests. One-way ANOVA with post test comparisons against controls (Dunnett’s test) showed significant differences compared with control group. #P < 0.0001, multiplicity adjusted. (F) Model for decitabine action at low dosage.
Discussion
Identification of biomarkers is critical for selecting patients who have a high likelihood of response to a given treatment. In breast cancer, ER expression and HER2 gene amplification/expression are targets for endocrine therapy and anti-HER2–based therapies, respectively (43–45). However, there is a lack of such targets in TNBC pointing to effective therapies. New drug targets are greatly needed. Because of the prominent role of epigenetic regulation in TNBC, targeting epigenetic regulators has shown promising benefit in a series of TNBC cell lines (46).
Demethylating agents have been shown to be effective in the treatment of hematological malignancies, and their efficacy in solid tumors was also evaluated in a variety of solid cancers (26, 47). Besides azacitidine and decitabine, which are the only demethylating drugs approved by the FDA for the treatment of MDS (48, 49), other DNMT inhibitors, such as guadecitabine, hydralazine, and MG98, have been investigated as anticancer agents (47). In patients with ovarian cancer, increased sensitivity was observed toward carboplatin after treatment with either azacitidine (50) or decitabine (51, 52). In patients with non–small cell lung carcinoma (NSCLC), increased efficacy of subsequent anticancer therapy was observed in patients who received prior azacitidine (53). However, a recent phase II clinical trial, including 13 patients with advanced TNBC treated with azacidine and the histone deacetylase inhibitor entinostat, reported by Connolly et al. showed no clinical response in these patients (54). All of these studies suggest that, in patients with solid tumors, more research is required to identify a subgroup of patients who might benefit from DNA demethylating agents.
Even in patients with MDS or acute myeloid leukemia (AML), only 50% have shown a clinical response to DNMT inhibitors. Prior studies have attempted to identify predictive biomarkers in patients with MDS or AML, but most found no or relatively weak correlation between variation in cancer genomes or epigenomes with response to demethylating drugs. Additionally, decitabine response was not shown to be associated with the methylation status of specific genes (55, 56). Similarly, genome-wide promoter DNA methylation studies have not yielded a robust predictive signature for response to these drugs (57). However, recent studies indicated that activation in the RAS/MEK/ERK pathway and DNMT1 mRNA expression are correlated with the cytotoxic activity of decitabine in ovarian cancer (29). More efforts are needed to identify molecular markers for prediction of decitabine response in cancers, including breast cancer.
In the current study, we demonstrated that in the TNBC PDX–derived organoids originating from primary tumors that were either responsive or resistant to chemotherapy (Supplemental Table 1) as well as tumors from after 20 weeks of NAC, DNMT protein levels, rather than mRNA levels, were highly correlated with decitabine sensitivity (Figure 1, D–F). Preclinical PDX models also showed greater effects of decitabine in tumors expressing higher level of DNMTs (Figure 2B). These findings suggest that DNMT protein levels may be biomarkers for decitabine response in TNBCs. For patients who might have developed resistance to standard chemotherapy and for whom the choices of alternative therapies are limited, decitabine could be very effective in a subset who have high levels of DNMTs. Although a few studies in solid tumors have shown that overexpression of DNMTs is associated with resistance to chemotherapy (58, 59), in our limited number of PDX-derived organoid models, we did not find a correlation between DNMT levels and NAC response using the corresponding clinical pathological complete response (pCR) for these organoids. However, in organoids that expressed high levels of DNMTs, decitabine could sensitize the organoids to paclitaxel, the first chemotherapy in the NAC regimen used in patients from whom PDX organoids were developed (Supplemental Figure 1A). This sensitization effect was not dependent on paclitaxel response. We understand that the PDX-derived organoids might be different from original human tumors at the molecular level. However, studies, including ours and many others, suggest that these preclinical models can faithfully represent human tumor biology and clinical treatment response (34, 60). Obviously, additional clinical and preclinical studies are required to validate all the findings.
Even though many mechanisms have been proposed for the antitumor activity of decitabine, the mechanisms underlying how decitabine suppresses tumor growth require more investigation. Decitabine was first thought to be incorporated into DNA, replacing cytosine in the DNA, leading to the formation of a covalent bond between DNMT1 and the 5-aza-cytosine ring that is trapped on the DNA. The replication fork progresses in the absence of DNMT1, resulting in passive loss of DNA methylation in the nascent strand, but not the DNA template (61). DNMT1 was also found to be involved in controlling cell growth (62). However, in this study, we found that a new mechanism involving the E3 ligase, TRAF6, mediated degradation of DNMTs following low concentration of decitabine treatment. A prior study has shown that decitabine induces proteosomal-dependent DNMT1 degradation by APC/CCdh1 in cervical cancer cells (HeLa) and the HCT116 colon cancer cell line (63, 64). In our study, we found that, not only DNMT1, but also DNMT3A and DNMT3B, were degraded following low-concentration decitabine treatment in multiple breast cancer cell lines (Figure 3C) and PDX tumor tissues (Figure 3A). MG132 only partially inhibited their degradation, but the lysosome inhibitor, concanamycin A, dramatically reversed the degradation process (Figure 3F). It is possible that the difference between our observations and previous studies might be due to the different cancer cells and the dose and duration of decitabine treatment. Low-dose decitabine has been used in a recent clinical trial for advanced lung cancer patients, where it led to a stable and durable response in some patients (53). Therefore, in our study, we treated the cells and PDX organoids with nanomolar concentrations of decitabine (27), and we observed a significant effect on growth inhibition in both breast cancer cell lines (Figure 3E) and PDX organoids (Figure 1D). Our findings identify a mechanism of action of DNMT inhibitors, which may be useful to elucidate resistance mechanisms for these drugs (65).
The ubiquitin system is composed of a ubiquitin-activating enzyme (E1), ubiquitin-carrier proteins (E2 enzymes; also known as ubiquitin-conjugating enzymes), and ubiquitin ligases (E3 enzymes) (66). The ubiquitin protein is a small protein, and its molecular weight is around 9 kd. Ubiquitin possesses 7 lysine residues (K6, K11, K27, K29, K33, K48, and K63) and an N-terminal methionine (M1) that may be utilized to form polyubiquitin chains. The human genome encodes about 800 E3 enzymes recognizing a large number of substrates, controlling different linkage-chain formation, and manipulating complicated signaling in the cells (67). The various types of linkage are usually associated with different cellular functions. K48 and K63 linkage ubiquitinations are the 2 main forms of polyubiquitination, responsible for 52% and 38% of all ubiquitination events in HEK293 cells, respectively (68). K48-linked polyubiquitin chains are involved in proteasome-dependent degradation, while K63-linked ubiquitination is a docking site for mediating protein-protein interactions or conformational changes. However, recent studies have also shown that K63-linked ubiquitination can mediate protein degradation. Sic1, dihydrofolate reductase, cyclin B1, and troponin 1 are all targeted for proteasome-dependent degradation mediated by the K63 linkage ubiquitination (69–72). STUB1 interacts with HIF1A and mediates its K63-linked ubiquitination, leading to the degradation of HIF1A through the chaperone-mediated autophagy pathway (73). K63-linked ubiquitination was also reported to selectively facilitate the clearance of inclusions associated with neurodegenerative diseases via autophagy (74).
TRAF6 is an E3 ubiquitin protein ligase that catalyzes lysine 63–linked (K63-linked) polyubiquitination and functions in Toll-like receptor signaling (75) and oncogenic activation of Akt (42). But it is also found to mediate KIF23 degradation through an autophagy pathway (76). The TRAF6-binding consensus sequence (PxExxAr/Ac) is a highly conserved protein–protein interaction motif (77, 78) that mediates the binding between TRAF6 and its substrates. We also found this conserved motif in DNMTs. Our experiments with endogenous immunopreciptation further confirmed the interaction of TRAF6 and DNMTs. TRAF6 contains a TRAF-C domain, a coiled-coil domain, 5 zinc fingers, and a RING domain (40). TRAF6 deletion constructs also showed TRAF6 binding to DNMTs through its TRAF-C domain, a domain that is known to bind to many TRAF6 substrates (78). The interactions led to TRAF6 ubiquitination of all 3 DNMTs (Figure 4, H–J) and promoted their degradation (Figure 5, A and B). However, the TRAF6 C70A mutant, lacking the E3 ligase activity, lost its capability to degrade DNMTs (Figure 4J and Figure 5C).
Our study has identified the DNMTs as new targets of K63-linked polyubiquitination through lysosome-mediated degradation (Figure 4, B–E, H, and I; and Figure 3F). In cancer cells, DNMT proteins are polyubiquitinated by the E3 ligase, TRAF6, thereby inducing lysosome-dependent protein degradation (Figure 3, F and G). Although several other proteins were also known to be degraded in this manner, such as GABAB1 and mind bomb-2 (79), the detailed mechanisms involved in K63-linked polyubiqutination–mediated lysosomal degradation need further investigation. In our study, decitabine treatment upregulated TRAF6 protein levels and mRNA levels (Figure 6, A–D), leading to enhanced degradation of DNMTs (Figure 5, D and E). However, how TRAF6 transcription is upregulated by decitabine remains unclear. The TRAF6 gene is not generally DNA methylated at promoter regions in breast cancer cells (Supplemental Table 2), indicating decitabine activation of TRAF6 may be through a pathway upstream of TRAF6. Kawai et al. has shown that TRAF6 activity was required for IFN regulatory factor 7 (IRF7) activation (80). Recent studies also indicated that decitabine treatment may induce the cancer cell endogenous retrovirus and activate IRF7 (81, 82), indicating that decitabine may activate endogenous retrovirus and antiviral responses that could upregulate TRAF6.
Taking these data together, our study suggests that decitabine induces degradation of DNMT1, DNMT3A, and DNMT3B by TRAF6 through a lysosome-dependent protein-degradation pathway (Figure 6F). This is one major mechanism by which decitabine inhibits tumor growth. We also found that growth in tumors with high DNMT protein levels was greatly suppressed by decitabine treatment, suggesting that DNMT protein levels might serve as a potential biomarker to guide drug selection in the clinic. Finally, TNBC PDX responded to decitabine regardless of chemotherapy response. Therefore, TNBC patients with high DNMT levels and resistance to standard chemotherapy may still benefit from decitabine. Future clinical studies are required to test decitabine or decitabine-like drugs in high-risk TNBC patients.
Methods
In vivo mouse maintenance and decitabine drug treatment.
Breast cancer PDX models were generated from baseline prechemotherapy percutaneous biopsy as described previously (33, 34). Liquid nitrogen–preserved tumor lines were thawed and injected in 6- to 8-week-old female NSG mice (Jackson Laboratory). When xenograft primary tumors reached 1 cm in diameter, mice were sacrificed and tissue fragments were transplanted to 6- to 8-week-old female NOD-SCID mice for the drug tests. Tumor growth was monitored twice weekly by a caliper. Once tumors reached 150 to 250 mm3, mice were randomized into control (vehicle, saline solution) or decitabine (5 mg/kg, i.p. 3 times weekly) groups, with each group consisting of 7 to 8 mice. The dose used in mice was converted from the low dose, 15 mg/m2/d, used in treating human hematopoietic malignancies. This dose in humans can achieve nanomolar plasma concentration (83, 84). The dose conversion and calculation were based on the FDA recommendation (https://www.fda.gov/downloads/drugs/guidances/ucm078932.pdf) and literature (85).
PDX tumor dissociation and human breast cancer cell isolation.
When tumors grew to 1 cm in diameter, tumors were dissected and weighed. For tissue dissociation, a portion of tumor tissue was dissected and cut into 2 mm3 sections in sterile PBS and incubated with 5 ml of the enzyme mix from a human Tumor Dissociation Kit (Miltenyi Biotech) prepared according to the manufacturer’s protocol. Tissue was digested at 37°C with rotation on a gentleMACS Dissociator (Miltenyi Biotech) program for “dissociation of tough tumors” according to the manufacturer’s protocol. The digested cell mixture was passed through a 70 μm MACS SmartStrainer, then a 40 μm MACS SmartStrainer to obtain the single-cell suspension. Dissociated cells were then spun down and washed with precooled washing buffer. Purified breast cancer tumor cells were obtained by removing the mouse cells using the Mouse Cell Depletion Purification Kit (Miltenyi Biotech) according to the manufacturer’s protocol.
Formation of spheroids from PDX tumors and evaluation of decitabine response.
For 3D cell culture, we used ultralow attachment NanoCulture Plate (NCP) (Scivax Corp), which is a scaffold-based 3D culture system that easily allows tumors to form spheroids (86). 50 μl MEF full medium (DMEM supplemented with 10% FBS, 1% glutamax [Life Technologies], 1% sodium pyruvate [Life Technologies], nonessential amino acids [Life Technologies], and 1% penicillin-streptomycin [Life Technologies]) with 5 μM ROCK inhibitor (Tocris) was placed into each well of a 96-well NanoCulture plate. Plates were spun at 300 g, and cells were then seeded at a final density of 2 × 104 cells in 90 μl medium. Growth medium was changed at day 7 using full MEF medium without adding ROCK inhibitor. Tumor spheroids were then incubated with various concentrations of decitabine for another 7 days. Fresh decitabine was added daily. Cell spheroids were counted using a Nikon Eclipse Ts100 (Nikon Instruments Inc.), and the susceptibility of the cells to decitabine was investigated using the percentage of viability.
Cell culture.
Human breast cancer Hs 578T (HTB-126), MDA-MB-231 (HTB-26), BT-549 (HTB-122), and HEK 293T cells were purchased from ATCC. Hs 578T and HEK 293T cells were cultured in DMEM (Gibco; Thermo Fisher Scientific) containing 10% FBS in 37°C with 5% CO2. BT-549 cells were cultured in RPMI-1640 (Gibco; Thermo Fisher Scientific) containing 10% FBS in 37°C with 5% CO2. MDA-MB-231 cells were cultured in Leibovitz’s L-15 medium (ATCC) containing 10% FBS in 37°C without CO2.
Breast cancer tumor spheroids were cultured in full MEF medium of DMEM (Gibco, Thermo Fisher Scientific) supplemented with 100 U/ml penicillin/streptomycin (Life Technologies), 2 mM Glutamax (Life Technologies), 1 × MEM NEAA (Thermo Fisher), and 1 mM sodium pyruvate (Corning) with or without 5 μM ROCK inhibitor (Tocris).
Chemicals, antibodies and plasmids.
S/FLAG/SBP-tagged TRAF6 construct was generated using TRAF6-WT plasmids purchased from Origen. All mutants were generated by site-directed mutagenesis (Stratagene), and sequences were verified by Sanger sequencing. Antibodies against DNMT1 (catalog 5032S) and DNMT3A (catalog 3598S, 2160S) were purchased from Cell Signaling Technology. Mouse monoclonal DNMT3B antibody (clone 52A1018) was purchased from Novus. Anti-FLAG (clone m2), anti-HA (clone HA-7), and anti–β-actin (clone AC-15) antibodies were purchased from MilliporeSigma. Anti-5mC antibody (catalog NA81) was purchased from MilliporeSigma. Purified recombinant protein of Myc-TRAF6 was purchased from Biocompare (catalog TP319528). Purified GST-DNMT1 was purchased from Active Motif (catalog 31335); prokaryotic-expressing Flag-tagged DNMT3A (catalog 31406) and His-tagged DNMT3B (catalog 31413) were purchased from Active Motif.
Immunoprecipitation and immunoblotting assays.
Methods used for performing immunoprecipitation and immunoblotting were described previously (87). Specifically, cells were lysed with sonication in NETN buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40) containing 50 mM β-glycerophosphate, 10 mM NaF, and protease inhibitor cocktail. Whole cell lysates were obtained by centrifugation and were then incubated with 4 μg antibodies and protein A/G sepharose beads (Amersham Biosciences) at 4°C overnight. The immunocomplexes were then washed 3 times with ice-cold NETN buffer. Proteins were eluted in 1× SDS loading buffer and then subjected to Western blot.
Dot blot assay for detecting global methylation.
The dot blot has been previously described (88). Briefly, isolated DNA was denatured in 0.1 M NaOH for 10 minutes at 95°C. Samples were neutralized with 1 M NH4OAC on ice, diluted by 2-fold, and spotted on the Immobilon-Ny+ Charged Nylon Membrane (MilliporeSigma). The blotted membrane was then washed in 2× SSC buffer, dried at 80°C for 5 minutes, and UV cross-linked at 120,000 μJ/cm2. The membrane was then blocked in Odyssey buffer (Li-Cor) diluted 1:1 in PBS overnight at 4°C. Mouse anti-5mC monoclonal antibody (Active Motif, 1:500) in Odyssey PBS was added for 3 hours at room temperature.
Colony-formation assay.
Cells were treated with the indicated concentrations of decitabine for 7 days and changed to fresh medium for another 5 to 10 days at 37°C, 5% CO2 to allow colony formation. Colonies were then stained with 5% GIEMSA and counted. Results were generated from 3 independent experiments and normalized for plating efficiencies.
CRISPR/Cas9-KO.
For CRISPR/Cas9-KO of human TRAF6 in MDA-MB-231 cells, the following small guide RNAs (sgRNAs) were used: sgTRAF6-1 (5′-AGTTGACAATGAAATACTGC-3′) and sgTRAF6-2 (5′-ACGTGAGATTCTTTCTCTGA-3′). The sgRNA sequences were cloned into the vector Lenti CRISPR-V2-puro. Cells were infected with Lenti-TRAF6-sgRNA-puro, followed by extensive selection with 2 μg/ml puromycin. Single colonies were obtained by serial dilution and amplification. Clones were identified by immunoblotting with anti-TRAF6 antibody and were verified by DNA sequencing.
mRNA extraction and qRT-PCR.
Total RNA was extracted from indicated cells using Quick-RNA MiniPrep Kit (Zymo Research) according to the manufacturer’s instructions. qRT-PCR was performed with the Power SYBR Green RNA-to-CT 1-Step Kit (AB) using the Stratagene Mx3005P Real-Time PCR Detection System (Stratagene). The primers for different genes tested were purchased from QIAGEN. All experiments were performed in triplicate.
For further information, see Supplemental Experimental Procedures.
Statistics.
The statistical data were generated from 3 independent experiments. Statistical analysis was performed by 2-tailed Student’s t test for 2-group comparisons and by 1-way ANOVA along with Dunnett’s multiple comparisons (comparing to control group) wherever appropriate.
Study approval.
All animal studies were reviewed and approved by the Mayo Clinic Institutional Animal Care and Use Committee (A17713, A43112, and A35715).
Author contributions
JY, BQ, JCB, MPG, and LW designed the study. JY, BQ, SQ, YZ, and SWL performed the experiments and collected the data. JY, BQ, AMM, SN, TL, DL, and KRK analyzed the data. AMM performed the pathology. JY, BQ, AMM, SN, and LW wrote and revised the manuscript. KRK, DWV, JAC, SAM, AMA, DWN, RJG, ZL, VJS, RW, JCB, MPG, and LW contributed to the interpretation of the results and supervised this project. All authors read and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
This work was supported by the Mayo Clinic Center for Individualized Medicine; Nadia’s Gift Foundation; John P. Guider; the Eveleigh Family; the Pharmacogenomics Research Network (grant U10GM 61388-15 to RW, LW, and MPG); the NIH (RO1 GM28157 to RW, CA 196648 to LW); the Mayo Clinic Cancer Center (grant CA15083-40A2 to MPG); the George M. Eisenberg Foundation for Charities; the Mayo Clinic Breast SPORE (grant P50CA 116201-9 to MPG, DWV, VJS, and KRK); the Prospect Creek Foundation; the Randy Shaver Cancer Research and Community Fund; and the Clinical and Translational Science Awards program (grant UL1 TR002377 from the National Center for Advancing Translational Sciences [NCATS], a component of the NIH).
Version 1. 04/30/2018
Electronic publication
Version 2. 06/01/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(6):2376–2388.https://doi.org/10.1172/JCI97924.
Contributor Information
Jia Yu, Email: yu.jia@mayo.edu.
Bo Qin, Email: qin.bo@mayo.edu.
Somaira Nowsheen, Email: nowsheen@uab.edu.
Tongzheng Liu, Email: Liutongzheng@jnu.edu.cn.
Sisi Qin, Email: qin.sisi@mayo.edu.
Yongxian Zhuang, Email: Zhuang.yongxian@mayo.edu.
Duan Liu, Email: Liu.duan@mayo.edu.
Alvaro Moreno-Aspitia, Email: MorenoAspitia.Alvaro@mayo.edu.
Zhenkun Lou, Email: lou.zhenkun@mayo.edu.
Richard Weinshilboum, Email: weinshilboum.richard@mayo.edu.
Judy C. Boughey, Email: boughey.judy@mayo.edu.
Liewei Wang, Email: wang.liewei@mayo.edu.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Gluz O, Liedtke C, Gottschalk N, Pusztai L, Nitz U, Harbeck N. Triple-negative breast cancer--current status and future directions. Ann Oncol. 2009;20(12):1913–1927. doi: 10.1093/annonc/mdp492. [DOI] [PubMed] [Google Scholar]
- 3.Lehmann BD, Pietenpol JA. Identification and use of biomarkers in treatment strategies for triple-negative breast cancer subtypes. J Pathol. 2014;232(2):142–150. doi: 10.1002/path.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) et al. Comparisons between different polychemotherapy regimens for early breast cancer: meta-analyses of long-term outcome among 100,000 women in 123 randomised trials. Lancet. 2012;379(9814):432–444. doi: 10.1016/S0140-6736(11)61625-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 6.Roll JD, Rivenbark AG, Jones WD, Coleman WB. DNMT3b overexpression contributes to a hypermethylator phenotype in human breast cancer cell lines. Mol Cancer. 2008;7:15. doi: 10.1186/1476-4598-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roll JD, et al. Dysregulation of the epigenome in triple-negative breast cancers: basal-like and claudin-low breast cancers express aberrant DNA hypermethylation. Exp Mol Pathol. 2013;95(3):276–287. doi: 10.1016/j.yexmp.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Silverman LR. DNA methyltransferase inhibitors in myelodysplastic syndrome. Best Pract Res Clin Haematol. 2004;17(4):585–594. doi: 10.1016/S1521-6926(04)00076-3. [DOI] [PubMed] [Google Scholar]
- 9.Kaminskas E, et al. Approval summary: azacitidine for treatment of myelodysplastic syndrome subtypes. Clin Cancer Res. 2005;11(10):3604–3608. doi: 10.1158/1078-0432.CCR-04-2135. [DOI] [PubMed] [Google Scholar]
- 10. National Comprehensive Cancer Network. NCCN clinical practice guidelines in oncology version 2.2013: acute myeloid leukemia. http://www.nccn.org/professionals/physician_gls/pdf/aml.pdf Accessed March 26, 2018.
- 11.Lee YG, et al. Comparative analysis between azacitidine and decitabine for the treatment of myelodysplastic syndromes. Br J Haematol. 2013;161(3):339–347. doi: 10.1111/bjh.12256. [DOI] [PubMed] [Google Scholar]
- 12.Fenaux P, et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol. 2009;10(3):223–232. doi: 10.1016/S1470-2045(09)70003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Creusot F, Acs G, Christman JK. Inhibition of DNA methyltransferase and induction of Friend erythroleukemia cell differentiation by 5-azacytidine and 5-aza-2’-deoxycytidine. J Biol Chem. 1982;257(4):2041–2048. [PubMed] [Google Scholar]
- 14.Christman JK, Mendelsohn N, Herzog D, Schneiderman N. Effect of 5-azacytidine on differentiation and DNA methylation in human promyelocytic leukemia cells (HL-60) Cancer Res. 1983;43(2):763–769. [PubMed] [Google Scholar]
- 15.Jeltsch A. Molecular enzymology of mammalian DNA methyltransferases. Curr Top Microbiol Immunol. 2006;301:203–225. doi: 10.1007/3-540-31390-7_7. [DOI] [PubMed] [Google Scholar]
- 16.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69(6):915–926. doi: 10.1016/0092-8674(92)90611-F. [DOI] [PubMed] [Google Scholar]
- 17.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 18.Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20(1):85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 19.Christman JK. 5-Azacytidine and 5-aza-2’-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21(35):5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 20.Jüttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2’-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci U S A. 1994;91(25):11797–11801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wijermans P, et al. Low-dose 5-aza-2’-deoxycytidine, a DNA hypomethylating agent, for the treatment of high-risk myelodysplastic syndrome: a multicenter phase II study in elderly patients. J Clin Oncol. 2000;18(5):956–962. doi: 10.1200/JCO.2000.18.5.956. [DOI] [PubMed] [Google Scholar]
- 22.Issa JP, et al. Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2’-deoxycytidine (decitabine) in hematopoietic malignancies. Blood. 2004;103(5):1635–1640. doi: 10.1182/blood-2003-03-0687. [DOI] [PubMed] [Google Scholar]
- 23.Kantarjian H, et al. Results of a randomized study of 3 schedules of low-dose decitabine in higher-risk myelodysplastic syndrome and chronic myelomonocytic leukemia. Blood. 2007;109(1):52–57. doi: 10.1182/blood-2006-05-021162. [DOI] [PubMed] [Google Scholar]
- 24.Issa JP, Kantarjian HM. Targeting DNA methylation. Clin Cancer Res. 2009;15(12):3938–3946. doi: 10.1158/1078-0432.CCR-08-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Momparler RL, Bouffard DY, Momparler LF, Dionne J, Belanger K, Ayoub J. Pilot phase I-II study on 5-aza-2’-deoxycytidine (Decitabine) in patients with metastatic lung cancer. Anticancer Drugs. 1997;8(4):358–368. doi: 10.1097/00001813-199704000-00008. [DOI] [PubMed] [Google Scholar]
- 26.Cowan LA, Talwar S, Yang AS. Will DNA methylation inhibitors work in solid tumors? A review of the clinical experience with azacitidine and decitabine in solid tumors. Epigenomics. 2010;2(1):71–86. doi: 10.2217/epi.09.44. [DOI] [PubMed] [Google Scholar]
- 27.Tsai HC, et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell. 2012;21(3):430–446. doi: 10.1016/j.ccr.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Girault I, Tozlu S, Lidereau R, Bièche I. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin Cancer Res. 2003;9(12):4415–4422. [PubMed] [Google Scholar]
- 29.Stewart ML, et al. KRAS genomic status predicts the sensitivity of ovarian cancer cells to decitabine. Cancer Res. 2015;75(14):2897–2906. doi: 10.1158/0008-5472.CAN-14-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simó-Riudalbas L, Melo SA, Esteller M. DNMT3B gene amplification predicts resistance to DNA demethylating drugs. Genes Chromosomes Cancer. 2011;50(7):527–534. doi: 10.1002/gcc.20877. [DOI] [PubMed] [Google Scholar]
- 31.Griffith LG, Swartz MA. Capturing complex 3D tissue physiology in vitro. Nat Rev Mol Cell Biol. 2006;7(3):211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 32.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5(9):675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 33.Goetz MP, et al. Tumor sequencing and patient-derived xenografts in the neoadjuvant treatment of breast cancer. J Natl Cancer Inst. 2017;109(7):djw306. doi: 10.1093/jnci/djw306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu J, et al. Establishing and characterizing patient-derived xenografts using pre-chemotherapy percutaneous biopsy and post-chemotherapy surgical samples from a prospective neoadjuvant breast cancer study. Breast Cancer Res. 2017;19(1):130. doi: 10.1186/s13058-017-0920-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borges S, et al. Pharmacologic reversion of epigenetic silencing of the PRKD1 promoter blocks breast tumor cell invasion and metastasis. Breast Cancer Res. 2013;15(2):R66. doi: 10.1186/bcr3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aparicio S, Hidalgo M, Kung AL. Examining the utility of patient-derived xenograft mouse models. Nat Rev Cancer. 2015;15(5):311–316. doi: 10.1038/nrc3944. [DOI] [PubMed] [Google Scholar]
- 37.Taylor SM, Jones PA. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell. 1979;17(4):771–779. doi: 10.1016/0092-8674(79)90317-9. [DOI] [PubMed] [Google Scholar]
- 38.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 39.Schneider M, et al. The innate immune sensor NLRC3 attenuates Toll-like receptor signaling via modification of the signaling adaptor TRAF6 and transcription factor NF-κB. Nat Immunol. 2012;13(9):823–831. doi: 10.1038/ni.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin Q, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nat Struct Mol Biol. 2009;16(6):658–666. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L, Xu M, Scotti E, Chen ZJ, Tontonoz P. Both K63 and K48 ubiquitin linkages signal lysosomal degradation of the LDL receptor. J Lipid Res. 2013;54(5):1410–1420. doi: 10.1194/jlr.M035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang WL, et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science. 2009;325(5944):1134–1138. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romond EH, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353(16):1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 44.Piccart-Gebhart MJ, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353(16):1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 45.Smith I, et al. 2-year follow-up of trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer: a randomised controlled trial. Lancet. 2007;369(9555):29–36. doi: 10.1016/S0140-6736(07)60028-2. [DOI] [PubMed] [Google Scholar]
- 46.Yang X, et al. Transcriptional activation of estrogen receptor alpha in human breast cancer cells by histone deacetylase inhibition. Cancer Res. 2000;60(24):6890–6894. [PubMed] [Google Scholar]
- 47.Linnekamp JF, Butter R, Spijker R, Medema JP, van Laarhoven HW. Clinical and biological effects of demethylating agents on solid tumours - A systematic review. Cancer Treat Rev. 2017;54:10–23. doi: 10.1016/j.ctrv.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 48.Issa JP, Kantarjian HM, Kirkpatrick P. Azacitidine. Nat Rev Drug Discov. 2005;4(4):275–276. doi: 10.1038/nrd1698. [DOI] [PubMed] [Google Scholar]
- 49.Kirkpatrick P. Decitabine. Nat Rev Drug Discov. 2006;5(11):891–892. doi: 10.1038/nrd2180. [DOI] [PubMed] [Google Scholar]
- 50.Fu S, et al. Phase 1b-2a study to reverse platinum resistance through use of a hypomethylating agent, azacitidine, in patients with platinum-resistant or platinum-refractory epithelial ovarian cancer. Cancer. 2011;117(8):1661–1669. doi: 10.1002/cncr.25701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matei D, et al. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res. 2012;72(9):2197–2205. doi: 10.1158/0008-5472.CAN-11-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fang F, et al. A phase 1 and pharmacodynamic study of decitabine in combination with carboplatin in patients with recurrent, platinum-resistant, epithelial ovarian cancer. Cancer. 2010;116(17):4043–4053. doi: 10.1002/cncr.25204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Juergens RA, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 2011;1(7):598–607. doi: 10.1158/2159-8290.CD-11-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Connolly RM, et al. Combination epigenetic therapy in advanced breast cancer with 5-azacitidine and entinostat: a phase II National Cancer Institute/Stand Up to Cancer Study. Clin Cancer Res. 2017;23(11):2691–2701. doi: 10.1158/1078-0432.CCR-16-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Traina F, et al. Impact of molecular mutations on treatment response to DNMT inhibitors in myelodysplasia and related neoplasms. Leukemia. 2014;28(1):78–87. doi: 10.1038/leu.2013.269. [DOI] [PubMed] [Google Scholar]
- 56.Bejar R, et al. TET2 mutations predict response to hypomethylating agents in myelodysplastic syndrome patients. Blood. 2014;124(17):2705–2712. doi: 10.1182/blood-2014-06-582809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fandy TE, et al. Early epigenetic changes and DNA damage do not predict clinical response in an overlapping schedule of 5-azacytidine and entinostat in patients with myeloid malignancies. Blood. 2009;114(13):2764–2773. doi: 10.1182/blood-2009-02-203547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang C, Mirkin BL, Dwivedi RS. DNA (cytosine) methyltransferase overexpression is associated with acquired drug resistance of murine neuroblastoma cells. Int J Oncol. 2001;18(2):323–329. doi: 10.3892/ijo.18.2.323. [DOI] [PubMed] [Google Scholar]
- 59.Segura-Pacheco B, et al. Global DNA hypermethylation-associated cancer chemotherapy resistance and its reversion with the demethylating agent hydralazine. J Transl Med. 2006;4:32. doi: 10.1186/1479-5876-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sachs N, et al. a living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172(1-2):373–386.e10. doi: 10.1016/j.cell.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 61.Wu JC, Santi DV. On the mechanism and inhibition of DNA cytosine methyltransferases. Prog Clin Biol Res. 1985;198:119–129. [PubMed] [Google Scholar]
- 62.Szyf M. The role of DNA methyltransferase 1 in growth control. Front Biosci. 2001;6:D599–D609. doi: 10.2741/szyf. [DOI] [PubMed] [Google Scholar]
- 63.Ghoshal K, et al. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol. 2005;25(11):4727–4741. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 64.Patel K, Dickson J, Din S, Macleod K, Jodrell D, Ramsahoye B. Targeting of 5-aza-2’-deoxycytidine residues by chromatin-associated DNMT1 induces proteasomal degradation of the free enzyme. Nucleic Acids Res. 2010;38(13):4313–4324. doi: 10.1093/nar/gkq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Treppendahl MB, Kristensen LS, Grønbæk K. Predicting response to epigenetic therapy. J Clin Invest. 2014;124(1):47–55. doi: 10.1172/JCI69737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258(13):8206–8214. [PubMed] [Google Scholar]
- 67.Adhikari A, Chen ZJ. Diversity of polyubiquitin chains. Dev Cell. 2009;16(4):485–486. doi: 10.1016/j.devcel.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 68.Dammer EB, et al. Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. J Biol Chem. 2011;286(12):10457–10465. doi: 10.1074/jbc.M110.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hofmann RM, Pickart CM. In vitro assembly and recognition of Lys-63 polyubiquitin chains. J Biol Chem. 2001;276(30):27936–27943. doi: 10.1074/jbc.M103378200. [DOI] [PubMed] [Google Scholar]
- 70.Kim HT, et al. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem. 2007;282(24):17375–17386. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]
- 71.Kirkpatrick DS, et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8(7):700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 72.Saeki Y, et al. Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J. 2009;28(4):359–371. doi: 10.1038/emboj.2008.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ferreira JV. K63 linked ubiquitin chain formation is a signal for HIF1A degradation by Chaperone-Mediated Autophagy. Sci Rep. 2015;5:10210. doi: 10.1038/srep10210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tan JM, et al. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum Mol Genet. 2008;17(3):431–439. doi: 10.1093/hmg/ddm320. [DOI] [PubMed] [Google Scholar]
- 75.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 76.Isakson P, Lystad AH, Breen K, Koster G, Stenmark H, Simonsen A. TRAF6 mediates ubiquitination of KIF23/MKLP1 and is required for midbody ring degradation by selective autophagy. Autophagy. 2013;9(12):1955–1964. doi: 10.4161/auto.26085. [DOI] [PubMed] [Google Scholar]
- 77.Pullen SS, Miller HG, Everdeen DS, Dang TT, Crute JJ, Kehry MR. CD40-tumor necrosis factor receptor-associated factor (TRAF) interactions: regulation of CD40 signaling through multiple TRAF binding sites and TRAF hetero-oligomerization. Biochemistry. 1998;37(34):11836–11845. doi: 10.1021/bi981067q. [DOI] [PubMed] [Google Scholar]
- 78.Ye H, et al. Distinct molecular mechanism for initiating TRAF6 signalling. Nature. 2002;418(6896):443–447. doi: 10.1038/nature00888. [DOI] [PubMed] [Google Scholar]
- 79.Zemoura K, Trümpler C, Benke D. Lys-63-linked ubiquitination of γ-aminobutyric acid (GABA), type B1, at multiple sites by the e3 ligase mind bomb-2 targets GABAB receptors to lysosomal degradation. J Biol Chem. 2016;291(41):21682–21693. doi: 10.1074/jbc.M116.750968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kawai T, et al. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat Immunol. 2004;5(10):1061–1068. doi: 10.1038/ni1118. [DOI] [PubMed] [Google Scholar]
- 81.Roulois D, et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162(5):961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chiappinelli KB, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162(5):974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cashen AF, Shah AK, Todt L, Fisher N, DiPersio J. Pharmacokinetics of decitabine administered as a 3-h infusion to patients with acute myeloid leukemia (AML) or myelodysplastic syndrome (MDS) Cancer Chemother Pharmacol. 2008;61(5):759–766. doi: 10.1007/s00280-007-0531-7. [DOI] [PubMed] [Google Scholar]
- 84.Derissen EJ, Beijnen JH, Schellens JH. Concise drug review: azacitidine and decitabine. Oncologist. 2013;18(5):619–624. doi: 10.1634/theoncologist.2012-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nair AB, Jacob S. A simple practice guide for dose conversion between animals and human. J Basic Clin Pharm. 2016;7(2):27–31. doi: 10.4103/0976-0105.177703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yoshii Y, et al. The use of nanoimprinted scaffolds as 3D culture models to facilitate spontaneous tumor cell migration and well-regulated spheroid formation. Biomaterials. 2011;32(26):6052–6058. doi: 10.1016/j.biomaterials.2011.04.076. [DOI] [PubMed] [Google Scholar]
- 87.Yu J, et al. Regulation of serine-threonine kinase Akt activation by NAD+-dependent deacetylase SIRT7. Cell Rep. 2017;18(5):1229–1240. doi: 10.1016/j.celrep.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Blaschke K, et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500(7461):222–226. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.