Abstract
DEP domain–containing 5 protein (DEPDC5) is a repressor of the recently recognized amino acid–sensing branch of the mTORC1 pathway. So far, its function in the brain remains largely unknown. Germline loss-of-function mutations in DEPDC5 have emerged as a major cause of familial refractory focal epilepsies, with case reports of sudden unexpected death in epilepsy (SUDEP). Remarkably, a fraction of patients also develop focal cortical dysplasia (FCD), a neurodevelopmental cortical malformation. We therefore hypothesized that a somatic second-hit mutation arising during brain development may support the focal nature of the dysplasia. Here, using postoperative human tissue, we provide the proof of concept that a biallelic 2-hit — brain somatic and germline — mutational mechanism in DEPDC5 causes focal epilepsy with FCD. We discovered a mutation gradient with a higher rate of mosaicism in the seizure-onset zone than in the surrounding epileptogenic zone. Furthermore, we demonstrate the causality of a Depdc5 brain mosaic inactivation using CRISPR-Cas9 editing and in utero electroporation in a mouse model recapitulating focal epilepsy with FCD and SUDEP-like events. We further unveil a key role of Depdc5 in shaping dendrite and spine morphology of excitatory neurons. This study reveals promising therapeutic avenues for treating drug-resistant focal epilepsies with mTORC1-targeting molecules.
Keywords: Genetics, Neuroscience
Keywords: Epilepsy, Molecular genetics, Neurodevelopment
Introduction
The human brain is genetically mosaic. Accumulation of somatic variants in postmitotic cells from healthy human cerebral tissues has recently been described, emphasizing that somatic mutational events are frequent in the brain (1). In neurodevelopmental disorders such as focal cortical dysplasia (FCD), brain somatic mutations in genes belonging to the mechanistic target of rapamycin complex 1 (mTORC1) pathway are increasingly reported (2, 3), especially activating somatic MTOR variants (4–7). FCD type II neuropathological hallmarks include cortical dyslamination, dysmorphic neurons (type IIa), and balloon cells (type IIb) (8). It is frequently associated with pediatric drug-resistant epilepsies and represents 9% of the epilepsy surgery population (9).
Loss-of-function mutations in DEP domain–containing 5 protein (DEPDC5) are the most common cause of familial focal epilepsies, in which FCD II cases are often reported (10), along with the tragic outcome of sudden unexpected death in epilepsy (SUDEP) (11, 12). DEPDC5 is a GTPase-activating protein that is part of the gap activity toward rags 1 (GATOR1) complex, and its inactivation leads to mTORC1 hyperactivity both in vitro (13) and in vivo (14, 15). mTORC1 is a master regulator of cell physiology, including neuronal growth. The specific brain function of DEPDC5 remains unknown. The fact that only a subset of patients among families with DEPDC5 mutations develop FCD, while other affected family members have apparently nonlesional epilepsy, led us to postulate the occurrence of Knudson’s 2-hit mechanism, which has not been definitively proven to date (16).
Since Depdc5 constitutive heterozygous–knockout rodents do not have an epileptic phenotype (14, 15), major hurdles remain in understanding the molecular and functional etiology of DEPDC5-related focal epilepsy. It is therefore crucial to generate a relevant model of the pathology and develop alternative therapeutic strategies to treat drug-resistant patients.
Results and Discussion
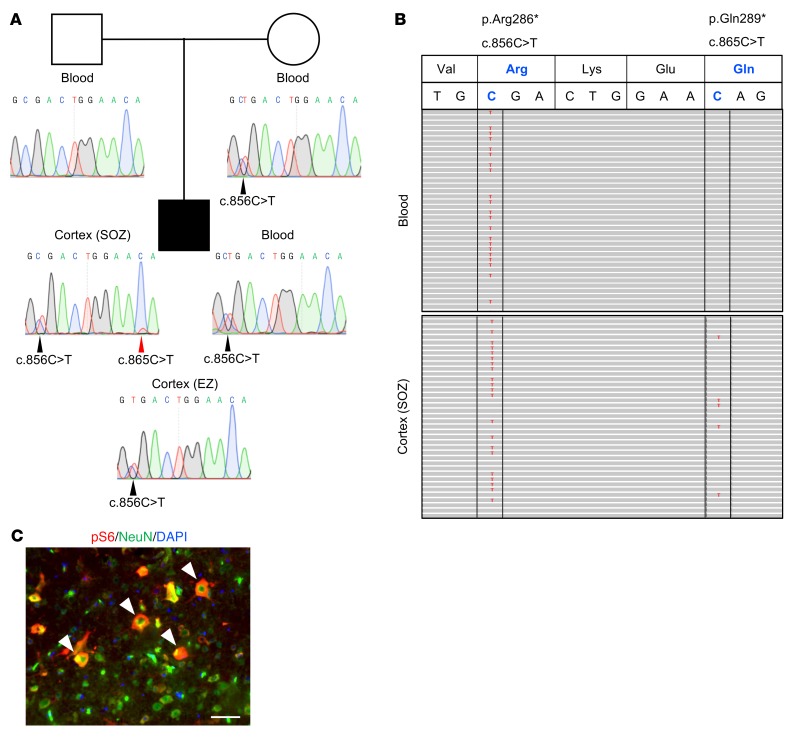
To pursue the 2-hit hypothesis, we searched for somatic variants in a panel of mTORC1 genes using deep sequencing of blood-brain paired DNA samples from 10 FCD II children who underwent surgery to treat their drug-resistant epilepsy. Capture sequencing identified a maternally inherited DEPDC5 heterozygous variant (NM_001242896: c.856C>T/p.Arg286*) in a 6-year-old child with sleep-related frontal lobe epilepsy and FCD IIa. We detected a second DEPDC5 somatic variant in trans configuration (c.865C>T/p.Gln289*; ~10% mosaic, 148/1505 reads) in the flash-frozen resected cortex from the seizure-onset zone (SOZ), but not in the blood (0 of 4,452 reads). Conventional Sanger sequencing detected the mosaic variant from the SOZ, but not from the surrounding epileptogenic zone (EZ) delimitated by stereoelectroencephalography, or from blood (Figure 1A). Ultra-deep amplicon sequencing detected both variants on distinct reads, indicating that they were located on different alleles and thus demonstrating a biallelic inactivation of DEPDC5 (Figure 1B). We further discovered a mutation gradient with a higher mosaic rate (~10%; 22,835 of 213,955 reads) in the SOZ than in the surrounding EZ (~0.3%; 485 of 154,962 reads). Both germline and somatic variants were nonsense, leading to loss of function, and were absent from the Genome Aggregation Database (http://gnomad.broadinstitute.org), and therefore considered to be pathogenic. To confirm that the DEPDC5 variants caused mTORC1 hyperactivation, we detected increased phosphorylation levels of the ribosomal protein S6, a well-known readout of mTORC1 activity, in enlarged NeuN+ neurons present in the resected brain specimen (Figure 1C).
Figure 1. DEPDC5 biallelic mutations in a patient.
(A) Family pedigree. The black square indicates the patient with FCD-associated epilepsy. Sanger sequence chromatograms show the germline nonsense variant (c.856C>T/p.Arg286*, black arrowheads) in the mother and child, and the somatic nonsense variant (c.865C>T/p.Gln289*, red arrowhead) that was only detected in the DNA extracted from the seizure-onset zone (SOZ), and not from the surrounding epileptogenic zone (EZ). (B) Integrative genomics viewer displays mutated bases in red in aligned reads: c.856C>T in blood (51%, 828 of 1,618 reads) and SOZ cortex DNA (48%, 748/1551 reads) and c.865C>T in SOZ cortex DNA only (10%, 148 of 1,505 reads). The 2 variants were systematically observed on different reads, indicating that they are in trans configuration. (C) Immunostaining against pS6 protein (red) and the neuronal marker NeuN (green) of a paraffin-embedded brain section from the patient. DAPI is shown in blue. White arrowheads indicate enlarged pS6+ neurons. Scale bar: 50 μm.
We subsequently generated a focal mosaic knockout of Depdc5 in the mouse brain, combining in utero electroporation (IUE) and CRISPR-Cas9 gene editing. The CRISPR-Cas9 system has been reported to introduce in vivo targeted indels leading to biallelic inactivation in postmitotic mouse neurons (17). Moreover, to circumvent the possible compensatory mechanisms of Depdc5 haploinsufficiency in rodents that account for the absence of spontaneous seizures (14, 15), we performed experiments on WT Swiss-Webster mice. We selected 2 guide RNAs (gRNA1 and gRNA2) targeting exon 16, which is common to all Depdc5 transcript isoforms, with minimal in silico predicted off-targets. We subsequently validated their high efficiency (>90%) to introduce CRISPR-Cas9 mosaic indels through DNA double-strand breaks in transfected HEK293T cells, with a restorable EGFP strategy (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI99384DS1). Both gRNAs were used for in vivo experiments and yielded similar results (see figure legends).
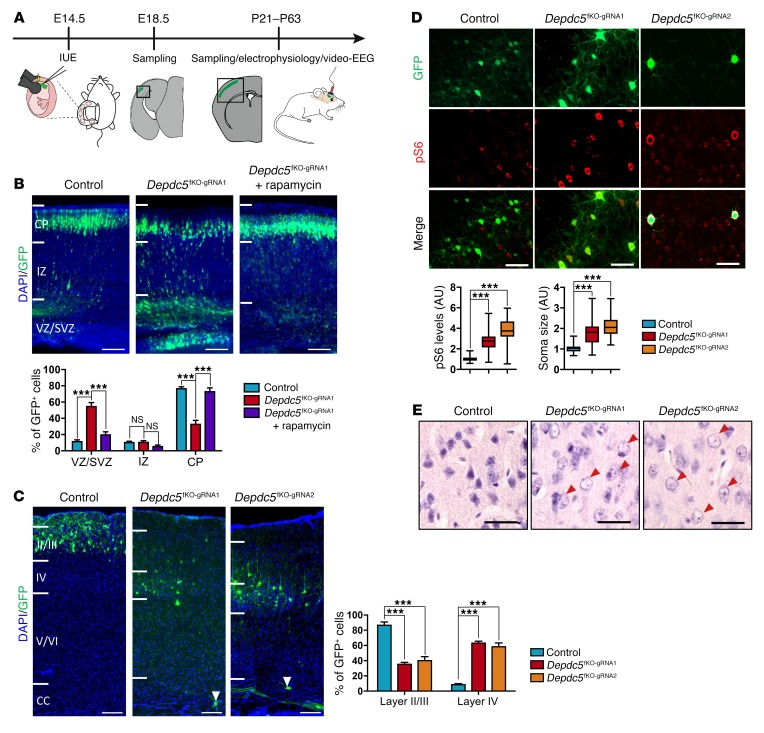
Swiss-Webster mouse embryos were electroporated in the ventricle at E14.5, a procedure that targets layer II/III–destined pyramidal neurons (18, 19). Both EGFP- and Cas9-expressing plasmids, with or without gRNA, were electroporated to generate Depdc5 focal knockout (Depdc5fKO) and control mice, respectively (Figure 2A). Site-specific deep sequencing of FACS-sorted GFP+ cells from Depdc5fKO embryos revealed an average indel rate of 20% (53,633 of 258,619 reads), validating our strategy to generate mosaic somatic mutations in vivo (Supplemental Figure 2A). We further estimated the proportion of these knockout progenitor cells to be approximately 4% of an embryonic brain hemisphere using FACS (Supplemental Figure 2B).
Figure 2. Delayed migration and increased mTORC1 activity in brain cortex of Depdc5fKO mice.
(A) Schema of IUE (in utero electroporation) procedure. (B) Representative coronal sections of control (n = 5), Depdc5fKO-gRNA1 (n = 6), and Depdc5fKO-gRNA1 plus rapamycin (n = 6) E18.5 embryo neocortices. DAPI, blue; GFP, green. Bottom: Histogram shows the distribution of electroporated GFP+ cells in neocortex. VZ, ventricular zone; SVZ, subventricular zone; IZ, intermediate zone; CP, cortical plate. ***P < 0.0001, by 2-way ANOVA. Scale bars: 100 μm. (C) Representative coronal sections of control (n = 5), Depdc5fKO-gRNA1 (n = 5), and Depdc5fKO-gRNA2 (n = 4) adult brain cortices. DAPI, blue; GFP, green. White arrowheads indicate ectopic neurons in the deeper cortical layers. CC, corpus callosum. Histogram shows the percentage of GFP+ cells in cortical layers II/III and IV. ***P = 0.0001, by 2-way-ANOVA. Scale bars: 100 μm. (D) Representative coronal sections of control (n = 5), Depdc5fKO-gRNA1 (n = 5), and Depdc5fKO-gRNA2 (n = 4) adult brain over the cortical layer III/IV boundary. GFP, green; pS6, red. Box and whisker plot shows fold changes in pS6 levels and soma size between ipsilateral GFP+ cells and contralateral GFP– cells in control, Depdc5fKO-gRNA1, and Depdc5fKO-gRNA2 adult brain cortices. ***P < 0.0001, by 1-way ANOVA (mean fold change in pS6: control = 1.05, Depdc5fKO-gRNA1 = 2.69, Depdc5fKO-gRNA2 = 3.82; mean fold change in soma size: control = 1.05, Depdc5fKO-gRNA1 = 1.8, Depdc5fKO-gRNA2 = 2.13; n = 30–70 cells per slice). Scale bars: 70 μm. (E) H&E staining of coronal brain sections from control (n = 3), Depdc5fKO-gRNA1 (n = 5), and Depdc5fKO-gRNA2 (n = 4) adult mice over cortical layers III/IV. Red arrowheads indicate balloon-like cells. Scale bars: 20 μm.
First, we examined the neuronal radial migration at E18.5. In contrast to the controls, most GFP+ cells from the Depdc5fKO embryos did not migrate to the cortical plate and were retained in ventricular and subventricular zones at E18.5 (Figure 2B). A single intraperitoneal injection of the mTORC1-specific inhibitor rapamycin into pregnant dams at E15 prevented this neuronal migratory defect in Depdc5fKO embryos, confirming an mTORC1-dependent mechanism (Figure 2B). Neuronal migratory abnormalities were still evident, even if less drastic, in adult mice. GFP+ cells from Depdc5fKO mice were distributed through layers III/IV, and we detected few ectopic, disorientated cells in deeper cortical layers (<1% of GFP+ cells), while we found, as expected, that they were distributed through layers II/III of the controls (Figure 2C). All GFP+ cells were confirmed to be neurons by counterstaining with NeuroTrace, a fluorescent Nissl marker (Supplemental Figure 2C). We assessed mTORC1 activity in adult Depdc5fKO animals to confirm Depdc5 inactivation by measuring the phosphorylation levels of S6 protein (Supplemental Figure 2D). PS6 immunostaining revealed increased phosphorylation levels of S6 (~3.5-fold) and an increased soma surface (~2-fold) in electroporated cells (Figure 2D), as observed in FCD II dysmorphic neurons. Moreover, H&E staining revealed numerous round, balloon-like cells with translucent cytoplasm and peripheral nuclei in the electroporated regions of Depdc5fKO mice, evocative of FCD IIb immature balloon cells (Figure 2E). By using 2 independent gRNAs, to avoid possible off-target effects, we were able to confirm that inactivation of Depdc5 during brain development causes FCD II cortical malformations.
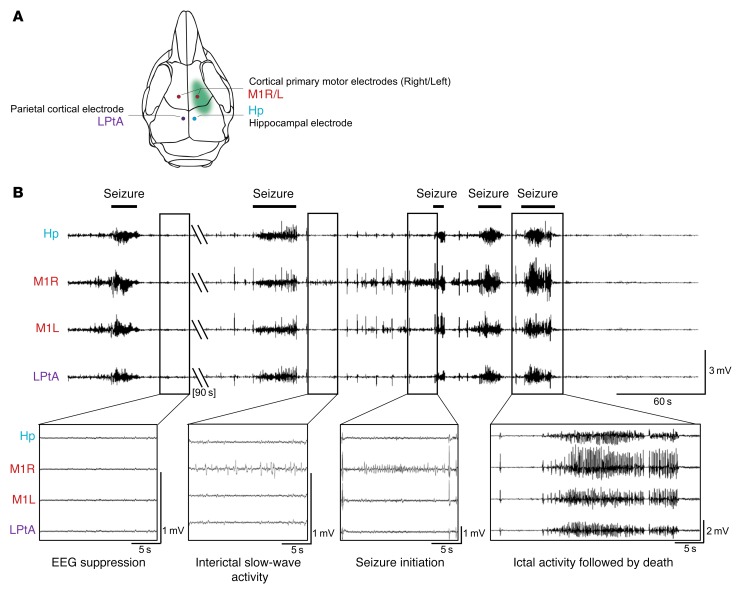
We next asked whether focal mosaic inactivation of Depdc5 in a small fraction of neurons is sufficient to cause spontaneous epileptic seizures. We performed continuous video recording combined with electroencephalography (video-EEG) in 13 Depdc5fKO and 8 control mice implanted with intracranial electrodes (Figure 3A). Four of thirteen (30%) Depdc5fKO mice displayed epileptic activity while wake, at the adult stage. In 1 mouse, electroclinical tonic seizures initiated from the electroporated cortical region between P46 and P68. In 3 other mice aged P37, P41, and P54, a single epileptic event occurred, followed by death, in a phenomenon resembling SUDEP. These epileptic episodes consisted of a severe cluster of tonic-clonic seizures, with alternate periods of EEG suppression and focal cortical 0.5-1 Hz slow-wave interictal activity. The focal origin of the seizures was further confirmed by ictal activity recordings initiated at the right cortical electrode near the electroporation site and accompanied by contralateral tonic limb movements (Figure 3B and Supplemental Video 1). Sudden death occurred after the last seizure, as shown by postictal electrocortical silence, and not during prolonged status epilepticus, consistent with the definition of SUDEP (Supplemental Figure 3 and Supplemental Video 2). We detected no spontaneous epileptic activity in the control mice, indicating that Cas9 protein has no epileptogenic effect.
Figure 3. Epileptic activity of adult Depdc5fKO mice.
(A) Schematic representation of implanted electrode locations. (B) EEG recordings of a cluster of seizures in a mouse aged P54. Hp, hippocampal; M1, primary motor cortex (R, right; L, left); LPtA, lateral parietal-temporal–associated cortex. Boxes below show different phases of EEG activity or suppression. A flat EEG after the last seizure was associated with the death of the animal.
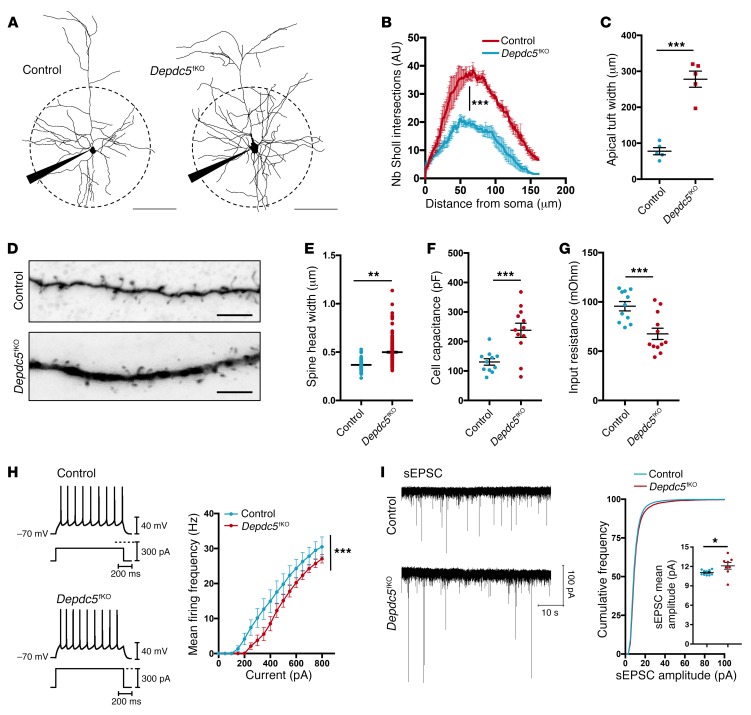
Finally, we searched for morphological and physiological changes in layer III crisperized pyramidal cells, which could contribute to this epileptic phenotype. We compared GFP+ neurons of control and Depdc5fKO mice aged P20–P24, before seizure onset, in order to avoid possible seizure-induced circuit damage. Neuronal reconstruction of biocytin-filled cells revealed a strikingly increased complexity of dendritic branching (Figure 4, A and B). Apical tufts were wider and dendritic thickness was increased in Depdc5fKO neurons compared with control neurons (Figure 4, C and D). Subsequent analysis showed hypertrophy of Depdc5fKO spines, characterized by a small but statistically significant increase in spine head width (Figure 4E), while spine density was unchanged (Supplemental Figure 4A). Thus, Depdc5 mediates dendrite and spine shaping, probably to affect synaptic transmission or dendritic integration in cortical pyramidal cells. Passive electrophysiological properties of layer III GFP+ pyramidal neurons revealed a significantly higher cell capacitance of Depdc5fKO neurons (Figure 4F), consistent with their increased membrane surface. Input resistance (Rm) was significantly reduced in Depdc5fKO neurons (Figure 4G) and was probably responsible for the shift toward larger inputs in the frequency-current curve (F-I curve) (Figure 4H). This shift indicates that a higher current is required to reach the voltage threshold for action potential activation. Nevertheless, above threshold, the gain of firing frequency (slope of the F-I curve) was significantly higher (Supplemental Figure 4B), suggesting an increased intrinsic responsiveness to current input changes, possibly due to alterations of ionic conductance. At the synaptic level, the mean amplitude of spontaneous excitatory postsynaptic currents (sEPSCs) was higher in Depdc5fKO neurons, while the mean frequency of sEPSCs was unchanged (Figure 4I and Supplemental Figure 4C), suggesting that Depdc5fKO spine hypertrophy might lead to altered expression of postsynaptic glutamate receptors. These data indicate that Depdc5 loss leads to enhanced excitatory synaptic activity that likely participates in the establishment of epileptic cortical activities.
Figure 4. Morphological and functional changes in Depdc5fKO neurons.
(A) Neuronal reconstruction of representative control (n = 5) and Depdc5fKO-gRNA1 (n = 5) recorded neurons. Scale bars: 100 μm. (B) Quantification of branching complexity by Sholl analysis. ***P < 0.0001, by 2-way-ANOVA for group effects. Nb, number. (C) Apical tuft width of control (n = 5) and Depdc5fKO-gRNA1 (n = 5) neurons. ***P = 0.0079, by Mann-Whitney U test. (D) Representative images of level-2 basal dendrite. Scale bars: 5 μm. (E) Dot plot showing the distribution of spine head widths (μm). **P = 0.0072, by unpaired t test (n = 4 neurons/group; n = 200 spines/neuron). (F) Capacitance (Cm) and (G) input resistance (Rin) for control (n = 11) and Depdc5fKO (n = 12) neurons. ***P = 0.002, by Mann-Whitney U test (mean Cm: controls = 130.5 pF, Depdc5fKO-gRNA1 = 238.1 pF; mean Rin: controls = 95.6 MOhm, Depdc5fKO-gRNA1 = 67.6 MOhm). (H) Left: Firing pattern of control Depdc5fKO-gRNA1 cells for 250 pA and 300 pA current input, respectively. Right: Mean frequency-current curve. ***P < 0.0001, by 2-way-ANOVA for group effects. (I) Left: Representative traces of sEPSC events over a 1-minute period. Right: Cumulative frequency distribution and dot plot of sEPSC mean amplitude. Mean amplitude: controls (n = 10) = 11.07 ± 0.12 pA; Depdc5fKO-gRNA1 (n = 8) = 12.1 ± 0.52 pA. *P = 0.0219, by Mann-Whitney U test.
In conclusion, our study highlights the emerging prospect that somatic mutations in a small proportion of neurons can cause neurodevelopmental disorders and that these, mutations underlie the cellular mosaicism observed in FCD brain specimens, i.e., dysmorphic neurons surrounded by normal neurons. Here, we report for the first time to our knowledge a definite case of biallelic — brain mosaic and germline — DEPDC5 mutations underlying FCD and proving the occurrence of Knudson’s 2-hit mechanism in focal epilepsy. This study further clarifies the intriguing issue of the wide spectrum of epilepsy severity among patients with germline DEPDC5 mutations and explains why only a subset of these patients have a refractory form of epilepsy-associated FCD, as previously suspected in a family (16). Because DEPDC5 is a negative regulator, loss of heterozygosity is necessary for the establishment of FCD along with a gradient of mosaicism, showing a different mutational mechanism than that in the MTOR gene, for which single, somatic activating mutations are sufficient to cause sporadic FCD (4–7).
We further demonstrate that biallelic inactivation in a Depdc5 brain somatic knockout mouse model accurately recapitulates the human condition. We could not distinguish between monoallelic or biallelic Cas9-mediated indels, nor could we show the absence of Depdc5 immunoreactivity (due to a lack of specific antibody) in GFP+ cells. However, we assumed that Cas9 cleavage produced homozygous knockout neurons as reported in other CRISPR-Cas9–engineered mouse models (17) and as suggested by the presence of an epileptic phenotype, which is absent in constitutive Depdc5+/– mice (15). We provide evidence that a small fraction of knockout neurons in the developing mouse cortex are sufficient to engender neuropathological hallmarks of FCD II (cortical dyslamination, pS6+ dysmorphic neurons, balloon-like cells) and trigger spontaneous focal epilepsy in the longer term. Our data therefore underline the concept that apparently nonlesional focal epilepsies may present developmental cytoarchitectural abnormalities undetectable by PET/MRI, eroding the dichotomous distinction between lesional and nonlesional epilepsy. We also report SUDEP-like episodes in mice following seizures, supporting clinical observations that DEPDC5 mutations may be associated with this tragic outcome for which the etiology remains elusive (11, 12). Human SUDEP genes are typically neurocardiac ion channel genes (SCN1A, SCN2A, SCN8A), and derived genetic SUDEP mouse models pointed out defective cardiac arrhythmias (20, 21). Besides, induced SUDEP mouse models highlighted the prime causality of cortical seizures in cardiorespiratory failure leading to SUDEP (22, 23). Here, we report that focal brain mosaic knockout of a non-channelopathy gene in mice results in spontaneous seizures and a phenotype that shares similarities with that of induced SUDEP mouse models in the course of electroclinical manifestations preceding the terminal outcome. Unraveling the physiology of DEPDC5-mediated SUDEP and the involvement of a possible predisposition to cardiac arrhythmia will have a critical impact on the clinical care of patients and on the implementation of preventive strategies.
An enigmatic question is why germline mutations of DEPDC5, a gene encoding a ubiquitously expressed signaling protein of the mTORC1 pathway, cause only neurological symptoms, while germline mutations of the other mTORC1 repressors TSC1 and TSC2 cause tuberous sclerosis (TSC), a multisystem disorder characterized by tumors in numerous organs. Here, we unveil the physiological function of Depdc5 in dendritic and spine modeling, which, when impaired, causes increased excitatory transmission, explaining how DEPDC5 mutations may lead to epileptogenesis. Neuronal shaping is involved in the pathogenesis of neuropsychiatric disorders (24) and is consistent with the psychiatric comorbidities and autistic features observed in patients with DEPDC5 mutations (25).
Finally, prenatal rapamycin prevented neuronal migratory defects in mice, proving that FCD due to somatic Depdc5 inactivation falls into the mTORopathy spectrum. Clinical trials with widespread rapamycin-derived mTORC1 inhibitors yielded promising results in controlling seizures in patients with TSC (26). Given the direct action of DEPDC5 in regulating mTORC1 activity, it has emerged as a promising therapeutic target to treat mTORC1-related focal epilepsy refractory to traditional antiepileptic medication and to avoid resective surgery. This mouse model therefore serves as a valuable functional platform for future clinical research directed toward the development of drugs that selectively enhance or mimic DEPDC5 activity (27).
Patients with FCD represent a growing proportion of today’s evolving epilepsy surgery population, a trend that poses crucial challenges for patient care and treatment and the allocation of resources among epilepsy centers (28). Along with the increasing attention focused on SUDEP as an underestimated cause of death in individuals with epilepsy (29), this disease represents a major public health issue. Altogether, our data provide essential and necessary scientific grounds for the identification of mTORC1-targeting, brain-specific molecules that (a) control seizures when antiepileptic drugs acting ion channels/neurotransmitters are ineffective, (b) prevent epileptogenesis, and (c) lead to preventive strategies against SUDEP occurrence in high-risk patients.
Methods
See the Supplemental Methods for a detailed description of the methods.
Study approval.
Human studies were approved by the ethics committee of CPP Ile de France II under registration no. ID-RCB/EUDRACT-2015-A00671-48 and the clinical trial identifier number NCT02890641 (ClinicalTrials.gov). Informed consent was obtained from all participants. Mouse studies were approved by the French Ministry of Research (no. 75-1319).
Author contributions
TR designed, performed, and analyzed most of the experiments. CD performed and analyzed all electrophysiology experiments. AB participated in the FACS and video-EEG experiments and revised the manuscript. S. Baldassari performed human genetic studies. EM performed immunostaining of human tissue and contributed to the genetic studies. GM performed in utero electroporation experiments. DR performed EEG implantations and analysis. VN supervised the EEG study. MC recruited the FCD cohort. EL and RM critically reviewed the manuscript. S. Baulac designed, supervised, and obtained funding for the study. TR and S. Baulac wrote the manuscript.
Supplementary Material
Acknowledgments
This work was funded by the European Research Council (682345, to S. Baulac and 322721, to RM); the program Investissements d’avenir ANR-10-IAIHU-06; the Fondation pour la Recherche Médicale (DEQ2015033, to S. Baulac; ECO20160736027, to TR; and FDT20160736468, to EM). We thank Sarah Ferrand-Sorbets and Georg Dorfmüller (Department of Pediatric Neurosurgery, Fondation Rothschild, Paris, France) for providing human brain specimens and Marc Polivka (Department of Anatomic Pathology, Hôpital Lariboisière, Paris, France) for neuropathology. We thank the ICM core facilities (sequencing, DNA bank, rodent behavior) for technical help, the Pitié-Salpétrière flow cytometry core facility, and Genosplice for bioinformatics.
Version 1. 04/30/2018
Electronic publication
Version 2. 06/01/2018
Print issue publication
Funding Statement
Advanced ERC grant to Richard Miles
Consolidator ERC grant to Stéphanie Baulac
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(6):2452–2458.https://doi.org/10.1172/JCI99384.
See the related Commentary at DEPDC5 takes a second hit in familial focal epilepsy.
Contributor Information
Théo Ribierre, Email: theo.ribierre@icm-institute.org.
Charlotte Deleuze, Email: charlotte.deleuze@icm-institute.org.
Alexandre Bacq, Email: alexandre.bacq@icm-institute.org.
Sara Baldassari, Email: sara.baldassari@icm-institute.org.
Elise Marsan, Email: elise.marsan@icm-institute.org.
Mathilde Chipaux, Email: mchipaux@for.paris.
Giuseppe Muraca, Email: giuseppe.muraca1975@gmail.com.
Delphine Roussel, Email: delphine.roussel@icm-institute.org.
Vincent Navarro, Email: vincent.navarro@aphp.fr.
Eric Leguern, Email: eric.leguern@aphp.fr.
Richard Miles, Email: richard.miles@upmc.fr.
References
- 1.Hoang ML, et al. Genome-wide quantification of rare somatic mutations in normal human tissues using massively parallel sequencing. Proc Natl Acad Sci U S A. 2016;113(35):9846–9851. doi: 10.1073/pnas.1607794113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poduri A, Evrony GD, Cai X, Walsh CA. Somatic mutation, genomic variation, and neurological disease. Science. 2013;341(6141):1237758. doi: 10.1126/science.1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marsan E, Baulac S. Review: Mechanistic target of rapamycin (mTOR) pathway, focal cortical dysplasia and epilepsy. Neuropathol Appl Neurobiol. 2018;44(1):6–17. doi: 10.1111/nan.12463. [DOI] [PubMed] [Google Scholar]
- 4.Lim JS, et al. Brain somatic mutations in MTOR cause focal cortical dysplasia type II leading to intractable epilepsy. Nat Med. 2015;21(4):395–400. doi: 10.1038/nm.3824. [DOI] [PubMed] [Google Scholar]
- 5.Nakashima M, et al. Somatic mutations in the MTOR gene cause focal cortical dysplasia type IIb. Ann Neurol. 2015;78(3):375–386. doi: 10.1002/ana.24444. [DOI] [PubMed] [Google Scholar]
- 6.Mirzaa GM, et al. Association of MTOR mutations with developmental brain disorders, including megalencephaly, focal cortical dysplasia, and pigmentary mosaicism. JAMA Neurol. 2016;73(7):836–845. doi: 10.1001/jamaneurol.2016.0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Møller RS, et al. Germline and somatic mutations in theMTORgene in focal cortical dysplasia and epilepsy. Neurol Genet. 2016;2(6):e118. doi: 10.1212/NXG.0000000000000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blümcke I, et al. The clinicopathologic spectrum of focal cortical dysplasias: a consensus classification proposed by an ad hoc Task Force of the ILAE Diagnostic Methods Commission. Epilepsia. 2011;52(1):158–174. doi: 10.1111/j.1528-1167.2010.02777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blumcke I, et al. Histopathological findings in brain tissue obtained during epilepsy surgery. N Engl J Med. 2017;377(17):1648–1656. doi: 10.1056/NEJMoa1703784. [DOI] [PubMed] [Google Scholar]
- 10.Baulac S. mTOR signaling pathway genes in focal epilepsies. Prog Brain Res. 2016;226:61–79. doi: 10.1016/bs.pbr.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 11.Nascimento FA, Borlot F, Cossette P, Minassian BA, Andrade DM. Two definite cases of sudden unexpected death in epilepsy in a family with a DEPDC5 mutation. Neurol Genet. 2015;1(4):e28. doi: 10.1212/NXG.0000000000000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bagnall RD, et al. Exome-based analysis of cardiac arrhythmia, respiratory control, and epilepsy genes in sudden unexpected death in epilepsy. Ann Neurol. 2016;79(4):522–534. doi: 10.1002/ana.24596. [DOI] [PubMed] [Google Scholar]
- 13.Bar-Peled L, et al. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340(6136):1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marsan E, et al. Depdc5 knockout rat: A novel model of mTORopathy. Neurobiol Dis. 2016;89:180–189. doi: 10.1016/j.nbd.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 15.Hughes J, et al. Knockout of the epilepsy gene Depdc5 in mice causes severe embryonic dysmorphology with hyperactivity of mTORC1 signalling. Sci Rep. 2017;7(1):12618. doi: 10.1038/s41598-017-12574-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baulac S, et al. Familial focal epilepsy with focal cortical dysplasia due to DEPDC5 mutations. Ann Neurol. 2015;77(4):675–683. doi: 10.1002/ana.24368. [DOI] [PubMed] [Google Scholar]
- 17.Platt RJ, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159(2):440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LoTurco J, Manent JB, Sidiqi F. New and improved tools for in utero electroporation studies of developing cerebral cortex. Cereb Cortex. 2009;19(Suppl 1):i120–i125. doi: 10.1093/cercor/bhp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taniguchi Y, Young-Pearse T, Sawa A, Kamiya A. In utero electroporation as a tool for genetic manipulation in vivo to study psychiatric disorders: from genes to circuits and behaviors. Neuroscientist. 2012;18(2):169–179. doi: 10.1177/1073858411399925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frasier CR, et al. Cardiac arrhythmia in a mouse model of sodium channel SCN8A epileptic encephalopathy. Proc Natl Acad Sci U S A. 2016;113(45):12838–12843. doi: 10.1073/pnas.1612746113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goldman AM, Buchanan G, Aiba I, Noebels JL. SUDEP animal Models. In: Pitkänen A, Buckmaster P, Galanopoulou A, Moshé S, eds. Models of Seizures and Epilepsy (Second edition). Amsterdam, Netherlands:Elsevier;2017:1007–1018. [Google Scholar]
- 22.Aiba I, Noebels JL. Spreading depolarization in the brainstem mediates sudden cardiorespiratory arrest in mouse SUDEP models. Sci Transl Med. 2015;7(282):282ra46. doi: 10.1126/scitranslmed.aaa4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aiba I, Wehrens XH, Noebels JL. Leaky RyR2 channels unleash a brainstem spreading depolarization mechanism of sudden cardiac death. Proc Natl Acad Sci U S A. 2016;113(33):E4895–E4903. doi: 10.1073/pnas.1605216113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14(3):285–293. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burger BJ, et al. Autistic siblings with novel mutations in two different genes: insight for genetic workups of autistic siblings and connection to mitochondrial dysfunction. Front Pediatr. 2017;5:219. doi: 10.3389/fped.2017.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.French JA, et al. Adjunctive everolimus therapy for treatment-resistant focal-onset seizures associated with tuberous sclerosis (EXIST-3): a phase 3, randomised, double-blind, placebo-controlled study. Lancet. 2016;388(10056):2153–2163. doi: 10.1016/S0140-6736(16)31419-2. [DOI] [PubMed] [Google Scholar]
- 27.Myers KA, Scheffer IE. DEPDC5 as a potential therapeutic target for epilepsy. Expert Opin Ther Targets. 2017;21(6):591–600. doi: 10.1080/14728222.2017.1316715. [DOI] [PubMed] [Google Scholar]
- 28.French J, Friedman D. The evolving landscape of epilepsy neuropathology. Lancet Neurol. 2018;17(3):202–203. doi: 10.1016/S1474-4422(17)30429-5. [DOI] [PubMed] [Google Scholar]
- 29.Devinsky O, Friedman D, Cheng JY, Moffatt E, Kim A, Tseng ZH. Underestimation of sudden deaths among patients with seizures and epilepsy. Neurology. 2017;89(9):886–892. doi: 10.1212/WNL.0000000000004292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.