Abstract
Many Toll-like receptors (TLRs) signal through TNF receptor–associated factor 6 (TRAF6) to activate innate immune responses. Here, we show that somatic nuclear autoantigenic sperm protein (sNASP) binds to TRAF6 to prevent TRAF6 autoubiquitination in unstimulated macrophages. Following LPS stimulation, a complex consisting of sNASP, TRAF6, IRAK4, and casein kinase 2 (CK2) is formed. CK2 phosphorylates sNASP at serine 158, allowing sNASP to dissociate from TRAF6. Free TRAF6 is then autoubiquitinated, followed by activation of downstream signaling pathways. In sNasp S158A knockin (S158A-KI) mice, LPS-treated macrophages could not phosphorylate sNASP, which remained bound to TRAF6. S158A-KI mice were more susceptible to sepsis due to a marked reduction in IL-1β, TNF-α, and IFN-γ production accompanied by an inability to clear bacteria and recruit leukocytes. Furthermore, phosphorylation-regulated release of sNASP from TRAF6 is observed following activation of TLR-1, -2, -4, -5, and -6. Thus, sNASP is a negative regulator of TLR signaling to modulate the innate immune response.
Keywords: Immunology, Inflammation
Keywords: Innate immunity
Introduction
The innate immune response is activated by pathogen-associated molecular patterns (PAMPs) through a family of Toll-like receptors (TLRs) (1, 2). TLR signaling can be classified into MyD88-dependent or MyD88-independent pathways. In the MyD88-dependent pathway, MyD88 is recruited to TLRs with interleukin 1 receptor–associated kinases (IRAKs) after detecting PAMPs to activate a ubiquitin E3 ligase, TNF receptor–associated factor 6 (TRAF6). Activated TRAF6 becomes modified by K63 polyubiquitin chains, which serve as scaffold to recruit TAK1 to activate multiple downstream signaling pathways (3, 4). The MyD88-independent pathway uses different intracellular adaptor molecules such as Toll/interleukin receptor 1–containing (TIR-containing) cytoplasmic adaptors TRIF and TRAM, but not MyD88.
TLR signaling is tightly regulated to maintain immune homeostasis because hyperactivation or hypoactivation of TLR signaling causes human diseases (5). Most studies have focused on negative regulation of TRAF6 activation. Indeed, several negative regulators are induced by TLR ligands to terminate TRAF6 activation (6). For example, A20 and CYLD are induced by TLR ligands to deubiquitinate polyubiquitinated TRAF6 to terminate downstream signaling events (7, 8). Turning off TRAF6 activation is necessary to avoid excessive immune response, as demonstrated by the development of spontaneous inflammation in A20-deficient mice (7). Ubiquitination and deubiquitination of TRAF6 are important in the regulation of TLR signaling pathways.
In contrast to the termination of TRAF6 signaling following activation, it is not known whether TRAF6 is regulated before activation. Here, we have shown that TRAF6 is bound by an inhibitory protein, somatic nuclear autoantigenic sperm protein (sNASP), in unstimulated cells. Following LPS stimulation, sNASP is phosphorylated and released from TRAF6 to allow downstream signaling. This is analogous to the release of IκB from NF-κB to allow NF-κB to transactivate proinflammatory cytokine genes. Using a dominant-negative sNASP knockin mouse model, we showed that the inability to release sNASP from TRAF6 prevented mice from mounting an effective immune response during sepsis. Thus, we have uncovered a critical control of TLR signaling that targets TRAF6, which is required for maintaining homeostasis of the innate immune response.
This is a new biological function of sNASP, which was previously shown to be a histone chaperone that binds core histones (H3 and H4) and linker histones (H1). sNASP transports linker histones into the nucleus and transfers H1 histones onto DNA to facilitate chromatin assembly (9, 10). Thus, sNASP is required for normal development (11). Overexpression or RNAi knockdown of NASP in human cells affected DNA damage repair, immune response, and cellular growth (12). In contrast to the nuclear activity of sNASP in histone regulation, sNASP regulates TRAF6 in the cytosol and identifies a new molecule for investigation in the innate immune response.
Results
sNASP inhibits TLR4-induced NF-κB activation through TRAF6.
NASP was shown to be a potential TRAF6 binding protein in 2 independent mass spectrometry screenings (11, 13). Of the 2 major NASP isoforms generated by alternative splicing, sNASP, but not testes-specific NASP (tNASP), is the major isoform that interacts with TRAF6 (Figure 1A). Reciprocal immunoprecipitation and Western blot analyses confirmed that endogenous sNASP binds to TRAF6 in human monocyte cell line THP-1 (Figure 1B). Similar results were obtained in a mouse macrophage cell line (Raw264.7) and primary murine macrophages (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI95720DS1). sNASP is also the predominant NASP isoform expressed in macrophage cells (Supplemental Figure 2). NASP was previously reported as a histone H1 binding protein that regulates histone transportation and cell cycle (11, 14). It is localized in both the nucleus and cytoplasm (10, 13). Cell fractionation experiments showed that sNASP binds to TRAF6 in the cytoplasm, but not in the nucleus (Supplemental Figure 3, A and B).
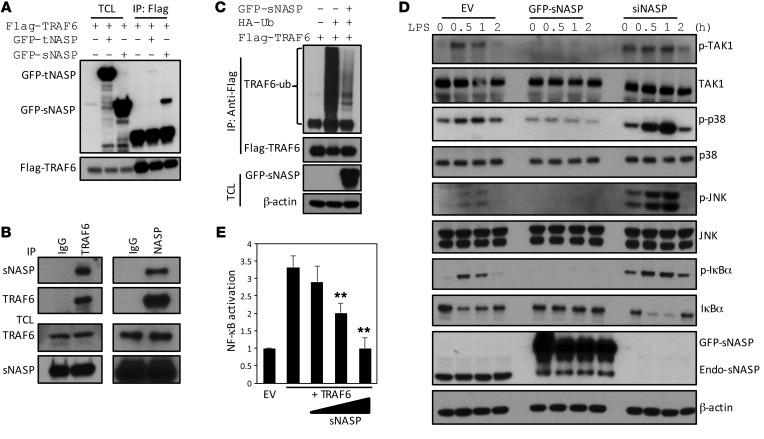
Figure 1. sNASP inhibits TLR4-induced NF-κB activation through TRAF6.
(A) Immunoprecipitation (IP) of Flag-TRAF6 (with anti-Flag agarose) from HEK293 cells transiently transfected with Flag-TRAF6 plus GFP-tNASP (+), GFP-tNASP (+), or empty vector (–), followed by immunoblotting (IB) with antibody against Flag or GFP. TCL IB was done with anti-Flag. TCL, total cell lysates. (B) IP of endogenous TRAF6 (with anti-TRAF6) or endogenous sNASP (with anti-NASP) or IgG from THP-1 cells, followed by IB with antibody against TRAF6 or NASP. TCL IB was done with anti-NASP and anti-TRAF6. (C) IP of Flag-TRAF6 (with anti-Flag agarose) in HEK293 cells transfected with Flag-tagged TRAF6 and hemagglutinin-tagged ubiquitin (HA-Ub) in the presence (+) or absence (–) of vector encoding GFP-tagged sNASP, and IB with anti-Flag followed by IB with antibody against Flag or Ub. TCL IB was done with anti-GFP and β-actin. (D) IB of indicated antibodies in LPS-stimulated Raw264.7 cells transduced with empty vector (EV) or GFP-tagged sNASP or siNASP. (E) Luciferase activity in HEK293 cells transfected with a luciferase reporter vector driven by an NF-κB–responsive promoter, plus EV or vector encoding TRAF6 and increasing concentrations of a vector encoding sNASP. Results were standardized to EV (set as 1). Data are mean ± SE for each group. **P < 0.01 (Student’s t test). Data represent a minimum of 3 independent experiments.
Overexpression of sNASP reduced autoubiquitination of TRAF6, but not TRAF3, in HEK293 cells (Figure 1C; Supplemental Figure 4, A and C; and Supplemental Figure 20A). Furthermore, sNASP specifically reduced K63-linked autoubiquitination (Supplemental Figure 4, B and D). LPS-induced phosphorylation of TAK1, p38 MAPK, JNK, and IκBα was decreased when sNASP was overexpressed in THP-1 cells. In contrast, phosphorylation of these proteins increased when sNASP was knocked down (Figure 1D and Supplemental Figure 20B). Similar results were obtained in Raw264.7 and bone marrow–derived macrophages (BMDMs) (Supplemental Figure 5, A–D). sNASP was found to inhibit TRAF6-mediated NF-κB activation in a dose-dependent manner (Figure 1E). To exclude potential sNASP effects in the nucleus, 2 sNASP deletion mutants that lacked nuclear localization signals, 1–233 and 1–348, were generated (Supplemental Figure 6A). Both deletion mutants were found in the cytoplasm only (Supplemental Figure 6B) and retained the ability to inhibit TRAF6-mediated NF-κB activation (Supplemental Figure 6C). Overexpression of GFP-sNASP led to downregulation of LPS-induced expression of IL-6 and TNF-α at the level of transcription, leading to diminished protein expression (Figure 2, A and B). Conversely, knockdown of NASP significantly increased the production of IL-6 and TNF-α at the level of both mRNA and protein (Figure 2, C and D, and Supplemental Figure 7). Western blot analysis confirmed appropriate overexpression or knocking down of sNASP (Supplemental Figure 5A). These findings suggest that sNASP negatively regulates TLR4-induced proinflammatory cytokine responses through TRAF6.
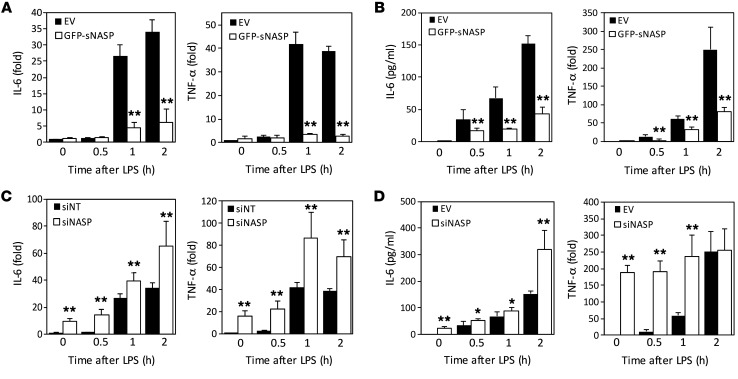
Figure 2. sNASP inhibits LPS-induced proinflammatory cytokine production.
Expression of TNF-α and IL-6 in Raw264.7 cell lines transduced with EV or GFP-tagged sNASP (A) or EV or siNASP (B) and stimulated with LPS. Results were normalized to the expression of ACTB (encoding β-actin) and are presented relative to those of untreated cells. (C and D) Production of TNF-α and IL-6 by Raw264.7 cells transduced as in A or B and stimulated with LPS. Data are mean ± SE for each group. *P < 0.05, **P < 0.01 (1-way ANOVA). Data represent a minimum of 3 independent experiments.
Phosphorylation of sNASP regulates its interaction with TRAF6 and cytokine production.
Thirty minutes after LPS treatment, sNASP was serine-phosphorylated, but not threonine-phosphorylated, in both Raw264.7 and THP-1 cells (Figure 3, A and B, and Supplemental Figure 20, C and D). Interestingly, endogenous sNASP dissociated from TRAF6 which correlated with increased serine-specific phosphorylation of sNASP 30 minutes after LPS stimulation (Figure 3B). These results suggest that serine phosphorylation of sNASP may regulate its interaction with TRAF6. Eight potential serine/threonine phosphorylation sites were found in sNASP from PhosphoSitePlus (PSP) (Supplemental Figure 8A). These predicted serine/threonine phosphorylation sites were individually substituted by alanine and expressed in THP-1 cells. Only substitution of serine 158 with alanine abolished LPS-induced serine phosphorylation (Supplemental Figure 8, B and C).
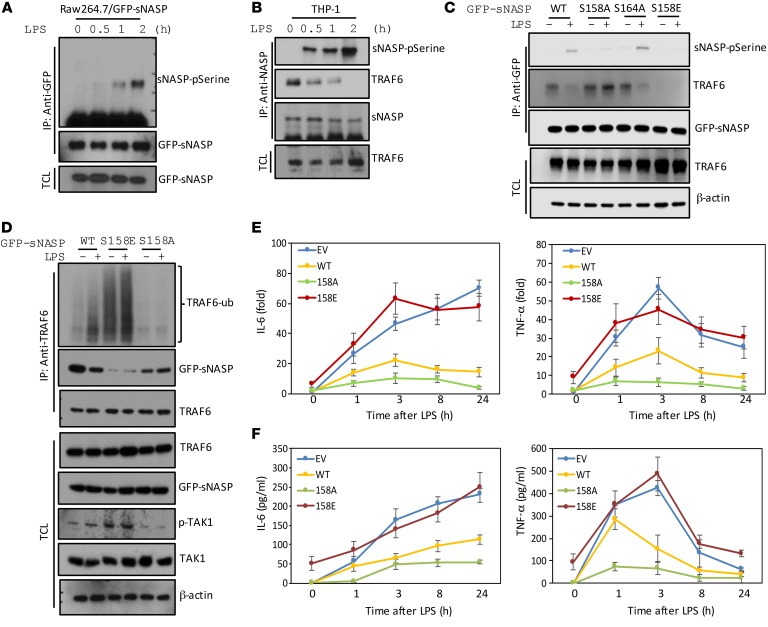
Figure 3. Phosphorylation of sNASP regulates its interaction with TRAF6 and affects cytokine production.
(A) Raw264.7 cells were transfected with GFP-tagged sNASP, stimulated with LPS, and assessed by IB with antibody against phosphorylated serine or GFP after IP with anti-GFP or by IB with anti-GFP in TCL. (B) Phosphorylation of the serine residue of endogenous sNASP in THP-1 cells following LPS stimulation, assessed by IB with antibody against phosphorylated serine (pSerine) or NASP after IP with anti-NASP. TCL IB was done with anti-TRAF6. (C) THP-1 cells were transfected with GFP-tagged WT sNASP or S158A, S164A, S158E mutants, followed by IB with antibody against phosphorylated serine, TRAF6, or GFP after IP with anti-GFP. TCL IB was done with anti-TRAF6 or anti–β-actin (below). (D) THP-1 cells were transfected with GFP-tagged WT sNASP or S158A, S158E mutants, followed by IB with antibody against Ub, TRAF6, or NASP after IP with anti-TRAF6. TCL IB was done with anti-TRAF6, anti-GFP, anti-pTAK1, anti-TAK1, or anti–β-actin. (E) Expression of TNF-α and IL-6 in Raw264.7 cell lines transfected with WT sNASP, S158A, S158E mutants, or EV and stimulated with LPS. Results were normalized to the expression of ACTB (encoding β-actin) and untreated cells. (F) Secretion of TNF-α and IL-6 by Raw264.7 cells transduced as in E and stimulated with LPS. Data are mean ± SE for each group. A–F represent a minimum of 3 independent experiments.
To further determine whether phosphorylation of serine 158 regulated sNASP association with TRAF6, we generated a GFP-sNASP S158E mutant, a constitutive active sNASP containing serine-to-glutamic acid substitution, to mimic phosphorylation. THP-1 cells were transfected with sNASP WT and various mutants and coimmunoprecipitated with TRAF6. There was a clear interaction between phosphorylation-deficient sNASP S158A mutant, but not phosphomimetic sNASP S158E mutant, and TRAF6 in unstimulated cells (Figure 3C). Furthermore, the nonphosphorylatable GFP-sNASP S158A mutant maintained its interaction with TRAF6 even after LPS treatment, whereas the phosphomimetic GFP-sNASP S158E mutant was unable to interact with TRAF6 in both unstimulated and LPS-stimulated cells (Figure 3C). Moreover, cells transfected with sNASP S158E mutant exhibited spontaneous TRAF6 autoubiquitination and TAK1 phosphorylation independent of LPS stimulation. Conversely, the sNASP S158A mutant lost LPS-induced TRAF6 autoubiquitination and TAK1 phosphorylation (Figure 3D and Supplemental Figure 20F). These results suggest that phosphorylation of sNASP affects its interaction with TRAF6 and downstream signaling.
Consistent with previous results, overexpression of WT sNASP resulted in a marked loss of LPS-induced IL-6 and TNF-α production (Figure 3, E and F). However, overexpression of phosphorylation-deficient sNASP S158A dramatically decreased LPS-triggered proinflammatory cytokines as compared with WT sNASP, suggesting that the nonphosphorylatable sNASP mutant competed with endogenous sNASP to exert a dominant-negative effect (Figure 3F). The phosphorylation-deficient sNASP S158A also markedly reduced cytokine RNA levels in Raw264.7 cells (Figure 3E). On the other hand, IL-6 and TNF-α productions were increased spontaneously without LPS stimulation in cells overexpressing the phosphomimetic mutant S158E (Figure 3, E and F). These results suggest that sNASP serine 158 phosphorylation is a major checkpoint of LPS-induced proinflammatory cytokine secretion.
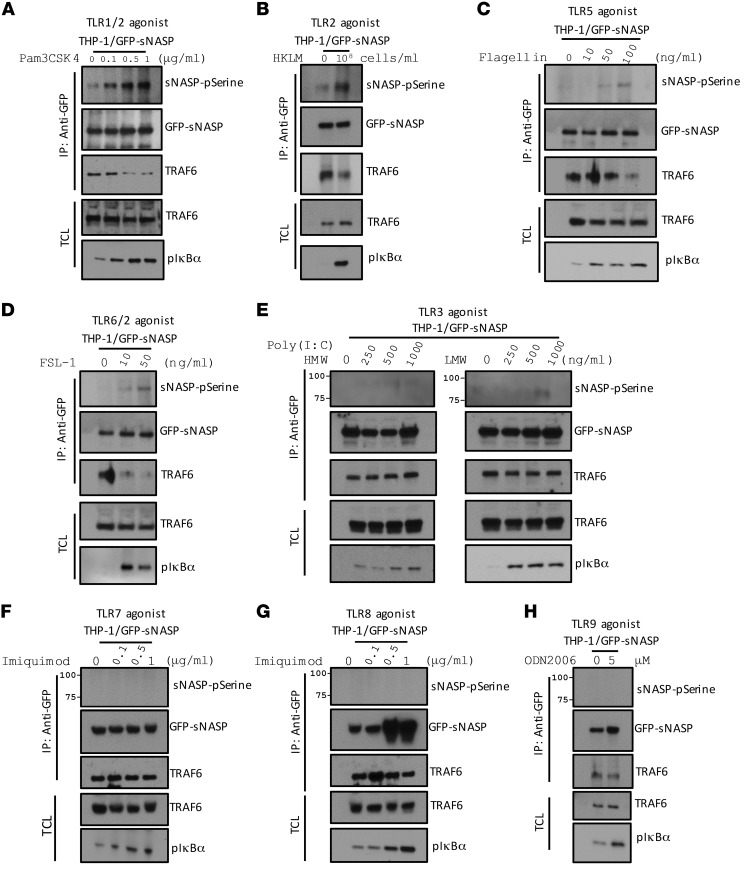
To test whether regulation of TLR4 signaling by phosphorylation of sNASP can be generalized to other TLRs, we treated THP-1 cells with various TLR ligands and studied the phosphorylation of sNASP. Phosphorylation of sNASP was increased in THP-1 cells stimulated by Pam3CSK4 (TLR1/2 agonist), HKLM (TLR2 agonist), flagellin (TLR5 agonist), and FSL-1 (TLR6/2 agonist) (Figure 4, A–D and Supplemental Figure 20, G–J). Consistent with our previous findings (Figure 3B), we also observed dissociation between TRAF6 and sNASP after stimulation by those agonists. However, cells treated with Poly (I:C) (TLR3 agonist), imiquimod (TLR7 agonist), ssRNA (TL8 agonist), and ODN2006 (TLR9 agonist) did not result in phosphorylation of sNASP or affect interaction between TRAF6 and sNASP (Figure 4, E–H). Thus, our data suggest that sNASP regulates selective TLR signaling initiated by bacterial outer surface components.
Figure 4. sNASP phosphorylation was observed following TLR-1, -2, -5, and -6 signaling.
THP-1 cells transfected with GFP-tagged WT sNASP were stimulated with Pam3CSK4 (A), HKML (B), flagellin (C), FSL-1 (D), Poly (I:C) HMW and LMW (E), imiquimod (F), ssRNA (G), and ODN2006 (H) assessed by IB with antibody against phosphorylated serine (pSerine), GFP, or TRAF6 after IP with anti-GFP. TCL IB was done with anti-TRAF6. Data represent at least 3 independent experiments.
CK2 phosphorylates sNASP in a complex containing IRAK4, TRAF6, and sNASP.
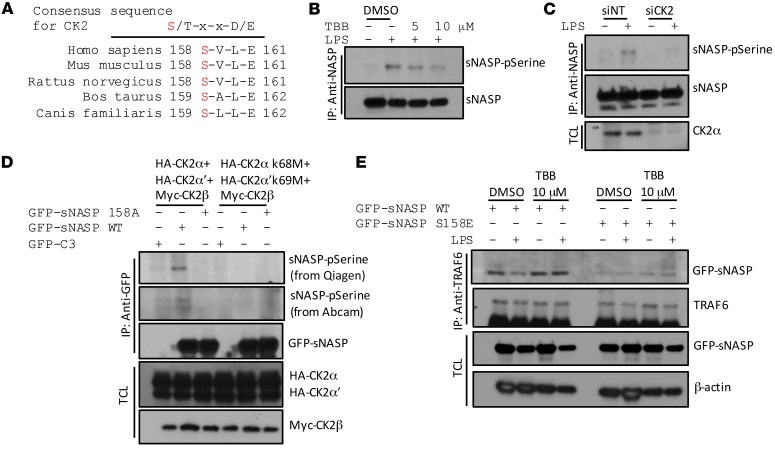
We next sought to identify the kinase responsible for phosphorylation of sNASP. A sequence alignment performed for sNASP showed that serine 158 is highly conserved in different species and the residues surrounding serine 158 (SVLE) conform to the putative CK2 phospho-motif (Figure 5A). To investigate the potential roles of CK2 in sNASP serine 158 phosphorylation, we tested whether CK2α phosphorylates the serine 158 residue in sNASP in an in vitro assay using WT and S158A mutant sNASP. Immunoprecipitated GFP-tagged sNASP was incubated with recombinant CK2 in vitro. Serine phosphorylation was detected in the WT sNASP but not S158A sNASP mutant (Supplemental Figure 9, A and B). Purified sNASP phosphorylated by CK2 was analyzed by liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis. Phosphorylated sNASP peptide 147–163 (ENDKTEEMPNDpSVLENK) was identified with high confidence, in full agreement with the mapping of serine 158 by in vitro mutagenesis (Supplemental Figure 9, C and D). These results also confirmed that CK2 can phosphorylate sNASP at serine 158 in vitro.
Figure 5. CK2 phosphorylates sNASP on serine 158.
(A) Alignment of sNASP sequence from multiple species revealed that serine 158 is highly conserved and contained in a motif recognized by CK2. X is any residue. (B) THP-1 cells stimulated with LPS in the absence (–) or presence (+) of TBB, an inhibitor of CK2, assessed by IB with antibody against phosphorylated serine (pSerine) or GFP after IP with anti-NASP. (C) THP-1 cells were stimulated with LPS in the presence of siNT or siCK2 and assessed by IB with antibody against phosphorylated serine (pSerine) or anti-GFP after IP with anti-NASP. TCL IB was done with anti-CK2α. (D) HEK293 cells transduced with GFP-tagged WT sNASP, S158A, or empty vector (C3) in the presence of Myc-tagged CK2β combined with HA-tagged CK2 catalytic subunits (CK2α and CK2α’) or CK2 kinase-dead mutants (CK2α K68M and CK2α’ K69M), assessed by IB with antibody against phosphorylated serine (pSerine) from Qiagen or Abcam or anti-GFP after IP with anti-GFP. TCL was IB with anti-HA or anti-Myc. (E) THP-1 cells transduced with GFP-tagged WT sNASP (WT) or S158A mutant in the absence (–) or presence (+) of TBB, assessed by IB with antibody against TRAF6 or GFP after IP with anti-TRAF6. TCL IB was done with anti-NASP and anti–β-actin. Data represent a minimum of 3 independent experiments.
To determine whether sNASP is a substrate of CK2 in cells, we used chemical and genetic tools to inhibit CK2 in THP-1 cells. TBB (tetrabromobenzimidazole), which has been reported as a cell-permeable selective inhibitor of CK2 (15), decreased serine phosphorylation of endogenous sNASP in a dose-dependent manner (Figure 5B and Supplemental Figure 20K). Moreover, depletion of the endogenous CK2α subunits in THP-1 cells also resulted in a reduction in sNASP phosphorylation compared with control cells (Figure 5C and Supplemental Figure 20L). CK2 is a protein kinase that in humans is composed of 2 catalytic subunits, CK2α and CK2α’, and 1 regulatory subunit, CK2β (16, 17). To further test whether CK2 kinase activity is important for sNASP phosphorylation, we transfected WT CK2α subunits or catalytically inactivated CK2α subunits (CK2α K68M and CK2α’ K69M) to HEK293 cells with sNASP WT or S158A mutant, and then performed coimmunoprecipitation experiments. HEK293 cells expressing WT CK2α and CK2α’, but not catalytically inactivated CK2α K68M and CK2α’ K69M mutants, had enhanced serine phosphorylation of sNASP (Figure 5D and Supplemental Figure 20M). Also, as shown in Figure 5D, overexpression with a combination of WT CK2 subunits, but not the kinase-dead mutants (CK2α K68M and CK2α’ K69M), caused phosphorylation of WT sNASP but not the S158A mutant. Taken together, these experiments indicate that sNASP is a bona fide substrate for CK2 in cells.
Because phosphorylation of sNASP regulates interaction between TRAF6 and sNASP, we asked whether CK2 also regulates interaction between TRAF6 and sNASP following LPS stimulation. We treated THP-1 cells expressing GFP-sNASP with CK2 inhibitor or siCK2, which inhibits serine phosphorylation, and found that treatment of cells with TBB or siCK2 retains sNASP interaction with TRAF6 following LPS stimulation (Supplemental Figure 10, A–D). Furthermore, when THP-1 cells expressing WT sNASP were treated with CK2 inhibitor (TBB), dissociation of TRAF6 with WT sNASP was blunted after LPS treatment (Figure 5E). However, CK2 inhibitor did not affect the interaction between TRAF6 and the phosphomimetic mutant S158E (Figure 5E), suggesting that phosphorylation of sNASP by CK2 regulates the interaction of TRAF6 and sNASP during TLR4 signaling.
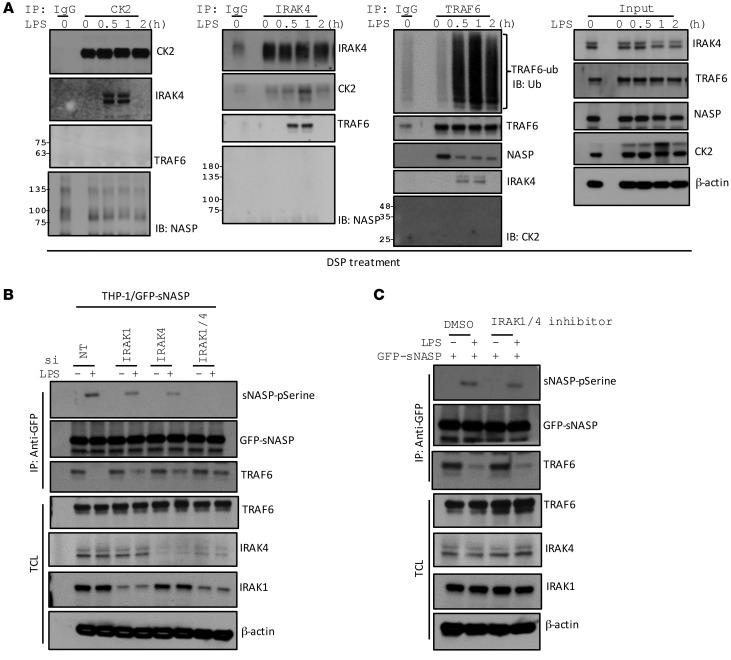
To investigate the mechanism whereby CK2 regulates sNASP phosphorylation, we treated THP-1 cells with LPS at different time points. Cell lysates were incubated with a cross-linker dithiobis(succinimidyl propionate) (DSP) and immunoprecipitated with different antibodies. CK2 bound to IRAK4 30 minutes after LPS stimulation, but did not directly associate with sNASP or TRAF6 (Figure 6A). On the other hand, IRAK4 associated with both CK2 and TRAF6 1 hour after LPS treatment, suggesting that IRAK4 is an adaptor for formation of a complex consisting of CK2 and TRAF6 following LPS stimulation of TLR4 (Figure 6A). Anti-TRAF6 immunoprecipitation showed that TRAF6 interacted with sNASP in unstimulated cells but dissociated from sNASP following binding to IRAK4 (Figure 6A). Depletion of both IRAK1 and IRAK4 completely blocked LPS-induced phosphorylation of sNASP, but retained the interaction between TRAF6 and sNASP (Figure 6B and Supplemental Figure 20N). However, IRAK1/4 kinase activity is not required for sNASP phosphorylation and interaction with TRAF6 (Figure 6C and Supplemental Figure 20O). Therefore, IRAK4 is an adaptor to bring together CK2 and TRAF6 for CK2-mediated phosphorylation of sNASP.
Figure 6. The IRAK1/4 complex is required for sNASP phosphorylation.
(A) THP-1 cells were stimulated with LPS for different time points and cross-linked with DSP for 30 minutes, and then IP with anti-CK2, anti-IRAK4, or anti-TRAF6 followed by IB with antibody against CK2, IRAK4, TRAF6, or NASP. TCL IB was done with antibody against CK2, IRAK4, TRAF6, NASP, or β-actin. (B) THP-1 cells were stimulated with LPS in the presence of siNT, siIRAK1, siIRAK4, or siIRAK1/4, and assessed by IB with antibody against phosphorylated serine (pSerine), anti-GFP, or TRAF6 after IP with anti-GFP. TCL IB was done with the indicated antibodies. (C) THP-1 cells were stimulated with LPS in the presence of DMSO or IRAK1/4 inhibitor, and assessed by IB with antibody against phosphorylated serine (pSerine), GFP, or TRAF6 after IP with anti-GFP. TCL IB was done with the indicated antibodies. Data represent a minimum of 3 independent experiments.
sNasp S158A-KI mice are more susceptible to sepsis due to defective immune response.
To address the physiologic relevance of sNASP/TRAF6 interaction, we generated sNasp S158A-KI mice using CRISPR/Cas9-mediated genome editing. To this end, we designed an sgRNA targeting exon 6 of sNasp and transcripted it to mRNA in vitro. To produce sNasp S158A-KI mice, the mRNA was injected into the embryos together with mutagenic oligonucleotides designed to introduce 1-bp substitutions (T to G) into the target locus (Supplemental Figure 11A). The validity of genetic substitution was confirmed by direct sequencing of tail DNAs (Supplemental Figure 11B). Loss of NASP led to early embryonic lethality in mice (11). In contrast, neither sNasp S158A/+ nor sNasp S158A/S158A mice showed any abnormalities of growth, survival, or behavior (data not shown). These findings suggest that the sNasp S158A allele is not required for embryonic development.
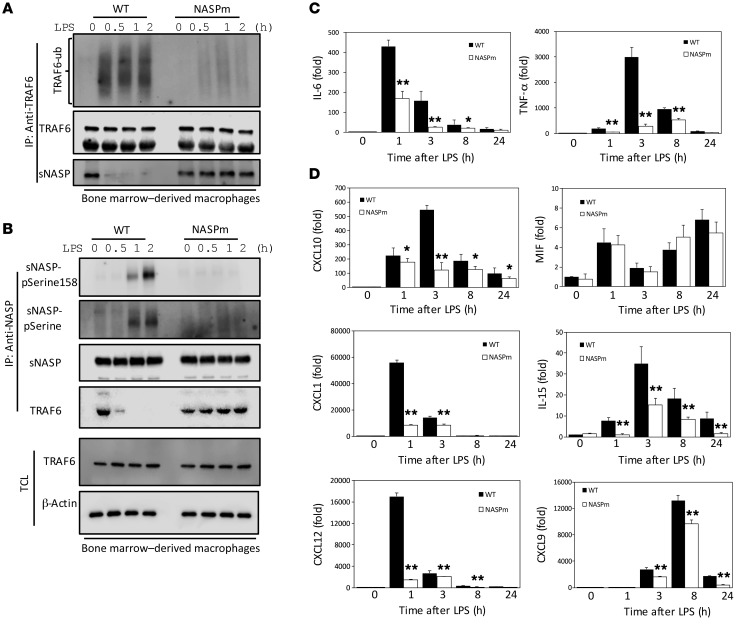
Next, we investigated whether LPS could increase sNASP S158 phosphorylation in BMDMs using a synthetic human antibody against phospho-sNASP S158 generated by a phage display system. In order to establish the specificity of this antibody, we transfected GFP-sNASP WT or phosphorylation mutants (S141A and S158A) into THP-1 cells. Phospho-sNASP S158 antibody detected LPS–induced sNASP phosphorylation in THP-1 cells transfected by sNASP-WT and S141A, but did not detect it in the sNASP S158A mutant (Supplemental Figure 12, A and B), confirming the mutant’s specificity for S158 phosphorylation. Because the phosphorylation-deficient sNASP S158A mutant resulted in defective TRAF6 signaling in THP-1 cells (Figure 3, C and D), we sought to assess if similar results also occurred in sNasp S158A macrophages treated with LPS. To test this possibility, we isolated BMDMs from WT and sNasp S158A-KI mice and examined sNASP phosphorylation and TRAF6 signaling after LPS treatment. Consistent with the results obtained with THP-1 and Raw264.7 cells, phosphorylation of sNASP and autoubiquitination of TRAF6 were markedly decreased in sNasp S158A-KI macrophages after LPS treatment (Figure 7, A and B, and Supplemental Figure 20, P and Q). Furthermore, the interaction between TRAF6 and sNASP was not affected by LPS in sNasp S158A-KI macrophages (Figure 7, A and B). LPS-induced sNasp S158A-KI, but not WT, primary macrophages also showed significant reduction of IL-6 and TNF-α (Figure 7C). These results indicate that the sNasp S158A-KI macrophage recapitulates the phenotype observed in cell lines. In addition to IL-6 and TNF-α, LPS-stimulated WT primary macrophages also secreted significantly higher levels of chemokines such as CXCL1, CXCL9, CXCL10, CXCL12, and IL-15 compared with LPS-stimulated sNasp S158A-KI primary macrophages (Figure 7D). It was shown that LPS-induced phosphorylation of CK2α at Thr360 and Ser362 enhanced CK2 activity (18). However, there was no difference in LPS-induced phosphorylation of CK2 between primary WT and sNasp S158A-KI macrophages (Supplemental Figure 13, A and B). Thus, TRAF6-sNASP signaling was regulated at a step distal to CK2.
Figure 7. sNASP S158A-KI macrophages have lower proinflammatory cytokine and chemokine production in response to LPS.
(A) BMDM cells from WT and NASP S158A-KI (NASPm) mice were stimulated with LPS and assessed by IB analysis with antibody against anti-ubiquitin, TRAF6, or NASP after IP with anti-TRAF6. (B) IP with anti-NASP followed by IB with phosphorylated serine (pSerine) or NASP in BMDM cells following LPS treatment. TCL IB was done with antibody against TRAF6, pSerine158-sNASP, or β-actin. (C and D) Expression of TNF-α and IL-6, CXCL10, MIF, CXCL1, IL-15, CXCL12, and CXCL9 in BMDM cells from WT and NASPm mice stimulated with LPS. Results were normalized to the expression of ACTB (encoding β-actin) and are presented relative to those of untreated cells. Data are mean ± SE for each group. *P < 0.05, **P < 0.01 (1-way ANOVA). Data represent a minimum of 3 independent experiments.
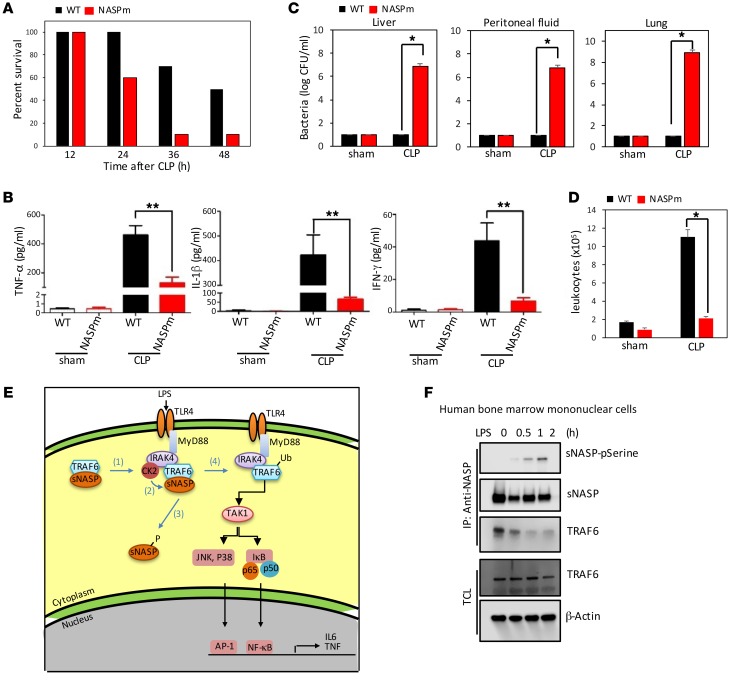
We next tested sNasp S158A-KI mice in a septic shock model. WT mice (C57BL/6J, n = 10) and sNasp S158A-KI mice (n = 10) underwent cecal ligation and puncture (CLP) to induce polymicrobial sepsis, and were monitored for survival. As shown in Figure 8A, more than 90 percent of the sNasp S158A-KI transgenic mice died within 36 hours, whereas 70 percent of WT mice lived. We did not observe an appreciable difference in survival rate between sNasp S158A-KI and WT mice after sham surgery (data not shown). To determine a possible cause for the increase in death rate in sNasp S158A-KI mice, we collected serum and assessed cytokine concentrations using ELISA assays. The serum of sNasp S158A-KI mice had a significantly lower concentration of TNF-α at 24 hours after CLP (Figure 8B). Additionally, the concentration of IL-1β and IFN-γ was also significantly lower in sNasp S158A-KI serum than in WT serum (Figure 8B). Therefore, our animal study suggested that the serine 158 substitution of sNASP markedly compromised the innate immune response to polymicrobial infection. In addition, bacterial counts in the lungs, liver, and peritoneal fluid after CLP were significantly higher in sNasp S158A-KI mice than in their WT littermates (Figure 8C). Flow cytometry indicated that the number of circulating CD11b+ leukocytes was much lower in the peritoneal fluid of sNasp S158A-KI mice than in WT mice after CLP challenge (Figure 8D). These results indicate that the serine 158 mutant of sNASP caused greater susceptibility to bacterial infection because of a defective innate immune response leading to ineffective clearance of bacteria.
Figure 8. sNasp S158A-KI mice are more susceptible to sepsis due to defective immune response.
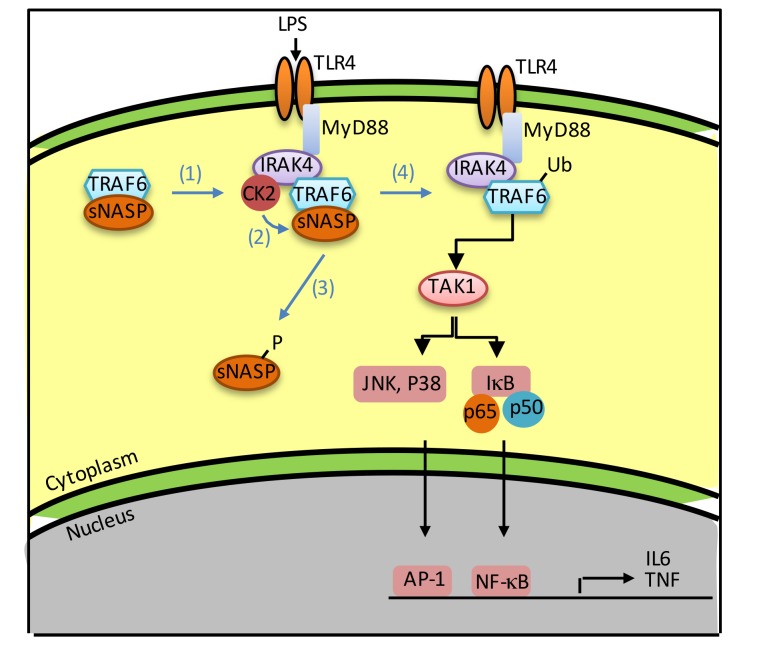
(A) Survival curves of WT and NASP S158A-KI (NASPm) mice following CLP (n = 10 per group per experiment). Bacterial load (as CFU) in the lung, liver, and peritoneal fluid (B), serum cytokines (C), and circulating leukocytes (CD11b+) (D) in WT and NASPm mice were measured 24 hours after sham treatment or CLP (n = 10 per group per experiment). (E) Model of TLR4 signaling regulated by sNASP. (1) Stimulation of TLR4 by LPS results in recruitment of IRAK4, CK2, and the TRAF6/sNASP complex. (2) CK2 phosphorylates sNASP. (3) p-sNASP is released from TRAF6. (4) TRAF6 autoubiquitination is followed by TAK-1 and IKK kinase activation and cytokine production. (F) IP with anti-NASP followed by IB with phosphorylated serine (pSerine), TRAF6, or NASP in human bone marrow mononuclear cells following LPS treatment. TCL IB was done with antibody against TRAF6 or β-actin. Results represent a minimum of 2 independent experiments with 10 mice each (A–D) or at least 3 (F) independent experiments. Data in B–D are mean ± SE for each group. *P < 0.05, **P < 0.01 (1-way ANOVA).
sNASP S158A-KI (NASPm) mice have a 2-fold reduction in neutrophil and eosinophil counts (Supplemental Figure 14). There was no significant change in lymphocyte, monocyte, basophil, or red blood cell counts. This difference could be due to a low-grade immune activation of the WT mice in a nonsterile environment. Next, we surveyed the requirement for sNASP in P selectin expression, chemotaxis, and phagocytosis. Following CLP-induced septic shock, P selection was induced only in the WT, but not mutant, endothelial cells in the liver (Supplemental Figure 15). However, MCP-1–induced macrophage chemotaxis was not affected in the mutant macrophage (Supplemental Figure 16). Thus, there appeared to be a defect in the early step of leukocyte recruitment.
MyD88-dependent activation of p38 has been shown to be involved in TLR-induced phagosome maturation (19, 20). Phagocytosed bacteria in phagosomes develop into phagolysosomes, and phagosomes and lysosomal markers can be used to study phagosome maturation. LysoTracker selectively labels late endosomes and lysosomes and colocalizes with lysosomal-associated membrane protein (LAMP) (21). Therefore, we monitored the maturation of phagosomes containing GFP–E. coli by their ability to colocalize with LysoTracker Red over time, using confocal fluorescence microscopy. The percentage of E. coli colocalized with LysoTracker Red at 1 hour reached 62% in WT macrophages, but only 10% in the mutant macrophages. The difference became more exaggerated at 90 minutes (Supplemental Figure 17, A and B). Taken together, the reduction of bacteria clearance in sNasp S158A-KI mice is multifactorial, including decreased basal white cell counts, poor adhesion molecule expression and leukocyte recruitment, insufficient cytokine/chemokine responses, and defective phagolysosome formation. We are planning to generate tissue-specific S158A-KI mice to further define the cause of early septic death.
We also tested WT and sNasp S158A-KI mice in lymphocytic choriomeningitis virus (LCMV) and influenza A virus (IAV) infection. LCMV and IAV both are single-stranded RNA viruses, which activate TLR7 but not TLR4 (22–24). WT mice (C57BL/6J, n = 10) and sNasp S158A-KI mice (n = 10) were infected with LCMV. The serum, spleen, kidney, and liver of LCMV-infected sNasp S158A-KI mice had similar viral titers compared with WT mice (Supplemental Figure 18A). The levels of TNF-α, IL-6, and IFN-γ were similar between LCMV-infected WT and sNasp S158A-KI mice (Supplemental Figure 18B). No significant differences were found between the survival rate and cytokine production in sNasp S158A-KI and WT mice after IAV infection (Supplemental Figure 19, A–C). These results demonstrate that sNASP regulates TLR4 but not TLR7 signaling, demonstrating the specificity of sNASP in TLR regulation (Figure 4F).
In conclusion, sNASP binds to TRAF6 in unstimulated cells to inhibit TRAF6 autoubiquitination and activation of downstream kinases. Upon LPS stimulation, a complex consisting of IRAK4, CK2, TRAF6, and sNASP was formed. CK2 phosphorylates sNASP at serine 158, releasing sNASP from TRAF6 and allowing TRAF6 to activate downstream kinases (Figure 8E). Similar responses were observed in human bone marrow mononuclear cells (Figure 8F and Supplemental Figure 20R). This inhibitory mechanism is biologically relevant because a dominant-negative mutant of sNASP prevents the mounting of an effective immune response during sepsis.
Discussion
NASP has a strong sequence identity and structural similarity to the SHNi-TPR superfamily, which is known as histone chaperone. Two major human NASP isoforms, sNASP and tNASP, were found in all dividing cells, including embryonic and malignant tissues. However, sNASP is the major isoform present in monocytes and macrophages (Supplemental Figure 2). Changes in NASP expression markedly affect cell growth and development, and loss of the NASP gene results in embryonic lethality (11, 14). Our interest in NASP was prompted by reports listing NASP as a TRAF6 binding partner in 2 independent mass spectrometry screenings (11, 13). To understand the role of NASP in the innate immune response, we manipulated the level of NASP in macrophage cell lines and examined the immune response following LPS stimulation. Our data reveal a new homeostatic mechanism using sNASP to regulate TLR-mediated NF-κB signaling through an inhibitory interaction with TRAF6. In resting macrophage cells, sNASP binds to TRAF6 to prevent its activation. However, after LPS stimulation, sNASP is phosphorylated by CK2 on serine 158 and dissociates from TRAF6 to allow downstream signaling. Therefore, sNASP serves as a critical checkpoint in TLR signaling and is a key regulator of innate immunity.
TLRs sense invading microbial pathogens to activate both innate and adaptive immunity. However, aberrant activation of TLRs can disturb immune hemostasis, leading to development of autoimmune and inflammatory diseases (3, 4, 25, 26). To prevent inappropriate activation of TLRs, multiple mechanisms, including dissociation of adaptor complexes, degradation of signal proteins, and transcriptional regulation, have been reported. For example, nucleotide binding oligomerization domain–like (NOD-like) receptor (NLR) family member X1 (NLRX1) binds to IKKβ and inhibits NF-κB activation; PDZ and LIM domain protein 2 (PDLIM2) was found to inhibit TLR signaling by degradation of the NF-kB component p65; and IkBNS is selectively recruited to the IL-6 promoter and regulates NF-kB activation (27–29). Since TRAF6 is a major signaling node in TLR activation, multiple regulators have been shown to directly target TRAF6, including A20, CYLD, USP4, SHP, SOCS3, TRIM38, and TANK (6, 30). A20, CYLD, and USP4 are deubiquitinating enzymes that remove the polyubiquitin chain to block TRAF6 signaling. TANK and SHP can bind to TRAF6 to inhibit its ubiquitination. SOCS3 and TRIM38 proteins are well-characterized E3 ubiquitin ligases that promote degradation of TRAF proteins. Hence, loss of these negative regulators of TRAF6 leads to hyperactivation of TLR signaling and inflammatory disorders (7, 8, 31). Since NASP-deficient mice are embryonic lethal (11) due to the role of NASP in histone regulation, we generated a dominant-negative form of sNASP that cannot be released from TRAF6 following LPS stimulation. In the sNasp S158A-KI mice, LPS cannot induce cytokine production and the mice are unable to clear bacteria or recruit leukocytes. Thus, sNASP is a bona fide inhibitor of TRAF6 activation.
Based on the CK2 consensus sequence motif, we postulated that CK2 could phosphorylate sNASP at serine 158. Indeed, biochemical, pharmacological, and genetic evidence has established that CK2 phosphorylates sNASP on serine 158. CK2 is a constitutively active kinase that is located in both the nuclear and cytoplasmic compartments (32). However, the majority of sNASP is not phosphorylated. Phosphorylation of sNASP is triggered by LPS-induced formation of a complex consisting of IRAK4, TRAF6, sNASP, and CK2. Thus, CK2 must be in the precise location to phosphorylate sNASP. CK2 activity can be regulated by proinflammatory cytokines. For example, LPS induces CK2 activity in murine Raw264.7 cells (33), TGF-β stimulates CK2 activity in murine mesangial cells and macrophages, and TNF-α activates CK2 in human cervical carcinoma HeLa and L929 cells (34–36). CK2 has been found to phosphorylate IκBα at multiple sites, including serine 283, 289, 293 and threonine 291, triggering IκBα degradation to activate NF-κB (37). CK2 also phosphorylates the p65 unit to regulate NF-κB–mediated inflammation (15). Thus, CK2 regulates multiple proteins involved in the inflammatory response, including TRAF6 signaling.
In addition to LPS, other TLR ligands such as Pam3CSK4 (a ligand of TLR1/2), HKLM (a ligand of TLR2), flagellin (a ligand of TLR5), and FSL-1 (a ligand of TLR6/2) also induced sNASP phosphorylation, but Poly (I:C) (a ligand of TLR3), imiquimod (a ligand of TLR7), ssRNA (a ligand of TL8), and ODN2006 (a ligand of TLR9) did not. Our result suggests that the subcellular localization of TLRs has a great influence on phosphorylation of sNASP. Phosphorylation of sNASP was only found in TLRs expressed on the cell surface and recognizes PAMPs in cell wall components and flagellin from both gram-positive and gram-negative bacteria, yeast, and fungi, but not in TLRs in intracellular compartments that detect PAMPs in nucleic acids derived from bacterial and viral pathogens. Specificity in different TLR signaling has been observed with different regulators. For instance, adaptor protein TIRAP, as part of the TLR4 downstream signaling molecule, interacts with TLR4 but not with TLR9. TIRAP-deficient mice respond normally to the TLR7 and TLR9 ligands but have defects in cytokine production and activation of the nuclear factor NF-κB in response to LPS (38).
Nasp-null mice are embryonic lethal, so sNasp S158A-KI mice were generated to further evaluate the physiologic function of sNASP in vivo (11). LPS-induced TRAF6 autoubiquitination and sNASP phosphorylation were impaired in the BMDMs of sNasp S158A-KI mice (Figure 7, A and B), consistent with results from cell line studies in which the sNASP level was manipulated (Figure 3, C and D). In a polymicrobial sepsis model, these mice were more susceptible to bacterial infection and failed to produce TNF-α and IL-1β. Thus, the sNasp S158A-KI mice displayed a primary immune deficiency phenotype following bacterial invasion. We have identified a previously unrecognized role for sNASP in the negative regulation of TLR signaling and established the mechanism whereby phosphorylation of sNASP by CK2 releases sNASP from TRAF6 to promote proinflammatory signaling. Our findings may allow for development of novel inhibitors to prevent CK2-mediated phosphorylation of sNASP to prevent TRAF6 activation to ameliorate harmful immune response.
Methods
Mice.
WT (C57BL/6) and sNASP S158A-KI mice were bred and maintained under pathogen-free conditions. The sNASP S158A-KI mice were generated with CRISPR/Cas9 technology, as illustrated in Supplemental Figure 11A. In brief, sNASP sgRNA was transcribed to mRNA in vitro, and mRNA guided the double-stranded DNA to be sheared by Cas9 at a specific site, then 120-bp donor oligo was integrated into the destination location by a homologous recombination. For genotyping, DNA was extracted from mouse tail samples using a DirectPCR Lysis Reagent (Viagen Biotech) according to the manufacturer’s guidelines. Tail DNA samples were subjected to PCR using the primers 5′-AGATCCCTTCAGAGCAGAT-3′ (forward) and 5′-TCCCTAAAGCCTACCAAAT-3 (reverse), and sequencing analysis (Supplemental Figure 11B).
CBC analysis.
Peripheral blood from WT and sNASP S158A-KI mice was collected via retro-orbital bleed into Microvettes coated with EDTA2K. Automated complete blood count with differential was performed using an IDEXX ProCyte Dx hematology analyzer (IDEXX Laboratories).
Cecal ligation puncture model.
Polymicrobial sepsis was induced by CLP with an 18-gauge needle as described (39). For survival studies, mice were monitored 2 times daily for up to 3 days. CFUs were measured by counting viable bacteria on agar plates in peritoneal fluid and lung and liver samples.
LCMV infection model.
The Armstrong strain of LCMV (LCMVarm) was propagated on BHK-21 cells and LCMV titers were determined by plaque assay on VeroE6 cells (40). Mice were infected by i.p. inoculation of 2 × 105 PFU LCMVarm cells. Uninfected mice were used in all in vivo experiments as background controls (data not shown). LCMV viral load was detected by quantitative PCR as previously described (41). For cytokine measurement, IL-6, TNF-α, and IFN-γ were quantified by BD cytometric bead array using an LSRFortessa flow cytometer and FCAP Array analysis software (BD Biosciences).
IAV infection model.
Influenza A/Puerto Rico/8/34 virus (IAV) infection and virus titer studies were performed as previously described (42). For the mortality studies, mice were infected by intranasal administration of 20,000 PFU IAV per mouse and monitored daily for up to 16 days. For the plaque and cytokine assays, mice were sacrificed 3 days after infection and lung tissue samples were harvested for analysis.
Cell culture and stimulation cells.
Peritoneal macrophages and BMDMs were cultured as previously described (43). The human monocyte THP-1 cell line (ATCC TIB-202, American Type Culture Collection [ATCC]) was cultured in RPMI-1640 medium (ATCC 30-2001, ATCC) supplemented with 2 mM glutamine, 10% FBS, and 1% penicillin and streptomycin. The embryonic kidney epithelial cell line HEK293 (ATCC CRL-1573, ATCC) and mouse macrophage Raw264.7 cell line (ATCC TIB-71, ATCC) were culture in DMEM supplemented with 10% FBS and 1% penicillin and streptomycin. Human bone marrow mononuclear cells were purchased from STEMCELL Technologies (catalog 70001) and were culture in RPMI-1640 medium supplemented with 10% FBS and 1% penicillin and streptomycin. All cell lines were maintained at 37°C at 5% CO2 levels. For cell stimulation, a Human TLR1-9 Agonist kit was purchased from InvivoGen (catalog tlrl-kit1hw). TBB (CK2 inhibitor, catalog T0826), IRAK1/4 inhibitor (catalog I5409), and MCP-1 (catalog SRP3109) were obtained from Sigma-Aldrich. DSP (catalog 22585) and LysoTracker Red (catalog L7528) were purchased from Thermo Scientific.
Chemotaxis assay.
Chemotaxis was estimated using a 24-well 6.5 mm Transwell (catalog 3422, Corning) according to the manufacturer’s instructions. A 24-well microchemotaxis plate in which the chambers were separated by an 8-μm pore size polycarbonate membrane was used. Serum-free medium was used as a control. Chemoattractant MCP-1 (20 ng/ml) was placed in the lower chamber and purified BMDM cells (2 × 106 cells/well) were placed in the upper chamber. Chambers were incubated at 37°C with 5% CO2 for 20 hours. The results are expressed as the mean number of BMDMs per field and are representative of triplicate measurements from 3 separate experiments.
Immunohistochemistry.
Twenty-four hours after CLP, WT and sNASP S158A-KI mice were euthanized (n = 8). Sham mice did not receive CLP. Before embedding in paraffin, liver specimens were fixed in 4% paraformaldehyde in PBS and dehydrated. Paraffin-embedded sections were deparaffinized with xylene and treated with gradually decreasing concentrations of ethanol. After washing 3 times with PBS, tissue sections were treated with 3% hydrogen peroxidase in methanol for 10 minutes to block endogenous peroxidase activity and then processed in citrate buffer and heated to 95°C for 25 minutes for antigen retrieval. Sections were incubated with blocking serum for 1 hour and later incubated with anti–P selectin (1:200, catalog ab202983, Abcam) overnight at 4°C. After washing in PBS 3 times, sections were first incubated with biotin-conjugated secondary antibody at 37°C for 45 minutes and then incubated with avidin-biotin complex at 37°C for another 30 minutes. After a final wash with PBS, the sections were used as DAB substrate to detect coloration.
Coimmunoprecipitation and immunoblotting.
IP and IB were performed essentially as described elsewhere (44). Cells were lysed by IP lysis buffer (20 mM Tris-HCl at pH 7.5, 0.5% NP-40, 150 mM NaCl, and protease inhibitors) (catalog P8340, Sigma). Lysates were sonicated and cleared of insoluble material by centrifugation at 15,700 g for 30 minutes. For immunoprecipitation, cell extracts were then incubated with the indicated antibody overnight at 4°C and subsequently with the respective Protein A/G beads (16-156/16-266, Millipore) for an additional 2 hours. After the beads were washed 3 times with lysis buffer, the immunoprecipitated products were eluted using 2× Laemmli sample buffer and boiled at 100°C for 5 minutes. Samples were subjected to 4–15% SDS-PAGE and transferred onto PVDF membrane (catalog IPVH00010, Millipore). Blocking was performed in 5% milk and membranes were incubated in primary antibodies overnight at 4°C. Membranes were incubated with HRP-conjugated secondary antibody for 1 hour and protein was visualized using SuperSignal West Pico Luminol Enhancer Solution (product 1859675, Thermo Scientific). For anti-Flag or anti-GFP immunoprecipitation, ANTI-FLAG M2 Affinity Gel (F2426, Sigma-Aldrich) or GFP-Trap_M (gtm-20, Chromotek) were used. The following antibodies were used for IP and IB (IB ratio noted unless otherwise specified): anti-NASP (IP 1:100, IB 1:1,000; SC-161915; Santa Cruz), anti-TRAF6 (IP 1:100, IB 1:1,000; SC-7221; Santa Cruz), anti-ubiquitin (1:1,000; SC-8017; Santa Cruz), anti-CK2α (IP 1:100, IB 1:1,000; SC-6479; Santa Cruz), anti–β-actin (1:5,000; SC-47778; Santa Cruz), anti-GAPDH (1:2,500; SC-3233; Santa Cruz), anti-LaminB (1:1,000; SC-6217; Santa Cruz), anti-IRAK1 (1:1,000; SC-7883; Santa Cruz), anti-IRAK4 (IP 1:100, IB 1:1,000; SC-99154; Santa Cruz), anti–phospho-TAK1 (1:1,000; 4531; Cell Signaling), anti-TAK1 (1:1,000; 4505; Cell Signaling), anti–phospho-p38 MAPK (1:1,000; 9211; Cell Signaling), anti–p38 MAPK (1:1,000; 9212; Cell Signaling), anti–phospho-JNK kinase (1:1,000; 4671; Cell Signaling), anti–JNK kinase (1:1,000; 9252; Cell Signaling), anti–phospho-IκBα Ser32 (1:1,000; 2859; Cell Signaling), anti-IκBα (1:1,000; 4812; Cell Signaling), anti-Myc (1:2,500; 631206; Clontech), anti-GFP (1:2,500; 632281; Clontech), anti–phosphothreonine Q7 (1:1,000; 37420; Qiagen), anti–phosphoserine Q5 (1:1,000; 37430; Qiagen), anti-phosphoserine (1:5,000; ab17465; Abcam), anti-HA (1:5,000; 901513; BioLegend), anti-Flag (1:5,000; F1804; Sigma-Aldrich), and anti–phospho-CK2α (pThr360/Ser362) (1:100; SAB4300628; Sigma-Aldrich). For phosphorylation IB, cells were lysed by RIPA buffer (BP-115D, Boston BioProducts), including protease inhibitors and Phosphatase Inhibitor Cocktail (P0044, Sigma-Aldrich), and PVDF membrane was incubated in Membrane Blocking Solution (000105, Life Technologies) for blocking and antibody incubation.
Quantitative reverse-transcriptase PCR.
RNA was extracted from cells using the RNeasy Mini Kit (74104, Qiagen) and the cDNA was synthesized by using the Advantage RT-PCR Kit (639506, Clontech) according to the manufacturer’s protocol. Quantitative real-time PCR was conducted using TaqMan Gene Expression Master Mix (4369016, Applied Biosystems) and IL-6, TNF-α, CXCL10, MIF, CXCL1, IL-15, CXCL12, and CXCL9 TaqMan Gene Expression Assay (Mm00446190_m1, Mm00443260_g1, Mm000445235_m1, Mm01611157_gH, Mm04207460_m1, Mm00434210_m1, Mm00445553_m1 and Mm00434946_m1, ThermoFisher Scientific). The results were normalized to expression of the gene encoding 18s and were quantified by the change-in-threshold method (ΔΔCT).
Cytokine measurement.
Cytokine measurement was assessed by ELISA. The serum and cell culture supernatants were collected and measured by ELISA according to the manufacturer’s protocol. Mouse Ready-Set-Go ELISA for IL-6 (88-7064), TNF-α (88-7324), IL-1β (88-7013), and IFN-γ (88-7314) were obtained from eBioscience.
siRNA knockdown.
Short interfering RNAs (siRNAs) were transfected by using DharmaFECT 1 reagent (T-2001-02, Dharmacon) according to the manufacturer’s protocol. Briefly, siRNAs and the DharmaFECT 1 reagent were diluted with serum-free DMEM separately and incubated for 5 minutes. These diluted reagents were then mixed, incubated for 20 minutes at room temperature to form an siRNA-DharmaFECT complex, and added dropwise to the cells. After 48 hours of transfection, cells were stimulated with LPS as described above. Sequences for siRNA are listed as follows: the siRNA specific for the gene encoding human NASP (siNASP, 5′-GGAACUGCUACCCGAAAUU-3′), CK2 (the pool of siCK2 no. 1, 5′-CUGGUCGCUUACAUCACUUUA-3′ and siCK2 no. 2, 5′-UCAAGAUGACUACCAGCUGU-3′), IRAK1(the pool of siIRAK1 no. 1, 5′-CGAAGAAAGUGAUGAAUUUUU-3′ and siIRAK1 no. 2, 5′- GAAAGACCUGGUGGAAGAUU-3′), IRAK4 (the pool of siIRAK4 no. 1, 5′-GCAGGACAGUGGUUAUUAAUU-3′ and siIRAK4 no. 2, 5′-GCCACCAUAUCAACACUUAUU-3′) and the siRNA specific for the gene encoding mouse NASP (the pool of siNASP no. 9, 5′-GGAUAUAAGUGAGCCUGAA-3′, siNASP no. 10, 5′-GCAGGAGAAUUACAGUUAUU-3′, siNASP no. 11, 5′-GGUAAGAAGUAUGGAGAAA-3′ and siNASP no. 12, 5′-GAUGAAAGAGGGUGAAGAA-3′). All siRNAs were synthesized from Sigma-Aldrich. siRNA Universal Negative control was purchased from Sigma-Aldrich (catalog S1C001).
Plasmid construction, cell transfection, and adenovirus transduction.
The plasmids encoding human GFP-tNASP and GFP-sNASP were gifts from Genevieve Almouzni (Centre de Recherche, Paris, France). The C-terminal fragment of sNASP was released by digestion of pGFP-sNASP with SbfI and ApaI (blunted with Klenow) or EcoNI and ApaI (blunted with Klenow) and then the remaining fragment of pEGFP-sNASP was ligated again to produce pGFP-sNASP 1–233 or pGFP-sNASP 1–348, respectively. PCR-based site-directed mutagenesis (45) was used to create mutant constructs of sNASP (GFP-sNASP, S123A, T127A, S138A, T141A, S158A, S158E, S164A, T361A, and S387A). The EcoRI-BamHI fragment from pGFP-sNASP was inserted into the same sites of p3XFlag-CMV (E4026, Sigma-Aldrich) to generate pFlag-sNASP. Adenovirus vectors including sNASP were subcloned into the pENTR vector (A10462, Thermo Scientific) using SalI-NotI sites, and then a recombinase reaction was performed according to the manufacturer’s instructions (K4930-00, Thermo Scientific) to get a pDEST-Flag–sNASP vector. All constructs were verified by DNA sequencing. The plasmids encoding Flag-TRAF6, HA-Ub k48, and HA-Ub k63 were obtained from Hui-Kuan Lin (The University of Texas MD Anderson Cancer Center, Houston, Texas, USA). The plasmids encoding HA-tagged TRAF3 were gifts from Shao-Cong Sun (The University of Texas MD Anderson Cancer Center, Houston, Texas, USA). The plasmids HA-CK2α, HA-CK2α K68M, HA-CK2α’, HA-CK2α’ K69M, and Myc-CK2β were obtained from the nonprofit Addgene Plasmid Repository (David Litchfield, University of Western Ontario, London, Ontario, Canada; Addgene plasmids 27086, 27089, 27087, 27090, and 27088). The plasmids HA-Ub and RGS-Ub have been described (46, 47). THP-1 cells were used for transient cell transfection, using X-tremeGene HP (06-365-752-001, Roche). HEK293 and Raw264.7 cells were transfected with expression plasmids by Lipofectamine 3000 (L3000015, Invitrogen) according to the manufacturer’s protocol. For adenovirus transduction, the construct pDEST-Flag–sNASP vector was transfected into 293A cells using Lipofectamine 3000. Virus-containing media were collected 48–72 hours later and passed through a 0.45 μm filter. BMDM cells were transduced with LacZ as control or Flag-sNASP–encoding adenovirus.
Luciferase assays.
NF-κB luciferase reporter assays were done according to our previous publication (48). Twenty-four hours before transfection, HEK293 cells were subcultured onto 24-well plates at a density of 2 × 105 cells/well. Cells were transfected with plasmids encoding NF-κB luciferase, pRL-TK Renilla luciferase, and different expression and control vectors. After 24 hours, cells were harvested and luciferase activities were determined using the Dual-Glo Luciferase Assay System (E2920, Promega). The results were normalized to internal Renilla luciferase activities. Data were obtained from 5 independent experiments.
Phagocytosis assay and fluorescence microscopy.
A total of 1 × 105 BMDMs was grown on glass cover slips in a 6-well plate overnight. LysoTracker Red (100 nM) was used to pretreat cells for 1 hour, and then cells were infected for 1 hour with GFP–E. coli (MOI = 20) and followed by several washes with PBS. Infected cells were further cultured in medium containing 10 μg/ml gentamicin. At the appropriate time points, the cells were washed in PBS and then fixed for 10 minutes at room temperature in 4% (vol/vol) paraformaldehyde. Slides were mounted with VECTASHIELD HardSet Antifade Mounting Medium with DAPI (H-1500, Vector Laboratories) and were imaged with a Nikon Eclipse Ti confocal microscope.
In vitro phosphorylation of sNASP by casein kinase II.
Recombinant GFP-sNASP (WT and sNASP158A) protein prepared by immunoprecipitation using GFP-Trap_M (gtm-20, Chromotek) was suspended in 30 μL of 1× NEBuffer for Protein Kinases supplemented with 1 μl casein kinase II (P6010, New England Biolabs) and 200 μM ATP (P0756, New England Biolabs) and incubated for 30 minutes at 30°C. The reaction was terminated by adding 30 μl of 2× SDS sample buffer and boiling for 5 minutes. Samples were analyzed on 10% SDS-PAGE. A standard Western blot was conducted and the membrane was probed with the indicated antibodies.
LC-MS/MS analysis.
Immunoprecipitated proteins were visualized by staining of SDS gels with GelCode Blue Stain Reagent (24590, Thermo Scientific). Proteins were excised from the gels and gel slices were submitted to the Harvard University Mass Spectrometry and Proteomics Resource Laboratory (Cambridge, MA) for protein identification and site-of-modification (phosphorylation) analysis.
Generation of human anti–phospho-sNASP S158.
A peptide corresponding to the amino acids 148–168 of human sNASP (Biotin-NPEGQ-NDKTEEMPNDS*VLENKSLQEN) was synthesized (Biomatik Inc.) with an amino-terminus biotin label in both phosphorylated and nonphosphorylated forms. A human anti–phospho-sNASP S158 single-chain variable fragment was generated against the phospho-peptide by phage display with a naive human phage display library constructed from peripheral blood lymphocytes (49).
Statistics.
SigmaPlot 13.0 software was used for data analysis. For statistical analyses of all quantifications (measurement of cytokine production and densitometric quantification of Western blots), statistical significance was determined by 2-tailed t test for 2 groups or 1-way ANOVA with multiple comparisons tests for 3 or more groups. Data are mean ± SEM. P < 0.05 was considered statistically significant. SigmaPlot 13.0 software was also used to assess the normality of distribution of investigated parameters. All parameters in our study were distributed normally and represented equal sample sizes. For Western blot quantification, gray bar graphs show the densitometric quantification of the indicated signals using ImageJ software. Signal values were normalized to unstimulated conditions (set at 1).
Study approval.
All mouse experiments were conducted in accordance with the Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011), and were approved by the Experimental Animal Ethical Committee at Shanghai Jiao Tong University School of Medicine and the IACUC of the University of Missouri.
Author contributions
FMY and ETHY developed the concepts, analyzed data, and designed experiments. FMY performed the experiments. JC developed the S158A-KI mice. YZ and WZ carried out the CLP and LCM virus studies. CX and BH carried out the influenza virus studies. MS generated the anti–phospho-sNASP S158 antibody. HMC established the laboratory in the University of Missouri, analyzed the data, and edited the manuscript. FMY and ETHY wrote the paper.
Supplementary Material
Acknowledgments
Supported by Strategic Hire of the University of Missouri (grant to ETHY); National Natural Science Foundation of China (grant 91229202), the National Basic Research Program of China (973 Program; grant 2013CB910902), the Specialized Research Fund for the Doctoral Program of Higher Education (grant 20120073110073), and Shanghai Committee of Science and Technology (grant 11DZ2260200) (to JC); and NIH R21 AI127404 (to BH). ETHY is the Frances T. McAndrew Chair of the University of Missouri. We would like to thank Runsheng Wang (MD Anderson Cancer Center) for technical assistance.
Version 1. 05/07/2018
Electronic publication
Version 2. 06/01/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(6):2459–2472.https://doi.org/10.1172/JCI95720.
Contributor Information
Feng-Ming Yang, Email: yangfeng@health.missouri.edu.
Yong Zuo, Email: zuoyong@shsmu.edu.cn.
Wei Zhou, Email: joesphchou@sjtu.edu.cn.
Chuan Xia, Email: xiac@health.missouri.edu.
Bumsuk Hahm, Email: hahmb@health.missouri.edu.
Jinke Cheng, Email: jkcheng@shsmu.edu.cn.
Hui-Ming Chang, Email: changhu@health.missouri.edu.
References
- 1.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449(7164):819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 2.Poltorak A, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282(5396):2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 3.Arthur JS, Ley SC. Mitogen-activated protein kinases in innate immunity. Nat Rev Immunol. 2013;13(9):679–692. doi: 10.1038/nri3495. [DOI] [PubMed] [Google Scholar]
- 4.O’Shea JJ, Murray PJ. Cytokine signaling modules in inflammatory responses. Immunity. 2008;28(4):477–487. doi: 10.1016/j.immuni.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cook DN, Pisetsky DS, Schwartz DA. Toll-like receptors in the pathogenesis of human disease. Nat Immunol. 2004;5(10):975–979. doi: 10.1038/ni1116. [DOI] [PubMed] [Google Scholar]
- 6.Kondo T, Kawai T, Akira S. Dissecting negative regulation of Toll-like receptor signaling. Trends Immunol. 2012;33(9):449–458. doi: 10.1016/j.it.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Turer EE, et al. Homeostatic MyD88-dependent signals cause lethal inflammation in the absence of A20. J Exp Med. 2008;205(2):451–464. doi: 10.1084/jem.20071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshida H, Jono H, Kai H, Li JD. The tumor suppressor cylindromatosis (CYLD) acts as a negative regulator for toll-like receptor 2 signaling via negative cross-talk with TRAF6 AND TRAF7. J Biol Chem. 2005;280(49):41111–41121. doi: 10.1074/jbc.M509526200. [DOI] [PubMed] [Google Scholar]
- 9.Richardson RT, et al. Characterization of the histone H1-binding protein, NASP, as a cell cycle-regulated somatic protein. J Biol Chem. 2000;275(39):30378–30386. doi: 10.1074/jbc.M003781200. [DOI] [PubMed] [Google Scholar]
- 10.Cook AJ, Gurard-Levin ZA, Vassias I, Almouzni G. A specific function for the histone chaperone NASP to fine-tune a reservoir of soluble H3-H4 in the histone supply chain. Mol Cell. 2011;44(6):918–927. doi: 10.1016/j.molcel.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 11.Richardson RT, et al. Nuclear autoantigenic sperm protein (NASP), a linker histone chaperone that is required for cell proliferation. J Biol Chem. 2006;281(30):21526–21534. doi: 10.1074/jbc.M603816200. [DOI] [PubMed] [Google Scholar]
- 12.Alekseev OM, Richardson RT, Alekseev O, O’Rand MG. Analysis of gene expression profiles in HeLa cells in response to overexpression or siRNA-mediated depletion of NASP. Reprod Biol Endocrinol. 2009;7:45. doi: 10.1186/1477-7827-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campos EI, et al. The program for processing newly synthesized histones H3.1 and H4. Nat Struct Mol Biol. 2010;17(11):1343–1351. doi: 10.1038/nsmb.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alekseev OM, Widgren EE, Richardson RT, O’Rand MG. Association of NASP with HSP90 in mouse spermatogenic cells: stimulation of ATPase activity and transport of linker histones into nuclei. J Biol Chem. 2005;280(4):2904–2911. doi: 10.1074/jbc.M410397200. [DOI] [PubMed] [Google Scholar]
- 15.Chantôme A, et al. Casein kinase II-mediated phosphorylation of NF-kappaB p65 subunit enhances inducible nitric-oxide synthase gene transcription in vivo. J Biol Chem. 2004;279(23):23953–23960. doi: 10.1074/jbc.M313731200. [DOI] [PubMed] [Google Scholar]
- 16.Basnet H, et al. Tyrosine phosphorylation of histone H2A by CK2 regulates transcriptional elongation. Nature. 2014;516(7530):267–271. doi: 10.1038/nature13736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulges A, et al. Protein kinase CK2 enables regulatory T cells to suppress excessive TH2 responses in vivo. Nat Immunol. 2015;16(3):267–275. doi: 10.1038/ni.3083. [DOI] [PubMed] [Google Scholar]
- 18.Kim KJ, et al. Platelet-activating factor enhances tumour metastasis via the reactive oxygen species-dependent protein kinase casein kinase 2-mediated nuclear factor-κB activation. Immunology. 2014;143(1):21–32. doi: 10.1111/imm.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blander JM, Medzhitov R. Regulation of phagosome maturation by signals from toll-like receptors. Science. 2004;304(5673):1014–1018. doi: 10.1126/science.1096158. [DOI] [PubMed] [Google Scholar]
- 20.Doyle SE, et al. Toll-like receptors induce a phagocytic gene program through p38. J Exp Med. 2004;199(1):81–90. doi: 10.1084/jem.20031237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang QQ, Li H, Oliver T, Glogauer M, Guo J, He YW. Integrin beta 1 regulates phagosome maturation in macrophages through Rac expression. J Immunol. 2008;180(4):2419–2428. doi: 10.4049/jimmunol.180.4.2419. [DOI] [PubMed] [Google Scholar]
- 22.Borrow P, Martínez-Sobrido L, de la Torre JC. Inhibition of the type I interferon antiviral response during arenavirus infection. Viruses. 2010;2(11):2443–2480. doi: 10.3390/v2112443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwasaki A, Pillai PS. Innate immunity to influenza virus infection. Nat Rev Immunol. 2014;14(5):315–328. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia C, Vijayan M, Pritzl CJ, Fuchs SY, McDermott AB, Hahm B. Hemagglutinin of influenza A virus antagonizes type I interferon (IFN) responses by inducing degradation of type I IFN receptor 1. J Virol. 2015;90(5):2403–2417. doi: 10.1128/JVI.02749-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider M, et al. The innate immune sensor NLRC3 attenuates Toll-like receptor signaling via modification of the signaling adaptor TRAF6 and transcription factor NF-κB. Nat Immunol. 2012;13(9):823–831. doi: 10.1038/ni.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu S, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347(6227):aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 27.Kuwata H, et al. IkappaBNS inhibits induction of a subset of Toll-like receptor-dependent genes and limits inflammation. Immunity. 2006;24(1):41–51. doi: 10.1016/j.immuni.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka T, Grusby MJ, Kaisho T. PDLIM2-mediated termination of transcription factor NF-kappaB activation by intranuclear sequestration and degradation of the p65 subunit. Nat Immunol. 2007;8(6):584–591. doi: 10.1038/ni1464. [DOI] [PubMed] [Google Scholar]
- 29.Xia X, et al. NLRX1 negatively regulates TLR-induced NF-κB signaling by targeting TRAF6 and IKK. Immunity. 2011;34(6):843–853. doi: 10.1016/j.immuni.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang XD, Sun SC. Targeting signaling factors for degradation, an emerging mechanism for TRAF functions. Immunol Rev. 2015;266(1):56–71. doi: 10.1111/imr.12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou F, Zhang X, van Dam H, Ten Dijke P, Huang H, Zhang L. Ubiquitin-specific protease 4 mitigates Toll-like/interleukin-1 receptor signaling and regulates innate immune activation. J Biol Chem. 2012;287(14):11002–11010. doi: 10.1074/jbc.M111.328187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyata Y, Nishida E. Evaluating CK2 activity with the antibody specific for the CK2-phosphorylated form of a kinase-targeting cochaperone Cdc37. Mol Cell Biochem. 2008;316(1-2):127–134. doi: 10.1007/s11010-008-9818-1. [DOI] [PubMed] [Google Scholar]
- 33.Lodie TA, Savedra R, Golenbock DT, Van Beveren CP, Maki RA, Fenton MJ. Stimulation of macrophages by lipopolysaccharide alters the phosphorylation state, conformation, and function of PU.1 via activation of casein kinase II. J Immunol. 1997;158(4):1848–1856. [PubMed] [Google Scholar]
- 34.Zdunek M, Silbiger S, Lei J, Neugarten J. Protein kinase CK2 mediates TGF-beta1-stimulated type IV collagen gene transcription and its reversal by estradiol. Kidney Int. 2001;60(6):2097–2108. doi: 10.1046/j.1523-1755.2001.00041.x. [DOI] [PubMed] [Google Scholar]
- 35.Singh NN, Ramji DP. Transforming growth factor-beta-induced expression of the apolipoprotein E gene requires c-Jun N-terminal kinase, p38 kinase, and casein kinase 2. Arterioscler Thromb Vasc Biol. 2006;26(6):1323–1329. doi: 10.1161/01.ATV.0000220383.19192.55. [DOI] [PubMed] [Google Scholar]
- 36.Van Lint J, et al. Tumor necrosis factor stimulates multiple serine/threonine protein kinases in Swiss 3T3 and L929 cells. Implication of casein kinase-2 and extracellular signal-regulated kinases in the tumor necrosis factor signal transduction pathway. J Biol Chem. 1992;267(36):25916–25921. [PubMed] [Google Scholar]
- 37.McElhinny JA, Trushin SA, Bren GD, Chester N, Paya CV. Casein kinase II phosphorylates I kappa B alpha at S-283, S-289, S-293, and T-291 and is required for its degradation. Mol Cell Biol. 1996;16(3):899–906. doi: 10.1128/MCB.16.3.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horng T, Barton GM, Flavell RA, Medzhitov R. The adaptor molecule TIRAP provides signalling specificity for Toll-like receptors. Nature. 2002;420(6913):329–333. doi: 10.1038/nature01180. [DOI] [PubMed] [Google Scholar]
- 39.Rittirsch D, Huber-Lang MS, Flierl MA, Ward PA. Immunodesign of experimental sepsis by cecal ligation and puncture. Nat Protoc. 2009;4(1):31–36. doi: 10.1038/nprot.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahmed R, Salmi A, Butler LD, Chiller JM, Oldstone MB. Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. J Exp Med. 1984;160(2):521–540. doi: 10.1084/jem.160.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCausland MM, Crotty S. Quantitative PCR technique for detecting lymphocytic choriomeningitis virus in vivo. J Virol Methods. 2008;147(1):167–176. doi: 10.1016/j.jviromet.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vijayan M, et al. Sphingosine 1-phosphate lyase enhances the activation of IKKε to promote type I IFN-mediated innate immune responses to influenza A virus infection. J Immunol. 2017;199(2):677–687. doi: 10.4049/jimmunol.1601959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heo KS, et al. ERK5 activation in macrophages promotes efferocytosis and inhibits atherosclerosis. Circulation. 2014;130(2):180–191. doi: 10.1161/CIRCULATIONAHA.113.005991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang FM, Lin YC, Hu MC. Identification of two functional nuclear localization signals mediating nuclear import of liver receptor homologue-1. Cell Mol Life Sci. 2011;68(7):1241–1253. doi: 10.1007/s00018-010-0521-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang FM, Pan CT, Tsai HM, Chiu TW, Wu ML, Hu MC. Liver receptor homolog-1 localization in the nuclear body is regulated by sumoylation and cAMP signaling in rat granulosa cells. FEBS J. 2009;276(2):425–436. doi: 10.1111/j.1742-4658.2008.06785.x. [DOI] [PubMed] [Google Scholar]
- 46.Gong L, Kamitani T, Millas S, Yeh ET. Identification of a novel isopeptidase with dual specificity for ubiquitin- and NEDD8-conjugated proteins. J Biol Chem. 2000;275(19):14212–14216. doi: 10.1074/jbc.275.19.14212. [DOI] [PubMed] [Google Scholar]
- 47.Kamitani T, Kito K, Nguyen HP, Yeh ET. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem. 1997;272(45):28557–28562. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- 48.Yang FM, Feng SJ, Lai TC, Hu MC. A calreticulin-dependent nuclear export signal is involved in the regulation of liver receptor homologue-1 protein folding. Biochem J. 2015;471(2):199–209. doi: 10.1042/BJ20150252. [DOI] [PubMed] [Google Scholar]
- 49.Shea C, Bloedorn L, Sullivan MA. Rapid isolation of single-chain antibodies for structural genomics. J Struct Funct Genomics. 2005;6(2-3):171–175. doi: 10.1007/s10969-005-5246-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.