Abstract
Painful diabetic neuropathy (PDN) is an intractable complication of diabetes that affects 25% of patients. PDN is characterized by neuropathic pain and small-fiber degeneration, accompanied by dorsal root ganglion (DRG) nociceptor hyperexcitability and loss of their axons within the skin. The molecular mechanisms underlying DRG nociceptor hyperexcitability and small-fiber degeneration in PDN are unknown. We hypothesize that chemokine CXCL12/CXCR4 signaling is central to this mechanism, as we have shown that CXCL12/CXCR4 signaling is necessary for the development of mechanical allodynia, a pain hypersensitivity behavior common in PDN. Focusing on DRG neurons expressing the sodium channel Nav1.8, we applied transgenic, electrophysiological, imaging, and chemogenetic techniques to test this hypothesis. In the high-fat diet mouse model of PDN, we were able to prevent and reverse mechanical allodynia and small-fiber degeneration by limiting CXCR4 signaling or neuronal excitability. This study reveals that excitatory CXCR4/CXCL12 signaling in Nav1.8-positive DRG neurons plays a critical role in the pathogenesis of mechanical allodynia and small-fiber degeneration in a mouse model of PDN. Hence, we propose that targeting CXCR4-mediated DRG nociceptor hyperexcitability is a promising therapeutic approach for disease-modifying treatments for this currently intractable and widespread affliction.
Keywords: Neuroscience
Keywords: Pain
Introduction
Painful diabetic neuropathy (PDN) is one of the most common and intractable complications of diabetes, affecting 25% of diabetic patients (1, 2). Given the increasing prevalence of type 2 diabetes mellitus (3), the incidence of PDN is expected to rise (4). Neuropathic pain associated with PDN substantially affects patients’ quality of life and health care costs (5) and is difficult to treat. Opiates are mostly ineffective for treating neuropathic pain and problematic for chronic use (2). Gabapentinoids and antidepressants produce limited relief in some patients but have many side effects and a low response rate for PDN (6–9). Thus, safer and more effective therapies based on mechanistic targets specific to PDN are urgently required.
The hallmarks of PDN are neuropathic pain and small-fiber degeneration (10, 11), particularly a “dying back” axonopathy that affects the smallest axons (12, 13) of the peripheral nervous system: the dorsal root ganglion (DRG) nociceptor axons. Acute pain is normally important for preventing tissue damage (14, 15). However, in conditions such as PDN, physiological pain transitions to pathological or neuropathic pain that does not serve any important physiological function. The complex pathophysiology underlying neuropathic pain in PDN (16) extends from primary afferent terminals to anatomical and functional changes in the brain and spinal cord that amplify nociceptive processing (16, 17). Diabetic patients (18) and experimental models of PDN (19, 20) have sensory neuron hyperexcitability, including spontaneous activity of DRG nociceptor axons and the terminals of C-fiber nociceptors (21, 22). The molecular pathways linking hyperexcitability to neuropathic pain and small-fiber degeneration in PDN are unknown. This gap in knowledge represents a critical barrier to progress in developing novel therapeutic approaches for PDN.
In our experiments, we identified DRG nociceptors via a molecular marker, the sodium channel Nav1.8 (23). Approximately 75% of DRG sensory neurons express Nav1.8, including more than 90% of C-nociceptors, a population of C–low-threshold mechanoreceptors and some Aδ-nociceptors and Aβ afferents (23). Thus, by focusing on the properties of Nav1.8-positive DRG neurons, we are likely to discover key changes in the behavior of DRG nociceptors in animal models of PDN.
One of the initial questions that must be addressed is what mechanisms trigger Nav1.8-positive DRG neuron hyperexcitability in diabetes. Promising hypotheses include altered gene expression and posttranslational modification of key ion channels (24, 25). For example, methylglyoxal, abundant during hyperglycemia (19, 20), induces posttranslational modifications in Nav1.8 sodium channels (26) that result in nociceptor hyperexcitability and mechanical allodynia in rodents. In addition, inflammatory mediators, including cytokines and chemokines, may increase Nav1.8-mediated currents by acutely activating Nav1.8 ion channels through second-messenger signaling or by enhancing channel expression (27–29). Consistent with this idea, we have shown that chemokines and their receptors are expressed by DRG neurons (30, 31) and that chemokine signaling is important in generating neuropathic pain in experimental models of PDN (30). However, the role of chemokines in generating Nav1.8-positive DRG neuron hyperexcitability, mechanical allodynia, and small-fiber degeneration in PDN remains unclear.
Although the causes of PDN are likely to be multifactorial, they include inflammatory processes (32). Inflammatory markers, such as IL-6, IL-2, and TNF-α, are elevated in hyperglycemia, suggesting a chronic, low-grade inflammatory state in diabetic patients (33, 34). Moreover, patients with higher plasma TNF-α have a greater risk of PDN (33, 35, 36). Expression of the chemokine receptor CXCR4, a G-protein–coupled, 7-span transmembrane receptor (GPCR), was elevated in a peripheral nerve microarray analysis of patients with progressive diabetic neuropathy (37). Consistent with this finding, we showed that, in the high-fat diet (HFD) mouse model of PDN (38), CXCR4 and its ligand, the chemokine CXCL12 (also known as stromal-derived factor 1), are crucial in the generation of mechanical allodynia, a pain hypersensitivity behavior associated with PDN in mice (30, 39) and humans (40, 41).
In light of these findings, we have now examined the mechanistic relationships between CXCL12/CXCR4 signaling, hyperexcitability in Nav1.8-positive DRG neurons, small-fiber degeneration, and mechanical allodynia in the HFD mouse model of PDN. We used electrophysiology, imaging, and chemogenetics to demonstrate that CXCL12/CXCR4 signaling is key to the development of Nav1.8-positive DRG neuron hyperexcitability, which is directly responsible for small-fiber degeneration and mechanical allodynia. Hence, therapies that target this mechanism represent a novel approach for PDN.
Results
Mechanical allodynia precedes small-fiber degeneration in diabetic mice.
Neuropathic pain and small-fiber neuropathy are well-recognized complications of type 2 diabetes, both in humans and animal models (10, 38). However, the temporal correlation between the onset of neuropathic pain behavior and small-fiber neuropathy has not been established. We set out to investigate this temporal relationship by measuring mechanical allodynia, a particular pain hypersensitivity behavior normally associated with PDN. We used the HFD mouse model of PDN. In this model, mice fed a diet high in fat content develop glucose intolerance, obesity, mechanical allodynia, and small-fiber degeneration over a period of 10 weeks (30, 38, 39). Hence, the key hallmarks of human PDN are recapitulated in this HFD model.
DRG neuron subtypes are identified using molecular markers (42–44). Because more than 90% of DRG nociceptors express Nav1.8 (23), we targeted our studies to this population. To investigate the onset of small-fiber degeneration, we used a molecular genetic strategy of crossing Nav1.8-Cre mice (45) with Ai9 (td-Tomato) mice (46). In the resulting Nav1.8-Cre;Ai9 mice, Nav1.8-positive DRG neurons were labeled red with td-Tomato reporter protein following Cre-dependent recombination, making it possible to visualize Nav1.8-positive neuron cell bodies in the DRG and their afferents in the dorsal horn of the spinal cord and in skin (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI92117DS1).
The Nav1.8-Cre;Ai9 mice were fed a HFD for 10 weeks. The mice gained weight (Supplemental Figure 1B) and became glucose intolerant 6 weeks after starting the HFD (Supplemental Figure 1C). We next used confocal microscopy to examine small-fiber degeneration in skin samples from Nav1.8-Cre;Ai9 mice. Starting at 8 weeks, HFD-fed mice displayed a dramatic reduction in intraepidermal nerve fiber (IENF) density, expressed as the number of nerves crossing the epidermal-dermal junction as a function of length, relative to control (regular diet [RD]) mice. There was no difference in IENF density between RD and HFD mice at 2, 4, or 6 weeks (Figure 1, A–C). We verified these results in skin samples from Nav1.8-Cre;Ai9 mice that had been on either a RD or HFD for 2 or 8 weeks by immunolabeling with an antibody against the protein gene product 9.5 (PGP 9.5), a pan-neuronal marker used for calculating IENF density and for diagnosing small-fiber neuropathies (13, 47). This independent verification excluded the possibility that the results reflected abnormal td-Tomato expression or transport in HFD mice (Supplemental Figure 1D).
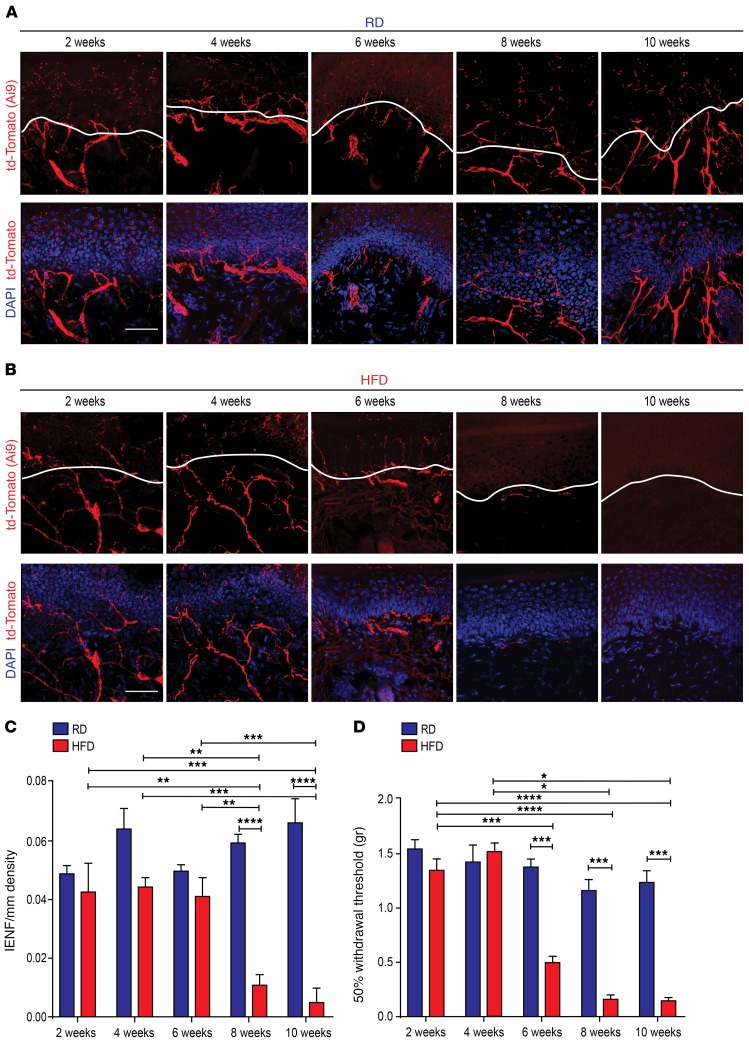
Figure 1. Onset of small-fiber degeneration and mechanical allodynia in mice fed a HFD.
(A) Confocal analysis of skin sections from Nav1.8-Cre;Ai9 mice fed a RD (blue) showed normal innervation. Nav1.8-positive fibers genetically labeled with td-Tomato are shown in red. Sections were stained with the nuclear marker DAPI (blue). Scale bar: 50 μm. (B) Skin sections from diabetic Nav1.8-Cre;Ai9 mice (HFD, red) had decreased innervation commencing 8 weeks after the start of the diet. Scale bar: 50 μm. (C) This effect was quantified using IENF density, and the epidermal-dermal junction is outlined in white in A and B. **P < 0.01, ***P < 0.001, and ****P < 0.0001 (n = 6 for all groups, with 3 noncontiguous sections analyzed per sample). (D) von Frey testing revealed the onset of mechanical allodynia in diabetic Nav1.8-Cre;Ai9 mice after 6 weeks on a HFD but not in RD-fed mice. *P < 0.05, ***P < 0.001, and ****P < 0.0001 (n = 7 mice/group). P values were calculated by 2-way ANOVA with Bonferroni’s multiple comparisons test. Values are expressed as the mean ± SEM.
We next determined the onset of mechanical allodynia by quantifying the withdrawal threshold of the hindpaw in response to stimulation with flexible von Frey filaments applied in order of ascending force. The von Frey experiments were conducted using random experimental group assignments by blinded investigators. Beginning at 6 weeks, Nav1.8-Cre;Ai9 mice fed a HFD had a significantly reduced withdrawal threshold compared with that of RD control mice, indicating the development of mechanical allodynia (Figure 1D). No statistically significant differences were noted between RD and HFD mice 2 or 4 weeks after commencement of the diet. Hence, HFD mice developed mechanical allodynia 2 weeks prior to small-fiber degeneration.
Intracellular calcium influx into DRG neurons increases in diabetic mice.
The molecular cascade linking neuropathic pain behavior to small-fiber degeneration in diabetes is incompletely understood. One phenomenon that could potentially explain both is enhanced Nav1.8-positive DRG neuron excitability. We applied a functional imaging technique using acutely isolated whole DRG explants to assess this hyperexcitability by measuring changes in internal calcium concentration ([Ca2+]i) in these neurons as PDN developed. We initially used a knockin mouse line that expressed the genetically encoded [Ca2+]i indicator protein GCaMP3 under the control of the PIRT promoter, which directs the expression of GCaMP3 in more than 95% of DRG neurons (48). Acutely excised DRG explants were isolated from Pirt-GCaMP3 mice 2, 4, 6, 8, 10, and 12 weeks after starting a HFD or RD. We measured the number of DRG neurons responding with [Ca2+]i transients to low and high concentrations of stimuli (i.e., capsaicin and high potassium buffer [HK]). In DRG explants from the mice that had been on a HFD for at least 6 weeks, the number of DRG neurons responding to a low concentration of capsaicin or HK was significantly higher than that in DRG explants isolated from RD-fed mice (Supplemental Figure 2A). In contrast, the number of DRG neurons responding to low capsaicin or HK after 2 or 4 weeks of a HFD or RD did not differ (Supplemental Figure 2B).
Given the cellular diversity and functional heterogeneity of DRG neurons (42–44, 49), we wanted to monitor [Ca2+]i in Nav1.8-positive DRG neurons. Therefore, we selectively expressed the [Ca2+]i indicator protein GCaMP6 in these neurons by crossing Nav1.8-Cre mice (45) with conditional reporter GCaMP6 mice (Ai96fl/fl;RCL-GCaMP6s) (50). We then performed imaging experiments on acutely isolated DRG explants from these mice 2 and 8 weeks after commencement of a HFD or RD (Supplemental Videos 1–4). We measured the number of Nav1.8-positive DRG neurons responding with [Ca2+]i transients to both low and high concentrations of capsaicin and HK. When mice had been on a HFD for 8 weeks, Nav1.8-positive DRG neurons were more likely to respond to lower concentrations of capsaicin and HK compared with neurons from RD mice (Figure 2, A–D). In contrast, the number of neurons responding to low capsaicin or HK after 2 weeks of a HFD or RD did not differ (Figure 2, E–H). These results demonstrate the development of Nav1.8-positive DRG neuron hyperexcitability in the HFD model of PDN, confirming the results that we obtained with the Pirt-GCamP3 mice.
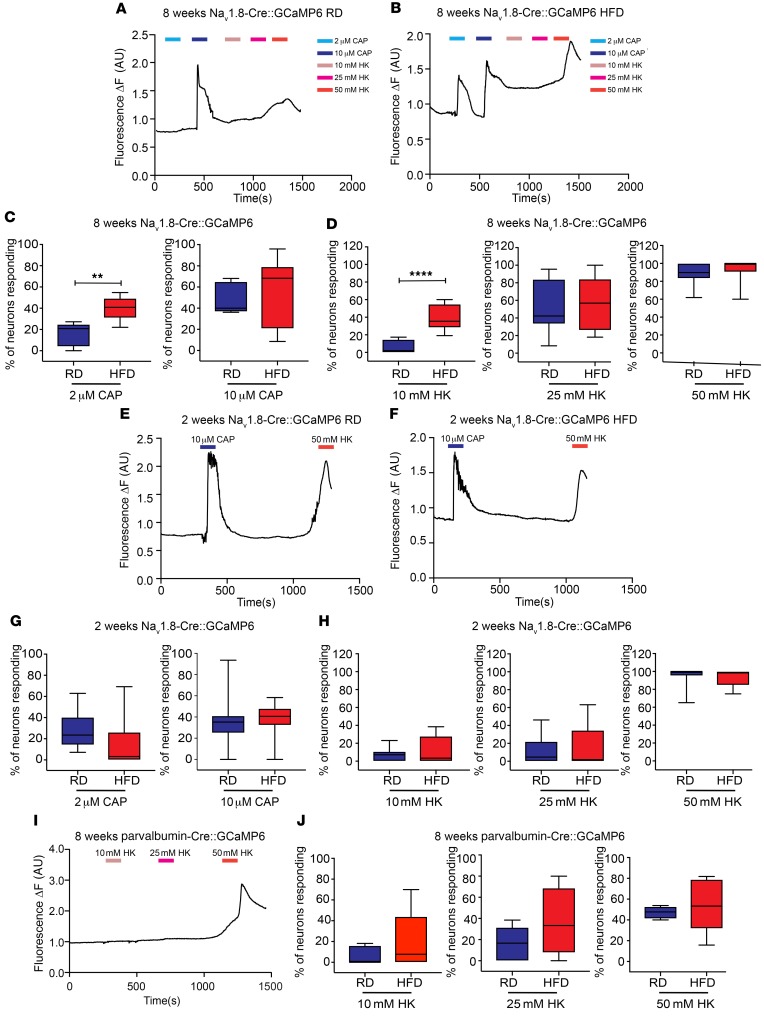
Figure 2. Nav1.8-positive DRG neurons show increased [Ca2+]i in mice fed a HFD.
(A and B) Representative traces of [Ca2+]i in acutely excised explants from Nav1.8-Cre GCaMP6 mice after 8 weeks on (A) a RD or (B) a HFD. The number of Nav1.8-positive neurons was quantified to assess the response to either (C) capsaicin (CAP) (2 μM or 10 μM) (**P < 0.01) (RD n = 381 neurons, 11 explants; HFD n = 519 neurons, 17 explants), or (D) 10 mM, 25 mM, or 50 mM HK (****P < 0.0001) (RD n = 381 neurons, 11 explants; HFD n = 519 neurons, 17 explants). Capsaicin- or HK-responsive DRG neurons are reported as a percentage of the total number of neurons that responded to 50 mM HK. (E and F) These same experiments were performed at 2 weeks in mice on (E) a RD or (F) a HFD. (G and H) No difference was found in the number of neurons responding to (G) capsaicin or (H) HK (RD n = 381 neurons, 11 explants; HFD n = 231 neurons, 10 explants). (I and J) In DRG explants from parvalbumin-Cre GCaMP6 mice, there were no significant differences between mice on a RD and those on a HFD after 8 weeks (RD n = 88 neurons, 6 explants; HFD n = 118 neurons, 9 explants). Values are expressed as the mean ± SEM. P values were calculated using a Mann-Whitney U test.
To determine whether this excitability was specific to Nav1.8-positive DRG neurons, we expressed GCaMP6 in proprioceptive DRG neurons by crossing parvalbumin-Cre mice, which have been used to study proprioceptor lineage (51–53), with GCaMP6 mice (Ai96fl/fl;RCL-GCaMP6s) (50). We then imaged acutely isolated DRG explants 2 and 8 weeks after starting the mice on a HFD or RD and measured the number of parvalbumin-positive DRG neurons responding with [Ca2+]i transients to low and high concentrations of capsaicin and HK. Parvalbumin-positive DRG neurons did not respond to either concentration of capsaicin, and their responses to HK after 2 or 8 weeks of a HFD or RD did not differ (Figure 2, I and J).
Diabetic Nav1.8-positive DRG neurons are hyperexcitable.
The increased sensitivity of Nav1.8-positive DRG neurons from HFD-fed mice to capsaicin and HK suggested hyperexcitability, which we tested directly in primary DRG cultures. We made current-clamp recordings of td-Tomato–labeled Nav1.8-positive neurons from Nav1.8-Cre;Ai9 mice fed a HFD or RD for 10 weeks. These neurons had a significantly lower rheobase compared with that of neurons from RD-fed mice (Figure 3, A–C). We observed no significant differences in the resting membrane potential (RMP) or action potential (AP) overshoot (Figure 3, D–F) but found that the firing frequency was increased in Nav1.8-positive neurons from HFD mice compared with those from RD mice (Figure 3, G–O). These electrophysiological properties support the conclusion that Nav1.8-positive DRG neurons from HFD mice become hyperexcitable.
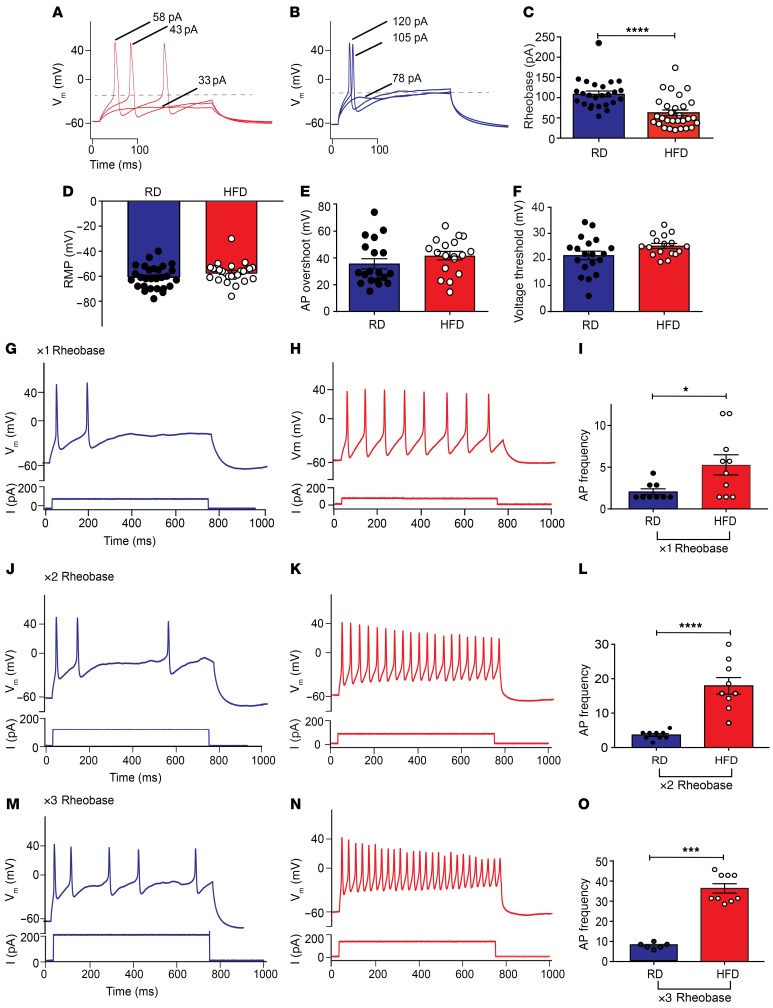
Figure 3. Nav1.8-positive DRG neurons display hyperexcitability in HFD-fed mice.
(A and B) Current-clamp recordings of DRG primary cultures from Nav1.8-Cre;Ai9 mice. Nav1.8-positive DRG neurons from HFD mice (A, red) (n = 29) had a lower rheobase than did neurons from RD mice (B, blue) (n = 25). (C) A significant decrease in rheobase was observed in HFD neurons (****P < 0.0001). (D) RMPs, (E) AP overshoot, and (F) voltage threshold for AP generation remained unchanged. (G–O) Representative current steps and associated voltage recordings are shown for DRG neurons from RD (blue) and HFD (red) mice, in which 700-ms rheobase current injections were done (G and H) 1× (n = 9; n = 10), (J and K) 2× (n = 9; n = 9), and (M and N) 3× (n = 6; n = 9), respectively. (H, K, and N) An increase in firing frequency was observed in neurons from HFD mice compared with (G, J, and M) neurons from RD mice. A significant increase was observed in the firing frequency in HFD DRG neurons compared with RD DRG neurons after (I) 1× (*P < 0.05), (L) 2× (****P < 0.0001), and (O) 3× (***P < 0.001) rheobase current injections, respectively. Values are expressed as the mean ± SEM. P values were calculated using a Mann-Whitney U test. Vm, voltage membrane.
CXCR4 chemokine receptor deletion from Nav1.8-positive DRG neurons prevents mechanical allodynia and small-fiber degeneration in diabetic mice.
What factors drive Nav1.8-positive DRG neuron hyperexcitability in PDN pathology? We previously reported that excitatory effects of chemokines are important in the development and maintenance of pain behaviors in neuropathic pain models (31, 54) and that CXCR4 signaling is important for the development of mechanical allodynia in HFD mice (30).
To extend these findings, we deleted CXCR4 receptors from Nav1.8-positive DRG neurons by crossing Nav1.8-Cre;Ai9 mice with CXCR4-floxed mice (CXCR4fl/fl) (55). This manipulation did not cause developmental defects (56), as the number of Nav1.8-positive DRG neurons labeled with td-Tomato was no different in Nav1.8-Cre;Ai9;CXCR4fl/+ heterozygous or Nav1.8-Cre;Ai9;CXCR4fl/fl homozygous mice (Supplemental Figure 3, A and B). Furthermore, we found no significant differences in the numbers of td-Tomato–positive DRG neurons that were also positive for IB4 (Supplemental Figure 3, A and B), which identifies nonpeptidergic nociceptive neurons (14, 57), demonstrating that these mice have normal segregation of peptidergic versus nonpeptidergic nociceptors after sensory neurogenesis (58, 59). These mice also had normal metabolic profiles. Like WT mice, both Nav1.8-Cre;Ai9;CXCR4fl/+ heterozygous and Nav1.8-Cre-Ai9;CXCR4fl/fl homozygous mice fed a HFD became obese (Supplemental Figure 3C) and glucose intolerant (Supplemental Figure 3D).
We tested for mechanical allodynia using the von Frey withdrawal threshold paradigm, as described above. In Nav1.8-Cre;Ai9;CXCR4fl/+ heterozygous HFD mice, the withdrawal threshold was significantly reduced compared with that of RD mice, indicating the development of mechanical allodynia (Figure 4A). In contrast, Nav1.8-Cre;Ai9;CXCR4fl/fl homozygous HFD mice showed normal withdrawal thresholds (Figure 4A), indicating that CXCR4 receptors in Nav1.8-positive DRG neurons are necessary for the establishment of mechanical allodynia in this model of PDN. We did not observe mechanical allodynia in RD mice with chemokine receptor CXCR4 deletion from Nav1.8-positive DRG neurons (Nav1.8-Cre;Ai9;CXCR4fl/fl homozygous) (Figure 4A), so CXCR4 deletion did not alter mechanical sensation.
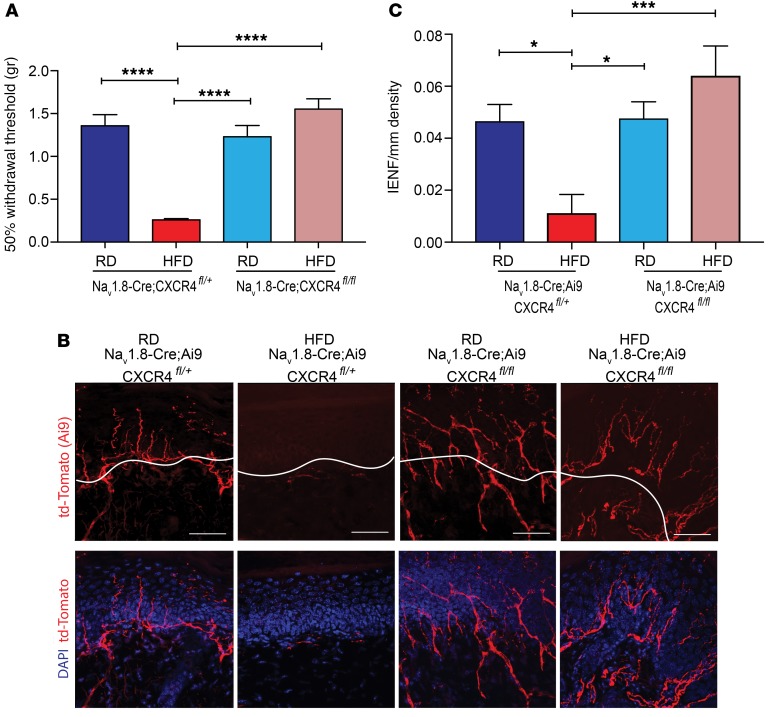
Figure 4. Selective chemokine receptor CXCR4 deletion from Nav1.8-postive DRG neurons prevents the development of mechanical allodynia and small-fiber degeneration in HFD-induced PDN.
(A) von Frey testing demonstrated that in HFD (red) Nav1.8-Cre;Ai9;CXCR4fl/+, which had a heterozygous deletion of CXCR4 from Nav1.8-postive DRG neurons, the withdrawal threshold was significantly reduced compared with that in Nav1.8-Cre;Ai9;CXCR4fl/+ mice on a RD (dark blue) and compared with the withdrawal threshold in mice with a homozygous deletion of CXCR4 (Nav1.8-Cre;Ai9;CXCR4fl/fl) on a RD (light blue). In contrast, Nav1.8-Cre;Ai9;CXCR4fl/fl (pink) mice on a HFD showed normalization of the withdrawal thresholds (****P < 0.0001) (n = 6/group). (B) Confocal analysis of Nav1.8-positive fibers from the skin of RD and HFD mice with heterozygous or homozygous deletions of CXCR4, showing td-Tomato (red) and merged images with the nuclear marker DAPI (blue). Nav1.8-Cre;Ai9;CXCR4fl/+ RD mice had normal skin innervation, whereas the same mice on a HFD had reduced innervation. However, selective homozygous deletion of CXCR4 in mice on a HFD prevented small-fiber degeneration. Scale bars: 50 μm. (C) This effect was quantified using IENF density, and the epidermal-dermal junction is outlined in white in B. *P < 0.05 and ***P < 0.001 (n = 7 for all groups, with 3 noncontiguous sections analyzed per sample). Values are expressed as the mean SEM. P values were calculated using 1-way ANOVA with a Bonferroni’s multiple comparisons test.
We next tested whether excitatory CXCL12/CXCR4 signaling in Nav1.8-positive neurons was necessary for small-fiber degeneration. Using confocal microscopy, we examined skin innervation in both Nav1.8-Cre;Ai9;CXCR4fl/+ heterozygous and Nav1.8-Cre;Ai9;CXCR4fl/fl homozygous mice fed a RD or HFD for 10 weeks. CXCR4 deletion from Nav1.8-positive DRG neurons significantly improved skin innervation in the diabetic mice (Figure 4, B and C). In contrast, heterozygous HFD mice had substantially depleted nerve terminals (Figure 4, B and C). These results were verified by immunolabeling using a PGP 9.5 antibody on the same skin samples, providing an independent verification for the fiber density measurements (Supplemental Figure 4). We found that skin innervation was normal in Nav1.8-Cre;Ai9;CXCR4fl/+ and Nav1.8-Cre;Ai9;CXCR4fl/fl mice fed a RD (Figure 4, B and C; and Supplemental Figure 4), demonstrating that CXCR4 deletion from Nav1.8-positive DRG neurons did not interfere with normal neurite outgrowth.
Excitatory CXCL12/CXCR4 signaling is enhanced in diabetic Nav1.8-positive DRG neurons.
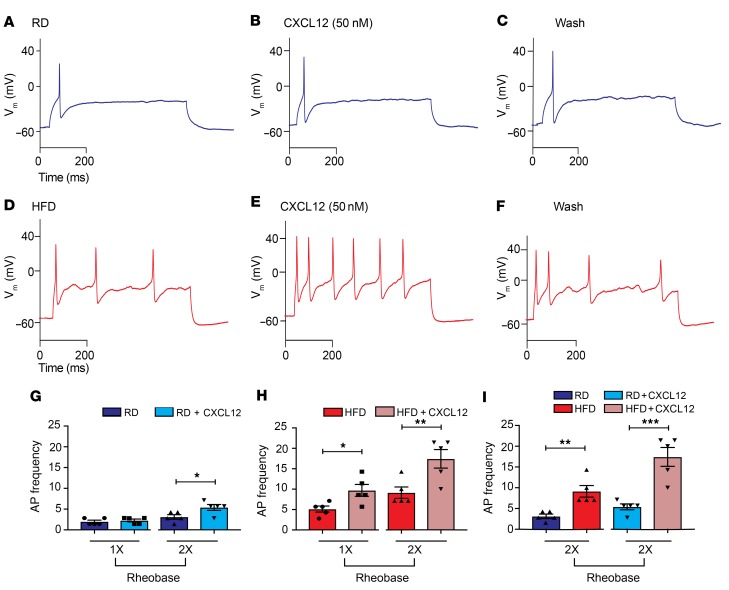
The above results demonstrate that CXCL12/CXCR4 signaling in Nav1.8-positive DRG neurons is necessary for mechanical allodynia and small-fiber degeneration in PDN. What is the mechanism for this effect? Our central hypothesis is that CXCL12/CXCR4 signaling triggers hyperexcitability and [Ca2+]i increases in Nav1.8-positive DRG neurons, which result in mechanical allodynia and axonal degeneration. To test this hypothesis, we performed current-clamp experiments on cultured DRG neurons from HFD and RD Nav1.8-Cre;Ai9 mice. Application of the chemokine CXCL12 (50 nM) increased the firing frequency of Nav1.8-positive neurons (Figure 5, A–F), and this increase was significantly greater in neurons from HFD mice (Figure 5, G–I). These results are consistent with a role for CXCL12/CXCR4 signaling in generating Nav1.8-positive DRG neuron hyperexcitability.
Figure 5. CXCL12/CXCR4 signaling produces increased firing frequencies in HFD-induced diabetic Nav1.8-positive DRG neurons.
(A) Current-clamp recordings of DRG primary cultures from Nav1.8-Cre;Ai9 mice. A typical illustration of APs generated using a depolarizing current injection from a RD Nav1.8-positive DRG neuron (blue) in response to a 700-ms input of ×1 rheobase current injection from the RMP (Vm) (–57 mV). (B) Application of CXCL12 (50 nM) produced no change in the firing of this neuron after current injection. (C) Results after a 5-minute wash. (D) Representative traces from a diabetic, HFD-fed Nav1.8-positive mouse DRG neuron (red) firing multiple APs in response to a 700-ms input of ×1 rheobase depolarizing current injections. (E and F) An increase in the firing frequency of HFD Nav1.8-positive neurons was observed after (E) CXCL12 (50 nM) application and (F) a wash. (G and I) The frequency of firing for each of these treatments was quantified. (G) A significant increase in AP frequency occurred after CXCL12 treatment in ×2 rheobase current pulses in RD Nav1.8-positive DRG neurons (*P < 0.05) (n = 5). (H) Significant increases were observed in firing frequencies following CXCL12 treatment of HFD Nav1.8-positive DRG neurons (red) after ×1 and ×2 rheobase depolarizing current injections from the RMPs. *P < 0.05 and **P < 0.01 (n = 5/group). (I) Comparison between RD and HFD DRG neurons after CXCL12 application showed significant increases in AP frequency in HFD DRG neurons. **P < 0.01 and ***P < 0.001 (n = 5/group). Values are expressed as the mean ± SEM. P values were calculated using a Mann-Whitney U test
Excitatory CXCL12/CXCR4 signaling was enhanced at 6 weeks of HFD treatment, around the time of onset of mechanical allodynia and preceding the onset of small-fiber degeneration. Indeed, [Ca2+]i transients in acutely excised DRG explants from Pirt-GCaMP3–transgenic mice showed that CXCL12 produced responses in a significantly greater number of neurons 6 weeks after starting a HFD (Supplemental Figure 5A). In contrast, we observed no difference in the number of DRG neurons responding to CXCL12 after mice had been on a HFD or RD for 2 or 4 weeks (Supplemental Figure 5B).
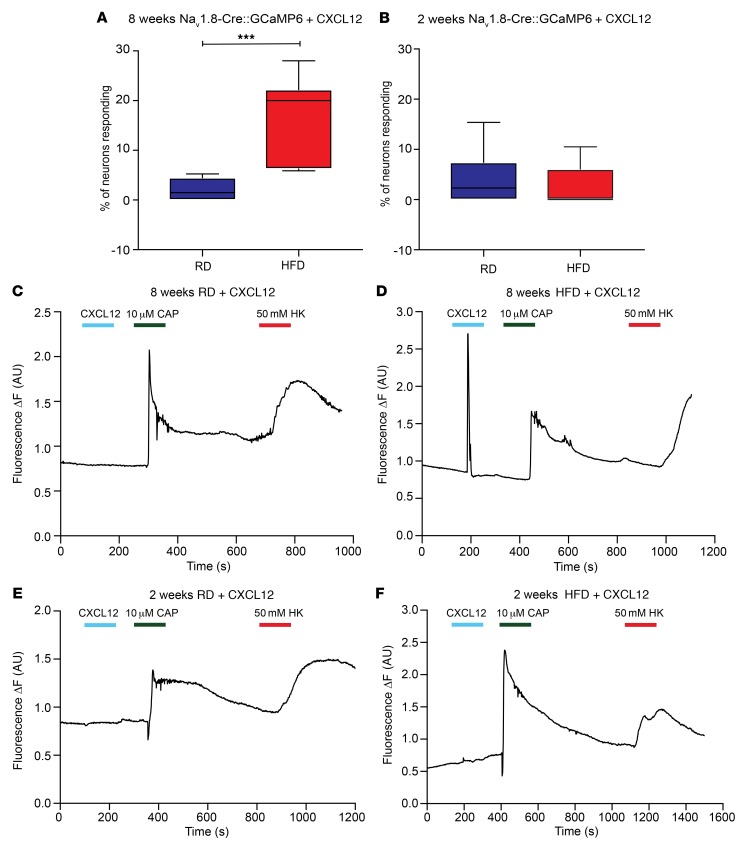
To demonstrate that this phenomenon was specific for Nav1.8-positive neurons, we performed similar [Ca2+]i imaging experiments on acutely excised DRG explants from Nav1.8-Cre::GCaMP6 mice 2 and 8 weeks after starting a HFD or RD (Supplemental Videos 5–8). A significantly greater number of Nav1.8-positive DRG neurons responded with increased [Ca2+]i after application of CXCL12 (100 nM) when mice had been on a HFD for 8 weeks versus DRG neurons from mice fed a RD (Figure 6, A, C, and D), but we found no difference after 2 weeks (Figure 6, B, E and F). Additionally, DRG explants from parvalbumin-Cre::GCaMP6 mice on a RD or HFD for 2 and 8 weeks did not respond with [Ca2+]i transients upon application of CXCL12 (Supplemental Table 2). These results are consistent with the idea that CXCL12/CXCR4 signaling may be important in the development of Nav1.8-positive DRG neuron hyperexcitability in PDN.
Figure 6. CXCR4 activation produces more frequent calcium responses in Nav1.8-positive DRG neurons from mice fed a HFD.
(A and B) [Ca2+]i responses of acutely excised DRG explants from RD (blue) and HFD (red) Nav1.8-Cre;GCaMP6 mice (A) 8 weeks and (B) 2 weeks after starting the diet. A significantly higher number of Nav1.8-positive DRG neurons responded with increased [Ca2+]i after application of CXCL12 (100 nM) when the mice had been on a HFD for 8 weeks compared with mice fed a RD (A). Data show capsaicin- or HK-responsive DRG neurons as a percentage of total neurons that responded to 50 mM HK. ***P < 0.001 (RD n = 333 neurons; 13 explants; HFD = 519 neurons, 17 explants). (C–F) Representative traces of [Ca2+]i transients in DRG explants from Nav1.8-Cre GCaMP6 mice. Explants were treated with capsaicin (10 μM) or HK (50 mM). (C and D) After 8 weeks, a greater number of neurons from HFD-fed mice responded to CXCL12 than did neurons from RD-fed mice. (E and F) Experiments were performed in mice after 2 weeks on a RD or HFD and showed no difference in responses to CXCL12 (RD n = 381 neurons, 11 explants; HFD n = 231 neurons, 10 explants). Values are expressed as the mean ± SEM. P values were calculated using a Mann-Whitney U test.
Reducing Nav1.8-positive DRG neuron excitability prevents and reverses mechanical allodynia and small-fiber degeneration in diabetic mice.
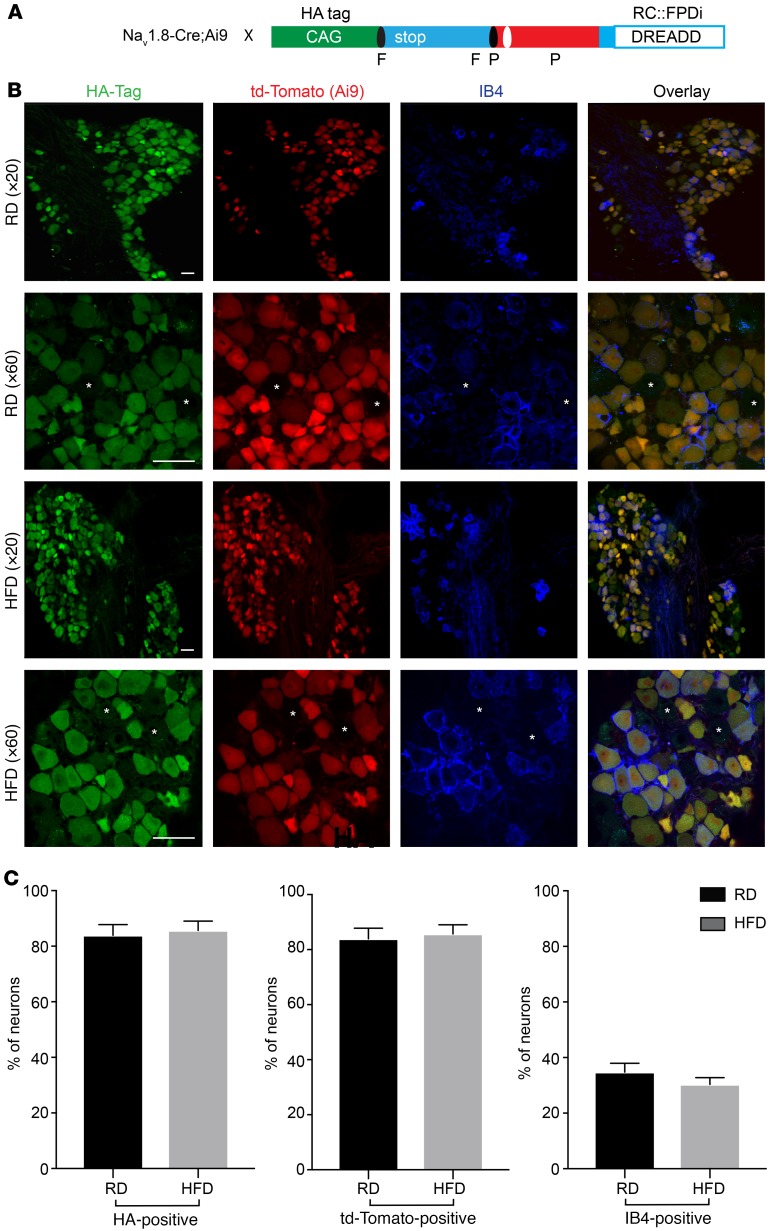
If this hyperexcitability is responsible for mechanical allodynia and small-fiber degeneration, then reducing hyperexcitability should have a significant impact on both phenomena. To reduce the excitability of Nav1.8-positive DRG neurons in vivo over the long term in freely behaving animals, we elected to use a chemogenetic platform by genetically introducing designer receptors exclusively activated by designer drugs (DREADDs) into Nav1.8-positive DRG neurons. We used an inhibitory DREADD receptor based on an engineered muscarinic acetylcholine receptor M4 (PDi), which works via activation of the inhibitory Gi/o protein pathway (60). Activation of this receptor with the small-molecule agonist clozapine-N-oxide (CNO) or its metabolite clozapine inhibits neuronal activity (reviewed in refs. 61–63). We expressed inhibitory hM4 DREADD (PDi) receptors in Nav1.8-positive DRG neurons by crossing Nav1.8-Cre;Ai9 mice with a mouse line that enables the conditional expression of DREADD receptors (62) (Figure 7A). We stained DRGs taken from the resulting Nav1.8-Cre;Ai9;RC::PDi mice and were able to visualize PDis with IHC using an antibody against HA, as in this construct, the inhibitory PDi DREADD contains an HA tag (62) (Figure 7A). To visualize nonpeptidergic neurons, we used the IB4 isolectin. Indeed, we found that PDis were expressed in all Nav1.8-positive DRG neurons and that the percentage of IB4-positive nonpeptidergic neurons (14, 57, 64) expressing PDis in mice on a RD or HFD did not differ (Figure 7, B and C).
Figure 7. Expression of the inhibitory DREADD receptor PDi in Nav1.8-positive DRG neurons.
(A) Breeding scheme and genetic constructs used to generate Nav1.8-Cre;Ai9;RC::PDi inhibitory DREADD mice; the inhibitory PDi DREADD receptor (PDi DREADDs) has an HA tag, and Nav1.8-positive DRG neurons are genetically labeled in red with td-Tomato. X, crossing (breeding mice); F, FRT-flanked transcriptional Stop; P, loxP-flanked-transcriptional Stop. (B) Confocal micrographs of DRGs from RD (top) and HFD (bottom) PDi DREADD–expressing mice (Nav1.8-Cre;Ai9;RC::PDi). Images show PDi DREADDs tagged with an HA epitope (green), Nav1.8 td-Tomato–expressing neurons (red), and IB4-positive neurons (blue). PDi DREADDs were found in small- and medium-diameter DRG neurons, some of which were IB4 positive and some IB4 negative. Large-diameter neurons (indicated by asterisks) did not express PDi DREADDs. Scale bars: 50 μm. Original magnification, ×20 and ×60. (C) Percentage of PDi DREADD–expressing neurons as determined by the HA tag, td-Tomato Nav1.8 neurons, and nonpeptidergic IB4-positive neurons. RD DRGs had 83.9% ± 3.4% HA- or td-Tomato–positive neurons versus 85.7% ± 3.8% for HFD DRGs. RD DRGs had 34.8% ± 3.2% IB4-positive neurons versus 35.4% ± 2.4% for HFD DRGs. There were no significant differences in the sizes of these cell populations between DRGs from RD and HFD PDi DREADD–expressing mice (n = 278 neurons [RD]; n = 227 [HFD]). Values are expressed as the mean ± SEM. P values were calculated using a Mann-Whitney U test.
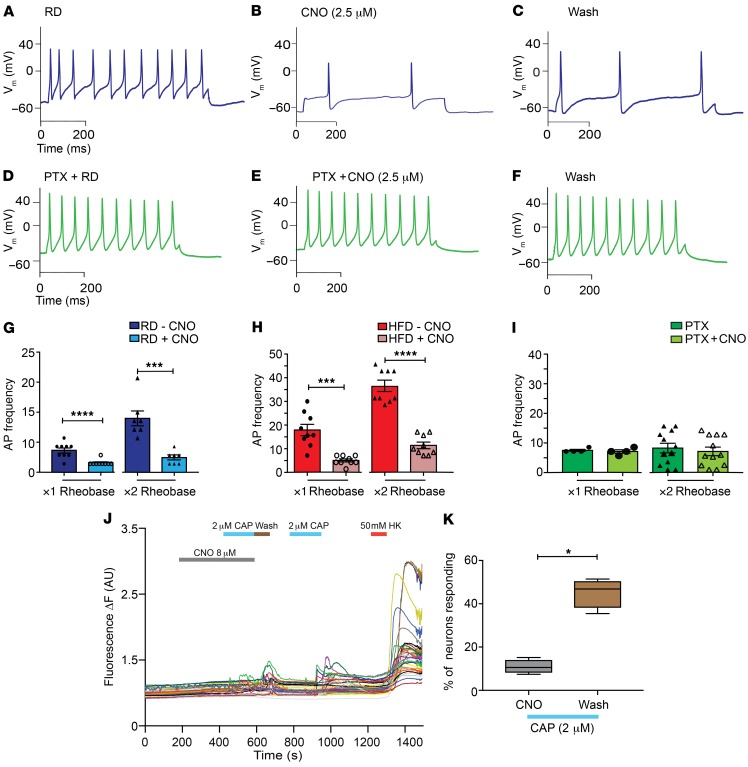
In vitro electrophysiology confirmed that CNO application reduced activity in Nav1.8-positive DRG neurons expressing inhibitory PDi receptors (Figure 8, A–C) as previously demonstrated in other types of neurons. Specifically, in current-clamp studies, CNO significantly reduced evoked AP frequency in cultured Nav1.8-positive DRG neurons from RD (Figure 8, A–C and G) and HFD Nav1.8-Cre;Ai9;RC::PDi mice (Figure 8H). When we incubated RD cultures overnight with pertussis toxin, CNO failed to produce any effects, indicating that the inhibitory effects observed were transduced through Gi/o, as expected (Figure 8, D–F, and I). Additionally, CNO reversibly reduced capsaicin-induced [Ca2+]i signals in DRG explants from mice encoding GCaMP6 together with PDis in Nav1.8-positive neurons (Nav1.8-Cre;RC::PDi GCaMP6 mice) (Figure 8, J and K).
Figure 8. Chemogenetic inhibition of Nav1.8-positive DRG neurons expressing the inhibitory DREADD receptor PDi is G-protein mediated.
(A) Current-clamp recordings from inhibitory PDi–expressing Nav1.8-positive neurons in primary cultures isolated from Nav1.8-Cre;Ai9;RC::PDi mice fed a RD (blue). (B) Application of CNO (2.5 μM) reduced the AP frequency, and (C) washing out the CNO partially restored the firing rate. (D–F) Overnight incubation of RD DRG cultures with pertussis toxin (PTX, green) abolished the inhibitory effect of CNO. (G) In RD Nav1.8-positive DRG neurons expressing DREADD receptors, a significant decrease in AP frequency after application of CNO at both the ×1 and ×2 rheobase was observed. ***P < 0.001 and ****P < 0.0001 (n = 7 and 9, respectively). (H) The same mice fed a HFD also showed a decrease in AP frequency after application of CNO.***P < 0.001 and ****P < 0.0001 (n = 9 for both groups). (I) Overnight incubation of DRG cultures with pertussis toxin abolished the inhibitory effects of CNO. There was no difference in AP frequency after preincubation with PTX and application of CNO at either the ×1 or ×2 rheobase (n = 4 and 12, respectively). (J) [Ca2+]i responses in DRG explants from Nav1.8-Cre;RC::PDi GCaMP6 mice showed that [Ca2+]i responses after addition of capsaicin (2 μM) were inhibited during incubation with CNO (8 μM for 5 min). After washing, Nav1.8-positive DRG neurons showed restored [Ca2+]i transients to capsaicin (2 μM) and HK (10 mM) (n = 120 neurons; 10 explants). (K) The responses to lower concentrations of capsaicin were quantified as the responses to capsaicin as a percentage of the total number of HK-responsive neurons. *P < 0.05. Values are expressed as the mean ± SEM. P values were calculated using a Mann-Whitney U test.
DREADD-independent effects of CNO have been reported (65), so we verified that CNO did not change the firing frequencies of DRG neurons from Nav1.8-Cre;Ai9 mice not expressing PDis in RD (Supplemental Figure 6, A–C and G) or HFD mice (Supplemental Figure 6, D–F and G). In summary, these results demonstrate that activating PDis in Nav1.8-positive DRG neurons had a reversible, CNO-dependent, inhibitory effect on the excitability of these neurons.
Additionally, CNO reversed mechanical allodynia in HFD Nav1.8-Cre;Ai9;RC::PDi mice expressing inhibitory DREADDs, but not in HFD Nav1.8-Cre;Ai9 mice not expressing inhibitory DREADDs in vivo. Indeed, using the von Frey pain behavioral assay, we observed that HFD Nav1.8-Cre;RC::PDi mice had a significantly lower withdrawal threshold for mechanical stimulation compared with animals on a RD (Supplemental Figure 7A). However, 1 hour after a single i.p. injection of CNO (10 mg/kg), the withdrawal threshold increased, returning to baseline 4 hours after injection (Supplemental Figure 7A). Injection of CNO did not reverse mechanical allodynia in diabetic Nav1.8-Cre;Ai9 mice not expressing PDis (Supplemental Figure 7B), indicating that CNO had no DREADD-independent effects. Both Nav1.8-Cre;Ai9:RC::PDi (Supplemental Figure 8, A and B) and Nav1.8-Cre;Ai9 (Supplemental Figure 8, C and D) mice fed a HFD had weight gain and glucose intolerance.
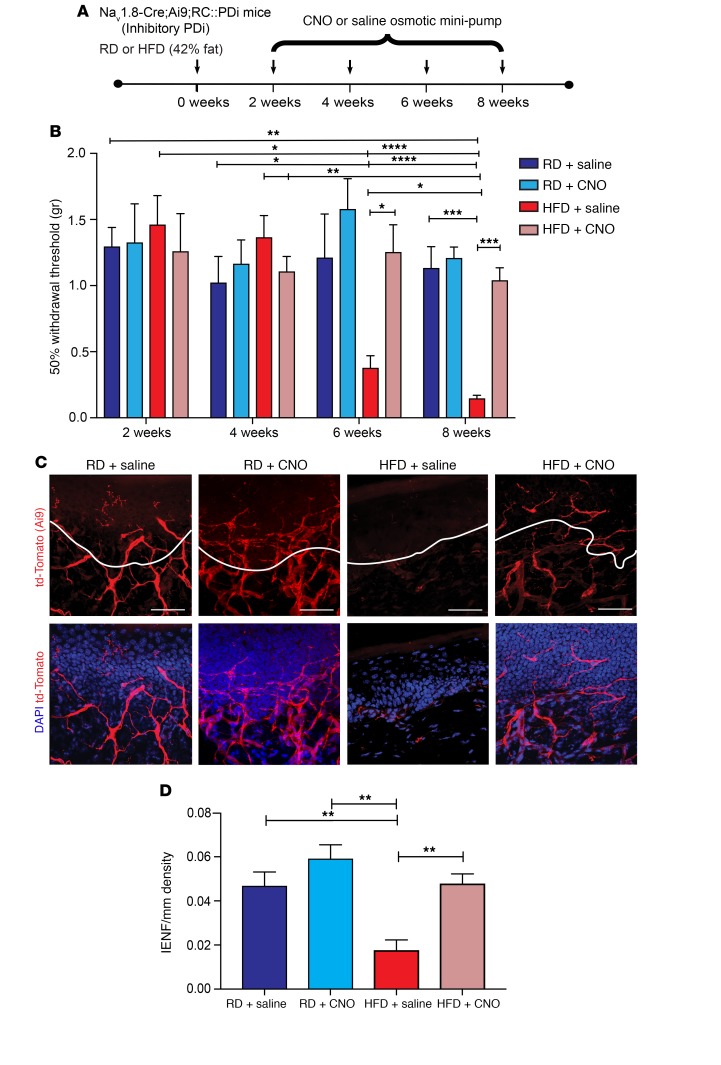
Our previous results suggested that small-fiber degeneration occurred 2 weeks after the onset of neuronal hyperexcitability (Figure 1, A–C, and Supplemental Figure 2). Thus, to evaluate the consequences of reducing Nav1.8-positive DRG neuronal hyperexcitability on small-fiber degeneration in PDN, we needed to achieve long-term activation of DREADD receptors in vivo. To do this, CNO was continuously delivered using osmotic minipumps implanted i.p. into Nav1.8-Cre;Ai9;RC::PDi mice between the second and eighth weeks (Figure 9A). Continuous CNO infusion did not alter the metabolic profile in mice expressing PDis, as a HFD induced obesity and glucose intolerance in Nav1.8-Cre;RC::PDi mice infused with either CNO or saline (Supplemental Figure 9, A–C). Additionally, we performed von Frey pain behavioral studies and found that when CNO was continuously infused from weeks 2–8, HFD-fed mice no longer developed mechanical allodynia (Figure 9B). However, mice continuously infused with saline over the same period developed mechanical allodynia after 6 weeks on a HFD (Figure 9B). These results are consistent with the idea that CXCL12/CXCR4-mediated hyperexcitability of Nav1.8-positive DRG neurons may be responsible for mechanical allodynia.
Figure 9. Long-term chemogenetic inhibition of Nav1.8-positive DRG neurons prevents mechanical allodynia and small-fiber degeneration in HFD-fed mice.
(A) Nav1.8-Cre;Ai9;RC::PDi mice were fitted with osmotic mini-pumps i.p. infusing either CNO (10 mg/kg/day) or saline between weeks 2 and 8 of either a RD or HFD. Each arrow represents a time point at which pain behavior was assessed. (B) von Frey testing was performed on Nav1.8-Cre;Ai9;RC::PDi mice 2, 4, 6, and 8 weeks after implantation of an osmotic mini-pump i.p. that delivered CNO (10 mg/kg/day) or saline into RD- or HFD-fed mice. Mice on a HFD showed a reduced withdrawal threshold starting at 6 weeks, which was reversed following CNO treatment. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 (n = 9/group). (C) Confocal micrographs of skin from these mice show td-Tomato in the Nav1.8 fibers (red) and merged images with the nuclear marker DAPI (blue). Mice on a RD given either saline or CNO showed normal skin innervation. In diabetic HFD-fed mice given saline, a reduction in skin innervation was observed, but it was reversed for mice on a HFD given CNO. Scale bars: 50 μm. (D) This effect was quantified using IENF density, and the epidermal-dermal junction is outlined in white in C, showing that CNO infusion prevented small-fiber degeneration in HFD-fed mice. **P < 0.01 (n = 6/group, with 3 noncontinuous sections analyzed per sample). Values are expressed as the mean ± SEM. P values were calculated using 2-way ANOVA with Bonferroni’s multiple comparisons test.
We next evaluated the consequences of reducing hyperexcitability on small-fiber degeneration. Long-term chemogenetic reduction of Nav1.8-positive DRG neuron hyperexcitability significantly improved skin innervation in HFD mice (Figure 9, C and D). Micrographs of skin from Nav1.8-Cre;Ai9;RC::PDi RD-fed control mice infused with saline or CNO mini-pumps showed normal skin innervation (Figure 9, C and D). In contrast, HFD Nav1.8-Cre;Ai9;RC::PDi mice with saline mini-pumps had greatly reduced innervation (Figure 9, C and D). However, HFD mice with CNO mini-pumps showed significantly improved innervation, which was not statistically different from that of RD mice (Figure 9, C and D). These results were verified by immunolabeling using a PGP 9.5 antibody on the same skin sample, providing an independent verification for the fiber density measurements (Supplemental Figure 10, A–C). These data demonstrate that reducing the hyperexcitability of Nav1.8-positive DRG neurons prevents small-fiber degeneration.
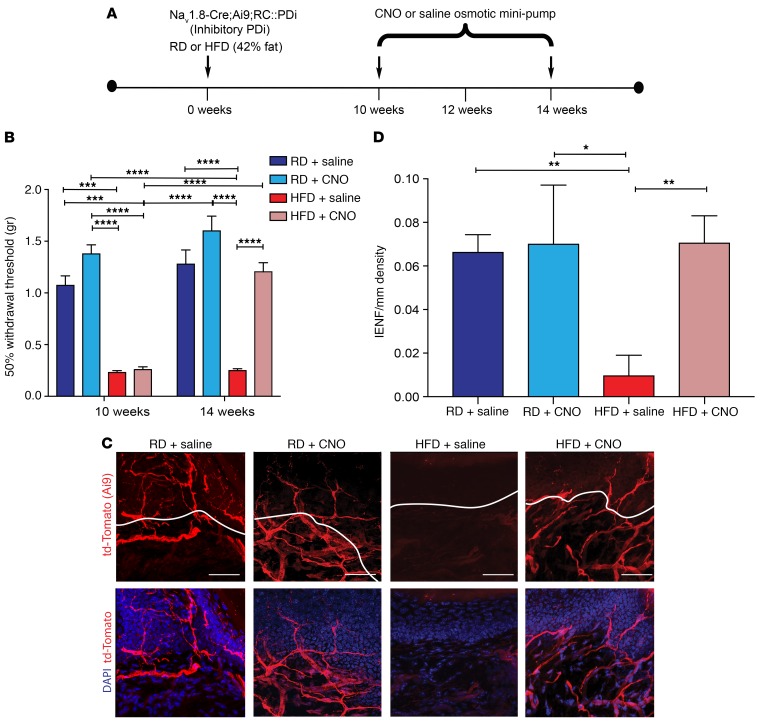
We next tested whether similar treatment could reverse these phenomena once they were established. We fitted Nav1.8-Cre;Ai9;RC::PDi mice with osmotic mini-pumps containing CNO or saline 10 weeks after the mice were started on a HFD (Figure 10A). By then, the mice had developed obesity, glucose intolerance, mechanical allodynia, and small-fiber degeneration. The obesity and glucose intolerance continued in HFD-fed mice (Supplemental Figure 9, D–F), but CNO infusion reversed their mechanical allodynia, while it persisted in saline-infused mice (Figure 10B). After 4 weeks of CNO infusion, small-fiber degeneration was completely reversed (Figure 10, C and D). These observations were confirmed with PGP 9.5 antibody immunolabeling on skin samples from the same mice (Supplemental Figure 11, A–C) as an independent verification for our fiber density measurements. We further established that CNO infusion did not affect mechanical allodynia or small-fiber degeneration in Nav1.8-Cre;Ai9 mice that were not expressing DREADDs, regardless of diet, at 2 to 8 weeks (Supplemental Figure 12, A–D) or 10 to 14 weeks (Supplemental Figure 13, A–D). Hence, these effects of CNO are DREADD dependent.
Figure 10. Chemogenetic inhibition of Nav1.8-positive DRG neurons can reverse small-fiber degeneration and mechanical allodynia in HFD-fed mice.
(A) Experimental protocol for osmotic mini-pump implantation in Nav1.8-Cre;Ai9;RC::PDi mice. Nav1.8-Cre;Ai9;RC::PDi mice were put on a RD or a HFD for 10 weeks and then implanted i.p. with an osmotic mini-pump delivering saline or CNO (10 mg/kg/day) for 4 weeks to determine whether CNO could reverse the effects of the HFD. Each arrow represents a time point at which pain behavior was assessed. (B) von Frey pain behavior testing demonstrated the presence of mechanical allodynia (reduction in withdrawal threshold) in mice after 10 weeks on a HFD. This mechanical allodynia was reduced after continuous treatment with CNO tested at the 14-week time point. ***P < 0.001 and ****P < 0.0001 (n = 6/group). (C and D) Confocal micrographs of skin from Nav1.8-Cre;Ai9;RC::PDi mice. td-Tomato–expressing Nav1.8 fibers (red) and merged images with the nuclear marker DAPI (blue). (C) Control mice on a RD with saline or CNO mini-pumps showed normal skin innervation. HFD mice implanted with a saline mini-pump showed reduced skin innervation. HFD mice fitted with CNO mini-pumps showed a significant improvement in skin innervation. Scale bars: 50 μm. (D) This effect was quantified using IENF density, and the intraepidermal-dermal junction is outlined in white in C. *P < 0.05 and **P < 0.01 (n = 6/group, with 3 noncontiguous sections analyzed per sample). Values are expressed as the mean ± SEM. P values were calculated using 2-way ANOVA with Bonferroni’s multiple comparisons test.
Increasing Nav1.8-positive DRG neuron excitability accelerates small-fiber degeneration in diabetic mice.
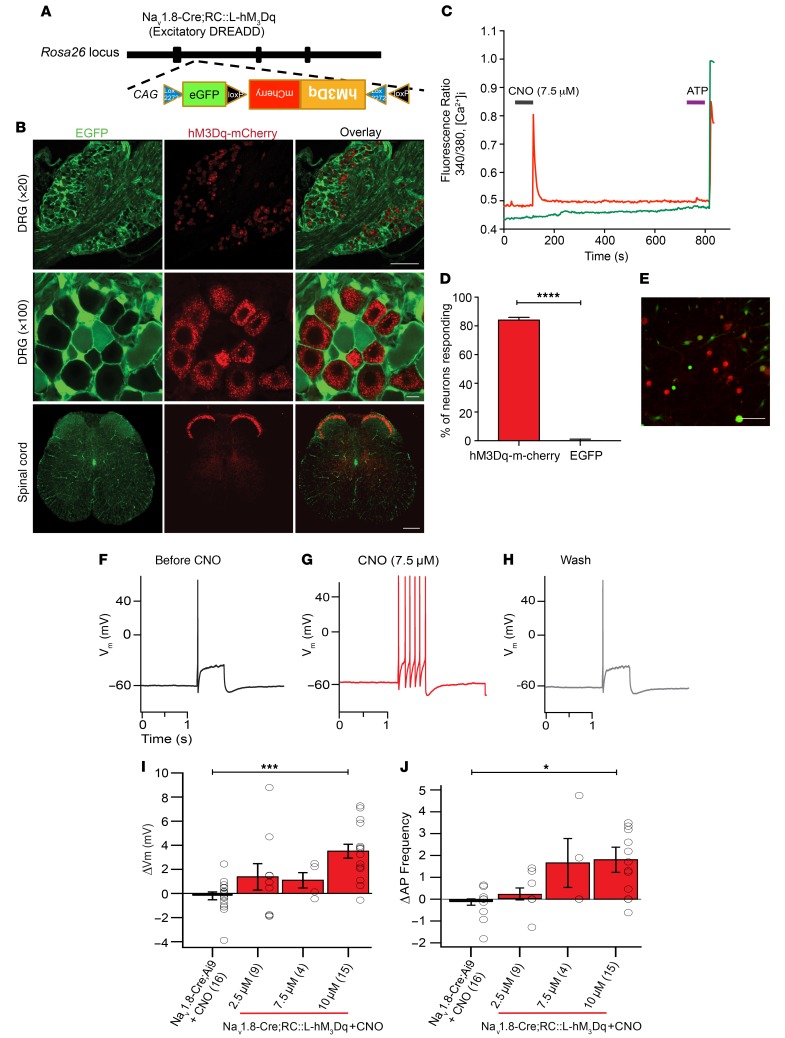
Next, we hypothesized that increasing neuronal excitability would accelerate mechanical allodynia and small-fiber degeneration. To test this hypothesis, we again used a chemogenetic approach, in which the expression of excitatory hM3Dq DREADDs (66) was induced in Nav1.8-positive DRG neurons. We used a mouse line with a Cre-responsive (Rosa-CAG=loxh M3Dq [RC::L-hM3Dq]) allele that also encodes EGFP and an hM3Dq-mCherry fusion protein. Cre activity inverts hM3Dq-mCherry, producing the proper orientation for transcription. RC::L-hM3Dq therefore expresses EGFP without recombinase activity and hM3Dq-mCherry after Cre-mediated recombination (Figure 11A). Using confocal microscopy, we confirmed the expression of hM3Dq DREADDs in Nav1.8-positive DRG neurons and the dorsal horn of the spinal cord (Figure 11B). Fura-2–based [Ca2+]i imaging of Nav1.8-positive DRG neurons cultured from Nav1.8-Cre;RC::L-hM3Dq mice revealed that CNO elicited robust [Ca2+]i signals in cells expressing the receptor (red), but not in cells without it (green) (Figure 11, C–E). Furthermore, in vitro current-clamp experiments showed that addition of CNO to Nav1.8-positive DRG neurons depolarized the membrane potential and increased the frequency of evoked APs in cultures from Nav1.8-Cre;RC::L-hM3Dq mice, but not from Nav1.8-Cre Ai9 control mice (Figure 11, F–J, and Supplemental Table 1).
Figure 11. Chemogenetic activation of hM3Dq excitatory DREADD receptors in Nav1.8-positive DRG neurons leads to increased neuronal excitability.
(A) The Nav1.8-Cre;RC::L-hM3Dq construct used in these experiments was designed so that Nav1.8-positive DRG neurons expressed m-Cherry–fused hM3Dq excitatory DREADD receptors, whereas all other cells expressed EGFP. (B) Representative images of DRGs (top and middle) and spinal cords (bottom) showing Nav1.8-positive DRG neurons expressing m-Cherry–fused hM3Dq excitatory DREADD receptors, whereas all other cells expressed EGFP. Original magnification, ×20, ×100, and ×10. Scale bars: 150 μm (top), 10 μm (middle), and 150 μm (bottom). (C–E) DRG neurons were cultured from hM3Dq excitatory DREADD mice and subjected to Fura-2–based [Ca2+]i imaging. Only cells expressing the hM3Dq DREADD receptors had [Ca2+]i responses to CNO (7.5 μM) (red), whereas all other EGFP-expressing cells did not respond (green). (D) Percentage of neurons responding to CNO (84.042% ± 1.9%). ****P < 0.0001, by Mann-Whitney U test (n = 94). (E) Representative image of the neurons used for [Ca2+]i imaging. Red indicates m-Cherry. Green indicates EGFP. Scale bar: 50 μm. (F–J) DRG primary cultures were prepared from these hM3Dq excitatory DREADD mice and m-Cherry–expressing cells were recorded. (G) Treatment with CNO (7.5 μM) along with a depolarizing current step led to increased AP frequency compared with (F) the current step alone or (H) after washing. (I) Changes in membrane voltage and (J) the AP frequency were quantified for various concentrations of CNO. These same experiments were performed in Nav1.8 td-Tomato DRG neurons that did not express DREADDs (Nav1.8-Cre;Ai9 mice). (I and J) Nav1.8-positve DRG neurons expressing the hM3Dq excitatory DREADD (Nav1.8-Cre;RC::L-hM3Dq) had significantly higher voltage membrane (Vm) and action potential frequencies compared to control Nav1.8-positve DRG neurons non-expressing the excitatory DREADD (Nav1.8-Cre;Ai9). *P < 0.05 and ***P < 0.001, by 1-way ANOVA with Tukey’s post hoc test (n = 16). Values are expressed as the mean ± SEM.
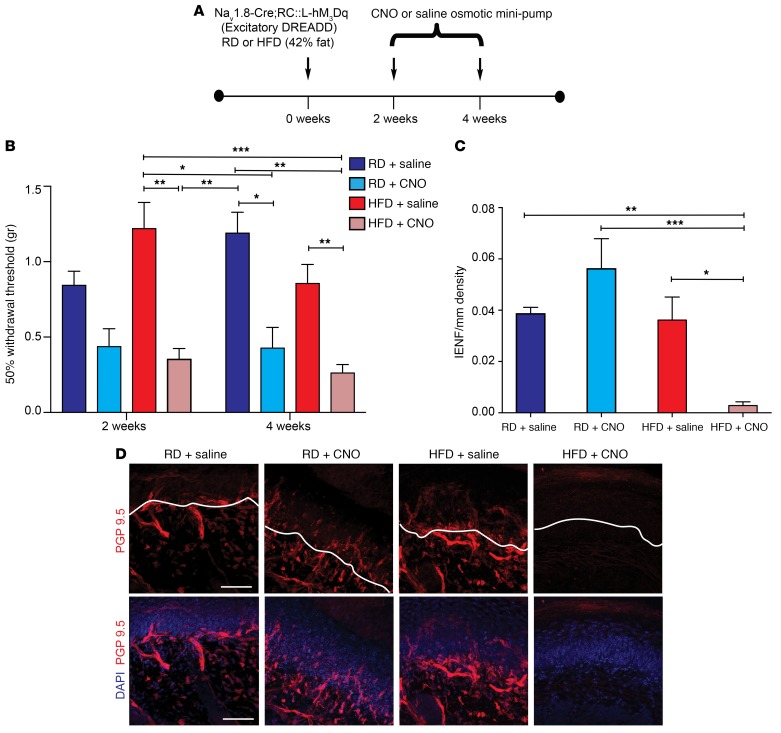
Next, we investigated the effects of long-term activation of hM3Dqs in vivo. We delivered CNO using osmotic mini-pumps placed i.p. into Nav1.8-Cre;RC::L-M3Dq mice 2 to 4 weeks after commencement of a HFD or RD (Figure 12A). After 4 weeks on the HFD, the mice had not yet developed glucose intolerance (Supplemental Figure 14, A–C). We found that mice fed either diet developed mechanical allodynia if CNO was continuously delivered from week 2 through week 4 (Figure 12B). Long-term chemogenetic activation of Nav1.8-positive DRG neurons also significantly accelerated small-fiber degeneration in HFD mice (Figure 12, C and D). Confocal micrographs from Nav1.8-Cre;RC::L-hM3Dq mice after 4 weeks on a RD or HFD with saline mini-pumps showed normal skin innervation. In contrast, Nav1.8-Cre;RC::L-hM3Dq mice on a HFD for 4 weeks with a CNO mini-pump had substantial depletion of nerve terminals (Figure 12, C and D), demonstrating accelerated pathology. In contrast, Nav1.8-Cre;RC::L-hM3Dq mice on a RD with CNO infusion did not develop small-fiber degeneration, at least after 4 weeks, the latest time point at which we examined these mice (Figure 12, C and D), indicating that increased excitability without diabetes was not sufficient to cause small-fiber degeneration.
Figure 12. Long-term chemogenetic activation of Nav1.8-positive DRG neurons results in significant acceleration of the development of mechanical allodynia and small-fiber degeneration in HFD-fed mice.
(A) Experimental setup of osmotic mini-pump implantation in Nav1.8-Cre;RC::L-hM3Dq mice. Nav1.8-Cre;RC::L-hM3Dq mice that expressed excitatory hM3Dq DREADD receptors were fed either a RD or a HFD and underwent i.p. implantation of an osmotic mini-pump, which administered either saline or CNO (10 mg/kg/day) for the period from 2 to 4 weeks following the commencement of a HFD or RD. (B) von Frey pain behavior testing demonstrated the onset of mechanical allodynia (reduction in withdrawal threshold) in HFD-fed mice (red) after 2 or 4 weeks following CNO administration. The RD mice (blue) also showed a reduction of their withdrawal threshold after 4 weeks of CNO administration. *P < 0.05, **P < 0.01, and ***P < 0.001 (n = 6/group). (C and D) Quantification (C) and confocal micrographs (D) of skin from Nav1.8-Cre;RC::L-hM3Dq mice on a RD for 4 weeks with saline mini-pumps showed normal skin innervation using PGP 9.5 (pseudo-colored red). Sections were colabeled with the nuclear marker DAPI (blue). In contrast, HFD mice with CNO mini-pumps had significant depletion of nerve terminals. Interestingly, in RD mice, increased excitability alone, produced by hM3Dq DREADD receptors, was not able to induce small-fiber degeneration in the absence of diabetes. Scale bar: 50 μm. This effect was quantified in C using IENF density, and the epidermal-dermal junction is outlined in white in D. *P < 0.05, **P < 0.01, and ***P < 0.001 (n = 6 from each group, with 3 noncontiguous sections analyzed per sample). P values were calculated using a 1-way ANOVA with Bonferroni’s multiple comparisons test. Values are expressed as the mean ± SEM.
Discussion
The results of our experiments demonstrated that excitatory CXCL12/CXCR4 signaling is a key factor in generating mechanical allodynia and small-fiber degeneration, two important features of PDN. We could prevent and reverse these phenomena by selective deletion of CXCR4 receptors or by chemogenetically limiting the excitability of Nav1.8-positive DRG neurons in the HFD mouse model of PDN. As activating CXCR4 receptors increased the excitability and [Ca2+]i of these neurons, we hypothesize that these effects may be responsible for the observed CXCR4-mediated mechanical allodynia and small-fiber degeneration. Therefore, these studies indicate that CXCR4-induced hyperexcitability of Nav1.8-positive DRG neurons represents a molecular pathway linking mechanical allodynia and axonal degeneration in diabetes and point to a potential new target for disease-modifying therapy, which is currently unavailable for PDN patients (6).
Painful symptoms vary among PDN patients (40), leading to different sensory phenotypes (40, 41) with different molecular mechanisms (25). In PDN patients, mechanical allodynia is commonly observed together with thermal hypoesthesia, particularly at later stages of the disease (40, 41). Similarly, in the HFD model, mice ultimately develop thermal hypoalgesia and mechanical allodynia, but not until 16 weeks after starting a HFD (38). After 10 weeks on a HFD, mice have mechanical allodynia without thermal hypoalgesia (30, 39). Given that sensory phenotypes are heterogeneous and vary with the disease stage, we decided to focus our study on mechanical allodynia rather than thermal pain behaviors. Mechanical allodynia is common in PDN patients (30, 39), though the relative contribution of its static and dynamic components, which are important in the clinic, may not be precisely duplicated in mouse models (25, 41). Nevertheless, our studies suggest that CXCR4 chemokine signaling is an important upstream mediator driving Nav1.8-positive DRG neuronal hyperexcitability, mechanical allodynia, and small-fiber degeneration in the HFD model. Thus, modulation of proalgesic chemokine signaling may provide an opportunity for disease modification. These results have the potential to transform the way small-fiber degeneration is treated and replace the largely ineffective approaches that are currently available for patients afflicted with PDN (6).
We demonstrated that the development of mechanical allodynia was inhibited following selective deletion of CXCR4 receptors and an associated reduction of hyperexcitability in Nav1.8-positive DRG neurons. The subtypes of DRG neurons traditionally linked to mechanical allodynia are C-fibers (67–70), low-threshold C-mechanoreceptors, and Aδ-mechanoreceptors (71–74). However, mechanical allodynia is also mediated by low-threshold Aβ-mechanoreceptors (71, 72). Given that all of these neuronal populations express Nav1.8 to some degree (23), our studies do not completely deconvolute the nature of the subtypes of neurons within the Nav1.8 population that are specifically associated with the occurrence of mechanical allodynia, which is something that could be achieved in future studies.
An additional limitation concerns the role of CXCR4-induced DRG hyperexcitability in the pathogenesis of axonal degeneration. One possibility is that blocking CXCR4 signaling protects against chronically increased [Ca2+]i, which produces axonal degeneration, as previously suggested (75) in the central (76) and peripheral neurons (77–79). In particular, increased [Ca2+]i is responsible for DRG neurite degeneration and contributes to nerve degeneration in a genetic model of small-fiber neuropathy (80). On the other hand, some reports have identified potentially beneficial effects of [Ca2+]i on axonal stability in a model of axon injury (81, 82). Therefore, the precise characteristics of [Ca2+]i in DRG neurons, including magnitude and acute or chronic signaling, may lead to different endpoints of axon structure and function.
Increased [Ca2+]i might contribute to axonal damage by altering mitochondrial function (83), including calcium homeostasis (84). Mitochondrial abnormalities occur in animal models of diabetes (32, 85). Specifically, DRG neurons show downregulation of mitochondrial respiratory chain complex proteins (86) and reduced respiratory chain activity (87). Thus, sustained CXCR4 signaling in Nav1.8-positive DRG neurons might initiate a cascade resulting in hyperexcitability and [Ca2+]i increases that could overwhelm the mitochondrial homeostatic mechanisms compromised by diabetes (32, 85), leading to small-fiber degeneration. Our observation that chemogenetic activation of Nav1.8-positive DRG neurons accelerated small-fiber degeneration only in HFD-fed mice (Figure 12D) supports this hypothesis.
Our chemogenetic approach revealed mechanisms underlying the development of mechanical allodynia and small-fiber degeneration in PDN. DREADDs are widely used to manipulate neural excitability (reviewed in refs. 61, 88), but they have some limitations. Inhibitory PDi expression in C-fibers using the TRPV1-Cre allele resulted in altered channel activity and second-messenger signaling, even without CNO, presumably because of the constitutive activity of overexpressed DREADD receptors in these experiments (89). Accordingly, we included saline controls to ensure that our findings were related to activation by CNO. We did not observe CNO-independent changes, perhaps because we used a different promoter to drive Cre expression (Nav1.8-Cre instead of TRPV1-Cre).
Recent reports have also suggested that there may be DREADD-independent effects of CNO (65) and have raised the possibility that CNO can rapidly convert to clozapine in vivo (63). To control for this possibility, we confirmed that CNO infusion did not affect mechanical allodynia or small-fiber degeneration in Nav1.8-Cre;Ai9 mice that were not expressing DREADDs, regardless of diet, at 2 to 8 weeks (Supplemental Figure 12, A–D) or 10 to 14 weeks (Supplemental Figure 13, A–D). Hence, all the effects of CNO we observed were DREADD dependent. All the controls for the chemogenetic platform listed above were essential for validating our experiments, as the data presented here represent the first time to our knowledge that any intervention has been shown to prevent and even reverse not only mechanical allodynia but also small-fiber degeneration in a diabetic model.
In summary, our results identify CXCL12/CXCR4 signaling as the initiator of a pathway linking hyperexcitability and increased [Ca2+]i in Nav1.8-positive DRG neurons to mechanical allodynia and small-fiber degeneration in PDN. From a translational perspective, we propose that blocking CXCR4 signaling or Nav1.8-positive DRG neuron hyperexcitability may represent a novel approach for the treatment of this intractable and widespread affliction. Indeed, a reduction of proalgesic CXCL12/CXCR4 signaling could abolish persistent excitability and increased [Ca2+]i, preventing not only neuropathic pain behavior but also the development of small-fiber degeneration. We also predict that drugs that reduce Nav1.8-expressing DRG neuronal hyperexcitability, such as specific sodium (90, 91), might effectively treat PDN. Moreover, the relationship between hyperexcitability, calcium overload, and axonal degeneration is likely to inform studies of other neurodegenerative diseases such as amyotrophic lateral sclerosis (ALS) (92) or Parkinson’s disease (PD) (93) that involve similar underlying events.
Methods
Animals.
Animals were housed on a 12-hour light/12-hour dark cycle with ad libitum access to food and water. We used the following mouse lines: Nav1.8-Cre;Ai9, Pirt-GCaMP3, Nav1.8-Cre::GCaMP6, parvalbumin-Cre::GCaMP6, and Nav1.8-Cre;RC::PDi mice; Nav1.8-Cre;Ai9;RC::PDi, Nav1.8-Cre;RC::PDi GCamP6, Nav1.8-Cre;RC::L-hM3Dq, and Nav1.8-Cre;Ai9;CXCR4fl/+ heterozygotes; and Nav1.8-Cre;Ai9;CXCR4fl/fl homozygotes.
HFD.
A HFD is a common rodent model of type 2 diabetes. Mice were fed 42% fat (Envigo TD88137) for 10 weeks. Control mice were fed a RD (11% fat). After 10 weeks on a RD or HFD, a glucose tolerance test was performed as described (39). To compare “diabetic” versus “nondiabetic” HFD mice, we set the cutoff for diabetes (≥140 mg/dl) at 2 SD above the mean for glucose 2 hours after glucose challenge in 129 WT littermate mice fed a RD (39, 94).
Detection of cutaneous innervation.
Skin samples were processed as previously described (39). Samples were imaged by confocal microscopy (Olympus fv10i, FluoView software) for confocal analysis. Composite Z-stack images were obtained and processed using Fiji software (NIH). The epidermal-dermal junction was outlined by a blinded observer who also noted its length. At least 3 other blinded reviewers counted the nerves crossing this line using the ImageJ Cell Counter plugin (NIH).
Behavioral testing.
von Frey behavioral studies were performed as previously described. von Frey experiments were conducted using random experimental group assignments (RD or HFD diet and treatment). Investigators who performed the von Frey tests and endpoint analyses were blinded to the experimental conditions. We have experience with randomized allocation and blinded analysis using this mouse model with sequenced numbering of mice at weaning (30, 39).
Calcium imaging in DRG explants.
L4 and L5 PirtGCaMP3 and Nav1.8-Cre;RC::PDi GCaMP6 mouse DRGs were dissected, incubated in artificial cerebrospinal fluid (ACSF) at room temperature, and mounted onto the stage of a Yokogawa CSU-X1 abd CSU-W1 upright spinning-disk confocal microscope (3i; Intelligent Imaging Innovations Inc.) equipped with an electron multiplication CCD camera (48).The activity of selected neurons of the explants expressing GCaMP3 or GCaMP6 (green fluorescence) was examined on the basis of peak amplitude of fluorescence change (ΔF/F0) for spontaneous activity compared with that of the stimulus. Analysis of [Ca2+]i imaging data was done with Fiji software using standard functions and a custom macro. Different concentrations of HK (K+) (10 and 50 mM) or capsaicin (1, 2, and 10 μM), CNO (8 μM), and CXCL12 (100 nM) were applied.
Preparation of primary cultures of DRG neurons.
DRG sensory neurons from diabetic Nav1.8-Cre;Ai9 mice, Nav1.8-Cre;Ai9;RC::PDi mice, and Nav1.8-Cre;RC::L-hM3Dq mice were dissociated as described (39)after 10 weeks on either a RD or HFD.
Electrophysiological recordings of DRG neurons.
For current-clamp recordings, patch electrodes with a resistance of 5 to 7 MΩ were filled with 140 mM KCl, 0.5 mM EGTA, 5 mM HEPES, and 3 Mg-ATP, pH 7.3 (300 mOsmol). The RMP was measured in each cell. Whole-cell, current-clamp recordings were obtained as previously described (95) using a MultiClamp patch-clamp amplifier (Molecular Devices). The data were captured with pClamp 10.0 software (Molecular Devices) and calculated with Clampfit (Molecular Devices), SigmaPlot (Systat Software), GraphPad Prism (GraphPad Software), and Igor (WaveMetrics). CNO (2.5, 7.5, or 10 μM) and CXCL12 (50 nM) were applied to culture.
Antibodies.
We used the following antibodies on DRG sections: HA-Tag (C29F4) rabbit monoclonal antibody (Cell Signaling Technology, catalog 3724, 1:250) and I-isolectin B4 (IB4 isolectin GS-IB4 Alexa Fluor 647 conjugate) (Invitrogen, Thermo Fisher Scientific, catalog I32450, 1:100). The secondary antibody Alexa Fluor 488 goat anti-rabbit antibody (Invitrogen, Thermo Fisher Scientific, 1:250) was used. Anti-PGP9.5 rabbit monoclonal antibody (MilliporeSigma, catalog AB1761-I, 1:250) was used on skin sections.
Immunohistochemical labeling.
Adult mice were deeply anesthetized with isoflurane and transcardially perfused with saline followed by 4% paraformaldehyde. DRG (lumbar levels 2–4) and spinal cord were processed as previously described (30). Tissue sections were analyzed by confocal microscopy.
CNO injection.
CNO (10 mg/kg, MilliporeSigma) in 200 ml of saline or saline alone was injected i.p. using a 25-gauge needle. Mice were tested for pain 1 hour and 4 hours after injection.
Chronic activation of DREADDs with CNO.
ALZET Osmotic Pumps were surgically implanted i.p. according to the manufacturer’s instructions in animals anesthetized with isoflurane. Pump models 2006 and 1004 were used for constant delivery (0.15 μl/h) of CNO (10 mg/kg/d) (VDM Biochemicals) or saline for 6 weeks and 4 weeks, respectively.
In vitro calcium imaging of DRG neurons.
Neurons from Nav1.8-Cre;RC::L-hM3Dq mice were cultured as described (39). Their responses to CNO (7.5 μM) were recorded using Fura-2–based [Ca2+]i imaging as previously described (39). For all experiments, capsaicin (100 nM), high K+ (25 mM), and ATP (100 μM) were added to the cells.
Statistics.
All statistical analysis was performed using GraphPad Prism 7.03 (GraphPad Software). For the measurement of blood glucose and behavioral testing, the significance of differences between the control and the various treatment groups, or between genotypes, was analyzed using a 1- or 2-way ANOVA with Bonferroni’s multiple comparisons tests. For calcium imaging experiments in vitro and in vivo, the data were tested for statistical significance using a Mann-Whitney U test. A 2-way ANOVA with Dunnett’s or Bonferroni’s multiple comparisons test was used to determine the IENF density. For the electrophysiological experiments, the data were tested for statistical significance using a Mann-Whitney U test or 1-way ANOVA with Tukey’s post hoc test. Student t tests were all 2 tailed. All values are expressed as the mean ± SEM, and a P value of less than 0.05 was considered statistically significant.
Study approval.
All methods involving animals were approved by the IACUC of Northwestern University.
Author contributions
NDJ performed von Frey behavioral studies, [Ca2+]i imaging studies, immunohistochemical labeling, and confocal analysis. Mouse breeding, diet administration, glucose tolerance testing, and IENF density counts were done by NDJ, CAR, BEH, and HRG. BJB and SH performed electrophysiological studies. DR and AAB performed [Ca2+]i imaging studies. DMM and NDJ performed statistical analysis. DMM and RJM supervised the project. DMM drafted the manuscript, which was edited by RJM. All authors read and approved the manuscript.
Supplementary Material
Acknowledgments
This work was supported by NIH grants K08 NS079482-01 (to DMM) and 5R01DA013141-14 (to RJM) and by NIH/Rush University Medical Center grant 1R01AR064251-01 (to RJM). All statistical analysis was reviewed by the Statistical Core at Northwestern University. We thank Rajeshwar Awatramani (Northwestern University) for helpful discussions. We thank Alexandra Fredrickson (Northwestern University) for helping with the evaluation of IENF density. We thank Susan Dymecki (Harvard Medical School), Xinzhong Dong (Johns Hopkins University), John Wood (University College London), Savio Chan (Northwestern University), and Patricia Jensen (NIH/NIEHS) for the generous gifts of mice (RC::PDi, Pirt-GCaMP3, Nav1.8-Cre, Parvalb-Cre, and RC::L-hM3Dq, respectively).
Version 1. 03/13/2018
In-Press Preview
Version 2. 04/23/2018
Electronic publication
Version 3. 06/01/2018
Print issue publication
Funding Statement
Submitted
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(6):2205–2225.https://doi.org/10.1172/JCI92117.
Contributor Information
Nirupa D. Jayaraj, Email: njayaraj@northwestern.edu.
Bula J. Bhattacharyya, Email: bulajha1@gmail.com.
Abdelhak A. Belmadani, Email: a-belmadni@northwestern.edu.
Dongjun Ren, Email: dongjren@gmail.com.
Craig A. Rathwell, Email: rathwell.craig@gmail.com.
Sandra Hackelberg, Email: shackelb@umich.edu.
Brittany E. Hopkins, Email: BrittanyHopkins2020@u.northwestern.edu.
Herschel R. Gupta, Email: hgupta1999@gmail.com.
Richard J. Miller, Email: r-miller10@northwestern.edu.
Daniela M. Menichella, Email: d-menichella@northwestern.edu.
References
- 1.American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2011;34 Suppl 1:S62–S69. doi: 10.2337/dc11-S062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spallone V, Lacerenza M, Rossi A, Sicuteri R, Marchettini P. Painful diabetic polyneuropathy: approach to diagnosis and management. Clin J Pain. 2012;28(8):726–743. doi: 10.1097/AJP.0b013e318243075c. [DOI] [PubMed] [Google Scholar]
- 3.Zimmet PZ, Magliano DJ, Herman WH, Shaw JE. Diabetes: a 21st century challenge. Lancet Diabetes Endocrinol. 2014;2(1):56–64. doi: 10.1016/S2213-8587(13)70112-8. [DOI] [PubMed] [Google Scholar]
- 4.Menke A, Casagrande S, Geiss L, Cowie CC. Prevalence of and Trends in Diabetes Among Adults in the United States, 1988-2012. JAMA. 2015;314(10):1021–1029. doi: 10.1001/jama.2015.10029. [DOI] [PubMed] [Google Scholar]
- 5.daCosta DiBonaventura M, Cappelleri JC, Joshi AV. A longitudinal assessment of painful diabetic peripheral neuropathy on health status, productivity, and health care utilization and cost. Pain Med. 2011;12(1):118–126. doi: 10.1111/j.1526-4637.2010.01012.x. [DOI] [PubMed] [Google Scholar]
- 6.Bril V, et al. Evidence-based guideline: Treatment of painful diabetic neuropathy: report of the American Academy of Neurology, the American Association of Neuromuscular and Electrodiagnostic Medicine, and the American Academy of Physical Medicine and Rehabilitation. Neurology. 2011;76(20):1758–1765. doi: 10.1212/WNL.0b013e3182166ebe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quilici S, et al. Meta-analysis of duloxetine vs. pregabalin and gabapentin in the treatment of diabetic peripheral neuropathic pain. BMC Neurol. 2009;9:6. doi: 10.1186/1471-2377-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Callaghan BC, Cheng HT, Stables CL, Smith AL, Feldman EL. Diabetic neuropathy: clinical manifestations and current treatments. Lancet Neurol. 2012;11(6):521–534. doi: 10.1016/S1474-4422(12)70065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finnerup NB, et al. Pharmacotherapy for neuropathic pain in adults: a systematic review and meta-analysis. Lancet Neurol. 2015;14(2):162–173. doi: 10.1016/S1474-4422(14)70251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Divisova S, et al. Intraepidermal nerve-fibre density as a biomarker of the course of neuropathy in patients with Type 2 diabetes mellitus. Diabet Med. 2016;33(5):650–654. doi: 10.1111/dme.12890. [DOI] [PubMed] [Google Scholar]
- 11.Latremoliere A, Woolf CJ. Central sensitization: a generator of pain hypersensitivity by central neural plasticity. J Pain. 2009;10(9):895–926. doi: 10.1016/j.jpain.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lauria G, Devigili G. Skin biopsy as a diagnostic tool in peripheral neuropathy. Nat Clin Pract Neurol. 2007;3(10):546–557. doi: 10.1038/ncpneuro0630. [DOI] [PubMed] [Google Scholar]
- 13.Sommer C, Lauria G. Skin biopsy in the management of peripheral neuropathy. Lancet Neurol. 2007;6(7):632–642. doi: 10.1016/S1474-4422(07)70172-2. [DOI] [PubMed] [Google Scholar]
- 14.Basbaum AI, Bautista DM, Scherrer G, Julius D. Cellular and molecular mechanisms of pain. Cell. 2009;139(2):267–284. doi: 10.1016/j.cell.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woolf CJ. Central sensitization: uncovering the relation between pain and plasticity. Anesthesiology. 2007;106(4):864–867. doi: 10.1097/01.anes.0000264769.87038.55. [DOI] [PubMed] [Google Scholar]
- 16.Feldman EL, Nave KA, Jensen TS, Bennett DLH. New Horizons in Diabetic Neuropathy: Mechanisms, Bioenergetics, and Pain. Neuron. 2017;93(6):1296–1313. doi: 10.1016/j.neuron.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tesfaye S, Boulton AJ, Dickenson AH. Mechanisms and management of diabetic painful distal symmetrical polyneuropathy. Diabetes Care. 2013;36(9):2456–2465. doi: 10.2337/dc12-1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ørstavik K, et al. Abnormal function of C-fibers in patients with diabetic neuropathy. J Neurosci. 2006;26(44):11287–11294. doi: 10.1523/JNEUROSCI.2659-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersson DA, et al. Methylglyoxal evokes pain by stimulating TRPA1. PLoS ONE. 2013;8(10):e77986. doi: 10.1371/journal.pone.0077986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bierhaus A, et al. Methylglyoxal modification of Nav1.8 facilitates nociceptive neuron firing and causes hyperalgesia in diabetic neuropathy. Nat Med. 2012;18(6):926–933. doi: 10.1038/nm.2750. [DOI] [PubMed] [Google Scholar]
- 21.Orstavik K, Jørum E. Microneurographic findings of relevance to pain in patients with erythromelalgia and patients with diabetic neuropathy. Neurosci Lett. 2010;470(3):180–184. doi: 10.1016/j.neulet.2009.05.061. [DOI] [PubMed] [Google Scholar]
- 22.Serra J, Duan WR, Locke C, Solà R, Liu W, Nothaft W. Effects of a T-type calcium channel blocker, ABT-639, on spontaneous activity in C-nociceptors in patients with painful diabetic neuropathy: a randomized controlled trial. Pain. 2015;156(11):2175–2183. doi: 10.1097/j.pain.0000000000000249. [DOI] [PubMed] [Google Scholar]
- 23.Shields SD, et al. Nav1.8 expression is not restricted to nociceptors in mouse peripheral nervous system. Pain. 2012;153(10):2017–2030. doi: 10.1016/j.pain.2012.04.022. [DOI] [PubMed] [Google Scholar]
- 24.Lauria G, et al. The role of sodium channels in painful diabetic and idiopathic neuropathy. Curr Diab Rep. 2014;14(10):538. doi: 10.1007/s11892-014-0538-5. [DOI] [PubMed] [Google Scholar]
- 25.von Hehn CA, Baron R, Woolf CJ. Deconstructing the neuropathic pain phenotype to reveal neural mechanisms. Neuron. 2012;73(4):638–652. doi: 10.1016/j.neuron.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Lera Ruiz M, Kraus RL. Voltage-Gated Sodium Channels: Structure, Function, Pharmacology, and Clinical Indications. J Med Chem. 2015;58(18):7093–7118. doi: 10.1021/jm501981g. [DOI] [PubMed] [Google Scholar]
- 27.Yang F, et al. SDF1-CXCR4 signaling contributes to persistent pain and hypersensitivity via regulating excitability of primary nociceptive neurons: involvement of ERK-dependent Nav1.8 up-regulation. J Neuroinflammation. 2015;12:219. doi: 10.1186/s12974-015-0441-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kao DJ, et al. CC chemokine ligand 2 upregulates the current density and expression of TRPV1 channels and Nav1.8 sodium channels in dorsal root ganglion neurons. J Neuroinflammation. 2012;9:189. doi: 10.1186/1742-2094-9-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang JG, et al. The chemokine CXCL1/growth related oncogene increases sodium currents and neuronal excitability in small diameter sensory neurons. Mol Pain. 2008;4:38. doi: 10.1186/1744-8069-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menichella DM, Abdelhak B, Ren D, Shum A, Frietag C, Miller RJ. CXCR4 chemokine receptor signaling mediates pain in diabetic neuropathy. Mol Pain. 2014;10:42. doi: 10.1186/1744-8069-10-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhangoo SK, et al. CXCR4 chemokine receptor signaling mediates pain hypersensitivity in association with antiretroviral toxic neuropathy. Brain Behav Immun. 2007;21(5):581–591. doi: 10.1016/j.bbi.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vincent AM, Callaghan BC, Smith AL, Feldman EL. Diabetic neuropathy: cellular mechanisms as therapeutic targets. Nat Rev Neurol. 2011;7(10):573–583. doi: 10.1038/nrneurol.2011.137. [DOI] [PubMed] [Google Scholar]
- 33.Kampoli AM, Tousoulis D, Briasoulis A, Latsios G, Papageorgiou N, Stefanadis C. Potential pathogenic inflammatory mechanisms of endothelial dysfunction induced by type 2 diabetes mellitus. Curr Pharm Des. 2011;17(37):4147–4158. doi: 10.2174/138161211798764825. [DOI] [PubMed] [Google Scholar]
- 34.Sjöholm A, Nyström T. Endothelial inflammation in insulin resistance. Lancet. 2005;365(9459):610–612. doi: 10.1016/S0140-6736(05)17912-4. [DOI] [PubMed] [Google Scholar]
- 35.Purwata TE. High TNF-alpha plasma levels and macrophages iNOS and TNF-alpha expression as risk factors for painful diabetic neuropathy. J Pain Res. 2011;4:169–175. doi: 10.2147/JPR.S21751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uçeyler N, Rogausch JP, Toyka KV, Sommer C. Differential expression of cytokines in painful and painless neuropathies. Neurology. 2007;69(1):42–49. doi: 10.1212/01.wnl.0000265062.92340.a5. [DOI] [PubMed] [Google Scholar]
- 37.Hur J, et al. The identification of gene expression profiles associated with progression of human diabetic neuropathy. Brain. 2011;134(Pt 11):3222–3235. doi: 10.1093/brain/awr228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Obrosova IG, et al. High-fat diet induced neuropathy of pre-diabetes and obesity: effects of “healthy” diet and aldose reductase inhibition. Diabetes. 2007;56(10):2598–2608. doi: 10.2337/db06-1176. [DOI] [PubMed] [Google Scholar]
- 39.Menichella DM, et al. Ganglioside GM3 synthase depletion reverses neuropathic pain and small fiber neuropathy in diet-induced diabetic mice. Mol Pain. 2016 doi: 10.1177/1744806916666284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baron R, Tölle TR, Gockel U, Brosz M, Freynhagen R. A cross-sectional cohort survey in 2100 patients with painful diabetic neuropathy and postherpetic neuralgia: Differences in demographic data and sensory symptoms. Pain. 2009;146(1-2):34–40. doi: 10.1016/j.pain.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 41.Themistocleous AC, et al. The Pain in Neuropathy Study (PiNS): a cross-sectional observational study determining the somatosensory phenotype of painful and painless diabetic neuropathy. Pain. 2016;157(5):1132–1145. doi: 10.1097/j.pain.0000000000000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiu IM, et al. Transcriptional profiling at whole population and single cell levels reveals somatosensory neuron molecular diversity. Elife. 2014;3.:e04660. doi: 10.7554/eLife.04660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Usoskin D, et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci. 2015;18(1):145–153. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- 44.Li CL, et al. Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity. Cell Res. 2016;26(8):967. doi: 10.1038/cr.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stirling LC, et al. Nociceptor-specific gene deletion using heterozygous NaV1.8-Cre recombinase mice. Pain. 2005;113(1-2):27–36. doi: 10.1016/j.pain.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 46.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13(1):133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lauria G, Lombardi R, Camozzi F, Devigili G. Skin biopsy for the diagnosis of peripheral neuropathy. Histopathology. 2009;54(3):273–285. doi: 10.1111/j.1365-2559.2008.03096.x. [DOI] [PubMed] [Google Scholar]
- 48.Han L, et al. A subpopulation of nociceptors specifically linked to itch. Nat Neurosci. 2013;16(2):174–182. doi: 10.1038/nn.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goswami SC, et al. Molecular signatures of mouse TRPV1-lineage neurons revealed by RNA-Seq transcriptome analysis. J Pain. 2014;15(12):1338–1359. doi: 10.1016/j.jpain.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499(7458):295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hippenmeyer S, et al. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3(5):e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niu J, et al. Modality-based organization of ascending somatosensory axons in the direct dorsal column pathway. J Neurosci. 2013;33(45):17691–17709. doi: 10.1523/JNEUROSCI.3429-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Nooij JC, Doobar S, Jessell TM. Etv1 inactivation reveals proprioceptor subclasses that reflect the level of NT3 expression in muscle targets. Neuron. 2013;77(6):1055–1068. doi: 10.1016/j.neuron.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.White FA, et al. Excitatory monocyte chemoattractant protein-1 signaling is up-regulated in sensory neurons after chronic compression of the dorsal root ganglion. Proc Natl Acad Sci U S A. 2005;102(39):14092–14097. doi: 10.1073/pnas.0503496102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mithal DS, Ren D, Miller RJ. CXCR4 signaling regulates radial glial morphology and cell fate during embryonic spinal cord development. Glia. 2013;61(8):1288–1305. doi: 10.1002/glia.22515. [DOI] [PubMed] [Google Scholar]
- 56.Belmadani A, Jung H, Ren D, Miller RJ. The chemokine SDF-1/CXCL12 regulates the migration of melanocyte progenitors in mouse hair follicles. Differentiation. 2009;77(4):395–411. doi: 10.1016/j.diff.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stucky CL, Rossi J, Airaksinen MS, Lewin GR. GFR alpha2/neurturin signalling regulates noxious heat transduction in isolectin B4-binding mouse sensory neurons. J Physiol (Lond) 2002;545(Pt 1):43–50. doi: 10.1113/jphysiol.2002.027656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Snider WD, McMahon SB. Tackling pain at the source: new ideas about nociceptors. Neuron. 1998;20(4):629–632. doi: 10.1016/S0896-6273(00)81003-X. [DOI] [PubMed] [Google Scholar]
- 59.Bennett DL, Averill S, Clary DO, Priestley JV, McMahon SB. Postnatal changes in the expression of the trkA high-affinity NGF receptor in primary sensory neurons. Eur J Neurosci. 1996;8(10):2204–2208. doi: 10.1111/j.1460-9568.1996.tb00742.x. [DOI] [PubMed] [Google Scholar]
- 60.Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc Natl Acad Sci U S A. 2007;104(12):5163–5168. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Urban DJ, Roth BL. DREADDs (designer receptors exclusively activated by designer drugs): chemogenetic tools with therapeutic utility. Annu Rev Pharmacol Toxicol. 2015;55:399–417. doi: 10.1146/annurev-pharmtox-010814-124803. [DOI] [PubMed] [Google Scholar]
- 62.Ray RS, et al. Impaired respiratory and body temperature control upon acute serotonergic neuron inhibition. Science. 2011;333(6042):637–642. doi: 10.1126/science.1205295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gomez JL, et al. Chemogenetics revealed: DREADD occupancy and activation via converted clozapine. Science. 2017;357(6350):503–507. doi: 10.1126/science.aan2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vulchanova L, et al. P2X3 is expressed by DRG neurons that terminate in inner lamina II. Eur J Neurosci. 1998;10(11):3470–3478. doi: 10.1046/j.1460-9568.1998.00355.x. [DOI] [PubMed] [Google Scholar]
- 65.MacLaren DA, et al. Clozapine n-oxide administration produces behavioral effects in long-evans rats: implications for designing DREADD experiments. eNeuro. 2016;3(5):ENEURO.0219-16.2016. doi: 10.1523/ENEURO.0219-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sciolino NR, et al. Recombinase-Dependent Mouse Lines for Chemogenetic Activation of Genetically Defined Cell Types. Cell Rep. 2016;15(11):2563–2573. doi: 10.1016/j.celrep.2016.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ossipov MH, Bian D, Malan TP, Lai J, Porreca F. Lack of involvement of capsaicin-sensitive primary afferents in nerve-ligation injury induced tactile allodynia in rats. Pain. 1999;79(2-3):127–133. doi: 10.1016/s0304-3959(98)00187-0. [DOI] [PubMed] [Google Scholar]
- 68.Minett MS, et al. Pain without nociceptors? Nav1.7-independent pain mechanisms. Cell Rep. 2014;6(2):301–312. doi: 10.1016/j.celrep.2013.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu CN, Wall PD, Ben-Dor E, Michaelis M, Amir R, Devor M. Tactile allodynia in the absence of C-fiber activation: altered firing properties of DRG neurons following spinal nerve injury. Pain. 2000;85(3):503–521. doi: 10.1016/S0304-3959(00)00251-7. [DOI] [PubMed] [Google Scholar]
- 70.King T, et al. Contribution of afferent pathways to nerve injury-induced spontaneous pain and evoked hypersensitivity. Pain. 2011;152(9):1997–2005. doi: 10.1016/j.pain.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abraira VE, Ginty DD. The sensory neurons of touch. Neuron. 2013;79(4):618–639. doi: 10.1016/j.neuron.2013.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li L, et al. The functional organization of cutaneous low-threshold mechanosensory neurons. Cell. 2011;147(7):1615–1627. doi: 10.1016/j.cell.2011.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seal RP, et al. Injury-induced mechanical hypersensitivity requires C-low threshold mechanoreceptors. Nature. 2009;462(7273):651–655. doi: 10.1038/nature08505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boada MD, et al. Fast-conducting mechanoreceptors contribute to withdrawal behavior in normal and nerve injured rats. Pain. 2014;155(12):2646–2655. doi: 10.1016/j.pain.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang JT, Medress ZA, Barres BA. Axon degeneration: molecular mechanisms of a self-destruction pathway. J Cell Biol. 2012;196(1):7–18. doi: 10.1083/jcb.201108111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Coleman MP, Perry VH. Axon pathology in neurological disease: a neglected therapeutic target. Trends Neurosci. 2002;25(10):532–537. doi: 10.1016/S0166-2236(02)02255-5. [DOI] [PubMed] [Google Scholar]
- 77.Lehning EJ, Doshi R, Isaksson N, Stys PK, LoPachin RM. Mechanisms of injury-induced calcium entry into peripheral nerve myelinated axons: role of reverse sodium-calcium exchange. J Neurochem. 1996;66(2):493–500. doi: 10.1046/j.1471-4159.1996.66020493.x. [DOI] [PubMed] [Google Scholar]
- 78.Persson AK, Hoeijmakers JGJ, Estacion M, Black JA, Waxman SG. Sodium Channels, Mitochondria, and Axonal Degeneration in Peripheral Neuropathy. Trends Mol Med. 2016;22(5):377–390. doi: 10.1016/j.molmed.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 79.Vargas ME, Yamagishi Y, Tessier-Lavigne M, Sagasti A. Live Imaging of Calcium Dynamics during Axon Degeneration Reveals Two Functionally Distinct Phases of Calcium Influx. J Neurosci. 2015;35(45):15026–15038. doi: 10.1523/JNEUROSCI.2484-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Estacion M, et al. Ca2+ toxicity due to reverse Na+/Ca2+ exchange contributes to degeneration of neurites of DRG neurons induced by a neuropathy-associated Nav1.7 mutation. J Neurophysiol. 2015;114(3):1554–1564. doi: 10.1152/jn.00195.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cho Y, Sloutsky R, Naegle KM, Cavalli V. Injury-induced HDAC5 nuclear export is essential for axon regeneration. Cell. 2013;155(4):894–908. doi: 10.1016/j.cell.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cho Y, Cavalli V. HDAC5 is a novel injury-regulated tubulin deacetylase controlling axon regeneration. EMBO J. 2012;31(14):3063–3078. doi: 10.1038/emboj.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bernardi P, Rasola A. Calcium and cell death: the mitochondrial connection. Subcell Biochem. 2007;45:481–506. doi: 10.1007/978-1-4020-6191-2_18. [DOI] [PubMed] [Google Scholar]
- 84.Rasola A, Bernardi P. The mitochondrial permeability transition pore and its involvement in cell death and in disease pathogenesis. Apoptosis. 2007;12(5):815–833. doi: 10.1007/s10495-007-0723-y. [DOI] [PubMed] [Google Scholar]
- 85.Vincent AM, et al. Mitochondrial biogenesis and fission in axons in cell culture and animal models of diabetic neuropathy. Acta Neuropathol. 2010;120(4):477–489. doi: 10.1007/s00401-010-0697-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Roy Chowdhury SK, et al. Impaired adenosine monophosphate-activated protein kinase signalling in dorsal root ganglia neurons is linked to mitochondrial dysfunction and peripheral neuropathy in diabetes. Brain. 2012;135(Pt 6):1751–1766. doi: 10.1093/brain/aws097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chowdhury SK, et al. Mitochondrial respiratory chain dysfunction in dorsal root ganglia of streptozotocin-induced diabetic rats and its correction by insulin treatment. Diabetes. 2010;59(4):1082–1091. doi: 10.2337/db09-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhu H, Roth BL. DREADD: a chemogenetic GPCR signaling platform. Int J Neuropsychopharmacol. 2014;18(1):pyu007. doi: 10.1093/ijnp/pyu007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Saloman JL, Scheff NN, Snyder LM, Ross SE, Davis BM, Gold MS. Gi-DREADD expression in peripheral nerves produces ligand-dependent analgesia, as well as ligand-independent functional changes in sensory neurons. J Neurosci. 2016;36(42):10769–10781. doi: 10.1523/JNEUROSCI.3480-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dib-Hajj SD, Yang Y, Black JA, Waxman SG. The Na(V)1.7 sodium channel: from molecule to man. Nat Rev Neurosci. 2013;14(1):49–62. doi: 10.1038/nrn3404. [DOI] [PubMed] [Google Scholar]
- 91.Eijkelkamp N, et al. Neurological perspectives on voltage-gated sodium channels. Brain. 2012;135(Pt 9):2585–2612. doi: 10.1093/brain/aws225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wainger BJ, et al. Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell Rep. 2014;7(1):1–11. doi: 10.1016/j.celrep.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lieberman OJ, et al. α-Synuclein-dependent calcium entry underlies differential sensitivity of cultured SN and VTA dopaminergic neurons to a Parkinsonian neurotoxin. eNeuro. 2017;4(6):ENEURO.0167-17.2017. doi: 10.1523/ENEURO.0167-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fenner D, et al. Generation of N-ethyl-N-nitrosourea (ENU) diabetes models in mice demonstrates genotype-specific action of glucokinase activators. J Biol Chem. 2011;286(45):39560–39572. doi: 10.1074/jbc.M111.269100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cummins TR, Dib-Hajj SD, Black JA, Akopian AN, Wood JN, Waxman SG. A novel persistent tetrodotoxin-resistant sodium current in SNS-null and wild-type small primary sensory neurons. J Neurosci. 1999;19(24):RC43. doi: 10.1523/JNEUROSCI.19-24-j0001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.