Abstract
In this work, we report the development of a general strategy for enhancing the efficiency of target capture in immunoassays, using a bifunctional fusion protein construct which incorporates a substrate-anchoring moiety for the high-abundance immobilization of an antigen-binding domain. This approach was informed by the development of a pseudo first-order rate constant model, and tested in a paper-based assay format using a fusion construct consisting of an rcSso7d binding module and a cellulose-binding domain. These rcSso7d-CBD fusion proteins were solubly expressed and purified from bacteria in high molar yields, and enable oriented, high-density adsorption of the rcSso7d binding species to unmodified cellulose within a 30-second incubation period. These findings were validated using two distinct, antigen-specific rcSso7d variants, which were isolated from a yeast surface display library via flow cytometry. Up to 1.6 micromoles of rcSso7d-CBD was found to adsorb per gram of cellulose, yielding a local binder concentration of up to 760 μM within the resulting active material. At this molar abundance, target antigens are captured from solution with nearly 100% efficiency, maximizing the attainable sensitivity for any given diagnostic system.
Keywords: Sso7d, cellulose-binding domain, paper-based diagnostics, functional material, antigen capture, affinity protein
Graphical abstract
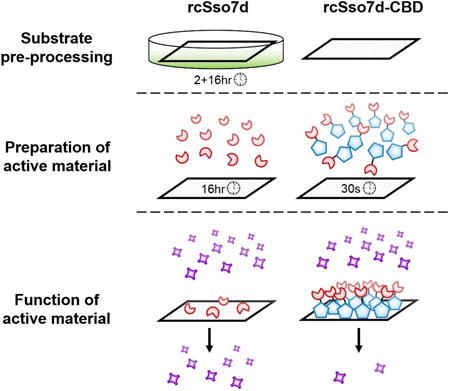
1. Introduction
Under the antigen-dilute conditions of a typical diagnostic assay, every target molecule that goes uncaptured represents a loss in potential binding signal, and directly diminishes diagnostic sensitivity. (Kelley et al., 2014; Rissin et al., 2013) Given that the signal-to-noise ratio for an immunoassay is directly proportional to the molar abundance of bound analyte, general strategies must be developed to enhance the efficiency of target capture, in order to boost the maximum achievable sensitivity for any given diagnostic platform.
According to the law of mass action, the stoichiometry and kinetics of a target-binding interaction can be favorably influenced via three general approaches: i) increasing the molar abundance and concentration of the soluble antigen, ii) increasing the abundance and concentration of its surface-immobilized binding partner, or iii) enhancing the affinity of this binding interaction under relevant assay conditions. (Esteban Fernández de Ávila et al., 2013) These guiding principles have been borne out in numerous experimental studies, which have demonstrated the advantageous impact of antigen pre-concentration (Ahmed et al., 2016; Giri et al., 2016; Tang et al., 2016) and enhanced binding affinity (Kaastrup et al., 2013; Ricci et al., 2016) upon target capture and assay sensitivity.
Previous studies have also explored the impact of the abundance of the surface-immobilized binding species upon the sensitivity of analyte detection. (Esteban Fernández de Ávila et al., 2013; Parsa et al., 2008; Peluso et al., 2003) However, while these studies confirmed improved diagnostic sensitivity for assays conducted at a higher abundance of immobilized binder, only modest densities of surface-bound species (e.g. picomoles/cm2) were achieved, and the target analyte was in molar excess of the immobilized binders. The implications of operating within the true target-depletion regime, wherein the binding protein is present in significant molar excess of the analyte, have not been thoroughly investigated.
In order to explore the consequences of enhanced binder immobilization upon target capture efficiency, we have developed a simplified binding model which employs a pseudo first-order rate constant (PFORC) to describe the antigen-binding interaction. This PFORC assumes a significant molar excess of the immobilized binding species, such that the abundance of available binder is effectively undiminished by the capture of soluble antigen. These modeling results indicate that within this high-abundance adsorption regime, the target antigen is rapidly and efficiently depleted from solution. Furthermore, this model suggests that at a large molar excess, the affinity of the immobilized binder has little effect upon the capture efficiency – so long as the local concentration of surface-bound species is at least ten-fold higher than the dissociation constant (KD), the binding reaction will proceed to near-completion. Thus, if this molar excess can be achieved, protein engineering efforts need not be invested into the affinity maturation of selected binders – depending on the specific immobilized abundance, a modest binding affinity in the high nanomolar or even low micromolar range could be sufficient for efficient target capture.
The predictions of this PFORC model were also validated experimentally, using a bifunctional fusion protein construct that combines a Type 3a cellulose-binding domain (CBD) with a modular binding scaffold based on the thermostable rcSso7d protein (Miller et al., 2016; Traxlmayr et al., 2016) (Figure 1). Previous studies have demonstrated the use of CBD fusion proteins for the bio-functionalization of cellulose substrates, in applications including protein purification, (Sugimoto et al., 2012; Tomme et al., 1998) textile manufacturing, (Levy and Shoseyov, 2002) and immunoassay development. (Dai et al., 2016; Holstein et al., 2016; Hussack et al., 2009; Kim et al., 2013) These studies have indicated that this CBD species adsorbs to cellulose in molar quantities which, in a standard diagnostic context, would yield a significant excess of immobilized protein relative to the soluble target. (Dai et al., 2016; Li et al., 2016) Our experimental studies have confirmed that the use of this substrate-anchoring domain in a paper-based assay format permits the rapid, oriented adsorption of the rcSso7d binding species on un-modified Whatman No. 1 chromatography paper, in sufficient abundance to completely capture up to 0.5 nanomoles of antigen from solution (e.g. depleting all antigen from 10 μL of a 50 μM solution). For the sample volumes and antigen concentrations observed in typical diagnostic assays (i.e. microliters, and concentrations in the picomolar to low nanomolar range), this binder abundance may represent greater than a 1000-fold molar excess relative to the soluble target. The high local concentration of this immobilized CBD fusion within the paper substrate (∼760 μM) also increases the rate of target capture, biasing the binding equilibrium toward the rapid depletion of the dilute antigen from solution.
Figure 1.
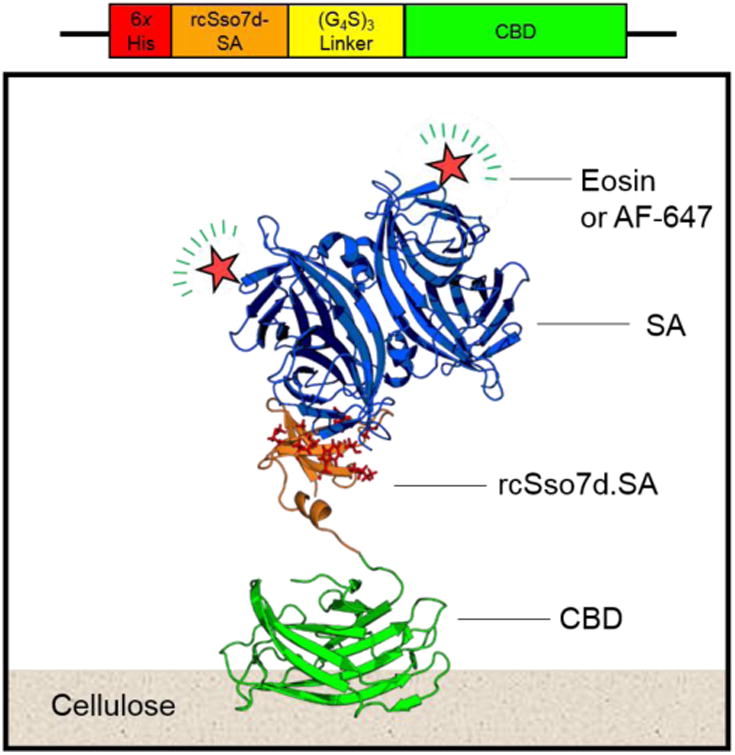
Schematic representation of the rcSso7d.SA-CBD genetic construct, and the relevant binding complexes for this immunoassay format. CBD: cellulose binding domain; rcSso7d: reduced charge protein Sso7d from Sulfolobus solfataricus; SA: streptavidin; AF-647: Alexa Fluor 647. PDB Structures: 4JO5 (CBD); (Yaniv et al., 2013) 1SSO (Sso7d); (Baumann et al., 1994) 1MEP (SA). (Hyre et al., 2006)
This surface-anchoring approach can be adapted to any substrate for which there is a known anchoring moiety, so long as the given bulk material features sufficient accessible surface area for the high-abundance immobilization of the binding construct, and is also structured so as to facilitate efficient transport of the antigen to the surface. For instance, solid-binding peptides have been used to immobilize biomolecules to a variety of substrates, ranging from metals and metal oxides to plastics, minerals, semiconductors, and carbon-based materials. (Care et al., 2017, 2015; Kumada, 2014; Seker and Demir, 2011) This strategy can also be extended to any immobilized target or class of binding domain which can interact with or be expressed as a genetic fusion to this anchoring moiety (e.g. antigens, antibodies and antibody fragments, non-antibody binding scaffolds, DNA oligonucleotides and aptamers, etc). (Holstein et al., 2016; Hussack et al., 2009; Rosa et al., 2014)
Lastly, this system has also been shown to be generalizable across varying soluble targets – the enhanced capture efficiency of the rcSso7d-CBD fusion was confirmed using two independent rcSso7d variants raised against the 52.8-kDa model antigen streptavidin, and a 33.1-kDa urine-based biomarker of active tuberculosis, Rv1656. (Napolitano et al., 2008) Though these distinct binding variants differ in target-specific affinity by a factor of ∼30, they perform with similar efficiency in the rcSso7d-CBD format, serving to validate the predictions of the PFORC model.
2. Materials and Methods
2.1 Yeast surface display selection and characterization of rcSso7d-based binding variants
The development and selection of rcSso7d.SA was described in our previous work. (Miller et al., 2016) The Rv1656-binding variant of rcSso7d was selected in similar fashion, from a yeast surface display library based on the reduced-charge Sso7d scaffold (rcSso7d). This yeast library was generated using trinucleotide oligo synthesis and in vivo homologous recombination with the linearized pCTcon2 plasmid. (Traxlmayr et al., 2016)
Both highly-avid magnetic bead sorting (Ackerman et al., 2009) (MBS) and fluorescence-activated cell sorting (FACS) (Chao et al., 2006) were used to select binders against a biotinylated Rv1656 target (Figure S2). The sorting stringency was increased over five rounds of FACS-based library screening, after which a sub-library was sequenced and rcSso7d.Rv1656 was selected for further characterization. The affinity of this species was assessed in a yeast surface display format, via a soluble titration of biotinylated Rv1656 against the displayed rcSso7d variant.
2.2 Recombinant protein expression, purification, and characterization
The genes for rcSso7d.SA and rcSso7d.Rv1656 were both cloned from the pCTcon2 yeast display plasmid into the pET28b(+) bacterial expression plasmid as previously described. (Miller et al., 2016) The rcSso7d.SA-CBD gene product was generated by Integrated DNA Technologies (IDT; Coralville, IA, USA) via gene synthesis, and traditional PCR cloning was used to integrate the rcSso7d.Rv1656 module into this rcSso7d-CBD fusion construct (see SI). All gene products were modified with an N-terminal hexahistidine tag for purification via immobilized metal affinity chromatography (IMAC). The pET14b-Rv1656 plasmid was provided by the lab of Dr. Antonio Campos-Neto at the Forsyth Institute. (Napolitano et al., 2008)
The heterologous expression of all protein species was conducted in a BL21(DE3) strain of E. coli, and induced via the addition of 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). Induced cells were lysed by ultrasonification, and the recombinant product was purified from the clarified lysate via IMAC. A 3-kDa Amicon Ultracentrifuge Filter cassette was used to buffer exchange the 9.24-kDa rcSso7d monomer 1,000-fold into the resuspension buffer (40 mM sodium acetate, pH 5.5). Products featuring a CBD fusion partner were buffer-exchanged using a 3.5kDa MWCO Slide-A-Lyzer Dialysis Cassette (Thermo Fisher Scientific, Waltham, MA, USA), in order to prevent the adsorption of the CBD fusion products to the cellulose acetate membrane of the spin filters.
Rv1656 was expressed in similar fashion using BL21(DE3) E. coli, and was resuspended in 50 mM HEPES buffer (pH 8.0) using a 10kDa MWCO Slide-A-Lyzer Dialysis Cassette. Purified Rv1656 was biotinylated using the EZ-Link Sulfo-NHS-LC-Biotin No-Weigh Format labeling kit from Thermo Fisher Scientific, and desalted using Micro G-25 Spin Columns from Santa Cruz Biotech (Dallas, TX, USA).
The concentrations of all purified proteins were assessed using a bicinchoninic acid (BCA) assay, and all standards and purified samples were tested in triplicate for greater accuracy. Protein purity was assessed using a freshly cast 15% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel, stained using Coomassie Brilliant Blue G-250.
2.3 Fabrication and testing of biofunctional cellulose test zones
Unmodified Whatman No. 1 chromatography paper was used as shipped for the immobilization of rcSso7d-CBD fusion proteins. In order to enable the covalent immobilization of rcSso7d variants lacking a CBD fusion partner, Whatman No. 1 chromatography paper was functionalized in 30 mM sodium metaperiodate solution as previously described. (Miller et al., 2016) This oxidized, aldehyde-functionalized cellulose was stored under vacuum in a desiccator until use, whereas non-functionalized paper was stored under ambient conditions. As previously described, a solid ink printer was used to produce test zone arrays, and this printed wax was melted through the paper thickness (0.18 mm) to yield test zones with an average area of 2.5 ± 0.1 mm2 (unless otherwise noted).
Stock solutions of purified rcSso7d and rcSso7d-CBD variants were diluted to the desired concentrations in resuspension buffer. For bare rcSso7d species, glycerol was also added to the solution at a final volumetric concentration of 10% in order to prevent evaporation during the extended initial incubation. Unless otherwise stated, all binding protein solutions were prepared at a final concentration of 30 μM. Negative controls for functionalized paper samples consisted of test zones contacted with 1 mg/mL bovine serum albumin (BSA). Bare paper test zones were used as the negative control for unmodified paper samples.
Functionalized test zones were modified with the bare rcSso7d variants, washed, and neutralized in Tris-buffered saline as described in our previous work. Both rcSso7d-CBD variants were contacted with unmodified paper in 6 μL aliquots for at least thirty seconds, and then washed twice in 20 μL of 1x phosphate-buffered saline (PBS; pH 7.4).
Protein-coated test zones were then contacted with 10 μL of the relevant antigen, diluted to the desired concentration in sterile-filtered 1x PBS/1% w/v BSA. rcSso7d.SA and rcSso7d.SA-CBD species were contacted with either streptavidin eosin (SA-E), prepared as previously described, (Miller et al., 2016) or streptavidin Alexa Fluor 647 (SA-AF647) sourced from Invitrogen (Carlsbad, CA, USA). rcSso7d.Rv1656-CBD was contacted with biotinylated recombinant Rv1656. All test zones were incubated with antigen solution for 30 minutes at room temperature, after which they were washed twice with PBS. Negative controls were incubated in PBS in the absence of soluble antigen during this period.
Assays incorporating rcSso7d.Rv1656 and rcSso7d.Rv1656-CBD were then subjected to an additional 30-minute incubation with SA-E/SA-AF647 at a concentration of 256 nM. SA-E samples were prepared in a citric acid-sodium phosphate buffer system (50mM citric acid, 90mM Na2HPO4, pH 4.5) containing 1% BSA, and washed in the same acidic buffer lacking BSA, in order to reduce non-specific binding of rcSso7d.Rv1656-CBD to the eosin reagent (Figure S8). Developed samples were blotted dry and stored in the dark in a freezer box until needed for fluorescence microscopy imaging.
2.4 Fluorescence microscopy
All samples were imaged as previously described (Miller et al., 2016), using an Olympus 1X81 Microscope. Unless otherwise noted, all samples developed with SA-E were exposed for 1000 ms using a Semrock TxRed-4040C filter set. Samples developed with SA-AF647 were exposed for either 80 ms or 100 ms (as noted) using a Semrock Cy5-4040C filter set. The ImageJ Auto Threshold function (Default algorithm) was used to identify the bounds of each sample zone, and the mean fluorescence intensity (MFI) of each sample was calculated by averaging the brightness of all pixels within the thresholded area. Four technical replicates were prepared for all experimental conditions, and the resultant MFI values were averaged for all replicates. Error bars represent one standard deviation from this mean intensity value.
2.5 Quantification of surface-immobilized CBD fusion proteins
A micro BCA assay (Thermo Fisher Scientific) was used to determine the immobilized surface density of the engineered rcSso7d.SA-CBD fusion protein on non-functionalized Whatman No. 1 chromatography paper. A series of standards was prepared by contacting test zones with known masses of rcSso7d.SA-CBD and allowing these solutions to evaporate in a vacuum chamber at room temperature for 30 minutes, yielding complete protein adsorption to the cellulosic substrate. Experimental samples were generated by applying a series of known soluble rcSso7d.SA-CBD concentrations to the test zones, followed by a PBS wash step.
All samples were excised from the test strips and deposited into the wells of a 96-well plate pre-filled with 150 μL of 40mM sodium acetate (pH 5.5). These test zones were vigorously stirred with clean pipette tips, and 150 μL of Working Reagent was then added to each sample well. The plate was incubated at 37°C for two hours, and after removing the paper test zones (wringing any entrained fluid back into the sample well), the absorbance at 562 nm was quantified for all samples.
The response curve for the evaporated standards was fit to a second-order polynomial, and this standard curve was used to determine the effective quantity of rcSso7d.SA-CBD immobilized on the washed samples. Proportional rcSso7d.SA-CBD retention was calculated by comparing these experimentally determined quantities to the known protein masses applied to the surface. In order to quantify the binding capacity of the cellulose substrate under these processing conditions, the density of Whatman No. 1 chromatography paper was measured in triplicate, and was found to be 0.088 ± 0.00016 mg/mm2. The area of the test zones was measured by determining the pixel density at 40× magnification (0.287 megapixels/mm2), and measuring the thresholded test zone area in ImageJ. For this micro BCA experiment, the average area of the test zones was found to be mm2, corresponding to a cellulose mass of 0.32 ± 0.021 mg.
3. Results and Discussion
3.1 Pseudo first-order rate constant model
In order to explore the effects of operating within the antigen-limited binding regime, a monovalent binding model based on the principles of mass-action kinetics was developed. This binding system can be described mathematically by a simple first-order differential equation:
Here, [L] and [R] represent the volumetric molar concentrations of free ligand and free receptor, respectively, and [LR] represents the concentration of the bound complex. By employing the law of molar conservation (e.g. [L] [L]0 – [LR] [R] [R]0 – [LR]), this monovalent binding system can be solved analytically (derivation in SI) to yield the expression:
However, when operating in the antigen-depletion regime, this relation can be simplified by noting that antigen capture does not significantly diminish the pool of free receptor, such that a constant concentration of available binding species can be assumed. This permits the use of a pseudo first-order rate constant (PFORC; units: s-1) which incorporates the initial receptor concentration:
By applying this PFORC in the first-order differential equation describing this binding system (derivation also in SI), we arrive at the following, more compact expression for the proportion of bound antigen (relative to the equilibrium value). Notably, this relation no longer depends upon the initial concentration of the soluble ligand, given that the receptor concentration alone determines the profile of the approach to binding equilibrium.
The binding regime affects not only the thermodynamics and stoichiometry of antigen capture, but also the binding kinetics. These basic models also enable the calculation of the time required for the system to reach 99% of equilibrium binding. The exact analytical expression for this value is:
In contrast, the PFORC model permits the calculation of a simplified, effective rate of reaction (kobs = koff + kon[R]), which can be incorporated into the following relation to evaluate t99:
By varying the initial receptor and ligand concentrations, proportional ligand capture at equilibrium and t99 values can be plotted for both the analytical solution and PFORC approximation (Figure 2), permitting direct comparison of the models and establishing bounds for the validity of the PFORC approach. We observe that the PFORC model is highly accurate throughout much of the regime where the immobilized receptor is in molar excess to the soluble ligand.
Figure 2.
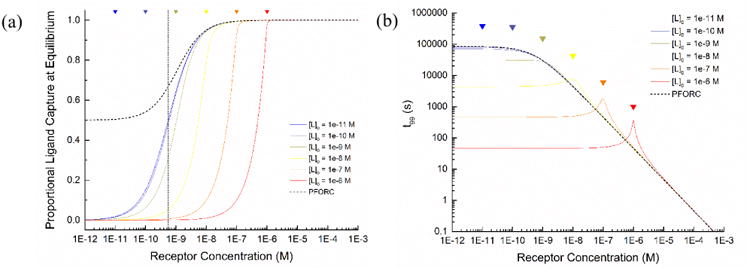
Comparison of analytical solution and PFORC model results. a) Ligand capture efficiency at equilibrium for the analytical and PFORC models. Curves represent the proportion of free ligand that is bound at equilibrium for varying initial concentrations of ligand and receptor. b) Calculated time required to achieve 99% of equilibrium binding in the analytical and PFORC models. All plots were generated using a KD of 5.5 × 10−10 M. Colored triangles denote the points where the receptor concentration is equivalent to the associated ligand concentration, to highlight the local changes near these values.
In fact, the PFORC model only appreciably deviates from the analytical solution as the initial receptor concentration either i) approaches the initial concentration of the free ligand, or ii) nears the dissociation constant of the binding pair, whichever value is greater (Figure S1). Generally, the proportional deviation between the analytical solution and the PFORC model only becomes significant for a ligand concentration or a KD within one order of magnitude of the local concentration of the immobilized receptor.
Note that this treatment assumes that all species are present in soluble form in the same volume, thereby establishing a direct link between molar concentration and molar abundance. In the context of a heterogeneous assay, the average local concentration of the immobilized binder within the test zone volume does not directly reflect its molar abundance relative to the soluble target. Thus we must consider the molar abundance of the immobilized species instead of its local concentration, and this quantity must be an order of magnitude greater than the abundance of the soluble target in order to yield antigen depletion as described by the PFORC model.
3.2 Selection and characterization of rcSso7d binding variants
In order to test the predictions of this basic binding model, we developed two distinct binding variants based on the thermostable rcSso7d scaffold. Both rcSso7d.SA and rcSso7d.Rv1656 were selected from a yeast surface display library of high initial diversity (∼1.4 billion library members) via magnetic bead sorting and flow cytometry. The amino acid sequence of these selected binding variants can be seen below (Table 1). As reported by Traxlmayr et al (2016), strong enrichment of the aromatic residues tyrosine and tryptophan was observed. This may serve to impart greater topological diversity and electron density upon the planar rcSso7d binding face, facilitating strong, conformal binding to the target antigen.
Table 1. Primary protein structure of selected rcSso7d binders.
| Protein Species | Primary Structure (N → C) (Variable AA residues) | Shorthand Tag |
|---|---|---|
| rcSso7d.SA | MATVKFTYQGEEKQVDISKIKIVARDGQYIDFKYDEGGGAYGYGWVSEKDAPKELLQMLEKQ | IADYDKYYW |
| rcSso7d. Rv1656 | MATVKFTYQGEEKQVDISKIKWVRRYGQYIGFSYDEGGGAWGKGYVSEKDAPKELLQMLEKQ | WRYYGSWKY |
The dissociation constants of the rcSso7d.SA and rcSso7d.Rv1656 modules were both measured in the yeast surface display format by titrating soluble, biotinylated antigen against monoclonal yeast populations expressing these binding species as surface-bound fusion proteins. The affinity of rcSso7d.SA was previously reported to be 556 ± 136 pM, (Miller et al., 2016) and the affinity of rcSso7d.Rv1656 was found to be 15.1 ± 7.0 nM (Figure S3).
In order to incorporate these binding proteins into the rcSso7d-CBD format, the gene encoding the type 3 cellulose binding domain of the CipA protein from Clostridium thermocellum (Genbank: HF912725.1, residues 364-522) was synthesized by IDT as a C-terminal fusion partner to the rcSso7d.SA species. This particular cellulose-binding domain was chosen for its high immobilization density and demonstrated activity in an immunoassay format, (Dai et al., 2016; Holstein et al., 2016; Hussack et al., 2009) as well for its thermal (McBee, 1954) and chemical stability. (Berdichevsky et al., 1999) The two fusion partners are joined by a flexible (G4S)3-linker sequence, and an internal BamHI site is included at the C-terminal end of the rcSso7d gene.
These rcSso7d-CBD constructs were expressed in BL21(DE3) E. coli and purified via a reusable IMAC column, yielding a product of electrophoretic purity within a single purification step (Figure S4). Protein concentration was quantified via a BCA assay, and the protein yield was determined to range from 131.4 mg/Lculture (14.28 mg/g wet cell mass (WCM)) for rcSso7d.SA-CBD to 105.5 mg/Lculture (8.55 mg/g WCM) for rcSso7d.Rv1656-CBD. Given a calculated cost basis for a single bacterial production run of $18.02 (see SI), and a conservative per-test usage of 5 micrograms, a single 36-hour production run at a 1000-mL scale can produce enough material for approximately 20,000 assays, at a cost of $0.0009/device. These favorable bio-manufacturing economics enable the high-throughput production of these paper-based assays, at a price point that is well-suited for low-cost biomedical applications in resource-limited settings.
3.3 Characterization of rcSso7d.SA-CBD cellulose-binding activity
In order to assess the capture efficiency of bioactive cellulose functionalized with the rcSso7d.SA-CBD fusion species, these binding proteins were immobilized in hydrophilic test zones and subsequently contacted with the soluble antigen, forming an immunocomplex. By using these half-sandwich assay formats, we are able to decouple the typical immunoassay binding steps, allowing each molecular interaction to be evaluated in isolation and engineered for optimal performance prior to re-integration into a full diagnostic format.
Fluorescence microscopy imaging of developed test zones indicates that the cellulose-binding domain strongly binds to unmodified Whatman No. 1 chromatography paper in high abundance (Figure 3), removing the need for substrate pre-processing steps. This represents a significant process improvement in the production of these paper-based assays, given that typical procedures require functionalization steps for the activation of inert cellulosic substrates, in order to immobilize diagnostic binding proteins in greater abundance. (Credou and Berthelot, 2014; Nery and Kubota, 2016; Shen et al., 2016; Yu et al., 2012; Zhao et al., 2016) These chemical pre-processing methods limit production throughput, and require efficient surface passivation steps following binder immobilization in order to prevent the non-specific adsorption of patient proteins and free detection reagents. (Vuoriluoto et al., 2016; Zhu et al., 2014) Additionally, stochastic chemical conjugation methods result in the non-oriented immobilization of the binding species, which can reduce the solvent accessibility of the target-binding paratope and result in an inactive sub-population of immobilized binder. (Song et al., 2012)
Figure 3.
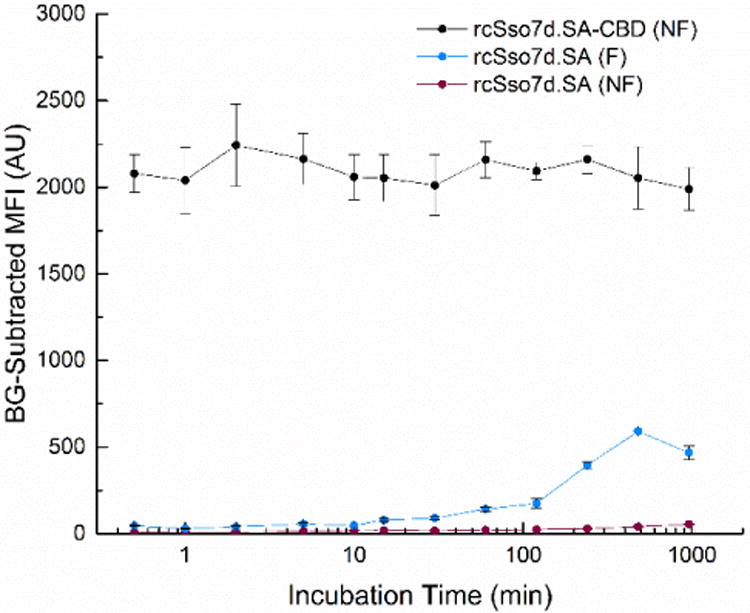
Time course of primary incubation. rcSso7d.SA-CBD was contacted with non-functionalized paper (NF) and rcSso7d.SA was contacted with both functionalized (F) and non-functionalized (NF) paper for periods of time ranging from 30 seconds to 16 hours, at soluble concentrations of 30 μM (180 picomoles of applied binder). Following washing and substrate neutralization, these samples were subsequently treated with 10 μL of SA-AF647 at a soluble concentration of 256 nM (2.56 picomoles of target). All samples were imaged in the Cy5 channel using an exposure time of 80 ms, and background-subtracted using the relevant negative control. Error bars represent the standard deviation of four independent replicates.
Furthermore, given the rate-dependent formation of the imine bond, an extended primary incubation is typically required in order for this covalent immobilization reaction to proceed to completion. Even following this incubation period in the functionalized paper format, this time-dependent process yielded sub-optimal antigen capture for the bare rcSso7d species (Figure 3).
In contrast, unmodified chromatography paper requires no special pre-treatment, can be stored under ambient conditions, and yields minimal nonspecific protein adsorption both prior to and during immunoassay development. The rcSso7d-CBD fusion also yields oriented display of the antigen-binding rcSso7d module, ensuring maximal paratope accessibility and surface activity. Finally, the CBD fusion rapidly binds to the cellulose substrate in high abundance. Regardless of whether the CBD fusion was contacted with the surface for a primary incubation period of 16 hours or 30 seconds, the binding signal was observed to be roughly equivalent, and significantly greater than that of the bare rcSso7d species (Figure 3). This drastically reduces the amount of time required to proceed from raw cellulose substrate to fully functional assays, from two days of processing time down to roughly ten minutes.
3.4 Characterization of assay sensitivity using cellulose-immobilized rcSso7d.SA-CBD
The greater surface density of the rcSso7d.SA-CBD species also results in the onset of discernible binding signal at lower concentrations of soluble antigen relative to the bare rcSso7d.SA species. Standard definitions of assay sensitivity establish a reliable detection threshold at three standard deviations above the average signal of the negative controls. By comparing the binding curves obtained by treating these species with a serial dilution of SA-E (Figure 4a), we find a conservative limit of detection (LOD) of 8.2 nM (IBG = 324.3 AU; σ = 41.7 AU) for the bare rcSso7d species, and 2.56 nM (IBG = 150.1 AU; σ = 5.9 AU) for rcSso7d.SA-CBD. The background signal due to non-specific SA-E binding was also lower for unmodified cellulose, yielding better discrimination of genuine binding signal from random fluctuations near the noise threshold. The binding curve for rcSso7d.SA-CBD is also seen to continue to rise at high nanomolar concentrations of the soluble antigen, whereas the binding signal appears to saturate for the rcSso7d.SA species. This suggests a significantly higher degree of rcSso7d.SA-CBD surface immobilization, which has implications for the more rapid and efficient capture of the target from solution.
Figure 4.
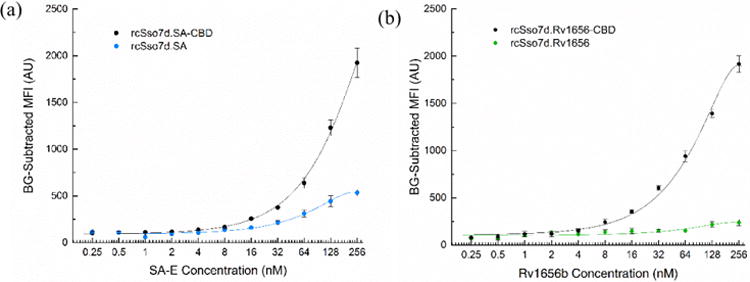
Comparison of antigen titration curves for a) rcSso7d.SA/rcSso7d.SA-CBD and b) rcSso7d.Rv1656/rcSso7d.Rv1656-CBD. rcSso7d and rcSso7d-CBD species were contacted with their associated substrates (functionalized and non-functionalized cellulose, respectively) for standard incubation times at a soluble concentration of 20 μM. Sample sets were treated with a serial dilution of a) SA-E or b) Rv1656b, at concentrations ranging from 256 nM to 0.25 nM. Samples contacted with Rv1656b were subsequently contacted with SA-E at a concentration of 256 nM. Samples were imaged in the Texas Red channel using an exposure time of 1000 ms. Datasets were fit with a second-order polynomial (rcSso7d.SA: −0.008362x2 + 3.851x + 100.0, r2 = 0.9904; rcSso7d.SA-CBD: −0.01059x2 + 9.899x + 100.0, r2 = 0.9986; rcSso7d.Rv1656: −0.002774x2 + 1.229x + 100.0, r2 = 0.8271; rcSso7d.Rv1656-CBD: −0.02791x2 +14.16x + 100.0, r2 = 0.9961). The baseline for these datasets was adjusted to an arbitrary value of 100AU in order to enable the comparison of signal onset. Error bars represent the standard deviation of four independent replicates.
These findings were also validated in a second, orthogonal binding system, using the rcSso7d.Rv1656 binding module (Figure 4b). In this system, too, a drastically improved binding response was observed with the rcSso7d-CBD fusion species, both in terms of its capture efficiency at high antigen concentrations, and its limit of detection (rcSso7d.Rv1656-CBD: LOD = 3.1 nM; IBG = 468.8 AU; σ = 17.3 AU; rcSso7d.Rv1656: LOD: 48.3 nM; IBG = 350.1 AU; σ = 32.2 AU). The background signal for the rcSso7d.Rv1656 species is significantly higher on unmodified cellulose, due to a limited degree of nonspecific binding to the aromatic eosin species (see Figure S8).
It should be noted that the effect of the 30-fold difference in affinity between these two binders can be observed qualitatively in the bare rcSso7d format. However, upon integration of these distinct binding species into the rcSso7d-CBD format, the binding curves are much more similar, suggesting that at higher immobilization densities, the binding affinity has little impact upon the ultimate capture efficiency.
3.5 Identification of the antigen-binding regime
While these observations demonstrate the benefits of incorporating the CBD fusion partner, it is necessary to characterize the binding regime directly in order to confidently validate the predictions of the PFORC model. Given the clear improvement in capture efficiency observed with the rcSso7d.SA-CBD species, we sought to determine whether this antigen binding could be further enhanced by contacting the cellulose substrate with greater molar quantities of rcSso7d.SA-CBD (Figure S6). A series of soluble rcSso7d.SA-CBD concentrations, ranging from 0.5 mg/mL to 7 mg/mL (18.3 μM to 256 μM), was applied to the paper test zones. These sample sets were incubated with a serial dilution of SA-E, ranging from 256 nM to 0.25 nM (Figure 5a).
Figure 5.
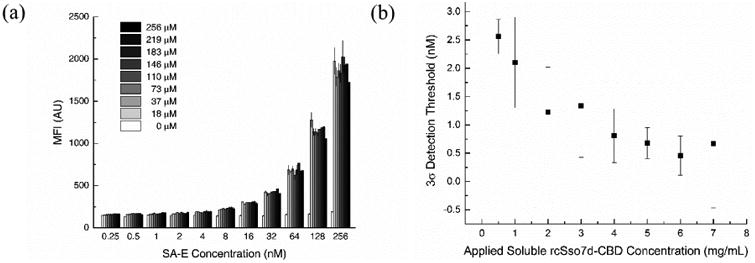
a) SA-E titration curves for various applied soluble concentrations of rcSso7d.SA-CBD. Sets of non-functionalized cellulose test zones were prepared with a range of soluble rcSso7d.SA-CBD concentrations. All test zones were contacted for 30s and washed, and were then treated for 30 minutes with a serial dilution of SA-E ranging from 256 nM to 0.25 nM. Samples were imaged in the Texas Red channel using an exposure time of 1000 ms. Datasets were fit with a second-order polynomial. Error bars represent the standard deviation of four independent replicates. b) Limits of detection for various applied concentrations of rcSso7d.SA-CBD. The measured MFI values for all negative control samples (with [SA-E] ranging from 256 nM to 0.25 nM) were averaged to calculate a conservative three-sigma detection threshold of 167.8 AU. Second-order polynomial lines of best fit were used to calculate the antigen concentration corresponding to this LOD for each sample set treated with a different applied rcSso7d.SA-CBD concentration. Second-order polynomial lines of best fit were also used to plot the upper and lower bounds of each data point (determined by the standard deviation), and these bounding trendlines were used to generate bounds on the limits of detection, represented by the error bars.
The resulting binding curves for each antigen titration are exceptionally regular, yielding an average r2 value of 0.9994 when fit with a second-order polynomial. These curves generally overlap, but while no large-scale trends are immediately apparent in these clustered data sets, we find that higher soluble concentrations of applied binder do yield greater capture efficiency at antigen concentrations in the low nanomolar range. Using the negative control dataset from all SA-E concentrations applied to bare cellulose (IBG = 150.1 AU; AU), we calculate a conservative three-sigma threshold MFI of Ith = 167.8 AU. Applying the second-order polynomial fit equations for each sample set, we find that as the applied concentration of rcSso7d.SA-CBD increases, the minimum detectable antigen concentration decreases (Figure 5b). This finding suggests that additional rcSso7d.SA-CBD binds to the cellulose substrate at higher applied concentrations, and indicates that this greater surface coverage yields improved capture efficiency at dilute antigen concentrations. Given that significantly higher MFI values are observed for more concentrated antigen solutions, this improvement in capture efficiency at low antigen concentrations is likely due to enhanced binding kinetics, rather than due to insufficient molar quantities of the immobilized binder at lower applied concentrations of rcSso7d.SA-CBD.
The general overlap of the rcSso7d.SA-CBD binding curves indicates that this binding system is operating in one of two regimes: either a) the assay is in fact within the antigen-depletion regime, such that there is no additional target to capture at a given soluble antigen concentration, or b) the cellulose substrate is saturated with immobilized rcSso7d.SA-CBD, preventing the adsorption of any additional binder. While these preliminary results suggest that the substrate is not saturated (namely the enhanced capture efficiency observed at dilute antigen concentrations with increased quantities of applied rcSso7d.SA-CBD), we sought to confirm this finding experimentally by directly quantifying the abundance of the immobilized rcSso7d.SA-CBD species on the cellulose substrate.
3.6 Direct quantification of rcSso7d-CBD surface abundance
In order to further verify the relevant binding regime for this binding system, a micro BCA assay was used to quantify the immobilized surface concentration of the rcSso7d.SA-CBD species. Known masses of rcSso7d.SA-CBD were evaporated onto test zones in order to generate a standard curve that was directly comparable to the washed experimental samples. A highly regular response curve was observed for all standard samples (r2 = 0.9978), and all washed samples fell within the bounds of this standard curve (Figure S7). A clear monotonic increase is observed for these experimental samples, indicating that the substrate is far from saturation under the binding conditions used at the standard concentration of 30 μM (Figure 6).
Figure 6.
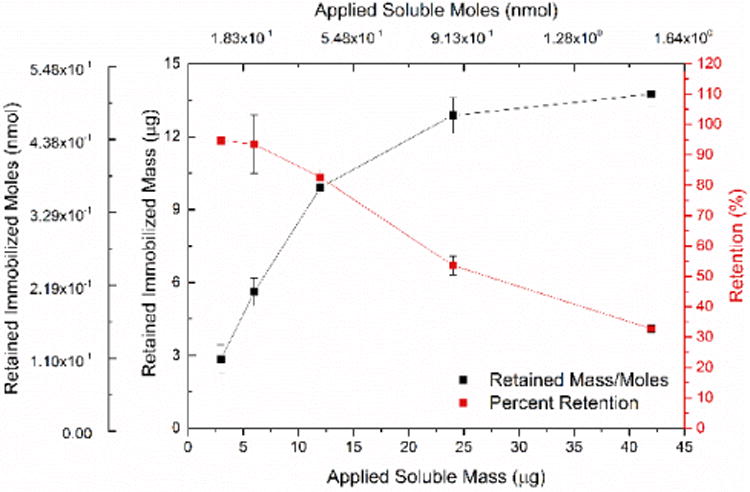
Micro BCA assay data indicating the adsorption efficiency of rcSso7d.SA-CBD on non-functionalized cellulose. A standard curve of known masses of adsorbed rcSso7d.SA-CBD was used to quantify the immobilization density of rcSso7d.SA-CBD on washed samples. Experimentally determined immobilized masses, assessed via this standard curve, are plotted against the known quantity of applied rcSso7d.SA-CBD, as is the percent retention. Error bars represent the standard deviation of four independent replicates.
This serves to confirm that antigen depletion is responsible for the similar response curves observed at varying soluble rcSso7d-CBD concentrations. The signal development observed for the washed samples indicates a molar abundance of rcSso7d-CBD that ranges from 0.1-0.5 nmol/test zone. Given an average test zone mass of 0.32 ± 0.021 mg, this equates to a surface density that varies from 0.32-1.56 μmol of rcSso7d-CBD/g cellulose, which agrees with previously reported values. (Dai et al., 2016; Li et al., 2016)
It should be noted that the efficiency of rcSso7d.SA-CBD immobilization decreases as higher soluble concentrations of protein are applied, indicating that substrate saturation can in fact occur at high immobilized surface density. Whereas the application of 0.1-0.2 nmol of rcSso7d-CBD to the surface results in an immobilized yield of ∼90%, this efficiency drops to ∼30% at an application of 1.5 nmol. Though higher densities of immobilized binder do allow enhanced capture efficiency at low antigen concentrations, these diminishing returns will necessarily impose practical and economic constraints on how near to saturation the surface coverage can be driven.
At a standard molar application of 180 picomoles (corresponding to a soluble concentration of 30 μM), the observed immobilization efficiency of 90% yields approximately 162 picomoles immobilized on the test substrate (corresponding to an average local concentration of ∼360 μM). At this molar abundance, immobilized rcSso7d.SA-CBD is present in 63.3-fold molar excess relative to the soluble antigen when contacted with a 10-μL sample at the highest titration concentration (256 nM). Under these conditions, the PFORC model predicts that rapid, complete depletion of the soluble ligand will occur (Figure S9). This approximation will remain valid for all dissociation constants and soluble target concentrations below 1.62 μM (for a 10 μL sample volume).
Finally, in order to directly test this prediction experimentally, the flow-through was collected following a 30-minute incubation of a 256 nM solution of SA-AF647 (10 μL) on rcSso7d.SA-CBD-coated test zones. This flow-through was applied directly to a second set of test zones coated with rcSso7d.SA-CBD, and following an additional 30-minute incubation, these sample sets were washed and imaged in the Cy5 channel. By using a standard curve of known concentrations of SA-AF647 applied to rcSso7d.SA-CBD-based assays (data not shown), the resultant fluorescence measurements can be correlated with their associated antigen concentration (Figure S10). These results indicate that following the initial depletion of SA-AF647 from a 256 nM solution, the concentration of the subsequent solution is 20.7 nM. This represents a capture efficiency of 92.2% during the initial incubation, confirming that rcSso7d.SA-CBD captures the available antigen with high efficiency.
4. Conclusions
In this study, we have considered the effects of operating within the antigen-depletion regime, using a simplified pseudo first-order rate constant model to predict the capture efficiency of immunoassays incorporating a molar excess of an immobilized binder. In order to test these predictions, we developed an rcSso7d-CBD fusion protein which can be readily expressed in bacteria and facilely purified in high molar yields. We have demonstrated that this species rapidly adsorbs to unmodified cellulose, resulting in a molar abundance of the binding species which is sufficient for the near-complete depletion of a soluble antigen from solution. These findings were validated with two distinct binding systems, and serve to validate the predictions of this simple PFORC model.
By operating within this antigen-depletion regime, we are able to maximize the analyte capture efficiency of the bioactive cellulose substrate. Given that this captured target is the biological signal which a diagnostic amplification method must render visually discernible, this enhanced capture efficiency guarantees that the maximum possible signal floor for a given biomarker can be achieved for every sample collected from a heterogeneous patient population. This general strategy, which uses a substrate-anchoring moiety for high-abundance surface adsorption of the target-binding species, is expected to be an applicable method of boosting diagnostic sensitivity in a broad array of assay formats.
Supplementary Material
Cloning protocols, yeast library screening, model derivations, cost-basis calculations, and additional results (PDF)
Highlights.
Molar excess of immobilized species yields complete target depletion from solution
Simple pseudo first-order rate constant model describes depletion regime binding
Surface-anchoring moiety fused to binding protein enhances substrate adsorption
rcSso7d-cellulose-binding domain (CBD) fusion allows efficient target capture
CBD allows rapid fabrication of paper-based assays without substrate preprocessing
Acknowledgments
We thank the staff of the Koch Institute Flow Cytometry core for sharing their expertise.
Funding Sources: Charles E. Reed Faculty Initiatives Fund supported this work. EAM acknowledges a training grant from the National Institutes of Health (2 T32 GM008334 26). DO and YJAM acknowledge support from MIT's UROP Program. This work was also partially supported by Cancer Center Support (Core) Grant P30CCA14051 from the National Cancer Institute.
Abbreviations
- CBD
cellulose-binding domain
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SA-E
streptavidin eosin
- SA-AF647
streptavidin Alexa Fluor 647
- PFORC
pseudo first-order rate constant
Footnotes
Associated Content: Supporting Information: The following files are available free of charge.
Author Contributions: The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ackerman M, Levary D, Tobon G, Hackel B, Orcutt KD, Wittrup KD. Biotechnol Prog. 2009;25:774–783. doi: 10.1002/btpr.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S, Bui MPPN, Abbas A. Biosens Bioelectron. 2016;77:249–263. doi: 10.1016/j.bios.2015.09.038. [DOI] [PubMed] [Google Scholar]
- Baumann H, Knapp S, Lundback T, Ladenstein R, Hard T. Nat Struct Biol. 1994;1:808–819. doi: 10.1038/nsb1194-808. [DOI] [PubMed] [Google Scholar]
- Berdichevsky Y, Lamed R, Frenkel D, Gophna U, Bayer EA, Yaron S, Shoham Y, Benhar I. Protein Expr Purif. 1999;17:249–259. doi: 10.1006/prep.1999.1125. [DOI] [PubMed] [Google Scholar]
- Care A, Bergquist PL, Sunna A. Trends Biotechnol. 2015;33:259–268. doi: 10.1016/j.tibtech.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Care A, Petroll K, Gibson ESY, Bergquist PL, Sunna A. Biotechnol Biofuels. 2017;10:1–16. doi: 10.1186/s13068-017-0715-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Nat Protoc. 2006;1:755–68. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- Credou J, Berthelot T. J Mater Chem B. 2014;2:4767–4788. doi: 10.1039/c4tb00431k. [DOI] [PubMed] [Google Scholar]
- Dai G, Hu J, Zhao X, Wang P. Sensors Actuators B Chem. 2016;238:138–144. [Google Scholar]
- Esteban Fernández de Ávila B, Watkins HM, Pingarrón JM, Plaxco KW, Palleschi G, Ricci F. Anal Chem. 2013;85:6593–6597. doi: 10.1021/ac4012123. [DOI] [PubMed] [Google Scholar]
- Giri B, Pandey B, Neupane B, Ligler FS. TrAC - Trends Anal Chem. 2016;79:326–334. [Google Scholar]
- Holstein CA, Chevalier A, Bennett S, Anderson CE, Keniston K, Olsen C, Li B, Bales B, Moore DR, Fu E, Baker D, Yager P. Anal Bioanal Chem. 2016;408:1335–1346. doi: 10.1007/s00216-015-9052-0. [DOI] [PubMed] [Google Scholar]
- Hussack G, Luo Y, Veldhuis L, Hall JC, Tanha J, MacKenzie R. Sensors. 2009;9:5351–5367. doi: 10.3390/s90705351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyre DE, Le Trong I, Merritt EA, Eccleston JF, Green NM, Stenkamp RE, Stayton PS. Protein Sci. 2006;15:459–467. doi: 10.1110/ps.051970306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaastrup K, Chan L, Sikes HD. Anal Chem. 2013;85:8055–8060. doi: 10.1021/ac4018988. [DOI] [PubMed] [Google Scholar]
- Kelley SO, Mirkin CA, Walt DR, Ismagilov RF, Toner M, Sargent EH. Nat Nanotechnol. 2014;9:969–980. doi: 10.1038/nnano.2014.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HD, Choi SL, Kim H, Sohn JH, Lee SG. Biotechnol Bioprocess Eng. 2013;18:575–580. [Google Scholar]
- Kumada Y. Biochim Biophys Acta - Proteins Proteomics. 2014;1844:1960–1969. doi: 10.1016/j.bbapap.2014.07.007. [DOI] [PubMed] [Google Scholar]
- Levy I, Shoseyov O. Biotechnol Adv. 2002;20:191–213. doi: 10.1016/s0734-9750(02)00006-x. [DOI] [PubMed] [Google Scholar]
- Li M, Yue Y, Zhang ZJ, Wang ZY, Tan TW, Fan LH. Bioconjug Chem. 2016;27:1579–1583. doi: 10.1021/acs.bioconjchem.6b00282. [DOI] [PubMed] [Google Scholar]
- McBee RH. J Bacteriol. 1954;67:505–6. doi: 10.1128/jb.67.4.505-506.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EA, Traxlmayr MW, Shen J, Sikes HD. Mol Syst Des Eng. 2016;1:377–381. doi: 10.1039/C6ME00032K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano DR, Pollock N, Kashino SS, Rodrigues V, Campos-Neto A. Clin Vaccine Immunol. 2008;15:638–43. doi: 10.1128/CVI.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nery EW, Kubota LT. J Pharm Biomed Anal. 2016;117:551–559. doi: 10.1016/j.jpba.2015.08.041. [DOI] [PubMed] [Google Scholar]
- Parsa H, Chin CD, Mongkolwisetwara P, Lee BW, Wang JJ, Sia SK. Lab Chip. 2008;8:2062. doi: 10.1039/b813350f. [DOI] [PubMed] [Google Scholar]
- Peluso P, Wilson DS, Do D, Tran H, Venkatasubbaiah M, Quincy D, Heidecker B, Poindexter K, Tolani N, Phelan M, Witte K, Jung LS, Wagner P, Nock S. Anal Biochem. 2003;312:113–24. doi: 10.1016/s0003-2697(02)00442-6. [DOI] [PubMed] [Google Scholar]
- Ricci F, Vallée-Bélisle A, Simon AJ, Porchetta A, Plaxco KW. Acc Chem Res. 2016;49:1884–1892. doi: 10.1021/acs.accounts.6b00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissin DM, Wilson DH, Duffy DC. The Immunoassay Handbook. Elsevier; 2013. Chapter 2.13: Measurement of Single Protein Molecules Using Digital ELISA; pp. 223–242. [Google Scholar]
- Rosa AMM, Louro AF, Martins SAM, Inácio J, Azevedo AM, Prazeres DMF. Anal Chem. 2014;86:4340–4347. doi: 10.1021/ac5001288. [DOI] [PubMed] [Google Scholar]
- Seker UOS, Demir HV. Molecules. 2011;16:1426–1451. doi: 10.3390/molecules16021426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M, Rusling J, Dixit CK. Methods. 2016;116:95–111. doi: 10.1016/j.ymeth.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HY, Zhou X, Hobley J, Su X. Langmuir. 2012;28:997–1004. doi: 10.1021/la202734f. [DOI] [PubMed] [Google Scholar]
- Sugimoto N, Igarashi K, Samejima M. Protein Expr Purif. 2012;82:290–296. doi: 10.1016/j.pep.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Tang R, Yang H, Choi JR, Gong Y, Hu J, Feng S, Pingguan-Murphy B, Mei Q, Xu F. Talanta. 2016;152:269–276. doi: 10.1016/j.talanta.2016.02.017. [DOI] [PubMed] [Google Scholar]
- Tomme P, Boraston A, McLean B, Kormos J, Creagh AL, Sturch K, Gilkes NR, Haynes CA, Warren RAJ, Kilburn DG. J Chromatogr B Biomed Appl. 1998;715:283–296. doi: 10.1016/s0378-4347(98)00053-x. [DOI] [PubMed] [Google Scholar]
- Traxlmayr MW, Kiefer JD, Srinivas RR, Lobner E, Tisdale AW, Mehta NK, Yang NJ, Tidor B, Wittrup KD. J Biol Chem. 2016;291:22496–22508. doi: 10.1074/jbc.M116.741314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuoriluoto M, Orelma H, Zhu B, Johansson LSS, Rojas OJ. ACS Appl Mater Interfaces. 2016;8:5668–5678. doi: 10.1021/acsami.5b11737. [DOI] [PubMed] [Google Scholar]
- Yaniv O, Morag E, Borovok I, Bayer EA, Lamed R, Frolow F, Shimon LJW. Acta Cryst. 2013;69:733–737. doi: 10.1107/S174430911301614X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A, Shang J, Cheng F, Paik BA, Kaplan J, Andrade RB, Ratner DM. Langmuir. 2012;28:11265–11273. doi: 10.1021/la301661x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Li H, Liu W, Guo Y, Chu W. Biosens Bioelectron. 2016;79:581–588. doi: 10.1016/j.bios.2015.12.099. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Xu X, Brault ND, Keefe AJ, Han X, Deng Y, Xu J, Yu Q, Jiang S. Anal Chem. 2014;86:2871–2875. doi: 10.1021/ac500467c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cloning protocols, yeast library screening, model derivations, cost-basis calculations, and additional results (PDF)


