Abstract
Purpose of review
HIV-1 infection is of global importance, and still incurs substantial morbidity and mortality. Although major pharmacologic advances over the past two decades have resulted in remarkable HIV-1 control, a cure is still forthcoming. One approach to a cure is to exploit natural mechanisms by which the host restricts HIV-1. Herein, we review past and recent discoveries of HIV-1 restriction factors, a diverse set of host proteins that limit HIV-1 replication at multiple levels, including entry, reverse transcription, integration, translation of viral proteins and packaging and release of virions.
Recent findings
Recent studies of intracellular HIV-1 restriction have offered unique molecular insights into HIV-1 replication and biology. Most recently, studies have revealed insights of how restriction factors drive HIV-1 evolution. Although HIV-1 restriction factors only partially control the virus, their importance is underscored by their effect on HIV-1 evolution and adaptation.
Summary
The list of host restriction factors that control HIV-1 infection is likely to expand with future discoveries. A deeper understanding of the molecular mechanisms of regulation by these factors will uncover new targets for therapeutic control of HIV-1 infection.
Keywords: HIV-1, Restriction factors, Interferon, Interferon stimulated genes
Introduction
HIV-1 infects 37 million people worldwide, killing roughly 1 million people per year. Life-saving antiretroviral therapy controls but does not eliminate the virus. Whereas an HIV-1 cure is necessary, a clearer understanding of the intracellular fate(s) of HIV-1 is essential before a potent cure can be designed. Indeed, the host cell has several mechanisms by which it can limit the burden of HIV-1, and an elucidation of these mechanisms offers keen insights into HIV-1 biology. In that context, we have reviewed recent advances describing intracellular genes that restrict HIV-1.
Innate Immunity: sensing and restriction
Innate immunity has received considerable attention recently for its role in host defense and inflammation. As a simplification, shared molecular patterns on invading pathogens can be recognized by intracellular sensors, triggering a downstream cascade of gene regulation in processes that have been extensively reviewed elsewhere [1, 2]. Many of the upregulated genes in innate immune cascades are devoted to ridding the host cell of the pathogen and/or recruiting a targeted adaptive response. With regards to viruses, viral nucleic acids and structural components are often recognized by pattern recognition receptors, whereas downstream antiviral effector molecules target replication within the cell [3–5]. A hallmark example of an innate immune pathway with potent antiviral effects is the type 1 interferon pathway: upon sensing of viral nucleic acids, type 1 interferons are released. These in turn trigger the transcription of hundreds of genes termed interferon-stimulated genes (ISGs), many of which directly constrain viral replication [6].
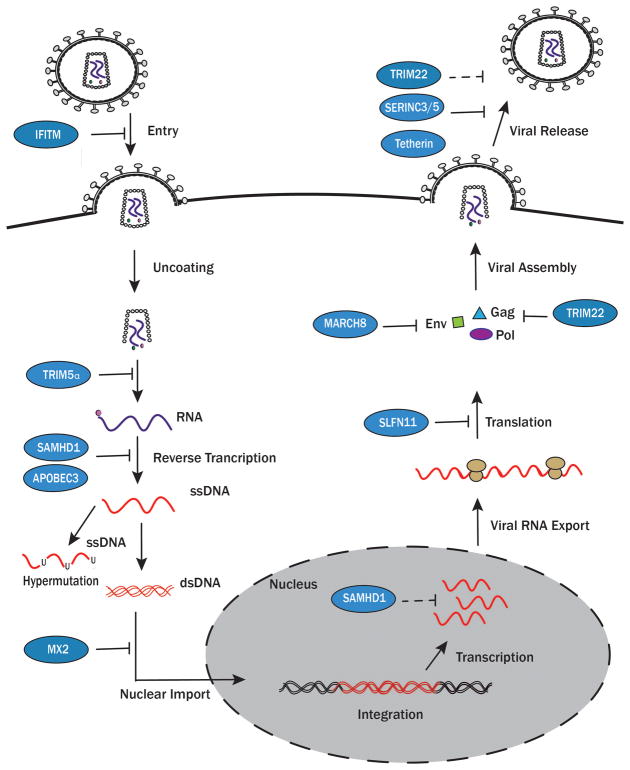
HIV-1 is a retrovirus: productive infection of host cells requires that uncoated HIV-1 RNA is reverse transcribed into a DNA provirus that is then integrated into the host cell’s genome. Conceptually, we have divided our review into restriction that occurs prior to HIV-1 integration and restriction that occurs post-integration (Figure). This conceptual division is to stress that pre-integration restriction necessarily occurs in cells that are not yet infected, whereas restriction post-integration occurs in cells that are already infected. We have also described the key adaptive measures that have evolved in HIV-1 to overcome restriction (Table).
Figure 1. Inhibition of HIV replication by known host restriction factors.
HIV-1 is inhibited by multiple restriction factors (blue) throughout its life cycle. IFITM proteins alter plasma membrane fluidity, inhibiting HIV-1 entry. TRIM5α prevents appropriate uncoating of the capsid. SAMHD1 inhibits cDNA synthesis by limiting the amount of free 2′-deoxynucleoside 5′-triphosphates (dNTPs) and possibly by its exonuclease activity (dashed lines). APOBEC3 proteins interfere with reverse transcription leading to uracilation of single-stranded DNA (ssDNA), a mechanism known as hypermutation. MX2 interferes with nuclear import, limiting the integration of proviral DNA. SLFN11 inhibits the translation of essential proteins needed to form HIV-1 virions. MARCH8 inhibits the incorporation of Env proteins. TRIM22 inhibits the production and incorporation of Gag proteins and inhibits LTR-dependent transcription. Tetherin, SERINC3/5, and TRIM22 prevent the virion from successfully budding. Dashed lines indicate proposed mechanisms. The pink circle denotes reverse transcriptase and the green circle denotes integrase, both packaged with the virion.
Table 1. Mechanisms of HIV-1 evasion.
Restriction factors are listed with the major adaptive strategy that HIV-1 employs to evade their restriction.
| Restriction Factors | HIV-1 Evasion Mechanism |
|---|---|
|
| |
| SAMHD1 [36] | Vpx inhibits SAMHD1 by reqcruting an E3 ligase leading to its ubinylation and degradation |
| APOBEC3 [67] | Vif inhibits APOBEC by leading to its polyubiquination and proteosomal degradation |
| MX2 [74] | Capsid mutates to evade interaction with MX2 binding loop |
| SERINC3/5 [90] | Nef inhibts SERINC3/5 by decreasing its incorporation into virions and redeistributing the proteins into an endocytic compartment |
| Tetherin [80] | Vpu inhibits trafficking of tetherin to the surface and promotes its ubinylation and degradation |
IFITM
The interferon-induced transmembrane gene family (IFITM), comprising IFITM1, IFITM2, and IFITM3, restrict a broad panoply of viruses from entering the host cell [7]. The IFITM family are ISGs. IFITM1 was the first in this group to be discovered in an RNA silencing screen of ISGs that conferred resistance to Influenza A, West Nile, and dengue viruses [8]. IFITM1 is mostly found on the plasma membrane and has a role in blocking entry of HIV-1 virions [9]. In contrast, IFITM2 and -3 are largely localized to the late endosome and lysosomes [10, 11]. The IFITM genes are thought to inhibit HIV-1 entry by changing the composition and curvature of the plasma membrane, perhaps reducing its fluidity, thereby interfering with a phenomenon known as hemifusion [12, 13]. Hemifusion is essential for the incorporation of an HIV-1 virion into a target cell [14]. Intriguingly, although restriction of HIV-1 entry occurs pre-integration, there are two reports suggesting a role post-integration: Compton et al. found that the presence of IFITM3 in HIV-1 infected cells decreased transmission of HIV-1 to target cells [15]. Moreover, time-lapse microscopy has shown that IFITM3 functions by decreasing fusion between the infected and uninfected target cell by affecting the virus membrane. Yu et al. confirmed that IFITMs were incorporated into HIV-1 virions and interacted with the envelope protein Env to restrict HIV-1 infection [16].
There is evidence that certain HIV-1 variants may be resistant to the restriction of IFITM proteins. In particular, transmitter/founder viruses have been found to be more resistant to IFITM restriction than viruses isolated from later times during infection; the later viruses may gain sensitivity as a result of escape from concomitant neutralizing antibody responses [17], although this in turn raises the question as to the relative relevance of IFITM as an in vivo restriction mechanism compared to antibody-driven selection.
TRIM5α and TRIM22
Tripartite motif 5α (TRIM5α), the longest splicing isoform of TRIM5 gene, was identified as a key molecule in old world monkeys that confers potent resistance against HIV-1 [18]. Among the large family of TRIM proteins consisting of over 70 family members in the human genome, TRIM5, TRIM11, TRIM15, TRIM19, TRIM22, TRIM28, and TRIM31 all have been described as having antiretroviral activity, although the human TRIM5α only confers modest resistance to HIV-1 [19–22]. It is thought, then, that TRIM proteins are required to prevent cross-species transmission of retroviruses [23].
TRIM proteins are multi-domain proteins defined by an N-terminal RING finger with E3 Ubiquitin ligase activity, one or two B-box domain(s), and a coiled-coil (RBCC) domain [19]. Some of the TRIM genes, including TRIM5α, also possess a C-terminal PRY/SPRY (SPla and the RYanodine Receptor) domain that is important for HIV-1 capsid recognition [24]. TRIM5α acts at multiple levels to restrict HIV-1 replication. It specifically binds the capsid (CA) lattice of HIV-1 and induces premature disassembly of viral particles, accompanied by proteosomal-degradation of viral components such as integrase [25]. TRIM5α has also been found to promote autophagic degradation of retroviruses by interacting with Atg8 proteins [26]. More recent studies show that TRIM5α inhibits HIV-1 infection by sensing the retrovirus capsid lattice, eliciting an innate immune response [27]. Interaction with the capsid lattice enhances TRIM5α RING-associated E3-Ub ligase, which in turn promotes the synthesis of free Lys63-linked Ub chains that bind to the TAK1 kinase complex. TAK1 ubiquitination leads to nuclear translocation of the transcription factors NF-κB and AP-1 that result in type 1 IFN and other pro-inflammatory cytokines [28] [29], broadly inducing an antiviral state.
Whereas the TRIM5α homolog in humans does not inhibit HIV-1 well, TRIM22, a human paralog of TRIM5α, is involved in type I IFN-mediated restriction of HIV-1 replication [30, 31]. TRIM22 inhibits HIV-1 replication by interfering with Tat-and NF-κB-independent LTR-driven transcription, and by preventing Sp1 binding to the HIV-1 promoter [32, 33]. TRIM22 may also interfere with virion assembly and release by preventing the trafficking and budding of Gag proteins and Gag-containing virus particles [30].
Notably, because human TRIM5α has only modest activity against HIV-1, it does not drive adaptation in the virus. However, several reports have suggested that HIV-1 could acquire adaptive mutations in the capsid gene, among others, in the setting of an engineered TRIM5α molecule that did have the capacity to restrict HIV-1 replication [34] [35].
SAMHD1
SAMHD1 (Sterile alpha motif and histidine-aspartate domain containing protein 1) was discovered in 2011 as a restriction factor by two independent groups using a mass spectrometry pull-down approach to identify proteins that co-immunoprecipitated with the viral protein x (Vpx) present in HIV-2, but not HIV-1 [36, 37]. SAMHD1 is expressed at high levels in myeloid-derived cells and in resting CD4+ T cells that are refractory to HIV-1 infection [36, 38, 39]. However, SAMHD1 is inducible by type I IFN in monocytes, but not in dendritic cells or resting CD4+ T cells [40]. SAMHD1 comprises three regions, the N-terminus sterile alpha motif (residues 1–109), a catalytic histidine aspartate core domain (residues 110–599), and the C-terminus (residues 600–626) [41]. The catalytic core domain has 2′-deoxynucleoside 5′-triphosphate triphosphohydrolase (dNTPase) activity that is believed to restrict HIV-1 by diminishing the intracellular pool of available dNTP in immune cells needed for cDNA synthesis during reverse transcription [42–44]. The anti-HIV-1 activity of SAMHD1 is negatively modulated by phosphorylation at residue Thr-592, and the phosphomimetic mutation T592E impairs dNTPase activity [45]. Indeed, dNTP levels are elevated in SAMHD1-deficient cells, whereas reduced dNTPs lead to an attenuation of reverse transcription [46–48].
Another feature of human SAMHD1 is that it exhibits metal-dependent 3′→5′ exonuclease activity against single-stranded DNAs and RNAs in vitro, suggesting that it may bind to and degrade HIV-1 RNA [49]. Ryoo and colleagues created several SAMHD1 mutants that exclusively retained either their exonuclease activity or their dNTPase activity and showed that the exonuclease alone was sufficient to partially restrict HIV-1 [50].
Restriction is overcome by Vpx in HIV-2 through interactions with the host Ub ligase adaptor DCAF1 that promotes SAMHD1 degradation [36, 37]. Whether HIV-1 has evolved adaptations to counter SAMHD1 is still not well understood, although recently Kyei et al. reported that HIV-1 might neutralize SAMHD1 in macrophages in concert with the cell cycle regulator cyclin L2 [51]. There is suggestive evidence for the clinical relevance of SAMHD1: the gene is mutated in a subset of patients suffering from Aicardi-Goutières syndrome (AGS), and monocytes from patients with AGS appear exquisitely susceptible to HIV-1 infection, in contrast to monocytes from healthy persons [52].
APOBEC3
Apolipoprotein B editing complex 3 (APOBEC3) family members are cytidine deaminases that include APOBEC3A-H [53, 54]. They are induced by Type I IFN and most play important roles in control of multiple retroviruses through RNA binding or through deamination of single-stranded DNA (ssDNA) [53]. APOBEC3 proteins are expressed variably in CD4+ and CD8+ T cell subsets, B cells, and myeloid cells [55]. APOBEC3G exerts a dominant antiviral effect in CD4+ T cell subsets [56], and in macrophages and primary CD4+ T cells shows more potent restriction of HIV-1 than the combined effect of APOBEC3F and APOBEC3DE [57]. The APOBEC3 family members restrict HIV-1 by hypermutating its genome, resulting in premature stop codons and defective proviruses that are incapable of propagating infection [58, 59]. APOBEC3G, for example, acts by temporarily inhibiting the reverse transcription step and deaminating C→U on the minus strand of the DNA, leading to a mismatch pairing of a G→A on the plus strand, thus causing genomic mutations [60]. Hypermutation leads to HIV-1 restriction in at least two ways: i) the host cell recognizes ssDNA as aberrant and degrades it [61]; and ii) proviral DNA is integrated as a defective provirus that is no longer capable of producing infectious virions, usually due to premature stop codons [62]. The restrictive activity of APOBEC3G depends on its encapsidation in the budding virion, mediated by the nucleocapsid domain of Gag molecules: upon virion fusion with a target cell, the viral RNA, reverse transcriptase, integrase, and APOBEC3G are released into the cytosol, allowing restriction to occur in the target cell [63, 64]. In addition, APOBEC3F and -G may exert antiretroviral activities that are independent of their deaminase functions by interfering with reverse transcription, either terminating the synthesis of the minus-strand or preventing tRNA molecules from binding HIV-1 nucleic acids.
HIV-1 has developed a significant adaptation to APOBEC3 restriction: the accessory protein Vif triggers the degradation of APOBEC3 proteins through polyubiquitination and proteasomal degradation [65–67]. Ubiquitination occurs by the recruitment of a Cullin 5–based E3 ubiquitin ligase complex consisting of elongin B, elongin C, and Rbx-1. The in vivo relevance of APOBEC3G is evident in recent reports showing that the HIV-1 latent reservoir in patients treated with antiretrovirals contains a large number of APOBEC3G hypermutated sequences [68, 69, 65]. Collectively, human studies suggest that the battle between HIV-1 and APOBEC3 is not concluded: whereas the latent reservoir is largely defective, there exist replication competent viruses that impede an HIV-1 cure. Similarly, Vif efficiently blocks APOBEC3 genes, but is insufficient in terminating all hypermutation. The latter point is promising for strategies aimed at enhancing inactivation of proviruses.
MX2
The IFN-inducible myxovirus resistance 2 (MX2/MXB) is a member of a family of dynamin-like GTPases that includes human MX1/MXA, another well-known IFN-inducible inhibitor of a broad range of viruses. In 2013, the anti-HIV-1 activity of MX2 was described by three separate groups [70–72]. MX2 appears to act at a late post-entry step prior to integration of proviral DNA, possibly through inhibition of nuclear import following reverse transcription, or by inhibiting the uncoating of HIV-1 [70, 71, 73]. It has also been proposed that MX2 could decrease integration of nascent viral DNA, leading to nuclear accumulation of 2′ LTR circles [70]. MX2 interacts with the HIV-1 capsid and may be dependent on CypA: substitutions in the CypA binding loop at positions 86–88 and 92 disrupt the binding of MX2 and capsid protein [74]. Similarly, treatment with cyclosporine, which blocks the CypA interaction with capsid, reduces the restrictive ability of MX2 [72].
There is little in vivo data to support the relevance of MX2 in HIV-1 restriction, although our group has found strong evidence that interferon administration to HIV-1 infected persons led to MX2 induction and proportional declines in plasma HIV-1 RNA in activated CD4+ T cells (unpublished). In addition, mutations in the HIV-1 capsid protein and in integrase confer resistance to MX2 [75]. It is yet to be understood how the potency of MX2 compares with other interferon-inducible HIV-1 restriction factors in humans.
SLFN11
SLFN11 belongs to the Schlafen family of mouse and human proteins implicated in the control of cell proliferation, immune responses, and the regulation of viral replication [76]. It has recently been identified as inducible by type 1 IFNs [77]. SLFN11 does not inhibit reverse transcription, DNA integration, production and nuclear export of viral RNA, or budding or release of viral particles. Its role in inhibition is one of the first to be associated with codon bias, inhibiting the expression of viral proteins. It binds to tRNAs, limiting their availability, thereby leading to a decrease in protein synthesis [77]. SLFN11 was also recently found to be significantly elevated in CD4+ T cells from elite HIV controllers as compared to non-controllers and ART-suppressed individuals, suggesting that SLFN11 may play a role in the suppression of HIV-1 in vivo [78]. However, it is not clear whether the effects of SLFN11 are sufficiently robust to promote adaptation in HIV-1 in vivo.
MARCH8
Recently, Tada et al. reported that the membrane-associated RING-CH8 (MARCH8) E3 ubiquitin ligase, a gene that is highly expressed in terminally differentiated myeloid cells such as macrophages and dendritic cells, possesses potent anti-HIV-1 activity in vitro [79]. The antiviral effect of MARCH8 was discovered serendipitously when the investigators noted that the transduction efficiency of lentiviruses produced from MARCH8-expressing cells was several-fold lower than that of control lentiviruses. MARCH8 blocks the incorporation of HIV-1 envelope glycoprotein into virus particles, resulting in a substantial reduction in the efficiency of virus entry, thus inhibiting its infectivity. Intriguingly, viruses are normally released, but are rendered non-infectious in the presence of MARCH8. Neither HIV-1 Vpr, Vpu nor Nef have detectable anti-MARCH8 activity, suggesting that HIV-1 lacks a counter-mechanism that dampens the effects of MARCH8. Studies are ongoing to determine the in vivo relevance of MARCH8, and whether HIV-1 can indeed adapt resistance to its effects.
Tetherin
Tetherin (Bone marrow stromal cell antigen-2 [BST-2]; CD317) is a restriction factor that was first identified in a microarray screen aimed at finding ISGs that are associated with the plasma membrane in cells that were not permissive for HIV-1 propagation [80]. Tetherin consists of an N-terminal cytoplasmic tail, a transmembrane helix, a coiled-coil ectodomain, and a C-terminal glycosylphosphatidylinositol (GPI) membrane anchor. It is localized in lipid rafts at the plasma membrane, in the trans-Golgi network, and in early recycling endosomes [81]. Tetherin inhibits the release of nascent HIV-1 particles by tethering the budding virions at the cell surface [82]. Nascent virions anchored to the membrane are then internalized and degraded in the lysosome. Interestingly, the topology of tetherin is unique; when a mimic of tetherin was designed from unrelated proteins, HIV-1 was inhibited, demonstrating that the topology of tetherin, not the sequence, is key to its restrictive function.
Another proposed role of tetherin in infection is its role as a viral sensor. It has been suggested that a budding virion could act as a pathogen-associated molecular pattern that is sensed by tetherin. As tetherin binds budding virions, clustering of tetherin dimers promotes the recruitment of TRAF2 and TRAF6, leading to activation of TAK1 and NF-κB and upregulation of proinflammatory cytokines such as CXCL10, IL-6 and IFN-β [83, 84]. The ability of tetherin to activate NF-κB requires multimerization [83].
HIV-1 has evolved several viral proteins have evolved to antagonize tetherin: HIV-1 Vpu (in groups M and N) and Nef (in group O) [85, 80, 86]. Indeed, the role of tetherin as a broad antiretroviral is evident in that HIV-2 and SIV have evolved resistance to their host versions of tetherin [87]. HIV-1 Vpu, a small transmembrane protein, interacts directly with tetherin at the trans-Golgi network and targets it for proteasomal or lysosomal degradation [88, 89]. Deletion of Vpu results in tetherin-mediated retention of virions at the plasma membrane [80]. In addition, Vpu also inhibits the activation of NF-κB by tetherin [84].
SERINC3/5
In a study structured to find constitutively expressed restriction factors that are suppressed by Nef, Usami et al., discovered that SERINC3 and SERINC5 were both able to restrict HIV-1 replication [90]. In Nef-deleted HIV-1 pseudotyped virus, the addition of SERINC5 significantly reduced infectivity. The amount of SERINC5 expression was directly associated with a quantitative loss of fusion of HIV-1 Δnef virions when co-expressed with a chimeric B-lactamase-Vpr. To further support these effects, SERINC5 mRNA level was inversely correlated with a requirement of Nef for HIV-1 replication in several cell lines (Jurkat, 293T, MT4).
SERINCs are cell surface proteins that function by incorporating serine into membrane lipids, most notably in sphingolipids and phosphatidylserine [91]. The lipid composition of the viral envelope is important for infectivity. Furthermore, SERINC5 inhibits the completion of reverse transcription of HIV-1 in the absence of Nef, which is an essential step prior to provirus integration [90]. Although the importance of SERINC3/5 can be inferred from their inhibition by Nef, more work is needed to understand the clinical relevance of these genes in HIV-1 infected persons.
Clinical Implications of HIV-1 Restriction Factors
Studies of restriction factors offer insights into the key molecular determinants of HIV-1 replication, and collectively outline a roadmap of HIV-1 vulnerabilities that could be targeted therapeutically. As an example, interferon alpha has been used to non-specifically target HIV-1 in several small trials of HIV-1 infected persons, and has been as effective as a single, modestly active antiretroviral drug [92]. Presumably, the actions of interferon alpha are at least partially in inducing ISGs that directly act on HIV-1 in the cells that are most susceptible to infection, but it is certainly possible that interferon alpha also recruits cellular immune responses. Although interferon alpha may not add benefit to HIV-1 infected patients because of its side effects, it is compelling to consider that genes and pathways downstream of interferon administration may be specifically targeted to produce similar effects on HIV-1 replication. We speculate that for HIV-1 infected patients who have suppressed viral replication with antiretrovirals, it may be possible to silence integrated proviruses by exploiting or mimicking the actions of post-integration restriction factors.
Another approach to HIV-1 treatment follows from its adaptation to host restriction factors by the use of accessory proteins. Since these genes appear to be essential for the virus to circumvent host innate immunity, enzymatic inhibition of the activities of these accessory proteins may restore the ability of the host to control HIV-1 using its suite of restriction factors. Indeed, there has been a push of small molecules into clinical trials that have been demonstrated to have high potency activity against HIV-1 accessory proteins [93,94].
Conclusion
To address the challenge of an HIV-1 cure, a better understanding is required of how HIV-1 persists despite host cell restriction. Presently, the science of restriction factors is moving from the bench to the bedside as we explore which restriction factors are active in HIV-1 infected persons, and what measures can be used to enhance restriction. Future HIV-1 cure trials may indeed exploit HIV-1 restriction or, alternatively, target the adaptive means by which HIV-1 evades restriction.
References
- 1.Iwasaki A, Medzhitov R. Control of adaptive immunity by the innate immune system. Nature immunology. 2015;16(4):343–53. doi: 10.1038/ni.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brubaker SW, Bonham KS, Zanoni I, Kagan JC. Innate immune pattern recognition: a cell biological perspective. Annual review of immunology. 2015;33:257–90. doi: 10.1146/annurev-immunol-032414-112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Montfoort N, Olagnier D, Hiscott J. Unmasking immune sensing of retroviruses: interplay between innate sensors and host effectors. Cytokine & growth factor reviews. 2014;25(6):657–68. doi: 10.1016/j.cytogfr.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Collins SE, Mossman KL. Danger, diversity and priming in innate antiviral immunity. Cytokine & growth factor reviews. 2014;25(5):525–31. doi: 10.1016/j.cytogfr.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Silvin A, Manel N. Innate immune sensing of HIV infection. Current opinion in immunology. 2015;32:54–60. doi: 10.1016/j.coi.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 6.McNab F, Mayer-Barber K, Sher A, Wack A, O’Garra A. Type I interferons in infectious disease. Nature reviews Immunology. 2015;15(2):87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nature reviews Immunology. 2013;13(1):46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139(7):1243–54. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu J, Pan Q, Rong L, He W, Liu SL, Liang C. The IFITM proteins inhibit HIV-1 infection. Journal of virology. 2011;85(5):2126–37. doi: 10.1128/JVI.01531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feeley EM, Sims JS, John SP, Chin CR, Pertel T, Chen LM, et al. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS pathogens. 2011;7(10):e1002337. doi: 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weston S, Czieso S, White IJ, Smith SE, Wash RS, Diaz-Soria C, et al. Alphavirus Restriction by IFITM Proteins. Traffic. 2016;17(9):997–1013. doi: 10.1111/tra.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chernomordik LV, Kozlov MM. Membrane hemifusion: crossing a chasm in two leaps. Cell. 2005;123(3):375–82. doi: 10.1016/j.cell.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 13.Li K, Markosyan RM, Zheng YM, Golfetto O, Bungart B, Li M, et al. IFITM proteins restrict viral membrane hemifusion. PLoS pathogens. 2013;9(1):e1003124. doi: 10.1371/journal.ppat.1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garg H, Viard M, Jacobs A, Blumenthal R. Targeting HIV-1 gp41-induced fusion and pathogenesis for anti-viral therapy. Current topics in medicinal chemistry. 2011;11(24):2947–58. doi: 10.2174/156802611798808479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15••.Compton AA, Bruel T, Porrot F, Mallet A, Sachse M, Euvrard M, et al. IFITM proteins incorporated into HIV-1 virions impair viral fusion and spread. Cell host & microbe. 2014;16(6):736–47. doi: 10.1016/j.chom.2014.11.001. A paper showing a mechanism in which IFITM incorporates itself into virions to block fusion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16••.Yu J, Li M, Wilkins J, Ding S, Swartz TH, Esposito AM, et al. IFITM Proteins Restrict HIV-1 Infection by Antagonizing the Envelope Glycoprotein. Cell reports. 2015;13(1):145–56. doi: 10.1016/j.celrep.2015.08.055. A paper showing how IFITM acts by inhibiting production and incrporation of envelope proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster TL, Wilson H, Iyer SS, Coss K, Doores K, Smith S, et al. Resistance of Transmitted Founder HIV-1 to IFITM-Mediated Restriction. Cell host & microbe. 2016 doi: 10.1016/j.chom.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–53. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 19.Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nature reviews Microbiology. 2005;3(10):799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 20.Han K, Lou DI, Sawyer SL. Identification of a genomic reservoir for new TRIM genes in primate genomes. PLoS genetics. 2011;7(12):e1002388. doi: 10.1371/journal.pgen.1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang F, Hatziioannou T, Perez-Caballero D, Derse D, Bieniasz PD. Antiretroviral potential of human tripartite motif-5 and related proteins. Virology. 2006;353(2):396–409. doi: 10.1016/j.virol.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 22.Battivelli E, Lecossier D, Matsuoka S, Migraine J, Clavel F, Hance AJ. Strain-specific differences in the impact of human TRIM5alpha, different TRIM5alpha alleles, and the inhibition of capsid-cyclophilin A interactions on the infectivity of HIV-1. Journal of virology. 2010;84(21):11010–9. doi: 10.1128/JVI.00758-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soll SJ, Wilson SJ, Kutluay SB, Hatziioannou T, Bieniasz PD. Assisted evolution enables HIV-1 to overcome a high TRIM5alpha-imposed genetic barrier to rhesus macaque tropism. PLoS pathogens. 2013;9(9):e1003667. doi: 10.1371/journal.ppat.1003667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Gold B, O’HUigin C, Diaz-Griffero F, Song B, Si Z, et al. Unique features of TRIM5alpha among closely related human TRIM family members. Virology. 2007;360(2):419–33. doi: 10.1016/j.virol.2006.10.035. [DOI] [PubMed] [Google Scholar]
- 25.Kutluay SB, Perez-Caballero D, Bieniasz PD. Fates of retroviral core components during unrestricted and TRIM5-restricted infection. PLoS pathogens. 2013;9(3):e1003214. doi: 10.1371/journal.ppat.1003214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandell MA, Kimura T, Jain A, Johansen T, Deretic V. TRIM proteins regulate autophagy: TRIM5 is a selective autophagy receptor mediating HIV-1 restriction. Autophagy. 2014;10(12):2387–8. doi: 10.4161/15548627.2014.984278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pertel T, Hausmann S, Morger D, Zuger S, Guerra J, Lascano J, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472(7343):361–5. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nepveu-Traversy ME, Berthoux L. The conserved sumoylation consensus site in TRIM5alpha modulates its immune activation functions. Virus research. 2014;184:30–8. doi: 10.1016/j.virusres.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 29.Campbell EM, Weingart J, Sette P, Opp S, Sastri J, O’Connor SK, et al. TRIM5alpha-Mediated Ubiquitin Chain Conjugation Is Required for Inhibition of HIV-1 Reverse Transcription and Capsid Destabilization. Journal of virology. 2016;90(4):1849–57. doi: 10.1128/JVI.01948-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barr SD, Smiley JR, Bushman FD. The interferon response inhibits HIV particle production by induction of TRIM22. PLoS pathogens. 2008;4(2):e1000007. doi: 10.1371/journal.ppat.1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh R, Gaiha G, Werner L, McKim K, Mlisana K, Luban J, et al. Association of TRIM22 with the type 1 interferon response and viral control during primary HIV-1 infection. Journal of virology. 2011;85(1):208–16. doi: 10.1128/JVI.01810-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kajaste-Rudnitski A, Marelli SS, Pultrone C, Pertel T, Uchil PD, Mechti N, et al. TRIM22 inhibits HIV-1 transcription independently of its E3 ubiquitin ligase activity, Tat, and NF-kappaB-responsive long terminal repeat elements. Journal of virology. 2011;85(10):5183–96. doi: 10.1128/JVI.02302-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33••.Turrini F, Marelli S, Kajaste-Rudnitski A, Lusic M, Van Lint C, Das AT, et al. HIV-1 transcriptional silencing caused by TRIM22 inhibition of Sp1 binding to the viral promoter. Retrovirology. 2015;12:104. doi: 10.1186/s12977-015-0230-0. A paper showing the mechanism in which TRIM22 inhibits HIV-1 LTR transcriptional activation by inhibition of Sp1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Setiawan LC, Kootstra NA. Adaptation of HIV-1 to rhTrim5alpha-mediated restriction in vitro. Virology. 2015;486:239–47. doi: 10.1016/j.virol.2015.09.017. [DOI] [PubMed] [Google Scholar]
- 35.Nakayama EE, Shioda T. Impact of TRIM5alpha in vivo. AIDS. 2015;29(14):1733–43. doi: 10.1097/QAD.0000000000000812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474(7353):654–7. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474(7353):658–61. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Descours B, Cribier A, Chable-Bessia C, Ayinde D, Rice G, Crow Y, et al. SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4(+) T-cells. Retrovirology. 2012;9:87. doi: 10.1186/1742-4690-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, et al. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nature medicine. 2012;18(11):1682–7. doi: 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.St Gelais C, de Silva S, Amie SM, Coleman CM, Hoy H, Hollenbaugh JA, et al. SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T-lymphocytes cannot be upregulated by interferons. Retrovirology. 2012;9:105. doi: 10.1186/1742-4690-9-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ahn J. Functional organization of human SAMHD1 and mechanisms of HIV-1 restriction. Biological chemistry. 2016;397(4):373–9. doi: 10.1515/hsz-2015-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480(7377):379–82. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- 43.Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nature immunology. 2012;13(3):223–8. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Powell RD, Holland PJ, Hollis T, Perrino FW. Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. The Journal of biological chemistry. 2011;286(51):43596–600. doi: 10.1074/jbc.C111.317628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang C, Ji X, Wu L, Xiong Y. Impaired dNTPase activity of SAMHD1 by phosphomimetic mutation of Thr-592. The Journal of biological chemistry. 2015;290(44):26352–9. doi: 10.1074/jbc.M115.677435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rehwinkel J, Maelfait J, Bridgeman A, Rigby R, Hayward B, Liberatore RA, et al. SAMHD1-dependent retroviral control and escape in mice. The EMBO journal. 2013;32(18):2454–62. doi: 10.1038/emboj.2013.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim B, Nguyen LA, Daddacha W, Hollenbaugh JA. Tight interplay among SAMHD1 protein level, cellular dNTP levels, and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. The Journal of biological chemistry. 2012;287(26):21570–4. doi: 10.1074/jbc.C112.374843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bonifati S, Daly MB, St Gelais C, Kim SH, Hollenbaugh JA, Shepard C, et al. SAMHD1 controls cell cycle status, apoptosis and HIV-1 infection in monocytic THP-1 cells. Virology. 2016;495:92–100. doi: 10.1016/j.virol.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beloglazova N, Flick R, Tchigvintsev A, Brown G, Popovic A, Nocek B, et al. Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. The Journal of biological chemistry. 2013;288(12):8101–10. doi: 10.1074/jbc.M112.431148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50••.Ryoo J, Choi J, Oh C, Kim S, Seo M, Kim SY, et al. The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nature medicine. 2014;20(8):936–41. doi: 10.1038/nm.3626. A paper showing that SAMHD1 also needs its RNase activity to inhibit HIV-1 and not just its ability to deplete DNTP levels. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kyei GB, Cheng X, Ramani R, Ratner L. Cyclin L2 is a critical HIV dependency factor in macrophages that controls SAMHD1 abundance. Cell host & microbe. 2015;17(1):98–106. doi: 10.1016/j.chom.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berger A, Sommer AF, Zwarg J, Hamdorf M, Welzel K, Esly N, et al. SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV-1 infection. PLoS pathogens. 2011;7(12):e1002425. doi: 10.1371/journal.ppat.1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harris RS, Dudley JP. APOBECs and virus restriction. Virology. 2015;479–480:131–45. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stavrou S, Ross SR. APOBEC3 Proteins in Viral Immunity. J Immunol. 2015;195(10):4565–70. doi: 10.4049/jimmunol.1501504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koning FA, Newman EN, Kim EY, Kunstman KJ, Wolinsky SM, Malim MH. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. Journal of virology. 2009;83(18):9474–85. doi: 10.1128/JVI.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gillick K, Pollpeter D, Phalora P, Kim EY, Wolinsky SM, Malim MH. Suppression of HIV-1 infection by APOBEC3 proteins in primary human CD4(+) T cells is associated with inhibition of processive reverse transcription as well as excessive cytidine deamination. Journal of virology. 2013;87(3):1508–17. doi: 10.1128/JVI.02587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chaipan C, Smith JL, Hu WS, Pathak VK. APOBEC3G restricts HIV-1 to a greater extent than APOBEC3F and APOBEC3DE in human primary CD4+ T cells and macrophages. Journal of virology. 2013;87(1):444–53. doi: 10.1128/JVI.00676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113(6):803–9. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 59.Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Current biology: CB. 2004;14(15):1392–6. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 60.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424(6944):99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 61.Casartelli N, Guivel-Benhassine F, Bouziat R, Brandler S, Schwartz O, Moris A. The antiviral factor APOBEC3G improves CTL recognition of cultured HIV-infected T cells. The Journal of experimental medicine. 2010;207(1):39–49. doi: 10.1084/jem.20091933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Russell RA, Moore MD, Hu WS, Pathak VK. APOBEC3G induces a hypermutation gradient: purifying selection at multiple steps during HIV-1 replication results in levels of G-to-A mutations that are high in DNA, intermediate in cellular viral RNA, and low in virion RNA. Retrovirology. 2009;6:16. doi: 10.1186/1742-4690-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang T, Zhang W, Tian C, Liu B, Yu Y, Ding L, et al. Distinct viral determinants for the packaging of human cytidine deaminases APOBEC3G and APOBEC3C. Virology. 2008;377(1):71–9. doi: 10.1016/j.virol.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Friew YN, Boyko V, Hu WS, Pathak VK. Intracellular interactions between APOBEC3G, RNA, and HIV-1 Gag: APOBEC3G multimerization is dependent on its association with RNA. Retrovirology. 2009;6:56. doi: 10.1186/1742-4690-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Desimmie BA, Delviks-Frankenberrry KA, Burdick RC, Qi D, Izumi T, Pathak VK. Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. Journal of molecular biology. 2014;426(6):1220–45. doi: 10.1016/j.jmb.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feng Y, Baig TT, Love RP, Chelico L. Suppression of APOBEC3-mediated restriction of HIV-1 by Vif. Frontiers in microbiology. 2014;5:450. doi: 10.3389/fmicb.2014.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nature medicine. 2003;9(11):1404–7. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 68.Jern P, Russell RA, Pathak VK, Coffin JM. Likely role of APOBEC3G-mediated G-to-A mutations in HIV-1 evolution and drug resistance. PLoS pathogens. 2009;5(4):e1000367. doi: 10.1371/journal.ppat.1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim EY, Lorenzo-Redondo R, Little SJ, Chung YS, Phalora PK, Maljkovic Berry I, et al. Human APOBEC3 induced mutation of human immunodeficiency virus type-1 contributes to adaptation and evolution in natural infection. PLoS pathogens. 2014;10(7):e1004281. doi: 10.1371/journal.ppat.1004281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70••.Goujon C, Moncorge O, Bauby H, Doyle T, Ward CC, Schaller T, et al. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature. 2013;502(7472):559–62. doi: 10.1038/nature12542. One of the three major papers released in 2013 first describing MX2 as an ISG that is also a restriction factor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71••.Kane M, Yadav SS, Bitzegeio J, Kutluay SB, Zang T, Wilson SJ, et al. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature. 2013;502(7472):563–6. doi: 10.1038/nature12653. One of the three major papers released in 2013 first describing MX2 as an ISG that is also a restriction factor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72••.Liu Z, Pan Q, Ding S, Qian J, Xu F, Zhou J, et al. The interferon-inducible MxB protein inhibits HIV-1 infection. Cell host & microbe. 2013;14(4):398–410. doi: 10.1016/j.chom.2013.08.015. One of the three major papers released in 2013 first describing MX2 as an ISG that is also a restriction factor. [DOI] [PubMed] [Google Scholar]
- 73.Fricke T, White TE, Schulte B, de Souza Aranha Vieira DA, Dharan A, Campbell EM, et al. MxB binds to the HIV-1 core and prevents the uncoating process of HIV-1. Retrovirology. 2014;11:68. doi: 10.1186/s12977-014-0068-x. doi: 10.1186/s12977-014-0068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu Z, Pan Q, Liang Z, Qiao W, Cen S, Liang C. The highly polymorphic cyclophilin A-binding loop in HIV-1 capsid modulates viral resistance to MxB. Retrovirology. 2015;12:1. doi: 10.1186/s12977-014-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matreyek KA, Wang W, Serrao E, Singh PK, Levin HL, Engelman A. Host and viral determinants for MxB restriction of HIV-1 infection. Retrovirology. 2014;11:90. doi: 10.1186/s12977-014-0090-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mavrommatis E, Fish EN, Platanias LC. The schlafen family of proteins and their regulation by interferons. Journal of interferon & cytokine research: the official journal of the International Society for Interferon and Cytokine Research. 2013;33(4):206–10. doi: 10.1089/jir.2012.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li M, Kao E, Gao X, Sandig H, Limmer K, Pavon-Eternod M, et al. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature. 2012;491(7422):125–8. doi: 10.1038/nature11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Abdel-Mohsen M, Raposo RA, Deng X, Li M, Liegler T, Sinclair E, et al. Expression profile of host restriction factors in HIV-1 elite controllers. Retrovirology. 2013;10:106. doi: 10.1186/1742-4690-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79••.Tada T, Zhang Y, Koyama T, Tobiume M, Tsunetsugu-Yokota Y, Yamaoka S, et al. MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins. Nature medicine. 2015;21(12):1502–7. doi: 10.1038/nm.3956. A study demonstrating how MARCH8 inhibits HIV-1 replication in macrophages by limiting envelopes incorporation into budding virions. [DOI] [PubMed] [Google Scholar]
- 80.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451(7177):425–30. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 81.Kupzig S, Korolchuk V, Rollason R, Sugden A, Wilde A, Banting G. Bst-2/HM1.24 is a raft-associated apical membrane protein with an unusual topology. Traffic. 2003;4(10):694–709. doi: 10.1034/j.1600-0854.2003.00129.x. [DOI] [PubMed] [Google Scholar]
- 82.Perez-Caballero D, Zang T, Ebrahimi A, McNatt MW, Gregory DA, Johnson MC, et al. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell. 2009;139(3):499–511. doi: 10.1016/j.cell.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Galao RP, Le Tortorec A, Pickering S, Kueck T, Neil SJ. Innate sensing of HIV-1 assembly by Tetherin induces NFkappaB-dependent proinflammatory responses. Cell host & microbe. 2012;12(5):633–44. doi: 10.1016/j.chom.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tokarev A, Suarez M, Kwan W, Fitzpatrick K, Singh R, Guatelli J. Stimulation of NF-kappaB activity by the HIV restriction factor BST2. Journal of virology. 2013;87(4):2046–57. doi: 10.1128/JVI.02272-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jia B, Serra-Moreno R, Neidermyer W, Rahmberg A, Mackey J, Fofana IB, et al. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS pathogens. 2009;5(5):e1000429. doi: 10.1371/journal.ppat.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kluge SF, Mack K, Iyer SS, Pujol FM, Heigele A, Learn GH, et al. Nef proteins of epidemic HIV-1 group O strains antagonize human tetherin. Cell host & microbe. 2014;16(5):639–50. doi: 10.1016/j.chom.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Serra-Moreno R, Evans DT. Adaptation of human and simian immunodeficiency viruses for resistance to tetherin/BST-2. Current HIV research. 2012;10(4):277–82. doi: 10.2174/157016212800792496. [DOI] [PubMed] [Google Scholar]
- 88.Dube M, Roy BB, Guiot-Guillain P, Mercier J, Binette J, Leung G, et al. Suppression of Tetherin-restricting activity upon human immunodeficiency virus type 1 particle release correlates with localization of Vpu in the trans-Golgi network. Journal of virology. 2009;83(9):4574–90. doi: 10.1128/JVI.01800-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McNatt MW, Zang T, Bieniasz PD. Vpu binds directly to tetherin and displaces it from nascent virions. PLoS pathogens. 2013;9(4):e1003299. doi: 10.1371/journal.ppat.1003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90••.Usami Y, Wu Y, Gottlinger HG. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature. 2015;526(7572):218–23. doi: 10.1038/nature15400. One of the first papers published on SERINC3/5 and how it restricts HIV-1 and is antagonized by nef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Inuzuka M, Hayakawa M, Ingi T. Serinc, an activity-regulated protein family, incorporates serine into membrane lipid synthesis. The Journal of biological chemistry. 2005;280(42):35776–83. doi: 10.1074/jbc.M505712200. [DOI] [PubMed] [Google Scholar]
- 92.Asmuth DM, Murphy RL, Rosenkranz SL, Lertora JJ, Kottilil S, Cramer Y, et al. Safety, tolerability, and mechanisms of antiretroviral activity of pegylated interferon Alfa-2a in HIV-1-monoinfected participants: a phase II clinical trial. The Journal of infectious diseases. 2010;201(11):1686–96. doi: 10.1086/652420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Adamson CS, Freed EO. Anti-HIV-1 Therapeutics: From FDA-approved Drugs to Hypothetical Future Targets. Molecular Interventions. 2009;9(2):70–74. doi: 10.1124/mi.9.2.5. http://doi.org/10.1124/mi.9.2.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Smithgall TE, Thomas G. Small molecule inhibitors of the HIV-1 virulence factor, Nef. Drug Discovery Today. Technologies. 2013;10(4):e451–e548. doi: 10.1016/j.ddtec.2013.07.002. http://doi.org/10.1016/j.ddtec.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]