Abstract
The heterotrimeric G-protein binding site on G-protein coupled receptors remains relatively unexplored regarding its potential as a new target of therapeutic intervention or as a secondary site of action by the existing drugs. Tauroursodeoxycholic acid bears structural resemblance to several compounds that were previously identified to specifically bind to the light-activated form of the visual receptor rhodopsin and to inhibit its activation of transducin. We show that TUDCA stabilizes the active form of rhodopsin, metarhodopsin II, and does not display the detergent-like effects of common amphiphilic compounds that share the cholesterol scaffold structure, such as deoxycholic acid. Computer docking of TUDCA to the model of light-activated rhodopsin revealed that it interacts using similar mode of binding to the C-terminal domain of transducin alpha subunit. The ring regions of TUDCA made hydrophobic contacts with loop 3 region of rhodopsin, while the tail of TUDCA is exposed to solvent. The results show that TUDCA interacts specifically with rhodopsin, which may contribute to its wide-ranging effects on retina physiology and as a potential therapeutic compound for retina degenerative diseases.
Keywords: Bile acid, Chemical biology, G protein, G protein-coupled receptor (GPCR), Molecular docking, Photoreceptor, Phototransduction, Retina, Rhodopsin, Signal transduction
1. Introduction
G-protein-coupled receptors (GPCRs) are versatile transmembrane proteins that are responsible for the detection of extracellular stimuli, such as hormones, neurotransmitters and light and for the transmission of that information inside the cell via interaction with the membrane-associated heterotrimeric G-proteins to regulate various intracellular second messenger pathways (Palczewski and Orban, 2013; Manglik and Kobilka, 2014). Consequently, GPCRs possess two distinct sites for the binding of ligands and for the interactions with G-proteins on its extracellular and intracellular interfaces respectively. The ligand binding sites of GPCRs have received the most attention due to their extreme importance in receptor pharmacology resulting in estimates that 30–50% of drugs currently on the market target GPCRs (Howard et al., 2001; Salon et al., 2011). The G-protein binding site is relatively less explored, but is quickly catching up with regard to the number of recent biochemical and structural studies (Kisselev et al., 2011; Preininger et al., 2013). Most notably, the NMR and X-ray structures of the C-terminal tail of the G-protein α-subunit, Gtα(340–350), one of the main protein domains on G-proteins that binds GPCRs and stabilizes its active conformation, provide solid structure-based framework for possible pharmacological exploration of this site (Kisselev et al., 1998; Choe et al., 2011).
We have used light receptor rhodopsin and its cognate G-protein transducin to identify a number of small molecule compounds that bind to and stabilize the active form of rhodopsin, metarhodopsin II (Meta II) (Taylor et al., 2008, 2010). These molecules can be used to modulate GPCR-G-protein interactions, which can potentially be helpful as therapeutics for various retinal diseases caused by constitutively active GPCRs, such as Leber Congenital Amaurosis (LCA), Congenital Stationary Night Blindness (CSNB) and some forms of Retinitis Pigmentosa (RP) (Park, 2014; Rao and Oprian, 1996; Fain, 2006; Dizhoor et al., 2008). One group of molecules that we identified via in-silico and biochemical screens belongs to natural products, sapogenins, which share triterpenoid scaffolds (Taylor et al., 2008). Sapogenins, such as madecassic acid, also share structural resemblance with bile acids. One of the bile acids that has recently been gaining significant interest due to broad medicinal properties is tauroursodeoxycholic acid (TUDCA), which is a natural hydrophilic molecule containing taurine conjugated with the ursodeoxycholic acid (UDCA). While a major constituent of bear bile (Luo et al., 2010), TUDCA is produced in humans at relatively low levels. Used as a therapeutic compound it has been shown to prevent hepatic cytotoxicity and have neuroprotective properties (Keene et al., 2002; Oveson et al., 2011; Romero-Ramirez et al., 2017). Interestingly, TUDCA has been in use since ancient times as part of the traditional Chinese medicine toolbox (Boatright et al., 2006). The mechanisms of TUDCA effects have been under active investigations. They are currently believed to be mediated via effects on mitochondrial toxicity and inhibition of apoptotic and anti-inflammatory pathways (Romero-Ramirez et al., 2017).
There is accumulating evidence that TUDCA has wide-ranging effects on retina physiology. Positive effects on the retina morphology and functions were noted for Diabetic Retinopathy (DR) (Gaspar et al., 2013; Wang et al., 2016; Fernandez-Sanchez et al., 2015), RP (Phillips et al., 2008; Drack et al., 2012; Fu and Zhang, 2014), and LCA (Fu and Zhang, 2014). Considering structural similarity to the small molecule compounds we identified in earlier studies, we investigated whether TUDCA may potentially exert some of its effects on retinal photo-receptors by binding to the transducin site on light-activated rhodopsin. We used an assay that quantitatively measures stabilization of the active form of rhodopsin, Meta II by UV–Visible spectroscopy. We also used computer simulations to determine whether TUDCA would dock at the transducin binding site on Meta II. Both methods strongly argue for the specific binding of TUDCA to light activated rhodopsin.
2. Experimental procedures
2.1. Isolation of rhodopsin
Dark adapted frozen bovine retinas are obtained from W.L. Lawson, Co. (NE). Rod outer segments (ROS) are prepared by the method of Papermaster and Dreyer (1974). Urea-washed ROS membranes (UM) are prepared using the procedure adapted from Yamazaki et al. (1982), and Willardson et al. (1993), essentially as we described earlier (Kisselev et al., 1999a, 2007). Rhodopsin concentration is measured as ΔA498 before and after bleaching in the presence of 20 mM hydroxylamine, based on the molar extinction coefficient at 498 nM of 42,700 M-1 cm-1 (Hong and Hubbell, 1972).
2.2. UV/Visible spectroscopy
The amount of extra Meta II was measured on a Cary-50 UV/Visible spectrophotometer (Varian, CA), at 4 °C, cuvette path-length 10 mm, essentially as we described before (Kisselev et al., 1994, 1999b). Specific temperatures were maintained using Peltier-controlled cuvette holder. The sample compartment was continuously infused with dry air. Photoactivation of rhodopsin was achieved by illumination of samples for 20 s with a 150-Watt fiber optic light source passed through a 490 ± 5 nm bandwidth interference filter. Samples contained 2.5 μM of urea-washed ROS membranes in buffer Meta II (20 mM Tris-HCL pH 8.0, 130 mM NaCl, 1 mM MgCl2, 1 mM EDTA) and various amounts of tauroursodeoxycholic acid or deoxycholic acid (Sigma, MO). 700 nm–250 nm spectra were recorded before and after activation of the sample with a 490 ± 5 nm light. The amount of Meta II was calculated as the absorbance difference A380–A417 before and after pho-toactivation. The amount of Meta II calculated in reaction buffer was taken as zero. Sample turbidity was measured as the absorbance difference A280–A700 after photoactivation. The data were processed off-line using KaleidaGraph 3.6.2. Full spectra scans were normalized to zero at 700 nm.
2.3. Acid trapping
Acid trapping was used to verify the Meta II state (Kisselev et al., 1998; Kito et al., 1968) of R*. Essentially, the extra MII-stabilization protocol was used with some minor modifications. The UV/Vis absorbance spectra of dark-adapted rhodopsin mixed with TUDCA was taken in the dark and then after light activation. The final concentration of TUDCA was 5 mM. Immediately following the light activation scan, 1% HCl (v/v) was added and mixed. The sample was incubated in the spectrophotometer at 4 °C for 5 min, then the absorbance spectra was scanned an additional time.
2.4. Generating conformations of TUDCA
TUDCA was drawn and minimized (Tripos forcefield) in Sybyl 7.3. Gasteiger-Huckel charges were added to TUDCA using Sybyl 7.3. Because Gtα(340–350) is a peptide, the charges were calculated by RosettaLigand, as RosettaLigand is optimized for calculating charges on proteins (Davis and Baker, 2009). The Omega package from Open Eye was used to generate a series of 200 low-energy conformations of TUDCA (OpenEye Scientific Software, 2005). These conformations were sampled by RosettaLigand to provide ligand flexibility during docking.
2.5. Preparing the receptor
A model of R* and the X-ray crystal structure of opsin bound to Gtα(340–350)K341L (3DQB) (Scheerer et al., 2008) were used for docking studies. The 3D intracellular (IC) loop model of R* resulted from a previous study (Taylor et al., 2007) in which experimental TrNOE structures of Gtα(340–350) and its analogs (Kisselev et al., 1998; Anderson et al., 2006a, 2006b) were docked onto the IC loops. These docked structures revealed a common binding mode with similar residue-residue interactions that were potentially important for complex formation between R* and Gtα(340–350). The other docking target used was the structure of opsin bound to Gtα(340–350) (Scheerer et al., 2008). The two different models were utilized for validation of Gtα(340–350) docking to Meta II. Both structures were repacked using ligand_rpkmin with default parameters in the RosettaLigand package. RosettaLigand repacks the side chains in a stochastic manner to remove any clashes that exist using RosettaLigand’s energy function. A total of 10 structures were output, and the minimum energy repacked structure was used for docking calculations. For the model of the R* loops, capping on loop termini had to be removed to run RosettaLigand.
2.6. Docking
RosettaLigand was used for all docking calculations. The standard flags were used for RosettaLigand as outlined in the Rosetta 3.0 software (Davis and Baker, 2009). A random perturbation of up to 5 Å in the X, Y, and Z dimensions from the center of mass was implemented. However, points outside a 5 Å sphere were not considered, yielding uniform sampling within the sphere. The starting position for the docking calculation was the center of mass of the Gtα(340–350) from either the X-ray crystal structure of opsin (for docking to opsin) or from the model of R*. For each compound docked, a total of 10,000 poses were generated.
As a proof-of-principle experiment, Gtα(340–350)K341L from the X-ray crystal structure was re-docked onto the opsin crystal structure to determine if the binding pose would recapitulate the crystal structure. The Gtα(340–350) conformation that resulted from a previous study (Taylor et al., 2007) on R* loops was re-docked onto the R* loops to determine if its pose could be recapitulated. TUDCA was docked onto the opsin crystal structure and onto the 3D IC loop model of rhodopsin in a conformation bound to Gtα(340–350).
The scoring scheme of Davis et al. in RosettaLigand was used (Davis and Baker, 2009). First, any pose where the ligand was not interacting with the receptor was discarded. The top 5% of poses ranked by total energy were isolated and then ranked by their interaction energy. The top 10 poses were further studied.
3. Results
3.1. TUDCA binding to R*
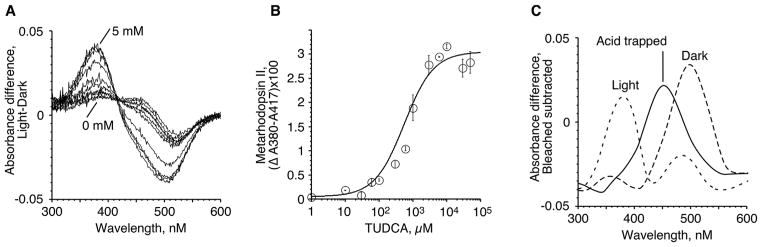
In order to test whether TUDCA specifically interacts with light activated rhodopsin, R*, we used an established spectroscopic assay that measures stabilization of the active R* intermediate, Meta II (Kisselev et al., 1999b). When rhodopsin is photoactivated at 4 °C, and pH 8.0, the dynamic equilibrium of two photointermediates, Meta I and Meta II is formed (Meta I, λmax = 480 nm and Meta II, λmax = 380 nm). Holo-transducin, Gt, and Gt-derived synthetic peptides form tight complex with Meta II and shift the Meta I-Meta II equilibrium towards Meta II. The extent of complex formation is reffected in the higher amplitude of the Meta II signal, which can be measured by the absorbance difference A380–A417 before and after photoactivation using UV/Visible spectroscopy. When TUDCA was added to the rhodopsin samples, we observed a concentration-dependent increase in the amount of Meta II, as seen as increase of absorbance at A380 nm, Fig. 1A. The light (L) minus dark (D) difference spectra also show parallel decrease of absorbance at A480, the spectral peak of the Meta I intermediate, Fig. 1A. The transition between Meta I and Meta II occurs via a well-defined isosbestic point at A417 nm. A380–A417 absorbance difference, a measure of Meta II in the sample, is plotted against TUDCA concentrations in Fig. 1B. The calculated EC50 of TUDCA in this assay is 450 μM.
Fig. 1.
Interaction of TUDCA with Meta II. A. UV/Visible difference spectra (L–D) of rhodopsin membranes in the presence of increasing concentrations of TUDCA (0–5 mM). B. Dose-dependent stabilization of Meta II by the indicated concentrations of TUDCA, n = 4. C. Smoothed UV/Visible difference spectra (L-bleached) of rhodopsin in the presence of 5 mM TUDCA: Dark, light activated, and acid-trapped spectra are shown.
In order to confirm identity of the Meta II photoproduct in R*-TUDCA complex we performed the acid-trapping test (Kisselev et al., 1998; Kito et al., 1968). Meta II contains a deprotonated Schiff base linkage between Lys296 and all-trans-retinal. In the strongly acidic environment Schiff base becomes re-protonated, which leads to the formation of the photoproduct at A440 nm. Contrary to the behavior of Meta II, the product of its decay, opsin + all-trans-retinal, has the covalent linkage between Lys296 and all-trans-retinal hydrolyzed. Thus, in the acid trapping test opsin + all-trans-retinal does not produce the A440 photoproduct. In the presence of 5 mM TUDCA light activated spectrum consists of the major photoproduct at A380 and a minor band at A480, which is likely Meta I due to incomplete Meta II stabilization by TUDCA. When hydrochloric acid was added to the R*-TUDCA reaction mixture, a distinct photoproduct absorbing at A450 was formed, Fig. 1C. Slight bathochromic spectral shift of the acid-denatured photoproduct (from A440 to A450) is because of the somewhat higher baseline absorbance at 400–440 nm in the fully bleached spectrum of rhodopsin in lipid membranes used to generate the difference spectrum (data not shown).
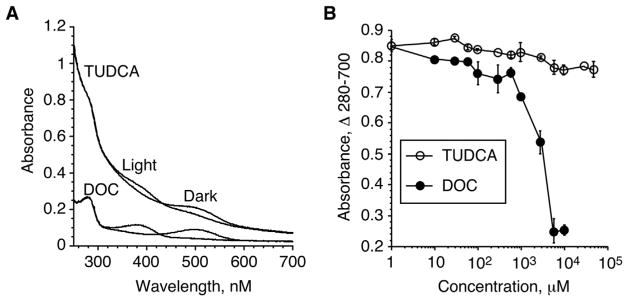
Analysis of the full spectra of R*-TUDCA samples also revealed that TUDCA appears to have little effect on the turbidity of the rhodopsin samples, and thus behave similarly to Gt, and Gt-derived synthetic peptides (Kisselev et al., 1998). The typical source of rhodopsin for the above spectroscopic experiments are Urea-washed ROS Membranes (UM), which contain highly enriched rhodopsin (99% purity) in its native membrane environment. Samples containing UM scatter light, which contribute to the steep slope of the full spectra, a phenomenon typical for membrane suspensions (Castanho et al., 1997). Addition of detergents, such as deoxycholate (DOC), reduce overall sample turbidity at concentrations above CMC, leading to flattening of the spectra (Goni and Alonso, 2000). Representative full UM spectra in the presence of 5 mM TUDCA or 5 mM DOC are shown in Fig. 2A. Despite structural similarities to detergents of the cholate group, such as DOC, Fig. 3, TUDCA had little effect on the turbidity of UM. The A280–A700 absorbance difference as measure of the overall slope of the full spectra after photoactivation remain essentially unaffected up to TUDCA concentration of 5 mM, and affected only slightly up to 50 mM, Fig. 2B. In strong contrast, DOC, a detergent often used to solubilize biological membranes shows precipitous decrease of sample turbidity with onset at 500 μM.
Fig. 2.
Differential effect of TUDCA and DOC on UM sample turbidity. A. Representative full Dark and Light-activated spectra of rhodopsin membranes in the presence of 5 mM TUDCA or 5 mM DOC. B. Dose-dependent effect on UM sample turbidity in the presence of increasing concentrations of TUDCA or DOC, n = 4.
Fig. 3.

Chemical structures of TUDCA and DOC.
3.2. Docking to opsin
In order to test whether TUDCA binds to the known G-protein site on R* we utilized two complimentary approaches: 1) Computer docking to the X-ray crystal structure of opsin stabilized by the high-affinity analogue of Gtα(340–350), Gtα(340–350)K341 (Scheerer et al., 2008), which offers the highest resolution of the transducin binding site on R*, but is inherently a static structure; and 2) docking to the molecular model of the R* intracellular loops (Taylor et al., 2007), that affords a more conformationally flexible interface which mimics dynamic R*-Gt interactions.
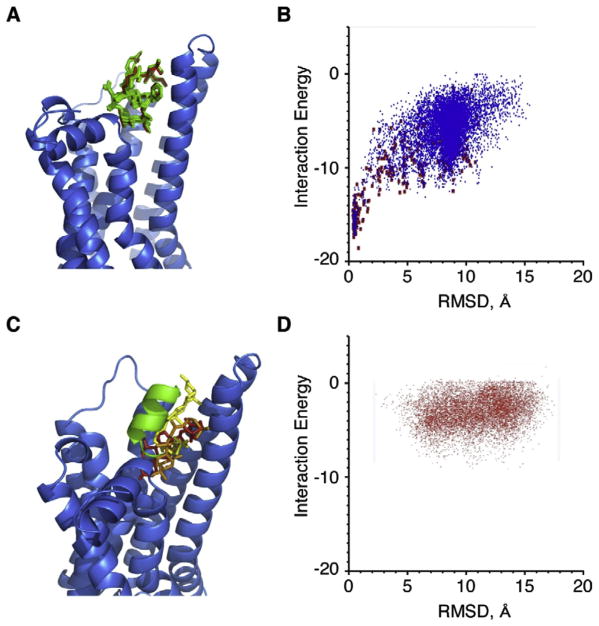
Before computer docking of TUDCA to R* was attempted, as a proof-of-principle for the utilized docking routine, Gtα(340–350)K341 was docked onto the X-ray crystal structure of opsin shown as a blue ribbon model. The top 10 poses of Gtα(340–350)K341 are conformationally very close to the X-ray crystal structure, with RMSDs ranging from 0.41 to 0.61 Å, Fig. 4A. The energy funnel that resulted from this calculation shows the minimum energy structures of the docked Gtα(340–350) K341 with the lowest RMSD values have converged well to the X-ray crystal structure of Gtα(340–350)K341, Fig. 4B. Next, TUDCA was docked onto the X-ray structure of opsin in a separate experiment. The top three TUDCA structures, corresponding to the three distinct energy minima, are shown docked to the X-ray crystal structure of opsin, Fig. 4C. The top ten TUDCA poses dock around the interface between Gtα(340–350)K341 and loop 3. The top two poses stretch along the reverse-turn region of Gtα(340–350)K341, and the third top pose docks along the hydrophobic helical region of Gtα(340–350)K341. The plot of TUDCA interaction energy versus RMSD from starting structure has three main minimal-energy regions, Fig. 4D. No other low energy docking sites of TUDCA on the X-ray structure of opsin were identified.
Fig. 4.
Computer docking of Gtα(340–350)K341L and TUDCA to R*, X-ray crystal structure of opsin. A. Gtα(340–350)K341L re-docked onto the X-ray crystal structure of opsin (blue ribbon). The docked poses are shown in green, and the X-ray crystal structure is shown in red as stick models. B. Plot of the energy funnel that resulted from the Gtα(340–350)K341L docking calculations. The blue dots represent all the docked poses, and the red squares are poses that scored in the top 5% of poses based on total energy. C. TUDCA docked onto the X-ray crystal structure of opsin (blue ribbon). The best scoring TUDCA pose is shown in red, followed by the second best in orange, and third best in yellow as stick models. The X-ray crystal structure of Gtα(340–350)K341L is shown in green as a ribbon. D. The plot of TUDCA interaction energy vs RMSD from the starting structure. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Docking to R* model
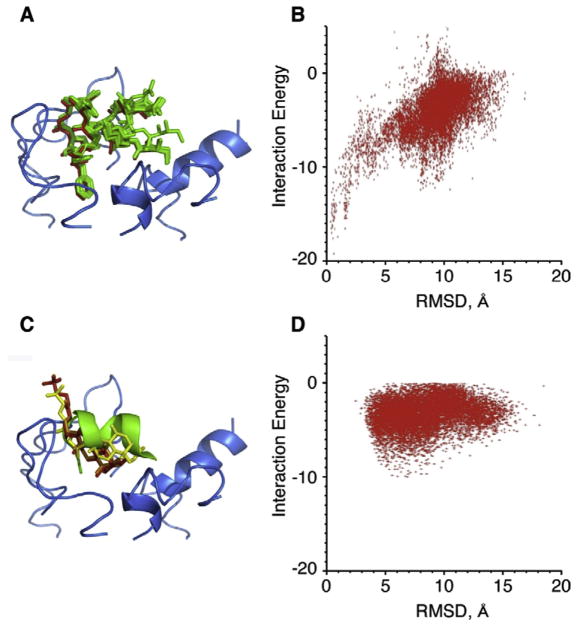
In order to explore R*-Gt interactions under conditions that closely resemble physiological, a dynamic model of R* intracellular loops was developed based on both experimental transferred nuclear Overhauser effect data from R* in complex with the native peptide Gtα(340–350) (Kisselev et al., 1998) and computational data (Taylor et al., 2007). The lowest energy pose of Gtα(340–350) from Taylor et al. (2007) was docked onto the loop model of R*. The top ten docked poses with lowest energy and RMSD values ranging from 0.48 to 1.01 Å, are shown in Fig. 5A. The energy funnel of Gtα(340–350) docked to R* loops, Fig. 5B, bares close resemblance to the energy funnel of Gtα(340–350)K341 docked to opsin, Fig. 4B.
Fig. 5.
Computer docking of native Gtα(340–350) and TUDCA to the model of the intracellular loops of R*. A. Ten lowest energy conformations of Gtα(340–350) are green stick models docked to the intracellular loops of R* (blue ribbon). The original model pose of Gtα(340–350) is in red. B. The energy funnel that resulted from the docking calculations showing convergence on the model of the complex. C. TUDCA shown as stick models docked onto the model of R* loops (blue ribbon). The best scoring solution that docked along the helical axis of Gtα(340–350) is shown in red, the second best in orange and the third best in yellow. The model of Gtα(340–350) is shown as green ribbon. D. The plot of interaction energy vs RMSD from the starting structure. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Finally, TUDCA was docked onto the R* loop model. The hydrophobic ring region of TUDCA docks similarly to Gtα(340–350), along the helical axis in this model. Out of the top ten best scoring poses, five poses stretched along the helical axis of Gtα(340–350). The top three poses that stretch along the helical axis are shown in Fig. 5C. Two of these poses are nearly identical and the third pose presents itself along slightly different regions of the helix location. Corresponding energy funnel is presented in Fig. 5D.
4. Discussion
Structural similarity of TUDCA’s triterpenoid scaffold to madecassic acid and some other small molecule compounds we identified in previous studies (Taylor et al., 2008) has prompted us to investigate whether reported positive effects of TUDCA on retinal pathophysiology may be related to its ability to bind to and stabilize the active intermediate of rhodopsin, Meta II. In the spectroscopic assays TUDCA demonstrates dose-dependent stabilization of Meta II, Fig. 1. Similar to the effect of Gt and Gt-derived synthetic peptides, TUDCA strongly shifts the Meta I – Meta II equilibrium towards Meta II with the spectroscopic transition proceeding via a single isosbestic point at A417 nm. Notably, increase of Meta II occurs at the expense of the Meta I intermediate. These results strongly argue for direct interactions and specific formation of the Meta II-TUDCA complex. Meta II identity in this complex was confirmed by the conversion of the Meta II photoproduct at A380 to the acid-trapped photoproduct absorbing at A450 under strongly acidic conditions, Fig. 1C. Calculated EC50 concentration of TUDCA in the Meta II stabilization assay is 450 μM, which is close to the EC50 for the native peptide Gtα(340–350) at 300 μM (Kisselev et al., 1998), (Downs et al., 2006). Thus, it appears that TUDCA binds to Meta II with approximately the same affinity as the native Gtα(340–350).
Additional support for the specificity of interactions comes from the analysis of TUDCA on the overall turbidity of the rhodopsin membranes, a sensitive test of membrane perturbation. Even at concentrations as high as 50 mM, which is ten-fold higher than its apparent EC50 in the Meta II assay, TUDCA has only slight effect on membrane turbidity, Fig. 2. In contrast, effect of the ionic detergent deoxycholate (DOC) that features similar structural scaffold to TUDCA on membrane turbidity is clearly observed by 1 mM. These results lead to the conclusion that TUDCA’s ability to bind to Meta II is specific, rather than related to its potential ability to solubilize lipid membranes and interfere in lipid-protein interactions.
To further substantiate TUDCA’s binding to Meta II, we examined whether TUDCA would dock to the known intracellular binding site for the Gtα subunit on Meta II. Using two independent docking models, one based on the static X-ray crystal structure of opsin stabilized by the high-affinity analogue of Gtα(340–350), Gtα(340–350)K341 (Scheerer et al., 2008), and the other based on the dynamic model of the R* intracellular loops (Taylor et al., 2007), we show that TUDCA successfully binds at the same site as Gtα(340–350) using similar mode of binding.
To verify the docking strategy, we tested our ability to recapitulate the docking model of native peptide Gtα(340–350) and its high-affinity analogue Gtα(340–350)K341 to the model of R* loops and opsin, respectively. Both peptides docked extremely well to their respective models with very small RMSDs when compared to the X-ray crystal structure of opsin and the R* model generated previously. The energy funnels for both compounds show the minimal energy poses also being the minimal RMSD from the native structure.
In both models, TUDCA docks along the crucial L344/L349/F350 hydrophobic patch of the Gtα(340–350) alpha helix (Kisselev et al., 1998) that makes contacts with loop 3 of rhodopsin (Scheerer et al., 2008) a GPCR site known for it’s importance in G-protein activation. The ring regions of TUDCA interact where the hydrophobic region of Gtα(340–350) is bound along the hydrophobic inner side of the cytoplasmic portion of TM5 and TM6 pair (Scheerer et al., 2008), and the hydrophilic tail of TUDCA wraps around so that it is more solvent exposed. TUDCA docks in much the same way other compounds found via a virtual screening approach, despite a different docking program being used (Taylor et al., 2008). These structurally similar compounds identified earlier, such as madecassic acid, successfully stabilized the Meta II state and also inhibited transducin activation. The ring structures on these compounds also bound along the hydrophobic portion of Gtα(340–350), parallel to the helical axis, which implies a common mode of recognition and binding. Future biochemical and in-vivo studies will determine whether TUDCA’s ability to allosterically regulate rhodopsin can be utilized to modulate signaling properties of retinal photoreceptors. It is of particular interest in this context that a small molecule quercetin was demonstrated to act as an allosteric modulator of mutated forms of rhodopsin, such as RP-associated G90V, validating the concept of this new methodology (Herrera-Hernández et al., 2017).
Strong evidence exists in published literature that such distinct retinal degenerations as RP and LCA may be related to the detrimental effects of constitutively active rhodopsin mutants or opsin, and that inhibiting this activity may be a valid therapeutic approach (Park, 2014; Rao and Oprian, 1996; Fain, 2006; Dizhoor et al., 2008). Meta II binding and stabilization data, as well as computer docking results presented here identify TUDCA as a compound that may exert at least some of its known anti-RD properties and effects on retinal photo-receptors by binding to the transducin site on light activated rhodopsin.
Acknowledgments
This work was supported by NIH Grants GM063203 and EY018107 (OGK), and by Career Development Award from Research to Prevent Blindness (OGK)
Abbreviations
- GPCR
G-protein coupled receptor
- G-protein
heterotrimeric GTP-binding protein
- R
dark-adapted rhodopsin
- R*
photoactivated rhodopsin
- Meta II
metarhodopsin II
- NMR
nuclear magnetic resonance
- TrNOE
transferred nuclear Overhauser effect
- IC
intracellular
- EC
extracellular
- TM
transmembrane
- UM
urea-washed rod outer-segment membranes
- ROS
rod outer segment
- RMSD
root mean square deviation
- Gt
transducin
- Gtα(340–350)
transducin alpha subunit C-terminal region
- TUDCA
tauroursodeoxycholic acid
- DOC
sodium deoxycholate
Footnotes
Conflicts of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Author contributions
OGK and GRM conceived and coordinated the study; EL, OGK and CMT wrote the paper. EL and OGK designed, performed and analyzed the experiments shown in Figs. 1 and 2. CMT designed, performed and analyzed the experiments shown in Figs. 4 and 5. All authors reviewed the results and approved the final version of the manuscript.
References
- Anderson MA, Ogbay B, Arimoto R, Sha W, Kisselev OG, Cistola DP, Marshall GR. Relative strength of cation-pi vs salt-bridge interactions: the Gtalpha (340–350) peptide/rhodopsin system. J Am Chem Soc. 2006a;128:7531–7541. doi: 10.1021/ja058513z. [DOI] [PubMed] [Google Scholar]
- Anderson MA, Ogbay B, Kisselev OG, Cistola DP, Marshall GR. Alternate binding mode of C-terminal phenethylamine analogs of G(t)alpha(340–350) to photoactivated rhodopsin. Chem Biol Drug Des. 2006b;68:295–307. doi: 10.1111/j.1747-0285.2006.00460.x. [DOI] [PubMed] [Google Scholar]
- Boatright JH, Moring AG, McElroy C, Phillips MJ, Do VT, Chang B, Hawes NL, Boyd AP, Sidney SS, Stewart RE, Minear SC, Chaudhury R, Ciavatta VT, Rodrigues CM, Steer CJ, Nickerson JM, Pardue MT. Tool from ancient pharmacopoeia prevents vision loss. Mol Vis. 2006;12:1706–1714. [PubMed] [Google Scholar]
- Castanho MA, Santos NC, Loura LM. Separating the turbidity spectra of vesicles from the absorption spectra of membrane probes and other chromophores. Eur Biophys J. 1997;26:253–259. [Google Scholar]
- Choe HW, Kim YJ, Park JH, Morizumi T, Pai EF, Krauss N, Hofmann KP, Scheerer P, Ernst OP. Crystal structure of metarhodopsin II. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- Davis IW, Baker D. RosettaLigand docking with full ligand and receptor flexibility. J Mol Biol. 2009;385:381–392. doi: 10.1016/j.jmb.2008.11.010. [DOI] [PubMed] [Google Scholar]
- Dizhoor AM, Woodruff ML, Olshevskaya EV, Cilluffo MC, Cornwall MC, Sieving PA, Fain GL. Night blindness and the mechanism of constitutive signaling of mutant G90D rhodopsin. J Neurosci. 2008;28:11662–11672. doi: 10.1523/JNEUROSCI.4006-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs MA, Arimoto R, Marshall GR, Kisselev OG. G-protein alpha and beta-gamma subunits interact with conformationally distinct signaling states of rhodopsin. Vis Res. 2006;46:4442–4448. doi: 10.1016/j.visres.2006.07.021. [DOI] [PubMed] [Google Scholar]
- Drack AV, Dumitrescu AV, Bhattarai S, Gratie D, Stone EM, Mullins R, Shefield VC. TUDCA slows retinal degeneration in two different mouse models of retinitis pigmentosa and prevents obesity in Bardet-Biedl syndrome type 1 mice. Invest Ophthalmol Vis Sci. 2012;53:100–106. doi: 10.1167/iovs.11-8544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fain GL. Why photoreceptors die (and why they don’t) Bioessays. 2006;28:344–354. doi: 10.1002/bies.20382. [DOI] [PubMed] [Google Scholar]
- Fernandez-Sanchez L, Lax P, Noailles A, Angulo A, Maneu V, Cuenca N. Natural compounds from saffron and bear bile prevent vision loss and retinal degeneration. Molecules. 2015;20:13875–13893. doi: 10.3390/molecules200813875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Zhang T. Pathophysilogical mechanism and treatment strategies for Leber congenital amaurosis. Adv Exp Med Biol. 2014;801:791–796. doi: 10.1007/978-1-4614-3209-8_99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar JM, Martins A, Cruz R, Rodrigues CM, Ambrosio AF, Santiago AR. Tauroursodeoxycholic acid protects retinal neural cells from cell death induced by prolonged exposure to elevated glucose. Neuroscience. 2013;253:380–388. doi: 10.1016/j.neuroscience.2013.08.053. [DOI] [PubMed] [Google Scholar]
- Goni FM, Alonso A. Spectroscopic techniques in the study of membrane solubilization, reconstitution and permeabilization by detergents. Biochim Biophys Acta. 2000;1508:51–68. doi: 10.1016/s0304-4157(00)00011-3. [DOI] [PubMed] [Google Scholar]
- Herrera-Hernández MG, Ramon E, Lupala CS, Tena-Campos M, Pérez JJ, Garriga P. Flavonoid allosteric modulation of mutated visual rhodopsin associated with retinitis pigmentosa. Sci Rep. 2017 Sep 11;7(1):11167. doi: 10.1038/s41598-017-11391-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K, Hubbell WL. Preparation and properties of phospholipid bilayers containing rhodopsin. Proc Natl Acad Sci USA. 1972;69:2617–2621. doi: 10.1073/pnas.69.9.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard AD, McAllister G, Feighner SD, Liu Q, Nargund RP, Van der Ploeg LH, Patchett AA. Orphan G-protein-coupled receptors and natural ligand discovery. Trends Pharmacol Sci. 2001;22:132–140. doi: 10.1016/s0165-6147(00)01636-9. [DOI] [PubMed] [Google Scholar]
- Keene CD, Rodrigues CM, Eich T, Chhabra MS, Steer CJ, Low WC. Tauroursodeoxycholic acid, a bile acid, is neuroprotective in a transgenic animal model of Huntington’s disease. Proc Natl Acad Sci U S A. 2002;99:10671–10676. doi: 10.1073/pnas.162362299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisselev OG, Ermolaeva MV, Gautam N. A farnesylated domain in the G protein gamma subunit is a specific determinant of receptor coupling. J Biol Chem. 1994;269:21399–21402. [PubMed] [Google Scholar]
- Kisselev OG, Kao J, Ponder JW, Fann YC, Gautam N, Marshall GR. Light-activated rhodopsin induces structural binding motif in G protein alpha subunit. Proc Natl Acad Sci U S A. 1998;95:4270–4275. doi: 10.1073/pnas.95.8.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisselev OG, Pronin AP, Gautam N. In: G-proteins: Techniques of Analysis. Manning DR, editor. CRC Press; 1999a. pp. 85–95. [Google Scholar]
- Kisselev OG, Meyer CK, Heck M, Ernst OP, Hofmann KP. Signal transfer from rhodopsin to the G-protein: evidence for a two- site sequential fit mechanism. Proc Natl Acad Sci U S A. 1999b;96:4898–4903. doi: 10.1073/pnas.96.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisselev OG. Photoactivation of rhodopsin and signal transfer to transducin. In: Fliesler SJ, Kisselev OG, editors. Signal Transduction in the Retina. CRC Press; 2007. pp. 33–54. [Google Scholar]
- Kisselev OG, Park JH, Choe H-W, Ernst OP. G Protein-coupled Receptors: from Structure to Function. Royal Society of Chemistry; 2011. pp. 54–74. [Google Scholar]
- Kito Y, Suzuki T, Azuma M, Sekoguti Y. Absorption spectrum of rhodopsin denatured with acid. Nature. 1968;218:955–957. doi: 10.1038/218955a0. [DOI] [PubMed] [Google Scholar]
- Luo Q, Chen Q, Wu Y, Jiang M, Chen Z, Zhang X, Chen H. Chemical constituents of bear bile. Zhongguo Zhongyao Zazhi. 2010;35:2416–2419. [PubMed] [Google Scholar]
- Manglik A, Kobilka B. The role of protein dynamics in GPCR function: insights from the beta2AR and rhodopsin. Curr Opin Cell Biol. 2014;27:136–143. doi: 10.1016/j.ceb.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oveson BC, Iwase T, Hackett SF, Lee SY, Usui S, Sedlak TW, Snyder SH, Campochiaro PA, Sung JU. Constituents of bile, bilirubin and TUDCA, protect against oxidative stress-induced retinal degeneration. J Neurochem. 2011;116:144–153. doi: 10.1111/j.1471-4159.2010.07092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, Orban T. From atomic structures to neuronal functions of g protein-coupled receptors. Annu Rev Neurosci. 2013;36:139–164. doi: 10.1146/annurev-neuro-062012-170313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papermaster DS, Dreyer WJ. Rhodopsin content in the outer segment membranes of bovine and frog retinal rods. Biochemistry. 1974;13:2438–2444. doi: 10.1021/bi00708a031. [DOI] [PubMed] [Google Scholar]
- Park PS. Constitutively active rhodopsin and retinal disease. Adv Pharmacol. 2014;70:1–36. doi: 10.1016/B978-0-12-417197-8.00001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MJ, Walker TA, Choi HY, Faulkner AE, Kim MK, Sidney SS, Boyd AP, Nickerson JM, Boatright JH, Pardue MT. Tauroursodeoxycholic acid preservation of photoreceptor structure and function in the rd10 mouse through postnatal day 30. Invest Ophthalmol Vis Sci. 2008;49:2148–2155. doi: 10.1167/iovs.07-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preininger AM, Meiler J, Hamm HE. Conformational flexibility and structural dynamics in GPCR-mediated G protein activation: a perspective. J Mol Biol. 2013;425:2288–2298. doi: 10.1016/j.jmb.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VR, Oprian DD. Activating mutations of rhodopsin and other G protein-coupled receptors. Annu Rev Biophys Biomol Struct. 1996;25:287–314. doi: 10.1146/annurev.bb.25.060196.001443. [DOI] [PubMed] [Google Scholar]
- Romero-Ramirez L, Nieto-Sampedro M, Yanguas-Casas N. Tauroursodeoxycholic acid: more than just a neuroprotective bile conjugate. Neural Regen Res. 2017;12:62–63. doi: 10.4103/1673-5374.198979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salon JA, Lodowski DT, Palczewski K. The significance of G protein-coupled receptor crystallography for drug discovery. Pharmacol Rev. 2011;63:901–937. doi: 10.1124/pr.110.003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, Hofmann KP, Ernst OP. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- Taylor CM, Nikiforovich GV, Marshall GR. Defining the interface between the C-terminal fragment of alpha-transducin and photoactivated rhodopsin. Biophys J. 2007;92:4325–4334. doi: 10.1529/biophysj.106.099242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CM, Barda Y, Kisselev OG, Marshall GR. Modulating G-protein coupled receptor/G-protein signal transduction by small molecules suggested by virtual screening. J Med Chem. 2008;51:5297–5303. doi: 10.1021/jm800326q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CM, Rockweiler NB, Liu C, Rikimaru L, Tunemalm AK, Kisselev OG, Marshall GR. Using ligand-based virtual screening to allosterically stabilize the activated state of a GPCR. Chem Biol Drug Des. 2010;75:325–332. doi: 10.1111/j.1747-0285.2009.00944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CF, Yuan JR, Qin D, Gu JF, Zhao BJ, Zhang L, Zhao D, Chen J, Hou XF, Yang N, Bu WQ, Wang J, Li C, Tian G, Dong ZB, Feng L, Jia XB. Protection of tauroursodeoxycholic acid on high glucose-induced human retinal microvascular endothelial cells dysfunction and streptozotocin-induced diabetic retinopathy rats. J Ethnopharmacol. 2016;185:162–170. doi: 10.1016/j.jep.2016.03.026. [DOI] [PubMed] [Google Scholar]
- Willardson BM, Pou B, Yoshida T, Bitensky MW. Cooperative binding of the retinal rod G-protein, transducin, to light-activated rhodopsin. J Biol Chem. 1993;268:6371–6382. [PubMed] [Google Scholar]
- Yamazaki A, Bartucca F, Ting A, Bitensky MW. Reciprocal effects of an inhibitory factor on catalytic activity and noncatalytic cGMP binding sites of rod phosphodiesterase. Proc Natl Acad Sci USA. 1982;79:3702–3706. doi: 10.1073/pnas.79.12.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]