Abstract
Circulating tumor-derived extracellular vesicles (EVs) have emerged as a promising source for identifying cancer biomarkers for early cancer detection. However, the clinical utility of EVs has thus far been limited by the fact that most EV isolation methods are tedious, nonstandardized, and require bulky instrumentation such as ultracentrifugation (UC). Here, we report a size-based EV isolation tool called ExoTIC (exosome total isolation chip), which is simple, easy-to-use, modular, and facilitates high-yield and high-purity EV isolation from biofluids. ExoTIC achieves an EV yield ~4–1000-fold higher than that with UC, and EV-derived protein and microRNA levels are well-correlated between the two methods. Moreover, we demonstrate that ExoTIC is a modular platform that can sort a heterogeneous population of cancer cell line EVs based on size. Further, we utilize ExoTIC to isolate EVs from cancer patient clinical samples, including plasma, urine, and lavage, demonstrating the device’s broad applicability to cancers and other diseases. Finally, the ability of ExoTIC to efficiently isolate EVs from small sample volumes opens up avenues for preclinical studies in small animal tumor models and for point-of-care EV-based clinical testing from fingerprick quantities (10–100 μL) of blood.
Keywords: exosome total isolation chip, ExoTIC, tumor-derived extracellular vesicles, isolation, lung cancer, biofluids
Graphical Abstract
Exosomes are nanometer-sized (30–180 nm) extracellular vesicles (EVs) that are actively shed from cells into body fluids.1–5 EVs that are released from tumor cells have recently received considerable attention,6,7 as they contain biomarkers such as tumor-specific proteins8,9 and nucleic acids (mRNA, microRNA, and DNA fragments) that are indicative of cancer stage and progression.5,6,10 Therefore, EVs in body fluids have emerged as a promising source of cancer biomarkers for diagnosis, prognostication, and treatment monitoring.7,11,12 However, the lack of technical standardized tools to consistently isolate high-yield and high-purity intact EVs poses a significant roadblock to their implementation for reliable biomarker discovery. Currently, ultracentrifugation (UC)13,14 is the most commonly used EV isolation method in the research community, despite being labor intensive, time-consuming, and resulting in poor EV quality. Other methods for EV isolation, such as multistep filtration15,16 still require a bulky centrifuge or vacuum system and large sample volumes (30–100 mL), yet deliver poor yields. Immuno-magnetic bead-based capture methods are expensive and rely heavily on specific antibodies,13,17,18 which vary from batch to batch and suffer from stability issues. Precipitation-based polyethylene glycol (PEG) methods are limited by small sample volumes and suffer from unacceptable purity10,19 for downstream analysis due to polymer contamination.19,20 Noncommercial microscale and nanoscale technologies sort EVs by size using acoustics, filtration, or by lateral displacement.21–26 However, these EV isolation methods are limited by the requirement of sophisticated fabrication technologies. Thus, a simple, inexpensive, and rapid EV isolation method that can process diverse biofluids is an essential but unmet need in many research and clinical settings.
To overcome these critical challenges, we have designed and implemented an EV isolation tool, the exosome total isolation chip (ExoTIC), which provides high-yield EVs for downstream analysis. We first demonstrate that ExoTIC can efficiently isolate intact EVs from culture media and healthy human plasma. We then show that ExoTIC is a highly modular tool that enables size-based EV sorting from heterogeneous EV populations. Further, we compare the microRNA and proteomic profiles from EVs isolated by ExoTIC and standard UC, demonstrating that results from our easy-to-use device are consistent with the labor-intensive UC method. Finally, we apply ExoTIC to EV isolation from clinical samples including plasma, urine, and lung bronchoalveolar lavage fluid. Our platform can process multiple types of complex biofluids and therefore has broad applicability to a whole host of cancers and other diseases.
RESULTS AND DISCUSSION
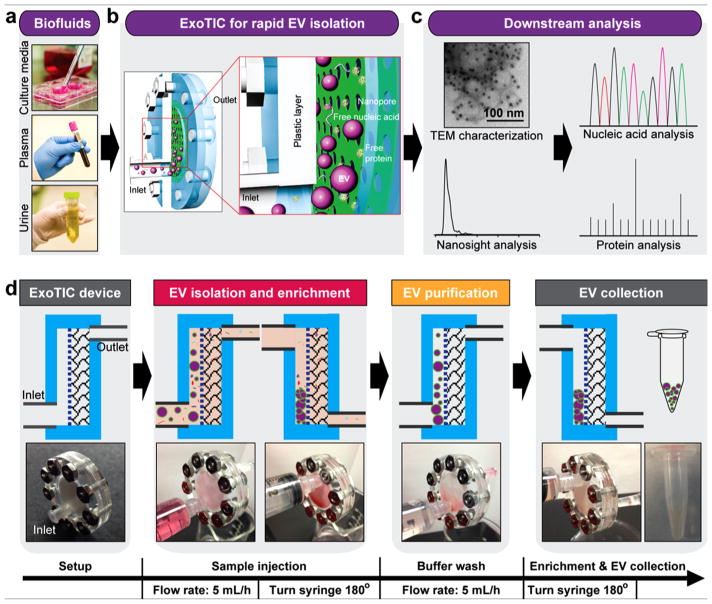
Working Principle of ExoTIC
ExoTIC was specifically designed to simplify EV isolation in the research and clinical point of care settings. ExoTIC uses a simple filtration approach in which EV-containing clinical samples, including culture media, plasma, and urine (Figure 1a), are passed through a nanoporous membrane to enrich and purify intact EVs in the 30–200 nm size range. Free nucleic acids, proteins, lipids, and other small fragments are flushed out (Figure 1b), and concentrated EVs are collected from the filter membrane using a standard pipet. The EVs are then characterized using nanoparticle tracking analysis (NTA) and transmission electron microscopy (TEM) for size, concentration, and morphology and then undergo downstream proteomic and transcriptomic analysis (Figure 1c). The detailed design and materials used for the ExoTIC device are shown in Figure S1. The fabrication process involves step-by-step assembly from laser-cut plastic layers, a polycarbonate track-etched nanoporous filter membrane, poly(ether sulfone) (PES) layer, and cellulose pad. The plastic housing is secured with metal screws and nuts, and a plastic ring-shaped gasket provides a leak-free seal (see Figure S2). The cellulose pad prevents deformation of the filter membrane under the pressures generated by the syringe pump. The workflow of isolating EVs from culture media is shown in Figure 1d. A 10 mL sample solution, prefiltered with a 0.22 μm PES syringe filter, is introduced continuously into the ExoTIC device via a 10 mL syringe using a syringe pump (multichannel, Figure S3) at a constant flow rate (5 mL/h). Once the sample has been concentrated down to 1 mL, the ExoTIC device is rotated by 180° so that the EVs are enriched at the end opposite to the inlet in order to minimize any potential sample loss. The concentrated EVs and any residual EVs bound to the filter membrane are recovered in the same tube, and the isolated EVs are then used for downstream physical characterization and molecular analysis.
Figure 1.
Schematic illustration of the ExoTIC device for extracellular vesicle isolation. (a) Various biofluids can be processed for EV isolation including culture media, plasma, and urine. (b) Schematic illustration of size-based EV isolation using the ExoTIC device. Intact EVs are enriched and purified at the filter, whereas the free proteins and nucleic acids are washed out. (c) Downstream analysis of EVs isolated from different clinical sample types for size, morphology, and molecular contents. (d) Schematic process of EV isolation from sample-in to EV-out. Device operation includes isolation of EVs from cell culture media (5 mL in 1 h), washing with PBS buffer (5 mL in 1 h), and collection of ~200 μL of EV solution for subsequent analysis. Total operation time for 5–10 mL of sample is under 3 h.
ExoTIC Provides Higher EV Yields Than UC and PEG Precipitation
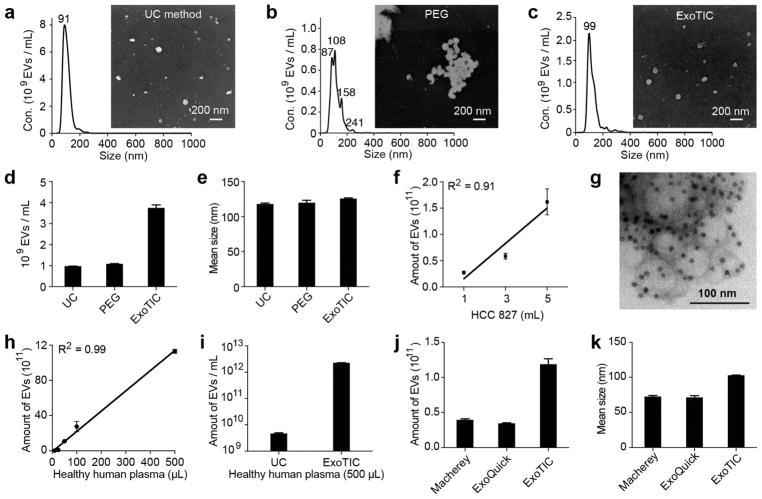
ExoTIC was compared with UC and polyethylene glycol (PEG)-based precipitation with respect to yield, size, and morphology of EVs isolated from culture media. ExoTIC isolated >90% of the EVs present in culture media (see Figure S4). The size distribution of EVs was measured using NTA,3 and their morphology and size were confirmed by scanning electron microscopy (SEM).27 As shown in Figure 2a, EVs isolated by UC had a unimodal peak centered at 91 nm by NTA and exhibited a size distribution of ~20–130 nm by SEM. The EVs purified by PEG precipitation formed aggregates and had multimodal peaks (Figure 2b). EVs isolated by ExoTIC had a unimodal peak at 99 nm by NTA (Figure 2c) with a size distribution of ~30–100 nm by SEM. ExoTIC also isolated EVs from culture media at a 4-fold higher yield compared to that with UC (Figure 2d). The mean size of the EVs purified by all three methods was ~120 nm (Figure 2e and Figure S5). Figure 2f provides a comparison of the total number of EVs purified from different volumes (from 1 to 5 mL) of HCC827 cell culture medium using ExoTIC, with the expected linear correlation between EV number and media volume. The TEM image in Figure 2g shows EVs labeled with immunogold for CD63.28,29 These results demonstrate that the ExoTIC device can efficiently isolate EVs over a wide size range while avoiding polymer contamination.
Figure 2.
Physical properties of EVs isolated by different methods. NTA and SEM analysis of EVs isolated by (a) UC, (b) PEG-based precipitation, and (c) ExoTIC. (d) Yield comparison of EVs purified from HCC827 lung adenocarcinoma cell culture medium by the ExoTIC device (5 mL), ultracentrifugation (60 mL), and PEG (5 mL). (e) Mean size of EVs purified by the three methods as determined by NTA (NanoSight NS300). (f) Total quantity of EVs purified from different volumes of HCC827 cell culture medium using the ExoTIC device. (g) TEM image of EVs isolated from cell culture media (HCC827 cell line) using the ExoTIC device, immuno-gold labeled for CD63 (dark spots). (h) Demonstration of the ExoTIC device’s ability to isolate EVs from plasma volumes as low as 10 μL up to 500 μL. (i) Yield comparison between UC and ExoTIC device of EVs isolated from 500 μL of plasma from healthy human donors. Comparison of three different isolation methods with respect to (j) yield and (k) mean size (as determined by NTA) of EVs isolated from 100 μL plasma (mean ± SD, n = 5). Mean size refers to the average size of the EVs in the size distribution. Mean size values are automatically generated in the NanoSight report.
We then compared the performance of ExoTIC with UC and PEG precipitation in isolating EVs from the plasma of healthy patients (see Figure S6).30 We first investigated the ability of the ExoTIC device to process low volumes (10 to 500 μL) of plasma (Figure 2h). When compared to UC purification of the same volume, the EV yield purified from 500 μL of healthy human plasma by the ExoTIC device was ~1000 times higher (Figure 2i). When compared with commercial PEG precipitation kits (ExoQuick and Macherey), the ExoTIC device achieved 3–4-fold higher EV yields (Figure 2j and Figure S7). The mean size of EVs isolated by ExoTIC was ~30% larger than that isolated by the other two commercial kits (Figure 2k), which may be due to the isolation methods.
Evaluation of EV MicroRNAs by ExoTIC and UC
EV microRNAs are a potential source of cancer biomarkers for early cancer diagnosis.31–35 The NanoString eCounter gene expression profiling technology has been successfully used to identify circulating microRNA signatures in patients with ulcerative colitis,36 Crohn’s Disease,37 and melanoma.38 Plasma and urine contain large amounts of ribonucleases (RNases). Therefore, a prerequisite for microRNAs and mRNAs to serve as biomarkers is that they must be resistant to degradation by RNases. These features make EV-derived microRNAs and mRNAs particularly suitable as biomarkers for diagnosis of cancer.
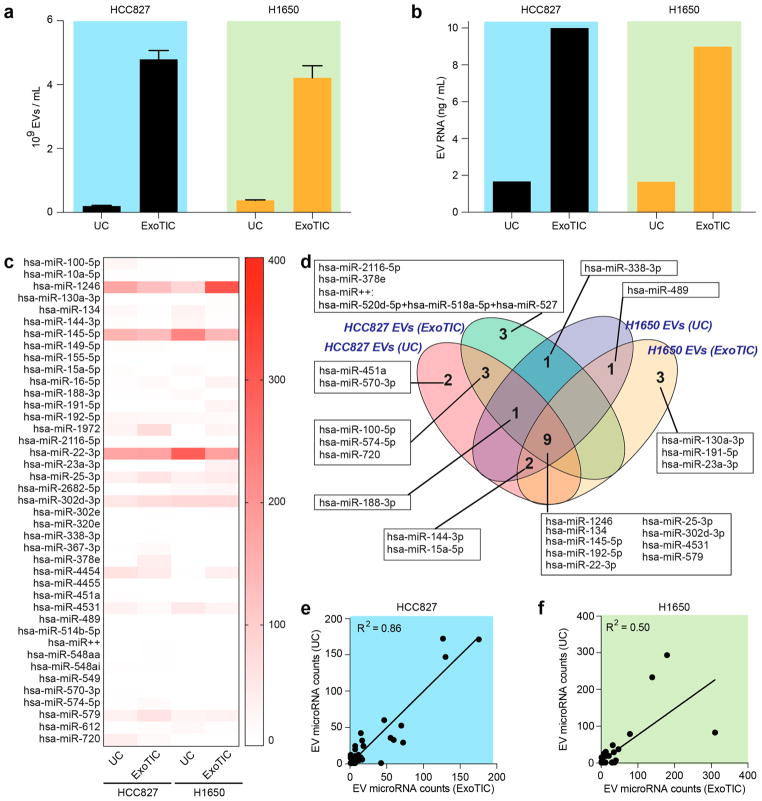
Using conditioned culture media from lung cancer cell lines (HCC827 and H1650) as a proof-of-concept, we compared ExoTIC- and UC-isolated EVs with respect to miRNA profiles. We first characterized the quantity and morphology of the EVs and subsequently analyzed their microRNA expression levels. ExoTIC again isolated >4-fold higher yields of EVs from conditioned culture media compared to UC (Figure 3a) and 6- fold higher yields of EV-derived microRNA compared to UC (Figure 3b and Table S1). MicroRNA expression levels between ExoTIC and UC were further analyzed by absolute quantitation of over 800 distinct microRNAs using fluorescent tags and digital imaging (Nanostring)39 (Figure 3c). The 40 most highly expressed microRNAs were common (100% overlap) to both EV isolation methods. The Venn diagram in Figure 3d for two lung cancer cell lines shows both overlapping and non-overlapping EV microRNAs (25 most highly expressed microRNAs) between ExoTIC and UC. For the HCC827 cell line, 13 microRNAs were found to be highly expressed in both UC and ExoTIC methods. Four microRNAs demonstrated high expression only in UC, and three microRNAs and one microRNA cluster demonstrated high expression only in ExoTIC. For the H1650 cell line, three microRNAs demonstrated high expression in ExoTIC, and 12 microRNAs showed high expression by both UC and ExoTIC. Nine of the highly expressed microRNAs were common to both cell lines and both isolation methods. Of these, hsa-miR-124640,41 and hsa-miR-13442,43 have been reported to be highly correlated with lung cancer (see Table S2). Figure 3e,f demonstrates the linear correlation of EV microRNA expression levels between the two methods in the two cell lines. The correlation coefficients (R2) between ExoTIC and UC microRNA counts are 0.86 and 0.50 for cell lines HCC827 and H1650, respectively. The difference in correlation coefficients between the two cell lines is attributable to cell-line-specific differences in EV size and density.
Figure 3.
Evaluation of microRNA extracted from EVs isolated by ExoTIC and UC. (a) EVs isolated from UC (60 mL) and ExoTIC (10 mL) in two cancer cell lines, HCC827 and H1650 (mean ± SD, n = 5). (b) RNA extracted from EVs that were isolated from the same culture media using UC and ExoTIC. (c) Heat-map of the top 40 EV microRNAs. (d) Venn diagram of EV microRNAs identified by UC and ExoTIC, indicating overlapping and non-overlapping microRNAs between the two methods in two cell lines. (e,f) Linear correlation of EV microRNA expression levels between the two methods in two cell lines. R2 represents Pearson correlation coefficient between two methods.
Although ExoTIC and UC methods showed similar micro-RNA profiles overall for the 40 most highly expressed EV microRNAs, we also observed that certain microRNAs were more highly expressed in EVs isolated by ExoTIC compared to UC and vice versa. These findings are supported by previous studies, which demonstrated stoichiometric differences in microRNA contents among different EV populations and between different methods.44–46 These differences in micro-RNA expression profiles can be attributed to the different separation mechanisms used by the two methods, which result in nonidentical EV populations, as UC separates EVs based on density while ExoTIC separates EVs based on size.
Evaluation of EV Protein Expression in ExoTIC and UC
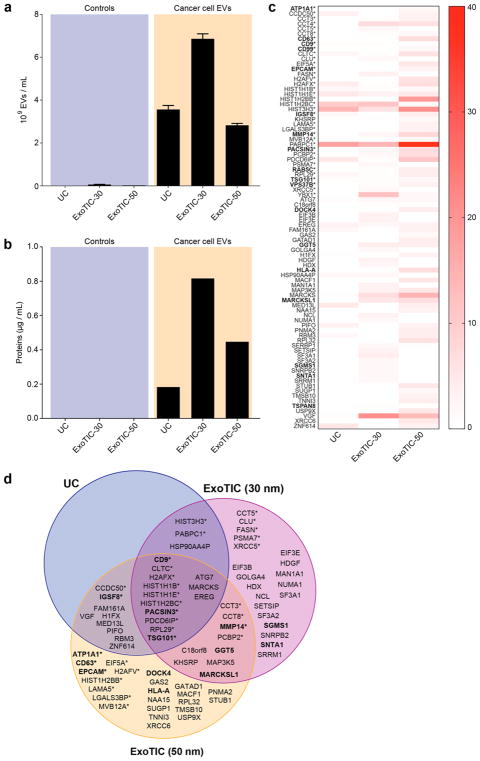
EVs derived from cancer cells carry a payload of proteins that reflects the types of proteins secreted and shed by the tumor.28,47 Isolated EV proteins are a rich source of biomarkers that could allow stratification of cancer patients into low-risk and high-risk groups for treatment. As a proof-of-concept, we demonstrated isolation of EVs and EV proteins from prostate cancer cell lines. Using liquid chromatography–mass spectrometry (LC/MS), we compared the ExoTIC and UC methods with respect to protein expression in EVs isolated from conditioned culture media from 22Rv1 prostate cancer cells. For 48 h prior to EV collection, we incubated cells in serum-free culture media to ensure that the proteins we detected were EV-specific and free of contaminating serum proteins and bovine EVs. To study the effect of EV size on protein profile, we isolated EVs using ExoTIC devices with two different pore sizes (30 and 50 nm) and compared with traditional UC. As with the lung cancer cell lines discussed previously, substantially more prostate cancer cell line EVs were isolated by either ExoTIC device than by UC (Figure 4a). Proteins were isolated from each of the EV preparations and measured, as shown in Figure 4b. Our data presented in Figure 4c,d shows that, of the 84 EV-derived proteins that we identified in total, UC, ExoTIC-30 nm, and ExoTIC-50 nm contributed 29% (25/84), 62% (52/84), and 75% (63/237 84), respectively. Thirty-seven of these proteins have previously been reported to be associated with this cancer cell line (Exocarta Database, www.exocarta.org) (see Table S3), of which 29% (11/37) were common to UC and ExoTIC-30 nm, 37% (14/37) were common to UC and ExoTIC-50 nm, and 54% (20/37) were common to ExoTIC-30 and ExoTIC-50. The 30 and 50 nm pore size ExoTIC devices accounted for most of the prostate cancer-associated EV proteins detected. Overall, our results show that we can detect a higher number of EV proteins using ExoTIC when compared to UC.
Figure 4.
Evaluation of EV protein composition. (a) Comparison of EV yield for EVs isolated from the serum-free culture media of 22Rv1 cells and controls (serum-free media without 22Rv1 cells) using UC, ExoTIC-30 (filter pore: 30 nm), and ExoTIC-50 (filter pore: 50 nm). (b) Comparison of EV protein yields for EV samples in (a). (c) Number of peptides identified by LC–MS/MS of EVs from 22Rv1 cells isolated by the three methods. (d) Venn diagram showing the EV proteins identified for each of the three EV isolation methods. For the proteins shown, no peptides were observed in control samples (media without EVs). *An asterisk indicates that the protein was previously identified in EVs from prostate cancer cell lines (www.exocarta.org). Membrane proteins are indicated in bold.
Different ExoTIC pore sizes (30 and 50 nm) may yield different EV populations, resulting in distinct protein contents. The lack of 100% overlap between the 30 and 50 nm isolations is likely due to differences in pressure drop across the filter membrane between the two ExoTIC devices and sampling error by the mass spectrometer. First, achieving the same flow rate (5 mL/h) in both devices requires ~5-fold higher operating pressure in the 30 nm device than in the 50 nm device (ΔP ~ 1/r3). These higher pressures may have changed overall filtering characteristics of the device for that specific flow rate range and pore size, which may have resulted in differences in isolating EVs. Therefore, the flow rate must be optimized independently for each filter pore size to account for the change in flow resistance. Second, the lack of overlap in this subset of proteins may be due to sampling error by the mass spectrometer, in that the instrument cannot comprehensively measure every protein in an LC–MS run (a limitation of MS). In a typical shotgun LC–MS/MS experiment, the mass spectrometer selects peptides as they elute from the LC for fragmentation in the MS. The number of peptides that can be fragmented in a complex mixture exceeds the processing speed of the mass spectrometer; therefore, not all of the same peptides are sampled from run to run.
ExoTIC as a Modular Platform for Size-Based Sorting of EVs
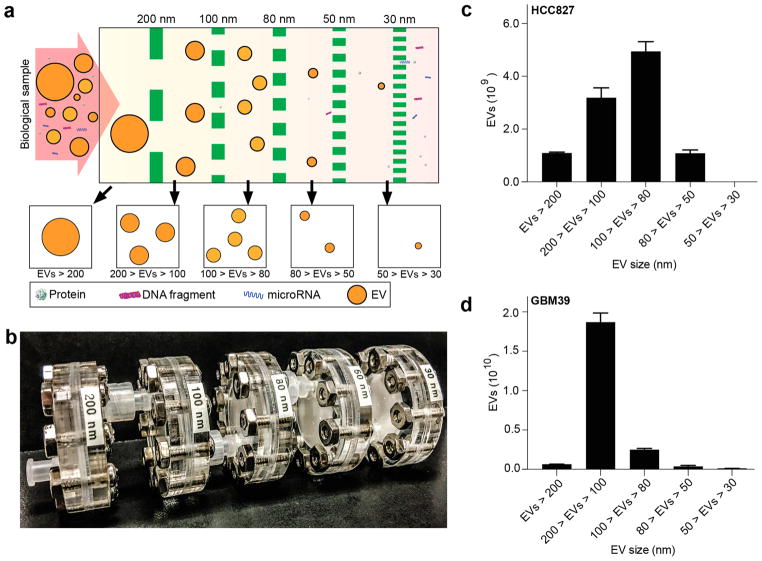
Cells release EVs in a broad range of sizes, with size-associated differences in biomolecular content.44,48 To address the need to sort different-sized EVs from the same sample, we designed ExoTIC as a modular unit such that several ExoTIC devices, each with a different membrane pore size (e.g., 200, 100, 80, 50, and 30 nm), can be connected in series to isolate EVs at several specific, narrow size ranges (Figure 5a,b and Figures S8 and S9). To validate our design, HCC 827 cell culture medium was injected into a series of ExoTIC modules with pore sizes of 200, 100, 80, and 50 nm. Retentates containing isolated EVs were separately collected from each ExoTIC module in the series and analyzed by NTA, confirming that progressively smaller EVs are captured at membranes of successively smaller pore sizes (see Figure S10). The modular ExoTIC arrangement was also used to compare differences in EV amount and size between two cancer cell lines, HCC 827 and GBM 39 (glioblastoma)49 (Figure 5c,d). We found distinctly different EV sizes and quantities between these two cell lines. The histogram of mode EV size at each filter cutoff corresponds well to the size distribution of EVs, showing that the modular system can sort different-sized EV subpopulations without affecting the overall size distribution, as might occur if there was preferential loss of EV yield at a particular filter size. The mode size of the EVs that pass through the filter decreased as the filter pore size became smaller. The mode size and number of EVs within a specific size range depended on the cell line studied. Thus, the modular ExoTIC system enables size fractionation of EVs for size-specific molecular analysis.
Figure 5.
Size-based isolation of EVs using the ExoTIC device. (a) Design schematic and (b) image of an actual modular ExoTIC device, comprised of 5 modules, each with a different membrane pore size, that connect in series for differential isolation of EVs from the same input sample. Quantity of EVs isolated at each size cutoff in the series from (c) HCC827 culture media and (d) GBM39 culture media (mean ± SD, n = 5).
EV Isolation from NSCLC Biological Fluids
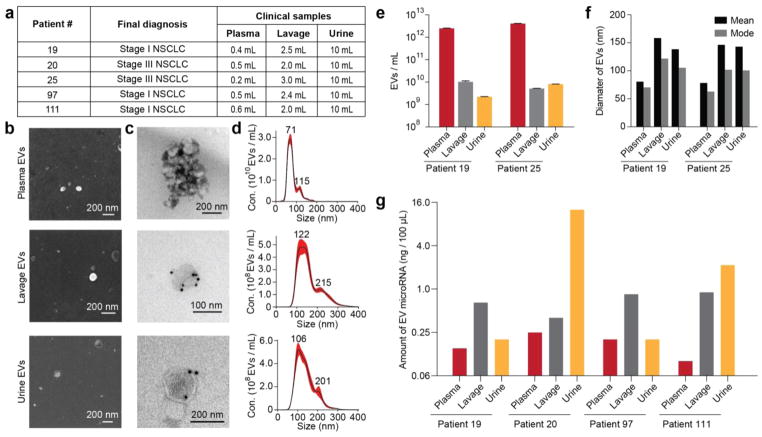
Next, we evaluated the efficiency and reproducibility of EV purification from limited clinical samples, including blood plasma, urine, and lung bronchoalveolar lavage (BAL) fluid from patients with non-small cell lung cancer (NSCLC) (Figure 6a). BAL is the recovery of fluid instilled into the airway during bronchoscopy and is routinely used to evaluate and diagnose lung cancer and other lung diseases. We successfully isolated EVs from these clinical samples and characterized their size and morphology by SEM (Figure 6b) and TEM (Figure 6c). The morphology and size distribution of the particles (see Figure S11), as well as the presence of the EV-specific surface marker CD63, confirmed that the particles we had isolated with the ExoTIC device were in fact EVs (Figure 6c,d and Figure S12). It is noteworthy that EVs from plasma were smaller and more abundant than EVs from BAL and urine (Figure 6e,f). Quantification was performed on total RNA isolated from EVs in the plasma, BAL, and urine samples of four patients with lung cancer. Although plasma yielded the most EVs, RNA quantities were in fact lowest in plasma EVs and highest in urine EVs (Figure 6g).
Figure 6.
Isolation and characterization of EVs from different biofluids of cancer patients. (a) Patient information and sample volume used for plasma, bronchoalveolar lavage, and urine to isolate EVs. EVs of patient 19 by fluid type and (b) SEM, (c) TEM with immuno-gold labeling for CD63 (GNP diameter 10 nm), and (d) nanoparticle tracking analysis of concentration. (e) Total quantity and (f) mean size and mode size of EVs isolated from the plasma, lavage, and urine of patients 19 and 25. (g) Amount of EV microRNAs extracted from the plasma, lavage, and urine of four lung cancer patients.
CONCLUSIONS
Despite the discovery of EVs 30 years ago, technical standardization of EV isolation and downstream RNA and protein analysis continues to be a major challenge in the field of cancer early detection and clinical translation. EV isolation in the research setting is still most commonly performed using UC, which has numerous limitations in terms of sample volume, yield, and the need for bulky instrumentation with longer turn-around time. Here, we present the ExoTIC device, which is simple, fast, cost-effective, and scalable and provides high-yield isolation of EVs from cell culture media and a variety of bodily fluids from cancer patients.
ExoTIC is a platform technology with four key innovative aspects over the commonly used UC method for EV isolation. First, ExoTIC isolates EVs from as little as 10 μL and up to 120 mL of sample in <3 h, whereas UC requires >50 mL of sample and twice the time. The ability to isolate EVs from small volumes is highly advantageous in cases where sample volumes are limited, such as in preclinical mouse models, where purification of EVs by UC would be extremely difficult as often only 10–20 μL of blood can be sampled at a given time. Moreover, small-volume processing can enable isolation of EVs from a fingerprick of blood, opening up avenues for EV testing at the point of care. Second, ExoTIC provides 4-fold and 1000- fold higher yields compared to UC in culture media and plasma, respectively, but in an automated and less labor-intensive manner which can easily be scaled for high-throughput processing. Third, ExoTIC can be implemented as a series of connected modular units, each with a different membrane pore size, to enable size-based sorting of heterogeneous EV populations. Size-selective isolation is a critical need in EV research because different cell types secrete EVs of different sizes4,29,44,48 and different-sized EVs, even from the same cell type, can differ markedly in molecular content.45,46,50,51
Furthermore, ExoTIC has a number of important design and technical advantages over EV isolation methods that are based on size exclusion,25,44 filtration,15,16,21 charge,30 microfluidics, 24,52,53 and acoustics23 (see Table S4). First, the robust mechanical design of ExoTIC prevents fluid leakage, enabling low device failure rates. Second, ExoTIC uses a standard female luer-lock that fits standard 5 and 10 mL syringes, allowing for easy adaptation to research and clinical lab workflows. Third, the ExoTIC device is designed to withstand high operating pressures as compared to existing filtration-based methods (the pressure applied in ExoTIC for EV isolation is 380 times lower than UC) (for calculations, see Supporting Information). Fourth, ExoTIC fabrication uses simple and rapid prototyping methods such as laser cutting and bolt assembly, thus readily enabling changes in overall design, including “swapping out” filters with different pore sizes and surface areas depending on the application (e.g., low vs high volume samples). Fourth, the material costs for ExoTIC include $0.70 for the filter membrane, $0.10 for plastics, and $0.10 for screws and nuts, such that a device can be made for under a dollar. The cost could be further lowered with large-scale production. Fifth, ExoTIC is completely enclosed and employs adhesive-free housing, reducing entry of environmental contaminants and dust. Sixth, a next-generation version of the device, in which the plastic components are replaced with metal (see Figures S13 and S14), can be sterilized by autoclave and individually packed for downstream analysis. Seventh, in low-resource settings or in the developing world, ExoTIC allows EV isolation to be performed without a syringe pump or any other machine simply by manually pushing the plunger of a syringe. Eighth, ExoTIC allows for easy collection of enriched EVs using a standard pipet at the inlet. The size of the filter, number of pores, and complexity of the sample determine how large of a sample volume can be processed with the ExoTIC device. For an ExoTIC device utilizing a 25 mm diameter filter membrane, the upper limit of sample volume for cell culture media processing can reach up to ~20 mL, whereas the upper limit is up to ~500 μL for plasma, which is a much more complex matrix. Finally, samples with volumes larger than 100 mL can be processed using multiple ExoTIC devices that can be run in parallel on the same syringe pump, as each pump can hold up to 6 syringes or more (Figure S3), and multiple syringe pumps can be run simultaneously. In principle, a 100 mL sample can be run with several devices in parallel, each containing 10–20 mL of sample, permitting processing within 2–3 h.
ExoTIC technology has several advantages over traditional methods for EV isolation for downstream point-of-care (POC) testing. Because ExoTIC is inexpensive, simple to use, rapid, and can be performed manually, this technology can enable POC testing of EVs in accordance with the ASSURED (affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free, and deliverable to end users) criteria as outlined by the World Health Organization (WHO) for disease diagnostics in resource limited settings. Furthermore, achieving high-yield isolation from small sample sizes enables sensitive exosome-based proteomic and transcriptomic biomarker detection at the POC.
In summary, we have isolated and characterized EVs from different types of clinical biofluid samples, potentially providing specific and distinct expression patterns of EV-specific micro-RNAs, mRNAs, genomic DNAs, and proteins. Future work will use ExoTIC-isolated EVs to investigate the genomic, transcriptomic, and proteomic signatures that discriminate cancer patients with high-risk disease from low-risk patients and healthy controls. In conclusion, this technology has the potential to enable accelerated EV-based biomarker discovery and molecular analysis that is simple, reliable, and quantitative with broad applicability to diagnosis, prognostication, and treatment monitoring in patients with cancer and other diseases.
MATERIALS AND METHODS
Please refer to Supporting Information for materials and methods related to the design, materials, and fabrication of the ExoTIC device and the downstream characterization of ExoTIC enriched EVs with respect to size, concentration, and morphology.
EV Isolation from Cell Culture Media Using the ExoTIC Device
After setup, the ExoTIC device was first flushed with 2 mL of 1X PBS buffer by manually pushing a 10 mL syringe or using a syringe pump (~5 min). Then, the EVs from culture media were isolated as follows: A five milliliter-volume of culture medium was drawn up in the same syringe and connected with the ExoTIC device. This syringe along with the ExoTIC device, was fixed onto a syringe pump. The device was oriented such that the outlet is in the “12 o’clock” position with respect to the inlet. A pump flow rate of 5 mL/h was applied to filter the culture media, concentrating EVs in front of the nanoporous membrane. Free proteins, nucleic acids, etc., which are smaller than the membrane pore size (~50 nm) pass through the filter pores. When ~500 μL of sample remained, the syringe and connected ExoTIC device are rotated 180° to orient the inlet of the device at the 12 o’clock position with respect to the outlet. The syringe pump continued to run at the same rate until the remaining medium was completely filtered. The EV-containing retentate was then washed by running 5 mL of 1X PBS through the device using the same syringe. The ExoTIC device was carefully disconnected from the syringe, and the purified EV solution was collected via the device inlet using a 200 μL pipet. The purified EV sample was stored at 4 °C for further molecular analysis.
EV Isolation by PEG Precipitation
A stock solution of polyethylene glycol was made using 20 g of PEG 8000 in 20 mL of 0.5 M NaCl PBS to a final PEG concentration of 40%. The solution was mixed by magnetic stirring until it turned clear. To isolate EVs, PEG solution was added to the filtered cell culture media at a ratio of 1:3. The mixture was then shaken to homogeneity and stored at 4 °C for 24 h. After incubation, centrifugation was performed at 1500g for 10 min to pellet the PEG-containing EVs. Aspiration of the supernatant left a white pellet at the bottom of the tube. The pellets were resuspended in 100 μL of PBS and stored at −80 °C for future EV analysis.
EV Isolation by Ultracentrifugation
Cell culture media (36 mL) was prepared as previously described and dispensed into two polycarbonate centrifugation tubes (26.3 mL capacity, Beckman Coulter). The tubes were then filled to the brim with PBS to prevent tube collapse during ultracentrifugation and were balanced to within 0.01 g of each other and placed within a Type 70ti rotor inside a Beckman Coulter XL–90 Ultracentrifuge. Tubes were labeled to indicate positioning within the rotor and to mark the expected location of the pellet. Samples were spun at 20000g (14000 rpm) for 30 min to pellet cell debris and other high-density particles. Supernatant was then placed back into clean sterile tubes, which were balanced and then secured into the rotor once more in the same orientations as before. The second ultracentrifugation step was performed at 100,000g (31200 rpm) for 1 h 30 min. EVs, in addition to particles of similar density, are pelleted in this step, while proteins and other molecules remain in suspension. The pellet may or may not be visible depending on cell type and EV concentration. Once the supernatant was aspirated, the pellet was resuspended in 100 μL of PBS and stored at −80 °C. Centrifuge tubes were sterilized and stored for later use.
EV Isolation from Human Plasma Using the ExoTIC Device
Blood from healthy human donors was collected at the Stanford Blood Center in tubes containing potassium EDTA anticoagulant, centrifuged at 1000g for 10 min at 4 °C to remove platelets, and 100 μL of the resulting plasma (see Figure S6) was used for subsequent steps (which collectively take ~5 min). Four hundred microliters of 1× PBS buffer was added to 100 μL of plasma and mixed. The 500 μL sample was then filtered using a low protein-binding filter (pore size: 200 nm), and the flow-through was collected in a 1.5 mL microtube. PBS buffer or DI water (1 mL) was used to wash the syringe filter, and the flow-through was collected in the same microtube.
EV isolation takes approximately 1 h
Approximately 1.5 mL of plasma PBS solution was withdrawn by a 10 mL syringe and connected with the ExoTIC device. After being fixed onto a syringe pump, a pumping rate of 1.5 mL/h was applied to enrich EVs in the ExoTIC device and remove free proteins, nucleic acids, and cell debris as depicted in panel (3) (see Figure S6). When about 500 μL was left, the syringe with the ExoTIC device was turned 180°, so that the inlet of the ExoTIC device was above the outlet as depicted in panel (4), to collect EVs in the chamber of the ExoTIC device. After turning, pumping continued at the same rate until the remaining media was completely filtered. Collecting the EV solution takes approximately 5 min. The ExoTIC device, containing EV solution, was carefully disconnected from the syringe. As depicted in Figure S6, a 200 μL size pipet was used to collect all of the EV solution through the inlet of the ExoTIC device. The collected EV sample was kept in a 1.5 mL microtube and stored at 4 °C for further analysis.
Extraction of microRNAs from EVs of Diverse Patient Biofluid Samples
All clinical samples were obtained with consent from patients being cared for clinically at Stanford Health Care after Institutional Review Board approval. After isolation of the putative exosomes through the ExoTIC device, miRNA isolation was performed using the miRCURY RNA Isolation Kit-Biofluids (cat no. 300112, Exiqon) for human serum. The miRCURY RNA Isolation Kit-Cell and Plant (cat no. 300110, Exiqon) was used to isolate miRNA from EVs in the urine and lavage. RNase-free pipet tips and RNase Zap (cat no. AM9780, Sigma-Aldrich) were used to prevent sample degradation and contamination.
Banked clinical samples of human serum, urine, and bronchoalveolar lavage (BAL), which had been stored at −80 °C, were thawed to initiate the process of miRNA extraction. Based on the product specifications, the recommended starting volume for human samples (serum, urine, and lavage) was 200 μL. In cases where the starting sample volume was less than 200 μL, RNase-free water (cat no. AM9937, Ambion) was used to bring the sample volume up to 200 μL. Cell debris were isolated from the sample by centrifugation at 3000g for 5 min. Proteinase K (cat no. 3115887001, Sigma-Aldrich) at a concentration of 3 μg/μL was added to the supernatant and incubated for 10 min at 37 °C to degrade proteins. After removal of the cell pellet, the supernatant containing the sample of interest was lysed with the lysis buffer provided in the kit. After vortexing the sample to ensure maximal mixing, the sample was incubated for 3 min prior to the addition of the protein precipitation buffer. Optimal precipitation was achieved by high-speed centrifugation at 11000g for 3 min. Furthermore, the addition of ~270 μL to the supernatant ensured the binding conditions were ideal for loading onto the microRNA Mini Spin Column. Upon loading onto the spin column, the sample was centrifuged at 11000g for 30 s and the subsequent flow-through was discarded. This step was repeated until the entire sample was spun through the column and dried to ensure maximal binding of the miRNA to the column. To prevent interference from DNA, recombinant DNase was added directly onto the membrane of the spin-column and incubated for 15 min at room temperature. The recommended wash buffer (prepared with 99% ethanol) was spun through the column to wash the sample. After complete drying of the membrane, the microRNA was eluted for downstream processing by adding RNase-free water directly onto the membrane, incubating for 1 minute at room temperature, and centrifuging at 11,000g for 1 min.
Proteomic Analysis of EV Samples
Protein concentrations in EV samples were measured by a Coomassie Bradford Assay according to the manufacturer’s instructions (ThermoFisher Scientific). Purified EV samples were lysed with 2% SDS to release EV proteins. Protein disulfide bonds were reduced by adding dithiothreitol (DTT, Sigma-Aldrich, St. Louis, MO) to a final concentration of 10 mM DTT. Samples were incubated at room temperature for 1.5 h. Iodoacetamide (Acros Organics, New Jersey) was added in 1.5-fold molar excess of DTT followed by a 1 h incubation at room temperature in the dark. Proteins were then precipitated with 100% cold acetone (Fisher Chemical, Pittsburgh, PA) overnight. Samples were then centrifuged at 15000 rpm for 10 min at 4 °C. Acetone was carefully removed without disturbing protein pellets. The protein pellets were then dried for 5 min at room temperature. 100 μL of 100% acetonitrile was added to each sample, followed by 15 min of sonication. Next, 30 μL of 50 mM ammonium bicarbonate containing 1 μg of trypsin (Promega, Sunnyvale, CA) was added to each sample, and proteins were digested at 37 °C for 1.5 h with shaking at 900 rpm. A second aliquot of 1 μg of trypsin was then added to each sample followed by another 1.5 h incubation period with 900 rpm shaking. Samples were dried by SpeedVac, reconstituted, and further desalted by C18 Zip-Tips (EMD Millipore, Darmstadt, Germany).
Tryptic peptides were loaded onto a C18 column (50 cm length, 2 μm particle size, Thermo Fisher Scientific, San Jose CA) and separated by reverse-phase chromatography using a nanoEasy nLC-1200 (Thermo Fisher Scientific, San Jose, CA). Eluted peptides were analyzed by online LC-MS analysis using a Q-Exactive Plus mass spectrometer (Thermo Fisher Scientific, San Jose, CA). Mobile phase A consisted of 0.1% formic acid in water, and the flow rate was 250 μL/min. Mobile phase B (0.1% formic acid in acetonitrile) was run at 2% for the first 5 min, ramped to 20% over 100 min, and rapidly increased to 95% for 21 min. The top 15 most abundant ions per MS1 scan were selected for higher energy collision induced dissociation (27 eV). MS1 resolution was set to 70000, AGC target was set to 1 × 106, and the m/z scan range was set to m/z = 375–1500. MS2 resolution was set to 17500 and AGC target to 2 × 105. Dynamic exclusion was enabled for 20 s. Data were searched by Byonic (Protein Metrics, San Carlos, CA) against the human UniProt database. Quantitation was performed by peptide count with a 1% false discovery rate. Proteins identified in control samples (serum-free media) were considered to be background proteins and were subtracted from the respective 22Rv1 experiments.
Supplementary Material
Acknowledgments
We thank Qiang Liu at Stanford University for his valuable comments and suggestions in this study. R.G.S. is supported by DOE Office of Basic Energy Sciences, Chemical Sciences Division, under Contract DE-AC02-76SF00515. The work was primarily supported by research funding provided by DoD grant (LC150650, 11976867, W81XWH-16-1-0200), Canary Foundation seed grant, NCI Center for Cancer Nanotechnology Excellence for Translational Diagnostics (NCIU54CA199075), and NIH grant (R00: HL117048).
Footnotes
Notes
The authors declare the following competing financial interest(s): Dr. U. Demirci is a founder of, and has an equity interest in: (i) DxNow Inc., a company that is developing microfluidic and imaging technologies for point-of-care diagnostic solutions, and (ii) Koek Biotech, a company that is developing microfluidic IVF technologies for clinical solutions, and iii) Levitas Inc., a company focusing on developing products for liquid biopsy. U.D.s interests were viewed and managed in accordance with the conflict of interest policies.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.7b04878.
Additional experimental methods, theoretical calculations, supporting figures and tables (PDF)
References
- 1.Trams EG, Lauter CJ, Salem N, Heine U. Exfoliation of Membrane Ecto-Enzymes in the Form of Micro – Vesicles. Biochim Biophys Acta, Biomembr. 1981;645:63–70. doi: 10.1016/0005-2736(81)90512-5. [DOI] [PubMed] [Google Scholar]
- 2.Pan BT, Teng K, Wu C, Adam M, Johnstone RM. Electron Microscopic Evidence for Externalization of the Transferrin Receptor in Vesicular Form in Sheep Reticulocytes. J Cell Biol. 1985;101:942–948. doi: 10.1083/jcb.101.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dragovic RA, Gardiner C, Brooks AS, Tannetta DS, Ferguson DJ, Hole P, Carr B, Redman CW, Harris AL, Dobson PJ, Harrison P, Sargent IL. Sizing and Phenotyping of Cellular Vesicles Using Nanoparticle Tracking Analysis. Nanomedicine. 2011;7:780–788. doi: 10.1016/j.nano.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colombo M, Raposo G, Thery C. Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 5.Raposo G, Stoorvogel W. Extracellular Vesicles: Exosomes, Microvesicles, and Friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Driscoll L. Expanding on Exosomes and Ectosomes in Cancer. N Engl J Med. 2015;372:2359–2362. doi: 10.1056/NEJMcibr1503100. [DOI] [PubMed] [Google Scholar]
- 7.Syn NL, Wang L, Chow EK, Lim CT, Goh BC. Exosomes in Cancer Nanomedicine and Immunotherapy: Prospects and Challenges. Trends Biotechnol. 2017;35:665–676. doi: 10.1016/j.tibtech.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, Hochberg FH, Breakefield XO, Weissleder R, Lee H. Protein Typing of Circulating Microvesicles Allows Real-Time Monitoring of Glioblastoma Therapy. Nat Med. 2012;18:1835–1840. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuld NJ, Hauser AD, Gastonguay AJ, Wilson JM, Lorimer EL, Williams CL. Smggds – 558 Regulates the Cell Cycle in Pancreatic, Non-Small Cell Lung, and Breast Cancers. Cell Cycle. 2014;13:941–952. doi: 10.4161/cc.27804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. Micrornas Are Transported in Plasma and Delivered to Recipient Cells by High-Density Lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheridan C. Exosome Cancer Diagnostic Reaches Market. Nat Biotechnol. 2016;34:359–360. doi: 10.1038/nbt0416-359. [DOI] [PubMed] [Google Scholar]
- 12.Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating Liquid Biopsies into the Management of Cancer. Nat Rev Clin Oncol. 2017;14:531–548. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 13.Thery C, Clayton A, Amigorena S, Raposo G. Curr Protoc Cell Biol. 2006. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids; pp. 3.22.1–3.22.29. [DOI] [PubMed] [Google Scholar]
- 14.Muller L, Hong CS, Stolz DB, Watkins SC, Whiteside TL. Isolation of Biologically-Active Exosomes from Human Plasma. J Immunol Methods. 2014;411:55–65. doi: 10.1016/j.jim.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinemann ML, Ilmer M, Silva LP, Hawke DH, Recio A, Vorontsova MA, Alt E, Vykoukal J. Benchtop Isolation and Characterization of Functional Exosomes by Sequential Filtration. J Chromatogr A. 2014;1371:125–135. doi: 10.1016/j.chroma.2014.10.026. [DOI] [PubMed] [Google Scholar]
- 16.Cheruvanky A, Zhou H, Pisitkun T, Kopp JB, Knepper MA, Yuen PS, Star RA. Rapid Isolation of Urinary Exosomal Biomarkers Using a Nanomembrane Ultrafiltration Concentrator. Am J Physiol Renal Physiol. 2007;292:F1657–F1661. doi: 10.1152/ajprenal.00434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS, Hochberg FH, Breakefield XO, Lee H, Weissleder R. Chip-Based Analysis of Exosomal Mrna Mediating Drug Resistance in Glioblastoma. Nat Commun. 2015;6:6999. doi: 10.1038/ncomms7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalra H, Adda CG, Liem M, Ang CS, Mechler A, Simpson RJ, Hulett MD, Mathivanan S. Comparative Proteomics Evaluation of Plasma Exosome Isolation Techniques and Assessment of the Stability of Exosomes in Normal Human Blood Plasma. Proteomics. 2013;13:3354–3364. doi: 10.1002/pmic.201300282. [DOI] [PubMed] [Google Scholar]
- 19.Taylor DD, Shah S. Methods of Isolating Extracellular Vesicles Impact Down-Stream Analyses of Their Cargoes. Methods. 2015;87:3–10. doi: 10.1016/j.ymeth.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 20.Taylor DD, Zacharias W, Gercel-Taylor C. Exosome Isolation for Proteomic Analyses and Rna Profiling. Methods Mol Biol. 2011;728:235–246. doi: 10.1007/978-1-61779-068-3_15. [DOI] [PubMed] [Google Scholar]
- 21.Davies RT, Kim J, Jang SC, Choi EJ, Gho YS, Park J. Microfluidic Filtration System to Isolate Extracellular Vesicles from Blood. Lab Chip. 2012;12:5202–5210. doi: 10.1039/c2lc41006k. [DOI] [PubMed] [Google Scholar]
- 22.He M, Crow J, Roth M, Zeng Y, Godwin AK. Integrated Immunoisolation and Protein Analysis of Circulating Exosomes Using Microfluidic Technology. Lab Chip. 2014;14:3773–3780. doi: 10.1039/c4lc00662c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee K, Shao H, Weissleder R, Lee H. Acoustic Purification of Extracellular Microvesicles. ACS Nano. 2015;9:2321–2327. doi: 10.1021/nn506538f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Wu HJ, Fine D, Schmulen J, Hu Y, Godin B, Zhang JX, Liu X. Ciliated Micropillars for the Microfluidic-Based Isolation of Nanoscale Lipid Vesicles. Lab Chip. 2013;13:2879–2882. doi: 10.1039/c3lc41343h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woo HK, Sunkara V, Park J, Kim TH, Han JR, Kim CJ, Choi HI, Kim YK, Cho YK. Exodisc for Rapid, Size-Selective, and Efficient Isolation and Analysis of Nanoscale Extracellular Vesicles from Biological Samples. ACS Nano. 2017;11:1360–1370. doi: 10.1021/acsnano.6b06131. [DOI] [PubMed] [Google Scholar]
- 26.Wunsch BH, Smith JT, Gifford SM, Wang C, Brink M, Bruce RL, Austin RH, Stolovitzky G, Astier Y. Nanoscale Lateral Displacement Arrays for the Separation of Exosomes and Colloids Down to 20 nm. Nat Nanotechnol. 2016;11:936–940. doi: 10.1038/nnano.2016.134. [DOI] [PubMed] [Google Scholar]
- 27.Lotvall J, Hill AF, Hochberg F, Buzas EI, Di Vizio D, Gardiner C, Gho YS, Kurochkin IV, Mathivanan S, Quesenberry P, Sahoo S, Tahara H, Wauben MH, Witwer KW, Théry C. Minimal Experimental Requirements for Definition of Extracellular Vesicles and Their Functions: A Position Statement from the International Society for Extracellular Vesicles. J Extracell Vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang K, Liu F, Fan J, Sun D, Liu C, Lyon CJ, Bernard DW, Li Y, Yokoi K, Katz MH, Koay EJ, Zhao Z, Hu Y. Nanoplasmonic Quantification of Tumour-Derived Extracellular Vesicles in Plasma Microsamples for Diagnosis and Treatment Monitoring. Nat Biomed Eng. 2017;1:0021. doi: 10.1038/s41551-016-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, Colás E, Cordeiro-da SA, Falcon-Perez JM, Ghobrial IM, Gimona M, Gursel I, Gursel M, Heegaard NH, Hendrix A, Kierulf P, et al. Biological Properties of Extracellular Vesicles and Their Physiological Functions. J Extracell Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in Exosome Isolation Techniques. Theranostics. 2017;7:789–804. doi: 10.7150/thno.18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skog J, Würdinger T, Van Rijn S, Meijer DH, Gainche L, Curry WT, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma Microvesicles Transport Rna and Proteins That Promote Tumour Growth and Provide Diagnostic Biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rana S, Malinowska K, Zöller M. Exosomal Tumor Microrna Modulates Premetastatic Organ Cells. Neoplasia. 2013;15:281IN14–295IN31. doi: 10.1593/neo.122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molino C, Mocerino C, Braucci A, Riccardi F, Trunfio M, Carrillo G, Vitale MG, Carteni G, De Sena G Breast Unit Cardarelli Hospital, N. I. Pancreatic Solitary and Synchronous Metastasis from Breast Cancer: A Case Report and Systematic Review of Controversies in Diagnosis and Treatment. World J Surg Oncol. 2014;12:2. doi: 10.1186/1477-7819-12-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh R, Pochampally R, Watabe K, Lu Z, Mo YY. Exosome-Mediated Transfer of Mir-10b Promotes Cell Invasion in Breast Cancer. Mol Cancer. 2014;13:256. doi: 10.1186/1476-4598-13-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao J, Liu R, Shi YJ, Yin LH, Pu YP. Exosome-Shuttling Microrna-21 Promotes Cell Migration and Invasion-Targeting Pdcd4 in Esophageal Cancer. Int J Oncol. 2016;48:2567–2579. doi: 10.3892/ijo.2016.3453. [DOI] [PubMed] [Google Scholar]
- 36.Polytarchou C, Oikonomopoulos A, Mahurkar S, Touroutoglou A, Koukos G, Hommes DW, Iliopoulos D. Assessment of Circulating Micrornas for the Diagnosis and Disease Activity Evaluation in Patients with Ulcerative Colitis by Using the Nanostring Technology. Inflamm Bowel Dis. 2015;21:2533–2539. doi: 10.1097/MIB.0000000000000547. [DOI] [PubMed] [Google Scholar]
- 37.Oikonomopoulos A, Polytarchou C, Joshi S, Hommes DW, Iliopoulos D. Identification of Circulating Microrna Signatures in Crohn’s Disease Using the Nanostring Ncounter Technology. Inflamm Bowel Dis. 2016;22:2063–2069. doi: 10.1097/MIB.0000000000000883. [DOI] [PubMed] [Google Scholar]
- 38.Pfeffer SR, Grossmann KF, Cassidy PB, Yang CH, Fan M, Kopelovich L, Leachman SA, Pfeffer LM. Detection of Exosomal Mirnas in the Plasma of Melanoma Patients. J Clin Med. 2015;4:2012–2027. doi: 10.3390/jcm4121957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, Ratcliffe AL, Webster PJ, Davidson EH, Hood L, et al. Direct Multiplexed Measurement of Gene Expression with Color-Coded Probe Pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 40.Huang W, Li H, Luo R. The Microrna-1246 Promotes Metastasis in Non-Small Cell Lung Cancer by Targeting Cytoplasmic Polyadenylation Element-Binding Protein. Diagn Pathol. 2015;10:127. doi: 10.1186/s13000-015-0366-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Kim G, An HJ, Lee MJ, Song JY, Jeong JY, Lee JH, Jeong HC. Hsa-Mir-1246 and Hsa-Mir-1290 Are Associated with Stemness and Invasiveness of Non-Small Cell Lung Cancer. Lung Cancer. 2016;91:15–22. doi: 10.1016/j.lungcan.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 42.Qin Q, Wei F, Zhang J, Wang X, Li B. Mir-134 Inhibits Non-Small Cell Lung Cancer Growth by Targeting the Epidermal Growth Factor Receptor. J Cell Mol Med. 2016;20:1974–1983. doi: 10.1111/jcmm.12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun C, Li S, Li D. Hsa-Mir-134 Suppresses Non-Small Cell Lung Cancer (NSCLC) Development through Down-Regulation of Ccnd1. Oncotarget. 2016;7:35960–35978. doi: 10.18632/oncotarget.8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caponnetto F, Manini I, Skrap M, Palmai-Pallag T, Di Loreto C, Beltrami AP, Cesselli D, Ferrari E. Size-Dependent Cellular Uptake of Exosomes. Nanomedicine. 2016;13:1011–1020. doi: 10.1016/j.nano.2016.12.009. [DOI] [PubMed] [Google Scholar]
- 45.Berckmans RJ, Sturk A, van Tienen LM, Schaap MC, Nieuwland R. Cell-Derived Vesicles Exposing Coagulant Tissue Factor in Saliva. Blood. 2011;117:3172–3180. doi: 10.1182/blood-2010-06-290460. [DOI] [PubMed] [Google Scholar]
- 46.Ogawa Y, Taketomi Y, Murakami M, Tsujimoto M, Yanoshita R. Small RNA Transcriptomes of Two Types of Exosomes in Human Whole Saliva Determined by Next Generation Sequencing. Biol Pharm Bull. 2013;36:66–75. doi: 10.1248/bpb.b12-00607. [DOI] [PubMed] [Google Scholar]
- 47.An T, Qin S, Xu Y, Tang Y, Huang Y, Situ B, Inal JM, Zheng L. Exosomes Serve as Tumour Markers for Personalized Diagnostics Owing to Their Important Role in Cancer Metastasis. J Extracell Vesicles. 2015;4:27522. doi: 10.3402/jev.v4.27522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willms E, Johansson HJ, Mager I, Lee Y, Blomberg KE, Sadik M, Alaarg A, Smith CI, Lehtio J, El Andaloussi S, Wood JAM, Vader P. Cells Release Subpopulations of Exosomes with Distinct Molecular and Biological Properties. Sci Rep. 2016;6:22519. doi: 10.1038/srep22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang E, Pohling C, Natarajan A, Witney TH, Kaur J, Xu L, Gowrishankar G, D’Souza AL, Murty S, Schick S, Chen L, Wu N, Khaw P, Mischel P, Abbasi T, Usmani S, Mallick P, Gambhir SS. G Ashwamax and Withaferin a Inhibits Gliomas in Cellular and Murine Orthotopic Models. J Neuro-Oncol. 2016;126:253–264. doi: 10.1007/s11060-015-1972-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gould SJ, Raposo G. As We Wait: Coping with an Imperfect Nomenclature for Extracellular Vesicles. J Extracell Vesicles. 2013;2:20389. doi: 10.3402/jev.v2i0.20389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ogawa Y, Miura Y, Harazono A, Kanai-Azuma M, Akimoto Y, Kawakami H, Yamaguchi T, Toda T, Endo T, Tsubuki M, Yanoshita R. Proteomic Analysis of Two Types of Exosomes in Human Whole Saliva. Biol Pharm Bull. 2011;34:13–23. doi: 10.1248/bpb.34.13. [DOI] [PubMed] [Google Scholar]
- 52.Liga A, Vliegenthart AD, Oosthuyzen W, Dear JW, Kersaudy-Kerhoas M. Exosome Isolation: A Microfluidic Road-Map. Lab Chip. 2015;15:2388–2394. doi: 10.1039/c5lc00240k. [DOI] [PubMed] [Google Scholar]
- 53.Liang LG, Kong MQ, Zhou S, Sheng YF, Wang P, Yu T, Inci F, Kuo WP, Li LJ, Demirci U, Wang S. An Integrated Double-Filtration Microfluidic Device for Isolation, Enrichment and Quantification of Urinary Extracellular Vesicles for Detection of Bladder Cancer. Sci Rep. 2017;7:46224. doi: 10.1038/srep46224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.