Abstract
This article provides a historical account of the discovery, chemistry, and biochemistry of two ubiquitous phosphoglycerolipids, phosphatidylserine (PS) and phosphatidylethanolamine (PE), including the ether lipids. In addition, the article describes the biosynthetic pathways for these phospholipids and how these pathways were elucidated. Several unique functions of PS and PE in mammalian cells in addition to their ability to define physical properties of membranes are discussed. For example, the translocation of PS from the inner to the outer leaflet of the plasma membrane of cells occurs during apoptosis and during some other specific physiological processes, and this translocation is responsible for profound life-or-death events. Moreover, mitochondrial function is severely impaired when the PE content of mitochondria is reduced below a threshold level. The discovery and implications of the existence of membrane contact sites between the endoplasmic reticulum and mitochondria and their relevance for PS and PE metabolism, as well as for mitochondrial function, are also discussed. Many of the recent advances in these fields are due to the use of isotope labeling for tracing biochemical pathways. In addition, techniques for disruption of specific genes in mice are now widely used and have provided major breakthroughs in understanding the roles and metabolism of PS and PE in vivo.
Keywords: mitochondria, membranes, phosphoglycerolipids, phospholipid trafficking, ether lipids
INTRODUCTION TO PHOSPHOLIPID BIOSYNTHESIS
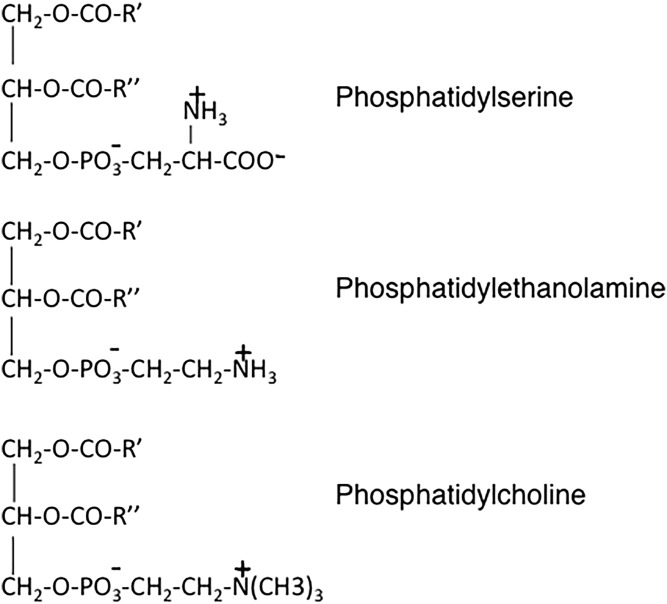
Biological membranes contain a vast array of different lipids: for example, membranes of mammalian cells contain more than a thousand different phosphoglycerolipids. In addition to the distinct polar headgroups at the sn-3 position of the phosphoglycerolipid molecules, a large part of the phospholipid diversity is due to the variety of acyl chains esterified at the sn-1 and sn-2 positions of the glycerol backbone. A large number of other lipids, including sterols and sphingolipids, also contribute to the unique properties of eukaryotic membranes. The principal lipid component of membranes in eukaryotic cells is phosphatidylcholine (PC) (Fig. 1), typically constituting 40–50% of total phosphoglycerolipids. In contrast, and with only a few exceptions, PC is absent from prokaryotic cells. Instead, phosphatidylethanolamine (PE) (Fig. 1) is usually the most abundant phospholipid in prokaryotic cells. In this article, I will provide some historical background on the discovery, characterization, and biosynthesis of two biologically important phospholipids, phosphatidylserine (PS) and PE (Fig. 1), with a focus on the biochemistry and physiological functions of these molecules in mammalian cells.
Fig. 1.
Chemical structures of PS, PE, and PC.
The concept that biological membranes serve as permeability barriers that separate cellular processes into distinct compartments was first suggested by Danielli and Davson (1) in 1935. This model was refined in 1972 by Singer and Nicolson (2) who introduced the “fluid mosaic model” in which they proposed that a biological membrane consists of a fluid two-dimensional lipid bilayer composed primarily of phospholipids into which proteins are embedded. This bilayer membrane model was later modified to include the presence of membrane microdomains that have specific protein and lipid compositions and that can further compartmentalize cellular processes (3). The (phospho)lipid composition of a membrane bilayer defines its fluidity and thereby regulates the functions of the proteins in that membrane. As well as having structural roles in membranes, many phosphoglycerolipids also play important regulatory roles in cells by being converted into “lipid second messengers,” such as diacylglycerol, arachidonic acid, the phosphatidylinositol phosphates, and lysophospholipids (4, 5).
PE is the second most abundant phospholipid in mammalian cells, typically providing 20–50% of total membrane phospholipids, whereas PS is usually considered to be a “minor” phospholipid, comprising only 2–15% of total phosphoglycerolipids in cellular membranes. Different types of mammalian tissues and cells have characteristic phospholipid compositions. For example, the two aminophospholipids, PS and PE, are more abundant in the brain than in other tissues. Moreover, in the brain and retina, the acyl chains of PS and PE are highly enriched in polyunsaturated fatty acids, primarily docosahexaenoic acid [22:6 (n-3)]. Thus, in human gray matter, greater than 36% of the acyl groups of PS consist of docosahexaenoyl acyl chains, suggesting a special function for these molecular species of PS in the nervous system. Indeed, the 22:6 (n-3) acyl chains of PS appear to be essential for the normal functioning and development of the nervous system [(6); reviewed in (7)].
Not only do different types of mammalian tissues and cells have distinct phospholipid compositions, but membranes of intracellular organelles also contain defined mixtures of phospholipids and other lipids. For example, the PE content of the mitochondrial inner membrane of mammalian cells is significantly higher than that of other organelles. In an additional level of complexity, the two leaflets of a membrane bilayer can also consist of different proportions of each phosphoglycerolipid class. The best characterized example of this topological arrangement is the plasma membrane bilayer in which PS and PE are distributed asymmetrically so that greater than 80% of PS and PE in this membrane is normally confined to the inner leaflet [(8); reviewed in (9)]. On the other hand, the choline-containing phospholipids, PC and sphingomyelin, are enriched in the outer, compared with the inner, leaflet of the plasma membrane bilayer.
Much of the following historical discussion of phospholipid chemistry and biochemistry was gleaned from the extensively researched three-volume book (1,966 pages) entitled Lipids that was published between 1951 and 1957 (10). All three volumes of this scholarly treatise were written (not edited!) by Harry J. Deuel (1887–1956), a professor of biochemistry at the University of Southern California (Fig. 2). Remarkably, these books were compiled and written without the benefit of a computer, PubMed, a photocopier, or a reference manager.
Fig. 2.
Harry Deuel, Jr. (1897–1956). Left: taken from (10). Right: the three volumes of H. J. Deuel’s treatise (10); from the author’s personal file.
THE DISCOVERY OF PS AND PE
The presence of phosphorus-containing lipids in animal tissues was probably first noted by Vauquelin (11) in 1812 in the fat-like material of the brain. Subsequently, in 1846, Gobley (12) reported that egg yolk contained a phosphorus-containing lipid that he called “lecithin,” derived from the Greek word for egg yolk. Later, Diakonow (13) concluded that choline and fatty acids were also components of the lecithin molecule. Remarkably, as early as 1874, Johann Ludwig Wilhelm Thudichum (14) showed that the products of the complete hydrolysis of lecithin were phosphoric acid, glycerol, fatty acids, and an organic base that contained a nitrogen atom. He coined the term “phosphatides” for this class of molecules (now known as the phosphoglycerolipids). In further studies reported in 1884, Thudichum separated two types of phosphatides from the brain on the basis of their solubility in alcohol: lecithin (now known as PC), which readily dissolved in alcohol, and “cephalin,” which was not soluble in alcohol (15). He suggested that instead of the choline constituent that was present in lecithin, cephalin contained an alternative nitrogenous base, originally called colamine (now known as ethanolamine). Thudichum (1829–1901) was a German biochemist and physician who, in 1884, published a book entitled A Treatise on the Chemical Constitution of the Brain (15). At the time of its publication, this book was widely criticized by many in the scientific community. Indeed, in Thudichum’s obituary in (16), his research on phospholipids was disrespectfully described as being “relatively insignificant”. Moreover, the venerable London newspaper, The Times, stated in its obituary of Thudichum on Sept. 10, 1901, that “the knowledge yielded by these researches was hardly commensurate with the time and cost at which it was obtained” (17). These sentiments apparently implied that nothing of medical or industrial value could be derived from Thudichum’s expensive studies. Sadly, similar shortsighted criticisms are often raised today to describe “discovery research” for which no commercial benefit is immediately evident.
As a sequel to Thudichum’s work, Taurog, Entemann, and Chaikoff (18) showed that lecithin and cephalin were widely distributed in animal tissues such as the liver, heart, and brain. They also found that the phospholipids in plasma from humans and dogs consisted almost entirely of the choline-containing phospholipids (lecithin and sphingomyelin), with only 5% of total plasma phospholipids being cephalin (18). These observations, made in 1944, are entirely consistent with today’s knowledge of plasma lipids.
PE was first purified more than one hundred years ago (1913) from cattle brain by Renall (19), who discovered that the two acyl chains of PE were typically different from one another, as was also the case for lecithin. As confirmation of the structural makeup of PE, the complete chemical synthesis of distearoyl-PE was achieved in 1924 by Levene and Rolf (20), and also by Grun and Limpacher (21). Subsequently, cephalin was chromatographically separated into two phosphoglycerolipids, PS and PE, by Folch (22). Remarkably, the “Folch” method for extraction and isolation of phospholipids (23) is still widely used today. The structural difference between PS and PE was revealed in 1945 when these two phospholipids were hydrolyzed by Artom (24). The hydrolysis products of PE included ethanolamine, whereas the hydrolysis of PS, surprisingly, yielded the amino acid, serine. Although the presence of PS in the cephalin mixture had been suspected by MacArthur (25) as early as 1914, the conclusion that an amino acid was a component of one of the phosphatides was completely unexpected and, for many years, was vigorously challenged. Eventually, however, the controversy was put to rest in 1941 when Folch and Schneider showed conclusively that a serine-containing phospholipid (PS) was, indeed, a component of ox brain cephalin (26). Subsequently, Folch isolated an almost pure (92–97%) preparation of PS from human brain and confirmed that the amino acid, serine, was a component of the molecule (22). In later studies, the presence of PC, PE, and PS in several rat tissues was conclusively demonstrated by Chargaff, Ziff, and Rittenberg (27) and by Artom (28).
During this era, in which the chemical structure of cephalin was being investigated, an interesting controversy arose concerning the origin of PE because no free ethanolamine could be detected in tissues. Thus, a prominent hypothesis for the origin of PE in cephalin was that the ethanolamine moiety of PE was not derived from free ethanolamine per se, but instead was an artifact that was generated by decarboxylation of the serine group of PS during the isolation procedure. This idea originated from a previous observation by Nord (29) that the direct decarboxylation of serine to ethanolamine occurred in putrefactive anaerobes. Thus, it was proposed that the PE in mammalian tissues was generated solely by the decarboxylation of PS, either during its isolation or during the postmortem changes that occurred in tissues. Nevertheless, Folch persevered in his contention that PE was a naturally occurring phospholipid because he had observed that the fatty acyl chain constituents of PS and PE were different. However, as was discovered later, and as is described in more detail in the PS decarboxylation section below, some PE and ethanolamine are, indeed, generated enzymatically from PS in vivo (30).
The historical information presented above on the isolation and basic chemical characterization of the phosphoglycerolipids is particularly noteworthy because much of this work was achieved almost 100 years ago at a time when the availability of experimental tools was extremely limited.
UNRAVELING PHOSPHOGLYCEROLIPID BIOSYNTHESIS
In the early 1900s, several investigators found that animals that had been fed a diet that contained no preformed phosphoglycerolipids developed normally, and that their tissues contained normal amounts of phospholipids [reviewed in (31)]. These observations implied that animals must be able to synthesize their own phospholipids in vivo. Additional evidence that phosphoglycerolipids were biosynthesized in animals came from some novel in vivo experiments executed by Sinclair (32), who added the “un-natural” fatty acid, elaidic acid (the trans isomer of oleic acid), to the diet of rats. He showed that elaidic acid became incorporated into phospholipids and fat, and was metabolized in a manner similar to that of its naturally occurring isomer, oleic acid. Around this time, also, a pivotal breakthrough in the development of methodology for elucidating the reactions of metabolic pathways was provided by the pioneering work of Schoenheimer and Rittenberg (33) in 1937 in which deuterium and other isotopes were used as metabolic tracers. This tracer technique was enthusiastically embraced by many biochemists, including one of Rittenberg’s colleagues, Bloch (34), as well as Cornforth and Gore (35). These renowned lipid biochemists used insightful isotopic labeling experiments to elucidate details of the cholesterol biosynthetic pathway. Isotopes were also used to delineate the biosynthetic pathways of the phospholipids. For example, Cavanagh and Raper (36) observed that deuterated fatty acids were incorporated in vivo into the phosphoglycerolipids of the liver, brain, kidney, and blood of rats. Similarly, when Stevens and Chaikoff (37) provided fatty acids containing [14C]-labeled carboxyl groups to rats, the 14C was incorporated into phosphoglycerolipids. During the late 1930s, the liver was considered to be the major site of phosphoglycerolipid synthesis in animals. However, by 1945, Artom (38) had also detected the biosynthesis of lecithin and cephalin in vivo in the intestine, kidney, muscle, and brain of rats using [32P]phosphate. In other experiments, Fishler et al. (39) showed in 1941 that phosphoglycerolipids were radiolabeled from 32Pi in vitro in liver slices, although at that time phospholipid synthesis could not be detected in liver homogenates. Furthermore, DeWitt Stetten (40) demonstrated that [15N]choline and [15N]ethanolamine were incorporated into PC and PE, respectively, in vivo in rats.
Thus, by the early 1950s, it had become clear that the phosphoglycerolipids, PC and PE, were biosynthesized from glycerol, fatty acids, phosphorus, and a nitrogenous base. The next major frontier was to dissect the biosynthetic pathways of these phospholipids in more detail. Some of the initial studies on PC biosynthesis were reported in 1953 by Kornberg and Pricer (41) who demonstrated that phosphocholine, radiolabeled with 14C and 32Pi, was converted by liver enzymes into a [14C]- and [32P]-containing lipid that they presumed was lecithin (i.e., PC); this conclusion was later confirmed by Rodbell and Hanahan (42). On the basis of these data, the following mechanism was proposed for the biosynthesis of lecithin:
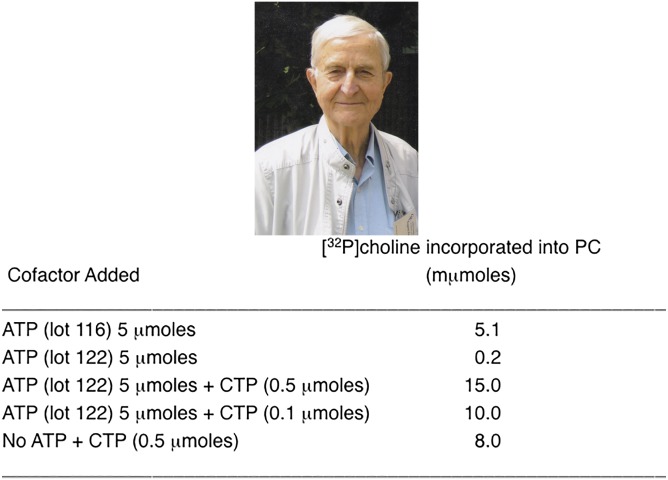
Kennedy and Weiss were intrigued by these findings and their curiosity led them to investigate in more detail the mechanism by which lecithin was formed from P-choline. In their initial studies, reported in 1956 (43), they concluded that ATP was required for the synthesis of lecithin from phosphocholine. However, when they employed some additional detective work, they made the surprising discovery that CTP, not ATP, was the ribonucleotide triphosphate involved in PC and PE biosynthesis (43). In these carefully performed studies, rat liver homogenates were incubated with [32P]phosphocholine in the presence of Mg2+ and ATP, with the result that [32P]-labeled phospholipids were produced. They found that addition of the cofactor ATP was essential for the reaction. Enigmatically, however, when instead of ATP, they added AMP plus a substrate (e.g., succinate) for the generation of ATP in situ via oxidative phosphorylation, no radioactive lecithin was produced. Importantly, as a positive control experiment, Kennedy and Weiss (43) demonstrated that the oxidative phosphorylation of AMP to ATP was occurring during these experiments. Thus, as an additional demonstration that ATP was, indeed, required for the biosynthesis of lecithin, a different preparation of ATP (Lot 122 in Fig. 3) was used in the reaction, but unexpectedly, [32P]phosphocholine did not become incorporated into lecithin (Fig. 3). One can readily imagine the discussions that occurred in the laboratory in response to these apparently contradictory findings! Consequently, several additional ATP preparations, as well as other nucleoside triphosphates (CTP, UTP, GTP, and ITP), were tested in the reaction. To their surprise, CTP was by far the most active of these nucleotides in promoting lecithin synthesis from P-choline (Fig. 3). Kennedy and Weiss (43) concluded, therefore, that the original ATP preparation (Lot 116 in Fig. 3) had been impure and contained a small amount of CTP (less than 1%). Thus, the activity of the original “ATP” in promoting the synthesis of lecithin was due to the contaminating CTP (Fig. 3). Thus, Kennedy and Weiss (43) concluded that CTP, not ATP, was the active component that was required for the conversion of phosphocholine to PC. This completely unanticipated result was the first demonstration that a cytidine nucleotide, CTP, could generate an “activated” form of a precursor (in this case CDP-choline) for use in a major biosynthetic reaction. From these observations, Kennedy and Weiss (43) proposed the following scheme to explain the role of CTP in the conversion of P-choline into lecithin:
Fig. 3.
Eugene P. Kennedy. Top: From the author’s personal file. Bottom: Results from an experiment showing that CTP, not ATP, is the nucleotide cofactor for PC biosynthesis [modified from (43)].
Similarly, Kennedy and Weiss (43) demonstrated that CTP was also required for the production of PE from CDP-ethanolamine by the following set of reactions:
Moreover, CDP-choline and CDP-ethanolamine were detected as naturally occurring nucleotides in rat liver and yeasts, and further experiments showed that the addition of highly purified CDP-choline or CDP-ethanolamine to the in vitro reactions also resulted in the synthesis of PC or PE, respectively. The possibility that the added CDP-choline had initially been converted to phosphocholine or choline, which was then incorporated into PC, was dismissed because the rate of conversion of CDP-choline to PC was much higher than when equivalent amounts of P-choline and CTP were used. Subsequent experiments on PC and PE synthesis demonstrated that two distinct enzymes mediated the conversion of P-choline and P-ethanolamine into the respective CDP-derivatives for the synthesis of PC and PE. In 1954, these two enzymes were designated “PC-cytidyl transferase” and “PE-cytidyl transferase” by Kalckar and Klenow (44) and the PC-cytidyl transferase from rat liver was further characterized by Kennedy, Smith, and Weiss (45).
For the final reaction of the CDP-choline pathway for PC synthesis, the enzyme that catalyzes the transfer of P-choline from CDP-choline to the sn-3 hydroxyl group of glycerol was originally called “PC-glyceride transferase” by Weiss, Smith, and Kennedy (46). These investigators established that this enzyme was specific for CDP-choline (46). The analogous enzymatic activity that transferred P-ethanolamine from CDP-ethanolamine to diacylglycerol to generate PE was called “PE-glyceride transferase”. Thus, by the late 1950s, the biosynthetic pathways of the major phospholipid classes (PC and PE) had been outlined, primarily as a result of the pioneering work of Gene Kennedy and his coworkers [reviewed in (47)]. More recent information on PE biosynthesis is provided in the Biosynthesis of PE section below.
HISTORY OF PS BIOSYNTHESIS
As indicated above (Unraveling Phosphoglycerolipid Biosynthesis), in 1941, Folch and colleague reported that one component of cephalin was a phosphoglycerolipid that contained the amino acid, serine (22, 26). Although many of his colleagues doubted that an amino acid could be a constituent of a lipid molecule, Folch subsequently confirmed his finding by isolating pure PS from cephalin of human brain and establishing that serine was, indeed, a component of the molecule (22, 48). The structure of PS (Fig. 1) (which, in contrast to the zwitterionic phospholipids, PC and PE, is negatively charged at cellular pH) was conclusively established by Baer and Maurukas (49) in 1955.
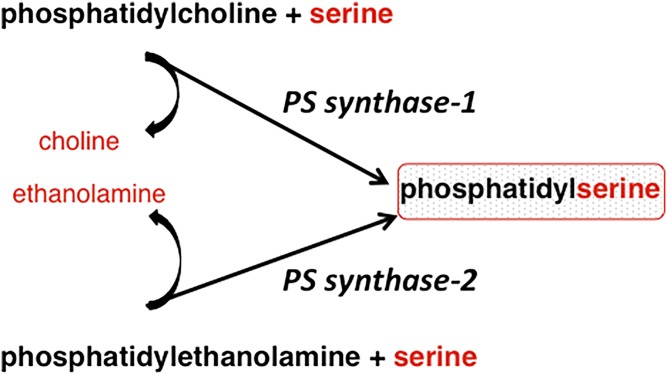
In mammalian cells, PS is synthesized by calcium-dependent base-exchange reactions in which the polar headgroup of a preexisting phospholipid (PC or PE) is exchanged for serine with the release of choline or ethanolamine, respectively (Fig. 4) (50). In contrast, in yeasts and prokaryotes, PS is synthesized by a completely different PS synthase that utilizes CDP-diacylglycerol and serine (51); the CDP-diacylglycerol biosynthetic pathway for PS synthesis has not been detected in mammalian cells. For many years, each type of cell was thought to make PS either by a base-exchange reaction or by the CDP-diacylglycerol pathway, but not by both pathways. However, in 1999, Gardner and Hampton (52) showed that, in plants, PS can be synthesized by both a base-exchange reaction and by a PS synthase that uses CDP-diacylglycerol. The latter enzyme is encoded by a cDNA that is 54% identical to the yeast PS synthase (52).
Fig. 4.
Base-exchange reactions for PS synthesis: from PC and PE, catalyzed by PS synthase-1 and PS synthase-2, respectively.
The base-exchange reaction for PS synthesis in mammalian cells is now known to be catalyzed by two distinct calcium-dependent PS synthases (base-exchange enzymes) (Fig. 4). One of these serine-exchange enzymes (now designated as PS synthase-2) uses PE as a substrate (53). This PS synthase was partially purified from brain in the 1980s by Spanner and Ansell (54), Baranska (55), and also by Suzuki and Kanfer (56). At that time, it was unclear whether the base-exchange reaction that catalyzed PS synthesis from PE could also operate as a phospholipase D. However, this base-exchange enzyme does not appear to possess any phospholipase D activity. The existence of an additional mammalian PS synthase (now known as PS synthase-1) was revealed in a series of chemical mutagenesis experiments in which a line of Chinese hamster ovary (CHO) cells was generated that lacked serine-exchange activity with PC, but retained the ability to synthesize PS from PE (i.e., PS synthase-2 activity) (57–59). The rate of PS synthesis in these cells, as monitored by in vitro radiolabeling, was 35–50% lower than in parental CHO cells, and the amount of cellular PS was also significantly lower (57–59). These observations implied that PS synthase-1 activity was defective in these mutant cells. Further mutagenesis of the PS synthase-1-deficient cells yielded a cell line in which total PS synthase activity was reduced by ∼95%, indicating that PS synthase-2 had been mutated in addition to PS synthase-1; moreover, these cells required exogenous PS for growth and survival. Thus, these data established that the two PS synthases together are responsible for essentially all PS synthesis in mammalian cells, and also showed that PS is required for cell viability (60). Moreover, these innovative mutagenesis experiments demonstrated that, in mammalian cells, PS synthase-1 preferentially uses PC as a substrate, whereas PS synthase-2 preferentially uses PE (Fig. 4). A cDNA encoding PS synthase-1 was subsequently cloned by complementation of the PS synthase-1-deficient CHO cells (61). A cDNA encoding PS synthase-2 was also identified by sequence alignment with the cDNA encoding PS synthase-1 in cDNA databases; the sequences of the two mammalian PS synthases are only ∼30% identical (62, 63).
Important questions that arise from these studies are: Why have mammalian cells evolved two distinct enzymes that produce PS (Fig. 4)? Is the duplication of PS synthase activity merely a compensatory mechanism in case one of the synthases is defective or does each PS synthase produce a unique pool of PS that is required for a distinct function? The tissue distribution of the mRNAs encoding PS synthase-1 and PS synthase-2 is different and varies during development, suggesting that the two PS synthases might produce pools of PS that are used for different purposes. PS is particularly abundant in the brain, comprising ∼15% of total phosphoglycerolipids, and PS appears to be crucial for normal functioning of the brain and the visual system (6, 7). In addition, PS synthase-1 mRNA is most abundant in the brain, but is also highly expressed in the kidney and liver and at lower levels in most other mouse tissues. On the other hand, PS synthase-2 mRNA is most highly expressed in Sertoli cells of the testis with much lower expression in other tissues, such as the brain and liver (62, 64, 65). As an additional indication that PS synthase-1 and PS synthase-2 might each be required for some individual functions, the two mammalian PS synthases appear to regulate phosphoglycerolipid homeostasis differently. For example, overexpression of PS synthase-1 in hepatoma cells markedly stimulated the incorporation of [3-3H]serine into PS, implying that the activity of PS synthase-1 can control the rate of PS synthesis (66); however, the amount of cellular PS was not increased. This apparent anomaly was explained by the observation that a compensatory increase in the conversion of PS to PE by PS decarboxylation had been induced (see the PS decarboxylation section below). Moreover, in the cells overexpressing PS synthase-1, an additional compensatory mechanism was implemented because the production of PE from ethanolamine via the CDP-ethanolamine pathway was attenuated (Fig. 5) (66). On the other hand, an equivalent level of overexpression of PS synthase-2 did not increase the incorporation of [3-3H]serine into PS or alter PE production by either the PS decarboxylation pathway or the CDP-ethanolamine pathway (62). Clearly, mammalian cells have the capacity to induce compensatory mechanisms to ensure that phospholipid homeostasis is maintained as far as possible. In addition, these studies suggest that the two PS synthases might produce distinct pools of PS to be used for specific purposes.
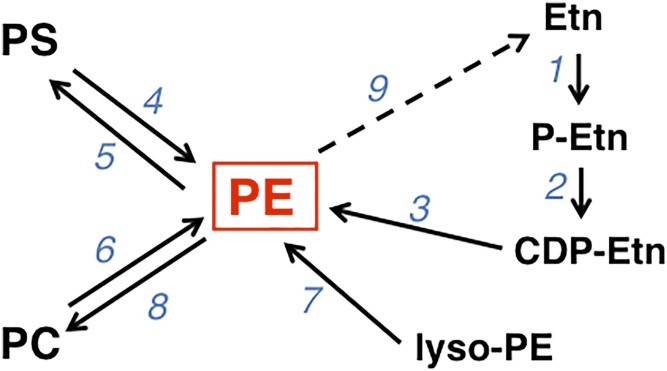
Fig. 5.
PE biosynthetic pathways. The enzyme catalyzing each reaction is indicated by the numbers 1–9. #1, ETNK; #2, ET; #3, CDP-ethanolamine:1,2-diacylglycerol ethanolaminephosphotransferase; #4, PSD; #5, PS synthase-2; #6, PS synthase-1; #7, lyso-PE acyltransferase; #8, PE N-methyltransferase; #9, release of ethanolamine from PE. Etn, ethanolamine.
The two mammalian PS synthases (Fig. 4) are integral membrane proteins that reside in the microsomal fraction of cell homogenates (67–69). Each enzyme requires calcium for its activity and the protein is predicted to contain several membrane-spanning domains (66, 70). As has also been shown for most of the enzymes that catalyze the terminal reactions of glycerolipid biosynthesis, the active sites of PS synthase-1 and PS synthase-2 are exposed on the cytosolic face of the endoplasmic reticulum (ER) (71). Thus, the major phosphoglycerolipids, PS, PC, and (some) PE, are generated on the cytosolic face of the ER membrane. This orientation of the enzymes that catalyze the terminal steps of phosphoglycerolipid synthesis raises an important question: how are phospholipids, such as PE, PC, and PS, translocated from the cytosolic to the luminal leaflet of the ER bilayer? Obviously, this movement must occur because both leaflets of the bilayer contain mixtures of these phospholipids. Yet, despite many attempts to define the transport protein that catalyzes the transbilayer movement of phosphoglycerolipids across the ER bilayer, the nature of this protein has not yet been unambiguously established.
In the early 1990s, a new technique by which a specific gene could be disrupted in mice (i.e., generation of “knockout mice”) was developed as a tool for understanding the physiological roles of proteins that are specified by particular genes (72, 73). This technique is now widely used and has radically changed the way in which the physiological functions of proteins can be investigated in vivo. In order to understand the roles of PS synthase-1 and -2 in vivo, my laboratory generated mice in which the genes encoding these enzymes were individually disrupted (Fig. 4) (65, 74, 75). Mice lacking PS synthase-2 appeared outwardly normal (75), indicating that PS synthase-2 is dispensable for mouse development and survival. Nevertheless, in male PS synthase-2-deficient mice, testis size was smaller than in wild-type mice. In addition, ∼10% of male offspring had atrophied testes and were infertile. Furthermore, the plasma level of follicle-stimulating hormone in male PS synthase-2 knockout mice was higher than in their wild-type littermates. The findings in the knockout mice are consistent with a defect in the function of Sertoli cells, the major cell type expressing PS synthase-2 in the testis. Although global elimination of PS synthase-2 markedly reduced total PS synthase activity in mouse tissues, the amounts of PS and PE were largely unchanged. A probable explanation for why the mass of PS was not reduced in the PS synthase-2 knockout mice is that PS synthase-1 activity (in primary hepatocytes isolated from these mice) was increased and PS degradation was attenuated (76). Thus, compensatory mechanisms had been induced for maintaining critical levels of PS in the absence of PS synthase-2, indicating that PS synthase-1 can, for the most part, substitute for a lack of PS synthase-2.
The gene encoding PS synthase-1 was also globally disrupted in mice (75). The PS synthase-1 knockout mice were viable and outwardly indistinguishable from their wild-type littermates. Despite an 85% reduction in total PS synthase activity in vitro, male and female mice lacking PS synthase-1 were fertile. Moreover, the amounts of PS in tissues of the knockout mice were normal, except for a modest reduction in the PS content of the liver. In marked contrast, however, and for reasons not completely understood, the viability of CHO cells lacking PS synthase-1 is severely compromised despite the presence of PS synthase-2 (68). Not unexpectedly, when mice lacking PS synthase-1 were crossed with mice lacking PS synthase-2, no viable offspring were generated that lacked both of the PS synthases (75). On the other hand, mice with three disrupted PS synthase alleles (i.e., Pss1−/−/Pss2+/− and Pss1+/−/Pss2−/− mice) were viable and retained some residual PS synthase activity with only a modest reduction in PS content of their tissues. These studies demonstrate that mice can tolerate the loss of either PS synthase-1 or PS synthase-2, and can survive with as little as 10% of normal PS synthase activity, whereas the complete elimination of PS synthase activity and the absence of PS are incompatible with survival.
Little information is available on how PS synthesis in mammalian cells is regulated by factors such as hormones or serine, or by the supply of energy, although some early studies indicated that PS synthesis in rat brain is regulated by phosphorylation via protein kinase C (77). In yeasts, an elegant system for regulation of PS synthesis by inositol at the level of gene expression has been described, primarily in the laboratories of Carman and Henry [reviewed in (78)]. However, as noted above, the synthesis of PS in mammalian cells and yeasts is mediated by completely different enzymatic reactions. It is not surprising, therefore, that the yeast-like system for regulation of gene expression of PS synthesis does not operate in mammalian cells. Because the tissue distribution of PS synthase-1 and PS synthase-2 in mammalian cells is distinct, it is possible that mechanisms underlying the transcriptional regulation of PS synthase-1 and PS synthase-2 are different in different types of mammalian cells and tissues. A preliminary characterization of transcriptional activation of the mammalian PS synthase-1 gene promoter in vitro and in vivo by Sp1 and N-Myc has been reported (79), but many more experiments are required before one can fully understand how PS synthesis is regulated at the level of gene expression in mammalian cells.
A completely different type of regulation of the PS biosynthetic pathway in mammalian cells was revealed in series of experiments performed by Nishijima, Kuge, and Akamatsu (80) in CHO cells. These studies showed that when the cellular content of PS increased, the rate of PS synthesis decreased, as measured by the rate of incorporation of 32Pi into PS. Thus, mammalian cells employ a mechanism by which end-product inhibition of PS synthesis by PS regulates the rate of PS biosynthesis. Subsequently, a mutant CHO cell line was generated in which the rate of PS synthesis was 2.5-fold higher than in parental CHO cells. In these mutant cells, PS synthesis was resistant to inhibition by exogenously added PS, suggesting that the mechanism by which PS attenuated PS synthesis had been abolished in the mutant cells (81). A point mutation at Arg-95 in PS synthase-1 was identified in these cells and shown to be responsible for the lack of end-product inhibition of PS synthesis (82). Similarly, a point mutation in Arg-97 of PS synthase-2 prevented the end-product inhibition of PS synthase-2 by PS (83). It is likely that the end-product inhibition of PS synthase-1 and PS synthase-2 activities is mediated by a direct binding of PS to the PS synthase proteins (63, 84).
Until recently, no example of a human disease caused by a mutation in PS synthase-1 or PS synthase-2 had been reported. However, whole-exome sequencing studies have identified a group of five patients with Lenz-Majewski syndrome (85) who had causative heterozygous mutations in the PS synthase-1 gene (86). This disorder is characterized by congenital abnormalities, dwarfism, hyperostosis, and intellectual impairment (85). Interestingly, the disease-causing mutations in the PS synthase-1 gene all lie within a highly conserved region of the protein that is responsible for the feedback regulation of PS synthase-1 activity by PS (84). Consequently, in fibroblasts derived from Lenz-Majewski patients, the rate of incorporation of [3H]serine into PS was markedly higher than in fibroblasts from control patients. Nevertheless, neither the amount of PS synthase-1 protein, nor the activity of PS synthase-2, was higher in the mutant cells than in the control cells. Instead, the mutations in the patients’ fibroblasts led to impaired end-product inhibition of PS synthase-1 activity. Consistent with this conclusion, the in vitro activity of PS synthase-1 in lysates of control fibroblasts, but not of the mutant fibroblasts, was strongly inhibited by exogenously added PS, but not by the addition of other phospholipids. Consequently, these studies show that the PS synthase-1 mutations in Lenz-Majewski syndrome patients are gain-of-function, rather than loss-of-function, mutations (86). In addition, despite the impaired end-product inhibition of PS synthesis, the mass of PS was not greater in the mutant fibroblasts than in the control fibroblasts. Presumably, compensatory mechanisms had been induced in the mutant fibroblasts to maintain normal phospholipid homeostasis. Additional cases of Lenz-Majewski syndrome have now also been reported to be due to mutations in the same region of the PS synthase-1 gene (87–89). However, the content of PS and PE in bone or other tissues and specific subcellular organelles was not measured in any of these studies. One can speculate that alterations (an increase?) in the PS content of specific tissues might be caused by the mutations and, thereby, contribute to the various clinical phenotypes. For example, an increased level of PS in osteoclasts has been proposed to accelerate the rate of bone formation without altering the rate of bone absorption (87–89). Thus, an increased amount of PS in bone cells might be responsible for the increased bone density in the patients. Nevertheless, it is not yet clear if, or how, the loss of end-product regulation of PS synthase-1 activity accounts for the multiple phenotypes of Lenz-Majewski patients, such as intellectual impairment, bone dysplasia, and cutis laxa (i.e., loose wrinkled skin) (88, 89).
PS IMPORT INTO MITOCHONDRIA
A key question frequently asked by cell biologists is: by what mechanisms are hydrophobic lipid molecules, such as PC, PE and PS, transported from their sites of synthesis (primarily on the ER) to other intracellular organelle membranes, such as those of mitochondria, plasma membrane, peroxisomes, lysosomes, and endosomes? Spontaneous diffusion of lipids through the aqueous milieu of the cytosol is energetically unfavorable and, therefore, is not likely to occur in most cases. Clearly, some inter-organelle lipid movement occurs via vesicles, but a vesicular transport mechanism probably does not mediate lipid import into mitochondria (69, 90–92). Nor do any of the known cytosolic “lipid transfer proteins” appear to be required for PS transfer from the ER through the cytosol to mitochondria because in permeabilized cells and reconstituted organelle systems, the transfer of PS to mitochondria for decarboxylation to PE (see the PS decarboxylation section below) occurs at the same rate as in intact cells (90–92). Although the mechanisms underlying inter-organelle lipid transport have, in most cases, not yet been established, some important clues have emerged in the case of PS transport from the ER to mitochondria.
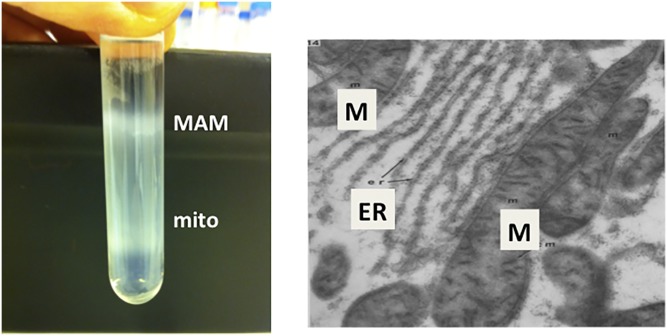
For the past half-century, numerous ultrastructural/electron microscopic studies of mammalian cells have revealed a close apposition between the ER and mitochondria (Fig. 6) that appears to be regulated by fasting and feeding (93–96). A close proximity has also been observed among the ER, mitochondria, and lipid droplets in hepatoma cells (97). Although the electron microscopic evidence suggested that physical connections might exist between the ER and mitochondria, these data were originally dismissed as being artifactual and lacking in biological importance. Nevertheless, in support of the idea that regions of the ER lie in close proximity to mitochondria, Shore and Tata (98) separated ER membranes into two subfractions that were associated to different degrees with mitochondria. Moreover, in the 1990s, some serendipitous cell fractionation studies in my laboratory revealed that mitochondria do, indeed, associate specifically and reversibly with elements of the ER, thereby providing a potential mechanism for the import of newly made PS into mitochondria (69). It is now known that approximately 20% of mitochondria in HeLa cells are normally in close juxtaposition with the ER, at a separation of 20–30 nm (99).
Fig. 6.
MAMs. Left: Percoll gradient isolation of MAMs and mitochondria from rat liver (from the author’s personal file). Right: Electron microscopic visualization of MAMs, shown as contacts/close proximity between ER and mitochondria (M) [From (95)].
In these subcellular fractionation experiments, we were investigating the subcellular location of enzymes involved in phospholipid synthesis in rat liver, and were aware that PS synthase activity had previously been localized to microsomal membranes (100, 101). We were, therefore, astonished and disappointed to discover that the mitochondria that we had isolated by a traditional subcellular fractionation technique (the pellet that sedimented upon a 10 min centrifugation of a postnuclear supernatant at 10,000 g) contained high levels of PS synthase activity. For example, in microsomes, PS synthase activity was 1.76 ± 0.69 nmol/h/mg protein, whereas in mitochondria, the activity was 1.82 ± 0.55 nmol/h/mg protein (69). We initially concluded that the mitochondria we had isolated were highly contaminated by microsomes. Rather than stopping at this point, we obtained an improved protocol for isolation of “pure” mitochondria from Ben de Kruijff (University of Utrecht, The Netherlands). This method involved ultracentrifugation of crude mitochondria (isolated as above) on a Percoll gradient (shown in Fig. 6). When we measured PS synthase activity in the lower light-brown band (mitochondria) from the Percoll gradient, we found that the activity was very low (0.10 ± 0.07 nmol/h/mg protein). In contrast, when we collected the diffuse white upper band from the Percoll gradient (Fig. 6) and centrifuged these membranes for 1 h at 100,000 g on a sucrose gradient, essentially all of the PS synthase activity (2.74 ± 0.9 nmol/h/mg protein) from the crude mitochondrial pellet was recovered in this “contaminating” membrane fraction. We initially called these membranes “fraction X” (69), but later changed their name to mitochondria-associated membranes (MAMs). Although MAMs contained many, but not all, properties of typical ER, the activities of several other lipid biosynthetic enzymes, including PS synthase-1 and -2, diacylglycerol acyltransferase, and acyl-CoA cholesterol acyltransferase, were several-fold enriched in MAMs compared with the bulk of ER [(69, 102–104); reviewed in (105)]. Proteomic analysis of MAMs isolated from mouse brain has now been reported (106, 107). Another surprising finding from our isolation of MAMs was that, according to immunoblotting, the PC-synthesizing enzyme, PE N-methyltransferase (which was the focus of the Dennis Vance laboratory next door!), was primarily localized to MAMs, rather than the bulk of ER (108); the bulk of ER did, however, contain some PE methyltransferase activity, but the protein was not detected by the antibody used for immunoblotting. Indeed, PE methyltransferase, which is expressed essentially only in hepatocytes, can be used as an excellent specific marker of MAMs in hepatocytes and liver. Because MAMs dissociate from mitochondria upon centrifugation on a Percoll gradient (Fig. 6) (69), we concluded that the association between MAMs and mitochondria was not the result of membrane fusion, but rather constituted a reversible contact event. ER-mitochondria contact sites (similar to the MAMs that were obtained from the livers of rats and mice) have now also been characterized from several other mammalian sources such as primary hepatocytes and CHO cells (92, 109–112), as well as plants (94, 113) and yeasts (114, 115) [reviewed in (105, 116)]. More recently, a similar, but slightly modified, protocol for isolation of MAMs has been reported (117).
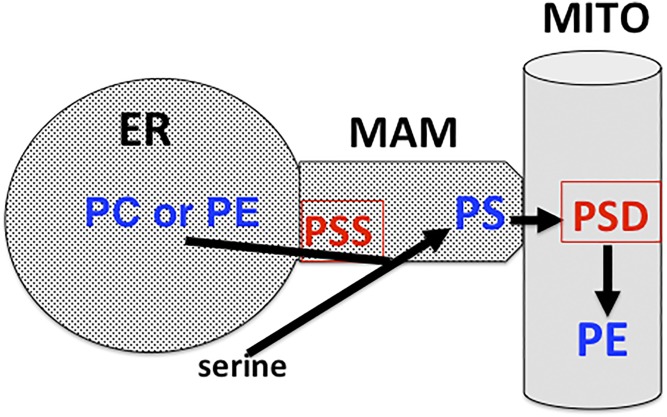
The finding that PS synthase-1 and -2 are enriched ∼4-fold in MAMs compared with the bulk of the ER (69, 92, 103) suggested that the majority of cellular PS might be produced in the MAMs (103). These intriguing observations led us, and others, to propose that the close juxtapositions, or contact sites, between the ER and mitochondria would provide a plausible mechanism by which a hydrophobic lipid, such as PS, is transported from its site of synthesis on the ER/MAM to mitochondria for decarboxylation to PE via PS decarboxylase (PSD) without moving directly through the aqueous milieu of the cytosol (Fig. 7) (69, 118, 119). Accordingly, when the crude mitochondrial fraction (containing MAMs + mitochondria) was incubated with [3H]serine in the absence of cytosolic proteins, radiolabeled PE was rapidly produced. Thus, the crude mitochondrial fraction contained all of the ingredients required for the synthesis of PS, its transport to mitochondria, and its decarboxylation to PE (Fig. 7). These studies indicated that the import of newly made PS into mitochondria occurs via MAMs. When these results were initially reported, there was profound and widespread skepticism about the role and biological relevance of MAMs in PS import into mitochondria. Nevertheless, numerous additional studies have now confirmed that MAM contact sites mediate the import of newly made PS from the ER/MAM to mitochondria for decarboxylation to PE by PSD (Fig. 7) (69, 90–92, 120–123).
Fig. 7.
Synthesis, translocation, and decarboxylation of PS. PS is synthesized in MAMs by serine-exchange via PS synthase (PSS) with either PC or PE. The newly made PS is subsequently imported via MAMs into mitochondria (MITO) and decarboxylated to PE therein via PSD.
The half-time for transport of newly made PS from its site of synthesis in the ER/MAM to the site of PSD on mitochondrial inner membranes (Fig. 7) is surprisingly long (∼6.5 h), and this step is rate limiting for the conversion of PS to PE in mammalian cells (91, 121). ATP is required for the transport of newly made PS to the site of PSD in intact mammalian cells, perhaps for orienting the ER and mitochondrial membranes appropriately (90, 91, 122). Moreover, the finding that newly made, rather than preexisting, PS is preferentially imported into mitochondria for decarboxylation to PE by PSD (122) indicates that a newly made pool of PS is channeled into mitochondria. More details of the mechanism by which PS is imported into mitochondria were revealed by some novel in vitro reconstitution experiments. Voelker (110) used a shearing process to disrupt “donor” cells in which PS had been previously radiolabeled. These disrupted labeled donor cells were then mixed with disrupted unlabeled “acceptor” cells. Interestingly, no radiolabeled PE was produced, demonstrating that the disrupted acceptor cells were unable to translocate the radiolabeled PS to mitochondria for decarboxylation to PE. On the other hand, PS that was made in the disrupted donor cells was efficiently decarboxylated to PE in the same set of disrupted cells. Thus, the translocation of PS to mitochondria for decarboxylation to PE occurred only in the same cells in which the PS was made, suggesting that newly made PS had been “channeled” into mitochondria (110). These data are consistent with the idea that PS is imported into mitochondria via close juxtaposition, or contact, between MAMs and mitochondria. In other supporting studies, the enrichment of donor membranes (MAMs) with PS (which appears to be primarily made in the MAMs) markedly increased the rate of PS import into mitochondria for decarboxylation to PE via PSD (124). Furthermore, an increased production of PS in MAMs increased the transport of PS from the ER/MAM to mitochondria (125). These data indicate that enrichment of MAMs with the negatively charged phospholipid, PS, promotes contact between MAMs and mitochondrial outer membranes.
After newly made PS reaches the mitochondrial outer membrane (Fig. 7), several additional hurdles must be overcome before the PS can access the catalytic site of PSD on mitochondrial inner membranes. First, upon arrival of PS at the cytosolic leaflet of the mitochondrial outer membrane, the PS must be translocated across this membrane. The molecular mechanism underlying the transbilayer movement of PS is currently not known. Next, the PS must traverse the inter-membrane space. This transfer appears to occur at the contact sites between mitochondrial outer and inner membranes (126), thereby eliminating the necessity for direct movement of PS through the hydrophilic environment of the inter-membrane space. It is likely that a triple contact site is formed between MAMs and the contact site between the outer and inner mitochondrial membranes (127, 128). Thus, the active site of PSD, which is exposed on the outer aspect of the mitochondrial inner membrane, would be able to access the newly imported PS. Clearly, additional information is required for elucidation of more details of the molecular mechanisms that mediate all of these steps for PS transport to the site of PSD on mitochondrial inner membranes.
The observed 20–30 nm separation between mitochondrial outer membranes and MAMs is consistent with the idea that a protein bridge reversibly tethers the two organelles (99). Studies that were performed primarily in yeasts (129) indicate that formation of contact sites between the ER and mitochondria is mediated by a proteinaceous tethering complex consisting of several components (130–136). Because several of these proteins that constitute the tethering complex of yeasts are not expressed in mammalian cells (137), more information is required about the protein constituents of a tethering complex in mammalian cells (138). One protein that has been suggested to be part of the mammalian ER/mitochondria tether is mitofusin-2, a protein that is required for mitochondrial fusion; mitofusin-2 has been detected in elements of the ER, mostly MAMs, as well as in mitochondrial outer membranes (134). A role for MAMs in mitochondrial function was also suggested by the finding that silencing mitofusin-2 expression in fibroblasts increased the distance between the ER and mitochondria and decreased [3H]serine incorporation into PE (139). Additional evidence for a role of mitofusin-2 in ER/mitochondria tethering is the observation that a mitochondrial ubiquitin ligase (MITOL) that ubiquitinates mitofusin-2 in vitro altered the number of ER-mitochondria contact sites (140). Furthermore, a defect in PS transport to mitochondria was corrected by decreasing the distance of tethering between the ER and mitochondria using artificial tethering proteins of specific lengths (125). Several other proteins have been implicated as components of the tethering complex between the ER/MAM and mitochondria of mammalian cells. For example, in neurons, the tethering of ER to mitochondria was reported to involve the PDZD8 protein as a mechanism for regulating calcium dynamics (141). Other proteins, such as the phosphofurin acidic cluster sorting protein, a cytosolic protein, also regulate the formation of MAM-mitochondria contact sites, but are not necessarily part of the tether (142). Thus, the identification of proteins that form a tethering complex between the ER/MAM and mitochondria and the precise biological functions of MAMs in mammalian cells are currently very active areas of investigation.
The import of PS into mitochondria for conversion to PE via PSD (Figs. 5, 7) is absolutely crucial for cell viability, as shown by the embryonic lethality caused by global elimination of PSD in mice (143). Moreover, reduction in the rate of production of PE in mitochondria via PSD profoundly impairs multiple mitochondrial functions, such as respiration, as well as mitochondrial morphology (143, 144). MAMs are required not only for PS import into mitochondria and the decarboxylation of PS to PE [reviewed in (105, 130, 145)], but also for regulation of calcium homeostasis because calcium transport between the ER and mitochondria occurs via MAMs (99, 131, 146). In addition, the inositol tris-phosphate receptor is highly enriched in MAMs (99, 109, 146, 147). Furthermore, the formation of inter-organelle contact sites between the ER/MAM and mitochondria appears to be critical for the regulation of apoptosis (133, 142), autophagosome formation (148), iron homeostasis (149), and mitochondrial DNA replication (150).
Several recent studies have suggested that MAMs play key roles in human disease processes. For example, an increased association of MAMs with mitochondria has been proposed to promote the progression of Alzheimer’s disease because the amyloid precursor protein and the protease (γ-secretase) that generate β-amyloid deposits are enriched in MAMs (139, 151–154). MAMs have also been implicated in familial Parkinson’s disease that is caused by mutations in α-synuclein, a protein that is linked with this disease and is located in MAMs. In these patients, MAM function is impaired (155). Recent studies have also implicated MAM dysfunction in type 2 diabetes (156). Nevertheless, despite these intriguing findings, many questions remain concerning the function of MAMs in human disorders.
The isolation and characterization of MAMs raised the possibility that similar membrane contact sites might be a feature that is common to other pairs of organelles, and might play an important role in inter-organelle communication as well as lipid transport. Although MAMs (i.e., the contacts between the ER and mitochondria) are the best-characterized membrane contact sites [reviewed in (105, 116, 157)], contact sites have now been detected between other pairs of organelles [reviewed in (158–160)]. For example, contact sites between the ER and plasma membrane are thought to be important for autophagosome formation (161). The tethering between the ER and the plasma membrane involves phosphatidylinositol-4-P and the oxysterol binding proteins, ORP5 and ORP8 (162). Membrane contact sites have also been observed between the ER and Golgi (163, 164), the ER and peroxisomes (165), mitochondria and lysosomes (166), and the ER and endosomes (159). Some of these inter-organelle contact sites have been proposed to mediate inter-organelle communication as well as lipid transfer [reviewed in (157)]. Nevertheless, it has not yet been established how generally membrane contact sites, including the MAMs, are used for the inter-organelle trafficking of lipids, such as sterols and other phospholipids.
BIOSYNTHESIS OF PE
The four PE biosynthetic pathways
PE can be synthesized in mammalian cells by four distinct pathways [reviewed in (167, 168)]: the CDP-ethanolamine pathway (reactions 1–3 in Fig. 5) (43), the PSD pathway (reaction 4 in Fig. 5) (30, 51), the base-exchange pathway (reactions 4 and 6 in Fig. 5) (169), and the acylation of lyso-PE (reaction 7 in Fig. 5) (170). It is noteworthy that the CDP-ethanolamine pathway is the only de novo pathway for PE synthesis, whereas the other three pathways involve transformation of a preformed phospholipid (Fig. 5). In general, as discussed in the Unraveling Phosphoglycerolipid Biosynthesis section above, the reactions of the CDP-ethanolamine pathway for PE biosynthesis parallel those of the CDP-choline pathway for PC synthesis (43).
The quantitative importance of each of these pathways of PE synthesis (Fig. 5) has not been adequately addressed, but appears to depend on the type of cell. In several types of cultured cells, including fibroblasts, more than 80% of PE appears to be derived from the PSD pathway, even when the cells are provided with adequate amounts of ethanolamine (171, 172). On the other hand, in some early studies, it was reported that, in hamster heart and rat hepatocytes, the CDP-ethanolamine pathway generated the majority of PE, whereas the PSD pathway accounted for only ∼5% of PE synthesis (173–175). According to one estimate, the synthesis of PE via the base-exchange reaction contributes only 8–9% of PE in rat hepatocytes at physiological concentrations of ethanolamine (∼20 μM) (169). The production of PE by the acylation of lyso-PE (reaction 7 in Fig. 5) is active in yeasts (170, 176) and probably also occurs in mammalian cells (144); interestingly, in yeasts, the majority of lyso-PE acyltransferase activity is localized to the MAMs (170). However, because radiolabeling was used in all of these calculations, and because the pool sizes, homogeneity of labeling, and the specific radioactivity of precursor molecules were not assessed, estimation of the relative contributions of the four pathways for PE synthesis is incomplete and requires further investigation (177).
An important question that arises is: why has more than one major pathway for PE synthesis evolved in mammalian cells? An obvious prediction is that alternative pathways would provide a backup for PE synthesis in case one pathway fails. This assumption does not, however, appear to represent the whole story because experiments in knockout mice show that both the PSD pathway (143) and the CDP-ethanolamine pathway (178, 179) for PE synthesis are required for mouse survival (see the PS decarboxylation and The CDP-ethanolamine pathway sections below).
PS decarboxylation
The enzymatic decarboxylation of PS to PE by PSD was first reported in 1964 (51). PSD decarboxylates the serine moiety of PS to generate the ethanolamine group of PE (Figs. 5, 7). The first PSD protein to be purified was from Escherichia coli in which this enzyme mediates the sole pathway for PE synthesis (51). In mammalian cells, PSD activity is restricted to mitochondrial inner membranes with the active site oriented toward the inter-membrane space (180, 181). Mammalian PSD belongs to a small family of decarboxylases that contain an unusual pyruvoyl prosthetic group (182). The enzymatically active form of PSD is generated from its precursor protein by an autocatalytic cleavage between the glycine and serine residues of an essential LGST motif. In mammalian cells, this proteolysis yields two subunits, α and β, which remain noncovalently attached to the mitochondrial inner membrane (183). During the proteolysis, the serine residue of the LGST motif is converted into the pyruvoyl group at the N terminus of the α subunit in which the active site of PSD resides (184). PSD activity in mammalian cells is derived from a single Pisd gene and the absolute requirement of the PSD pathway for PE synthesis in mammals was established by the demonstration that global disruption of the Pisd gene in mice is embryonically lethal (143). In contrast, yeasts express two distinct PSD activities: Psd1, which is similar to the mammalian PSD protein and is localized to mitochondria, and Psd2, which is encoded by a completely different gene and resides in the Golgi/vacuole (185, 186). Yeasts that lack both Pisd1 and Pisd2 activities are ethanolamine auxotrophs (185, 186). Interestingly, and in contrast to previous reports, the Psd1 protein in yeasts has recently been reported to have dual locations: the ER and mitochondria. Moreover, the PE made by Psd1 in the ER was proposed to have a function that was distinct from the PE made by Psd1 in mitochondria (187). It will be interesting to determine whether mammalian PSD has similar dual localizations and functions in mammalian cells.
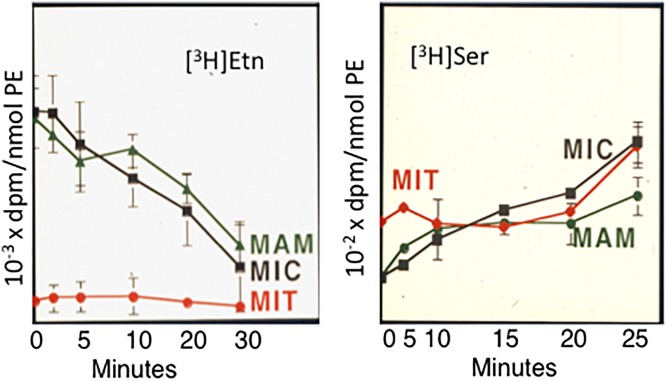
It is noteworthy that the final steps of the CDP-ethanolamine pathway (Fig. 5) and the PSD pathway for PE synthesis occur in different subcellular locations. Whereas the final reaction of the CDP-ethanolamine pathway for PE synthesis operates on the ER, PE synthesis via PSD occurs in mitochondria. Thus, it is possible that two distinct pools of PE are produced by these two biosynthetic pathways. In support of this possibility, in CHO cells and hepatocytes, mitochondrial PE is derived preferentially from PE that is made in situ via PSD in mitochondria rather than being made by the CDP-ethanolamine pathway in the ER and subsequently imported into mitochondria (122). This conclusion was reached from experiments in CHO cells in which PE was pulse-labeled with either [3H]serine (as a marker of PE derived from PSD) or [3H]ethanolamine (as a marker of PE made from the CDP-ethanolamine pathway) (Fig. 8). After various time periods, subcellular fractions (microsomes, MAMs, and mitochondria) were isolated over a time course and PS and PE were obtained from the fractions. The specific radioactivity of [3H]serine-derived PE in mitochondria rapidly exceeded that in microsomes and MAMs. On the other hand, essentially all of the [3H]ethanolamine-labeled PE remained in the microsomal and MAM fractions and was not significantly imported into mitochondria (Fig. 8), even after 24 h (92). In addition, these, and other, studies showed that PE made in mitochondria via PSD is efficiently exported from mitochondria and transported to other organelle membranes, such as the ER and plasma membrane (122, 188). Parallel studies, with similar conclusions, were performed in yeasts by Daum and Voelker and their colleagues (189, 190). Thus, the PE in mitochondrial membranes is primarily made in situ in mitochondria via PSD, rather than being made from the CDP-ethanolamine pathway in the ER.
Fig. 8.
Origin of mitochondrial PE. Incorporation of [3H]ethanolamine (Etn) (left) and [3H]serine (Ser) (right) into PE from subcellular fractions of rat hepatocytes [from (92)]. MIC, microsomes; MIT, mitochondria.
The findings described above are consistent with the mitochondrial defects and lethality that occur in PSD knockout mice (143). Because Pisd+/− heterozygous mice appeared to be normal in all respects, whereas complete elimination of PSD was 100% embryonically lethal, we reduced PSD activity in CHO cells by ∼85% using siRNA silencing so that we could determine the impact of reducing mitochondrial PE content. The mitochondrial PE content was thereby reduced by 25–30%, causing mitochondrial fragmentation and dramatically impairing mitochondrial functions, including oxidative phosphorylation (144). Importantly, as discussed in The CDP-ethanolamine pathway section below, genetic elimination of the CDP-ethanolamine pathway for PE synthesis in mice is also embryonically lethal (178). Consequently, PE synthesis via the CDP-ethanolamine pathway and PE derived from the PSD pathway are both essential for mouse viability. It appears, therefore, that the pool of PE made by PSD (in mitochondria) and the pool of PE made by the CDP-ethanolamine pathway (in the ER) are at least partially compartmentalized so that each pool is used for some specific functions.
The CDP-ethanolamine pathway
Surprisingly little is known about the origin of the ethanolamine that is used for PE synthesis via the CDP-ethanolamine pathway (Fig. 5). Nor has the mechanism by which ethanolamine is imported into cells been unequivocally established. The normal plasma concentration of free ethanolamine in rodents and humans is ∼20 μM (191). Some (probably the majority of) ethanolamine in animals is supplied from dietary sources. On the other hand, in plants such as Arabidopsis thalania (192) and in some microorganisms, serine can be directly decarboxylated to ethanolamine. In contrast, the production of ethanolamine by direct decarboxylation of serine has not been observed in mammalian cells. Another important, and often ignored, source of ethanolamine for PE synthesis is the mitochondrial decarboxylation of the serine moiety of PS by PSD (Fig. 5) (30, 181) (see the PS decarboxylation section above). The product, PE, can subsequently be degraded to release free ethanolamine that acts as a substrate for PE synthesis via the CDP-ethanolamine pathway (Fig. 5). A completely different endogenous source of ethanolamine for PE synthesis in mammalian cells is from the degradation of sphingosine-P by the enzyme, sphingosine-P lyase (193, 194). This reaction is thought to provide only a small fraction of the total ethanolamine that is required for PE synthesis via the CDP-ethanolamine pathway (195, 196). Thus, the majority of ethanolamine incorporated into PE via the CDP-ethanolamine pathway appears to be derived from dietary sources, as well as PS decarboxylation.
In some studies performed in 1956, Kennedy, Smith, and Weiss (45) identified two enzymatic activities that participate in PE synthesis via the CDP-ethanolamine pathway: CTP:phosphoethanolamine cytidylyltransferase (ET) and CDP-ethanolamine:1,2-diacylglycerol phosphoethanolamine transferase (Fig. 5). Mammalian genes encoding the three enzymes of this pathway have now been identified. In the first step of this pathway, ethanolamine is phosphorylated by ethanolamine kinase (ETNK) to produce phosphoethanolamine (Fig. 5). Four related genes encode enzymes that exhibit ETNK activity; all of these kinases are soluble cytosolic proteins. Two of the kinases primarily phosphorylate choline, but also have low activity toward ethanolamine. The two other ETNKs, ETNK1 and ETNK2, appear to be specific for ethanolamine (197–199). The two ethanolamine-specific kinases have distinct tissue distributions. Whereas ETNK2 is primarily expressed in liver and reproductive tissues, ETNK1 is widely expressed in most tissues, with highest expression in liver and kidney. Homozygous disruption of the Etnk2 gene in mice reduced hepatic ETNK activity by ∼80%, but did not alter phospholipid levels in tissues and the male mice developed normally. However, in female Etnk2−/− mice, litter size was reduced and perinatal death occurred in ∼20% of the pups (198). These findings suggest that there is at least some, although not complete, functional redundancy of the two ETNK isoforms.
In the second step of the CDP-ethanolamine pathway for PE synthesis (Fig. 5), ET converts phosphoethanolamine to CDP-ethanolamine in a reaction analogous to the cytidylyltransferase reaction of the CDP-choline pathway for PC synthesis. The ET protein was first purified from rat liver cytosol by Sundler and Akesson (200) in 1975. ET (encoded by the Pcyt2 gene) is distinct from the cytidylyltransferase of the PC biosynthetic pathway because ET specifically utilizes P-ethanolamine, but not P-choline (201). Moreover, unlike the amphitropic enzyme, CTP:phosphocholine cytidylyltransferase-α (202–204), ET is not activated by lipids and there is no conclusive evidence that ET is activated upon binding to membranes. Furthermore, in contrast to CTP:phosphocholine cytidyltransferase-α, ET is not present in the nucleus of mammalian cells. The ET protein is encoded by a single gene (Pcyt2) that generates two distinct ET isoforms produced by differential splicing events (205, 206). An unusual feature of ET is the presence of two catalytic motifs separated by a short linker region derived from an alternatively spliced exon; both catalytic motifs are required for ET activity (205, 206).
The absolute requirement of the CDP-ethanolamine pathway of PE synthesis for mouse development was demonstrated by global disruption of the Pcyt2 gene in mice, which resulted in lethality around embryonic day 8.5 (178). It is noteworthy that in Pcyt2−/− mice, despite the apparently normal activity of PSD for PE synthesis (see the PS decarboxylation section above), PSD was unable to compensate for the loss of ET activity. The Pcyt2+/− heterozygous mice were outwardly normal and the amount of PE in tissues was normal despite the attenuated rate of PE synthesis via the CDP-ethanolamine pathway, according to radiolabeling studies with [3H]glycerol (178, 179). Interestingly, diacylglycerols and triacylglycerols accumulated in the livers of Pcyt2+/− mice causing hepatic steatosis, presumably because the utilization of diacylglycerols for PE synthesis was diminished (179, 207). In other studies, the Pcyt2 gene was disrupted specifically in the livers of mice. The liver-specific Pcyt2−/− mice appeared outwardly normal, but the amount of triacylglycerols in the liver was 10-fold higher than in Pcyt+/+ mice (208). Skeletal muscle-specific ET knockout mice were also generated (209). In the muscles of these mice, the PE content was lower than in the muscles of wild-type mice, with a concomitant increase in diacylglycerol content that was not associated with insulin resistance. However, mitochondrial function was improved, not impaired, in the muscles of the skeletal muscle-specific ET knockout mice (209).
In the final reaction of the CDP-ethanolamine pathway, PE is formed by the transfer of phosphoethanolamine from CDP-ethanolamine to diacylglycerol in a reaction catalyzed by CDP-ethanolamine:1,2-diacylglycerol ethanolaminephosphotransferase (Fig. 5). This enzymatic activity is contributed by integral membrane proteins that reside primarily on ER membranes, with some activity on nuclear membranes (210). More than one protein has been identified that exhibits ethanolaminephosphotransferase activity, but none of these proteins has been purified to homogeneity. One of these transferases, CEPT1, has dual activity with both CDP-ethanolamine and CDP-choline (211). This protein exhibits a striking preference for diacylglycerol species that contain a palmitoyl chain at the sn-1 position and a docosahexaenoyl chain at the sn-2 position. Correspondingly, in hepatocytes, nearly 50% of PE made by the CDP-ethanolamine pathway consists of this molecular species, although the function, if any, of this selectivity is not yet understood. In other studies, the muscle-specific elimination of CEPT1 in high-fat diet-fed mice altered the phospholipid composition of the sarcoplasmic reticulum and increased insulin sensitivity in muscle (212). In addition to the dual specificity ethanolaminephosphotransferase, a second ethanolaminephosphotransferase activity, EPT-1 (also called selenoprotein-1), that is specific for CDP-ethanolamine (213), has more recently been discovered. Selenoprotein-1 is most highly expressed in the brain, placenta, and liver. Recently, a human with the severe motor neuron disorder, hereditary spastic paraplegia, has been reported to have a mutation in selenoprotein-1 (214, 215).
In contrast to the abundance of information available on mechanisms by which PC synthesis is regulated by CTP:phosphocholine cytidylyltransferase [reviewed in (216)], little is known about regulation of the CDP-ethanolamine pathway for PE synthesis. In the 1970s, Sundler demonstrated that, in rats and cultured rat hepatocytes, the reaction catalyzed by ET is rate limiting for PE synthesis under most metabolic conditions (191). In these experiments, freshly prepared hepatocytes were incubated with increasing concentrations of ethanolamine, and a corresponding increase in the amount of P-ethanolamine was observed with no change in the concentration of CDP-ethanolamine. These data indicated that the cytidylyltransferase, ET, of the CDP-ethanolamine pathway catalyzes the slowest, and therefore rate-limiting, step for PE synthesis, as was the case for the cytidylyltransferase in the CDP-choline pathway for PC synthesis,. However, under some metabolic conditions, the supply of diacylglycerol, a substrate for the final reaction of the CDP-ethanolamine pathway, appears to limit the rate of PE synthesis by this pathway (174, 217).
The mechanisms by which the CDP-ethanolamine pathway is regulated at the level of expression of the Pcyt2 gene remain incompletely understood, although basal transcription of ET is mediated by the transcription factor, NF-Y (218). Information on regulation of the CDP-ethanolamine pathway for PE synthesis by hormones and other metabolic factors is largely lacking. However, ET is activated by protein kinase C-mediated phosphorylation in the linker region of the protein and in the C-terminal catalytic domain (219). Moreover, the mRNA encoding ET and the amount of ET protein increase during differentiation of muscle cells (220). In addition, the two major pathways of PE synthesis are coordinately regulated under certain metabolic conditions because the amount of ET protein and its activity are ∼40% and 100%, respectively, higher in tissues of heterozygous PSD knockout mice than in wild-type control mice (143).
PE ether lipids
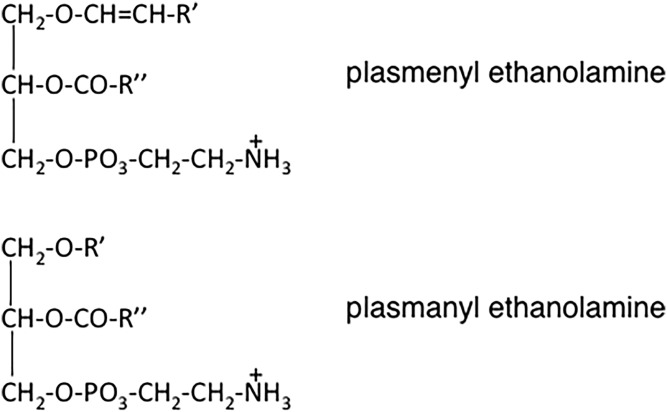
The existence of a unique class of phosphorus-containing lipids that contain an O-alkyl group was first proposed by Doree (221) in 1909 in England. Later, in 1929, Feulgen, Imhauser, and Behrens (222) discovered that when a phosphatide fraction was stained with fuchsin sulfurous acid, a purple color developed that was characteristic of an aldehyde; the presence of the aldehyde was apparently due to the degradation of the alkyl or alkenyl bond of this novel class of lipids. Subsequently, in the 1950s, the nature of the chemical linkage of the alkyl group at the sn-1 position in a subset of PE species was established by several research groups who also showed that some PE species alternatively contained an alk-1-enyl (i.e., a vinyl ether) linkage at the sn-1 position (Fig. 9). Confirmation that ether lipids were constituents of animal tissues was provided by chemical studies and by demonstration by Snyder and colleagues in 1969 that the cell-free synthesis of the alkyl ether bond occurs (223, 224). A historical review of this discovery and of the chemistry and biochemistry of the ethanolamine plasmalogens can be found in (225).
Fig. 9.
Structures of the ether lipids: plasmenylethanolamine (top) and plasmanylethanolamine (bottom).
The PE ether lipids are widely distributed throughout the animal kingdom and are also present in small amounts in plants. In mammals, ∼20% of total phospholipids contain either an O-alkyl or an O-alkenyl linkage at the sn-1 position (Fig. 9); the remainder of the PE consists of the diacyl species. Thus, PE molecules can be subclassified into the diacyl, alkylacyl (i.e., plasmanyl), and the alkenylacyl (i.e., plasmenyl) classes. In liver, plasmenyl-PE, with the O-alk-1′-enyl linkage, accounts for only 0.8% of total phospholipids (226, 227), whereas PE in the brain contains a much larger proportion of plasmenyl PE, up to 70% of total ethanolamine phospholipids. Moreover, in neuronal cell membranes, ∼30% of total phospholipids and ∼90% of the ethanolamine phospholipids consist of plasmenyl-PE. PE ether lipids are also abundant in inflammatory cells and tumor cells in which the ether lipids comprise up to 70% of the ethanolamine phosphoglyceride pool. The O-alkyl and O-alkenyl chains at the sn-1 position of the ether lipids generally consist of 16:0, 18:0, or 18:1 chains, whereas arachidonoyl (20:4) and docosahexanoyl (20:6) acyl chains are the most common at the sn-2 position. The PC ether lipids, which are usually the alkyl species, are usually far less abundant than the PE ether lipids, except in the heart of some animals, such as hamsters, where plasmanylcholines are a prominent phospholipid species (228, 229).
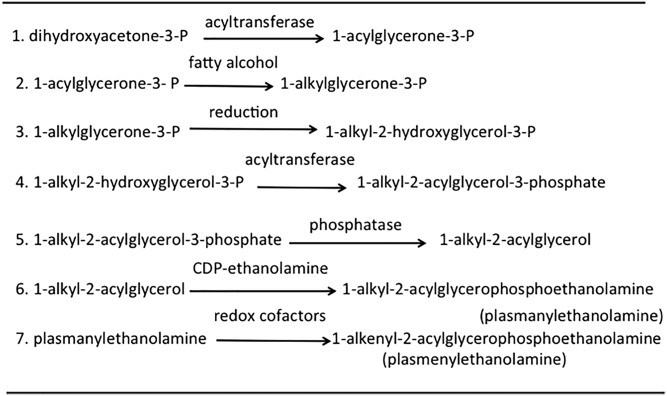
The biosynthetic pathway for the PE ether lipids is complex (outlined in Fig. 10) [reviewed in (226, 227)]. The first two steps (steps 1 and 2 in Fig. 10), the formation of 1-alkylglycerone-3-phosphate (also called 1-alkyldihydroxyacetone phosphate) from dihydroxyacetone phosphate, occur exclusively in peroxisomes. The first two sequential reactions for the production of 1-alkylglycerone-3-phosphate from dihydroxyacetone phosphate are catalyzed by dihydroxyacetone-3-phosphate O-acyltransferase and 1-alkylglycerone-3-phosphate synthase (reactions 1 and 2 in Fig. 10); the latter reaction uses a fatty alcohol as a substrate. The 1-alkylglycerone-3-phosphate is then converted into 1-alkyl-2-acylglycerol in the ER by sequential reduction, acylation, and dephosphorylation steps (reactions 3–5 in Fig. 10). In step 6 of Fig. 10, 1-alkyl-2-acylglycerol reacts with CDP-ethanolamine, using either the dual function choline/ethanolamine phosphotransferase or the CDP-ethanolamine-specific phosphotransferase, selenoprotein-1 (both of which are also used for diacyl PE synthesis; see The CDP-ethanolamine pathway section above), yielding plasmanyl-PE (plasmanylethanolamine) (reaction 6 in Fig. 10). Finally, the alkyl group of plasmanyl-PE is desaturated to the 1-alkenyl group in an unusual reaction catalyzed by the plasmanyl-PE-specific enzyme, plasmanyl-PE desaturase, which uses redox factors (reaction 7 in Fig. 10). Interestingly, the fatty aldehyde derived from the degradation of sphinganine (dihydrosphingosine) has also been reported to serve as an effective donor of the alk-1-enyl chain of plasmenylethanolamine (230).
Fig. 10.
Enzymes and their subcellular locations for plasmenylethanolamine biosynthesis. #1, Dihydroxyacetone-3-P acyltransferase (peroxisomes); #2, ether bond formation by 1-alkylglycerone-3-P synthase (peroxisomes); #3, 1-alkylglycerone-3-P reductase (ER); #4, acyl-CoA-1-alkyl-2-acylglycerol-3-P acyltransferase (ER); #5, 1-alkyl-2-acylglycerol-3-phosphate phosphohydrolase (ER); #6, CDP-ethanolamine 1-alkyl-2-acylglycerol ethanolaminephosphotransferase (ER); #7, plasmanylethanolamine 1-alkyldesaturase (ER).
Ether lipids appear to play important structural roles in animal cell membranes, although the precise functions of these lipids are still not entirely clear. Nevertheless, there is strong evidence that alkenyl ether lipids function as antioxidants due to the presence of the vinyl ether (231, 232), and also regulate intracellular cholesterol trafficking (233). Humans with inborn errors of ether lipid synthesis exhibit severe pathologies, particularly in the brain. For example, mutations in either of the two peroxisomal enzymes, dihydroxyacetone phosphate acyltransferase or 1-alkylglycerone-3-phosphate synthase (steps 1 and 2 in Fig. 10), cause the syndrome, rhizomelic chondrodysplasia punctata (226, 234). Furthermore, in the inherited disorder, Zellweger syndrome (235, 236), peroxisome maturation is prevented, thereby abolishing the first two steps in formation of the ether bond of PE ether lipids (226). Moreover, a recent study reported that selenoprotein-1 (the ethanolaminephosphotransferase that catalyzes reaction 6 in Fig. 10) is mutated in a patient with hereditary spastic paraplegia, a disorder in which myelination and neurodevelopment are profoundly impaired (215). In skin fibroblasts from this patient, levels of plasmenylethanolamine are significantly reduced, revealing a role for selenoprotein-1 in maintaining homeostasis of the ether-linked phospholipids in humans.
FUNCTIONS OF PS AND PE
Topological arrangement of phospholipids in the plasma membrane
The concentration of PS in mammalian cells is highest in the plasma membrane and endosomes, but very low in mitochondria, particularly in mitochondrial inner membranes, the site at which PS is decarboxylated to PE (see The CDP-ethanolamine pathway section above). Several novel functions of PS in mammalian cells have recently been revealed. For example, PS is enriched on the lumenal, compared with the cytosolic, leaflet of the ER membrane (237), and is highly enriched on the inner, compared with the outer, leaflet of the plasma membrane [(8); reviewed in (9)]. This asymmetric transbilayer distribution of PS in the plasma membrane is established and maintained by the unidirectional inward transbilayer movement of PS in a process that requires ATP. The “floppase” responsible for this transbilayer translocation of PS has been identified as ATP8A1, a member of the P-type ATPase protein family [reviewed in (238)]. In addition, the bi-directional transbilayer movement of PS and other phospholipids in the plasma membrane has been proposed to be mediated by scramblase-1, a protein that randomizes the distribution of phospholipids, including PS and PE, within the plasma membrane bilayer of mammalian cells, without the need for ATP (239, 240). However, the involvement of scramblase-1 in the transbilayer movement of PS is still controversial because deletion of scramblase-1 in mice did not significantly alter PS distribution in the plasma membrane (241). The translocation of PS from the inner to the outer leaflet of the plasma membrane (probably mediated by a calcium-induced inactivation of ATP8A1 and activation of scramblase) plays an important biological role during apoptosis. At the onset of apoptosis, an increase in the intracellular calcium concentration promotes PS translocation across the plasma membrane so that more PS becomes exposed on the outer surface of the cell (242–244). This alteration in transbilayer distribution of PS in the plasma membrane represents one of the signals by which apoptotic cells are recognized and removed by phagocytes (242–244) via a PS receptor (244–246). PS receptors, such as Tim4 (245) and the so-called PS receptor PSR-1 (247), on phagocytic cells have been proposed to recognize and bind to PS when it becomes exposed on the outside of the apoptotic cell, resulting in the eventual engulfment of the apoptotic cell. Nevertheless, the identity of the PS receptor involved in removal of the apoptotic cells remains controversial [reviewed in (248, 249)]. PS also becomes exposed on the outside of the plasma membrane of activated platelets during initiation of the blood-clotting cascade (250–255), and on the outside of sperm during their maturation (256, 257).
In addition, PE is also normally enriched on the inner, compared with the outer, leaflet of the plasma membrane. In the canalicular membrane of hepatocytes, the floppase responsible for the inward translocation of PE across the membrane has been identified as ATP8B1, a protein related to ATP8A1. Mutations in ATP8B1, and reduced activity of this protein, impair canalicular bile acid export and cause retention of bile salts in the liver, resulting in the human disorder, progressive familial intrahepatic cholestasis (PFIC1). It has been proposed that the deficiency in ATP8B1 induces membrane instability, thereby reducing the function of bile acid exporters and causing the accumulation of bile salts in the hepatocytes (258).
Binding of anionic PS to signaling proteins
In addition to the biological functions that are activated in response to the redistribution of PS across the plasma membrane bilayer, PS is required for numerous other crucial intracellular processes, perhaps because of its anionic nature (237, 259–261). For example, positively charged proteins (e.g., Src and members of the Ras and Rho GTPase family) can bind to PS and direct the protein to the plasma membrane. PS also binds to the C2 domain of specific proteins and targets them to phagosomes (260, 262, 263). Furthermore, PS increases the activity of several signaling proteins that contain a C2 domain, including synaptotagmin and dynamin (264) as well as protein kinase C (265) and Akt (via its PH domain) (266). In addition, PS binds to the heat shock protein, Hsp70, thereby inducing formation of ion channels in the plasma membrane (267). Recently, PS was also shown to be essential for the formation of caveolae, invaginations in the plasma membrane that are involved in signaling events (268).
Mitochondrial PE and mitochondrial function
The content of PE in mitochondrial membranes is higher than that in other organelle membranes. Indeed, PE is particularly enriched in mitochondrial inner membranes (40% of total phospholipids). As already discussed above in the PS decarboxylation section, PS is the precursor of the PE that is produced by PSD in mitochondrial inner membranes. These observations are consistent with the finding that the majority of mitochondrial PE is synthesized in situ in mitochondria via PSD (92) [see the PS decarboxylation section above and Fig. 5). Interestingly, although mitochondrial PE content and mitochondrial function are markedly reduced in PSD-deficient cells (144), mitochondrial biogenesis and oxidative capacity are enhanced in muscle cells lacking ET (209). These observations further support the idea that, in several types of mammalian cells, mitochondrial PE is primarily synthesized via PSD in mitochondria, not by the CDP-ethanolamine pathway in the ER.
A decrease in mitochondrial PE content profoundly alters mitochondrial morphology and functions not only in mammalian cells (143, 144), but also in yeasts (269) and Trypanosoma brucei (270). In mammalian cells, silencing of PSD expression by 85% reduced the amount of mitochondrial PE by ∼30% and caused extensive mitochondrial fragmentation (144). Similarly, in embryos of mice lacking PSD, the mitochondria are grossly misshapen and exhibit a punctate distribution, suggesting defective mitochondrial fusion (143). Because the majority of ATP in mammalian cells is generated in mitochondria by oxidative phosphorylation via the electron transport chain, mitochondrial PE deficiency impairs the activities of proteins of the electron transport chain (144). These observations are consistent with studies showing that mitochondrial dysfunction contributes to cardiovascular disease (271), diabetes (272), neurodegeneration (273), and cancer progression (274, 275). Interestingly, in the brain, PE is required for the propagation and infectivity of prions, although the mechanism underlying this requirement remains unknown (276).
Changes in the phospholipid molar ratio in cells can also profoundly alter cell functions. For example, elimination of PE N-methyltransferase (a hepatic enzyme that synthesizes ∼30% of hepatic PC via methylation of PE) in mice by gene targeting decreases the molar ratio of PC/PE (by increasing PE and reducing PC) in mitochondria and stimulates mitochondrial respiration, as well as activities of proteins of the electron transport chain (277). In addition, in primary hepatocytes isolated from these knockout mice, the amount of ATP was double that in wild-type control mice (277). Thus, the amount of PE in mitochondrial membranes can profoundly alter mitochondrial morphology and functions, including energy production (144).
Importance of PE for membrane structure and function
Mammalian cells contain a large number of distinct molecular species of PE in which the acyl chains at the sn-1 and sn-2 positions are different. This acyl-chain diversity is, at least in part, due to a remodeling process in which phospholipases and lyso-phospholipid acyltransferases participate [reviewed in (278)]. Both the acyl-chain composition and headgroup constituents of membrane phospholipids define the physical properties of the membrane. For example, the cone-shape of the PE molecule promotes membrane curvature (279, 280), and the ability of PE to enhance membrane fusion is related to its ability to form hexagonal II phases in membranes (279, 281–283). PE is also required for contractile ring disassembly at the cleavage furrow of mammalian cells during cytokinesis (284), probably because of the physical properties of PE. Furthermore, in some novel studies, Dowhan and colleagues showed that in E. coli, PE markedly influences the topology of membrane proteins, such as lactose permease, and assists in their folding by acting as a “lipid chaperone” (285–287).
Phospholipids are also important components of plasma lipoproteins, such as VLDLs, low density lipoproteins, and high density lipoproteins. The phospholipids reside in the outer monolayer that surrounds the neutral lipid core (primarily triacylglycerols and cholesteryl esters) of the lipoprotein particles, and are required for formation and stability of the particles (288). PC is the most abundant phospholipid in the surface monolayer of VLDLs, but small amounts of PE (∼5% of total phospholipids) and other phospholipids are also incorporated into circulating VLDLs. Interestingly, however, the PE content of newly secreted VLDLs, and of VLDLs isolated from the Golgi lumen (289), is much higher than in circulating VLDLs (290), suggesting that PE might play a role in the assembly and/or secretion of VLDLs [reviewed in (291)].
Precursor functions of PE
Other important functions of PE in mammalian cells include its role in autophagy. During the process of autophagy, PE, primarily that derived from PSD (292), becomes covalently attached to the autophagy protein, LC3. Consequently, LC3 is recruited to membranes and enhances autophagosome formation (292). PE also provides the ethanolamine moiety of the glycerophosphatidylinositol anchors that attach many cell surface signaling proteins to the plasma membrane, thereby activating these proteins (293). In addition, PE is a precursor of anandamide (N-arachidonoylethanolamine) (294), a ligand for cannabinoid receptors in the brain.
CONCLUSION
In this article, I have focused on the remarkable historical background on the discovery and chemistry of two phosphoglycerolipids, PS and PE, and have also provided an account of the step-wise elucidation of the biosynthetic pathways for PS and PE in mammalian cells. In addition, the extensive use of knockout mice in understanding many of the physiological functions of PS and PE was discussed. This article also includes a discussion of the mechanism by which newly made PS is transported from the MAM/ER to mitochondria for decarboxylation to PE. In the final section of this article, the impact of a deficiency of PS or PE on key biological processes in mammalian cells was described. Clearly, one can predict that, in the future, many of the unanswered questions regarding the metabolism of PS and PE, as well as the consequences of depletion of these two phospholipids from mammalian cells, will be addressed using newly developed tools.
Acknowledgments
The author would like to sincerely thank all the people who have worked in the J. Vance laboratory, visiting scientists, postdoctoral fellows, graduate students, and technicians, as well as many other collaborators who have contributed over the years to much of the work discussed in this article.
Footnotes
Abbreviations:
- CHO
- Chinese hamster ovary
- ER
- endoplasmic reticulum
- ET
- CTP:phosphoethanolamine cytidylyltransferase
- ETNK
- ethanolamine kinase
- MAM
- mitochondria-associated membrane
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PS
- phosphatidylserine
- PSD
- phosphatidylserine decarboxylase
REFERENCES
- 1.Danielli J. F., and Davson H.. 1935. A contribution to the theory of permeability of thin films. J. Cell. Comp. Physiol. 5: 495–547. [Google Scholar]
- 2.Singer S. J., and Nicolson G. L.. 1972. The fluid mosaic model of the structure of cell membranes. Science. 175: 720–731. [DOI] [PubMed] [Google Scholar]
- 3.Simons K., and Ikonen E.. 1997. Functional rafts in cell membranes. Nature. 387: 569–572. [DOI] [PubMed] [Google Scholar]
- 4.Berridge M. J., and Irvine R. F.. 1984. Inositol trisphosphate, a novel second messenger in cellular signal transduction. Nature. 312: 315–321. [DOI] [PubMed] [Google Scholar]
- 5.Nishizuka Y. 1992. Intracellular signaling by hydrolysis of phospholipids and activation of protein kinase C. Science. 258: 607–614. [DOI] [PubMed] [Google Scholar]
- 6.Kimura A. K., and Kim H-Y.. 2013. Phosphatidylserine synthase-2: high efficiency for synthesizing phosphatidylserine containing docosahexaenoic acid. J. Lipid Res. 54: 214–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim H. Y. 2007. Novel metabolism of docosahexaenoic acid in neural cells. J. Biol. Chem. 282: 18661–18665. [DOI] [PubMed] [Google Scholar]
- 8.Schick P. K., Kurica K. B., and Chacko G. K.. 1976. Location of phosphatidylethanolamine and phosphatidylserine in the human platelet plasma membrane. J. Clin. Invest. 57: 1221–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Op den Kamp J. A. F. 1979. Lipid asymmetry in membranes. Annu. Rev. Biochem. 48: 47–71. [DOI] [PubMed] [Google Scholar]
- 10.Deuel H. J. 1957. The Lipids: Their Chemistry and Biochemistry. Interscience Publishers, Inc., New York. [Google Scholar]
- 11.Vauquelin M. 1812. Analyse de la matière cérébrale de l’homme et de quelques animaux. Ann. Chim. 81: 37–51. [Google Scholar]
- 12.Gobley M. 1846. Recherches chimiques sur le jaune d’oed. J. Pharm. Chim. 9: 1–15, 81–91, 161–174. [Google Scholar]
- 13.Diakonow C. 1868. Ueber die chemische Constitution des Lecithin [On the chemical constitution of lecithin]. Zentr. Med. Wiss. 6: 2–3, 97–99. [Google Scholar]
- 14.Thudichum J. L. W. 1874. Researches on the chemical constitution of the brain. Report of the Medical Officer of the Privy Council and Local Government Board (n.s.) London. No. 3, append. 5, 113–247. [Google Scholar]
- 15.Thudichum J. L. W. 1884. A Treatise on the Chemical Constitution of Brain. Balliere. Tindall, and Cox, London. [Google Scholar]
- 16.Dr. J. L. W. Thudicum [obituary]. Nature. 1901. 64: 527. [Google Scholar]
- 17. J. L. W. Thudicum [obituary]. The Times. September 10, 1901. London. [Google Scholar]
- 18.Taurog A., Entemann C., and Chaikoff I. L.. 1944. The choline- and non-choline-containing phospholipids of plasma. J. Biol. Chem. 156: 385–391. [Google Scholar]
- 19.Renall M. H. 1913. Über den stickstoffhältigen Bestandteil des Kephalins. Biochem. Zeitschr. 55: 296–300. [Google Scholar]
- 20.Levene P. A., and Rolf I. P.. 1924. Synthetic lecithins. J. Biol. Chem. 60: 677–683. [Google Scholar]
- 21.Grun R., and Limpacher A.. 1927. Synthese der Kephaline. Eur. J. Inorg. Chem. 60: 151–156. [Google Scholar]
- 22.Folch J. 1941. Isolation of phosphatidylserine from brain cephalin, and identification of the serine component. J. Biol. Chem. 139: 973–974. [Google Scholar]
- 23.Folch J., Lees M., and Sloane-Stanley G. H.. 1959. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226: 495–509. [PubMed] [Google Scholar]
- 24.Artom C. 1945. A quantitative method for ethanolamine and serine as a basis for the determination of phosphatidyl ethanolamine and phosphatidyl serine in tissues. J. Biol. Chem. 157: 585–594. [Google Scholar]
- 25.MacArthur C. G. 1914. Brain cephalin: I. Distribution of the nitrogeneous hydrolysis products of cephalin. J. Am. Chem. Soc. 36: 2397–2401. [Google Scholar]
- 26.Folch J., and Schneider H. A.. 1941. An amino acid constituent of ox brain cephalin. J. Biol. Chem. 137: 51–62. [Google Scholar]
- 27.Chargaff E., Ziff M., and Rittenberg D.. 1942. A study of the nitrogenous constitutents of tissue phosphatides. J. Biol. Chem. 144: 343–352. [Google Scholar]
- 28.Artom C. 1945. Some data on the distribution of individual phospholipids in rat tissues and in human plasma. J. Biol. Chem. 157: 595–599. [Google Scholar]
- 29.Nord F. F. 1919. Ueber die katalytische Hydrierung von Cholesterin und Cholesterylen. Biochem. Zeitschr. 99: 281–285. [Google Scholar]
- 30.Borkenhagen L. F., Kennedy E. P., and Fielding L.. 1961. Enzymatic formation and decarboxylation of phosphatidylserine. J. Biol. Chem. 236: PC28–PC32. [Google Scholar]
- 31.Chaikoff I. L. 1942. The application of labeling agents to the study of phospholipid metabolism. Physiol. Rev. 22: 291–317. [Google Scholar]
- 32.Sinclair R. G. 1935. The metabolism of the phospholipids: VIII. the passage of elaidic acid into tissue phospholipids. Evidence of the intermediary role of liver phospholipid into fat metabolism. J. Biol. Chem. 111: 515–526. [Google Scholar]
- 33.Schoenheimer R., and Rittenberg D.. 1937. Deuterium as an indicator in the study of intermediary metabolism: XI. Further studies on the biological uptake of deuterium into organic substances, with special reference to fat and cholesterol formation. J. Biol. Chem. 121: 235–236. [Google Scholar]
- 34.Bloch K. 1965. The biological synthesis of cholesterol. Science. 150: 19–28. [DOI] [PubMed] [Google Scholar]
- 35.Cornforth J. W., and Youhotsky Gore I.. 1957. Studies on the biosynthesis of cholesterol. IV. Degradation of rings C and D. Biochem. J. 65: 94–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cavanagh B., and Raper H. S.. 1939. A study of the passage of fatty acids of food into lipins and glycerides of the body using deuterium as an indicator. Biochem. J. 33: 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stevens B. P., and Chaikoff I. L.. 1951. Incorporation of short chain fatty acids into phospholipides by the rat. J. Biol. Chem. 193: 465–472. [PubMed] [Google Scholar]
- 38.Artom C. 1945. Some data on the distribution of individual phospholipids in rat tissues and human plasma. J. Biol. Chem. 157: 601–612. [Google Scholar]
- 39.Fishler M. C., Taurog A., Perlman I., and Chaikoff I. L.. 1941. The synthesis and breakdown of liver phospholipids in vitro with radioactive phosphorus as indicator. J. Biol. Chem. 141: 809–818. [Google Scholar]
- 40.Stetten D. 1941. Biological relationships of choline, ethanolamine, and related compounds. J. Biol. Chem. 138: 437–438. [Google Scholar]
- 41.Kornberg A., and Pricer W. E. Jr. 1953. Enzymatic esterification of alpha-glycerophosphate by long chain fatty acids. J. Biol. Chem. 204: 345–357. [PubMed] [Google Scholar]
- 42.Rodbell M., and Hanahan D. J.. 1955. Lecithin synthesis in liver. J. Biol. Chem. 214: 607–618. [PubMed] [Google Scholar]
- 43.Kennedy E. P., and Weiss S. B.. 1956. The function of cytidine coenzymes in the biosynthesis of phospholipides. J. Biol. Chem. 222: 193–214. [PubMed] [Google Scholar]
- 44.Kalckar H. M., and Klenow H.. 1954. Nonoxidative and nonproteolytic enzymes; biosynthesis and metabolism of phosphorus compounds. Annu. Rev. Biochem. 23: 527–586. [DOI] [PubMed] [Google Scholar]
- 45.Kennedy E. P., Smith S. W., and Weiss S. B.. 1956. New synthesis of lecithin in an isolated enzyme system. Nature. 178: 594–595. [DOI] [PubMed] [Google Scholar]
- 46.Weiss S. B., Smith S. W., and Kennedy E. P.. 1958. The enzymatic formation of lecithin from cytidine diphosphate choline and D-1,2-diglyceride. J. Biol. Chem. 231: 53–64. [PubMed] [Google Scholar]
- 47.Kennedy E. P. 1957. Metabolism of lipides. Annu. Rev. Biochem. 26: 119–148. [DOI] [PubMed] [Google Scholar]
- 48.Folch J. 1948. The chemical structure of phosphatidyl serine. J. Biol. Chem. 174: 439–450. [PubMed] [Google Scholar]
- 49.Baer E., and Maurukas J.. 1955. Phosphatidylserine. J. Biol. Chem. 212: 25–38. [PubMed] [Google Scholar]
- 50.Hubscher H. G., Dils R. R., and Pover W. F.. 1959. Studies on the biosynthesis of phosphatidylserine. Biochim. Biophys. Acta. 36: 518–528. [DOI] [PubMed] [Google Scholar]
- 51.Kanfer J., and Kennedy E. P.. 1964. Metabolism and function of bacterial lipids. II. Biosynthesis of lipids in Escherichia coli. J. Biol. Chem. 239: 1720–1726. [PubMed] [Google Scholar]
- 52.Gardner R. G., and Hampton R. Y.. 1999. A highly conserved signal controls degradation of 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase in eukaryotes. J. Biol. Chem. 274: 31671–31678. [DOI] [PubMed] [Google Scholar]
- 53.Ansell G. B., and Chojnacki T.. 1966. The incorporation of the phosphate esters of N-substituted aminoethanols into the phospholipids of brain and liver. Biochem. J. 98: 303–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spanner S., and Ansell G. B.. 1982. Activation of glycerophosphocholine phosphodiesterase in rat forebrain by Ca2+. Biochem. J. 208: 845–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barańska J. 1982. Biosynthesis and transport of phosphatidylserine in the cell. Adv. Lipid Res. 19: 163–184. [DOI] [PubMed] [Google Scholar]
- 56.Suzuki T. T., and Kanfer J. N.. 1985. Purification and properties of an ethanolamine-serine base exchange enzyme of rat brain microsomes. J. Biol. Chem. 260: 1394–1399. [PubMed] [Google Scholar]
- 57.Kuge O., Nishijima M., and Akamatsu Y.. 1985. Isolation of a somatic cell mutant defective in phosphatidylserine biosynthesis. Proc. Natl. Acad. Sci. USA. 82: 1926–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Voelker D. R., and Frazier J. L.. 1986. Isolation and characterization of a Chinese hamster ovary cell line requiring ethanolamine or phosphatidylserine for growth and exhibiting defective phosphatidylserine synthase activity. J. Biol. Chem. 261: 1002–1008. [PubMed] [Google Scholar]
- 59.Kuge O., and Nishijima M.. 1997. Phosphatidylserine synthase I and II of mammalian cells. Biochim. Biophys. Acta. 1348: 151–156. [DOI] [PubMed] [Google Scholar]
- 60.Kuge O., Saito K., and Nishijima M.. 1997. Cloning of a Chinese hamster ovary (CHO) cDNA encoding phosphatidylserine synthase (PSS) II, overexpression of which suppresses the phosphatidylserine biosynthetic defect of a PSS I-lacking mutant of CHO-K1 cells. J. Biol. Chem. 272: 19133–19139. [DOI] [PubMed] [Google Scholar]
- 61.Kuge O., Nishijima M., and Akamatsu Y.. 1991. A Chinese hamster cDNA encoding a protein essential for phosphatidylserine synthase I activity. J. Biol. Chem. 266: 24184–24189. [PubMed] [Google Scholar]
- 62.Stone S. J., and Vance J. E.. 1999. Cloning and expression of murine liver phosphatidylserine synthase (PSS)-2: differential regulation of phospholipid metabolism by PSS1 and PSS2. Biochem. J. 342: 57–64. [PMC free article] [PubMed] [Google Scholar]
- 63.Kuge O., Hasegawa K., Ohsawa T., Saito K., and Nishijima M.. 2003. Purification and characterization of Chinese hamster phosphatidylserine synthase 2. J. Biol. Chem. 278: 42692–42698. [DOI] [PubMed] [Google Scholar]
- 64.Sturbois-Balcerzak B., Stone S. J., Sreenivas A., and Vance J. E.. 2001. Structure and expression of the murine phosphatidylserine synthase-1 gene. J. Biol. Chem. 276: 8205–8212. [DOI] [PubMed] [Google Scholar]
- 65.Bergo M. O., Gavino B. J., Steenbergen R., Sturbois B., Parlow A. F., Sanan D. A., Skarnes W. C., Vance J. E., and Young S. G.. 2002. Defining the importance of phosphatidylserine synthase 2 in mice. J. Biol. Chem. 277: 47701–47708. [DOI] [PubMed] [Google Scholar]
- 66.Stone S. J., Cui Z., and Vance J. E.. 1998. Cloning and expression of mouse liver phosphatidylserine synthase-1 cDNA: overexpression in rat hepatoma cells inhibits the CDP-ethanolamine pathway for phosphatidylethanolamine biosynthesis. J. Biol. Chem. 273: 7293–7302. [DOI] [PubMed] [Google Scholar]
- 67.Jelsema C. L., and Morré D. J.. 1978. Distribution of phospholipid biosynthetic enzymes among cell components of rat liver. J. Biol. Chem. 253: 7960–7971. [PubMed] [Google Scholar]
- 68.Saito K., Kuge O., Akamatsu Y., and Nishijima M.. 1996. Immunochemical identification of the pssA gene product as phosphatidylserine synthase I of Chinese hamster ovary cells. FEBS Lett. 395: 262–266. [DOI] [PubMed] [Google Scholar]
- 69.Vance J. E. 1990. Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 265: 7248–7256. [PubMed] [Google Scholar]
- 70.Saito K., Nishijima M., and Kuge O.. 1998. Genetic evidence that phosphatidylserine synthase II catalyzes the conversion of phosphatidylethanolamine to phosphatidylserine in Chinese hamster ovary cells. J. Biol. Chem. 273: 17199–17205. [DOI] [PubMed] [Google Scholar]
- 71.Bell R. M., Ballas L. M., and Coleman R. A.. 1981. Lipid topogenesis. J. Lipid Res. 22: 391–403. [PubMed] [Google Scholar]
- 72.Breslow J. L. 1993. Transgenic mouse models of lipoprotein metabolism and atherosclerosis. Proc. Natl. Acad. Sci. USA. 90: 8314–8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maeda N., Li H., Lee D., Oliver P., Quarfordt S. H., and Osada J.. 1994. Targeted disruption of the apolipoprotein C–III gene in mice results in hypotriglyceridemia and protection from postprandial hypertriglyceridemia. J. Biol. Chem. 269: 23610–23616. [PubMed] [Google Scholar]
- 74.Vance D. E., and Vance J. E.. 2009. Physiological consequences of disruption of mammalian phospholipid biosynthetic genes. J. Lipid Res. 50: S132–S137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Arikketh D., Nelson R., and Vance J. E.. 2008. Defining the importance of phosphatidylserine synthase-1 (PSS1): unexpected viabliity of PSS1-deficient mice. J. Biol. Chem. 283: 12888–12897. [DOI] [PubMed] [Google Scholar]
- 76.Steenbergen R., Nanowski T. S., Nelson R., Young S. G., and Vance J. E.. 2006. Phospholipid homeostasis in phosphatidylserine synthase-2-deficient mice. Biochim. Biophys. Acta. 1761: 313–323. [DOI] [PubMed] [Google Scholar]
- 77.Kanfer J. N., McCartney D., and Hattori H.. 1988. Regulation of the choline, ethanolamine and serine base exchange enzyme activities of rat brain microsomes by phosphorylation and dephosphorylation. FEBS Lett. 240: 101–104. [DOI] [PubMed] [Google Scholar]
- 78.Carman G. M., and Henry S. A.. 1999. Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Prog. Lipid Res. 38: 361–399. [DOI] [PubMed] [Google Scholar]
- 79.Tasseva G., Cole L., and Vance J. E.. 2011. N-Myc and SP regulate phosphatidylserine synthase-1 expression in brain and glial cells. J. Biol. Chem. 286: 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nishijima M., Kuge O., and Akamatsu Y.. 1986. Phosphatidylserine biosynthesis in cultured Chinese hamster ovary cells. I. Inhibition of de novo phosphatidylserine biosynthesis by exogenous phosphatidylserine and its efficient incorporation. J. Biol. Chem. 261: 5784–5789. [PubMed] [Google Scholar]
- 81.Hasegawa K., Kuge O., Nishijima M., and Akamatsu Y.. 1989. Isolation and characterization of a Chinese hamster ovary cell mutant with altered regulation of phosphatidylserine biosynthesis. J. Biol. Chem. 264: 19887–19892. [PubMed] [Google Scholar]
- 82.Kuge O., Hasegawa K., Saito K., and Nishijima M.. 1998. Control of phosphatidylserine biosynthesis through phosphatidylserine-mediated inhibition of phosphatidylserine synthase I in Chinese hamster ovary cells. Proc. Natl. Acad. Sci. USA. 95: 4199–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kuge O., Saito K., and Nishijima M.. 1999. Control of phosphatidylserine synthase II activity in Chinese hamster ovary cells. J. Biol. Chem. 274: 23844–23849. [DOI] [PubMed] [Google Scholar]
- 84.Ohsawa T., Nishijima M., and Kuge O.. 2004. Functional analysis of Chinese hamster phosphatidylserine synthase 1 through systematic alanine mutagenesis. Biochem. J. 381: 853–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lenz W. D., and Majewski F.. 1974. A generalized disorder of the connective tissues with progeria, choanal atresia, symphalangism, hypoplasia of dentine and craniodiaphyseal hypostosis. Birth Defects Orig. Artic. Ser. 10: 133–136. [PubMed] [Google Scholar]
- 86.Sousa S. B., Jenkins D., Chanudet E., Tasseva G., Ishida M., Anderson G., Docker J., Ryten M., Sa J., Saraiva J. M., et al. 2014. Gain-of-function mutations in the phosphatidylserine synthase 1 (PTDSS1) gene cause Lenz-Majewski syndrome. Nat. Genet. 46: 70–76. [DOI] [PubMed] [Google Scholar]
- 87.Whyte M. P., Blythe A., McAlister W. H., Nenninger A. R., Bijanki V. N., and Mumm S.. 2015. Lenz-Majewski hyperostotic dwarfism with hyperphosphoserinuria from a novel mutation in PTDSS1 encoding phosphatidylserine synthase 1. J. Bone Miner. Res. 30: 606–614. [DOI] [PubMed] [Google Scholar]
- 88.Sohn M., Ivanova P., Brown H. A., Toth D. J., Varnai P., Kim Y. J., and Balla T.. 2016. Lenz-Majewski mutations in PTDSS1 affect phosphatidylinositol 4-phosphate metabolism at ER-PM and ER-Golgi junctions. Proc. Natl. Acad. Sci. USA. 113: 4314–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Piard J., Lespinasse J., Vlckova M., Mensah M. A., Iurian S., Simandlova M., Malikova M., Bartsch O., Rossi M., Lenoir M., et al. 2018. Cutis laxa and excessive bone growth due to de novo mutations in PTDSS1. Am. J. Med. Genet. 176: 668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Voelker D. R. 1989. Phosphatidylserine translocation to the mitochondrion is an ATP-dependent process in permeabilized animal cells. Proc. Natl. Acad. Sci. USA. 86: 9921–9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Voelker D. R. 1989. Reconstitution of phosphatidylserine import into rat liver mitochondria. J. Biol. Chem. 264: 8019–8025. [PubMed] [Google Scholar]
- 92.Shiao Y-J., Lupo G., and Vance J. E.. 1995. Evidence that phosphatidylserine is imported into mitochondria via a mitochondria-associated membrane and that the majority of mitochondrial phosphatidylethanolamine is derived from decarboxylation of phosphatidylserine. J. Biol. Chem. 270: 11190–11198. [DOI] [PubMed] [Google Scholar]
- 93.Bernhard W., Haguenau F., Gautier A., and Oberling C.. 1952. Submicroscopical structure of cytoplasmic basophils in the liver, pancreas and salivary gland; study of ultrafine slices by electron microscope. [Article in German] Z. Zellforsch. Mikrosk. Anat. 37: 281–300. [PubMed] [Google Scholar]
- 94.Morré D. J., Merritt W. D., and Lembi C. A.. 1971. Connections between mitochondria and endoplasmic reticulum in rat liver and onion stem. Protoplasma. 73: 43–49. [DOI] [PubMed] [Google Scholar]
- 95.Fawcett D. W. 1955. Observations on the cytology and electron microscopy of hepatic cells. J. Natl. Cancer Inst. 15: 1475–1503. [PubMed] [Google Scholar]
- 96.Franke W. W., and Kartenbeck J.. 1971. Outer mitochondrial membrane continuous with endoplasmic reticulum. Protoplasma. 73: 35–41. [DOI] [PubMed] [Google Scholar]
- 97.Farese R. V. Jr., and Walther T. C.. 2009. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell. 139: 855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shore G. C., and Tata J. R.. 1977. Two fractions of rough endoplasmic reticulum from rat liver. J. Cell Biol. 72: 714–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rizzuto R., Pinton P., Carrington W., Fay F. S., Fogarty K. E., Lifshitz L. M., Tuft R. A., and Pozzan T.. 1998. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 280: 1763–1766. [DOI] [PubMed] [Google Scholar]
- 100.van Golde L. M. G., Raben J., Batenburg J. J., Fleischer B., Zambrano F., and Fleischer S.. 1974. Biosynthesis of lipids in Golgi complex and other subcellular fractions from rat liver. Biochim. Biophys. Acta. 360: 179–192. [DOI] [PubMed] [Google Scholar]
- 101.Vance J. E. 1988. Compartmentalization of phospholipids for lipoprotein assembly on the basis of molecular species and biosynthetic origin. Biochim. Biophys. Acta. 963: 70–81. [DOI] [PubMed] [Google Scholar]
- 102.Rusiñol A. E., Cui Z., Chen M. H., and Vance J. E.. 1994. A unique mitochondria-associated membrane fraction from rat liver has a high capacity for lipid synthesis and contains pre-Golgi secretory proteins including nascent lipoproteins. J. Biol. Chem. 269: 27494–27502. [PubMed] [Google Scholar]
- 103.Stone S. J., and Vance J. E.. 2000. Phosphatidylserine synthase-1 and -2 are localized to mitochondria-associated membranes. J. Biol. Chem. 275: 34534–34540. [DOI] [PubMed] [Google Scholar]
- 104.Stone S. J., Levin M. C., Zhou P., Han J., Walther T. C., and Farese R. V. Jr. 2009. The endoplasmic reticulum enzyme DGAT2 is found in mitochondria-associated membranes and has a mitochondrial targeting signal that promotes its association with mitochondria. J. Biol. Chem. 284: 5352–5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vance J. E. 2014. MAM (mitochondria-associated membranes) in mammalian cells: lipids and beyond. Biochim. Biophys. Acta. 1841: 595–609. [DOI] [PubMed] [Google Scholar]
- 106.Poston C. N., Krishnan S. C., and Bazemore-Walker C. R.. 2013. In-depth proteomic analysis of mammalian mitochondria-associated membranes (MAM). J. Proteomics. 79: 219–230. [DOI] [PubMed] [Google Scholar]
- 107.Lin Q., and Liou Y. C.. 2017. Fishing for key players in ER-mitochondrial contacts. J. Biol. Chem. 292: 16393–16394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cui Z., Vance J. E., Chen M. H., Voelker D. R., and Vance D. E.. 1993. Cloning and expression of a novel phosphatidylethanolamine N-methyltransferase. J. Biol. Chem. 268: 16655–16663. [PubMed] [Google Scholar]
- 109.Meier P. J., Spycher M. A., and Meyer U. A.. 1981. Isolation and characterization of rough endoplasmic reticulum associated with mitochondria from normal rat liver. Biochim. Biophys. Acta. 646: 283–297. [DOI] [PubMed] [Google Scholar]
- 110.Voelker D. R. 1993. The ATP-dependent translocation of phosphatidylserine to the mitochondria is a process that is restricted to the autologous organelle. J. Biol. Chem. 268: 7069–7074. [PubMed] [Google Scholar]
- 111.Ardail D., Privat J-P., Egret-Charlier M., Levrat C., Lerme F., and Louisot P.. 1990. Mitochondrial contact sites. Lipid composition and dynamics. J. Biol. Chem. 265: 18797–18802. [PubMed] [Google Scholar]
- 112.Loewen C. J., and Levine T. P.. 2005. A highly conserved binding site in vesicle-associated membrane protein-associated protein (VAP) for the FFAT motif of lipid-binding proteins. J. Biol. Chem. 280: 14097–14104. [DOI] [PubMed] [Google Scholar]
- 113.Wang Z., and Benning C.. 2012. Chloroplast lipid synthesis and lipid trafficking through ER-plastid membrane contact sites. Biochem. Soc. Trans. 40: 457–463. [DOI] [PubMed] [Google Scholar]
- 114.Zinser E., Sperka-Gottlieb C. D., Fasch E. V., Kohlwein S. D., Paltauf F., and Daum G.. 1991. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote saccharomyces cerevisiae. J. Bacteriol. 173: 2026–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schumacher M. M., Choi J. Y., and Voelker D. R.. 2002. Phosphatidylserine transport to the mitochondria is regulated by ubiquitination. J. Biol. Chem. 277: 51033–51042. [DOI] [PubMed] [Google Scholar]
- 116.Herrera-Cruz M. S., and Simmen T.. 2017. Over six decades of discovery and characterization of the architecture at mitochondria-associated membranes (MAMs). Adv. Exp. Med. Biol. 997: 13–31. [DOI] [PubMed] [Google Scholar]
- 117.Area-Gomez E. 2014. Assessing the function of mitochondria-associated ER membranes. Methods Enzymol. 547: 181–197. [DOI] [PubMed] [Google Scholar]
- 118.Holthuis J. C., and Levine T. P.. 2005. Lipid traffic: floppy drives and a superhighway. Nat. Rev. Mol. Cell Biol. 6: 209–220. [DOI] [PubMed] [Google Scholar]
- 119.Shiao Y-J., Balcerzak B., and Vance J. E.. 1998. A mitochondrial membrane protein is required for translocation of phosphatidylserine from mitochondria-associated membranes to mitochondria. Biochem. J. 331: 217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Voelker D. R. 1985. Disruption of phosphatidylserine translocation to the mitochondria in baby hamster kidney cells. J. Biol. Chem. 260: 14671–14676. [PubMed] [Google Scholar]
- 121.Voelker D. R. 1990. Characterization of phosphatidylserine synthesis and translocation in permeabilized animal cells. J. Biol. Chem. 265: 14340–14346. [PubMed] [Google Scholar]
- 122.Vance J. E. 1991. Newly made phosphatidylserine and phosphatidylethanolamine are preferentially translocated between rat liver mitochondria and endoplasmic reticulum. J. Biol. Chem. 266: 89–97. [PubMed] [Google Scholar]
- 123.Simbeni R., Pon L., Zinser E., Paltauf F., and Daum G.. 1991. Mitochondrial membrane contact sites of yeast. Characterization of lipid components and possible involvement in intramitochondrial translocation of phospholipids. J. Biol. Chem. 266: 10047–10049. [PubMed] [Google Scholar]
- 124.Wu W. I., and Voelker D. R.. 2004. Reconstitution of phosphatidylserine transport from chemically defined donor membranes to phosphatidylserine decarboxylase 2 implicates specific lipid domains in the process. J. Biol. Chem. 279: 6635–6642. [DOI] [PubMed] [Google Scholar]
- 125.Kannan M., Lahiri S., Liu L. K., Choudhary V., and Prinz W. A.. 2017. Phosphatidylserine synthesis at membrane contact sites promotes its transport out of the ER. J. Lipid Res. 58: 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Voelker D. R. 1991. Adriamycin disrupts phosphatidylserine import into the mitochondria of permeabilized CHO-K1 cells. J. Biol. Chem. 266: 12185–12188. [PubMed] [Google Scholar]
- 127.Ardail D., Gasnier F., Lermé F., Simonot C., Louisot P., and Gateau-Roesch O.. 1993. Involvement of mitochondrial contact sites in the subcellular compartmentalization of phospholipid biosynthetic enzymes. J. Biol. Chem. 268: 25985–25992. [PubMed] [Google Scholar]
- 128.Ardail D., Lerme F., and Louisot P.. 1991. Involvement of contact sites in phosphatidylserine import into liver mitochondria. J. Biol. Chem. 266: 7978–7981. [PubMed] [Google Scholar]
- 129.Kornmann B., Currie E., Collins S. R., Schuldiner M., Nunnari J., Weissman J. S., and Walter P.. 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 325: 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rowland A. A., and Voeltz G. K.. 2012. Endoplasmic reticulum-mitochondria contacts: function of the junction. Nat. Rev. Mol. Cell Biol. 13: 607–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hayashi T., Rizzuto R., Hajnoczky G., and Su T. P.. 2009. MAM: more than just a housekeeper. Trends Cell Biol. 19: 81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Flis V. V., and Daum G.. 2013. Lipid transport between the endoplasmic reticulum and mitochondria. Cold Spring Harb. Perspect. Biol. 5: a013235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Csordás G., Renken C., Várnai P., Walter L., Weaver D., Buttle K. F., Balla T., Mannella C. A., and Hajnóczky G.. 2006. Structural and functional features and significance of the physical linkage between ER and mitochondria. J. Cell Biol. 174: 915–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.de Brito O. M., and Scorrano L.. 2008. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 456: 605–610. [DOI] [PubMed] [Google Scholar]
- 135.de Brito O. M., and Scorrano L.. 2010. An intimate liaison: spatial organization of the endoplasmic reticulum-mitochondria relationship. EMBO J. 29: 2715–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Levine T. P., and Patel S.. 2016. Signalling at membrane contact sites: two membranes come together to handle second messengers. Curr. Opin. Cell Biol. 39: 77–83. [DOI] [PubMed] [Google Scholar]
- 137.Nguyen T. T., Lewandowska A., Choi J. Y., Markgraf D. F., Junker M., Bilgin M., Ejsing C. S., Voelker D. R., Rapoport T. A., and Shaw J. M.. 2012. Gem1 and ERMES do not directly affect phosphatidylserine transport from ER to mitochondria or mitochondrial inheritance. Traffic. 13: 880–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Voelker D. R. 2009. Genetic and biochemical analysis of non-vesicular lipid traffic. Annu. Rev. Biochem. 78: 827–856. [DOI] [PubMed] [Google Scholar]
- 139.Area-Gomez E., Del Carmen Lara Castillo M., Tambini M. D., Guardia-Laguarta C., de Groof A. J., Madra M., Ikenouchi J., Umeda M., Bird T. D., Sturley S. L., et al. 2012. Upregulated function of mitochondria-associated ER membranes in Alzheimer disease. EMBO J. 31: 4106–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sugiura A., Nagashima S., Tokuyama T., Amo T., Matsuki Y., Ishido S., Kudo Y., McBride H. M., Fukuda T., Matsushita N., et al. 2013. MITOL regulates endoplasmic reticulum-mitochondria contacts via Mitofusin2. Mol. Cell. 51: 20–34. [DOI] [PubMed] [Google Scholar]
- 141.Hirabayashi Y., Kwon S. K., Paek H., Pernice W. M., Paul M. A., Lee J., Erfani P., Raczkowski A., Petrey D. S., Pon L. A., et al. 2017. ER-mitochondria tethering by PDZD8 regulates Ca(2+) dynamics in mammalian neurons. Science. 358: 623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Simmen T., Aslan J. E., Blagoveshchenskaya A. D., Thomas L., Wan L., Xiang Y., Feliciangeli S. F., Hung C. H., Crump C. M., and Thomas G.. 2005. PACS-2 controls endoplasmic reticulum-mitochondria communication and Bid-mediated apoptosis. EMBO J. 24: 717–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Steenbergen R., Nanowski T. S., Beigneux A., Kulinski A., Young S. G., and Vance J. E.. 2005. Disruption of the phosphatidylserine decarboxylase gene in mice causes embryonic lethality and mitochondrial defects. J. Biol. Chem. 280: 40032–40040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tasseva G., Bai H. D., Davidescu M., Haromy A., Michelakis E., and Vance J. E.. 2013. Phosphatidylethanolamine deficiency in mammalian mitochondria impairs oxidative phosphorylation and alters mitochondrial morphology. J. Biol. Chem. 288: 4158–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tatsuta T., Scharwey M., and Langer T.. 2014. Mitochondrial lipid trafficking. Trends Cell Biol. 24: 44–52. [DOI] [PubMed] [Google Scholar]
- 146.Rizzuto R., Brini M., Murgia M., and Pozzan T.. 1993. Microdomains with high Ca2+ close to IP3-sensitive channels that are sensed by neighboring mitochondria. Science. 262: 744–747. [DOI] [PubMed] [Google Scholar]
- 147.Csordás G., Várnai P., Golenár T., Roy S., Purkins G., Schneider T. G., Balla T., and Hajnóczky G.. 2010. Imaging interorganelle contacts and local calcium dynamics at the ER-mitochondrial interface. Mol. Cell. 39: 121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., Oomori H., Noda T., Haraguchi T., Hiraoka Y., et al. 2013. Autophagosomes form at ER-mitochondria contact sites. Nature. 495: 389–393. [DOI] [PubMed] [Google Scholar]
- 149.Xue Y., Schmollinger S., Attar N., Campos O. A., Vogelauer M., Carey M. F., Merchant S. S., and Kurdistani S. K.. 2017. Endoplasmic reticulum-mitochondria junction is required for iron homeostasis. J. Biol. Chem. 292: 13197–13204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lewis S. C., Uchiyama L. F., and Nunnari J.. 2016. ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science. 353: aaf5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Area-Gomez E., de Groof A. J., Boldogh I., Bird T. D., Gibson G. E., Koehler C. M., Yu W. H., Duff K. E., Yaffe M. P., Pon L. A., et al. 2009. Presenilins are enriched in endoplasmic reticulum membranes associated with mitochondria. Am. J. Pathol. 175: 1810–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Schon E. A., and Przedborski S.. 2011. Mitochondria: the next (neurode)generation. Neuron. 70: 1033–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zampese E., Fasolato C., Kipanyula M. J., Bortolozzi M., Pozzan T., and Pizzo P.. 2011. Presenilin 2 modulates endoplasmic reticulum (ER)-mitochondria interactions and Ca2+ cross-talk. Proc. Natl. Acad. Sci. USA. 108: 2777–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pera M., Larrea D., Guardia-Laguarta C., Montesinos J., Velasco K. R., Agrawal R. R., Xu Y., Chan R. B., Di Paolo G., Mehler M. F., et al. 2017. Increased localization of APP-C99 in mitochondria-associated ER membranes causes mitochondrial dysfunction in Alzheimer disease. EMBO J. 36: 3356–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Guardia-Laguarta C., Area-Gomez E., Rub C., Liu Y., Magrane J., Becker D., Voos W., Schon E. A., and Przedborski S.. 2014. α-Synuclein is localized to mitochondria-associated ER membranes. J. Neurosci. 34: 249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tubbs E., Theurey P., Vial G., Bendridi N., Bravard A., Chauvin M. A., Ji-Cao J., Zoulim F., Bartosch B., Ovize M., et al. 2014. Mitochondria-associated endoplasmic reticulum membrane (MAM) integrity is required for insulin signaling and is implicated in hepatic insulin resistance. Diabetes. 63: 3279–3294. [DOI] [PubMed] [Google Scholar]
- 157.Vance J. E. 2015. Phospholipid synthesis and transport in mammalian cells. Traffic. 16: 1–18. [DOI] [PubMed] [Google Scholar]
- 158.Prinz W. A. 2014. Bridging the gap: membrane contact sites in signaling, metabolism, and organelle dynamics. J. Cell Biol. 205: 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Phillips M. J., and Voeltz G. K.. 2016. Structure and function of ER membrane contact sites with other organelles. Nat. Rev. Mol. Cell Biol. 17: 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Levine T., and Loewen C.. 2006. Inter-organelle membrane contact sites: through a glass, darkly. Curr. Opin. Cell Biol. 18: 371–378. [DOI] [PubMed] [Google Scholar]
- 161.Nascimbeni A. C., Giordano F., Dupont N., Grasso D., Vaccaro M. I., Codogno P., and Morel E.. 2017. ER-plasma membrane contact sites contribute to autophagosome biogenesis by regulating local PI3P synthesis. EMBO J. 36: 2018–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chung J., Torta F., Masai K., Lucast L., Czapla H., Tanner L. B., Narayanaswamy P., Wenk M. R., Nakatsu F., and De Camilli P.. 2015. Intracellular transport. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER-plasma membrane contacts. Science. 349: 428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Goto A., Charman M., and Ridgway N. D.. 2016. Oxysterol-binding protein activation at endoplasmic reticulum-golgi contact sites reorganizes phosphatidylinositol 4-phosphate pools. J. Biol. Chem. 291: 1336–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Liu L. K., Choudhary V., Toulmay A., and Prinz W. A.. 2017. An inducible ER-Golgi tether facilitates ceramide transport to alleviate lipotoxicity. J. Cell Biol. 216: 131–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hua R., Cheng D., Coyaud E., Freeman S., Di Pietro E., Wang Y., Vissa A., Yip C. M., Fairn G. D., Braverman N., et al. 2017. VAPs and ACBD5 tether peroxisomes to the ER for peroxisome maintenance and lipid homeostasis. J. Cell Biol. 216: 367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wong Y. C., Ysselstein D., and Krainc D.. 2018. Mitochondria-lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature. 554: 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Vance J. E. 2008. Phosphatidylserine and phosphatidylethanolamine in mammalian cells: two metabolically-related aminophospholipids. J. Lipid Res. 49: 1377–1387. [DOI] [PubMed] [Google Scholar]
- 168.Vance J. E., and Tasseva G.. 2013. Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochim. Biophys. Acta. 1831: 543–554. [DOI] [PubMed] [Google Scholar]
- 169.Sundler R., Akesson B., and Nilsson A.. 1974. Quantitative role of base exchange in phosphatidylethanolamine synthesis in isolated rat hepatocytes. FEBS Lett. 43: 303–307. [DOI] [PubMed] [Google Scholar]
- 170.Riekhof W. R., Wu J., Jones J. L., and Voelker D. R.. 2007. Identification and characterization of the major lysophosphatidylethanolamine acyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 282: 28344–28352. [DOI] [PubMed] [Google Scholar]
- 171.Voelker D. R. 1984. Phosphatidylserine functions as the major precursor of phosphatidylethanolamine in cultured BHK-21 cells. Proc. Natl. Acad. Sci. USA. 81: 2669–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Miller M. A., and Kent C.. 1986. Characterization of the pathways for phosphatidylethanolamine biosynthesis in Chinese hamster ovary mutant and parental cell lines. J. Biol. Chem. 261: 9753–9761. [PubMed] [Google Scholar]
- 173.Zelinski T. A., and Choy P. C.. 1982. Phosphatidylethanolamine biosynthesis in isolated hamster heart. Can. J. Biochem. 60: 817–823. [DOI] [PubMed] [Google Scholar]
- 174.Tijburg L. B., Geelen M. J., and Van Golde L. M.. 1989. Biosynthesis of phosphatidylethanolamine via the CDP-ethanolamine route is an important pathway in isolated rat hepatocytes. Biochem. Biophys. Res. Commun. 160: 1275–1280. [DOI] [PubMed] [Google Scholar]
- 175.Bleijerveld O. B., Brouwers J. F., Vaandrager A. B., Helms J. B., and Houweling M.. 2007. The CDP-ethanolamine pathway and phosphatidylserine decarboxylation generate different phosphatidylethanolamine molecular species. J. Biol. Chem. 282: 28362–28372. [DOI] [PubMed] [Google Scholar]
- 176.Riekhof W. R., and Voelker D. R.. 2006. Uptake and utilization of lyso-phosphatidylethanolamine by Saccharomyces cerevisiae. J. Biol. Chem. 281: 36588–36596. [DOI] [PubMed] [Google Scholar]
- 177.Bjerve K. S. 1985. The biosynthesis of phosphatidylserine and phosphatidylethanolamine from L-[3–14C]serine in isolated rat hepatocytes. Biochim. Biophys. Acta. 833: 396–405. [DOI] [PubMed] [Google Scholar]
- 178.Fullerton M. D., Hakimuddin F., and Bakovic M.. 2007. Developmental and metabolic effects of disruption of the mouse CTP:phosphoethanolamine cytidylyltransferase gene (Pcyt2). Mol. Cell. Biol. 27: 3327–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Fullerton M. D., Hakimuddin F., Bonen A., and Bakovic M.. 2009. The development of a metabolic disease phenotype in CTP:phosphoethanolamine cytidylyltransferase-deficient mice. J. Biol. Chem. 284: 25704–25713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Percy A. K., Moore J. F., Carson M. A., and Waechter C. J.. 1983. Characterization of brain phosphatidylserine decarboxylase: localization in the mitochondrial inner membrane. Arch. Biochem. Biophys. 223: 484–494. [DOI] [PubMed] [Google Scholar]
- 181.Zborowski J., Dygas A., and Wojtczak L.. 1983. Phosphatidylserine decarboxylase is located on the external side of the inner mitochondrial membrane. FEBS Lett. 157: 179–182. [DOI] [PubMed] [Google Scholar]
- 182.Snell E. E. 1977. Pyruvate-containing enzymes. Trends Biochem. Sci. 2: 131–135. [Google Scholar]
- 183.Onguka O., Calzada E., Ogunbona O. B., and Claypool S. M.. 2015. Phosphatidylserine decarboxylase 1 autocatalysis and function does not require a mitochondrial-specific factor. J. Biol. Chem. 290: 12744–12752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Li Q-X., and Dowhan W.. 1988. Structural characterization of Escherichia coli phosphatidylserine decarboxylase. J. Biol. Chem. 263: 11516–11522. [PubMed] [Google Scholar]
- 185.Trotter P. J., Pedretti J., Yates R., and Voelker D. R.. 1995. Phosphatidylserine decarboxylase 2 of Saccharomyces cerevisiae. Cloning and mapping of the gene, heterologous expression and creation of the null allele. J. Biol. Chem. 270: 6071–6080. [DOI] [PubMed] [Google Scholar]
- 186.Trotter P. J., and Voelker D. R.. 1995. Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 270: 6062–6070. [DOI] [PubMed] [Google Scholar]
- 187.Friedman J. R., Kannan M., Toulmay A., Jan C. H., Weissman J. S., Prinz W. A., and Nunnari J.. 2018. Lipid homeostasis is maintained by dual targeting of the mitochondrial PE biosynthesis enzyme to the ER. Dev. Cell. 44: 261–270.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Shiao Y-J., and Vance J. E.. 1995. Evidence for an ethanolamine cycle: differential recycling of the ethanolamine moiety of phosphatidylethanolamine derived from phosphatidylserine and ethanolamine. Biochem. J. 310: 673–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Achleitner G., Zweytick D., Trotter P. J., Voelker D. R., and Daum G.. 1995. Synthesis and intracellular transport of aminoglycerophospholipids in permeabilized cells of the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 270: 29836–29842. [DOI] [PubMed] [Google Scholar]
- 190.Osman C., Voelker D. R., and Langer T.. 2011. Making heads or tails of phospholipids in mitochondria. J. Cell Biol. 192: 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sundler R. 1975. Ethanolaminephosphate cytidylyltransferase. Purification and characterization of the enzyme from rat liver. J. Biol. Chem. 250: 8585–8590. [PubMed] [Google Scholar]
- 192.Rontein D., Wu W. I., Voelker D. R., and Hanson A. D.. 2003. Mitochondrial phosphatidylserine decarboxylase from higher plants. Functional complementation in yeast, localization in plants, and overexpression in Arabidopsis. Plant Physiol. 132: 1678–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zhou J., and Saba J. D.. 1998. Identification of the first mammalian sphingosine phosphate lyase gene and its functional expression in yeast. Biochem. Biophys. Res. Commun. 242: 502–507. [DOI] [PubMed] [Google Scholar]
- 194.Van Veldhoven P. P., Gijsbers S., Mannaerts G. P., Vermeesch J. R., and Brys V.. 2000. Human sphingosine-1-phosphate lyase: cDNA cloning, functional expression studies and mapping to chromosome 10q22. Biochim. Biophys. Acta. 1487: 128–134. [DOI] [PubMed] [Google Scholar]
- 195.Saba J. D., Nara F., Bielawska A., Garrett S., and Hannun Y. A.. 1997. The BST1 gene of Saccharomyces cerevisiae is the sphingosine-1-phosphate lyase. J. Biol. Chem. 272: 26087–26090. [DOI] [PubMed] [Google Scholar]
- 196.Hannun Y. A., Luberto C., and Argraves K. M.. 2001. Enzymes of sphingolipid metabolism: from modular to integrative signaling. Biochemistry. 40: 4893–4903. [DOI] [PubMed] [Google Scholar]
- 197.Lykidis A., Wang J., Karim M. A., and Jackowski S.. 2001. Overexpression of a mammalian ethanolamine-specific kinase accelerates the CDP-ethanolamine pathway. J. Biol. Chem. 276: 2174–2179. [DOI] [PubMed] [Google Scholar]
- 198.Tian Y., Jackson P., Gunter C., Wang J., Rock C. O., and Jackowski S.. 2006. Placental thrombosis and spontaneous fetal death in mice deficient in ethanolamine kinase 2. J. Biol. Chem. 281: 28438–28449. [DOI] [PubMed] [Google Scholar]
- 199.Gustin S. E., Western P. S., McClive P. J., Harley V. R., Koopman P. A., and Sinclair A. H.. 2008. Testis development, fertility, and survival in ethanolamine kinase 2-deficient mice. Endocrinology. 149: 6176–6186. [DOI] [PubMed] [Google Scholar]
- 200.Sundler R., and Akesson B.. 1975. Biosynthesis of phosphatidylethanolamines and phosphatidylcholines from ethanolamine and choline in rat liver. Biochem. J. 146: 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Tijburg L. B. M., Vermeulen P. S., and van Golde L. M. B.. 1992. Ethanolamine phosphate cytidylyltransferase. Methods Enzymol. 209: 258–263. [DOI] [PubMed] [Google Scholar]
- 202.Cornell R. B., and Northwood I. C.. 2000. Regulation of CTP:phosphocholine cytidylyltransferase by amphitropism and relocalization. Trends Biochem. Sci. 25: 441–447. [DOI] [PubMed] [Google Scholar]
- 203.Lykidis A., Jackson P., and Jackowski S.. 2001. Lipid activation of CTP: phosphocholine cytidylyltransferase alpha: characterization and identification of a second activation domain. Biochemistry. 40: 494–503. [DOI] [PubMed] [Google Scholar]
- 204.Cornell R. B., and Ridgway N. D.. 2015. CTP:phosphocholine cytidylyltransferase: Function, regulation, and structure of an amphitropic enzyme required for membrane biogenesis. Prog. Lipid Res. 59: 147–171. [DOI] [PubMed] [Google Scholar]
- 205.Poloumienko A., Cote A., Quee A-T., Zhu L., and Bakovic M.. 2004. Genomic organization and differential splicing of the mouse and human Pcyt2 genes. Gene. 325: 145–155. [DOI] [PubMed] [Google Scholar]
- 206.Tie A., and Bakovic M.. 2007. Alternative splicing of CTP:phosphoethanolamine cytidylyltransferase produces two isoforms that differ in catalytic properties. J. Lipid Res. 48: 2172–2181. [DOI] [PubMed] [Google Scholar]
- 207.Fullerton M. D., and Bakovic M.. 2010. Complementation of the metabolic defect in CTP:phosphoethanolamine cytidylyltransferase (Pcyt2)-deficient primary hepatocytes. Metabolism. 59: 1691–1700. [DOI] [PubMed] [Google Scholar]
- 208.Leonardi R., Frank M. W., Jackson P. D., Rock C. O., and Jackowski S.. 2009. Elimination of the CDP-ethanolamine pathway disrupts hepatic lipid homeostasis. J. Biol. Chem. 284: 27077–27089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Selathurai A., Kowalski G. M., Burch M. L., Sepulveda P., Risis S., Lee-Young R. S., Lamon S., Meikle P. J., Genders A. J., McGee S. L., et al. 2015. The CDP-ethanolamine pathway regulates skeletal muscle diacylglycerol content and mitochondrial biogenesis without altering insulin sensitivity. Cell Metab. 21: 718–730. [DOI] [PubMed] [Google Scholar]
- 210.Henneberry A. L., Wright M. M., and McMaster C. R.. 2002. The major sites of cellular phospholipid synthesis and molecular determinants of fatty acid and lipid head group specificity. Mol. Biol. Cell. 13: 3148–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Henneberry A. L., and McMaster C. R.. 1999. Cloning and expression of a human choline/ethanolaminephosphotransferase: synthesis of phosphatidylcholine and phosphatidyethanolamine. Biochem. J. 339: 291–298. [PMC free article] [PubMed] [Google Scholar]
- 212.Funai K., Lodhi I. J., Spears L. D., Yin L., Song H., Klein S., and Semenkovich C. F.. 2016. Skeletal muscle phospholipid metabolism regulates insulin sensitivity and contractile function. Diabetes. 65: 358–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Horibata Y., and Hirabayashi Y.. 2007. Identification and characterization of human ethanolaminephosphotransferase1. J. Lipid Res. 48: 503–508. [DOI] [PubMed] [Google Scholar]
- 214.Ahmed M. Y., Al-Khayat A., Al-Murshedi F., Al-Futaisi A., Chioza B. A., Pedro Fernandez-Murray J., Self J. E., Salter C. G., Harlalka G. V., Rawlins L. E., et al. 2017. A mutation of EPT1 (SELENOI) underlies a new disorder of Kennedy pathway phospholipid biosynthesis. Brain. 140: 547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Horibata Y., Elpeleg O., Eran A., Hirabayashi Y., Savitzki D., Tal G., Mandel H., and Sugimoto H.. Ethanolamine phosphotransferase 1 (selenoprotein I) is critical for the neural development and maintenance of plasmalogen in human. J. Lipid Res. Epub ahead of print. March 2, 2018; doi:10.1194/jlr.P081620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Lagace T. A., and Ridgway N. D.. 2013. The role of phospholipids in the biological activity and structure of the endoplasmic reticulum. Biochim. Biophys. Acta. 1833: 2499–2510. [DOI] [PubMed] [Google Scholar]
- 217.Tijburg L. B., Houweling M., Geelen M. J., and Van Golde L. L. M.. 1989. Inhibition of phosphatidylethanolamine synthesis by glucagon in isolated rat hepatocytes. Biochem. J. 257: 645–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ando H., Aoyama C., Horibata Y., Satou M., Mitsuhashi S., Itoh M., Hosaka K., and Sugimoto H.. 2015. Transcriptional suppression of CTP:phosphoethanolamine cytidylyltransferase by 25-hydroxycholesterol is mediated by nuclear factor-Y and Yin Yang 1. Biochem. J. 471: 369–379. [DOI] [PubMed] [Google Scholar]
- 219.Pavlovic Z., Zhu L., Pereira L., Singh R. K., Cornell R. B., and Bakovic M.. 2014. Isoform-specific and protein kinase C-mediated regulation of CTP:phosphoethanolamine cytidylyltransferase phosphorylation. J. Biol. Chem. 289: 9053–9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Zhu L., Michel V., and Bakovic M.. 2009. Regulation of the mouse CTP: phosphoethanolamine cytidylyltransferase gene Pcyt2 during myogenesis. Gene. 447: 51–59. [DOI] [PubMed] [Google Scholar]
- 221.Dorée C. 1909. The occurrence and distribution of cholesterol and allied bodies in the animal kingdom. Biochem. J. 4: 72–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Feulgen R., Imhauser K., and Behrens M.. 1929. Zur Kenntnis des Plasmalogens. I. Mitteilung. Eigenschaften des Plasmalogens, Darstellung und Natur des Plasmals. Z. Physiol. Chem. 180: 161–179. [Google Scholar]
- 223.Snyder F., Malone B., and Wykle R. L.. 1969. The biosynthesis of alkyl ether bonds in lipids by a cell-free system. Biochem. Biophys. Res. Commun. 34: 40–47. [DOI] [PubMed] [Google Scholar]
- 224.Snyder F., Wykle R. L., and Malone B.. 1969. A new metabolic pathway: biosynthesis of alkyl ether bonds from glyceraldehyde-3-phosphate and fatty alcohols by microsomal enzymes. Biochem. Biophys. Res. Commun. 34: 315–321. [DOI] [PubMed] [Google Scholar]
- 225.Snyder F. 1999. The ether lipid trail: a historical perspective. Biochim. Biophys. Acta. 1436: 265–278. [DOI] [PubMed] [Google Scholar]
- 226.Brites P., Waterham H. R., and Wanders R. J.. 2004. Functions and biosynthesis of plasmalogens in health and disease. Biochim. Biophys. Acta. 1636: 219–231. [DOI] [PubMed] [Google Scholar]
- 227.Braverman N. E., and Moser A. B.. 2012. Functions of plasmalogen lipids in health and disease. Biochim. Biophys. Acta. 1822: 1442–1452. [DOI] [PubMed] [Google Scholar]
- 228.Paltauf F. 1994. Ether lipids in biomembranes. Chem. Phys. Lipids. 74: 101–139. [DOI] [PubMed] [Google Scholar]
- 229.Xu Y. F., O K., and Choy P. C.. 1994. Plasmenylcholine (1-O-alk-1′-enyl-2-acyl-sn-glycero-3-phosphocholine) biosynthesis in guinea-pig heart and liver: cholinephosphotransferase is a bifunctional enzyme for the synthesis of phosphatidylcholine and plasmenylcholine. Biochem. J. 301: 131–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Stoffel W., LeKim D., and Heyn G.. 1970. Metabolism of sphingosine bases. XIV. Sphinganine (dihydrosphingosine), an effective donor of the alk-1-enyl chain of plasmalogens. Hoppe Seylers Z. Physiol. Chem. 351: 875–883. [DOI] [PubMed] [Google Scholar]
- 231.Zoeller R. A., Morand O. H., and Raetz C. R. H.. 1988. A possible role for plasmalogens in protecting animal cells against photosensitized killing. J. Biol. Chem. 263: 11590–11596. [PubMed] [Google Scholar]
- 232.Zoeller R. A., Lake A. C., Nagan N., Gaposchkin D. P., Legner M. A., and Lieberthanl W.. 1999. Plasmalogens as endogenous antioxidants: somatic cell mutants reveal the importance of the vinyl ether. Biochem. J. 338: 769–776. [PMC free article] [PubMed] [Google Scholar]
- 233.Munn N. J., Arnio E., Liu D., Zoeller R. A., and Liscum L.. 2003. Deficiency in ethanolamine plasmalogen leads to altered cholesterol transport. J. Lipid Res. 44: 182–192. [DOI] [PubMed] [Google Scholar]
- 234.de Vet E. C. J. M., Ijist L., Oostheim W., Wanders R. J. A., and van den Bosch H.. 1998. Alkykldihydroxyacetonephosphate synthase: fate in peroxisome biogenesis disorders and identification of the point mutation underlying a single enzyme deficiency. J. Biol. Chem. 273: 10296–10301. [DOI] [PubMed] [Google Scholar]
- 235.Schrakamp G., Roosenboom C. F. P., Schutgens R. B. H., Wanders R. J. A., Heymans H. S. A., Tager J. M., and van den Bosch H.. 1985. Alkyl dihydroxyacetone phosphate synthase in human fibroblasts and its deficiency in Zellweger syndrome. J. Lipid Res. 26: 867–873. [PubMed] [Google Scholar]
- 236.Schutgens R. B. H., Romeyn G. J., Wanders R. J. A., van den Bosch H., Schrakamp G., and Heymans H. S. A.. 1984. Deficiency of acyl-CoA dihydroxyacetone phosphate acyltransferase in patients with Zellweger (cerebro-hepato-renal) syndrome. Biochem. Biophys. Res. Commun. 120: 179–184. [DOI] [PubMed] [Google Scholar]
- 237.Fairn G. D., Schieber N. L., Ariotti N., Murphy S., Kuerschner L., Webb R. I., Grinstein S., and Parton R. G.. 2011. High-resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. J. Cell Biol. 194: 257–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Graham T. R. 2004. Flippases and vesicle-mediated protein transport. Trends Cell Biol. 14: 670–677. [DOI] [PubMed] [Google Scholar]
- 239.Frasch S. C., Henson P. M., Kailey J. M., Richter D. A., Janes M. S., Fadok V. A., and Bratton D. L.. 2000. Regulation of phospholipid scramblase activity during apoptosis and cell activation by protein kinase Cd. J. Biol. Chem. 275: 23065–23073. [DOI] [PubMed] [Google Scholar]
- 240.Hankins H. M., Baldridge R. D., Xu P., and Graham T. R.. 2015. Role of flippases, scramblases and transfer proteins in phosphatidylserine subcellular distribution. Traffic. 16: 35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Zhou Q., Zhao J., Wiedmer T., and Sims P. J.. 2002. Normal hemostasis but defective hematopoietic response to growth factors in mice deficient in phospholipid scramblase 1. Blood. 99: 4030–4038. [DOI] [PubMed] [Google Scholar]
- 242.Fadok V. A., Voelker D. R., Campbell P. A., Cohen J. J., Bratton D. L., and Henson P. M.. 1992. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J. Immunol. 148: 2207–2216. [PubMed] [Google Scholar]
- 243.Fadok V. A., de Cathelineau A., Daleke D. L., Henson P. M., and Bratton D. L.. 2001. Loss of phospholipid asymmetry and surface exposure of phosphatidylserine is required for phagocytosis of apoptotic cells by macrophages and fibroblasts. J. Biol. Chem. 276: 1071–1077. [DOI] [PubMed] [Google Scholar]
- 244.Balasubramanian K., Mirnikjoo B., and Schroit A. J.. 2007. Regulated externalization of phosphatidylserine at the cell surface: implications for apoptosis. J. Biol. Chem. 282: 18357–18364. [DOI] [PubMed] [Google Scholar]
- 245.Miyanishi M., Tada K., Koike M., Uchiyama Y., Kitamura T., and Nagata S.. 2007. Identification of Tim4 as a phosphatidylserine receptor. Nature. 450: 435–439. [DOI] [PubMed] [Google Scholar]
- 246.Park D., Hochreiter-Hufford A., and Ravichandran K. S.. 2009. The phosphatidylserine receptor TIM-4 does not mediate direct signaling. Curr. Biol. 19: 346–351. [DOI] [PubMed] [Google Scholar]
- 247.Yang H., Chen Y. Z., Zhang Y., Wang X., Zhao X., Godfroy J. I. 3rd, Liang Q., Zhang M., Zhang T., Yuan Q., et al. 2015. A lysine-rich motif in the phosphatidylserine receptor PSR-1 mediates recognition and removal of apoptotic cells. Nat. Commun. 6: 5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Henson P. M., and Bratton D. L.. 2013. Antiinflammatory effects of apoptotic cells. J. Clin. Invest. 123: 2773–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Bratton D. L., and Henson P. M.. 2008. Apoptotic cell recognition: will the real phosphatidylserine receptor(s) please stand up? Curr. Biol. 18: R76–R79. [DOI] [PubMed] [Google Scholar]
- 250.Zwaal R. F., Comfurius P., and Bevers E. M.. 1998. Lipid-protein interactions in blood coagulation. Biochim. Biophys. Acta. 1376: 433–453. [DOI] [PubMed] [Google Scholar]
- 251.Bevers E. M., Comfurius P., van Rijn J. L., Hemker H. C., and Zwaal R. F.. 1982. Generation of prothrombin-converting activity and the exposure of phosphatidylserine at the outer surface of platelets. Eur. J. Biochem. 122: 429–436. [DOI] [PubMed] [Google Scholar]
- 252.Elliott J. I., Surprenant A., Marelli-Berg F. M., Cooper J. C., Cassady-Cain R. L., Wooding C., Linton K., Alexander D. R., and Higgins C. F.. 2005. Membrane phosphatidylserine distribution as a non-apoptotic signalling mechanism in lymphocytes. Nat. Cell Biol. 7: 808–816. [DOI] [PubMed] [Google Scholar]
- 253.Shaw A. W., Pureza V. S., Sligar S. G., and Morrissey J. H.. 2007. The local phospholipid environment modulates the activation of blood clotting. J. Biol. Chem. 282: 6556–6563. [DOI] [PubMed] [Google Scholar]
- 254.Majumder R., Quinn-Allen M. A., Kane W. H., and Lentz B. R.. 2008. A phosphatidylserine binding site in factor Va C1 domain regulates both assembly and activity of the prothrombinase complex. Blood. 112: 2795–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Williamson P., Bevers E. M., Smeets E. F., Comfurius P., Schlegel R. A., and Zwaal R. F. A.. 1995. Continuous analysis of the mechanism of activated transbilayer lipid movement in platelets. Biochemistry. 34: 10448–10455. [DOI] [PubMed] [Google Scholar]
- 256.Gadella B. M., Rathi R., Brouwers J. F., Stout T. A., and Colenbrander B.. 2001. Capacitation and the acrosome reaction in equine sperm. Anim. Reprod. Sci. 68: 249–265. [DOI] [PubMed] [Google Scholar]
- 257.Flesch F. M., Brouwers J. F., Nievelstein P. F., Verkleij A. J., van Golde L. M., Colenbrander B., and Gadella B. M.. 2001. Bicarbonate stimulated phospholipid scrambling induces cholesterol redistribution and enables cholesterol depletion in the sperm plasma membrane. J. Cell Sci. 114: 3543–3555. [DOI] [PubMed] [Google Scholar]
- 258.van der Woerd W. L., Wichers C. G., Vestergaard A. L., Andersen J. P., Paulusma C. C., Houwen R. H., and van de Graaf S. F.. 2016. Rescue of defective ATP8B1 trafficking by CFTR correctors as a therapeutic strategy for familial intrahepatic cholestasis. J. Hepatol. 64: 1339–1347. [DOI] [PubMed] [Google Scholar]
- 259.Yeung T., Gilbert G. E., Shi J., Silvius J., Kapus A., and Grinstein S.. 2008. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 319: 210–213. [DOI] [PubMed] [Google Scholar]
- 260.Yeung T., Heit B., Dubuisson J. F., Fairn G. D., Chiu B., Inman R., Kapus A., Swanson M., and Grinstein S.. 2009. Contribution of phosphatidylserine to membrane surface charge and protein targeting during phagosome maturation. J. Cell Biol. 185: 917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Leventis P. A., and Grinstein S.. 2010. The distribution and function of phosphatidylserine in cellular membranes. Annu. Rev. Biophys. 39: 407–427. [DOI] [PubMed] [Google Scholar]
- 262.Sigal C. T., Zhou W., Buser C. A., McLaughlin S., and Resh M. D.. 1994. Amino-terminal basic residues of Src mediate membrane binding through electrostatic interaction with acidic phospholipids. Proc. Natl. Acad. Sci. USA. 91: 12253–12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Finkielstein C. V., Overduin M., and Capelluto D. G.. 2006. Cell migration and signaling specificity is determined by the phosphatidylserine recognition motif of Rac1. J. Biol. Chem. 281: 27317–27326. [DOI] [PubMed] [Google Scholar]
- 264.Powell K. A., Valova V. A., Malladi C. S., Jensen O. N., Larsen M. R., and Robinson P. J.. 2000. Phosphorylation of dynamin I on Ser-795 by protein kinase C blocks its association with phospholipids. J. Biol. Chem. 275: 11610–11617. [DOI] [PubMed] [Google Scholar]
- 265.Verdaguer N., Corbalan-Garcia S., Ochoa W. F., Fita I., and Gomez-Fernandez J. C.. 1999. Ca(2+) bridges the C2 membrane-binding domain of protein kinase Calpha directly to phosphatidylserine. EMBO J. 18: 6329–6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Huang B. X., Akbar M., Kevala K., and Kim H. Y.. 2011. Phosphatidylserine is a critical modulator for Akt activation. J. Cell Biol. 192: 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Arispe N., Doh M., Simakova O., Kurganov B., and De Maio A.. 2004. Hsc70 and Hsp70 interact with phosphatidylserine on the surface of PC12 cells resulting in a decrease of viability. FASEB J. 18: 1636–1645. [DOI] [PubMed] [Google Scholar]
- 268.Hirama T., Das R., Yang Y., Ferguson C., Won A., Yip C. M., Kay J. G., Grinstein S., Parton R. G., and Fairn G. D.. 2017. Phosphatidylserine dictates the assembly and dynamics of caveolae in the plasma membrane. J. Biol. Chem. 292: 14292–14307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Joshi A. S., Thompson M. N., Fei N., Huttemann M., and Greenberg M. L.. 2012. Cardiolipin and mitochondrial phosphatidylethanolamine have overlapping functions in mitochondrial fusion in Saccharomyces cerevisiae. J. Biol. Chem. 287: 17589–17597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Signorell A., Gluenz E., Rettig J., Schneider A., Shaw M. K., Gull K., and Butikofer P.. 2009. Perturbation of phosphatidylethanolamine synthesis affects mitochondrial morphology and cell-cycle progression in procyclic-form Trypanosoma brucei. Mol. Microbiol. 72: 1068–1079. [DOI] [PubMed] [Google Scholar]
- 271.Ren J., Pulakat L., Whaley-Connell A., and Sowers J. R.. 2010. Mitochondrial biogenesis in the metabolic syndrome and cardiovascular disease. J. Mol. Med. (Berl.). 88: 993–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Supale S., Li N., Brun T., and Maechler P.. 2012. Mitochondrial dysfunction in pancreatic beta cells. Trends Endocrinol. Metab. 23: 477–487. [DOI] [PubMed] [Google Scholar]
- 273.Johri A., and Beal M. F.. 2012. Mitochondrial dysfunction in neurodegenerative diseases. J. Pharmacol. Exp. Ther. 342: 619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Baysal B. E., Ferrell R. E., Willett-Brozick J. E., Lawrence E. C., Myssiorek D., Bosch A., van der Mey A., Taschner P. E., Rubinstein W. S., Myers E. N., et al. 2000. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 287: 848–851. [DOI] [PubMed] [Google Scholar]
- 275.Keckesova Z., Donaher J. L., De Cock J., Freinkman E., Lingrell S., Bachovchin D. A., Bierie B., Tischler V., Noske A., Okondo M. C., et al. 2017. LACTB is a tumour suppressor that modulates lipid metabolism and cell state. Nature. 543: 681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Deleault N. R., Piro J. R., Walsh D. J., Wang F., Ma J., Geoghegan J. C., and Supattapone S.. 2012. Isolation of phosphatidylethanolamine as a solitary cofactor for prion formation in the absence of nucleic acids. Proc. Natl. Acad. Sci. USA. 109: 8546–8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.van der Veen J. N., Lingrell S., da Silva R. P., Jacobs R. L., and Vance D. E.. 2014. The concentration of phosphatidylethanolamine in mitochondria can modulate ATP production and glucose metabolism in mice. Diabetes. 63: 2620–2630. [DOI] [PubMed] [Google Scholar]
- 278.Lands W. E. 2000. Stories about acyl chains. Biochim. Biophys. Acta. 1483: 1–14. [DOI] [PubMed] [Google Scholar]
- 279.Verkleij A. J., Leunissen-Bijvelt J., de Kruijff B., Hope M., and Cullis P. R.. 1984. Non-bilayer structures in membrane fusion. Ciba Found. Symp. 103: 45–59. [DOI] [PubMed] [Google Scholar]
- 280.Martens S., and McMahon H. T.. 2008. Mechanisms of membrane fusion: disparate players and common principles. Nat. Rev. Mol. Cell Biol. 9: 543–556. [DOI] [PubMed] [Google Scholar]
- 281.Cullis P. R., de Kruijff B., Hope M., Verkleij A. J., Nayar R., Farren S. B., Tildock C., Madden T. D., and Bally M. B.. 1983. Structural properties of lipids and their functional role in biological membranes. In Membrane Fluidity in Biology. R. C. Aloia, editor. Academic Press, New York. 39–81. [Google Scholar]
- 282.Epand R. M., Fuller N., and Rand R. P.. 1996. Role of the position of unsaturation on the phase behavior and intrinsic curvature of phosphatidylethanolamines. Biophys. J. 71: 1806–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Dowhan W., and Bogdanov M.. 2002. Functional roles of lipids in membranes. In Biochemistry of Lipids, Lipoproteins and Membranes. 4th edition. D. E. Vance and. J. E. Vance, editors. Elsevier Science, Amsterdam. 599. [Google Scholar]
- 284.Emoto K., Kobayashi T., Yamaji A., Aizawa H., Yahara I., Inoue K., and Umeda M.. 1996. Redistribution of phosphatidylethanolamine at the cleavage furrow of dividing cells during cytokinesis. Proc. Natl. Acad. Sci. USA. 93: 12867–12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Bogdanov M., Sun J., Kaback H. R., and Dowhan W.. 1996. A phospholipid acts as a chaperone in assembly of a membrane transport protein. J. Biol. Chem. 271: 11615–11618. [DOI] [PubMed] [Google Scholar]
- 286.Bogdanov M., Umeda M., and Dowhan W.. 1999. Phospholipid-assisted refolding of an integral membrane protein. Minimum structural features for phosphatidylethanolamine to act as a molecular chaperone. J. Biol. Chem. 274: 12339–12345. [DOI] [PubMed] [Google Scholar]
- 287.Dowhan W., and Bogdanov M.. 2012. Molecular genetic and biochemical approaches for defining lipid-dependent membrane protein folding. Biochim. Biophys. Acta. 1818: 1097–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Yao Z. M., and Vance D. E.. 1988. The active synthesis of phosphatidylcholine is required for very low density lipoprotein secretion from rat hepatocytes. J. Biol. Chem. 263: 2998–3004. [PubMed] [Google Scholar]
- 289.Hamilton R. L., and Fielding P. E.. 1989. Nascent very low density lipoproteins from rat hepatocytic Golgi fractions are enriched in phosphatidylethanolamine. Biochem. Biophys. Res. Commun. 160: 162–173. [DOI] [PubMed] [Google Scholar]
- 290.Skipski V. P. 1972. Quantitation, composition and metabolism of blood lipids. In Blood Lipids and Lipoproteins. G. Nelson, editor. Wiley-Interscience, New York. 471–483. [Google Scholar]
- 291.van der Veen J. N., Kennelly J. P., Wan S., Vance J. E., Vance D. E., and Jacobs R. L.. 2017. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochim. Biophys. Acta. 1859: 1558–1572. [DOI] [PubMed] [Google Scholar]
- 292.Hailey D. W., Rambold A. S., Satpute-Krishnan P., Mitra K., Sougrat R., Kim P. K., and Lippincott-Schwartz J.. 2010. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 141: 656–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Menon A. K., and Stevens V. L.. 1992. Phosphatidylethanolamine is the donor of the ethanolamine residue linking a glycosylphosphatidylinositol anchor to protein. J. Biol. Chem. 267: 15277–15280. [PubMed] [Google Scholar]
- 294.Jin X. H., Okamoto Y., Morishita J., Tsuboi K., Tonai T., and Ueda N.. 2007. Discovery and characterization of a Ca2+-independent phosphatidylethanolamine N-acyltransferase generating the anandamide precursor and its congeners. J. Biol. Chem. 282: 3614–3623. [DOI] [PubMed] [Google Scholar]