Abstract
Ethanolamine phosphotransferase (EPT)1, also known as selenoprotein 1 (SELENOI), is an enzyme that transfers phosphoethanolamine from cytidine diphosphate-ethanolamine to lipid acceptors to produce ethanolamine glycerophospholipids, such as diacyl-linked phosphatidylethanolamine (PE) and ether-linked plasmalogen [1-alkenyl-2-acyl-glycerophosphoethanolamine (plasmenyl-PE)]. However, to date there has been no analysis of the metabolomic consequences of the mutation of EPT1 on the concentration of ethanolamine glycerophospholipids in mammalian cells. We studied a patient with severe complicated hereditary spastic paraplegia, sensorineural-deafness, blindness, and seizures. Neuroimaging revealed hypomyelination, followed by brain atrophy mainly in the cerebellum and brainstem. Using whole exome sequencing, we identified a novel EPT1 mutation (exon skipping). In vitro EPT activity, as well as the rate of biosynthesis of ethanolamine glycerophospholipids, was markedly reduced in cultures of the patient’s skin fibroblasts. Quantification of phospholipids by LC-MS/MS demonstrated reduced levels of several PE species with polyunsaturated fatty acids, such as 38:6, 38:4, 40:6, 40:5, and 40:4. Notably, most plasmenyl-PE species were significantly decreased in the patient’s cells, whereas most plasmanylcholine [1-alkyl-2-acyl-glycerophosphocholine (plasmanyl-PC)] species were increased. Similar findings regarding decreased plasmenyl-PE and increased plasmanyl-PC were obtained using EPT1-KO HeLa cells. Our data demonstrate for the first time the indispensable role of EPT1 in the myelination process and neurodevelopment, and in the maintenance of normal homeostasis of ether-linked phospholipids in humans.
Keywords: phospholipids/phosphatidylethanolamine, phospholipids/phosphatidylcholine, phospholipids/biosynthesis, phospholipids/metabolism, ethanolamine phosphotransferase 1, neurodegenerative disease, hereditary spastic paraplegia, whole exome sequencing
Phosphatidylethanolamine (PE) is the second most abundant diacyl-linked phospholipid after phosphatidylcholine (PC) in mammals, generally constituting approximately 20–40% of cellular phospholipids (1). In addition to diacyl-linked phospholipids, approximately 20% of phospholipids in mammalian cells are ether-linked phospholipids containing an alkenyl or alkyl group at the sn-1 position, which distinguishes them from diacyl-linked phospholipids. In general, the 1-alkenyl group is present in plasmenylethanolamine [1-alkenyl-2-acyl-glycerophosphoethanolamine (plasmenyl-PE; also known as plasmalogen)], whereas the 1-alkyl group is present in plasmanylcholine [1-alkyl-2-acyl-glycerophosphocholine (plasmanyl-PC)]. The amounts of these ether-linked lipids vary from tissue to tissue. In humans, the highest concentration of plasmenyl-PE is found in the brain, followed by heart, lung, kidney, spleen, skeletal muscle, and testis. In the liver, plasmenyl-PE accounts for only 0.8% of the total phospholipids. The concentration of plasmanyl-PC is highest in heart and skeletal muscle and is low in other organs (2, 3).
In mammals, PE is synthesized by two major pathways: the Kennedy [or cytidine diphosphate (CDP)-ethanolamine] pathway in the endoplasmic reticulum (ER) and the phosphatidylserine (PS) decarboxylation pathway by PS decarboxylase (PSD) in mitochondria (1). In the Kennedy pathway, phosphoethanolamine is transferred from CDP-ethanolamine to diacylglycerol with the release of CMP and the production of PE by ethanolamine phosphotransferase (EPT) or choline/EPT (CEPT). For the biosynthesis of plasmenyl-PE, plasmanyl-PE is first synthesized from CDP-ethanolamine and 1-alkyl-2-acyl-glycerol by EPT or CEPT (supplemental Fig. S1). The 1-alkyl group is then desaturated to form the 1-alkenyl group by plasmanyl-PE desaturase.
Plasmanyl-PC is synthesized from CDP-choline and 1-alkyl-2-acyl-glycerol by choline phosphotransferase (CPT) or CEPT. In contrast to plasmanyl-PE, there is no specific desaturase for plasmanyl-PC. Therefore, to synthesize plasmenyl-PC, plasmenyl-PE is hydrolyzed by phospholipase C to form free 1-alkenyl-2-acyl-glycerol, which is then modified by CPT or CEPT to produce plasmenyl-PC (2, 3). It is also reported that plasmenyl-PC can be synthesized by methylation of the ethanolamine group in plasmenyl-PE (4–6).
Biosynthesis of the 1-alkyl group requires peroxisomes. The 1-alkyl-dihydroxyacetonephosphate (DHAP), which is responsible for introduction of the characteristic ether-bond, is synthesized from DHAP by glyceronephosphate O-acyltransferase (GNPAT), also named DHAP acyltransferase (DHAPAT), and alkylglycerone phosphate synthase (AGPS), also named alkyl-DHAP synthase (alkyl-DHAPS) in the peroxisome. The 1-alkyl-DHAP is then converted to 1-alkyl-2-acyl-glycerol in the ER by three enzymatic reactions, for the reduction, acylation, and removal of the phosphate group (2, 3).
In a previous study, we showed that human selenoprotein I (SELENOI) (7) exhibits EPT activity and termed this enzyme, EPT1 (8). In contrast to CEPT1, which has dual specificity and can use both CDP-ethanolamine and CDP-choline to produce PE and PC, respectively (9), EPT1 specifically uses CDP-ethanolamine as the phosphobase donor. We also showed that EPT1 is expressed most abundantly in brain, placenta, liver, and pancreas, followed by heart, skeletal muscle, lung, and kidney in humans. However, to date, no study has elucidated the in vivo outcome of EPT1 deficiency on the content of ethanolamine glycerophospholipids in mammalian cells.
In the present study, we identified a novel EPT1 mutation (exon skipping) in a patient with severe complicated hereditary spastic paraplegia (HSP), sensorineural deafness, and blindness associated with a severe conduction abnormality along the visual pathway. Follow-up by neuroimaging revealed neonatal hypomyelination followed by brain atrophy affecting mainly the cerebellum and the midbrain. Metabolomic study of the patient’s fibroblasts revealed markedly reduced levels of several diacyl-linked PE molecular species with polyunsaturated fatty acids, such as 38:6, 38:4, 40:6, 40:5, and 40:4. Interestingly, the level of plasmenyl-PE was more dramatically decreased than that of PE. In contrast, the level of plasmanyl-PC was increased, whereas there was no change in the level of PC. Similar results, showing a decrease in plasmenyl-PE and an increase in plasmanyl-PC, were obtained following analysis of the phospholipid profile of EPT1-KO HeLa cells. The present data demonstrate for the first time the major role of EPT1 in myelin formation, in normal brain development, and in the maintenance of ether-linked phospholipids in humans.
MATERIALS AND METHODS
Patient, standard protocol, and approvals
The patient was born to a consanguineous Arab Muslim family residing in northern Israel and was clinically, biochemically, and genetically investigated. The study protocol was performed according to the principles of the Helsinki Declaration and approved by the Institutional Ethics Review Committee, the National Committee for Genetics Studies of the Israeli Ministry of Health, and the Independent Ethics Committee of Dokkyo Medical University Hospital (ID:29002). Informed consent was obtained from the parents.
Clinical and laboratory evaluation
The patient was clinically evaluated by pediatric neurologists, a specialist in metabolic disorders, and by geneticists. Biochemical evaluation using standard laboratory techniques included measurement of plasma pH and plasma concentrations of glucose, electrolytes, liver and muscle enzymes, blood urea nitrogen, creatinine, lactate, amino acids, very-long-chain fatty acids, and phytanic acid. Isoelectric focusing of plasma transferrin was conducted, and urinary amino acids, organic acids, and amino acid levels were measured. Cerebrospinal fluid lactate levels were also measured. Electrophysiological tests, including brain-stem auditory evoked potentials and visual evoked potentials, were performed. Brain imaging using a 1.5 T system (GE Signa 9.1) was performed and evaluated by conventional T1-weighted and T2-weighted images. Fluid-attenuation inversion recovery, diffusion-weighted images, and single-voxel proton magnetic resonance spectroscopy images were obtained.
Whole exome sequencing
Whole exome sequencing (WES) of the patient was performed on exon targets captured using a SureSelect Human All Exon 50 Mb kit V4 (Agilent Technologies, Santa Clara, CA). Sequences were determined using a HiSeq2500 system (Illumina, San Diego, CA). The full sequencing methodology and variant interpretation protocol were previously described (10).
Cell culture
Skin fibroblasts isolated from a healthy control and the patient were cultured in RPMI-1640 supplemented with 20% FBS and penicillin/streptomycin. HeLa cells were cultured in DMEM (high glucose) with 10% FBS. Cells were maintained at 37°C in a humidified incubator containing 5% CO2.
Enzyme activity
EPT activity was measured as described previously (8). Briefly, cells were lysed in 20 mM Tris-HCl (pH 8.0) containing 5 μg/ml each of leupeptin, pepstatin, and chymostatin, in an ultrasonic bath sonicator for 10 s. The protein content was measured using a BCA protein assay kit (Thermo Fisher Scientific, Waltham, MA). The reaction mixture (25 μl) contained 50 mM Tris-HCl buffer (pH 8.0), 5 mM MnCl2, 1 mM EGTA, 0.5 mM 1,2-dioleoyl-sn-glycerol, 0.002% (w/v) Tween 20, cell lysate, and 20 μM CDP-ethanolamine (ethanolamine 1,2-14C, 50 mCi/mmol). After incubation at 37°C for 10 min, the reaction was stopped by adding 60 μl of chloroform-methanol (1:1, v/v). After centrifugation at 8,000 g for 5 min, the organic phase was applied to precoated Silica Gel 60 TLC plates (Merck, Darmstadt, Germany), which were then developed with chloroform-methanol-water (65:35:8, v/v/v). Radioactive phospholipids were analyzed using a FLA-7000 imaging analyzer and quantified using ImageQuant TL version 8.1 (GE Healthcare UK Ltd., Amersham, UK). Results were normalized per total amount of protein.
Metabolic labeling of ethanolamine glycerophospholipids
Cells were cultured in medium containing 0.2 μCi/ml of ethanolamine hydrochloride (1,2-14C, 110 mCi/mmol) for 12 (skin fibroblasts) or 2 h (HeLa cells) at 37°C in a humidified incubator containing 5% CO2. Phospholipids were extracted from the cells using the Bligh and Dyer method (11) and analyzed by TLC. To separate plasmenyl-PE and PE, TLC plates were exposed to HCl vapor for 3 min at room temperature before development to hydrolyze the 1-alkenyl group in plasmenyl-PE. The amount of PE and the lyso-form of PE derived from plasmenyl-PE were analyzed using an imaging analyzer as described above. The amount of protein was measured using a BCA protein assay kit, and the amount of labeled lipids was normalized per total amount of protein.
Generation of EPT1-KO HeLa cells by genome editing using CRISPR-Cas9
To generate EPT1-KO HeLa cells using CRISPR/Cas9 technology, guide RNA targeting exon 1 of human EPT1 was designed using the CRISPR design tool (available at: https://www.atum.bio/eCommerce/cas9/input). Guide RNA (sense, 5′-caccgAGTTTTCGGGTCGTCATGGC3′ antisense, 5′-aaacGCCATGACGACCCGAAAACTc3′) was cloned into the pSpCas9(BB)-2A-Puro (pX459) vector (Addgene, Cambridge, MA). HeLa cells were transfected with this vector using Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. Forty-eight hours after transfection, the cells were cultured in the presence of 2 μg/ml puromycin for 10 days, and then surviving cells were seeded as single colonies in 96-well plates. Single clones were expanded and screened for EPT1 expression by measuring EPT activity. The genomic DNA was isolated using a standard proteinase K/phenol method, then the DNA fragments for the target regions were amplified by PCR using one set of primers (hEPT1exon1-S: 5′-AGCGCGGCTCTCCTACCTTCTCGGGCAGC-3′ and hEPT1exon1-A: 5′-AAATAGGAAGGGAATTCCCTCGCTCCATG-3′). PCR products were purified using a QIAquick gel extraction kit (Qiagen, Hilden, Germany) and sequenced directly. To identify the mutated site of each allele, the DNA fragments obtained above were subcloned into pcDNA3.1 vector and several DNA inserts were sequenced.
Quantification of phospholipids by LC-MS/MS
Cells were lysed in 20 mM Tris-HCl buffer (pH 8.0) and the amount of protein was measured using a BCA protein assay kit. Phospholipids were extracted from cell lysate containing 300–400 μg protein using the Bligh and Dyer method in the presence of 2 μg of internal standards (1,2-dipentadecanoyl PC, 1,2-diheptadecanoyl PE, 1,2-diheptadecanoyl PS, and N-heptadecanoyl SM; Avanti Polar Lipids, Alabaster, AL). To eliminate phospholipid containing the 1-alkenyl group, lipids were treated with 0.15 N HCl at room temperature for 2 h. Lipids were analyzed by reverse-phase ultra-high-pressure LC using an Acquity UPLC BEH C18 column (1.7 μm, 2.1 × 50 mm) (Waters, Milford, MA) coupled to a 5500 QTRAP mass spectrometer (Sciex Inc., Framingham, MA). A binary gradient consisting of solvent A (acetonitrile:methanol:water, 1:1:3, v/v/v, containing 5 mM ammonium acetate) and solvent B (2-propanol containing 5 mM ammonium acetate) was used. The gradient profile was as follows: 0–1 min, 95% A; 1–9 min, 5–95% B linear gradient; 9–13 min, 95% B. The flow rate was 0.3 ml/min and the column temperature was 40°C. Each phospholipid species containing ethanolamine was detected in multiple reaction monitoring (MRM) mode by selecting the m/z of the phospholipid species at Q1 and the precursor ion of m/z 196 at Q3 in negative ion mode (12). For choline-containing phospholipids, the precursor ion of m/z 184 at Q3 was monitored in positive ion mode. To identify plasmenyl-PE containing 16:0, 18:0, and 18:1 at the sn-1 position, the precursor ions of m/z 364, 392, and 390 at Q3, respectively, were detected as described in (13). For quantification of PS, phosphatidylinositol (PI), and SM, neutral loss of m/z 87 in negative mode, the precursor ion of m/z 241 in negative mode, and the precursor ion of m/z 184 in positive mode, respectively, were monitored. Lipids were quantified using MultiQuant version 2.0 (Sciex) and normalized against the internal standards and amount of protein.
Expression of patient’s EPT1 and immunoblotting
Total RNA was isolated from cells with an RNeasy Mini kit (Qiagen). cDNA was synthesized with ReverTra Ace reverse transcriptase (Toyobo, Osaka, Japan). To obtain DNA fragments encoding EPT1, PCR was performed using a forward primer (F: 5′-ATGGCTGGCTACGAATACGTGAGCCC-3′) and a reverse primer (R: 5′-CAGGCCAATATTCTTTTCTTCCATTCC-3′). After being purified from the gel, PCR products were cloned into pcDNA3.1/V5-His TOPO vector (Thermo Fisher Scientific). DNA fragments encoding WT or the patient’s EPT1 fused with a Flag-tag at the N terminus were amplified by PCR and cloned into pCAG vector (Wako Pure Chemical Industries, Osaka Japan). Cells were transfected with the expression vectors using Lipofectamine 2000 and incubated for 24 h. Cells were then treated with 30 μM of MG-132 (a potent cell-permeable proteasome and calpain inhibitor; Enzo Life Sciences, Farmingdale, NY) for 6 h. Proteins were separated with SDS-PAGE and transferred to nitrocellulose membranes (Amersham Protran, GE Healthcare, Chicago, IL) using a Trans-Blot SD semi-dry transfer blotter (Bio-Rad, Hercules, CA), then incubating the membranes with 5% (w/v) skim milk in TBS for 1 h and washing three times with T-TBS (TBS containing 0.02% Tween 20). Membranes were then incubated with anti-DYKDDDDK (Flag) Tag antibody (Cell Signaling Technology, Danvers, MA) overnight at 4°C, washed three times with T-TBS, and then incubated with horseradish peroxidase-conjugated IgGs for 1 h at room temperature. Membranes were washed three times with T-TBS and stained with Clarity Western ECL substrate (Bio-Rad) according to the manufacturer’s instructions and visualized using a ChemiDoc Touch (Bio-Rad).
Statistical analysis
Quantitative data are presented as mean ± SD. The statistical significance was assessed using the Student’s t-test. P < 0.05 was considered statistically significant.
RESULTS
Clinical and biochemical studies
The patient is a 4-year-old boy, the second child of an Israeli Arab couple who are first cousins and reside in northern Israel. Their firstborn daughter, who is 6 years old, has moderate intellectual disability, speaks only a few words, and achieved all motor milestones later than normal. Her slow development was attributed to asphyxia, low APGAR score (5 in the first minute and 7 at 5 minutes).
The patient was born after 39 weeks of gestation in a normal pregnancy by elective cesarean section. His APGAR score was 9/9, despite the presence of meconium amniotic fluid. Physical examination at age 2 h revealed hypertonia alternating with hypotonia, followed by tonic-clonic seizures, which responded to phenobarbital. His head circumference was 35 cm, his weight was 3.3 kg, and no dysmorphic features or shortening of proximal limbs were present. Routine biochemical studies, including plasma glucose, electrolytes, liver and kidney function, and screening for intrauterine and bacterial infections, were all normal. Cerebrospinal fluid glucose, protein, cells, culture, lactate, and amino acids were all normal. The extended metabolic work up, including plasma ammonia, lactate, amino acids, isoelectric focusing of plasma transferrin, very-long-chain fatty acids, and urinary amino and organic acids, was within the normal range.
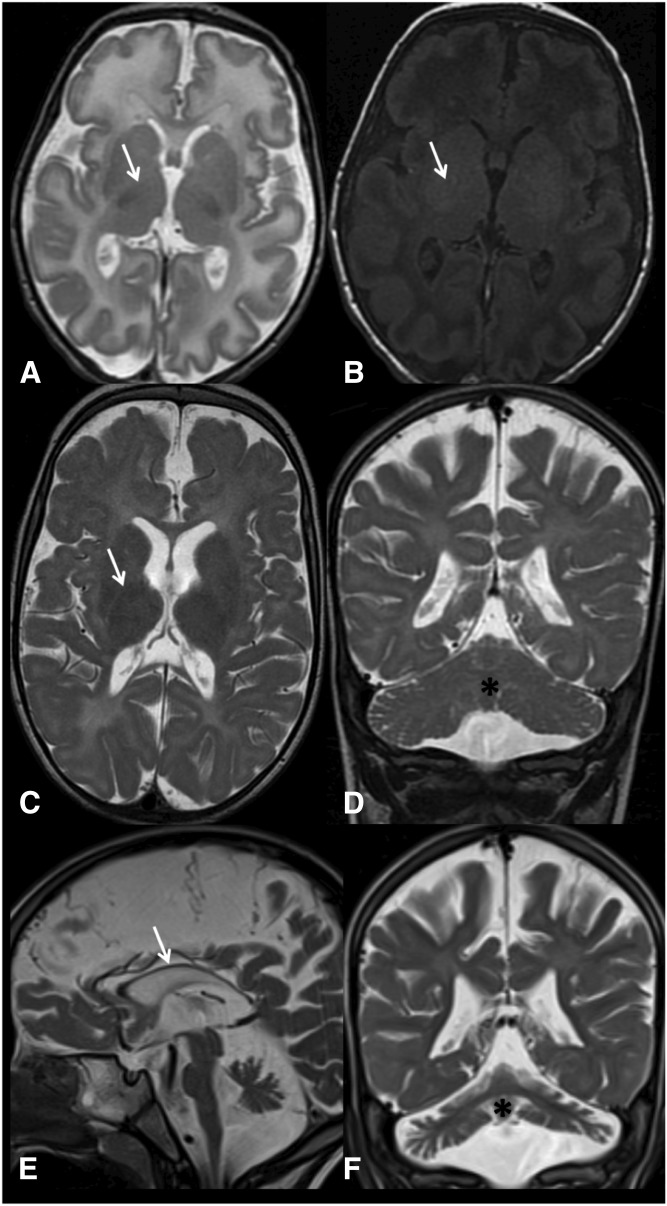
Brain MRI studies (Fig. 1) revealed progressive white matter abnormalities. At age 3 weeks, hypomyelination was present with no signs of atrophy. A year later, there was a continued delayed myelination pattern and diffuse atrophy, including in the cerebellum. At 4 years of age, increased white matter diminution was evident, with hypoplasia of the optic nerves, extremely thin corpus callosum, and continued cerebellar and midbrain atrophy. Magnetic resonance spectroscopy did not reveal lactate or any other abnormal peaks. Between birth and age 4 years, the patient did not achieve any developmental milestones, except for smiling when he is held in his mother’s arms. Clinical seizures recurred since the age of 3 years. Currently, at 4 years of age, his head circumference is at the third centile (progressive microcephaly). There are no dysmorphic features other than roving eye movements (searching nystagmus) and no social eye contact. He has a high arched palate with a bifid uvula, pronounced head lag, marked spasticity of the lower limbs, spontaneous clonus, and moderate spasticity of the upper limbs. Auditory brainstem response analysis indicated severe sensorineural hearing impairment and prolonged central conduction times along the auditory pathway. He had a normal electroretinography scan, but no visual evoked potentials could be elicited. These last findings indicate a severe conduction abnormality along the visual pathway pointing to central impairment of the visual system. The EEG showed a disorganized background with multifocal generalized epileptic discharge. The patient’s progressive spasticity necessitated repeated orthopedic surgeries.
Fig. 1.
MRI images of the patient’s brain. MRI images at the age of 3 weeks (A, B), 1 year (C, D), and 4 years (E, F). At 3 weeks of age, there is a delayed myelination pattern, as evident by the signal of the posterior limb of internal capsule (PLIC) (arrow). At this age, normal myelination should be evident in the PLIC by a low T2 signal (A) and a high T1 signal (B). Additionally, there is an overall increase in the brain water content in the T2 image (A). No signs of atrophy were evident at 3 weeks of age. At 1 year of age, there is a continued delayed myelination pattern [PLIC signal, arrow (C)]; additionally, there is a mild increase in the white matter T2 signal and diffuse atrophy, including the cerebellum [asterisk (D)]. At 4 years of age, atrophy and white matter diminution are evident, with an extremely thin corpus callosum [(E) arrow] and continued cerebellar atrophy [(F) asterisk].
WES analysis
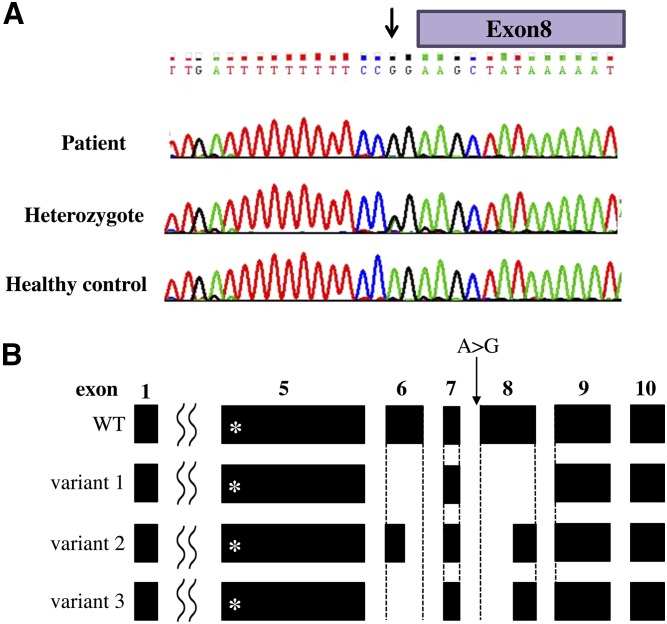
Given the consanguinity in the family, we performed WES under the hypothesis of a recessively inherited rare causal allele. WES of the patient yielded 42.0 million mapped reads with a mean coverage of ×57. Following alignment and variant calling, we performed a series of filtering steps. These included removing variants that were less than ×8, were off-target, heterozygous, synonymous, had MAF >0.5% or homozygotes at the ExAC [Exome Aggregation Consortium, Cambridge, MA (http://exac.broadinstitute.org/) database], or MAF >4% in the Hadassah in-house database (∼800 ethnic-matched exome analyses). Only 13 variants survived this filtering (Table 1), but given the conservation score, predicted pathogenicity, and genotyping of the family members, we focused on Chr2:26607825 A>G, NM_033505:exon8:c.732-2A>G in EPT1 (Fig. 2A, supplemental Fig. S2A). This variant was calculated to markedly reduce the splicing score, and is not carried by the ∼125,000 healthy individuals whose WES data are deposited in the gnomAD website (http://gnomad.broadinstitute.org/) database. Furthermore, no homozygous loss-of-function variants in the EPT1 gene were found in this cohort, suggesting that homozygosity for a deleterious mutation in the human EPT1 gene is not tolerated.
TABLE 1.
Homozygous variants surviving after exome filtering
| Chr | Base Pairs (Hg19) | WT | Mut | AA | Gene | GERP | Exac Het | rs Number |
| 2 | 26534681 | T | C | R-440-G | ADGRF3 | — | 0 | — |
| 2 | 26607825 | A | G | ACCEPTOR | EPT1 | 5.8 | 0 | — |
| 3 | 128598673 | A | C | K-47-Q | ACAD9 | 3 | 0 | — |
| 3 | 184041709 | A | G | I-806-V | EIF4G1 | 5.9 | 44 | rs62287499 |
| 4 | 170498175 | A | C | I-308-M | NEK1 | 4.2 | 52 | rs10034957 |
| 9 | 120176823 | C | T | A-132-T | ASTN2 | 3.1 | 2 | — |
| 9 | 139571511 | T | G | I-132-L | AGPAT2 | — | 2 | — |
| 9 | 140652341 | C | T | S-460-L | EHMT1 | — | 0 | — |
| 11 | 116658693 | C | T | G-5-E | ZPR1 | 4.2 | 0 | — |
| 12 | 104350450 | C | A | A-23-S | C12ORF73 | 4.4 | 8 | rs201697510 |
| 14 | 24769850 | — | GAG | [ins]162-E | NOP9 | — | 0 | rs113258190 |
| 17 | 7220663 | G | C | P-1449-A | NEURL4 | 5 | 0 | rs374832287 |
| 17 | 47246239 | G | A | R-491-H | B4GALNT2 | — | 143 | rs141826857 |
Fig. 2.
Summary of the EPT1 mutation in the patient. A: Sequence chromatogram of exon/intron 8 around the mutation site (arrow). Patient (upper panel), heterozygote (middle panel), and healthy control (lower panel). B: Splicing variants of EPT1 in the patient. Variant 1 was isolated from lymphocytes and variants 2 and 3 were from fibroblasts. The asterisks in exon 5 indicate the location of CDP-alcohol phosphatidyltransferase motif. Note the partial or full deletion of exon 6 and 8 in splicing variants.
To demonstrate the functional importance of the EPT1 c.732-2A>G variant, we generated cDNA from fresh lymphocytes from the patient and found that it was markedly shortened compared with the fragment generated from a healthy control. Sequence determination revealed a splicing variant having skipped exons 6 and 8, with the predicted loss of 290 bp associated with a frameshift. We named this EPT1 variant as variant 1 (Fig. 2B). Then, we also analyzed cDNA for EPT1 using the patient’s skin fibroblasts. When RT-PCR was performed with one set of primers encoding the N and C termini of EPT1 ORF [forward (F) and reverse (R), respectively, in Supplemental Fig. S2A]), smeared DNA bands were obtained from the patient’s cDNA, whereas a single 1158 bp band corresponding to a full-length size of EPT1 was obtained from a healthy control (Supplemental Fig. S2B). Then, the smeared bands were totally extracted, subcloned, and sequenced, and we found that they contained at least two EPT1 splicing variants, variants 2 and 3 (Fig. 2B). In variant 2, both exon 6 and exon 8 were partially deleted. In variant 3, exon 6 was fully deleted and exon 8 was partially deleted at the same position of variant 2. These results suggest that the A>G mutation causes generation of several splicing variants with truncation in exons 6 and 8.
EPT activity and de novo synthesis of ethanolamine-containing phospholipids is impaired in the patient
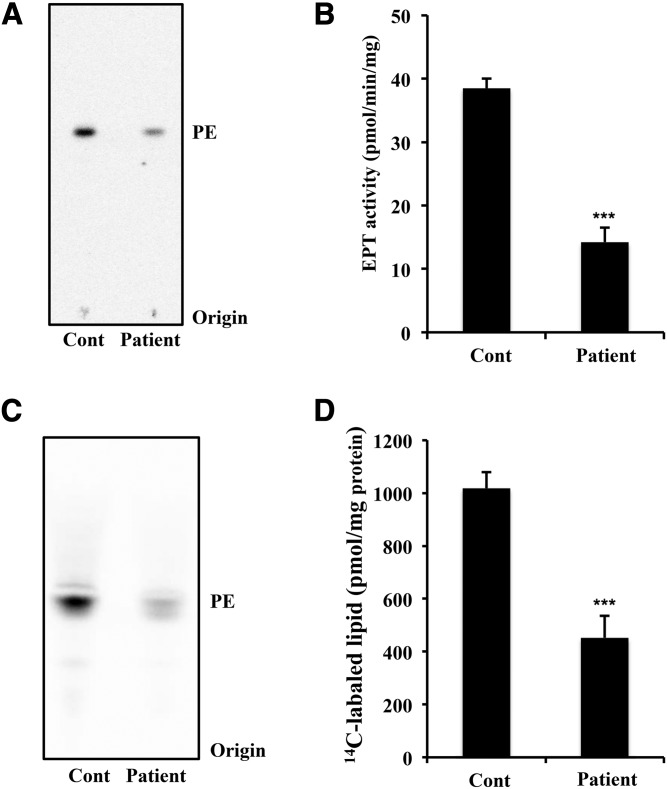
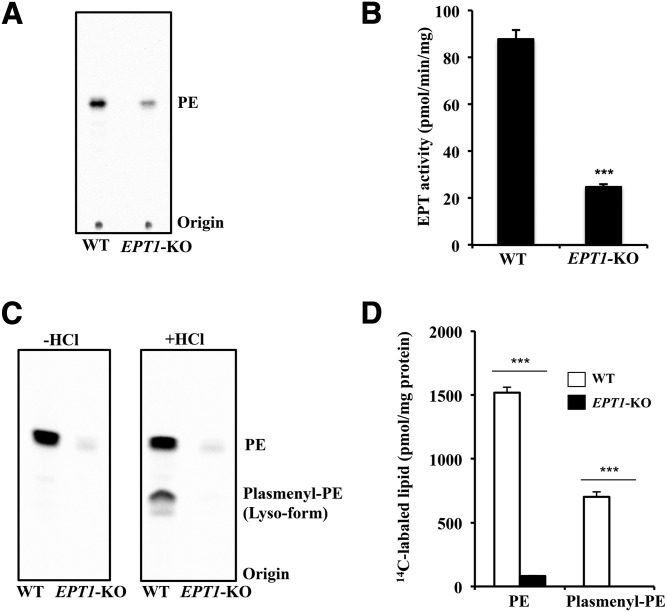
To confirm that EPT1 mutation affects the enzymatic activity of EPT, we cultured skin fibroblasts from a healthy control and from the patient and measured the in vitro EPT activities. Cell lysates were incubated with diacylglycerol in the presence of 14C-radiolabeled CDP-ethanolamine. After the lipids were extracted from the reaction mixture, the radiolabeled lipids were separated and analyzed by TLC. As shown in Fig. 3A and B, the activity for production of PE in the patient was significantly lower than that of the control. Even though the patient has an EPT1 mutation, about one-third of the enzymatic activity remained, possibly due to CEPT1 activity.
Fig. 3.
EPT activity and de novo synthesis of ethanolamine glycerophospholipids in the skin fibroblasts. A, B: EPT activity in the cell lysates of a healthy control (Cont) and of the patient. Activity was measured using radiolabeled CDP-ethanolamine as the substrate in the presence of diacylglycerol and Mn2+ for 10 min. After extraction, radioactive phospholipids were analyzed by TLC (A) and quantified using ImageQuant TL version 8.1 (B). Data are expressed as the mean ± SD from four independent reactions. C, D: Metabolic labeling of ethanolamine glycerophospholipids. Skin fibroblasts were cultured in the presence of radiolabeled ethanolamine for 12 h. After extraction, radioactive phospholipids were analyzed by TLC (C) and quantified (D). The values shown are the mean ± SD from five independent culture dishes. Each experiment was repeated twice with similar results. ***P < 0.001 as compared with control cells.
We next analyzed the de novo synthesis of ethanolamine glycerophospholipids. Skin fibroblasts were cultured in the presence of 14C-ethanolamine for 12 h for metabolic labeling of the ethanolamine glycerophospholipids. After extraction from the cells, the lipids were analyzed by TLC. As shown in Fig. 3C and D, 14C-ethanolamine-labeled lipids with the same Rf value as PE were significantly reduced in the patient compared with the control. These results suggest that the biosynthesis of ethanolamine glycerophospholipids is markedly impaired in the patient.
Phospholipid profiling of skin fibroblasts by LC-MS/MS
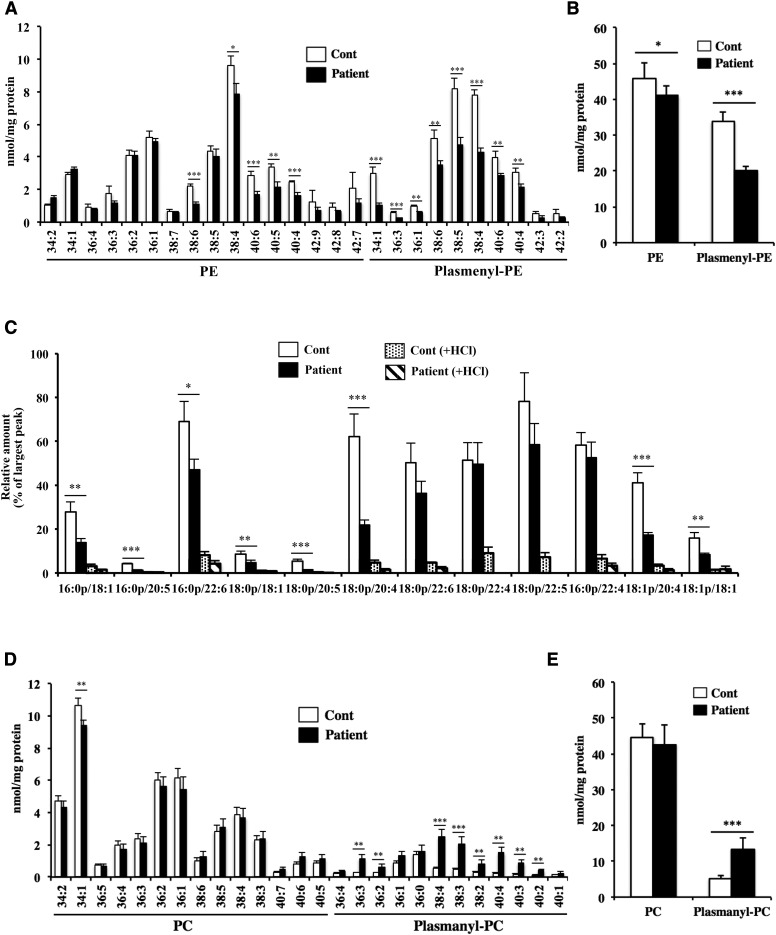
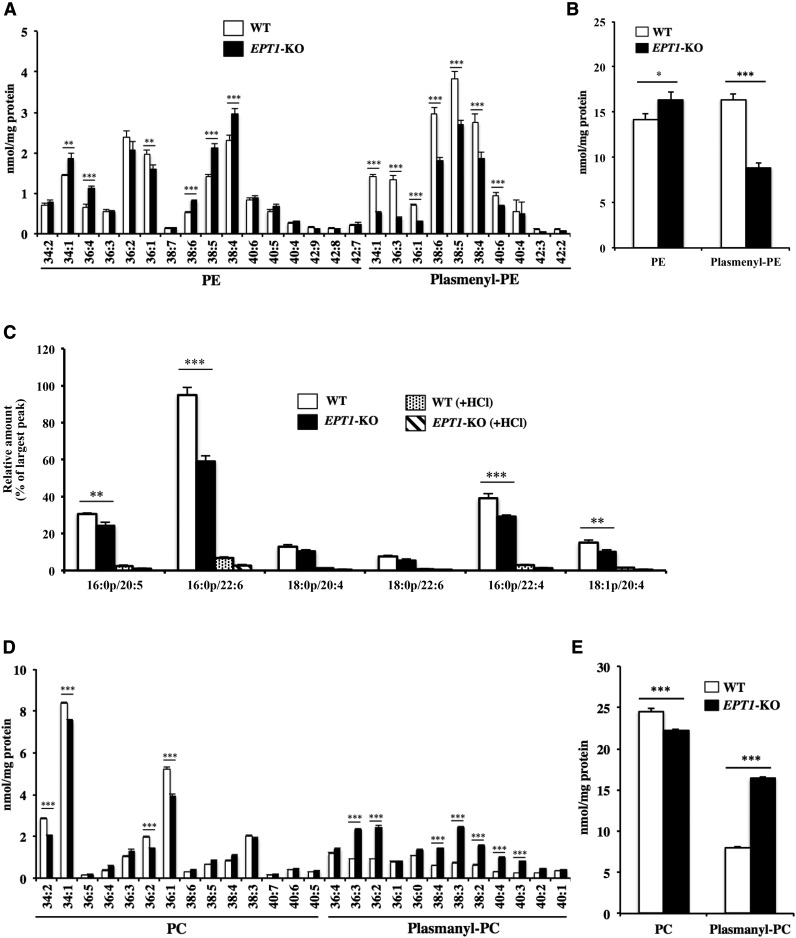
We next analyzed the composition of the phospholipids in the fibroblasts. The phospholipids were extracted from the cell and quantitatively measured using LC-MS/MS. We measured the ethanolamine glycerophospholipids by monitoring the precursor ion of m/z 196 in negative ion mode, which corresponds to the m/z of glycerol phosphoryl ethanolamine-water (12). As shown in Fig. 4A, PE molecular species with polyunsaturated fatty acids, such as 38:6, 38:4, 40:6, 40:5, and 40:4, were markedly reduced in the patient. Notably, most plasmenyl-PE species were significantly reduced in the patient. As shown in Fig. 4B, the total amount of plasmenyl-PE obtained from the patient’s fibroblasts was much decreased, and the total amount of PE was also slightly decreased in the patient compared with the healthy control. To confirm that the reduced lipids were surely plasmenyl-PE rather than plasmanyl-PE species, we measured the same sample following treatment with HCl. The alkenyl group in plasmenyl-PE is degraded by HCl, whereas neither PE nor plasmanyl-PE is sensitive to HCl (14). Plasmenyl-PE, but not PE, was dramatically reduced after HCl treatment (supplemental Fig. S3A), indicating that the decreased lipids in the patient’s fibroblasts were mainly plasmenyl-PE. We also measured plasmenyl-PE species with another MRM transition in positive ion mode by monitoring the characteristic fragments resulting from collisions at the sn-1 position, as reported in (13). As shown in Fig. 4C, this MRM transition also confirmed the reduced level of plasmenyl-PE in the patient.
Fig. 4.
Quantification of ethanolamine and choline glycerophospholipids in the skin fibroblasts. Total lipids were extracted from the skin fibroblasts using the Bligh-Dyer method in the presence of internal standard, and the amount of each phospholipid was determined using LC-MS/MS. PE and plasmenyl-PE species (A) and total amount of ethanolamine phospholipids (B) scanned by monitoring the precursor ion of m/z 196 in negative mode. C: Plasmenyl-PE scanned by monitoring the precursor ions of m/z 364, 392, and 390 (fragment ions derived from plasmenyl-PE with 16:0, 18:0, and 18:1 at the sn-1 position, respectively) in positive mode. To eliminate plasmenyl-PE, samples were treated with HCl. PC and plasmanyl-PC species (D) and total amount of choline phospholipids (E) scanned by monitoring the precursor ion of m/z 184 in positive mode. Values were normalized against the amount of protein in the cells. Data are expressed as the mean ± SD from quadruplet analyses in one experiment. Each experiment was repeated at least three times with similar results *P < 0.05, **P < 0.01, and ***P < 0.001 as compared with control (Cont) cells.
We also measured choline glycerophospholipids by monitoring the precursor ion of m/z 184 in positive ion mode, which corresponds to the m/z of phosphoryl choline. As shown in Fig. 4D, most diacyl-linked PC species were similar in both the control and the patient. In contrast, many plasmanyl-PC species were significantly increased in the patient. As shown in Fig. 4E, the total amount of plasmanyl-PC was significantly increased in the patient compared to the healthy control. We also analyzed the same sample after treatment with HCl, and confirmed that the lipids that increased in the patient were surely derived from plasmanyl-PC, not from plasmenyl-PC (supplemental Fig. S3B).
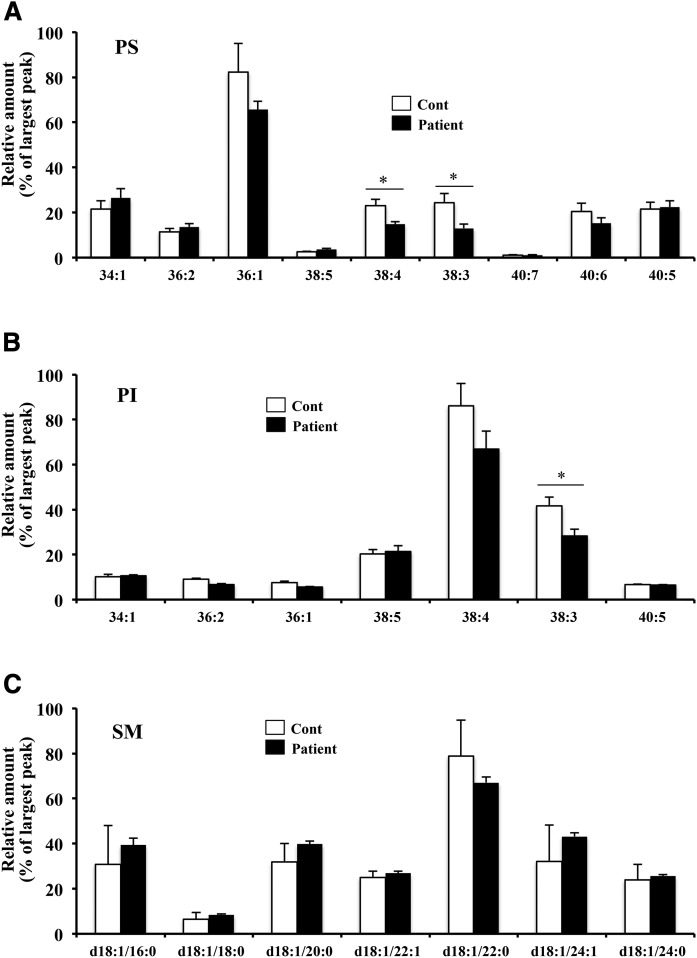
We compared other phospholipids, such as PS, PI, and SM, but could not find marked changes between the control and the patient except the decrease in PS molecular species with 38:4 and 38:3, and PI molecular species with 38:3 (Fig. 5). Taken together, these results suggest that the EPT1 mutation in the patient causes a decrease in plasmenyl-PE and an increase in plasmanyl-PC.
Fig. 5.
Quantification of PS, PI, and SM in skin fibroblasts. PS (A), PI (B), and SM (C) species were determined using LC-MS/MS with MRM scanning of the neutral loss of m/z 87 in negative mode, the precursor ion of m/z 241 in negative mode, and the precursor ion of m/z 184 in positive mode, respectively. Values were normalized against the amount of protein in the cells. Data are the mean ± SD from triple analyses in one experiment. Each experiment was repeated at least three times with similar results. *P < 0.05 as compared with control (Cont) cells.
Generation of EPT1-KO HeLa cells
To confirm the results obtained from the above analysis of the patient’s fibroblasts, we generated a line of stable HeLa cells in which the EPT1 gene was ablated using the CRISPR/Cas9 system. After transfection of a Cas9 plasmid with gRNA targeting the EPT1 exon 1, puromycin-selected cells were cloned. We obtained a clone with a heterozygous biallelic mutation (supplemental Fig. S4A). Alleles 1 and 2 showed a 1 bp insertion and a 5 bp deletion around the start codon, respectively (supplemental Fig. S4B), indicating that the open reading frame of both EPT1 alleles is frame-shifted. We found that the KO cells showed a significant reduction of in vitro EPT activity (Fig. 6A, B). Next, we analyzed the de novo synthesis of ethanolamine glycerophospholipids by metabolic labeling using 14C-ethanolamine. As shown in Fig. 6C, 14C-labeled ethanolamine glycerophospholipids with the same Rf value as PE were significantly reduced in the KO cells. To separate plasmenyl-PE from PE, the TLC plate was exposed to HCl vapor before development to hydrolyze the alkenyl-group, resulting in formation of the lyso-form (2-acyl) derived from plasmenyl-PE. The levels of both plasmenyl-PE and PE were markedly decreased in the KO cells (Fig. 6D). These results suggest that the biosynthesis of ethanolamine glycerophospholipids is impaired by the EPT1 mutation.
Fig. 6.
EPT activity and de novo synthesis of ethanolamine glycerophospholipids in EPT1-KO HeLa cells. A, B: EPT activity in the cell lysate of WT and EPT1-KO HeLa cells was measured using radiolabeled CDP-ethanolamine as the substrate in the presence of diacylglycerol and Mn2+ for 10 min. After extraction, the radioactive phospholipids were analyzed by TLC (A) and quantified using ImageQuant TL version 8.1 (B). Data are the mean ± SD from four independent reactions. C, D: Metabolic labeling of ethanolamine glycerophospholipids. Cells were cultured in the presence of radiolabeled ethanolamine for 2 h. After extraction and application onto TLC plates, radioactive lipids were treated with HCl vapor to degrade plasmenyl-PE and produce the lyso-form of the lipid. Lipids were analyzed by TLC (C) and quantified (D). Values shown are the mean ± SD from five independent culture dishes. Each experiment was repeated at least three times with similar results. ***P < 0.001 as compared with WT.
Phospholipid profiling of EPT1-KO HeLa cells with LC-MS/MS
We measured ethanolamine glycerophospholipids using LC-MS/MS in the EPT1-KO cells. The amount of glycerophospholipids containing the alkenyl or alkyl group was confirmed by HCl treatment (supplemental Fig. S5A, B). As shown in Fig. 7A and C, most plasmenyl-PE species were significantly reduced in the KO cells, whereas the levels of PE species, such as 34:1, 36:4, 38:6, 38:5, and 38:4, were increased in the KO cells. As shown in Fig. 7B, the total amount of plasmenyl-PE in the KO cells was decreased to about half of that in WT. In contrast, the total PE amount was slightly increased in the KO cells compared to WT. Similar to the results obtained using the patient’s fibroblasts, most plasmanyl-PC species were markedly increased in the KO cells (Fig. 7D). As shown in Fig. 7E, the total amount of plasmanyl-PC in the KO cells was increased to about twice the amount in WT. These results support our conclusion that an EPT1 mutation results in a decrease in plasmenyl-PE and an increase in plasmanyl-PC.
Fig. 7.
Quantification of ethanolamine and choline glycerophospholipids in EPT1-KO HeLa cells. After extraction from WT or EPT1-KO HeLa cells using the Bligh-Dyer method in the presence of internal standard, the amount of each phospholipid was determined using LC-MS/MS. PE and plasmenyl-PE species (A) and total amount of ethanolamine phospholipids (B) were scanned by monitoring the precursor ion of m/z 196 in negative mode. C: Plasmenyl-PE was scanned by monitoring the precursor ions of m/z 364, 392, and 390 in positive mode. To eliminate plasmenyl-PE, samples were treated with HCl. PC and plasmanyl-PC species (D) and total amount of choline phospholipids (E) were scanned by monitoring the precursor ion of m/z 184 in positive mode. Values were normalized against the amount of protein in the cells. Data are the mean ± SD from quadruplet analyses in one experiment. Each experiment was repeated at least three times with similar results. **P < 0.01 and ***P < 0.001 as compared with WT cells.
Expression of the patient’s EPT1 in EPT1-KO HeLa cells
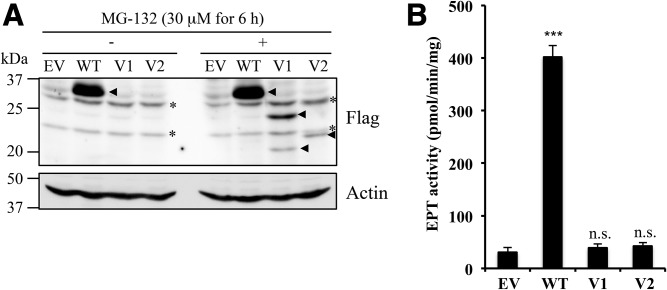
Human EPT1 has been predicted to contain seven transmembrane helices and the CDP-alcohol phosphatidyltransferase motif, an amino acid sequence essential for EPT catalytic activity (supplemental Fig. S2C) (8, 15). The splicing variants found in the patient still contained the CDP-alcohol phosphatidyltransferase motif (Fig. 2B, supplemental Fig. S2C). Therefore, we tried to investigate the EPT enzyme activities of these variants. As the ORF of variant 3 is the same as variant 1, we prepared expression vectors containing WT, variant 1, and variant 2 fused with a Flag tag at the N terminus, and those constructs were transfected into the EPT1-KO cells. The expressions of the recombinant proteins were confirmed by immunoblotting using anti-Flag antibody. The estimated molecular masses of WT, variant 1, and variant 2 were 44, 23, and 27 kDa, respectively. As shown in Fig. 8A (left panel), a 35 kDa protein band was detected in cells transfected with WT vector. However, no protein expression was detected in cells transfected with variant 1 or 2 vectors. Therefore, we treated the cells with MG-132, a proteasome inhibitor, after transfection. As in Fig. 8A (right panel), protein bands of 24 and 22 kDa could be detected in variants 1 and 2 transfected cells with MG-132, respectively. In the lane of variant 1 with MG-132, a smaller protein band, possibly cleaved product of expressed variant 1 protein, was also detected around 20 kDa. Next, we measured EPT1 enzyme activities in the cell lysates treated with MG-132. As shown in Fig. 8B, clearly increased EPT enzyme activity was observed in WT-expressing cells, whereas no increase was detected in variant-expressing cells. These results suggest that variant enzymes are not functional because the truncated EPT1 variants were expressed only when cells were treated with MG-132. The endogenously expressed patient’s variant enzymes may be constantly eliminated by ubiquitin/proteasome-mediated protein quality control in steady state conditions.
Fig. 8.
Expression of EPT1 variants in EPT1-KO HeLa cells. A: The EPT1-KO HeLa cells were transfected with empty vector (EV), vector containing Flag-tag fused WT EPT1 (WT), variant 1 (V1), and variant 2 (V2). After 24 h, cells were treated with 30 μM of MG-132 for 6 h (right panel). After cell lysates were separated by SDS-PAGE, Flag-tagged proteins were analyzed by immunoblotting using anti-Flag antibody. Actin was used as a protein loading control. Arrowheads and asterisks indicate Flag-specific and nonspecific signals, respectively. (B) EPT enzyme activity of the transfected cells treated with MG-132 was measured using radiolabeled CDP-ethanolamine as the substrate in the presence of diacylglycerol and Mn2+ for 10 min. ***P < 0.001 as compared with EV. n.s., not significant. Each experiment was repeated twice with similar results.
DISCUSSION
PE is the second most abundant phospholipid in the lipid bilayer of organelle membranes in mammals and has various functions, such as membrane fusion (16, 17), lipidation of the autophagy component (18, 19), and providing a supply of phosphoethanolamine for the GPI anchor (20, 21) and for use in the synthesis of anandamide (22). Although the physiological roles of plasmenyl-PE are not fully understood, it is believed to play roles as an antioxidant and as a regulator of cholesterol trafficking (2).
EPT1 is the final enzyme that synthesizes these ethanolamine glycerophospholipids, but little is known about the contribution of EPT1 to the homeostasis of ethanolamine glycerophospholipids. Furthermore, there are no reports of EPT1-KO mice, and the biological function of EPT1 in animals has not been clarified. We herein describe a patient featuring severe complicated autosomal recessive HSP, including blindness, deafness, and seizures. Neuroimaging showed neonatal hypomyelination followed by progressive brain atrophy, with striking involvement of the cerebellum and brain stem. WES revealed an EPT1 mutation. We demonstrated for the first time that plasmenyl-PE is decreased and plasmanyl-PC is increased in both this patient’s cells and in EPT1-KO HeLa cells. These results suggest that EPT1 is indispensable for neuronal development and myelin synthesis, and plays an important role in the maintenance of ether-linked phospholipids in human cells.
In the human central nervous system, plasmenyl-PE constitutes 30% of the total phospholipids and 90% of the ethanolamine glycerophospholipids in neuronal cell membranes (2, 23). It is widely recognized that plasmenyl-PE protects the neuronal membrane and myelin sheath from oxidative damage. Several neurodevelopmental deficits are observed in plasmenyl-PE deficiency diseases, such as Zellweger syndrome and rhizomelic chondrodysplasia punctata, including hypomyelination (24–27). Several studies have also demonstrated that the level of plasmenyl-PE is decreased in several brain disorders, such as Alzheimer’s disease (28, 29), Parkinson’s disease (30), Down syndrome (31), and schizophrenia (32). Thus, our results indicate that plasmenyl-PE is an important constituent for myelin formation and neuronal development and function.
Myelin formation is a process involved in normal brain development and maturation. Numerous genes are involved in controlling the events required for assuring normal myelination. Various inborn errors of metabolism are associated with hypomyelination, in which myelin is not properly formed, and demyelinating leukodystrophies, in which myelin is initially formed, but is later destroyed (33). Pelizaeus-Merzbacher disease (PMD) is a prototype of hypomyelination [PMD (MIM 312080), an X-linked hypomyelinating disorder caused by mutations in the proteolipid protein 1 gene (PLP1)]. Targeted metabolomic analyses of PMD patients’ fibroblasts revealed significantly decreased concentrations of ethanolamine plasmalogens (34). The authors suggested that defects in peroxisomal function, coupled with the PLP1 gene of PMD, result in changes in plasmalogen metabolism. However, the finding of hypomyelination and decreased cellular content of plasmenyl-PE in EPT1-deficient human fibroblasts indicates that it is not the peroxisomes but rather the contribution of the ER enzyme that orchestrates the myelin biosynthesis process.
Recently, another EPT1 mutation was reported in patients with a complex neurodegenerative phenotype (35). The clinical phenotypes in the family are similar to our report herein in that they are characterized by neurodegenerative complicated HSP with hypomyelination upon neuroimaging. In these patients, Arg112, which is a highly conserved amino acid in the CDP-alcohol phosphotransferase motif of EPT1, is substituted to Pro. Yeast expressing the mutant EPT1Arg112Pro showed only 3% of the de novo PE synthesis of WT. These authors also reported that, despite the dramatic reduction in EPT activity, the level of PE in the blood, which may reflect PE synthesis in liver, was not affected in the patients. We also found that, in contrast to plasmenyl-PE, the total amount of PE is not notably changed in either the patient’s fibroblasts or EPT1-KO HeLa cells (Figs. 4, 7). Clarification of this point will require taking an alternative pathway of PE synthesis into consideration. It is widely accepted that in many types of cultured cells, a large amount of PE is made from the decarboxylation of PS via mitochondrial PSD (36–38). Importantly, it was reported that PE molecular species generated by PS decarboxylation are different from those generated by the CDP-ethanolamine pathway, i.e., PSD preferentially synthesizes PE with polyunsaturated fatty acids (39). In EPT1-KO HeLa cells, the amount of PE with polyunsaturated fatty acids, such as 36:4, 38:6, 38:5, and 38:4, is increased compared with WT (Fig. 7A). In the patient’s fibroblasts, the amount of PS with 38:4 and 38:3 are decreased (Fig. 5A), suggesting that PS species with polyunsaturated fatty acids are used to compensate for the loss of PE production in EPT1-deficient cells. Thus, the most likely reason that the total amount of PE was not greatly affected by EPT1 deficiency may be due to the compensational PE production by PSD. PE can also be synthesized by CEPT1 or two minor pathways: PS synthase 2 (PSS2), which can exchange the serine group of PS with ethanolamine (38, 40, 41), and lysoPE acyltransferase (LPEAT), which converts lysoPE to PE (42, 43). However, PSD, PSS2, and LPEAT cannot compensate for the reduction in plasmenyl-PE because they are not believed to be involved in the synthesis of this lipid. Of the enzymes mentioned above, only CEPT1 can synthesize plasmenyl-PE. In Figs. 3 and 6, we demonstrated adequate EPT activity and de novo synthesis of ethanolamine glycerophospholipids in both the patient and in EPT1-KO cells, possibly due to CEPT1 activity. Because the CEPT1 activities of both the fibroblasts and the HeLa cells were lower than their EPT1 activities (one-third and one-fourth of the total EPT activity, respectively), CEPT1 activity may be insufficient to rescue the level of decreased plasmenyl-PE. Further study on CEPT1-KO cells will clarify the contribution of this enzyme to plasmenyl-PE synthesis. We also demonstrated that plasmanyl-PC is increased in both the patient and KO cells, possibly because EPT1 mutation causes a surplus of 1-alkyl-2-acyl-glycerol in the ER, which is used for the synthesis of plasmanyl-PE. CPT and CEPT use this extra 1-alkyl-2-acyl-glycerol to produce plasmanyl-PC in the cells (supplemental Fig. S1).
In summary, this study demonstrated that EPT1 is indispensable for the maintenance of plasmenyl-PE, a critical component of both neuronal membranes and myelin sheaths, and is essential for normal neurodevelopment in humans.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Takashi Namatame of the Clinical Research Center for DNA sequencing.
Footnotes
Abbreviations:
- CDP
- cytidine diphosphate
- CEPT
- choline/ethanolamine phosphotransferase
- CPT
- choline phosphotransferase
- DHAP
- dihydroxyacetonephosphate
- EPT
- ethanolamine phosphotransferase
- ER
- endoplasmic reticulum
- HSP
- hereditary spastic paraplegia
- MRM
- multiple reaction monitoring
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PI
- phosphatidylinositol
- plasmanyl-PC
- 1-alkyl-2-acyl-glycerophosphocholine
- plasmenyl-PE
- 1-alkenyl-2-acyl-glycerophosphoethanolamine
- PLIC
- posterior limb of internal capsule
- PMD
- Pelizaeus-Merzbacher disease
- PS
- phosphatidylserine
- PSD
- phosphatidylserine decarboxylase
- WES
- whole exome sequencing
This work was supported in part by Japan Society for the Promotion of Science Grant 17K08642. The authors declare no competing financial interests.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Vance J. E. 2015. Phospholipid synthesis and transport in mammalian cells. Traffic. 16: 1–18. [DOI] [PubMed] [Google Scholar]
- 2.Braverman N. E., and Moser A. B.. 2012. Functions of plasmalogen lipids in health and disease. Biochim. Biophys. Acta. 1822: 1442–1452. [DOI] [PubMed] [Google Scholar]
- 3.Brites P., Waterham H. R., and Wanders R. J.. 2004. Functions and biosynthesis of plasmalogens in health and disease. Biochim. Biophys. Acta. 1636: 219–231. [DOI] [PubMed] [Google Scholar]
- 4.Mogelson S., and Sobel B. E.. 1981. Ethanolamine plasmalogen methylation by rabbit myocardial membranes. Biochim. Biophys. Acta. 666: 205–211. [DOI] [PubMed] [Google Scholar]
- 5.Mozzi R., Gramignani D., Andriamampandr C., Freysz L., and Massarelli R.. 1989. Choline plasmalogen synthesis by the methylation pathway in chick neurons in culture. Neurochem. Res. 14: 579–583. [DOI] [PubMed] [Google Scholar]
- 6.Lee T. C., Qian C. G., and Snyder F.. 1991. Biosynthesis of choline plasmalogens in neonatal rat myocytes. Arch. Biochem. Biophys. 286: 498–503. [DOI] [PubMed] [Google Scholar]
- 7.Kryukov G. V., Castellano S., Novoselov S. V., Lobanov A. V., Zehtab O., Guigo R., and Gladyshev V. N.. 2003. Characterization of mammalian selenoproteomes. Science. 300: 1439–1443. [DOI] [PubMed] [Google Scholar]
- 8.Horibata Y., and Hirabayashi Y.. 2007. Identification and characterization of human ethanolaminephosphotransferase1. J. Lipid Res. 48: 503–508. [DOI] [PubMed] [Google Scholar]
- 9.Henneberry A. L., and McMaster C. R.. 1999. Cloning and expression of a human choline/ethanolaminephosphotransferase: synthesis of phosphatidylcholine and phosphatidylethanolamine. Biochem. J. 339: 291–298. [PMC free article] [PubMed] [Google Scholar]
- 10.Ta-Shma A., Zhang K., Salimova E., Zernecke A., Sieiro-Mosti D., Stegner D., Furtado M., Shaag A., Perles Z., Nieswandt B., et al. 2017. Congenital valvular defects associated with deleterious mutations in the PLD1 gene. J. Med. Genet. 54: 278–286. [DOI] [PubMed] [Google Scholar]
- 11.Bligh E. G., and Dyer W. J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 12.Taguchi R., Houjou T., Nakanishi H., Yamazaki T., Ishida M., Imagawa M., and Shimizu T.. 2005. Focused lipidomics by tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 823: 26–36. [DOI] [PubMed] [Google Scholar]
- 13.Zemski Berry K. A., and Murphy R. C.. 2004. Electrospray ionization tandem mass spectrometry of glycerophosphoethanolamine plasmalogen phospholipids. J. Am. Soc. Mass Spectrom. 15: 1499–1508. [DOI] [PubMed] [Google Scholar]
- 14.Frosolono M. F., and Rapport M. M.. 1969. Reactivity of plasmalogens: kinetics of acid-catalyzed hydrolysis. J. Lipid Res. 10: 504–506. [PubMed] [Google Scholar]
- 15.Nogly P., Gushchin I., Remeeva A., Esteves A. M., Borges N., Ma P., Ishchenko A., Grudinin S., Round E., Moraes I., et al. 2014. X-ray structure of a CDP-alcohol phosphatidyltransferase membrane enzyme and insights into its catalytic mechanism. Nat. Commun. 5: 4169. [DOI] [PubMed] [Google Scholar]
- 16.Epand R. M., Fuller N., and Rand R. P.. 1996. Role of the position of unsaturation on the phase behavior and intrinsic curvature of phosphatidylethanolamines. Biophys. J. 71: 1806–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Umeda M., and Emoto K.. 1999. Membrane phospholipid dynamics during cytokinesis: regulation of actin filament assembly by redistribution of membrane surface phospholipid. Chem. Phys. Lipids. 101: 81–91. [DOI] [PubMed] [Google Scholar]
- 18.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., and Yoshimori T.. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19: 5720–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanida I., Sou Y. S., Ezaki J., Minematsu-Ikeguchi N., Ueno T., and Kominami E.. 2004. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J. Biol. Chem. 279: 36268–36276. [DOI] [PubMed] [Google Scholar]
- 20.Kinoshita T., and Fujita M.. 2016. Biosynthesis of GPI-anchored proteins: special emphasis on GPI lipid remodeling. J. Lipid Res. 57: 6–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Homans S. W., Ferguson M. A., Dwek R. A., Rademacher T. W., Anand R., and Williams A. F.. 1988. Complete structure of the glycosyl phosphatidylinositol membrane anchor of rat brain Thy-1 glycoprotein. Nature. 333: 269–272. [DOI] [PubMed] [Google Scholar]
- 22.Ueda N., Tsuboi K., and Uyama T.. 2013. Metabolism of endocannabinoids and related N-acylethanolamines: canonical and alternative pathways. FEBS J. 280: 1874–1894. [DOI] [PubMed] [Google Scholar]
- 23.Panganamala R. V., Horrocks L. A., Geer J. C., and Cornwell D. G.. 1971. Positions of double bonds in the monounsaturated alk-1-enyl groups from the plasmalogens of human heart and brain. Chem. Phys. Lipids. 6: 97–102. [DOI] [PubMed] [Google Scholar]
- 24.Heymans H. S., Schutgens R. B., Tan R., van den Bosch H., and Borst P.. 1983. Severe plasmalogen deficiency in tissues of infants without peroxisomes (Zellweger syndrome). Nature. 306: 69–70. [DOI] [PubMed] [Google Scholar]
- 25.Heymans H. S., vd Bosch H., Schutgens R. B., Tegelaers W. H., Walther J. U., Müller-Höcker J., and Borst P.. 1984. Deficiency of plasmalogens in the cerebro-hepato-renal (Zellweger) syndrome. Eur. J. Pediatr. 142: 10–15. [DOI] [PubMed] [Google Scholar]
- 26.Bams-Mengerink A. M., Majoie C. B., Duran M., Wanders R. J., Van Hove J., Scheurer C. D., Barth P. G., and Poll-The B. T.. 2006. MRI of the brain and cervical spinal cord in rhizomelic chondrodysplasia punctata. Neurology. 66: 798–803, discussion 789. [DOI] [PubMed] [Google Scholar]
- 27.Dorninger F., Brodde A., Braverman N. E., Moser A. B., Just W. W., Forss-Petter S., Brugger B., and Berger J.. 2015. Homeostasis of phospholipids - The level of phosphatidylethanolamine tightly adapts to changes in ethanolamine plasmalogens. Biochim. Biophys. Acta. 1851: 117–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kou J., Kovacs G. G., Hoftberger R., Kulik W., Brodde A., Forss-Petter S., Honigschnabl S., Gleiss A., Brugger B., Wanders R., et al. 2011. Peroxisomal alterations in Alzheimer’s disease. Acta Neuropathol. 122: 271–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodenowe D. B., Cook L. L., Liu J., Lu Y., Jayasinghe D. A., Ahiahonu P. W., Heath D., Yamazaki Y., Flax J., Krenitsky K. F., et al. 2007. Peripheral ethanolamine plasmalogen deficiency: a logical causative factor in Alzheimer’s disease and dementia. J. Lipid Res. 48: 2485–2498. [DOI] [PubMed] [Google Scholar]
- 30.Fabelo N., Martin V., Santpere G., Marin R., Torrent L., Ferrer I., and Diaz M.. 2011. Severe alterations in lipid composition of frontal cortex lipid rafts from Parkinson’s disease and incidental Parkinson’s disease. Mol. Med. 17: 1107–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy E. J., Schapiro M. B., Rapoport S. I., and Shetty H. U.. 2000. Phospholipid composition and levels are altered in Down syndrome brain. Brain Res. 867: 9–18. [DOI] [PubMed] [Google Scholar]
- 32.Kaddurah-Daouk R., McEvoy J., Baillie R., Zhu H., Yao J. K., Nimgaonkar V. L., Buckley P. F., Keshavan M. S., Georgiades A., and Nasrallah H. A.. 2012. Impaired plasmalogens in patients with schizophrenia. Psychiatry Res. 198: 347–352. [DOI] [PubMed] [Google Scholar]
- 33.van der Knaap M. S., Breiter S. N., Naidu S., Hart A. A., and Valk J.. 1999. Defining and categorizing leukoencephalopathies of unknown origin: MR imaging approach. Radiology. 213: 121–133. [DOI] [PubMed] [Google Scholar]
- 34.Wood P. L., Smith T., Pelzer L., and Goodenowe D. B.. 2011. Targeted metabolomic analyses of cellular models of pelizaeus-merzbacher disease reveal plasmalogen and myo-inositol solute carrier dysfunction. Lipids Health Dis. 10: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahmed M. Y., Al-Khayat A., Al-Murshedi F., Al-Futaisi A., Chioza B. A., Pedro Fernandez-Murray J., Self J. E., Salter C. G., Harlalka G. V., Rawlins L. E., et al. 2017. A mutation of EPT1 (SELENOI) underlies a new disorder of Kennedy pathway phospholipid biosynthesis. Brain. 140: 547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voelker D. R. 1984. Phosphatidylserine functions as the major precursor of phosphatidylethanolamine in cultured BHK-21 cells. Proc. Natl. Acad. Sci. USA. 81: 2669–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller M. A., and Kent C.. 1986. Characterization of the pathways for phosphatidylethanolamine biosynthesis in Chinese hamster ovary mutant and parental cell lines. J. Biol. Chem. 261: 9753–9761. [PubMed] [Google Scholar]
- 38.Vance J. E. 2008. Phosphatidylserine and phosphatidylethanolamine in mammalian cells: two metabolically related aminophospholipids. J. Lipid Res. 49: 1377–1387. [DOI] [PubMed] [Google Scholar]
- 39.Bleijerveld O. B., Brouwers J. F., Vaandrager A. B., Helms J. B., and Houweling M.. 2007. The CDP-ethanolamine pathway and phosphatidylserine decarboxylation generate different phosphatidylethanolamine molecular species. J. Biol. Chem. 282: 28362–28372. [DOI] [PubMed] [Google Scholar]
- 40.Sundler R., Akesson B., and Nilsson A.. 1974. Quantitative role of base exchange in phosphatidylethanolamine synthesis in isolated rat hepatocytes. FEBS Lett. 43: 303–307. [DOI] [PubMed] [Google Scholar]
- 41.Bjerve K. S. 1984. Phospholipid substrate-specificity of the L-serine base-exchange enzyme in rat liver microsomal fraction. Biochem. J. 219: 781–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao J., Shan D., Revett T., Li D., Wu L., Liu W., Tobin J. F., and Gimeno R. E.. 2008. Molecular identification of a novel mammalian brain isoform of acyl-CoA:lysophospholipid acyltransferase with prominent ethanolamine lysophospholipid acylating activity, LPEAT2. J. Biol. Chem. 283: 19049–19057. [DOI] [PubMed] [Google Scholar]
- 43.Shindou H., Hishikawa D., Harayama T., Eto M., and Shimizu T.. 2013. Generation of membrane diversity by lysophospholipid acyltransferases. J. Biochem. 154: 21–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.