Abstract
In this study, we present the clinicopathological features associated with PD-L1 protein and mRNA expression in a large Asian cohort of patients with non-small cell lung cancer (NSCLC) and assessed the prognostic implications of PD-L1 expression, particularly in early stage NSCLC. We retrospectively analyzed 687 NSCLC specimens (476 adenocarcinoma and 211 squamous cell carcinoma) using tissue microarray. PD-L1 immunohistochemistry (IHC) was performed using Dako 22C3 pharmDx assay and PDL1 mRNA was measured using RNA in situ hybridization (RISH). The overall prevalence of PD-L1 protein expression was 25.2% in tumor cells and PDL1 mRNA expression was 11.9%. There was a strong positive correlation between PD-L1 IHC and RISH results (Spearman’s rho = 0.6, p<0.001). In adenocarcinoma, PD-L1 protein and mRNA expressions significantly correlated with poorly differentiated histologic subtype (p<0.001 and p = 0.002, respectively). PD-L1 expression was also associated with genetic alteration in adenocarcinoma. High PD-L1 expression level was associated with EGFR-naïve and KRAS-mutant subgroup (p = 0.001 and p = 0.017, respectively). With a 1% cut-off value, PD-L1 protein expression showed a short overall survival duration in early stage adenocarcinoma with marginal significance (p = 0.05, Hazard ratio = 1.947). Our study revealed that PD-L1 expression varied with histologic subtype and genomic alteration status in lung adenocarcinoma, and activation of the PD-L1 pathway may be a poor prognostic factor especially in early stage lung adenocarcinoma. In addition, PDL1 RISH showed promising results in predicting PD-L1 protein expression in NSCLC.
Introduction
Researchers have recently become interested in developing immunotherapies for the treatment of non-small cell lung cancer (NSCLC), particularly monoclonal antibodies targeting the programmed cell death-1 (PD-1) receptor and its ligand (PD-L1) [1, 2]. Interaction of PD-1 with PD-L1 inhibits T-cell activation, allowing tumor cells to bypass immune surveillance. Therefore, blockade of the PD-1/PD-L1 axis may enhance the active immune response against tumors. Currently, different types of monoclonal antibodies targeting PD-1 or PD-L1, including nivolumab for NSCLC with squamous cell histology [3] and non-squamous cell histology [4] in the second-line setting, pembrolizumab for NSCLC with high PD-L1 expression (≥ 50%) in the first-line setting [5] or in the second-line setting for tumors with 1–49% PD-L1 expression [6], and atezolizumab for all subtypes of NSCLC in the second-line setting [7], are available. Responses to PD-1/PD-L1 inhibitors are improved in patients with high tumor PD-L1 expression compared with those exhibiting low or no PD-L1 [4–6,8]. Therefore, PD-L1 protein expression is the only biomarker that can predict which patients are more likely to respond to anti-PD-1/PD-L1 therapy in the clinical setting. However, the correlations between PD-L1 expression in tumor cells and treatment response to anti-PD-1 or anti-PD-L1 therapy is still unclear because almost 10% of patients with PD-L1-negative tumors also responded to PD-1/PD-L1 inhibitors in the above clinical trials [4,6].
Besides acting as a predictive biomarker, PD-L1 shows inconsistent results among various studies as a prognostic biomarker. Studies investigating the prognostic role of PD-L1 and its association with clinicopathological features and driver mutations in NSCLC have yielded quite different results [9–13]. This discrepancy may be attributed to differences in ethnicity, heterogeneous histological subtypes, and stages. Furthermore, clinical trials with checkpoint inhibitors have focused on advanced, inoperable tumors; thus, data reporting the predictive and prognostic roles of PD-L1 expression in early-stage NSCLC are limited.
The variety of PD-L1 immunohistochemical (IHC) assays, involving the use of different antibodies and interpretation criteria, may also contribute to the lack of consistent results [14]. Given the difficulties associated with PD-L1 IHC, an alternative method for accurately evaluating PD-L1 expression is needed. An antibody-independent assay for RNA in situ hybridization (RISH) in formalin-fixed, paraffin-embedded (FFPE) tumor tissues using an RNAscope assay has been favored for its specificity and interpretative objectivity. In gastric cancer and small cell lung cancer, PDL1 mRNA exhibited a positive nonlinear relationship with PD-L1 protein using this assay, suggesting the potential applications of the RNAscope assay in future clinical studies [15,16].
In this study, we evaluated the clinicopathological features associated with PD-L1 protein and mRNA expression in a large Asian cohort of patients with NSCLC and investigated the prognostic implications of PD-L1 expression, particularly in early stage NSCLC.
Materials and methods
Patients and samples
Our cohort consisted of 687 patients with NSCLC, including 476 with adenocarcinoma (ADC) and 211 with squamous cell carcinoma (SqCC) who underwent surgical resection between May 2003 and December 2012 at Seoul National University Bundang Hospital. None received pre-operative chemotherapy or radiation therapy. Clinicopathological information was obtained from clinical records and pathology reports. The pathologic staging was based on the 7th edition of the American Joint Committee on Cancer staging manual [17]. The study protocol was approved by the Institutional Review Board of Seoul National University Bundang Hospital (B-1704/393-303).
Histological analyses
All resected tumor specimens were fixed with formalin and then stained with hematoxylin and eosin (H&E). All H&E slides were carefully reviewed by two of the authors (H. Kim and J.H. Chung) to confirm the original diagnosis and classify the histological subtype. ADC in situ and minimally invasive ADC samples were excluded from the study. All other invasive ADC samples were categorized as lepidic, papillary, acinar, micropapillary, solid, or invasive mucinous according to the 2015 World Health Organization Classification of Lung Tumors [18]. These histological subtypes were used to determine tumor grade (lepidic, well differentiated; acinar and papillary, moderately differentiated; and micropapillary and solid, poorly differentiated).
Construction of the tissue microarray (TMA)
The slides were independently reviewed by two pathologists (H. Kim and J.H. Chung) to select the most representative sections. The most representative tumor area was carefully marked on the H&E-stained slide of each sample tissue. A TMA was constructed using 2-mm-diameter cores derived from the representative tumor areas selected at random of the FFPE tissue blocks from each case by SuperBioChips Laboratories (Seoul, Korea).
IHC analysis of PD-L1 protein
TMAs were sectioned at a thickness of 4-μm and stained using the Dako pharmDx assay. Briefly, the slides were stained with anti-PD-L1 22C3 mouse monoclonal primary antibodies with the EnVision FLEX visualization system on a Dako Autostainer Link 48 instrument (Carpinteria, CA, USA), along with negative control reagents and cell line run controls, as per the manufacturer’s instructions. The IHC slides were scored independently by two pathologists (H.J. Kwon and H. Kim). PD-L1 was considered positive in tumor cells only in cases of at least 100 viable tumor cells, if membranous staining alone or membranous and cytoplasmic staining together was present. Membranous staining in tumor cells directly adjacent to immune cells was not considered positive if the surface touching immune cells was the only stained part. The percentage of stained cells in the overall area of the tumor (Tumor Proportion Score) was scored regardless of intensity [6]. Cases were then classified by two different cut-off values, 1% and 50%, based on the published association of this cut-off with anti-PD-1 therapeutic response [6].
RNA in situ hybridization of PDL1 mRNA
PDL1 mRNAs were measured using RNAscope assays (Advanced Cell Diagnostics [ACD], Hayward, CA, USA) following the manufacturer’s instructions [19]. Briefly, 5-μm-thick sections were deparaffinized; incubated with pretreatment reagents 1, 2, and 3 at room temperature for 10 min; boiled for 15 min; and incubated at 40°C for 30 min. TMA sections were then hybridized with Hs-CD274-probes (ACD) at 40°C for 2 h. Hybridization signals were amplified and visualized with an RNAscope 2.0 HD detection kit (Red). RNAscope results were examined under a standard bright field microscope at 200–400× magnification. Positive signals presented as red punctuate dots. PPIB and DapB were used as positive and negative probes, respectively, to control tissue RNA conditions and nonspecific hybridization.
PD-L1 mRNA signals were in the tumor compartment or mesenchyme, as visualized by red dotted or clustered patterns. No standard scoring criteria for PD-L1 mRNA expression in NSCLC had been determined; therefore, we adopted the RNAscope system scoring guidelines (“RNA scope score”): 0 (no staining or < 1 dot per 10 cells); 1 (1–3 dots per cell); 2 (4–9 dots per cell); 3 (10–15 dots per cell); and 4 (> 15 dots per cell and > 10% dots in clusters) [19]. We also evaluated the tumor proportion that showed at least 1 dot. We classified signals according to the proportion as follows: 0 (0 and < 1%); 1 (1–9%); 2 (10–49%); and 3 (50–100%), which was defined as the “RNA proportion score”. Because PD-L1 RNA scope and proportion scores showed a linear correlation (r = 0.83, p < 0.01, data not shown), cases showing either an RNA scope score of 1 or more or an RNA proportion score of 1 or more were designated as PD-L1 mRNA positive.
Detection of mutations in EGFR and KRAS and rearrangement of the ALK gene
Polymerase chain reaction and DNA sequencing with FFPE tissue samples were used to analyze EGFR mutations in exons 18–21 and KRAS mutations at codons 12, 13, and 61, as described previously [20]. Rearrangement of the ALK gene was assessed using fluorescence in-situ hybridization with an ALK probe (Vysis LSI ALK Break Apart Rearrangement probe; Abbott Molecular, Park, IL, USA) and a 15% cut-off value, as described previously [20].
Statistical analysis
Statistical analysis was carried out using Stata Statistical Software version 14 (Stata Corp., College Station, TX, USA) and R program (R Foundation for Statistical Computing, Vienna, Austria). Spearman’s test and logistic regression were performed to compare assays and determine appropriate cut-off values. Cohen’s ĸ coefficient of agreement was obtained to cross-check the results. A Kaplan-Meier analysis was performed to construct survival curves, and statistical significance was assessed using log-rank tests. A multivariate analysis was performed by Cox proportional hazards regression modeling. All statistical tests were two sided, and statistical significance was accepted for p values of less than 0.05.
Results
Clinicopathological characteristics
Clinicopathological characteristics are summarized in Table 1. Briefly, there were a total of 429 men (62.4%) and 258 women (37.6%) with a median age of 64 years (range: 21–85 years). Approximately half of the patients were never smokers (n = 297; 43.2%). This may be the reason that ADC was the most prevalent histological subtype (n = 476; 69.3%). The pathological stage was I in 359 patients (52.2%), II in 162 patients (23.6%), III in 141 patients (20.6%), and IV in 25 patients (3.6%).
Table 1. Clinicopathological characteristics.
| Adenocarcinoma | Squamous cell carcinoma | Total | |||||
|---|---|---|---|---|---|---|---|
| Characteristic | Number of cases | % | Number of cases | % | Number of cases | % | |
| Age (year) | |||||||
| Median (range) | 64 (21–83) | 68 (31–85) | 64 (21–85) | ||||
| Sex | |||||||
| Male | 229 | 48.1 | 200 | 94.8 | 429 | 62.4 | |
| Female | 247 | 51.9 | 11 | 5.2 | 258 | 37.6 | |
| Smoking status¶ | |||||||
| Never smoker | 284 | 59.7 | 13 | 6.2 | 297 | 43.2 | |
| Current smoker | 91 | 19.1 | 122 | 57.8 | 213 | 31.0 | |
| Ex-smoker | 101 | 21.2 | 76 | 36.0 | 177 | 25.8 | |
| Tumor size (cm) | |||||||
| Mean (range) | 3.1 (0.5–16.0) | 4.0 (0.8–14.5) | 3.4 (0.5–16.0) | ||||
| Pleural invasion | |||||||
| Absent | 272 | 57.1 | 149 | 70.6 | 421 | 61.3 | |
| Present | 204 | 42.9 | 62 | 29.4 | 266 | 38.7 | |
| Venous invasion | |||||||
| Absent | 361 | 75.8 | 168 | 79.6 | 529 | 77.0 | |
| Present | 115 | 24.2 | 43 | 20.4 | 158 | 23.0 | |
| Lymphatic invasion | |||||||
| Absent | 248 | 52.1 | 130 | 61.6 | 378 | 55.0 | |
| Present | 228 | 47.9 | 81 | 38.4 | 309 | 45.0 | |
| Pathologic stage | |||||||
| I | 271 | 56.9 | 88 | 41.7 | 359 | 52.5 | |
| II | 90 | 18.9 | 72 | 34.1 | 162 | 23.6 | |
| III | 96 | 20.2 | 45 | 21.3 | 141 | 20.6 | |
| IV | 19 | 4.0 | 6 | 2.9 | 25 | 3.6 | |
| PD-L1 protein expression | |||||||
| < 1% | 399 | 83.8 | 115 | 54.5 | 514 | 74.8 | |
| 1–49% | 48 | 10.1 | 57 | 27.0 | 105 | 15.3 | |
| ≥ 50% | 29 | 6.1 | 39 | 18.5 | 68 | 9.9 | |
| PDL1 mRNA expression | |||||||
| Negative | 447 | 93.9 | 158 | 74.9 | 605 | 88.1 | |
| Positive | 29 | 6.1 | 53 | 25.1 | 82 | 11.9 | |
| Total | 476 | 69.3 | 211 | 30.7 | 687 | 100 | |
PD-L1, programmed cell death ligand-1; mRNA, messenger RNA
¶ smoking status was defined as follows: never smoker (<100 cigarettes per lifetime); current smoker (≥100 cigarettes per lifetime and smoked at the time of lung cancer diagnosis or quit ≤1 year prior to the diagnosis); ex-smoker (≥100 cigarettes per lifetime and quit >1 year prior to the diagnosis)
PD-L1 protein and mRNA expression
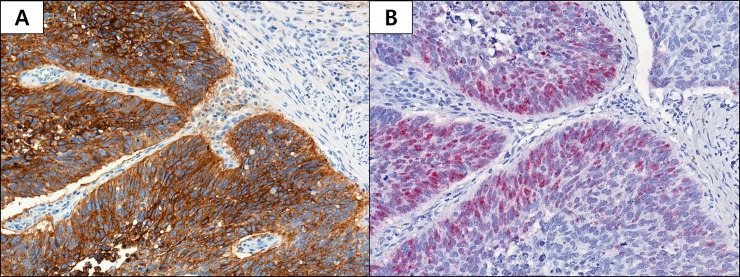
The overall prevalence of PD-L1 protein expression in tumor cells was 25.2% (173/687; Table 1). With a 1% cut-off, PD-L1 was positive in 16.2% (77/476) and 45.5% (96/211) of ADC and SqCC, respectively. With a 50% cut-off, the positive rates were 6.1% (29/476) for ADC and 18.5% (39/211) for SqCC. Thus, PD-L1 protein appeared to be present in a higher percentage of SqCC samples than ADC samples (p < 0.05). PD-L1 mRNA expression was detected in 11.9% (82/687) of patients. In subgroup analysis, SqCC showed higher mRNA positivity than ADC (25.1% versus 6.1%; p < 0.05). Fig 1 shows representative images of PD-L1 protein (Fig 1A) and mRNA expression (Fig 1B) based on IHC and RNAscope, respectively. Membranous expression of PD-L1 IHC can be readily seen in A, while in B, red dotted or clustered PDL1 mRNA signals can be noted.
Fig 1.
Programmed Death Ligand 1 (PD-L1) Protein (A) and mRNA (B) Expression in Non-small Cell Lung Cancer. (A) Membranous expression of PD-L1 protein in tumor cells (20× magnification). (B) PD-L1 mRNA signals located in the nucleus and mesenchyme within tumor compartments are denoted by red dotted or clustered patterns (20× magnification).
Correlation between PD-L1 protein and mRNA expression
PD-L1 protein expression showed a strong positive correlation with PD-L1 mRNA expression (Spearman’s rho = 0.6, p < 0.001). As the TPS of PD-L1 protein expression increased, PDL1 mRNA expression was observed frequently (Table 2).
Table 2. Correlation between PD-L1 mRNA and protein expression.
| IHC TPS (%) | mRNA expression (number, %) | Total | |
|---|---|---|---|
| Negative | Positive | ||
| <1 | 500 (97.3) | 14 (2.7) | 514 (74.8) |
| 1–49 | 85 (80.9) | 20 (19.1) | 105 (15.3) |
| ≥50 | 20 (29.4) | 48 (70.6) | 68 (9.9) |
| Total | 605 (88.1) | 82 (11.9) | 687 (100) |
IHC, immunohistochemistry; TPS, tumor proportion score
We calculated the overall percentage agreement (OPA) pairwise between assays at two PD-L1 IHC cut-off values (1% and 50%). OPA with 1% and 50% IHC cut-off values were 80.1% and 91.1%, respectively (Table 3). Positive and negative percentage agreement (PPA and NPA) were calculated for mRNA assays against the IHC (1% and 50% cut-off values). With a 1% cut-off, the PPA and NPA of mRNA assays were 78.1% and 80.3%, respectively. Applying a 50% cut-off, PPA was decreased (46.9%), whereas NPA was increased (95.7%; Table 3).
Table 3. Agreement between PD-L1 protein and mRNA expression results.
| PD-L1 IHC 1% cutoff | PD-L1 IHC 50% cutoff | |||||
|---|---|---|---|---|---|---|
| OPA | PPA | NPA | OPA | PPA | NPA | |
| PDL1 mRNA | 80.1% | 78.1% | 80.3% | 91.1% | 46.9% | 95.7% |
PD-L1, Programmed cell death ligand-1; IHC, immunohistochemistry; mRNA, messenger RNA; OPA, overall percentage agreement; PPA, positive percentage agreement; NPA, negative percentage agreement
Association between PD-L1 status and clinicopathological parameters
Next, we investigated the associations between PD-L1 status and clinicopathological parameters in ADC and SqCC. In ADC, both PD-L1 protein and mRNA expression were correlated with histologic subtype (p < 0.001 and p = 0.002, respectively; Table 4). PD-L1 showed higher expression in the poorly differentiated (solid and micropapillary predominant) histologic subgroup than in the well-differentiated (lepidic predominant) subgroup (Fig 2). PD-L1 expression was also associated with genetic alterations. Although PD-L1 protein and mRNA expression levels were lower in the EGFR-mutated group than in the EGFR-negative group (p = 0.001 and p = 0.016, respectively), PD-L1 protein expression was higher in the KRAS-mutated group than in the KRAS-negative group (p = 0.017). Smoking history, pathological stage, and ALK status were not associated with PD-L1 status. In SqCC, only tumor size was associated with PD-L1 protein expression with marginal significance (p = 0.049; data not shown).
Table 4. Association between PD-L1 status and clinicopathologic variables in lung adenocarcinoma.
| PD-L1 IHC (number (%)) | PD-L1 RNA scope (number (%)) | |||||||
|---|---|---|---|---|---|---|---|---|
| Total | Negative | Weak positive | Strong positive | p value | Negative | Positive | p value | |
| Smoking history | ||||||||
| Yes | 192 | 156 (81.3%) | 22 (11.5%) | 14 (7.3%) | > 0.05 | 180 (93.8%) | 12 (6.2%) | > 0.05 |
| No | 284 | 243 (85.6%) | 26 (9.2%) | 15 (5.3%) | 267 (94.0%) | 17 (6.0%) | ||
| Histologic subtype ¶ | ||||||||
| WD | 42 | 41 (97.6%) | 1 (2.4%) | 0 | <0.001 | 41 (97.6%) | 1 (2.4%) | 0.002 |
| MD | 360 | 309 (85.8%) | 33 (9.2%) | 18 (5.0%) | 343 (95.3%) | 17 (4.7%) | ||
| PD | 68 | 43 (62.7%) | 14 (20.9%) | 11(16.4%) | 57 (83.6%) | 11 (16.4%) | ||
| Mucinous | 6 | 6 (100%) | 0 | 0 | 6 (100%) | 0 | ||
| Pathologic stage | ||||||||
| IA-IIA | 340 | 286 (84.1%) | 33 (9.7%) | 21 (6.2%) | > 0.05 | 320 (94.1%) | 20 (5.9%) | > 0.05 |
| IIB-IV | 136 | 113 (83.1%) | 15 (11.0%) | 8 (5.9%) | 127 (93.4%) | 9 (6.6%) | ||
| EGFR mutation | ||||||||
| Present | 223 | 201 (90.1%) | 17 (7.6%) | 5 (2.2%) | 0.001 | 216 (96.9%) | 7 (3.1%) | 0.016 |
| Absent | 229 | 179 (78.2%) | 27 (11.8%) | 23 (10.0%) | 209 (91.3%) | 20 (8.7%) | ||
| KRAS mutation | ||||||||
| Present | 23 | 15 (65.2%) | 4 (17.4%) | 4 (17.4%) | 0.017 | 20 (87.0%) | 3 (13.0%) | > 0.05 |
| Absent | 237 | 203 (85.7%) | 23 (9.7%) | 11 (4.6%) | 223 (94.1%) | 14 (5.9%) | ||
| ALK rearrangement | ||||||||
| Present | 24 | 21 (87.5%) | 1 (4.2%) | 2 (8.3%) | > 0.05 | 24 (100%) | 0 | > 0.05 |
| Absent | 181 | 161 (89.0%) | 12 (6.6%) | 8 (4.4%) | 177 (97.8%) | 4 (2.2%) | ||
PD-L1, Programmed cell death ligand-1; IHC, immunohistochemistry; WD, well differentiated; MD, moderately differentiated; PD, poorly differentiated; EGFR, Epidermal growth factor receptor; KRAS, Kirsten rat sarcoma 2 viral oncogene homolog; ALK, Anaplastic lymphoma kinase.
¶ lepidic, well differentiated; acinar and papillary, moderately differentiated; and micropapillary and solid, poorly differentiated.
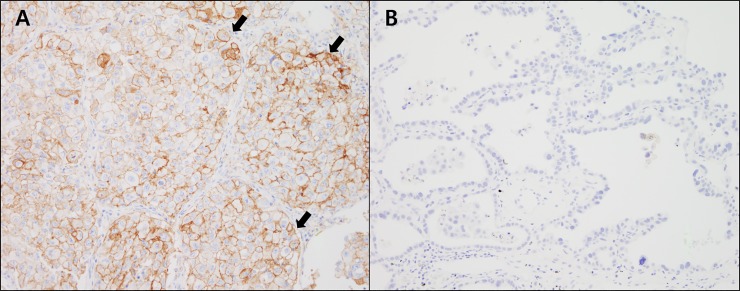
Fig 2. Microphotographs of representative examples of PD-L1 protein expression according to histological subtypes of lung adenocarcinoma.
(A) PD-L1 protein is expressed in tumor cell membranes (>50%) in 37.3% of solid predominant ADC (arrowheads). (B) In contrast, PD-L1 was not expressed in most lepidic predominant ADC (97.6%). (A and B, 20× magnification).
Survival analysis
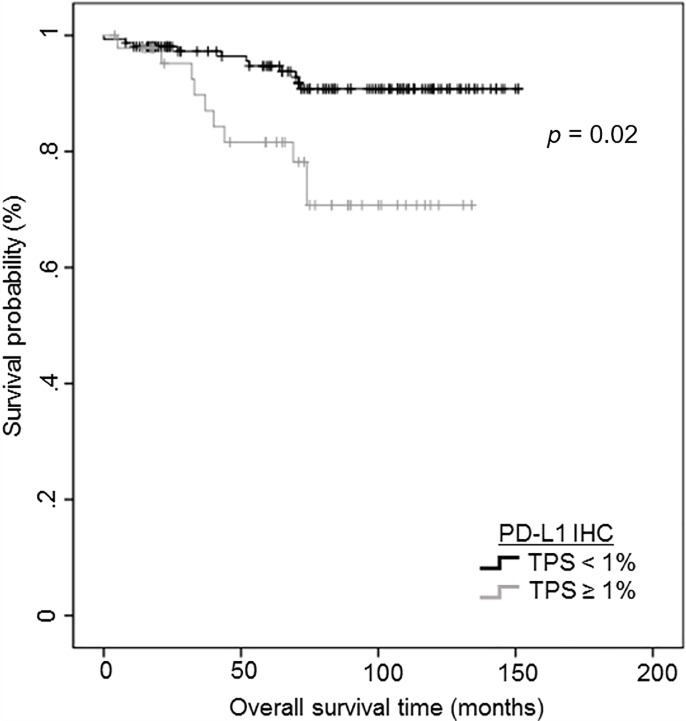
We performed survival analysis to investigate the prognostic role of PD-L1 protein and mRNA expression in ADC and SqCC. In ADC, lymphovascular/perineural invasion and pathologic TNM stage were independent poor prognostic factors for disease-free survival (DFS) and overall survival (OS; Table 5A). PD-L1 expression was not associated with patient survival in the full cohort of patients with ADC. To investigate the prognostic significance of PD-L1 expression, we performed survival analysis in the early stage (I and IIA) ADC subgroup containing 340 patients. In subgroup analysis, PD-L1 protein expression over 1% was associated with shorter OS using univariate analysis (p = 0.02) (Fig 3) and tended to show poor prognosis with marginal significance (p = 0.05, hazard ratio: 1.947) in multivariate analysis after adjusting for the conventional clinicopathological covariates (Table 5B). For PD-L1 mRNA, there were no significant survival differences in both the full cohort and early stage ADC subgroup. In SqCC, PD-L1 protein and mRNA expression levels were not associated with survival (data not shown).
Table 5. Survival analysis in full cohort (A) and early-stage subgroup (B) of lung adenocarcinoma.
| A (n = 476) | |||||||
| Disease-free survival | Overall survival | ||||||
| Univariate | Multivariate | Univariate | Multivariate | ||||
| Clinicopathologic variables | Category | p value | p value | HR (95% CI) | p value | p value | HR (95% CI) |
| sex | Male vs female | 0.947 | - | 0.028 | 0.669 | ||
| age | ≥66 vs <66 | 0.038 | 0.07 | < 0.001 | < 0.001 | 2.340 (1.556–3.497) | |
| smoking history | ever vs never | 0.934 | 0.019 | 0.068 | |||
| histologic subtype | PD vs. WD/MD | 0.003 | 0.42 | 0.002 | 0.412 | ||
| pleural invasion | Present vs. absent | < 0.001 | < 0.001 | 0.031 | 1.575 (1.042–2.379) | ||
| vascular invasion | Present vs. absent | < 0.001 | 0.026 | 1.444 (1.045–1.995) | < 0.001 | 0.693 | |
| lymphatic invasion | Present vs. absent | < 0.001 | 0.003 | 1.667 (1.214–2.372) | < 0.001 | 0.006 | 1.885 (1.197–2.967) |
| Perineural invasion | Present vs. absent | 0.04 | 0.665 | 0.009 | 0.301 | ||
| pTNM stage | IIB, III and IV vs I, IIA | <0.001 | <0.001 | 2.129 (1.564–2.899) | < 0.001 | < 0.001 | 2.684 (1.788–4.029) |
| PD-L1 protein expression | >1% vs <1% | 0.196 | 0.054 | ||||
| >50% vs <50% | 0.382 | 0.381 | |||||
| PDL1 mRNA expression | positive vs negative | 0.488 | 0.127 | ||||
| B (n = 340) | |||||||
| Disease-free survival | Overall survival | ||||||
| Univariate | Multivariate | Univariate | Multivariate | ||||
| Clinicopathologic variables | Category | p value | p value | HR (95% CI) | p value | p value | HR (95% CI) |
| sex | Male vs female | 0.475 | 0.13 | ||||
| age | ≥66 vs <66 | 0.749 | 0.021 | 0.043 | 1.830(1.020–3.284) | ||
| smoking history | ever vs never | 0.501 | 0.01 | 0.017 | 2.026(1.133–3.624) | ||
| histologic subtype | PD vs. WD/MD | 0.015 | 0.255 | 0.086 | |||
| pleural invasion | Present vs. absent | 0.001 | 0.25 | 0.006 | 0.032 | 1.908(1.057–3.443) | |
| vascular invasion | Present vs. absent | 0.006 | 0.352 | 0.204 | |||
| lymphatic invasion | Present vs. absent | <0.001 | 0.069 | 0.001 | 0.02 | 2.038(1.120–3.709) | |
| Perineural invasion | Present vs. absent | 0.11 | 0.037 | 0.074 | |||
| pTNM stage | IIA vs. I | <0.001 | 0.008 | 2.323(1.361–3.966) | 0.001 | 0.164 | 2.194(1.140–4.001) |
| PD-L1 protein expression | >1% vs <1% | 0.129 | 0.02 | 0.05 | 1.947(1.000–3.791) | ||
| >50% vs <50% | 0.416 | 0.064 | |||||
| PD-L1 mRNA expression | positive vs negative | 0.887 | 0.909 | ||||
PD-L1, Programmed cell death ligand-1; mRNA, messenger RNA; HR, hazard ratio; CI, confidence interval; WD, well differentiated; MD, moderately differentiated; PD, poorly differentiated
Fig 3. Kaplan–Meier curves depicting the prognostic impact of programmed cell death ligand-1 (PD-L1) protein expression on overall survival in early stage (I and IIA) non-small cell lung cancer subgroup.
Cases with PD-L1 protein expression over 1% showing a shorter overall survival duration (p = 0.02).
Discussion
In this study, we demonstrated that PD-L1 expression was significantly associated with histologic grade and genetic alteration status in lung ADC. We also found that PD-L1 protein expression was an adverse prognostic marker for OS in patients with early stage lung ADC. We also assessed PD-L1 mRNA expression and compared PD-L1 protein and mRNA expression; the results showed that PD-L1 mRNA was a potential surrogate marker, with a positive correlation with protein expression.
Our study showed that PD-L1 protein and mRNA expression levels were significantly associated with high histologic grade and solid subtype of ADC. Our results are in line with the results of several studies which reported the correlation between PD-L1 expression in tumor cells and poor differentiation and solid histology [9–12]. This finding may be clinically useful when small biopsies from patients show negative PD-L1 expression, but only the lepidic component was biopsied. Because small biopsies may miss the region of the tumor with high PD-L1 expression due to the heterogeneity issue [21], re-biopsy could be considered in solid tumor areas to ensure that the patient is a candidate. From our experience, PD-L1 was found to be strongly expressed in poorly differentiated cells but negative in papillary and lepidic components in a small biopsy specimen (not published data). In particular, in the case of pembrolizumab, which has received FDA approval as first line therapy for metastatic NSCLC, accurate evaluation of PD-L1 expression in advanced stage patients, which can only be performed with biopsy, is crucial for identifying patients to be a candidate to anti-PD-1 therapy [5, 22].
From biological point of view, elevated expression of PD-L1 poorly differentiated lung ADCs compared with well-differentiated ADCs might account the inactivation of effector-immune cells through PD-1 receptor signaling which could ultimately enhance tumor progression. This relationship supports the results of our study that ADCs with high expression of PD-L1 are associated with poorly differentiated histology and poor prognosis.
The relationship between EFGR mutation status and PD-L1 expression in NSCLC is still unclear. Although preclinical studies have suggested that EGFR-driven NSCLC inhibits antitumor immunity through activation of the PD-1/PD-L1 pathway in an intrinsic manner, epidemiological studies have suggested that EGFR-mutant NSCLC is more likely to exhibit decreased PD-L1 expression. Two recent pooled analyses have provided further support for this inverse relationship. In one study, patients harboring EGFR mutations were more likely to have decreased PD-L1 expression (odds ratio: 1.79, 95% confidence interval: 1.10–2.93) [23], and in another study, PD-L1 expression was associated with EGFR wild-type status (odds ratio: 0.61, 95% confidence interval: 0.42–0.90, P = 0.01) [24]. One reason for these conflicting results between EGFR mutations and PD-L1 expression could be the variability, including the assessment of biomarkers from a single lesion site at a single time point, which often provides poor insights into spatiotemporal dynamics. For example, PD-L1 expression has been shown to fluctuate during EGFR tyrosine kinase inhibitor (TKI) and post-progression [25]. In our study, none of the patients received EGFR TKI treatment, which could exclude the temporal heterogeneity of PD-L1 status. PD-L1 protein and mRNA expression were lower in the EGFR-mutated group than in the EGFR-negative group. Low PD-L1 expression in EGFR-mutated ADC may be related to the lower prevalence of PD-L1 in the Asian population than in Western populations. In our cohort, PD-L1 expression was observed in 16.2% of patients with a 1% cut-off and in only 6.1% of patients with a 50% cut-off. Several reports have shown low PD-L1 prevalence in lung ADC of Asian patients [26]. Thus, the difference in EGFR mutation prevalence may be one of the causes of ethnic differences in PD-L1 expression.
The prognostic impact on PD-L1 expression in NSCLC is still controversial [2,27], but our results along with several reports addressed the association of PD-L1 expression and poor clinical outcomes[11,28]. Koh et al. demonstrated that PD-L1 expression is a poor prognostic factor of DFS in patients with pulmonary ADC [11]. They suggested that PD-L1 expression in tumor cells and infiltration of PD-1+/CD8+ tumor infiltrating lymphocytes may not only induce T-cell exhaustion but also inhibit tumor cell death. A meta-analysis with 1,550 patients with NSCLC from nine studies has also demonstrated that PD-L1 protein expression in NSCLC is associated with poor prognosis [28]. Another meta-analysis reported that PD-L1 expression was associated with poor patient outcome in only Asian NSCLC subgroup, suggesting that ethnic difference might be associated with the prognostic implication of PD-L1 [29]. These discrepancies may be due to differences in the PD-L1 assay method, heterogeneity according to the NSCLC subtype, and various stages and treatment modalities [29]. To minimize these issues, we investigated the associations between PD-L1 expression and survival in a large cohort of patients with NSCLC, including many early stage tumors that had been resected with curative intention. Our study demonstrated that PD-L1 protein expression was a poor prognostic factor affecting overall survival in patients with early stage lung ADC, but had no prognostic value in patients with SqCC histology.
In this study, PD-L1 expression was found as frequently in stages I and II (24.8% and 27.8%, respectively) as in stages III and IV (23.4% and 24.0%, respectively), indicating that aberrant expression of this ligand may be an early event. Patients with early stage NSCLC may have a more intact immune system and the potential for long-lasting immune priming against micrometastases [30]. Therefore, immunotherapy for early stage cancer could increase the cure rate, reduce tumor burden, and enable local approaches (such as surgery) in additional patients, taking advantage of minimal residual disease. Ongoing clinical trials have been investigating the effects of neoadjuvant or adjuvant immunotherapy for resectable early stage lung cancer, and several studies have shown promising results. PD-L1 expression may be important not only as a prognostic factor but also as a predictive biomarker for early stage lung cancer immunotherapy. Thus, further studies are needed to determine whether PD-L1 may be related to drug response in early stage disease.
Finally, we assessed PD-L1 expression using IHC and RISH. Many studies have attempted to assess PD-L1 expression using different techniques, including RISH [15,16]. The OPA of RISH was over 80% compared with the FDA-approved IHC method, and our study illustrated the possible application of RISH as a complementary diagnostic test, providing accurate detection of PD-L1 in NSCLC. However, there was discrepancy in the expression of PD-L1 protein and mRNA. Among 68 cases with PD-L1 protein expression over 50% of tumor cells, 20 cases (29.4%) were confirmed no PD-L1 mRNA expression on RISH. There were several consideration of the discrepancy. First of all, RNA is a weaker molecule compared with DNA or protein, so RNA is more sensitive to procedures of fixation and deproteinization. Secondly, RISH is a semi-automatic procedure including the incubation step with the probe as a manual procedure, that could also affect the results. Although the novel RISH assay used in our study provide a higher level of target sensitivity and specificity when compared with many IHC protocols achieved using by Z-pairs oligonucleotides [19], there might be analytic variables, especially during manual procedures. Lastly, PD-L1 protein expression may be affect by post-transcriptional modification. Emerging evidence supports that PD-L1 expression is regulated on a post-transcriptional and translational level by various intracellular pathway [31,32]. These molecules can induce or suppress PD-L1 protein expression via PI3K/Akt signaling pathway or IFNγ pathway without PDL1 mRNA expression. Further investigation of the mechanism of PD-L1 protein expression bypassing mRNA expression, and minimizing the analytic variables of RISH assay is necessary to reduce discrepancy between the two assay.
The lack of response data to anti-PD-1/PD-L1 drugs is the major limitation of our study. The issue on utilizing PD-L1 mRNA assay should be made based on data from clinical trials of the drug under consideration and further studies are needed with the therapeutic responses.
In conclusion, elevated PD-L1 expression was associated with poorly differentiated histology and EGFR-naïve status in lung ADC. In addition, PD-L1 expression may be a prognostic marker in patients with early stage lung cancer.
Supporting information
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Korea Healthcare Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (HI17C1290) and the National Research Foundation of Korea (NRF) Grant funded by the Korean Government (MSIT) (No. 2017R1A5A1015626) to JHC, and the National Research Foundation of Korea (NRF-2017R1C1B1011620) to HK. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Postow MA, Callahan MK, Wolchok JD. Immune checkpoint blockade in cancer therapy. J Clin Oncol. 2015;33: 1974–1982. doi: 10.1200/JCO.2014.59.4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366: 2443–2454. doi: 10.1056/NEJMoa1200690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brahmer J, Reckamp KL, Baas P, Crinò L, Eberhardt WE, Poddubskaya E, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373: 123–135. doi: 10.1056/NEJMoa1504627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373: 1627–1639. doi: 10.1056/NEJMoa1507643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375: 1823–1833. doi: 10.1056/NEJMoa1606774 [DOI] [PubMed] [Google Scholar]
- 6.Herbst RS, Baas P, Kim DW, Felip E, Pérez-Gracia JL, Han JY, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387: 1540–1550. doi: 10.1016/S0140-6736(15)01281-7 [DOI] [PubMed] [Google Scholar]
- 7.Rittmeyer A, Barlesi F, Waterkamp D, Park K, Ciardiello F, von Pawel J, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 2017;389: 255–265. doi: 10.1016/S0140-6736(16)32517-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372: 2018–2028. doi: 10.1056/NEJMoa1501824 [DOI] [PubMed] [Google Scholar]
- 9.Yang CY, Lin MW, Chang YL, Wu CT, Yang PC. Programmed cell death-ligand 1 expression in surgically resected stage I pulmonary adenocarcinoma and its correlation with driver mutations and clinical outcomes. Eur J Cancer. 2014;50: 1361–1369. doi: 10.1016/j.ejca.2014.01.018 [DOI] [PubMed] [Google Scholar]
- 10.Cha YJ, Kim HR, Lee CY, Cho BC, Shim HS. Clinicopathological and prognostic significance of programmed cell death ligand-1 expression in lung adenocarcinoma and its relationship with p53 status. Lung Cancer. 2016;97: 73–80. doi: 10.1016/j.lungcan.2016.05.001 [DOI] [PubMed] [Google Scholar]
- 11.Koh J, Go H, Keam B, Kim MY, Nam SJ, Kim TM, et al. Clinicopathologic analysis of programmed cell death-1 and programmed cell death-ligand 1 and 2 expressions in pulmonary adenocarcinoma: comparison with histology and driver oncogenic alteration status. Mod Pathol. 2015;28: 1154–1166. doi: 10.1038/modpathol.2015.63 [DOI] [PubMed] [Google Scholar]
- 12.Takada K, Okamoto T, Shoji F, Shimokawa M, Akamine T, Takamori S, et al. Clinical significance of PD-L1 protein expression in surgically resected primary lung adenocarcinoma. J Thorac Oncol. 2016;11: 1879–1890. doi: 10.1016/j.jtho.2016.06.006 [DOI] [PubMed] [Google Scholar]
- 13.Scheel AH, Ansén S, Schultheis AM, Scheffler M, Fischer RN, Michels S, et al. PD-L1 expression in non-small cell lung cancer: correlations with genetic alterations. Oncoimmunology. 2016;5: e1131379 doi: 10.1080/2162402X.2015.1131379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirsch FR, McElhinny A, Stanforth D, Ranger-Moore J, Jansson M, Kulangara K, et al. PD-L1 immunohistochemistry assays for lung cancer: results from phase 1 of the Blueprint PD-L1 IHC Assay Comparison Project. J Thorac Oncol. 2017;12: 208–222. doi: 10.1016/j.jtho.2016.11.2228 [DOI] [PubMed] [Google Scholar]
- 15.Erber R, Stöhr R, Herlein S, Giedl C, Rieker RJ, Fuchs F, et al. Comparison of PD-L1 mRNA expression measured with the CheckPoint Typer(R) Assay with PD-L1 protein expression assessed with immunohistochemistry in non-small cell lung cancer. Anticancer Res. 2017;37: 6771–6778. doi: 10.21873/anticanres.12137 [DOI] [PubMed] [Google Scholar]
- 16.Kim HR, Cha YJ, Hong MH, Gandhi M, Levinson S, Jung, et al. Concordance of programmed death-ligand 1 expression between primary and metastatic non-small cell lung cancer by immunohistochemistry and RNA in situ hybridization. Oncotarget. 2017;8: 87234–87243. doi: 10.18632/oncotarget.20254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edge S, Byrd DR, Compton CC, Fritz AG, Greene F, Trotti A, eds. AJCC cancer staging handbook New York: Springer; 2009. [Google Scholar]
- 18.Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, et al. The 2015 World Health Organization Classification of Lung Tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10: 1243–1260. doi: 10.1097/JTO.0000000000000630 [DOI] [PubMed] [Google Scholar]
- 19.Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14: 22–29. doi: 10.1016/j.jmoldx.2011.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim H, Yang JM, Jin Y, Jheon S, Kim K, Lee CT, et al. MicroRNA expression profiles and clinicopathological implications in lung adenocarcinoma according to EGFR, KRAS, and ALK status. Oncotarget. 2017;8: 8484–8498. doi: 10.18632/oncotarget.14298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ilie M, Long-Mira E, Bence C, Butori C, Lassalle S, Bouhlel L, et al. Comparative study of the PD-L1 status between surgically resected specimens and matched biopsies of NSCLC patients reveal major discordances: a potential issue for anti-PD-L1 therapeutic strategies. Ann Oncol. 2016;27: 147–153. doi: 10.1093/annonc/mdv489 [DOI] [PubMed] [Google Scholar]
- 22.Miller RA, Cagle PT, Bernicker EH. First-line immune therapy-implications for pathologists. Arch Pathol Lab Med. 2016;140: 739–740. doi: 10.5858/arpa.2016-0904-ED [DOI] [PubMed] [Google Scholar]
- 23.Dong ZY, Zhang JT, Liu SY, Su J, Zhang C, Xie Z, et al. EGFR mutation correlates with uninflamed phenotype and weak immunogenicity, causing impaired response to PD-1 blockade in non-small cell lung cancer. Oncoimmunology. 2017;6: e1356145 doi: 10.1080/2162402X.2017.1356145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang M, Li G, Wang Y, Wang Y, Zhao S, Haihong P, et al. PD-L1 expression in lung cancer and its correlation with driver mutations: a meta-analysis. Sci Rep. 2017;7: 10255 doi: 10.1038/s41598-017-10925-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen N, Fang W, Zhan J, Hong S, Tang Y, Kang S, et al. Upregulation of PD-L1 by EGFR activation mediates the immune escape in EGFR-driven NSCLC: implication for optional immune targeted therapy for NSCLC patients with EGFR mutation. J Thorac Oncol. 2015;10: 910–923. doi: 10.1097/JTO.0000000000000500 [DOI] [PubMed] [Google Scholar]
- 26.Pan Y, Zheng D, Li Y, Cai X, Zheng Z, Jin Y, et al. Unique distribution of programmed death ligand 1 (PD-L1) expression in East Asian non-small cell lung cancer. J Thorac Dis. 2017;9: 2579–2586. doi: 10.21037/jtd.2017.08.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366: 2455–2465. doi: 10.1056/NEJMoa1200694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun JM, Zhou W, Choi YL, Choi SJ, Kim SE, Wang Z, et al. Prognostic significance of PD-L1 in patients with non-small cell lung cancer: a large cohort study of surgically resected cases. J Thorac Oncol. 2016;11: 1003–1011. doi: 10.1016/j.jtho.2016.04.007 [DOI] [PubMed] [Google Scholar]
- 29.Pan ZK, Ye F, Wu X, An HX, Wu JX. Clinicopathological and prognostic significance of programmed cell death ligand1 (PD-L1) expression in patients with non-small cell lung cancer: a meta-analysis. J Thorac Dis. 2015;7: 462–470. doi: 10.3978/j.issn.2072-1439.2015.02.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lavin Y, Kobayashi S, Leader A, Amir ED, Elefant N, Bigenwald C, et al. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell. 2017;169: 750–765.e17. doi: 10.1016/j.cell.2017.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang L, Huang F, Mei J, Wang X, Zhang Q, Wang H, et al. Posttranscriptional control of PD-L1 expression by 17β-Estradiol via PI3K/AKT signaling pathway in ERα-positive cancer cell lines. Int J Gynecol Cancer. 2017;27: 196–205. doi: 10.1097/IGC.0000000000000875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yee D, Shah KM, Coles MC, Sharp TV, Lagos D. MicroRNA-155 induction via TNF-α and IFN-γ suppresses expression of programmed death ligand-1 (PD-L1) in human primary cells. J Biol Chem. 2017;292: 20683–20693. doi: 10.1074/jbc.M117.809053 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.