Abstract
Osteoporosis has been associated with pulmonary silicosis in California horses exposed to soils rich in cytotoxic silica dioxide crystals, a syndrome termed silicate associated osteoporosis (SAO). The causal mechanism for the development of osteoporosis is unknown. Osteoporotic lesions are primarily located in bone marrow-rich sites such as ribs, scapula and pelvis. Gene transcription patterns within bone marrow and pulmonary lymph nodes of affected horses may offer clues to disease pathobiology. Bone marrow core and tracheobronchial lymph node tissue samples harvested postmortem from affected and unaffected horses were examined histologically and subjected to RNA sequencing (RNA-seq). Sequenced data were analyzed for differential gene expression and gene ontology. Metatranscriptomic and metagenomic assays evaluated samples for infectious agents. Thirteen of 17 differentially expressed transcripts in bone marrow were linked to bone and cartilage formation such as integrin binding bone sialoprotein (log2FC = 3.39, PFDR = 0.013) and chondroadherin (log2FC = 4.48, PFDR = 0.031). Equus caballus solute carrier family 9, subfamily A2 (log2FC = 3.77, PFDR = 0.0034) was one of the four differentially expressed transcripts linked to osteoclast activity. Osteoblasts were hyperplastic and hypertrophic in bone marrow from affected horses. Biological pathways associated with skeletal morphogenesis were significantly enriched in affected horses. The 30 differentially expressed genes in affected lymph nodes were associated with inflammatory responses. Evidence of infectious agents was not found. The SAO affected bone marrow molecular signature demonstrated increased transcription and heightened activation of osteoblasts. Increased osteoblastic activity could be part of the pathological mechanism for osteoporosis or a compensatory response to the accelerated osteolysis. Transcriptome data offer gene targets for inquiries into the role of osteocytes and osteoblasts in SAO pathogenesis. Viral or bacterial infectious etiology in SAO is less likely based on metatranscriptomic and metagenomic data but cannot be completely ruled out.
Introduction
Progressive osteoporosis is a comorbid condition in horses that have chronic pulmonary silicosis.[1] Clinical manifestations of osteoporosis include debilitating skeletal deformities, cervical osteoarthritis, and pathological fractures.[1, 2] Lytic bone lesions are typically found in ribs, vertebrae, scapulae, and the pelvis.[1, 3] Osteoporosis is more strongly associated with silicotic lymphadenitis in the nodes that drain the lung parenchyma (tracheobronchial lymph nodes (tLNs)), than with pulmonary silicosis.[1] The condition is termed silicate associated osteoporosis (SAO) when the skeletal and thoracic (lung and tLN) lesions are present simultaneously.[1] Cases of SAO are geographically clustered in regions with soil containing high levels of cytotoxic silicates (e.g., cristobalite—SiO2 polymorph), which are aerosolized and inhaled as fine crystalline silicate particles. [2, 4] Other causes of chronic granulomatous pulmonary disease have not been found in affected horses.[1] The pathogenic link between silicosis and osteoporosis remains putative.
Abnormal bone remodeling and anomalous osteoclast morphology (hypertrophy, hyperplasia), coupled with excessive osteolysis in SAO, are histologically similar to the lytic phase of Paget’s disease of bone (PDB) in people.[5] However, the chronic pulmonary inflammation induced by cytotoxic silicates, age of onset, skeletal distribution, and progressive osteolysis set SAO apart from PDB [1, 3, 5] (reviewed by Nebot Valenzuela et al. [6]). The roles of immunological background and other contributing factors for the development of SAO, such as dose and duration of exposure to the silicate crystals warrant further investigation. A previous investigation did not reveal viral or bacterial infections as cofactors in SAO through culture and microscopic examination of affected tissues, however broad metatranscriptomic and metagenomic assays to look for novel infectious agents have not been performed.[1] Though bone lesions have been well characterized grossly and morphologically in SAO, the underlying cause of the pathologic osteolysis is unknown.
SAO has only been reported in horses. However, singular cases of similar comorbidities have been suspected to occur in humans. [7–9] Osteoporosis has been associated with pulmonary diseases such as cystic fibrosis [10] and chronic obstructive pulmonary disease in people.[11, 12] Other extra-skeletal inflammatory diseases associated with osteoporosis include inflammatory bowel disease [13] and systemic lupus erythematosus. [14] Hyper-activation of osteoclasts during systemic inflammation suggests that a subset of pro-inflammatory signals may induce osteoporosis.[15]
Merging of osteoclast activation and pro-inflammatory signals is in part supported by molecular studies. Osteoclast differentiation and activation are regulated mainly through an interaction between “receptor activator of nuclear factor κ-β” (RANK) and its ligand (RANKL).[16] Pro-inflammatory cytokines like IL1, TNFα and IL6 potentiate RANK downstream signaling cascades, or increase paracrine production of RANKL. Thus, these pro-inflammatory cytokines can increase osteoclast activity.[17] Therefore, osteolysis in SAO may be derived from chronic cytotoxic silicate-induced inflammation within the respiratory tract. We hypothesized that the inflammatory trigger initiates aberrant osteoclastogenesis in the bone marrow (BM), where osteoclast precursors receive systemic signals, resulting in accelerated osteolysis and osteoporosis in affected horses.
To test our hypothesis we generated differential gene expression (DGE) profiles of the main tissue sites affected by SAO, the rib BM and tLN, in affected and unaffected horses. Because of the hypothesized systemic effect of pro-osteoclastogenic trigger, BM samples were collected away from gross lesions of osteoporosis with markedly altered cell populations. This tissue collection protocol was designed to capture the pattern of cellular signals before significant tissue heterogeneity would dominate the transcriptional differences. The presence of transcripts from infectious agents in the target tissues was also evaluated.
Material and methods
Animals
Samples from the two tissue targets, BM cores and tLNs, were collected post mortem from sex and age-matched SAO+ and SAO- horses (S1 Table). There is no known breed predisposition for SAO.[1] To prevent inclusion of subclinically affected horses in the control group, inclusion criteria for control horses included absence of exposure to SAO endemic regions (i.e., exposure to cytotoxic silica dioxide particles) and absence of granulomatous pulmonary disease on post mortem examination. Study animals consisted of 8 SAO-affected, (3 females: Appaloosa, Quarter Horse and Thoroughbred, and 5 males: Holsteiner, Quarter Horse cross, Quarter Horse, Paint and Arabian), and 8 unaffected horses, (4 females: Hanoverian, Thoroughbred, Paint and Quarter Horse, and 4 males: Quarter Horse, Standardbred/Arabian cross, Mustang cross, and Morgan/Arabian cross). The median age of SAO+ horses in BM tissue group (n = 8, 20 years, range 12 to 30) was not significantly different (Mann-Whitney test, p = 0.46) from the median age of unaffected horses (n = 8, 17.5 years, range 8 to 26). Similarly, the median age of SAO+ horses in tLN tissue group (n = 7, 19 years, range 12 to 30) was not significantly different (Mann-Whitney test, p = 0.48) from the median age of the unaffected horses (n = 6, 17 years, range 11 to 26). Clinical disease in SAO horses was categorized as mild or severe based on the absence or presence, respectively, of gross skeletal deformities. Formal consent was obtained from owners for horses donated to the study. All horses were euthanized with an intravenous overdose of pentobarbital sodium (FATAL-PLUS—Pentobarbital Sodium 390 mg/mL, Vortech Pharmaceuticals, Dearborn, MI) solution (>100 mg/kg) due to poor quality of life (SAO horses) or neurologic, chronic focal musculoskeletal or behavioral abnormalities, or trauma (S1 Table). The project was approved by the University of California Davis Institutional Animal Care and Use Committee.
SAO clinical phenotyping
The medical history included known exposure to SAO endemic regions. Musculoskeletal conditions including history of skeletal deformities, lameness, fracture(s), decreased range of motion, and exercise intolerance, as well as abnormalities of the respiratory system were investigated and documented. Ante mortem clinical evaluation included physical examination and complete blood cell count and serum biochemistry including ionized calcium concentrations. All horses in the study received non-steroidal anti-inflammatory (NSAIDs) medications (e.g. flunixin meglumine, firocoxib, or phenylbutazone) at standard dosing regiments for pain management prior to euthanasia. Horses treated with bisphosphonates, including zoledronic acid (Zometa), clodronate (Osphos) and tiludronate (Sigma), within the past twelve months were excluded from the study. The diagnosis of SAO was confirmed by post mortem evidence of silicosis in the tLN and osteoporosis.[1]
Post mortem evaluation
The skull, scapulae, humeri, ribs, pelvis, lungs, lymph nodes (cervical, thoracic and abdominal), endocrine organs (pituitary, thyroid, and adrenal glands; parathyroid glands were examined in 3 of the affected and 2 control horses due to the difficulty in finding the parathyroid glands in this specie), skeletal muscles, major abdominal organs and reproductive organs (when present) were evaluated grossly and histologically. Sections of the 7th, 8th, 9th, or 10th rib and other bones were evaluated microscopically to confirm SAO status when bone deformity, porosity, fracture, or discoloration were present.
Histology
All tissues were fixed in 10% buffered formalin. Rib BM cores specimens were decalcified with 10% ethylenediaminetetraacetic acid (EDTA; Amresco, Solon, OH) at 4°C for 2 weeks prior to trimming and sectioning. Trimmed tissues underwent routine histological processing resulting in 4μm sections stained with hematoxylin and eosin using routine protocols.
Microscopic features used to confirm silicosis in tLNs included necrotizing and fibrosing granulomas with intralesional refractile crystals. Features used to confirm osteoporosis included atypically large and numerous intralesional osteoclasts with increased numbers of nuclei, and abnormal bone tissue remodeling characterized by haphazard, anastomosing, thin, woven and lamellar bone trabeculae with mosaic cement lines in areas of prior lysis.
Bone formation and resorption ratios were calculated using the length (in μm) of bone surface covered by osteoblasts or scalloped sites of resorption (Howship's lacunae), respectively, to the total length of bone surface (bone perimeter) using digital images of histological sections obtained at 100x magnification (image area = 877x666μm). Image software (Adobe Photoshop CS6, Adobe Systems Incorporated, San Jose, CA) was used for the analysis of images with sufficient morphological detail for cell and tissue identification (2–6 per specimen). The measuring scale was set to the calibration bar embedded in the images. Bone formation surface was defined as bone surface length in μm lined by osteoblasts. Bone resorption surfaces were defined by length in μm of scalloped bone surface contours. Osteoblast height, as a reflection of osteoblast hypertrophy and activity, was defined as the average apical to basilar distance in μm of 3 to 5 osteoblasts per image. Bone surfaces with artifacts and/or indistinct cellular morphology as well as surfaces created at the image edges were excluded from analysis. The calculated ratios were compared between SAO+ and unaffected horses using a two-sided t-test with p<0.05 considered statistically significant.
Tissue targets for transcriptome sequencing and DGE analysis
Rib BM cores and tLN are characteristically affected in SAO and, therefore, were the target sites for in situ transcription profiling of SAO+ and unaffected horses. Rib BM core samples were collected away from grossly visible rib lesions.
The interval from euthanasia to the time of BM collection ranged from 20 min to 2 hours. However, one sample (SAO6) was preserved at 4°C for 12hrs post mortem and included in the study after passing RNA quality assessment (defined in the next paragraph). BM was collected aseptically by removing a segment of cortical bone with handsaw (Bosch PS50-2B, Mt. Prospect, IL) at the middle of the rib (7, 8, 9 or 10). The BM sample included both trabecular bone and inter-trabecular tissues. Transverse segments of tLN, including the cortex and medulla, were collected during necropsy, within 1–4 hours after euthanasia. Approximately 5 mm cubes of BM and tLN samples were placed into individual 1.8 ml cryogenic tubes (Nunc CryoTubes Sigma-Aldrich, Roskilde, Denmark) filled with 1.5mL of RNA–preserving solution (RNAlater, Vilnius, Lithuania) and immediately cryopreserved in liquid nitrogen. Samples were stored at -80°C until RNA extraction. Next, a second set of samples of the BM and tLNs from the same site were fixed in 10% buffered formalin for microscopic examination.
RNA extraction and quality validation
The tissues stored in RNAlater were thawed on ice and sectioned using sterile instruments into ~0.3 cm cubes. The tissue cubes were individually crushed within liquid nitrogen to a fine powder using a sterile and UV-light-treated ceramic mortar and pestle also cleaned with RNase inhibiting solution (RNaseZAP wipes, Sigma-Aldrich, Saint Louis, MO). Total RNA was extracted using a commercial kit (RNeasy Plus Mini Kit, Qiagen, Hilden, Germany) following manufacturer’s protocol. Any contaminating genomic DNA was degraded enzymatically (RNase-Free DNase I and RNeasy Mini Elute Clean Up kits, Qiagen). An aliquot of the resulting total RNA with absorbency ratio (A260nm/280nm) of >1.9 was quantified spectrophotometrically (NanoDrop 1000, ThermoFisher Scientific, Waltham, MA) at 260nm wavelength. The quality of the RNA was assessed using a capillary electrophoresis technique (Agilent 2100 Bioanalyzer, Agilent Technologies, Santa Clara, CA, (S1 Table)).
RNA-seq library construction and RNA sequencing
Samples of total RNA (500ng) with RIN of ≥ 7.5 were submitted to the DNA Technologies Core (University of California Davis Genome Center) and used for construction of strand-specific, poly-A enriched libraries. Following stranded mRNA-Seq Kit (KAPA Stranded mRNA-Seq Kit, Illumina platform, Cape Town, South Africa) manufacturer’s protocol, poly-A enrichment followed by cDNA synthesis and A tailing adapter ligation was performed. Library amplification was completed in 10 PCR cycles. Capillary electrophoresis technique (Agilent 2100 Bioanalyzer, Agilent Technologies, Santa Clara, CA) assessed distribution of the library fragments. Libraries were pooled and sequenced across two lanes with specification of 100-bp paired-end reads (100PE), (Illumina HiSeq 3000, San Diego, CA).
Sequence reads quality assessment, trimming, and alignment
Raw reads were processed with expHTS [18] to trim low quality reads and adapter contamination, and to remove PCR duplicates. Trimmed reads for each sample were aligned to the EquCab2 horse genome with EquCab2.83 annotation (Ensembl), using STAR v. 2.5.1a aligner, [19] which also generated raw read counts per gene that were the input for the statistical analysis.
Differential gene expression analyses
Differential gene expression analyses were conducted using the statistical limma-voom pipeline (version 3.28.17) [20] [21] and ANOVA statistical model that factors in presence of disease (affected/ unaffected), osteoporosis phenotype (mild, severe), tissue type (BM, tLN), and RNA isolation batch. Statistical significance of the genes was associated with an adjusted p value of <0.05. GO enrichment analysis was conducted (Bioconductor package topGO: Enrichment Analysis for Gene Ontology. R package version 2.24.0) to identify biological functions enriched in SAO+ tissues transcriptome.[22]
PANTHER pathway analysis
The files of genes from the DGE analysis with ENSEMBL gene IDs and fold-change expressed in log2 for BM and LN tissues were loaded into Gene List Analysis (http://www.pantherdb.org, https://doi.org/10.1093/nar/gkw1138). The settings included statistical enrichment test defaults and Equus Caballus (EquCab2.0) as the reference species. Statistical significance was set as p-value below the Bonferroni correction was used to accounted for multiple comparisons in the analysis. PANTHER Pathways and GO-Slim Biological Processes were selected as the Annotation Data Sets for analysis.
BRAKEN (Bayesian Re-estimation of Abundance after Classification with KrakEN) analysis
Because the pathogenesis of SAO is unknown, non-equine reads were evaluated for similarity to sequences of infectious agents that could be attributed to viruses or bacteria. Trimmed reads were evaluated with KrakEN, a k-mer based metagenomic classification tool, using a combined horse, viral, and bacterial database, followed by the BRAKEN metagenomic classification pipeline for transcription frequency of non-equine genetic material, and reported as fractions relative to the Equus Caballus taxonomy labels assigned by KrakEN.[23–25]
Metagenomic analysis, viral discovery
Metagenomic analysis evaluated SAO+ tissues for viral transcripts. Samples of SAO+ BM and tLN SAO1, SAO7, SAO3, SAO9 (not used in transcriptome study)), as well as buffy coat (SAO7, SAO9) and lung (SAO1, SAO3) tissues were pooled. Viral capsid-protected nucleic acid was extracted and enriched according to an established protocol.[26] An Illumina MiSeq library was constructed using random RT-PCR followed by use of the Illumina Nextera kit and sequenced on an Ilumina MiSeq platform using 250 bases paired end. Sequence reads were de novo assembled using the Ensemble program and contigs and singletons then translated into hypothetical protein sequences, which were compared to all viral proteins in the NCBI virus RefSeq database using BLASTx. Detailed descriptions of the methods used in this analysis are included as supplemental material (S1 Protocol).
RNA mapping statistics
Sufficient depth of sequencing and an adequate mapping rate were achieved for the DGE analysis. The average reads across BM and tLN samples were ~25.7 million reads/sample. Following trimming adapter contamination and removal of low quality reads and PCR duplicates with expHTS software, the average mapping rate to EquCab2.0 was 92.63%, where 54.6% reads uniquely aligned to an annotated horse gene and 32.4% aligned to regions of genome without annotation. 5.28% of reads aligned to multiple regions in the genome and 0.35% aligned with some annotation overlap, therefore, these reads were not assigned to a specific gene.
Samples segregate based on tissue type and disease phenotype
The biologically relevant segregation of the sampled tissues in the MDS plots demonstrated high quality of starting genetic material, sequencing, and mapping techniques (S1–S3 Figs). The MDS plots illustrated that the clustering of SAO+ and SAO- transcriptomes in BM and tLN tissues were most influenced by the tissue origin (S1 Fig), and the disease phenotype (S2 and S3 Figs), rather than biological variations such as breed or age (S5 and S6 Figs) among horses. SAO+ BM samples with mild osteoporosis phenotype clustered with unaffected samples (S2 Fig).
Results
Differential gene expression in BM and tLN
The ANOVA statistical model that accounted for the osteoporosis phenotype resulted in 17 significantly differentially expressed genes in BM and 36 significantly differentially expressed genes in tLN tissue (list of selected genes in Tables 1 and 2 and S2 and S3 Tables).
Table 1. Seventeen differentially expressed genes in SAO+ BM tissue listed in tissue function groups with base 2 logarithmic fold change and false discovery adjusted p-value.
| Tissue Function | Gene | Specific Function | Log2FC, (PFDR) |
|---|---|---|---|
| Bone resorption | Equus caballus solute carrier family 9, subfamily A2 (SLC9A2) | Na+/H+ exchanger, cation proton antiporter; expressed in osteoclasts [27] | 3.77 (0.034) |
| Bone resorption, formation, and maintenance | Osteomodulin, Osteoadherin (OMD) | Bone homeostasis and mineralization couples activity of Oc and Ob [28, 29] | 3.92 (0.013) |
| Distal-less homeobox3 (DLX3) | Homeobox gene embryonic bone development, Ob differentiation and Os function; Oc differentiation [30, 31] Higher expression in Os compared to Ob [32] | 4.49 (0.033) | |
| *Transient receptor potential cation channel, subfamily V, member 4 (TRPV4) | Skeletal development at the growth plate; cartilage homeostasis [33–35] and osteoclast differentiation [36] | 3.77 (0.030) | |
| Bone homeostasis and formation | Collagen type XXIV, alpha 1 (COL24A1) | Precedes collagen I deposition in ossification centers [37] | 4.18 (0.013) |
| Sp7 transcription factor (SP7), Osterix (Osx) | Ob and chondrocyte differentiation from mesenchyme [38–40]; functions downstream of Runx2, BMP, Wnt [41] | 3.32 (0.031) | |
| Integrin binding sialoprotein, bone sialoprotein (BSP) | Mineralization of bone and cartilage during bone development [42] | 3.93 (0.013) | |
| Osteocalcin; bone gamma-carboxyglutamate (gla) protein (BGLAP) | Bone mineralization; regulated by Runx2 and Dlx3 and produced by in BM pluripotent cells during osteogenesis [31, 43, 44] | 2.44 (0.029) | |
| Sphingomyelin phosphodiesterase 3 (SMPD3) | Endochondral bone formation in skeletal development [45, 46] | 2.70 (0.021) | |
| Collagen type XI, alpha 2 (COLXIA2) | Skeletal development as cartilage ECM, embryonic development of the skeleton [47] | 3.37 (0.029) | |
| Chondroadherin (CHAD) | Embryonic development of the skeleton, [48] minor component of bone matrix, [49] higher expression in Ob than Os [32] | 4.48 (0.031) | |
| Anoctamin 5 (ANO5) | Embryonic development developing somites; Growth-plate chorndrocytes; Ob [50] | 3.94 (0.037) | |
| Hydroxysteroid (17-beta) dehydrogenase 6 (HSD17B6) | Epimerization / interconversion estrone (E1) and estradiol (E2) [51] | -2.99 (0.013) | |
| WAP four-disulfide core domain 1 (WFDC1) | Secreted protease inhibitor with higher expression in Ob than Os [32] | 2.67 (0.038) | |
| Cadherin 15 (CDH15) | Cell adhesion molecule with higher expression in Os than Ob [32] | 3.97 (0.034) | |
| Inflammation | Death-associated protein kinase 2 (DAPK2) | Granulocytes migration with activation and differentiation [52] | 2.36 (0.031) |
*Genes with functional overlap in Ob (osteoblast), Os (osteocyte) and Oc (osteoclast)
Table 2. Differentially expressed genes in tLN and base 2 logarithmic fold change with false discovery adjusted p-value.
The selected three of the 36 differentially expressed genes are listed for their relevance to SAO.
| Gene (Names and symbol) | Function | Log2FC, (PFDR) |
|---|---|---|
| Pentraxin 3 (PTX3) | Macrophages, dendritic cells and stromal cells soluble pattern recognition receptor (response to insults like silicates, infection and trauma);[53] identified as a promoter in TNF-α and IL-1β production through binding NF-κB elements in vitro [54] | 3.9 (0.029) |
| Equus caballus Fc fragment of IgA receptor (FCAR) | Mediates immunological response in myeloid cells including alveolar macrophages that interact with IgA engaged with its antigen [55] | 2.20 (0.030) |
| Dentin matrix acidic phosphoprotein 1 (DMP1) | Tumor suppressor gene from calmodulin kinase family with broad function in cell signaling;[52] extracellular matrix protein important for biomineralization;[56, 57] expressed in cancer with predilection for bone metastases (eg. human lung cancer)[58] | 3.01 (0.032) |
Bone marrow differential gene expression
Overall 12 of 17 differentially expressed genes (relative to unaffected animals) in SAO+ BM were found to be associated with skeletal formation (Table 1). Sixteen of the transcripts were increased in SAO, with decreased expression only found in hydroxysteroid (17-beta) dehydrogenase 6, (HSD17B6). Among the genes with increased transcripts, Equus caballus solute carrier family 9, subfamily A2 (SLC9A2) expression is linked to osteoclast differentiation and survival.[27] Transient receptor potential cation channel, subfamily V, member 4 is a marker of osteoclast differentiation (TRPV4).[36] Osteomodulin (OMD) [28] and distal-less homeobox 3 (DLX3)[30] gene products are transcriptional indicators of osteoclast activity and differentiation, respectively. Death-associated protein kinase 2 (DAPK2),[41, 52] was the only genetic footprint of response to pro-inflammatory stimulus in BM of SAO+ horses. Five of the differentially expressed genes associated with the developing skeleton include DLX3,[59] Sp7 transcription factor (SP7),[40, 41] collagen type XXIV, alpha 1 (COL24A1),[37] collagen type XI, alpha 2 (COLXIA2),[47] and chondroadherin (CHAD). [48] Increased transcription of bone formation markers included integrin binding sialoprotein (BSP),[60, 61] osteocalcin (OC),[62] and sphingomyelin phosphodiesterase 3 (SMPD3).[45, 46]
Tracheobronchial lymph node differential gene expression
Three out of 36 differentially expressed genes in the tLNs from SAO+ horses were included for their functional relevance to the silicosis induced inflammatory process (Table 2). Six of the 36 genes were uncharacterized proteins. The other 27 genes were broadly associated with immune functions and demonstrated a nonspecific pattern of differential expression (S3 Table).
Gene ontology (GO) enrichment analysis of bone marrow and tracheobronchial lymph node transcriptomes
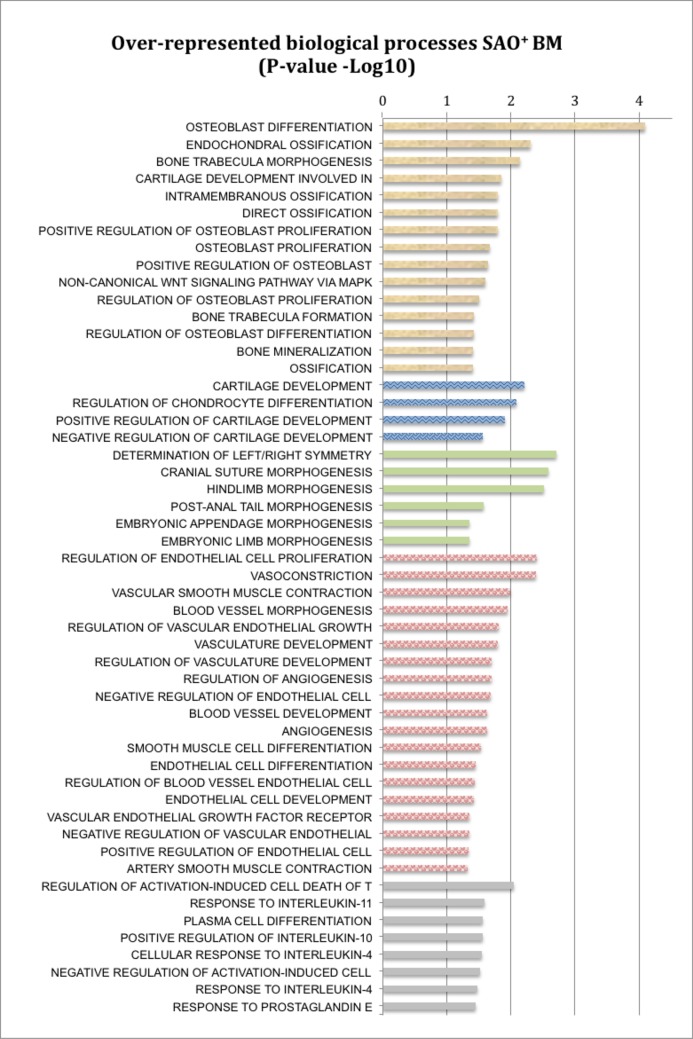
GO analysis probed the entire transcriptome of SAO+ and unaffected tissues against the structured dataset of biological functions to capture the significant differences in gene signaling and interactions relative to biological processes. The data is presented in raw p-value for the Kolmogorov-Smirnov test for the over-represented GO terms (S4 and S5 Tables). (Raw p-values are reported because there is no multiple testing correction method for hierarchically arranged GO terms). Differences in biological functions were identified in BM and tLN tissues. The order (P-value) revealed the statistical significance in the enriched biological function (S4 and S5 Tables). In BM transcriptome, 326 enriched biologic functions associated with SAO had P values <0.05 (S4 Table). The smallest p-value associated with enriched biological function was osteoblast differentiation (p = 8.20E-05) with other top functions related to skeletal and vascular systems development (Fig 1 and S4 Table). In the affected tLN, 448 enriched biological functions (P value <0.05) were broadly divided into immunological activation and few indicators of infection (S5 Table, S4 Fig).
Fig 1. The selected, significantly over-represented biological processes in SAO+ BM sample (based on raw p value <0.05).
The biological processes are listed in arbitrary categories associated with formation of bone (tan), cartilage (blue), embryonic skeleton (green), vessels (pink), and immune system responses (gray). Bars represent negative Log 10 conversion of the p value to illustrate the confidence in their over-representation in SAO+ BM. S4 Table includes the entire list of all the significant biological functions for BM GO enrichment analysis.
Panther analysis: PANTHER enrichment test
Increased signaling through the integrin (P00034, p = 5.91E-03), PDGF (P00047, p = 3.44E-02) and cadherin (P00012, p = 6.58E-03) pathways were found in the SAO+ BM samples compared to unaffected samples. Down-regulation of T cell activation (P00053, p = 1.51E-03), increased coagulation (P00011, p = 7.87E-03), and decreased signaling through PDGF (P00047, p = 7.92E-03) were over-represented pathways in affected tLN samples.
Morphological link to transcriptome patterns informed through DGE and GO
BM and tLN sampled from the same site as tissues harvested for RNA were evaluated microscopically to assess the biological relevance of the transcriptome data.
Bone marrow
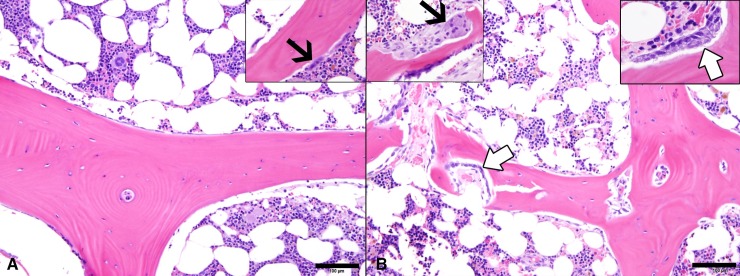
Bone formation, resorption, and total perimeters were quantified in SAO+ and unaffected BM samples. Mean (± SD) ratio of bone forming surface to total bone surface perimeter was greater (p<0.001) in SAO+ horses (0.31 ± 0.14) than unaffected horses (0.05 ± 0.04). Osteoblast hypertrophy, captured as apical to basilar height, was significantly greater (p = 0.002) in SAO+ horses (11.6 ± 2.2 μm) than unaffected horses (5.4 ± 3.8 μm). Mean ratio of bone resorption surface to total bone surface perimeter was greater (p = 0.016) in SAO+ horses (0.15 ± 0.12) than unaffected horses (0.03 ± 0.04). However, few osteoclasts were present in sections (ranging from 0 to 6 per image area) compared to the other cell populations and numbers per total bone perimeter were not statistically different (p = 0.401) between affected and unaffected horses. However, osteoclasts were subjectively more ubiquitous in SAO+ horses with greater cell volume and nuclei numbers (Fig 2). Cartilaginous tissue was not identified in the rib sections.
Fig 2.
Representative photomicrographs of decalcified unaffected (A) and SAO+ (B) BM sections. Inter-trabecular BM stroma contained similarly mixed cell populations. A thin layer of bone lining cells cover the trabecular bone surfaces in the control BM sample (A). Rows of osteoblasts (white arrows) line some of the bone surfaces in SAO+ (B) and fewer surfaces in control (not shown) BM specimens. Rare osteoclasts (black arrows) are noted in the specimens but the osteoclasts and associated resorption bays appear larger in SAO+ BM. [H&E stain; Scale bars 100 μm].
Tracheobronchial lymph nodes
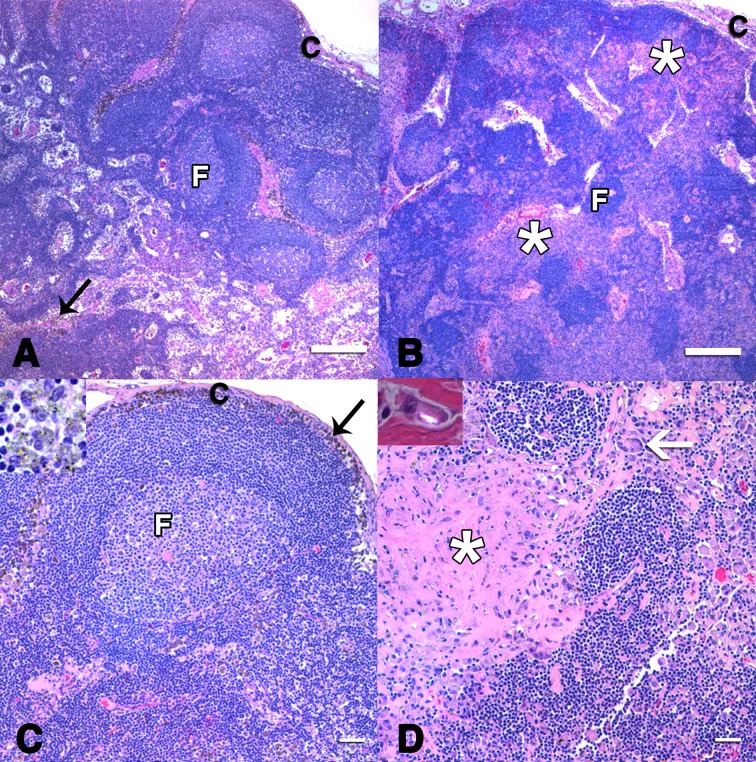
The lymphoid follicles in SAO+ horses were atrophied compared to the well-defined follicular architecture present in unaffected tLN samples (Fig 3). Mild to moderate granulomatous inflammation and fibrosis obscured large regions of the tLN medullary cords and cortex in the SAO+ group (Fig 3). Variable numbers of thin, refractile crystals (<2 μm long) were identified within regions of inflammation and fibrosis under polarized light in tLNs only from affected horses. These crystals were non-pigmented and indistinct under transmitted white light. Different refractile, brown pigmented crystals were observed in 10–30% of the sinusoidal macrophages of all tLNs. Necrosis, fibrosis, multinucleated macrophages, and inflammation were not associated with these crystals in tissues.
Fig 3.
Photomicrographs of unaffected (A, C) and SAO+ tLNs (B, D). The affected tLN had regions of pallor in the cortex and medullary cords (asterisks) subjacent to atrophied lymphoid follicles (“F”, B). Abundant mature and smudged collagen deposits (fibrosis) co-localized with numerous macrophages, giant cells (arrow, D) and refractile, sharp crystalline particles (inset—high magnification of macrophage under polarized light, D). Granular, pigmented crystals (black arrows and inset, A, C) consistent with incidental anthracosilicosis were noted in the macrophages of both unaffected and affected tLNs (not shown in B, D) without inflammation, necrosis and fibrosis characteristic of reaction to cytotoxic silicate crystals. Infectious organisms were not found in any lymph nodes. (c–lymph node capsule) [H&E stain; Scale bars: (A,B) 500μm; (C,D) 50μm].
Metatranscriptomic analysis
To determine if the presence of infectious agents was associated with the differential transcriptome data from SAO+ and unaffected horses, metatranscriptomic analysis was conducted. In all samples, reads assigned to Equus caballus comprised greater than 99.8% of all reads or read pairs. The most abundant, non-equine species present in affected and control BM and tLN tissues included Choristoneura occidentalis granulovirus (an arthropod virus, <0.09% of reads) and Alteromonas mediterranea (a marine bacterium, <0.02% of reads). One SAO+ BM sample had Mycobacterium tuberculosis as the second most abundant species (0.00017% of reads) without microscopic evidence of granulomatous lesions.
Viral discovery
Metagenomic analysis was conducted on selected tissues from 3 SAO+ horses to determine if viral genetic material could be detected. No viral RNA, despite the findings of granulovirus in the metatranscriptomic analysis, was identified in pooled tissues (BM, tLN, blood, and lung).
Discussion
The cardinal lesions of SAO in tLN and bone marrow-rich skeletal sites were present in all affected horses, supporting circumstantial evidence for their interconnected role in the pathobiology of SAO.[1] The molecular signature within the microenvironment of affected pulmonary and skeletal tissues was explored to enhance the understanding of SAO disease pathogenesis. RNA-seq was used in the investigation of BM and tLN transcriptomes. To compare the pattern of molecular signatures with the biological processes within BM and tLN tissues, transcriptome results were assessed in light of the histologic features in the sites used for molecular analysis. Several transcripts associated with bone formation (e.g., increased osteoblast-associated activity) and a lesser number for bone resorption were overrepresented in SAO+ BM transcriptome relative to unaffected animals. DGE and GO enrichment analyses demonstrated pro-inflammatory activity in SAO+ tLNs, but inherent tissue heterogeneity may have diluted a more distinct transcriptome pattern. There was no evidence of infectious co-factors in SAO etiology. This study provides a novel viewpoint of a dominant molecular signature of bone formation (increased osteoblastic activity) in a condition that has been morphologically defined by osteoclastic hyperactivity and clinically significant osteoporosis.[1] This data introduced an unexpected conundrum.
Although unexpected, the paucity of transcripts associated with osteoclast differentiation in SAO+ BM tissue suggests a number of pathogenic possibilities. Lack of anticipated transcriptome pattern associated with increased signaling through pro-osteoclastogenic RANK may reflect the low frequency of these transcripts within the complex cellular milieu of BM. Alternatively, the lack of evidence for a dominant osteoclastogenic signal may reflect the brief temporal nature of such a signal. Regardless, within lesions characterized by abundant bone removal, histologic evidence of overt osteoblastic activity is concurrently present. By design, tissue RNA samples were obtained from grossly normal bone, anatomically distant from osteoporotic lesions.
DGE and GO enrichment analyses of the SAO+ BM transcriptome revealed predilection for skeletal formation transcription. This finding was supported by morphologically increased osteoblast activity in affected BM samples compared to quiescent bone surfaces of unaffected tissues. Osterix (Sp7) is one of the differentially increased transcripts indicative of enhanced osteoblast differentiation.[41] Expression of osteoblast activity markers downstream of Sp7 like IBSP, BGLAP and COL24A1 provided further transcriptional evidence of increased osteoblast differentiation in SAO+ BM.[43, 63, 64]
Marked osteoblastic activity coupled with hyperactive osteoclasts is characteristic of SAO lesions and other bone diseases like fibrous osteodystrophy. However, overrepresented activation of osteoblasts away from regions of intense osteolysis was not anticipated. Activation of osteoblasts as a systemic reparative or compensatory mechanism despite minimal osteoclastic activity within tested sample is a potential explanation. The results could be considered in the context of increased mechanical loading distributed along weakened bone and increased differentiation of BM stromal cells toward osteo-chondral lineage.[65] The induction of osteoblast differentiation by pro-inflammatory cytokines like Il-1β was also a new consideration.[66] Trabecular osteocytes interact with BM stroma and may add to the pool of differentially expressed genes like DLX3 and CDH15 shown to have a higher transcription level than osteoblasts in one study.[32]
Mechanisms involved in fetal bone formation are rekindled during endochondral ossification in skeletal fracture repair and potentially activated in SAO+ BM. Increased transcripts in our data are downstream of the master regulators of skeletal formation like bone morphogenetic protein (BMP) (reviewed by V.S. Salazar) [67] and runt related transcription factor 2 (Runx2) [68], (reviewed by T. Komori).[69] Transcriptional pattern of osteo-chondroprogenitor cell differentiation is reflected in the DGE signature and enriched biological functions associated with chondrocyte development and endochondral ossification despite histological absence of cartilage in BM samples. For example, genes such as COLXIA2, SMPD3, TRPV4, CHAD, and ANO5 are associated with cartilage tissue functions [45, 47] although cartilage and endochondral ossification have not been reported in SAO osteolytic lesions. Skeletal tissue-stereotypic genetic regulations of repair and embryonic pathways of the skeletal formation likely overlap regardless of the mechanism of injury. The roles for the genes associated with endochondral ossification and skeletal morphogenesis might be expanded as transcription patterns within SAO bone lesions and appendicular skeletal sites rarely affected by osteoporosis are elucidated in the future.
Increased osteoclast differentiation and activity in SAO+ BM was speculatively linked to the increased transcription of SLC9A2, DLX3, TRPV4 and OMD.[27, 30, 36] The latter gene is up regulated in an osteoblast response to osteoclastic activity.[28] Results from GO analysis did not demonstrate biological functions associated with osteoclasts. The sensitivity of the transcriptome analysis might not be sufficient to detect differential expression of osteoclasts outnumbered in BM samples by other cell populations. Alternatively, osteoclast transcriptome may be sample-dependent and thus indicative of multifocal rather than generalized nature of the pro-osteoclastic differentiation and osteolysis in SAO.
Our results suggest a multifocal rather than systemic activation of an osteolytic trigger in SAO. The multifocal distribution of osteolysis has been noted in post mortem examination and on nuclear scintigraphic examination of SAO+ horses, but the potential significance has not been realized.[1, 3] The putative differential transcriptome expression pattern may only be captured within osteolytic lesions and would be sample site dependent. Additionally, the histogenic nature of the cytologically atypical, multinucleated, osteolytic cells in SAO may need to be defined beyond microscopic phenotype together with the histogenesis of their precursors. Nevertheless, our results bring attention to osteoblasts and their multidimensional role in bone tissue formation, regulation of their microenvironment, and implication in pathogenesis of bone fragility diseases.
The role of the only down-regulated transcript, HSD17B6, in SAO+ BM is unclear because function of the equine isoenzyme is not defined in the literature. HSD17B6 might have an important role in bone as some isoenzymes catalyze the interconversion of estrone (E1) to “osteoprotective” estradiol E2).[51] In human and mice prostate tissues, HSD17B6 has been identified as a mediator of the conversion of androgen substrates into inactive forms.[70] Depending on the substrate and the product of the hydroxysteroid dehydrogenase, depletion of HSD17B6 in SAO+ horses could be a response to accelerated bone loss, or part of the disease pathobiology resulting in excessive osteolysis.
Compared to GO and DGE analysis, PANTHER pathway evaluation of the BM transcriptome did not explicitly demonstrate osteoblast-associated functions. Pathway analysis showed increased signaling through the integrin and cadherin pathways broadly used by cells in the BM tissue.
Both DGE and GO enrichment analyses showed evidence for nonspecific inflammation in the tLNs of SAO+ horses. DGE analysis did not reveal a transcription pattern to explain the association between the skeleton and lung conditions in SAO. The 30 functionally defined differentially expressed genes within the SAO+ tLNs showed variably regulated transcripts broadly related to immune system functions. Theoretically, connections between silicosis and osteoporosis could be made with some transcripts. For example, an increase in DMP1 transcription echoes BM tropism of an osteolytic trigger in SAO because it was up-regulated in bone tropic metastatic lung cancers.[58] Physiologic expression of DMP1 in non-mineralized tissues may indicate a wider purpose for extra-skeletal regulation of mineralization.[71] PTX3 up-regulation may have some connection with silicosis as a promoter in TNF-α and IL-1β production through binding NF-κB elements, as demonstrated in vitro.[54] Increased production of IL-1 and TNF-α has been implicated in the pathobiology of silicosis.[72, 73] Interestingly, PTX3 is increased in cultured human bronchial epithelial cells exposed to cytotoxic silicates.[74] Inherent cell population variability within affected and unaffected tLN tissues may have distorted the signal pattern of the silicosis and diluted the significant differentially expressed genes. Heterogeneity included regions where more than 40% of affected tLN had been replaced by fibrosis, necrosis, and granulomatous inflammation in response to intralesional refractile crystals (presumed cristobalite).
Modification of the tissue target, sample collection, and RNA preparation protocols, as well as identification of SAO clinical markers would curtail some of the limitations for the prospective investigation of this condition. Because necropsy and histopathology remain the gold standard diagnostic tools to confirm SAO, only horses in terminal condition were recruited for the study. Concerns over transportation of severely affected SAO+ horses with advanced osteoporosis resulted in challenging barriers for patient recruitment. Isolation of the specific cell targets from BM and blood of live animals, such as osteoblasts or osteoclasts using laser dissection, and monocytes with a cell sorting technique may prove useful for future studies. The implication of the results from the metatranscriptomic analysis in our study was unclear since the low frequency transcripts of an arthropod virus and marine bacteria were present in all the samples. Misclassification of the non-equine transcripts was considered as a possibility. The finding of low frequency transcripts of Mycobacterium tuberculosis in one SAO+ BM sample was not supported by clinical and pathological examination. Elimination of the poly A RNA selection step would potentially resolve limitations of the metatranscriptomic analysis in including infectious agents that do not transcribe their genetic material in this RNA form.
In conclusion, the data revealed an SAO associated transcriptome profile in BM tissue skewed toward skeletal formation which is also supported by microscopic findings (at bone sites distinct from lesions of osteolysis). The hypothesized molecular signal of a pathogenic link connecting the pulmonary and skeletal lesions was not elucidated by our results, and increased transcripts associated with osteoclast differentiation and activation were relatively fewer than osteoblast related transcripts. A clear transcriptome pattern was not identified between affected and unaffected animals due to the cellular heterogeneity in the tLN samples. Specific cell targets from the BM and tLN tissues would address the potential dilution of the disease-specific transcriptome pattern. Transcriptome profile of osteoclasts, osteoclast precursors or macrophages from osteolytic SAO lesions may provide better understanding of osteoclast dysregulation. The effect of ongoing pulmonary inflammation and silicosis on osteoblasts and regulation of bone formation warrants investigation in light of our findings. This is the first study that uses RNA-seq technology in investigation of disease mechanism in the equine co-morbid condition though RNA-seq has been used in research of other equine diseases.[75–79] SAO may provide an opportunity for a unique understanding of mechanisms in an environmentally induced condition with pulmonary/tLN and skeletal comorbidity.
Supporting information
The sequenced samples transcript patterns cluster based on tissue in this MDS plot that simultaneously evaluated BM and tLN transcriptomes together.
(TIF)
BM MDS plot demonstrates clustering of cases (red) based on bone phenotype. The group with mild osteoporosis (encircled in black) co-localized with control cases.
(TIF)
tLN MDS plot demonstrates clustering of control cases and wider dispersal of the cases transcriptome profiles.
(TIF)
The most enriched biological processes related infectious agents like virus and bacterial elements in the SAO+ tLN with raw p-value <0.05 are included in the bar chart. Bars represent -Log 10 converted p values. S5 Table includes the entire list of all the significant biological functions for tLN GO enrichment analysis.
(TIF)
(TIF)
(TIF)
Animal signalment and quality of RNA extracted from BM (BM RIN) and tLN (LN RIN) for SAO+ and Control horses.
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
Acknowledgments
We thank Dr. Coral Armstrong and Dr. Durham for their clinical guidance and expertise in SAO that helped to shape this study. We thank Dr. Joseph Fass, Dr. Matt Settles, Nik Joshi for consultations on study design and Metatranscriptomic analysis and its interpretation. We are grateful to Dr. Patricia Pesavento for consultation and interpretation of the Metagenomic analysis. Dr. Nicole Baumgarth and Dr. Clare Yellowley critically reviewed the paper. Ashley Nola and Tiffany Lee were instrumental in assisting with necropsies and collecting tissues for histopathology and RNA extraction.
Abbreviations
- BM
bone marrow
- DGE
differential gene expression
- GO
Gene ontology
- M
mild SAO+ phenotype
- MDS
Multidimensional scaling plot
- RIN
RNA (ribonucleic acid) integrity score
- RNA-seq
RNA sequencing
- S
severe SAO+ phenotype
- SAO
Silicate Associated Osteoporosis
- SAO+
tissues and samples from horses positive for SAO phenotype
- SAO-
tissues and samples from horses negative for SAO phenotype, and without exposure to the SAO endemic regions
- tLN
tracheobronchial lymph node
Data Availability
All sequence data is available from the SRA database (accession: SRP133539). Other relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was financially supported by the Beijing Genomics Institute at UC Davis Signature Research in Genomics Grant Program (SRGESAO 69240 to SMS); the Center for Equine Health UC Davis (V431U81 to RZ) with funds provided by the State of California satellite wagering fund and contributions by private donors; National Institutes of Health (NIH), National Center for Research Resources as a Ruth L. Kirschstein National Research Service (NRSA) Award: Comparative Medical Science Training Program: T32 OD 011147 to RZ. Support for C.J.F. provided by the NIH: National Institute of Neurological Disorders and Stroke (1K01OD015134-01A1 and L40 TR001136). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Arens AM, Barr B, Puchalski SM, Poppenga R, Kulin RM, Anderson J, et al. Osteoporosis associated with pulmonary silicosis in an equine bone fragility syndrome. Veterinary pathology. 2011;48(3):593–615. doi: 10.1177/0300985810385151 . [DOI] [PubMed] [Google Scholar]
- 2.Durham M AC, editor Fractures and bone deformities in 18 horses with silicosis in Proceedings. 52nd Annual Convention of the American Association of Equine Practitioners; 2006; San Antonio, TX.
- 3.Anderson JD, Galuppo LD, Barr BC, Puchalski SM, Macdonald MM, Whitcomb MB, et al. Clinical and scintigraphic findings in horses with a bone fragility disorder: 16 cases (1980–2006). Journal of the American Veterinary Medical Association. 2008;232(11):1694–9. doi: 10.2460/javma.232.11.1694 . [DOI] [PubMed] [Google Scholar]
- 4.Schwartz LW, Knight HD, Whittig LD, Malloy RL, Abraham JL, Tyler NK. Silicate pneumoconiosis and pulmonary fibrosis in horses from the Monterey-Carmel peninsula. Chest. 1981;80(1 Suppl):82–5. . [DOI] [PubMed] [Google Scholar]
- 5.Seitz S, Priemel M, Zustin J, Beil FT, Semler J, Minne H, et al. Paget's disease of bone: histologic analysis of 754 patients. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research. 2009;24(1):62–9. doi: 10.1359/jbmr.080907 . [DOI] [PubMed] [Google Scholar]
- 6.Nebot Valenzuela E, Pietschmann P. Epidemiology and pathology of Paget's disease of bone—a review. Wien Med Wochenschr. 2016. doi: 10.1007/s10354-016-0496-4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yildizgoren MT, Ekiz T, Nadir Ozis T, Baki AE, Tutkun E, Ozgirgin N. Osteoporosis: can it be related to silicosis? Tuberk Toraks. 2014;62(1):98–9. . [DOI] [PubMed] [Google Scholar]
- 8.Yildizgoren MT, Ozis TN, Baki AE, Tutkun E, Yilmaz H, Tiftik T, et al. Evaluation of bone mineral density and 25-hydroxyvitamin D levels in subjects with silica exposure. Environ Health Prev Med. 2016;21(3):149–53. doi: 10.1007/s12199-016-0509-3 ; PubMed Central PMCID: PMCPMC4823218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balgairies E, Amoudru C. [Spontaneous fractures of the ribs in silicotics]. J Radiol Electrol Arch Electr Medicale. 1957;38(11–12):1112–5. . [PubMed] [Google Scholar]
- 10.Sheikh S, Gemma S, Patel A. Factors associated with low bone mineral density in patients with cystic fibrosis. Journal of bone and mineral metabolism. 2014. doi: 10.1007/s00774-014-0572-z . [DOI] [PubMed] [Google Scholar]
- 11.Jorgensen NR, Schwarz P, Holme I, Henriksen BM, Petersen LJ, Backer V. The prevalence of osteoporosis in patients with chronic obstructive pulmonary disease: a cross sectional study. Respiratory medicine. 2007;101(1):177–85. doi: 10.1016/j.rmed.2006.03.029 . [DOI] [PubMed] [Google Scholar]
- 12.Chen SJ, Liao WC, Huang KH, Lin CL, Tsai WC, Kung PT, et al. Chronic obstructive pulmonary disease is a strong independent risk factor for osteoporosis and pathologic fractures: a population-based cohort study. QJM: monthly journal of the Association of Physicians. 2015. doi: 10.1093/qjmed/hcv062 . [DOI] [PubMed] [Google Scholar]
- 13.Adriani A, Pantaleoni S, Luchino M, Ribaldone DG, Reggiani S, Sapone N, et al. Osteopenia and osteoporosis in patients with new diagnosis of inflammatory bowel disease. Panminerva medica. 2014;56(2):145–9. . [PubMed] [Google Scholar]
- 14.Garcia-Carrasco M, Mendoza-Pinto C, Escarcega RO, Jimenez-Hernandez M, Etchegaray Morales I, Munguia Realpozo P, et al. Osteoporosis in patients with systemic lupus erythematosus. The Israel Medical Association journal: IMAJ. 2009;11(8):486–91. . [PubMed] [Google Scholar]
- 15.McLean RR. Proinflammatory cytokines and osteoporosis. Curr Osteoporos Rep. 2009;7(4):134–9. . [DOI] [PubMed] [Google Scholar]
- 16.Yasuda H, Shima N, Nakagawa N, Yamaguchi K, Kinosaki M, Mochizuki S, et al. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(7):3597–602. ; PubMed Central PMCID: PMCPMC19881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takayanagi H. New immune connections in osteoclast formation. Ann N Y Acad Sci. 2010;1192:117–23. doi: 10.1111/j.1749-6632.2009.05303.x . [DOI] [PubMed] [Google Scholar]
- 18.Streett DP, KR; Gerritsen, AT; Hunter, SS; Settles, ML., editor expHTS: analysis of high throughput sequence data in an experimental framework. Proceedings of the 6th ACM Conference on Bioinformatics, Computational Biology and Health Informatics; 2015 September 09–12, 2015 Atlanta, Georgia: ACM New York, NY, USA.
- 19.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635 ; PubMed Central PMCID: PMCPMC3530905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. doi: 10.1093/bioinformatics/btp616 ; PubMed Central PMCID: PMCPMC2796818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47 doi: 10.1093/nar/gkv007 ; PubMed Central PMCID: PMCPMC4402510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22(13):1600–7. doi: 10.1093/bioinformatics/btl140 . [DOI] [PubMed] [Google Scholar]
- 23.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15(3):R46 doi: 10.1186/gb-2014-15-3-r46 ; PubMed Central PMCID: PMCPMC4053813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu J BF, Thielen P, Salzberg SL. Bracken: estimating species abundance in metagenomics data. PeerJ Computer Science. 2017;(3:e104). https://doi.org/10.7717/peerj-cs.104. [Google Scholar]
- 25.David A. Streett KRP, Alida T. Gerritsen, Samuel S. Hunter, and Matthew L. Settles., editor expHTS: analysis of high throughput sequence data in an experimental framework. ACM Conference on Bioinformatics, Computational Biology and Health Informatics (BCB '15); 2015; ACM, New York, NY, USA.
- 26.Victoria JG, Kapoor A, Li L, Blinkova O, Slikas B, Wang C, et al. Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J Virol. 2009;83(9):4642–51. doi: 10.1128/JVI.02301-08 ; PubMed Central PMCID: PMCPMC2668503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Battaglino RA, Pham L, Morse LR, Vokes M, Sharma A, Odgren PR, et al. NHA-oc/NHA2: a mitochondrial cation-proton antiporter selectively expressed in osteoclasts. Bone. 2008;42(1):180–92. doi: 10.1016/j.bone.2007.09.046 ; PubMed Central PMCID: PMCPMC3593247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ninomiya K, Miyamoto T, Imai J, Fujita N, Suzuki T, Iwasaki R, et al. Osteoclastic activity induces osteomodulin expression in osteoblasts. Biochemical and biophysical research communications. 2007;362(2):460–6. doi: 10.1016/j.bbrc.2007.07.193 . [DOI] [PubMed] [Google Scholar]
- 29.Tashima T, Nagatoishi S, Sagara H, Ohnuma S, Tsumoto K. Osteomodulin regulates diameter and alters shape of collagen fibrils. Biochemical and biophysical research communications. 2015;463(3):292–6. doi: 10.1016/j.bbrc.2015.05.053 . [DOI] [PubMed] [Google Scholar]
- 30.Choi SJ, Roodman GD, Feng JQ, Song IS, Amin K, Hart PS, et al. In vivo impact of a 4 bp deletion mutation in the DLX3 gene on bone development. Dev Biol. 2009;325(1):129–37. doi: 10.1016/j.ydbio.2008.10.014 ; PubMed Central PMCID: PMCPMC4617241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hassan MQ, Javed A, Morasso MI, Karlin J, Montecino M, van Wijnen AJ, et al. Dlx3 transcriptional regulation of osteoblast differentiation: temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene. Mol Cell Biol. 2004;24(20):9248–61. doi: 10.1128/MCB.24.20.9248-9261.2004 ; PubMed Central PMCID: PMCPMC517873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paic F, Igwe JC, Nori R, Kronenberg MS, Franceschetti T, Harrington P, et al. Identification of differentially expressed genes between osteoblasts and osteocytes. Bone. 2009;45(4):682–92. doi: 10.1016/j.bone.2009.06.010 ; PubMed Central PMCID: PMCPMC2731004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamande SR, Yuan Y, Gresshoff IL, Rowley L, Belluoccio D, Kaluarachchi K, et al. Mutations in TRPV4 cause an inherited arthropathy of hands and feet. Nat Genet. 2011;43(11):1142–6. doi: 10.1038/ng.945 . [DOI] [PubMed] [Google Scholar]
- 34.Rock MJ, Prenen J, Funari VA, Funari TL, Merriman B, Nelson SF, et al. Gain-of-function mutations in TRPV4 cause autosomal dominant brachyolmia. Nat Genet. 2008;40(8):999–1003. doi: 10.1038/ng.166 ; PubMed Central PMCID: PMCPMC3525077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guilak F, Leddy HA, Liedtke W. Transient receptor potential vanilloid 4: The sixth sense of the musculoskeletal system? Ann N Y Acad Sci. 2010;1192:404–9. doi: 10.1111/j.1749-6632.2010.05389.x ; PubMed Central PMCID: PMCPMC3580043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masuyama R, Vriens J, Voets T, Karashima Y, Owsianik G, Vennekens R, et al. TRPV4-mediated calcium influx regulates terminal differentiation of osteoclasts. Cell Metab. 2008;8(3):257–65. doi: 10.1016/j.cmet.2008.08.002 . [DOI] [PubMed] [Google Scholar]
- 37.Koch M, Laub F, Zhou P, Hahn RA, Tanaka S, Burgeson RE, et al. Collagen XXIV, a vertebrate fibrillar collagen with structural features of invertebrate collagens: selective expression in developing cornea and bone. J Biol Chem. 2003;278(44):43236–44. doi: 10.1074/jbc.M302112200 . [DOI] [PubMed] [Google Scholar]
- 38.Koga T, Matsui Y, Asagiri M, Kodama T, de Crombrugghe B, Nakashima K, et al. NFAT and Osterix cooperatively regulate bone formation. Nat Med. 2005;11(8):880–5. doi: 10.1038/nm1270 . [DOI] [PubMed] [Google Scholar]
- 39.Peng Y, Shi K, Wang L, Lu J, Li H, Pan S, et al. Characterization of Osterix protein stability and physiological role in osteoblast differentiation. PLoS One. 2013;8(2):e56451 doi: 10.1371/journal.pone.0056451 ; PubMed Central PMCID: PMCPMC3574093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao Y, Jheon A, Nourkeyhani H, Kobayashi H, Ganss B. Molecular cloning, structure, expression, and chromosomal localization of the human Osterix (SP7) gene. Gene. 2004;341:101–10. doi: 10.1016/j.gene.2004.05.026 . [DOI] [PubMed] [Google Scholar]
- 41.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108(1):17–29. . [DOI] [PubMed] [Google Scholar]
- 42.Vincent K, Durrant MC. A structural and functional model for human bone sialoprotein. J Mol Graph Model. 2013;39:108–17. doi: 10.1016/j.jmgm.2012.10.007 . [DOI] [PubMed] [Google Scholar]
- 43.Jazayeri M, Shokrgozar MA, Haghighipour N, Mahdian R, Farrokhi M, Bonakdar S, et al. Evaluation of Mechanical and Chemical Stimulations on Osteocalcin and Runx2 Expression in Mesenchymal Stem Cells. Mol Cell Biomech. 2015;12(3):197–213. . [PubMed] [Google Scholar]
- 44.Stein GS, Lian JB, van Wijnen AJ, Stein JL, Montecino M, Javed A, et al. Runx2 control of organization, assembly and activity of the regulatory machinery for skeletal gene expression. Oncogene. 2004;23(24):4315–29. doi: 10.1038/sj.onc.1207676 . [DOI] [PubMed] [Google Scholar]
- 45.Li J, Manickam G, Ray S, Oh CD, Yasuda H, Moffatt P, et al. Smpd3 Expression in both Chondrocytes and Osteoblasts Is Required for Normal Endochondral Bone Development. Mol Cell Biol. 2016;36(17):2282–99. doi: 10.1128/MCB.01077-15 ; PubMed Central PMCID: PMCPMC4985927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khavandgar Z, Murshed M. Sphingolipid metabolism and its role in the skeletal tissues. Cell Mol Life Sci. 2015;72(5):959–69. doi: 10.1007/s00018-014-1778-x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lui VC, Ng LJ, Sat EW, Cheah KS. The human alpha 2(XI) collagen gene (COL11A2): completion of coding information, identification of the promoter sequence, and precise localization within the major histocompatibility complex reveal overlap with the KE5 gene. Genomics. 1996;32(3):401–12. doi: 10.1006/geno.1996.0135 . [DOI] [PubMed] [Google Scholar]
- 48.Shen Z, Gantcheva S, Mansson B, Heinegard D, Sommarin Y. Chondroadherin expression changes in skeletal development. Biochem J. 1998;330 (Pt 1):549–57. ; PubMed Central PMCID: PMCPMC1219172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou HY. Proteomic analysis of hydroxyapatite interaction proteins in bone. Ann N Y Acad Sci. 2007;1116:323–6. doi: 10.1196/annals.1402.023 . [DOI] [PubMed] [Google Scholar]
- 50.Mizuta K, Tsutsumi S, Inoue H, Sakamoto Y, Miyatake K, Miyawaki K, et al. Molecular characterization of GDD1/TMEM16E, the gene product responsible for autosomal dominant gnathodiaphyseal dysplasia. Biochemical and biophysical research communications. 2007;357(1):126–32. doi: 10.1016/j.bbrc.2007.03.108 . [DOI] [PubMed] [Google Scholar]
- 51.Luu-The V, Zhang Y, Poirier D, Labrie F. Characteristics of human types 1, 2 and 3 17 beta-hydroxysteroid dehydrogenase activities: oxidation/reduction and inhibition. J Steroid Biochem Mol Biol. 1995;55(5–6):581–7. . [DOI] [PubMed] [Google Scholar]
- 52.Geering B, Stoeckle C, Rozman S, Oberson K, Benarafa C, Simon HU. DAPK2 positively regulates motility of neutrophils and eosinophils in response to intermediary chemoattractants. J Leukoc Biol. 2014;95(2):293–303. doi: 10.1189/jlb.0813462 . [DOI] [PubMed] [Google Scholar]
- 53.Mantovani A, Valentino S, Gentile S, Inforzato A, Bottazzi B, Garlanda C. The long pentraxin PTX3: a paradigm for humoral pattern recognition molecules. Ann N Y Acad Sci. 2013;1285:1–14. doi: 10.1111/nyas.12043 . [DOI] [PubMed] [Google Scholar]
- 54.Basile A, Sica A, d'Aniello E, Breviario F, Garrido G, Castellano M, et al. Characterization of the promoter for the human long pentraxin PTX3. Role of NF-kappaB in tumor necrosis factor-alpha and interleukin-1beta regulation. J Biol Chem. 1997;272(13):8172–8. . [DOI] [PubMed] [Google Scholar]
- 55.Maliszewski CR, March CJ, Schoenborn MA, Gimpel S, Shen L. Expression cloning of a human Fc receptor for IgA. J Exp Med. 1990;172(6):1665–72. ; PubMed Central PMCID: PMCPMC2188749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.D'Souza RN, Cavender A, Sunavala G, Alvarez J, Ohshima T, Kulkarni AB, et al. Gene expression patterns of murine dentin matrix protein 1 (Dmp1) and dentin sialophosphoprotein (DSPP) suggest distinct developmental functions in vivo. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research. 1997;12(12):2040–9. doi: 10.1359/jbmr.1997.12.12.2040 . [DOI] [PubMed] [Google Scholar]
- 57.He G, Dahl T, Veis A, George A. Dentin matrix protein 1 initiates hydroxyapatite formation in vitro. Connect Tissue Res. 2003;44 Suppl 1:240–5. . [PubMed] [Google Scholar]
- 58.Chaplet M, De Leval L, Waltregny D, Detry C, Fornaciari G, Bevilacqua G, et al. Dentin matrix protein 1 is expressed in human lung cancer. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research. 2003;18(8):1506–12. doi: 10.1359/jbmr.2003.18.8.1506 . [DOI] [PubMed] [Google Scholar]
- 59.Li H, Marijanovic I, Kronenberg MS, Erceg I, Stover ML, Velonis D, et al. Expression and function of Dlx genes in the osteoblast lineage. Dev Biol. 2008;316(2):458–70. doi: 10.1016/j.ydbio.2008.01.001 ; PubMed Central PMCID: PMCPMC2679944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ingram RT, Clarke BL, Fisher LW, Fitzpatrick LA. Distribution of noncollagenous proteins in the matrix of adult human bone: evidence of anatomic and functional heterogeneity. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research. 1993;8(9):1019–29. doi: 10.1002/jbmr.5650080902 . [DOI] [PubMed] [Google Scholar]
- 61.Fisher LW, Whitson SW, Avioli LV, Termine JD. Matrix sialoprotein of developing bone. J Biol Chem. 1983;258(20):12723–7. . [PubMed] [Google Scholar]
- 62.Hoang QQ, Sicheri F, Howard AJ, Yang DS. Bone recognition mechanism of porcine osteocalcin from crystal structure. Nature. 2003;425(6961):977–80. doi: 10.1038/nature02079 . [DOI] [PubMed] [Google Scholar]
- 63.Mizuno M, Imai T, Fujisawa R, Tani H, Kuboki Y. Bone sialoprotein (BSP) is a crucial factor for the expression of osteoblastic phenotypes of bone marrow cells cultured on type I collagen matrix. Calcified tissue international. 2000;66(5):388–96. . [DOI] [PubMed] [Google Scholar]
- 64.Matsuo N, Tanaka S, Yoshioka H, Koch M, Gordon MK, Ramirez F. Collagen XXIV (Col24a1) gene expression is a specific marker of osteoblast differentiation and bone formation. Connect Tissue Res. 2008;49(2):68–75. doi: 10.1080/03008200801913502 . [DOI] [PubMed] [Google Scholar]
- 65.David V, Martin A, Lafage-Proust MH, Malaval L, Peyroche S, Jones DB, et al. Mechanical loading down-regulates peroxisome proliferator-activated receptor gamma in bone marrow stromal cells and favors osteoblastogenesis at the expense of adipogenesis. Endocrinology. 2007;148(5):2553–62. doi: 10.1210/en.2006-1704 . [DOI] [PubMed] [Google Scholar]
- 66.Sonomoto K, Yamaoka K, Oshita K, Fukuyo S, Zhang X, Nakano K, et al. Interleukin-1beta induces differentiation of human mesenchymal stem cells into osteoblasts via the Wnt-5a/receptor tyrosine kinase-like orphan receptor 2 pathway. Arthritis Rheum. 2012;64(10):3355–63. doi: 10.1002/art.34555 . [DOI] [PubMed] [Google Scholar]
- 67.Salazar VS, Gamer LW, Rosen V. BMP signalling in skeletal development, disease and repair. Nat Rev Endocrinol. 2016;12(4):203–21. doi: 10.1038/nrendo.2016.12 . [DOI] [PubMed] [Google Scholar]
- 68.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89(5):755–64. . [DOI] [PubMed] [Google Scholar]
- 69.Komori T. Runx2, a multifunctional transcription factor in skeletal development. J Cell Biochem. 2002;87(1):1–8. doi: 10.1002/jcb.10276 . [DOI] [PubMed] [Google Scholar]
- 70.Biswas MG, Russell DW. Expression cloning and characterization of oxidative 17beta- and 3alpha-hydroxysteroid dehydrogenases from rat and human prostate. J Biol Chem. 1997;272(25):15959–66. . [DOI] [PubMed] [Google Scholar]
- 71.Terasawa M, Shimokawa R, Terashima T, Ohya K, Takagi Y, Shimokawa H. Expression of dentin matrix protein 1 (DMP1) in nonmineralized tissues. Journal of bone and mineral metabolism. 2004;22(5):430–8. doi: 10.1007/s00774-004-0504-4 . [DOI] [PubMed] [Google Scholar]
- 72.Li X, Hu Y, Jin Z, Jiang H, Wen J. Silica-induced TNF-alpha and TGF-beta1 expression in RAW264.7 cells are dependent on Src-ERK/AP-1 pathways. Toxicol Mech Methods. 2009;19(1):51–8. doi: 10.1080/15376510802354201 . [DOI] [PubMed] [Google Scholar]
- 73.Mossman BT, Churg A. Mechanisms in the pathogenesis of asbestosis and silicosis. American journal of respiratory and critical care medicine. 1998;157(5 Pt 1):1666–80. doi: 10.1164/ajrccm.157.5.9707141 . [DOI] [PubMed] [Google Scholar]
- 74.Perkins TN, Peeters PM, Shukla A, Arijs I, Dragon J, Wouters EF, et al. Indications for distinct pathogenic mechanisms of asbestos and silica through gene expression profiling of the response of lung epithelial cells. Hum Mol Genet. 2015;24(5):1374–89. doi: 10.1093/hmg/ddu551 ; PubMed Central PMCID: PMCPMC4402341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tessier L, Cote O, Clark ME, Viel L, Diaz-Mendez A, Anders S, et al. Impaired response of the bronchial epithelium to inflammation characterizes severe equine asthma. BMC Genomics. 2017;18(1):708 doi: 10.1186/s12864-017-4107-6 ; PubMed Central PMCID: PMCPMC5591550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Finno CJ, Bordbari MH, Valberg SJ, Lee D, Herron J, Hines K, et al. Transcriptome profiling of equine vitamin E deficient neuroaxonal dystrophy identifies upregulation of liver X receptor target genes. Free Radic Biol Med. 2016;101:261–71. doi: 10.1016/j.freeradbiomed.2016.10.009 ; PubMed Central PMCID: PMCPMC5154892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Semik E, Gurgul A, Zabek T, Ropka-Molik K, Koch C, Mahlmann K, et al. Transcriptome analysis of equine sarcoids. Vet Comp Oncol. 2017;15(4):1370–81. doi: 10.1111/vco.12279 . [DOI] [PubMed] [Google Scholar]
- 78.Tallmadge RL, Shen L, Tseng CT, Miller SC, Barry J, Felippe MJ. Bone marrow transcriptome and epigenome profiles of equine common variable immunodeficiency patients unveil block of B lymphocyte differentiation. Clin Immunol. 2015;160(2):261–76. doi: 10.1016/j.clim.2015.05.005 ; PubMed Central PMCID: PMCPMC4581901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McIlwraith CW, Kawcak CE, Frisbie DD, Little CB, Clegg PD, Peffers MJ, et al. Biomarkers for equine joint injury and osteoarthritis. Journal of orthopaedic research: official publication of the Orthopaedic Research Society. 2017. doi: 10.1002/jor.23738 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The sequenced samples transcript patterns cluster based on tissue in this MDS plot that simultaneously evaluated BM and tLN transcriptomes together.
(TIF)
BM MDS plot demonstrates clustering of cases (red) based on bone phenotype. The group with mild osteoporosis (encircled in black) co-localized with control cases.
(TIF)
tLN MDS plot demonstrates clustering of control cases and wider dispersal of the cases transcriptome profiles.
(TIF)
The most enriched biological processes related infectious agents like virus and bacterial elements in the SAO+ tLN with raw p-value <0.05 are included in the bar chart. Bars represent -Log 10 converted p values. S5 Table includes the entire list of all the significant biological functions for tLN GO enrichment analysis.
(TIF)
(TIF)
(TIF)
Animal signalment and quality of RNA extracted from BM (BM RIN) and tLN (LN RIN) for SAO+ and Control horses.
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
Data Availability Statement
All sequence data is available from the SRA database (accession: SRP133539). Other relevant data are within the paper and its Supporting Information files.