Abstract
Drought stress negatively affects plant growth and development. An increasing number of reports have revealed the involvement of APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors (TFs) in biotic and abiotic stress regulation in plants. However, research on these TFs in the peanut plant (Arachis hypogaea) has been limited. Here, we isolated a full-length coding sequence (CDS) of the AP2/ERF family gene AhDREB1 from the peanut plant and showed that its expression was induced by Polyethylene Glycol (PEG) 6000 and exogenous abscisic acid (ABA) treatment. When overexpressed in Arabidopsis, AhDREB1 increased both ABA levels and ABA sensitivity, affected the ABA signaling pathway and increased the expression of downstream drought stress-related genes RD29A, P5CS1, P5CS2 and NCED1. These results demonstrate that AhDREB1 can improve tolerance to drought via the ABA-dependent pathway in Arabidopsis. In the peanut plant, the specific histone deacetylases (HDACs) inhibitor trichostatin A (TSA) promotes AhDREB1 transcription and the enrichment level of H3ac was increased in regions of the AhDREB1 gene during TSA and PEG treatment. In summary, histone acetylation can affect the expression of AhDREB1 under osmotic stress conditions, thereby improving plant drought resistance.
Keywords: APETALA2/Ethylene Responsive Factor, AhDREB1, peanut, abscisic acid, drought stresses, histone acetylation
1. Introduction
Plants are sessile and constantly subjected to various abiotic stresses, including drought, cold, high salinity, heat and alkalinity, which seriously affect their growth, development and productivity [1]. Of these stresses, drought is one of main causes of loss in production of agricultural crops across the world [2]. Drought can result in plant water loss and leaf wilting, and can lead to irreversible, damaging changes including, ultimately, plant death [3].
Over a long period of evolution, plants have developed diverse survival strategies and complex mechanisms to cope with drought stress [4]. Plants respond to drought stress by altering molecular and physiological processes at the molecular, cellular, tissue and whole-plant levels [5,6]. At the molecular level, plants can optimize their growth and development by regulating the expression of numerous stress responses via two main pathways, one of which is abscisic acid (ABA)-dependent, while the other is not [7]. However, several studies suggest that, in fact, there may be cross-talk between the two types of signaling pathway; thus, they may be interdependent [8]. The phytohormone ABA is a pivotal regulator of plant growth and development and of plant responses to drought stress [3]. The content of ABA in plants increases rapidly under drought stress conditions, and returns to its original level when the stress is removed. Because it correlates with drought resistance, the level of ABA is an important index of drought resistance in plants [9]. Multiple processes are involved in the biosynthesis of ABA, but the main factor during drought is the increase in expression of 9-cis-epoxycarotenoid dioxygenase (NCED), which encodes a key enzyme in the ABA biosynthetic pathway [10,11]. In addition, plants regulate the expression of drought-resistance genes via relevant components of ABA signaling pathway, thus protecting plants from drought stress. ABA can change the transcription level of drought-related genes via the ABA-responsive element (ABRE) binding protein [12,13].
In the ABA-independent pathway, dehydration-responsive element binding protein (DREB), plays the main role in regulating many drought-responsive genes. Other transcription factors such as NAC (NAM, ATAF, CUC), MYB (v-myb avian myeloblastosis viral oncogene homolog), MYC (Myelocytomatosis proteins), WRKY and nuclear factor-Y (NF-Y) are also involved in drought response and tolerance [14]. DREB activates drought-associated genes, such as RD29A, P5CS1, SODs, by binding to dehydration responsive element (DRE) motifs [15,16], which comprise nine conserved bases (TACCGACAT), in their promoter regions, thereby increasing the plant’s drought resistance [14,17]. DREB transcription factors form a subfamily of the APETALA2/Ethylene Responsive Factor (AP2/ERF) protein family [18,19].
AP2/ERFs are a large family of plant-specific transcription factors (TFs) and the rapid development of genome-wide sequencing technology and bioinformatics tools has led to the identification of AP2/ERF genes in many species; for example, 139 and 122 AP2/ERF genes have been found in rice and Arabidopsis thaliana, respectively [20]. TFs from the AP2/ERF family have either one or two highly conserved DNA-binding AP2/ERF domains, each of 58 or 59 amino acids residues [21]. Based on DNA binding domain (DBD) and sequence similarity, AP2/ERF family members have been divided into four major subfamilies, comprising DREB factors, ERFs, AP2 factors and a subfamily called Related to ABI3 and VP1 (RAV) [18,19]. The DREB and ERF subfamilies, both containing a single AP2 domain, are distinguished from each other by differences in the conserved residues of their DBDs. In DREB proteins, the 14th and 19th positions of the DBD are valine (V14) and glutamic acid (E19), whereas in ERFs, the corresponding positions are alanine (A14) and aspartic acid (D19) [18]. Members of the AP2 subfamily have two AP2/ERF domains, while members of the RAV subfamily have a B3 DBD in addition to an AP2/ERF domain [18,19].
In recent years, an increasing number of reports have revealed AP2/ERF TFs to be involved in multiple biological processes that regulate plant growth and development, and especially in abiotic stress tolerance. Among the AP2/ERF family, DREB and ERF subfamily proteins have been well characterized in abiotic stress responses involving drought, salt, cold and other stresses [20]. Thus, overexpression of GsERF71, which encodes an AP2/ERF TF from wild soybean, improves plant alkaline stress tolerance by upregulating H+-ATPase and by modifying the accumulation of auxin [22]. In Arabidopsis, overexpressing AtERF019 increases tolerance to water deficiency, and also results in phenotypes that delay flowering and senescence [23]. In Arabidopsis, overexpression of DREB1 can enhance tolerance to cold stress, promote the accumulation of proline and result in phenotypes of plant dwarfing and delay of flowering [24]. Overexpression of ScDREB8 significantly improves salt tolerance of Arabidopsis at the seedling stage by upregulating the expression of downstream stress-related genes and improving reactive oxygen species (ROS) scavenging ability [25]. Overexpression of OsDREB genes enhances drought tolerance in rice [26]. In Arabidopsis, strains overexpressing AtDREB2C are highly sensitive to ABA during seed germination and show enhanced tolerance to cold and heat stress [8].
It has been shown that epigenetic regulation plays an important role in plant growth and development [27]. Epigenetic regulation mainly involves two aspects of DNA and histone. At the level of DNA, it is mainly the methylation of genomic DNA, while at the histone levels are mainly histone acetylation, methylation, glycosylation, phosphorylation, ubiquitination, and so on [28,29]. Histone acetylation levels are controlled by the dynamic equilibrium between histone acetyltransferases (HATs) and histone deacetylases (HDACs) [30]. Thus, acetylation of histone lysine residues by HATs can relax chromatin structure and thus facilitate gene activation. Conversely, HDACs remove acetyl groups from acetylated histones, and this activity is generally associated with transcriptional repression and gene silencing [31]. Recently, some studies have shown that histone acetylation can affect the development of plant seeds, the formation and opening of flowers, the elongation of roots, gametophyte development and other biological processes. [32,33]. In maize, mutation of the HDAC gene hda101 results in a smaller kernel than that of wild type, and in defects of basal endosperm transfer layer (BETL) cells, which suggests that HDA101 can affect the development of seed in maize [34]. In A. thaliana, AtHDA5 affects flowering time, and the hda5 mutant exhibits a late-flowering phenotype [35]. An increasing number of reports have shown that histone acetylation affects ABA and other phytohormone signaling pathways, and is also important for the epigenetic regulation of plant responses to external stress [36,37]. However, the specific mechanisms are not clear. Recently, it has been reported that the MSI1-HDA19 complex affects ABA signaling pathway by binding to the chromatin of ABA receptor genes PYL2, PYL4 and PYL6, where it maintains low levels of acetylation of histone H3 at lysine 9, thereby regulating expression levels of these genes [38]. In A. thaliana, AtHDA9 negatively affects plant sensitivity to drought and salt stresses by modifying histone acetylation levels of a large number of stress-responsive genes [27]. In maize, HDACs can affect the level of histone acetylation in chromatin, which in turn promotes the expression of ZmDREB1 under cold stress [39].
Peanut (Arachis hypogaea L.) is one of the most important oil crops and food sources of protein, and is cultivated in the semi-arid tropical and subtropical regions of the world [40]. Peanut plants are constantly subjected to various biotic and abiotic stresses during growth and development. Drought stress is one of the main factors that limit the growth, yield and quality of peanuts [36]. Therefore, it is meaningful to study how peanut plants respond to drought stress, which has potential applications in the breeding of drought-resistant strains. Previously, in the peanut transcriptome database, we found a DREB-like gene (comp63385_c0) that contained a key AP2/ERF domain and was upregulated by water deficit and exogenous ABA [41]. This gene was named AhDREB1 (GenBank accession No. KU143745.1). In this paper, the full-length coding sequence (CDS) of AhDREB1 was isolated from peanut leaves and its transcription was found to be induced by Polyethylene Glycol (PEG) 6000 and exogenous ABA treatment. Overexpression of AhDREB1 in Arabidopsis increased ABA sensitivity during seed germination, affected the expression of ABA signaling pathway-related genes and enhanced drought resistance. Furthermore, the H3ac enrichment level was increased in various regions of AhDREB1 chromatin during osmotic stress and treatment with the specific HDAC inhibitor trichostatin A (TSA). In summary, histone acetylation can affect the expression of AhDREB1 under drought conditions, and may be important in further improving drought resistance of peanut plants.
2. Results
2.1. Isolation and Characterization of Peanut AhDREB1 Gene
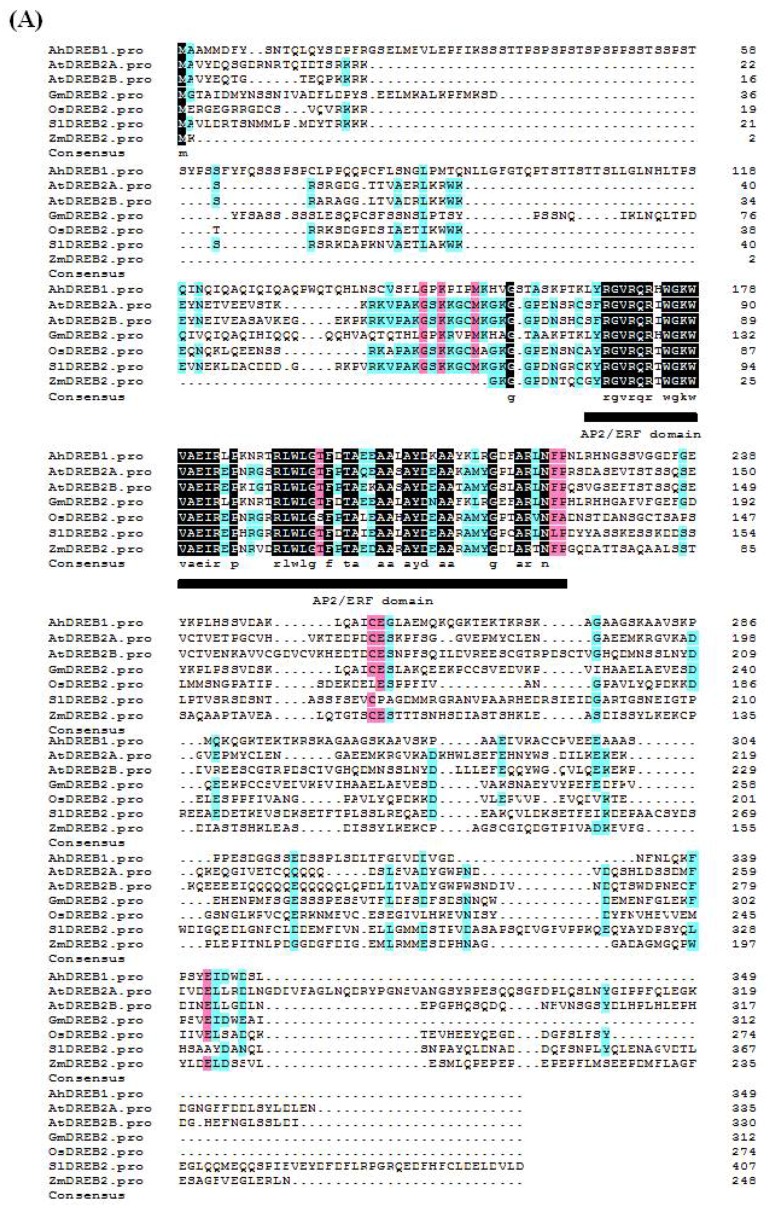
The full-length CDS of AhDREB1 gene was isolated from peanut leaf DNA consists of a 1050-bp open reading frame (ORF) that encodes a polypeptide of 349 amino acid residues and predicted molecular mass ~37.8 kDa (Figure 1A). Sequence analysis showed that AhDREB1 contains one conserved AP2/ERF DBD (residues 167–227), including the two conserved residues V14 and L19 specific to DREB TFs [18]. Multiple sequence alignment using DNAMAN8.0 software suggested that AhDREB1 also has a low degree of similarity to the known DREB genes from A. thaliana, Glycine max, Oryza sativa, Solanum lycopersicum and Zea mays outside the AP2/ERF domain (Supplementary Figure S1). The AhDREB1 promoter sequence contains many putative stress response-related cis-elements, including examples of the ABRE element (6 hits), CE3 recognition site (1 hit), MYB recognition site (1 hit) and heat shock response element (HSE) recognition site (3 hits) (Supplementary Figure S2). Secondary structure prediction suggested AhDREB1 has no transmembrane structure (Figure 1B). To determine the subcellular localization of AhDREB1, we engineered a plasmid construct in which the AhDREB1 sequence without the stop codon was inserted into the 5′ end of the eGFP gene in the p35S-eGFP vector. The p35S::AhDREB1-eGFP recombinant plasmid was transformed into Arabidopsis protoplast cells, with the empty p35s-eGFP vector used as a control. Confocal microscopy imaging showed that, while the eGFP (enhanced Green Fluorescent Protein) control protein was distributed throughout the cell, the AhDREB1-eGFP fusion protein was only visible in the nucleus (Figure 1C). These results suggest that AhDREB1 encodes a nuclear protein containing one conserved AP2/ERF DBD, and that AhDREB1 belongs to the DREB TF subfamily.
Figure 1.
AhDREB1 is a member of the APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factor family in peanut. (A) Alignment of amino acid sequences of AhDREB1 with six dehydration-responsive element binding protein (DREB) subfamily proteins. AhDREB1: GenBank number API65088.1; AtDREB2A: GenBank number BAA33794.1; AtDREB2B: GenBank number BAA33795.1; GmDREB2: GenBank number AAQ57226.1; OsDREB2: GenBank number AAN02487.2; SlDREB2: GenBank number ADZ15315.1; ZmDREB2: GenBank number AFI71287.1. Identical amino acid residues are shaded in black. Conservative AP2/ERF domain are marked with a black underline. (B) AhDREB1 has no transmembrane domain, transmembrane structure was analyzed using online TMHMM 2.0 (http://www.cbs.dtu.dk/services/). (C) The intracellular localization of AhDREB1 in Arabidopsis protoplasts. Image showed the eGFP protein was observed in the whole Arabidopsis protoplasts cells, which the AhDREB1-eGFP fusion protein was localized in nucleus in Arabidopsis protoplasts cells, determined by confocal and bright-field microscopy.
2.2. Expression of AhDREB1 Is Induced by Osmotic Stress and ABA Treatment
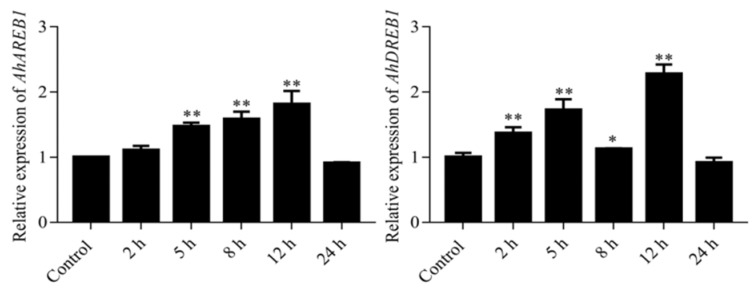
The expression profile of AhDREB1 was examined in peanut leaves treated with 20% PEG 6000 or 100 μM ABA. AhDREB1 expression was increased at 2 h of PEG treatment, slightly declined at 5 h, then significantly increased and maintained a relatively high level from 8 to 24 h. The drought-related genes AhAREB1 and AhNCED1 were also expressed in response to PEG. AhAREB1 levels did not change significantly at time points up to 5 h, but then were significantly upregulated at 8 h, and continuously increased from 12 to 24 h. The expression of AhNCED1 was significantly increased at 2 h, slightly decreased at 5 to 8 h, although it remained higher than the control level, then slowly increased from 12 to 24 h (Figure 2). The expression of AhDREB1 could be induced by osmotic stress caused by PEG treatment suggests that AhDREB1 itself is a drought-responsive gene. Consistent with this, we found that 100 μM ABA treatment produced a remarkable increase in AhDREB1 expression level at 2 to 5 h, which slightly declined at 8 h, then significantly increased at 12 h, and finally declined again at 24 h. In comparison, AhAREB1 expression continuously increased from 2 to 12 h, and then declined 24 h after ABA treatment (Figure 3). Thus, AhDREB1 expression can be increased by both osmotic stress and treatment with exogenous ABA.
Figure 2.
Expression analyses of AhDREB1, AhNCED1 and AhAREB1 following 20% PEG treatment in peanut leaves by real-time quantitative PCR (RT-qPCR). Time points of 2, 5, 8, 12, and 24 h were sampled to observe the expression changing trend. The untreated group was used as the control. Mean and SD were obtained from more than three biological replicates. Asterisks indicate significant differences from control (Student’s t test p values, ** p < 0.01).
Figure 3.
Expression analyses of AhDREB1 and AhAREB1 following 100 μM abscisic acid (ABA) treatment in peanut leaves by RT-qPCR. Time points of 2, 5, 8, 12, and 24 h were sampled to observe the expression changing trend. The untreated group was used as the control. Mean and SD were obtained from more than three biological replicates. Asterisks indicate significant differences from control (Student’s t test p values, * p < 0.05 and ** p < 0.01).
2.3. Overexpression of AhDREB1 in Arabidopsis Enhances Plant Tolerance to Drought Stress
AhDREB1 overexpression (AhDREB1-OX) Arabidopsis lines were obtained to understand the role of AhDREB1 in the drought stress response. Of more than 20 independent lines produced, three independent T3 homozygous AhDREB1-OX lines (OX-1, OX-2, OX-3) with a relatively high level of AhDREB1 transcription were selected for further studies (Figure 4A). We first examined whether AhDREB1-OX lines at the seedling stage display tolerant phenotypes under drought stress conditions. When three-week-old Col and AhDREB1-OX lines were drought stressed for two weeks, OX-2 and OX-3 plants exhibited strong drought tolerance, while Col and OX-1 plants showed severe wilting (Figure 4C). After 2 d rehydration, almost all wild-type and OX-1 plants failed to recover, while OX-2 and OX-3 plants recovered rapidly (Figure 4C). After recovery, 68% of OX-2 and 77% of OX-3 plants survived, in contrast to only 30% of Col and 24% of OX-1 plants (Figure 4B). We found that water content in AhDREB1-OX lines was not significantly different to that of Col plants during dehydration treatment (Figure 4D), but the stomatal aperture in leaves of OX-2 plants was smaller than in Col plants under stress conditions (Figure 4E) (Supplementary Figure S3). The endogenous ABA levels in the rosette leaves of all three independent T3 homozygous transgenic lines were significantly higher than Col under normal growth conditions. Intriguingly, however, ABA content of OX-2 and OX-3 plants was higher than that in Col plants under drought stress conditions, although OX-1 ABA levels were not different to controls (Figure 4F).
Figure 4.
Overexpression of AhDREB1 in Arabidopsis enhances plant tolerance to drought stress. (A) The expression levels of AhDREB1 in AhDREB1-OX lines. (B) Survival rate of 3-week-old wild-type and AhDREB1-OX lines during the drought stress test. (C) Drought tolerance phenotype of 3-week-old wild-type and AhDREB1-OX lines were drought stress for 2 weeks and rehydration 2days. (D) The change in water content in overexpression Arabidopsis thaliana. (E) Stomatal opening in the leaves of 3-week-old wild-type and AhDREB1-OX lines under control conditions or after drought stress for 2 weeks; n = 180. ‘*’ indicates a significant difference at the level of p < 0.05 between AhDREB1-OX lines and Col plants under control treatment or drought stress conditions. (F) ABA content in the leaves of 3-week-old wild-type and AhDREB1-OX lines under control conditions or after drought stress for 2 weeks. All experiments, mean and SD were obtained from more than three biological replicates. Asterisks in (A) to (F), indicate significant differences from Col (Student’s t test P values, * p < 0.05 and ** p < 0.01).
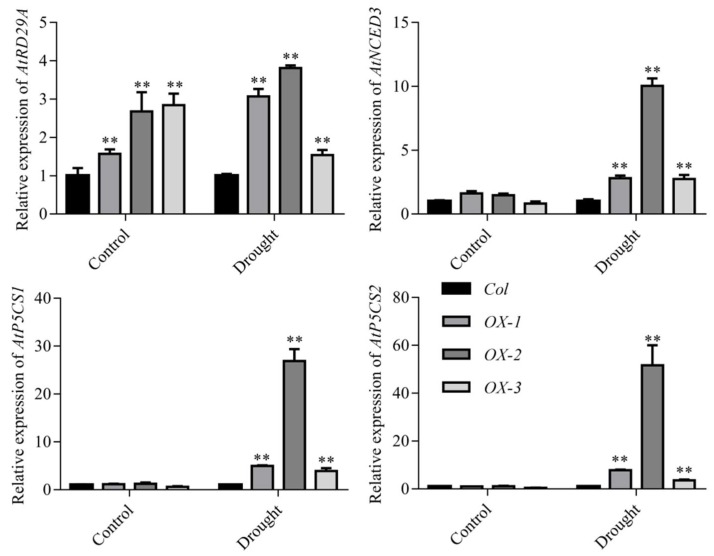
Given that AhDREB1-OX plants showed enhanced drought tolerance, we investigated whether the transcription of drought stress-related marker genes was altered compared to Col control plants. We found that transcript levels of the drought stress-related genes AtRD29A, AtNCED3, AtP5CS1 and AtP5CS2 were significantly higher than Col in all three transgenic lines after drought treatment, especially in OX-2 plants (Figure 5). At the same time, the transcriptional level of AhDREB1 was also significantly increased during PEG treatment. These results indicate that overexpression of AhDREB1 can promote downstream drought-related gene expression. In summary, overexpression of AhDREB1 can enhance drought resistance of the vegetative stages of A. thaliana by increasing ABA levels and promoting the expression of drought-resistance genes.
Figure 5.
Expression analyses of drought stress-related marker genes: AtRD29A, AtNCED3, AtP5CS1, AtP5CS2 in the wild type and transgenic plants under drought stress. The untreated group was used as the control. Mean and SD were obtained from more than three biological replicates. Asterisks indicate significant differences from control (Student’s t test P values, ** p < 0.01).
2.4. AhDREB1 Increases ABA Sensitivity and Affects ABA Signaling Pathway in AhDREB1-OX Arabidopsis Plants
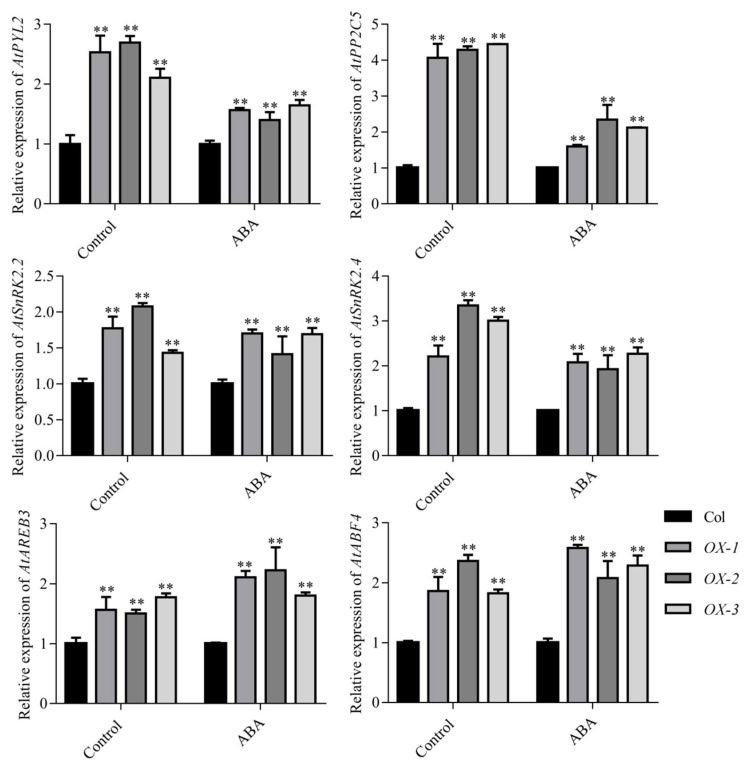
To investigate whether AhDREB1 affects plant drought tolerance via the ABA-dependent pathway, we checked ABA sensitivity of AhDREB1-OX transgenic plants at the germination stage. Seeds of the three overexpression lines, together with Col controls, were germinated in one-half-strength Murashige and Skoog (1/2 MS) medium containing ABA at one of three different concentrations (0, 0.5, 2 μM). AhDREB1-OX seeds exhibited a significantly slower germination rate than wild type following 0.5 and 2 μM ABA treatment, but there were no differences between AhDREB1-OX and Col plants without ABA treatment (Figure 6A–D). Thus, ABA sensitivity of AhDREB1-OX plants was increased. Furthermore, the transcription levels of ABA signaling pathway-related genes AtPYL2, AtPP2C5, AtSnRK2.2, AtSnRK2.4, AtAREB3 and AtABF4 were significantly higher than in Col plants under both normal growth conditions and after exogenous application of 10 μM ABA (Figure 7). In conclusion, AhDREB1 increases ABA sensitivity and affects the ABA signaling pathway in AhDREB1-OX Arabidopsis plants.
Figure 6.
AhDREB1 increases ABA sensitivity in Arabidopsis. (A–C) Seed germination rate of AhDREB1-OX lines and Col in response to different concentrations of ABA. Numbers of germinated seedlings were recorded from 0 to 192 h after stratification on one-half-strength Murashige and Skoog (1/2 MS) agar plates containing 0, 0.5, or 2 μM ABA, respectively. (D) Photographs of seed germination on agar plates containing 0, 0.5 or 2 μM ABA. Mean and SD were obtained from more than three biological replicates.
Figure 7.
Expression changes of ABA signaling pathway related gene: AtPYL2, AtPP2C5, AtSnRK2.2, AtSnRK2.4, AtAREB3, AtABF4 following 10 μM ABA treatment. The untreated group was used as the control. Mean and SD were obtained from more than three biological replicates. Asterisks indicate significant differences from control (Student’s t test p values, ** p < 0.01).
2.5. Histone Acetylation Is Involved in AhDREB1 Transcriptional Regulation
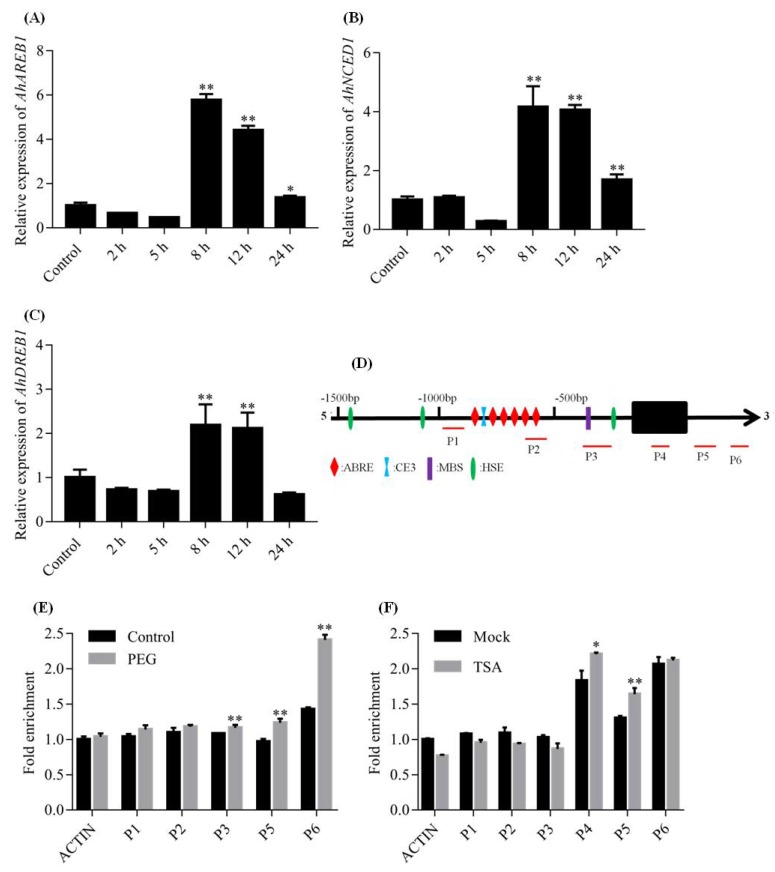
Trichostatin A (TSA) is a small-molecule histone deacetylase (HDAC) inhibitor, that is, it represses histone deacetylation. We found that TSA treatment of peanut leaves slightly reduced the expression of AhDREB1 at 2 to 5 h, while a relatively high transcription level of AhDREB1 was detected at 8 to 12 h, followed by a decrease in expression at 24 h (Figure 8A). When we checked the drought-related genes AhAREB1 and AhNCED1, we observed that their expression profiles were both essentially consistent with that of AhDREB1 during TSA treatment (Figure 8B,C). These results suggest that TSA can promote the expression of AhDREB1 and, therefore, that histone acetylation modification may be involved in its transcriptional regulation. To clarify this, chromatin immunoprecipitation (ChIP) assays were performed using antibodies that specifically recognize histone H3ac modifications. H3ac levels in chromatin of peanut leaves treated with TSA and PEG were analyzed in various regions of the AhDREB1 gene using ChIP-quantitative PCR (qPCR). TSA treatment resulted in relatively high H3ac enrichment levels in the P4 and P5 regions of AhDREB1 chromatin, whereas no statistically significant changes in H3ac were detected in other regions (Figure 8E). Treatment with 20% PEG also enriched levels of H3ac, but only in the P3, P5, P6 regions (Figure 8F). In contrast, the application of 100 μM ABA (Supplementary Figure S4) produced no significant changes in H3ac levels in any region of AhDREB1 chromatin. In conclusion, histone acetylation is involved in AhDREB1 transcriptional regulation and the level of the histone acetylation in AhDREB1 chromatin can be modulated by both TSA and 20% PEG treatment.
Figure 8.
Histone acetylation is involved in AhDREB1 transcriptional regulation. (A–C) Expression analyses of AhDREB1 and stress resistance genes following 1 μΜ treatment in peanut leaves by RT-qPCR. Time points of 2, 5, 8, 12, and 24 h were sampled to observe the expression changing trend. The untreated group was used as the control. Mean and SD were obtained from more than three biological replicates. Asterisks indicate significant differences from control (Student’s t test p values, * p < 0.05 and ** p < 0.01). (D) The structure of the AhDREB1 promoter. PCR amplification (P1–P6) for chromatin immunoprecipitation (ChIP) assays are indicated. (E,F) H3ac levels in chromatin of peanut leaves under 1 μM TSA and 20% PEG treatment, respectively. The untreated group was used as the control. Mean and SD were obtained from more than three biological replicates. Asterisks indicate significant differences from control (Student’s t test p values, * p < 0.05 and ** p < 0.01).
3. Discussion
AP2/ERFs are plant-specific transcription factors. Increasing evidence shows that AP2/ERF TF genes are induced in response to various stresses, including drought, abscisic acid, salt, cold, heat and alkalinity [42]. Here, we isolated full-length CDS of AhDREB1 gene, which encodes an AP2/ERF TF of the DREB subfamily (Figure 1A). The expression of AhDREB1 is induced by PEG 6000 and ABA (Figure 2 and Figure 3), which suggests that AhDREB1 might play an important role in the plant drought response and the ABA signaling pathway. A large number of studies have revealed that AP2/ERF TFs can enhance the stress tolerance when overexpressed in transgenic plants [20,43]. For instance, overexpression of ERF1-V in wheat improves resistance to powdery mildew, salt and drought stress [44]. Similarly, overexpression of SpERF1 enhances drought tolerance of transgenic A. thaliana [45]. It has been reported that overexpression of OsERF71 can enhance the drought stress tolerance of rice by changing the morphological structure of roots and by regulating the expression of genes related to lignin synthesis and drought-related genes [46]. In the current paper, overexpression of AhDREB1 significantly enhanced drought resistance in A. thaliana (Figure 4C) in the vegetative stages of at least two OX lines (Figure 4B).
It is well known that ABA is a pivotal regulator of plant responses to drought stress. The level of ABA increases rapidly under drought stress and this is a function of its biosynthesis, catabolism and transport [10]. NCED is a key enzyme in the ABA synthesis pathway [10,11] and, in Arabidopsis, the respective gene AtNCED3, which is induced by drought stress, controls the level of endogenous ABA under drought-stress conditions. Overexpression of AtNCED3 causes an increase in endogenous ABA levels and promotes transcription of drought- and ABA-inducible genes, which improve drought tolerance in Arabidopsis [47]. In peanut plants, the expression of AhNCED1 is significantly upregulated by dehydration and high salinity, while overexpression of AhNCED1 in Arabidopsis plants leads to the accumulation of endogenous ABA and improved water-stress tolerance [48]. ABA transport is also an important factor affecting ABA levels and plant responses to drought stress. Previous studies have shown that mutation of the Arabidopsis abcg40 gene, which encodes an ABA transporter, causes stomata to close more slowly in response to ABA, and strongly delays the upregulation of ABA-responsive genes, resulting in reduced drought tolerance [49]. In peanut, AhATL1, an ABCG transporter subfamily gene, is upregulated by water stress and exogenous ABA treatment. Overexpression of AhATL1 decreases ABA sensitivity and drought resistance by influencing ABA transport in Arabidopsis [50]. These studies show that the level of ABA is closely related to drought tolerance and thus ABA levels can be used as an important index of drought resistance in plants [9]. Consistent with their drought-resistant phenotype, we showed here that ABA contents of the Arabidopsis AhDREB1-OX lines OX-2 and OX-3 were significantly higher than in Col under drought stress, although line OX-1 showed no significant difference to the control (Figure 4F). We suspected that the phenomena may be related to the expression of AhDREB1 in Arabidopsis thaliana. Indeed, AhDREB1 expression in OX-1 line is relatively low compared to the other two lines (Figure 4A), which may affect the level of ABA under drought stress and affect the drought resistance of Arabidopsis. It is interesting that the ABA levels of AhDREB1-OX lines were also significantly higher than Col under normal growth conditions. Previous studies have shown that ABA is an important factor in regulating seed dormancy and germination [51]. In Arabidopsis, strains overexpressing AtDREB2C are ABA hypersensitive during germination and show enhanced plant tolerance to cold and heat stress [8]. The overexpression of GhERF38 increased ABA sensitivity in seed germination and seedling period in Arabidopsis [52]. Our AhDREB1-OX lines were also ABA hypersensitive during seed germination and seedling development. Thus we speculated that AhDREB1 may affect ABA signaling pathway in Arabidopsis. To verify this hypothesis, we checked expression levels of ABA signaling pathway-related genes in AhDREB1-OX lines and Col plants subjected to 10 μM ABA treatment. All of these genes showed significantly higher expression than the wild-type under both normal conditions and during ABA treatment (Figure 7). These results suggest AhDREB1 increases ABA sensitivity and affects ABA signaling pathway in Arabidopsis.
It has been reported that ERF and DREB subfamily TFs can directly interact with GCC boxes (GCCGCC) or DRE elements in the promoters of downstream stress- and defence-related genes to regulate multiple stress responses [20]. Overexpression of SpERF1 enhances drought tolerance of transgenic A. thaliana after binding to DRE elements in the promoters of the abiotic stress-responsive genes P5CS1, RD29A, HSP101, LEA4-5, HSP70 and COR47 [45]. ThCRF1 binds DRE and CGG-box motifs and induces the expression of various genes, including P5CS, SOD and POD, thereby enhancing tolerance to salt stress [17]. In this paper, to further investigate the molecular mechanism by which AhDREB1 enhances drought tolerance, RT-qPCR was carried out to quantify transcription levels of RD29A, P5CS1, P5CS2 and NCED1. All four genes were significantly upregulated in AhDREB1-OX lines subjected to drought stress (Figure 5). This result was consistent with the drought-resistant phenotype described above. In summary, AhDREB1 affects ABA sensitivity and ABA signaling by increasing the level of endogenous ABA and the expression of downstream drought stress-related genes, which in turn increase A. thaliana drought resistance in overexpressing lines.
Histone acetylation is one of the most important histone modifications and can affect chromatin structure and gene expression. TSA, a small molecule HDAC inhibitor, can cause transient increases in acetylation of histone H2B, H4, and H3 in chromatin [53]. It has been reported that RAB18, RD29B and HSP70, together with four late embryogenesis abundant protein (LEA) genes, are upregulated by TSA during seed germination [54]. TSA also suppresses cold-induced transcription of the ZmDREB1 gene in maize [39]. In our studies, the expression of AhDREB1 was significantly upregulated by TSA (Figure 8A), implying that histone acetylation may be involved in the transcriptional regulation of AhDREB1. We investigated the effect of TSA treatment on the enrichment level of H3ac in AhDREB1 chromatin using ChIP-qPCR assays. The levels of H3ac were significantly increased in the P4 and P5 regions of AhDREB1 (Figure 8E), consistent with our AhDREB1 gene expression results following TSA treatment. We therefore conclude that histone acetylation is involved in the transcriptional regulation of AhDREB1.
Many studies have shown that histone acetylation plays an important role in the regulation of plant growth and development, and in the responses to abiotic stress. Thus, in Arabidopsis, H3K4me3 and H3K9ac levels increase in the coding regions of RD29A, RD29B, RD20 and RAP2.4 genes during drought stress [55]. HDACs positively affect the expression of the cold-induced ZmDREB1 gene, and this affects plant tolerance to cold stress [39]. Acetylation of H3K9 and K3K14 is increased under PEG and ABA treatment in A. hypogaea [36], while in A. thaliana, AtHDA9 negatively affects plant sensitivity to drought and salt stresses by modulating histone acetylation levels of a large number of stress-responsive genes [27]. In this paper, we found that overexpression of AhDREB1 enhances plant tolerance to drought stress and that AhDREB1 transcription is affected by histone acetylation. Accordingly, we hypothesised that AhDREB1 is affected by histone acetylation under drought conditions, and that this affects drought resistance in whole plants. To verify the hypothesis, we tested H3ac levels following treatment with 20% PEG and found these to increase in the P3, P5, P6 regions of the AhDREB1 gene (Figure 8F). However, treatment with 100 μM ABA yielded no significant change in H3ac levels (Supplementary Figure S4). In conclusion, histone acetylation is involved in the transcriptional regulation of AhDREB1 under osmotic stress conditions by increasing H3ac levels, thereby further improving plant drought resistance.
In summary, we isolated a full-length CDS of the AP2/ERF family gene AhDREB1 from peanut leaves and showed that AhDREB1 overexpression can improve the tolerance of Arabidopsis to drought via the ABA-dependent pathway. At the same time, histone acetylation is involved in the transcriptional regulation of AhDREB1 during osmotic stress, further improving drought resistance. This is the first report that histone acetylation is involved in the transcriptional regulation of a DREB subfamily gene in peanut. The findings provide new insight into how AP2/ERF TFs enhance plant tolerance to drought stress, which may lead to successful breeding of drought resistant strains of peanuts and other crops.
4. Material and Methods
4.1. Peanut Plant Material and Growth Conditions
Seeds of peanut (Arachis hypogaea L. cv. Yueyou 7) were provided by the Crop Research Institute, Guangdong Academy of Agricultural Sciences, China. Seeds were soaked in water for 12 h, and then transferred to moist filter paper in an artificial climate incubator with a cycle of 16 h light from fluorescent and incandescent lamps (200 μmol·m−2·s−1) at 26 °C followed by 8 h darkness at 22 °C for 48 h until germination. Germinated seeds were planted in a potting mixture of vermiculite, perlite and soil (1:1:1) in the artificial climate incubator. Plants were watered with half-strength Murashige and Skoog nutrient solution every other day [36].
4.2. Isolation and Sequence Analysis of AhDREB1 from Arachis hypogaea L.
The total RNA of peanut leaves was extracted. Reverse transcription is performed by using 200 units Superscript III Reverse Transcriptase (Invitrogen, catalog no. 18080, Shanghai, China) and 500 ng oligo-dT primer and then the first strand of cDNA was synthesized. Reverse transcribed (RT) conditions were: 70 °C for 10 min (min), 42 °C for 1 h, and 15 min at 70 °C [36]. The cDNA was used as a template to amplify AhDREB1 open reading frame (ORF) by PCR using the following primers: AhDREB1 ORF forward primer 5′-ATGGCAGCAATGATGGATTTCTACA-3′, AhDREB1 ORF reverse primer 5′-TCACAGAGAATCCCAATCAATCTCA-3′. PCR amplification was performed as follows: 94 °C for 7 min, then 35 cycles of 94 °C for 30 s (s), 55 °C for 30 s and 72 °C for 1 min 30 s, with a final extension step at 72 °C for 10 min. PCR products were purified with an Agarose Gel DNA Purification Kit (TaKaRa, catalog no. DV805A, Dalian, China) and were ligated into the pMD19 T-vector (TaKaRa, catalog no. 6013, Dalian, China). Plasmids were sequenced on both strands.
Sequence analysis was performed using Lasergene 7.0 software (DNAStar, Madison, WI, USA). On-line BLAST analysis of DNA and amino acid sequences was performed at the National Center for Biotechnology Information Services website (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple sequence alignment was performed using DNAMAN8.0 software [56]. The AhDREB1 promoter sequence was identified using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). AhDREB1 was assessed for the presence of a transmembrane domain using online TMHMM 2.0 (http://www.cbs.dtu.dk/services/) [50]. Promoter elements were analyzed using PlantCARE [57,58].
4.3. Subcellular Localization Analysis
The expression vectors for subcellular localization were constructed as follows: the AhDREB1 coding region was prepared by PCR using the primer set forward primer 5′-ATGGCAGCAATGATGGATTTCTACA-3′ and reverse primer 5′-TCACAGAGAATCCCAATCAATCTCA-3′. The amplified fragment was cloned into pMD19 T-vector and sequenced on both strands. Subsequently, the AhDREB1 ORF was cloned into the p35S::eGFP vector [59]. The fusion construct p35S::AhDREB1-eGFP was driven by the 35S promoter. The recombinant plasmid vector was transformed into Arabidopsis protoplasts. After incubation for 12–18 h in the dark, eGFP fluorescence signals in protoplasts were observed under the confocal microscope (Carl Zeiss LSM 710; Carl Zeiss, Germany).
4.4. Peanut Plant Treatments
Peanut seedlings were removed from the soil at the four-leaf stage, and after the roots were carefully rinsed with deionized water, they were maintained in 1/8 MS medium for up to 2 h. Seedlings were treated with 20% PEG or 100 μM ABA for 2, 5, 8, 12 and 24 h. Trichostatin A (TSA) was used to treat seedlings at 1 μM for 2, 5, 8, 12 and 24 h [36]. Untreated seedlings planted in the soil were used as control plants. Seedlings placed in an equivalent volume of deionized water instead of ABA or TSA solutions were used as mock treatments. Seedlings were transferred to an artificial climate incubator (26 °C, 60% moisture) under continuous light. Peanut leaves were harvested and frozen at −80 °C prior to use.
4.5. Real-Time Quantitative PCR (RT-qPCR)
Total RNA was extracted from peanut leaves or Arabidopsis thaliana seedlings [36]. For first-strand cDNA synthesis, 1 μg high-quality total RNA was reverse transcribed (RT) using a Prime Script TM RT Reagent Kit (Perfect Real Time, TaKaRa, Dalian, China). The cDNA sample was used as the template for RT-qPCR. Relative transcript levels of AhDREB1 were evaluated in an ABI 7500 system using the ChamQ SYBR qPCR Master Mix (Low ROX Premixed, Vazyme Biotech Co., Ltd., Nanjing, China). Each reaction contained 5 μL 2 × ChamQ SYBR qPCR Master Mix, 10 ng cDNA, 0.2 μL 0.05 mM stock solution of each primer in a final volume of 10 μL. The PCR thermal cycles were as follows: 95 °C for 30 s, followed by 40 cycles at 95 °C for 5 s and 60 °C for 34 s. The AhACTIN and AtACTIN2 genes were used as internal controls for A. hypogaea and A. thaliana, respectively, as described [36,59]. Relative expression levels were calculated using the relative 2−ΔΔCt method [60]. The relevant primers for the analysis of gene transcription levels are listed one by one in Supplementary Table S1.
4.6. Plant Transformation and Growth Conditions
The recombinant vector p35S::AhDREB1-eGFP was transformed into Agrobaterium tumefaciens strain EHA105. Positive A. tumefaciens colonies containing the vector were employed for Arabidopsis Col transformation by the floral dip method [59]. T0 generation seeds were harvested and sowed on 1/2 MS medium containing 50 mg/L kanamycin for selection, with repeat screening until T3. Individual plants were genotyped and homozygous plants were selected by PCR. Subsequent experiments were perform using T3 seeds. Arabidopsis growing conditions: light (a daily cycle of 16 h light and 8 h dark), temperature (22 ± 2 °C), relative humidity (60–70%).
4.7. Drought Stress Treatment of Arabidopsis
The seeds of AhDREB1 overexpression lines and wild type Arabidopsis were grown in 1/2 MS medium and were vernalized for 48 h at 4 °C. Then seeds were placed in a greenhouse with a daily cycle of 16 h light and 8 h dark at 20 ± 2 °C. After germination, the seedlings were planted in peat soil and grown for three weeks under the same light and temperature conditions. Three-week-old Arabidopsis thaliana was treated by soil drought for approximately 2 weeks under the same greenhouse conditions as above. The survival rate of Arabidopsis thaliana was counted after 2 days of rehydration. The survival rate was determined in three independent experiments [60].
4.8. Stomatal Movement Assay
To observe the stomatal opening of wild-type A. thaliana and AhDREB1-OX lines under normal growth and soil drought conditions, rosette leaves were detached from three-week-old plants. The detached rosette leaves were soaked in open stomatal buffer (10 mM MES, 5 mM KCl, 50 mM CaCl2, pH 6.15) in a growth chamber under incandescent lamps (200 μmol·m−2·s−1) at 20 °C for 3 h. Stomatal apertures were measured as described previously [49]. Digimizer software was used to measure stomatal apertures.
4.9. Relative Water Content Determination
Arabidopsis rosette leaves under normal growth and drought stress conditions were cut and collected to determine the fresh weight (mf). Leaf material was placed at 105 °C for 0.5 h, then dried to constant weight at 80 °C, giving the dry weight (md). Water content (%) = (mf − md)/mf × 100%. The relative water content was determined in triplicate for each sample [61].
4.10. Determination of ABA Content
As previously described, ABA was extracted from Arabidopsis rosette leaves under soil drought or control conditions [48]. Extraction in 80% (v/v) aqueous methanol, and high performance liquid chromatography fractionation in a SinoChrom ODS AP C18 column (250 × 4.6 mm, 5 μm, Dalian, China), were conducted as reported previously [62]. The level of ABA was determined in triplicate for each sample.
4.11. ABA Sensitivity
Sensitivity of AhDREB1-OX and wild-type seeds to ABA during seeds germination was assessed. To test the effects of ABA on germination, the seeds (80 seeds each, three repeats) were germinated on 1/2 MS medium with 0.8% agar containing a gradient concentration of ABA (0, 0.5, 2 μM), and the germination rate of the treated seeds was calculated every 12 h until 192 h [59].
4.12. ChIP-Quantitative PCR
For the ChIP assay, leaves (500 mg) of four-leaf-stage (two weeks after planting) peanut plants were fixed with cold MC buffer (10 mM potassium phosphate pH 7.0, 50 mM NaCl, 0.1 M sucrose, 1% formaldehyde) for 20 min by vacuum concentration. Then add glycine to make its final concentration of 125 mM, vacuum incubation 10 min to terminate cross-linking. After washing leaves in MC buffer, leaves were collected and frozen in liquid nitrogen, and stored at −80 °C. The ChIP assay was performed as described previously [63]. One microgram of anti-H3ac and rabbit IgG (Millipore, Shanghai, China) were used for immunoprecipitation. Specifically, precipitated DNA was recovered and analyzed by ChIP-qPCR using SYBR Premix ExTaq Mix (Takara Bio, Dalian, China). Peanut ACTIN genes were used to calculate the relative fold-enrichment of target DNA fragments. The ChIP-qPCR primers are listed in Supplemental Table S2.
4.13. Statistical Analysis
Quantitative data were expressed as mean ± SD. The statistical significance of experimental data was assessed by Student t-test or ANOVA (one-way analysis of variance with a Least Significant Difference (LSD) post-hoc test), as appropriate, using the SPSS17.0 statistical package (Chicago, IL, USA).
Acknowledgments
We are grateful to Alan Tunnacliffe (Chief Editor) at Cambridge Academic Manuscripts, for technical support and critical reading of the manuscript. This work was supported by Natural Science Foundation of Guangdong Province (8151063101000011 to Ling Li), Innovation Project of Graduate School of South China Normal University (2017LKXM060 to Baihong Zhang).
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/19/5/1441/s1.
Author Contributions
B.Z., L.S. and L.L. designed the research. B.Z. performed the research. B.Z., L.S. and B.H. analyzed the data and prepared figures. B.Z. wrote the manuscript in consulation with L.S. and L.L. L.L. and B.H. contributed reagents/materials/analysis tools.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Agarwal P.K., Gupta K., Lopato S., Agarwal P. Dehydration responsive element binding transcription factors and their applications for the engineering of stress tolerance. J. Exp. Bot. 2017;68:2135–2148. doi: 10.1093/jxb/erx118. [DOI] [PubMed] [Google Scholar]
- 2.Vinocur B., Altman A. Recent advances in engineering plant tolerance to abiotic stress: Achievements and limitations. Curr. Opin. Biotechnol. 2005;16:123–132. doi: 10.1016/j.copbio.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Gonzalez-Villagra J., Rodrigues-Salvador A., Nunes-Nesi A., Cohen J.D., Reyes-Diaz M.M. Age-related mechanism and its relationship with secondary metabolism and abscisic acid in Aristotelia chilensis plants subjected to drought stress. Plant Physiol. Biochem. 2018;124:136–145. doi: 10.1016/j.plaphy.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 4.Liu J., Wang R., Liu W., Zhang H., Guo Y., Wen R. Genome-Wide Characterization of Heat-Shock Protein 70s from Chenopodium quinoa and Expression Analyses of Cqhsp70s in Response to Drought Stress. Genes. 2018;9:35. doi: 10.3390/genes9020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohnert H.J., Jensen R.G. Strategies for engineering water-stress tolerance in plants. Trends Biotechnol. 1996;14:89–97. doi: 10.1016/0167-7799(96)80929-2. [DOI] [Google Scholar]
- 6.Wang W.X., Vinocur B., Altman A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta. 2003;218:1–14. doi: 10.1007/s00425-003-1105-5. [DOI] [PubMed] [Google Scholar]
- 7.Yamaguchi-Shinozaki K., Shinozaki K. Gene regulatory network in drought and cold stress responses. Plant Cell Physiol. 2005;46:S33. [Google Scholar]
- 8.Lee S.J., Kang J.Y., Park H.J., Kim M.D., Bae M.S., Choi H.I., Kim S.Y. DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity. Plant Physiol. 2010;153:716–727. doi: 10.1104/pp.110.154617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roychoudhury A., Paul S., Basu S. Cross-talk between abscisic acid-dependent and abscisic acid-independent pathways during abiotic stress. Plant Cell Rep. 2013;32:985–1006. doi: 10.1007/s00299-013-1414-5. [DOI] [PubMed] [Google Scholar]
- 10.Finkelstein R. Abscisic Acid synthesis and response. Arabidopsis Book. 2013;11:e0166. doi: 10.1199/tab.0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maruyama K., Urano K., Yoshiwara K., Morishita Y., Sakurai N., Suzuki H., Kojima M., Sakakibara H., Shibata D., Saito K., et al. Integrated Analysis of the Effects of Cold and Dehydration on Rice Metabolites, Phytohormones, and Gene Transcripts. Plant Physiol. 2014;164:1759–1771. doi: 10.1104/pp.113.231720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakashima K., Ito Y., Yamaguchi-Shinozaki K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009;149:88–95. doi: 10.1104/pp.108.129791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujita Y., Fujita M., Shinozaki K., Yamaguchi-Shinozaki K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 2011;124:509–525. doi: 10.1007/s10265-011-0412-3. [DOI] [PubMed] [Google Scholar]
- 14.Singh D., Laxmi A. Transcriptional regulation of drought response: A tortuous network of transcriptional factors. Front. Plant Sci. 2015;6:895. doi: 10.3389/fpls.2015.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamaguchi-Shinozaki K., Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamaguchi-Shinozaki K., Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005;10:88–94. doi: 10.1016/j.tplants.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 17.Qin L., Wang L., Guo Y., Li Y., Umut H., Wang Y. An ERF transcription factor from Tamarix hispida, ThCRF1, can adjust osmotic potential and reactive oxygen species scavenging capability to improve salt tolerance. Plant Sci. 2017;265:154–166. doi: 10.1016/j.plantsci.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 18.Sakuma Y., Liu Q., Dubouzet J.G., Abe H., Shinozaki K., Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- 19.Riechmann J.L., Heard J., Martin G., Reuber L., Jiang C., Keddie J., Adam L., Pineda O., Ratcliffe O.J., Samaha R.R., et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 20.Phukan U.J., Jeena G.S., Tripathi V., Shukla R.K. Regulation of Apetala2/Ethylene Response Factors in Plants. Front. Plant Sci. 2017;8:150. doi: 10.3389/fpls.2017.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohme-Takagi M., Shinshi H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell. 1995;7:173–182. doi: 10.1105/tpc.7.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu Y., Duan X., Ding X., Chen C., Zhu D., Yin K., Cao L., Song X., Zhu P., Li Q., et al. A novel AP2/ERF family transcription factor from Glycine soja, GsERF71, is a DNA binding protein that positively regulates alkaline stress tolerance in Arabidopsis. Plant Mol. Biol. 2017;94:509–530. doi: 10.1007/s11103-017-0623-7. [DOI] [PubMed] [Google Scholar]
- 23.Scarpeci T.E., Frea V.S., Zanor M.I., Valle E.M. Overexpression of AtERF019 delays plant growth and senescence, and improves drought tolerance in Arabidopsis. J. Exp. Bot. 2017;68:673–685. doi: 10.1093/jxb/erw429. [DOI] [PubMed] [Google Scholar]
- 24.Gilmour S.J., Sebolt A.M., Salazar M.P., Everard J.D., Thomashow M.F. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000;124:1854–1865. doi: 10.1104/pp.124.4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang Y., Li X., Zhang D., Gao B., Yang H., Wang Y., Guan K., Wood A.J. ScDREB8, a novel A-5 type of DREB gene in the desert moss Syntrichia caninervis, confers salt tolerance to Arabidopsis. Plant Physiol. Biochem. PPB. 2017;120:242–251. doi: 10.1016/j.plaphy.2017.09.014. [DOI] [PubMed] [Google Scholar]
- 26.Chen J.Q., Meng X.P., Zhang Y., Xia M., Wang X.P. Over-expression of OsDREB genes lead to enhanced drought tolerance in rice. Biotechnol. Lett. 2008;30:2191–2198. doi: 10.1007/s10529-008-9811-5. [DOI] [PubMed] [Google Scholar]
- 27.Zheng Y., Ding Y., Sun X., Xie S., Wang D., Liu X., Su L., Wei W., Pan L., Zhou D.X. Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis. J. Exp. Bot. 2016;67:1703–1713. doi: 10.1093/jxb/erv562. [DOI] [PubMed] [Google Scholar]
- 28.Luo M., Liu X., Singh P., Cui Y., Zimmerli L., Wu K. Chromatin modifications and remodeling in plant abiotic stress responses. Biochim. Biophys. Acta. 2012;1819:129–136. doi: 10.1016/j.bbagrm.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 29.Swindell W.R., Huebner M., Weber A.P. Plastic and adaptive gene expression patterns associated with temperature stress in Arabidopsis thaliana. Heredity. 2007;99:143–150. doi: 10.1038/sj.hdy.6800975. [DOI] [PubMed] [Google Scholar]
- 30.Liu X., Yang S., Zhao M., Luo M., Yu C.W., Chen C.Y., Tai R., Wu K. Transcriptional repression by histone deacetylases in plants. Mol. Plant. 2014;7:764–772. doi: 10.1093/mp/ssu033. [DOI] [PubMed] [Google Scholar]
- 31.Courey A.J., Jia S. Transcriptional repression: The long and the short of it. Genes Dev. 2001;15:2786–2796. doi: 10.1101/gad.939601. [DOI] [PubMed] [Google Scholar]
- 32.Hu Y., Qin F., Huang L., Sun Q., Li C., Zhao Y., Zhou D.X. Rice histone deacetylase genes display specific expression patterns and developmental functions. Biochem. Biophys. Res. Commun. 2009;388:266–271. doi: 10.1016/j.bbrc.2009.07.162. [DOI] [PubMed] [Google Scholar]
- 33.Varotto S., Locatelli S., Canova S., Pipal A., Motto M., Rossi V. Expression profile and cellular localization of maize Rpd3-type histone deacetylases during plant development. Plant Physiol. 2003;133:606–617. doi: 10.1104/pp.103.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang H., Liu X., Xin M., Du J., Hu Z., Peng H., Rossi V., Sun Q., Ni Z., Yao Y. Genome-Wide Mapping of Targets of Maize Histone Deacetylase HDA101 Reveals Its Function and Regulatory Mechanism during Seed Development. Plant Cell. 2016;28:629–645. doi: 10.1105/tpc.15.00691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo M., Tai R., Yu C.W., Yang S., Chen C.Y., Lin W.D., Schmidt W., Wu K. Regulation of flowering time by the histone deacetylase HDA5 in Arabidopsis. Plant J. 2015;82:925–936. doi: 10.1111/tpj.12868. [DOI] [PubMed] [Google Scholar]
- 36.Su L.C., Deng B., Liu S., Li L.M., Hu B., Zhong Y.T., Li L. Isolation and characterization of an osmotic stress and ABA induced histone deacetylase in Arachis hygogaea. Front. Plant Sci. 2015;6:512. doi: 10.3389/fpls.2015.00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo M., Wang Y.Y., Liu X., Yang S., Lu Q., Cui Y., Wu K. HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J. Exp. Bot. 2012;63:3297–3306. doi: 10.1093/jxb/ers059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mehdi S., Derkacheva M., Ramstrom M., Kralemann L., Bergquist J., Hennig L. The WD40 Domain Protein MSI1 Functions in a Histone Deacetylase Complex to Fine-Tune Abscisic Acid Signaling. Plant Cell. 2016;28:42–54. doi: 10.1105/tpc.15.00763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu Y., Zhang L., Zhao L., Li J., He S., Zhou K., Yang F., Huang M., Jiang L., Li L. Trichostatin A selectively suppresses the cold-induced transcription of the ZmDREB1 gene in maize. PLoS ONE. 2011;6:e22132. doi: 10.1371/journal.pone.0022132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao X., Li C., Wan S., Zhang T., Yan C., Shan S. Transcriptomic analysis and discovery of genes in the response of Arachis hypogaea to drought stress. Mol. Biol. Rep. 2018;45:119–131. doi: 10.1007/s11033-018-4145-4. [DOI] [PubMed] [Google Scholar]
- 41.Li X.Y., Lu J.B., Liu S., Liu X., Lin Y.Y., Li L. Identification of rapidly induced genes in the response of peanut (Arachis hypogaea) to water deficit and abscisic acid. BMC Biotechnol. 2014;14:58. doi: 10.1186/1472-6750-14-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rong W., Qi L., Wang A., Ye X., Du L., Liang H., Xin Z., Zhang Z. The ERF transcription factor TaERF3 promotes tolerance to salt and drought stresses in wheat. Plant Biotechnol. J. 2014;12:468–479. doi: 10.1111/pbi.12153. [DOI] [PubMed] [Google Scholar]
- 43.Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta. 2012;1819:86–96. doi: 10.1016/j.bbagrm.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 44.Xing L., Di Z., Yang W., Liu J., Li M., Wang X., Cui C., Wang X., Wang X., Zhang R. Overexpression of ERF1-V from Haynaldia villosa Can Enhance the Resistance of Wheat to Powdery Mildew and Increase the Tolerance to Salt and Drought Stresses. Front. Plant Sci. 2017;8:1948. doi: 10.3389/fpls.2017.01948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Y., Dong C., Li X., Du J., Qian M., Sun X., Yang Y. A novel AP2/ERF transcription factor from Stipa purpurea leads to enhanced drought tolerance in Arabidopsis thaliana. Plant Cell Rep. 2016;35:2227–2239. doi: 10.1007/s00299-016-2030-y. [DOI] [PubMed] [Google Scholar]
- 46.Lee D.K., Jung H., Jang G., Jeong J.S., Kim Y.S., Ha S.H., Do Choi Y., Kim J.K. Overexpression of the OsERF71 Transcription Factor Alters Rice Root Structure and Drought Resistance. Plant Physiol. 2016;172:575–588. doi: 10.1104/pp.16.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iuchi S., Kobayashi M., Taji T., Naramoto M., Seki M., Kato T., Tabata S., Kakubari Y., Yamaguchi-Shinozaki K., Shinozaki K., et al. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001;27:325–333. doi: 10.1046/j.1365-313x.2001.01096.x. [DOI] [PubMed] [Google Scholar]
- 48.Wan X.R., Li L. Regulation of ABA level and water-stress tolerance of Arabidopsis by ectopic expression of a peanut 9-cis-epoxycarotenoid dioxygenase gene. Biochem. Biophys. Res. Commun. 2006;347:1030–1038. doi: 10.1016/j.bbrc.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 49.Kang J., Hwang J.U., Lee M., Kim Y.Y., Assmann S.M., Martinoia E., Lee Y. PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl. Acad. Sci. USA. 2010;107:2355–2360. doi: 10.1073/pnas.0909222107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ge K., Liu X., Li X.Y., Hu B., Li L. Isolation of an ABA Transporter-Like 1 Gene from Arachis hypogaea That Affects ABA Import and Reduces ABA Sensitivity in Arabidopsis. Front. Plant Sci. 2017;8:1150. doi: 10.3389/fpls.2017.01150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sah S.K., Reddy K.R., Li J.X. Abscisic Acid and Abiotic Stress Tolerance in Crop Plants. Front. Plant Sci. 2016;7:571. doi: 10.3389/fpls.2016.00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma L.F., Hu L.X., Fan J.B., Amombo E., Khaldun A.B.M., Zheng Y., Chen L. Cotton GhERF38 gene is involved in plant response to salt/drought and ABA. Ecotoxicology. 2017;26:841–854. doi: 10.1007/s10646-017-1815-2. [DOI] [PubMed] [Google Scholar]
- 53.Waterborg J.H. Plant histone acetylation: In the beginning …. BBA-Gene Regul. Mech. 2011;1809:353–359. doi: 10.1016/j.bbagrm.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 54.Tai H.H., Tai G.C.C., Beardmore T. Dynamic histone acetylation of late embryonic genes during seed germination. Plant Mol. Biol. 2005;59:909–925. doi: 10.1007/s11103-005-2081-x. [DOI] [PubMed] [Google Scholar]
- 55.Kim J.M., To T.K., Ishida J., Morosawa T., Kawashima M., Matsui A., Toyoda T., Kimura H., Shinozaki K., Seki M. Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol. 2008;49:1580–1588. doi: 10.1093/pcp/pcn133. [DOI] [PubMed] [Google Scholar]
- 56.Wan X., Li L. Molecular cloning and characterization of a dehydration-inducible cDNA encoding a putative 9-cis-epoxycarotenoid dioxygenase in Arachis hygogaea L. DNA Seq. 2005;16:217–223. doi: 10.1080/10425170500129785. [DOI] [PubMed] [Google Scholar]
- 57.Lescot M., Dehais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., Rouzé P., Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X., Li X., Li M., Yan Y., Liu X., Li L. Dual Function of NAC072 in ABF3-Mediated ABA-Responsive Gene Regulation in Arabidopsis. Front. Plant Sci. 2016;7:1075. doi: 10.3389/fpls.2016.01075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li X.Y., Liu X., Yao Y., Li Y.H., Liu S., He C.Y., Li J.M., Lin Y.Y., Li L. Overexpression of Arachis hypogaea AREB1 gene enhances drought tolerance by modulating ROS scavenging and maintaining endogenous ABA content. Int. J. Mol. Sci. 2013;14:12827–12842. doi: 10.3390/ijms140612827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim J.M., To T.K., Matsui A., Tanoi K., Kobayashi N.I., Matsuda F., Habu Y., Ogawa D., Sakamoto T., Matsunaga S., et al. Acetate-mediated novel survival strategy against drought in plants. Nat. Plants. 2017;3:17097. doi: 10.1038/nplants.2017.97. [DOI] [PubMed] [Google Scholar]
- 61.Hu B., Liu X., Hong L., Li L., Luo G.Y. Expression and localization of Arachis hypogaea 9-cis epoxycarotenoid dioxygenase 1 (AhNCED1) of peanut under water stress. Biotechnol. Biotechnol. Equip. 2010;24:1562–1568. doi: 10.2478/V10133-010-0005-7. [DOI] [Google Scholar]
- 62.Liu S., Lv Y., Wan X.R., Li L.M., Hu B., Li L. Cloning and expression analysis of cDNAs encoding ABA 8′-hydroxylase in peanut plants in response to osmotic stress. PLoS ONE. 2014;9:e97025. doi: 10.1371/journal.pone.0097025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu S., Li M.J., Su L.C., Ge K., Li L.M., Li X.Y., Liu X., Li L. Negative feedback regulation of ABA biosynthesis in peanut (Arachis hypogaea): A transcription factor complex inhibits AhNCED1 expression during water stress. Sci. Rep. 2016;6:37943. doi: 10.1038/srep37943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.