Abstract
Neuroprotective peptides represent an attractive pharmacological strategy for the prevention or treatment of age-related diseases, for which there are currently few effective therapies. Lactoferrin (LF)-derived peptides (PKHs) and a set of six rationally-designed tryptophan (W)-containing heptapeptides (PACEIs) were characterized as prolyl endopeptidase (PEP) inhibitors, and their effect on β-amyloid peptide (Aβ) toxicity in a Caenorhabditis elegans model of Alzheimer’s disease (AD) was evaluated. Two LF-derived sequences, PKH8 and PKH11, sharing a W at the C-terminal end, and the six PACEI heptapeptides (PACEI48L to PACEI53L) exhibited significant in vitro PEP inhibition. The inhibitory peptides PKH11 and PACEI50L also alleviated Aβ-induced paralysis in the in vivo C. elegans model of AD. Partial or total loss of the inhibitory effect on PEP was achieved by the substitution of W residues in PKH11 and PACEI50L and correlated with the loss of protection against Aβ toxicity, pointing out the relevance of W on the neuroprotective activity. Further experiments suggest that C. elegans protection might not be mediated by an antioxidant mechanism but rather by inhibition of Aβ oligomerization and thus, amyloid deposition. In conclusion, novel natural and rationally-designed W-containing peptides are suitable starting leads to design effective neuroprotective agents.
Keywords: neurodegenerative diseases, amyloid β peptide, Caenorhabditis elegans, prolyl endopeptidase inhibition, lactoferrin-derived peptides, rationally-designed peptides, tryptophan, molecular docking
1. Introduction
Neurodegenerative diseases account for a significant proportion of morbidity and mortality in the developed countries. Moreover, these disorders are becoming more frequent due to the increased life expectancy [1]. Alzheimer’s disease (AD), the most common type of senile dementia, is a neurodegenerative disorder with enormous social and economic impact [2]. Although the cause(s) of AD is still controversial, it is accepted that the accumulation of amyloid β peptide (Aβ) in brain plaques triggers downstream neurotoxic events, leading to neuronal dysfunction, cell death, and neurodegeneration [3]. Aβ is a 38–43 amino acid peptide which derives from the β-amyloid precursor protein (βAPP) through proteolytic processing by β- and γ-secretases. Aβ 1–42 (Aβ1–42) appears to be the species that first forms deposits in AD [2], and its role in the oxidative damage in AD brain has also been established [4]. These findings suggest amyloid plaques or Aβ production as targets for drug development [1]. In addition, compounds that exhibit antioxidant activity can be considered potential therapeutic agents for AD [5]. In this context, the nematode Caenorhabditis elegans engineered to express human Aβ1–42 is a convenient in vivo model that has been used in drug screening for potential AD therapeutics. The resulting transgenic strains develop a concomitant progressive paralysis phenotype, being a well-suited model for correlating Aβ expression and toxicity [6,7,8].
Prolyl endopeptidase (PEP; E.C. 3.4.21.26) also called prolyl oligopeptidase (POP) is a highly conserved serine protease enzyme that cleaves peptide bonds at the carboxyl side of proline residues (P) in peptides up to 30 amino acids long. It has been found in a wide range of species and tissues, especially in the human brain. PEP can degrade biologically active P-containing peptides, including peptide-like hormones, neuroactive peptides, and various cellular factors [9]. Moreover, levels of PEP activity are altered in many degenerative conditions and psychiatric disorders, such as AD, amnesia, depression, and schizophrenia, and therefore, the enzyme is a potential therapeutic target for these diseases [10]. PEP was suggested to be a putative γ-secretase and accordingly PEP inhibitors abolished the formation of Aβ in neuroblastoma cells and prevented amyloid deposition in a mouse model of accelerated senescence [11]. However, PEP is not only involved in cleaving off physiologically active peptides, and it has been speculated that its physiological role results from its direct interaction with partner proteins [12].
The potential therapeutic use of peptides derived from natural sources has been extensively discussed during the last decades [13,14,15]. Dietary peptides have been claimed to have positive effects on weight loss and glycemia management [16] and also in the prevention and treatment of cancer, cardiovascular and infective diseases, and mental health disorders [17,18]. Moreover, many peptides seem to act through more than a single mechanism of action and, hence, they can be considered multifunctional sequences [19,20]. Natural peptides also represent an excellent starting point as leading candidates for the rational design of synthetic sequences with improved biological activity, specificity, and stability [21].
Natural sources of PEP-inhibitory peptides mainly include meat and fish by-products [22,23,24,25,26], cereals [27], and milk proteins [25,28,29,30]. By contrast, peptides showing protection against the toxicity caused by the accumulation of Aβ are scarce. These potential neuroprotective peptides were isolated from a cocoa by-product [31], maize [32], and scorpion venom [33]. Furthermore, small rationally-designed peptides based on specific Aβ motifs able to interact with Aβ, modifying its kinetics of aggregation, and reducing its toxicity have been described as an attractive pharmacological strategy [34,35].
Lactoferrin (LF), a well-characterized component of milk whey, is a multifunctional iron glycoprotein that exhibits a diverse range of biological activities including antimicrobial, antiviral, antioxidant, and immunomodulatory activities [36]. LF-derived peptides share some biological activities with the intact protein and possess antihypertensive properties [20]. Recently, we have shown the inhibitory effect of an LF-based product on Aβ toxicity [37], but there is no information about the neuroprotective effects of LF-derived peptides.
In the present study, we investigated the PEP-inhibitory activity of LF-derived peptides and of a set of sequence-related synthetic heptapeptides. Peptides exhibiting PEP-inhibitory activity in vitro were examined in a transgenic C. elegans model of AD to evaluate their in vivo protective effects against Aβ toxicity. With the aim of characterizing the mechanism(s) mediating C. elegans protection, we further examined their in vivo antioxidant effect and their in silico molecular interactions with Aβ. Finally, the role of tryptophan residues (W) on the neuroprotective activity of peptides was investigated.
2. Results
2.1. Lactoferrin Hydrolysates Inhibit Prolyl Endopeptidase Activity
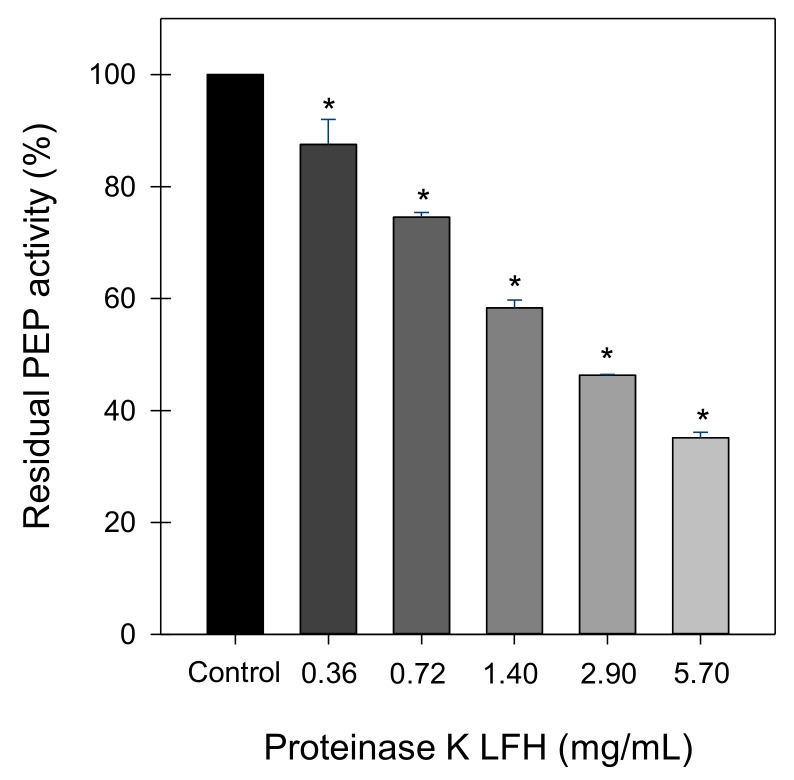
LF hydrolysates (LFH) generated by pepsin, proteinase K, or trypsin and subjected to ultrafiltration through a 3 kDa cut-off membrane showed PEP inhibitory activity (Table 1). At the maximum concentration tested (2 mg/mL), proteinase K LFH provoked the highest inhibitory effect (41% of inhibition) while pepsin and trypsin hydrolysates inhibited PEP by approximately 20%. The proteinase K LFH (0.36–5.7 mg/mL) provoked significant concentration-dependent inhibition of PEP (Figure 1), reaching 65% inhibition at the highest concentration assayed (5.7 mg/mL). Non-hydrolysed LF (0.7 and 1.4 mg/mL) did not show any inhibitory effect on PEP activity.
Table 1.
Effects of lactoferrin hydrolysates (LFH) on prolyl endopeptidase (PEP) activity.
| LFH | Concentration (mg/mL) | PEP Residual Activity (%) 1 |
|---|---|---|
| Pepsin LFH | 1 | 94.7 ± 0.1 |
| 2 | 84.1 ± 2.9 ** | |
| Proteinase K LFH | 1 | 76.6 ± 0.3 ** |
| 2 | 59.4 ± 1.2 ** | |
| Trypsin LFH | 1 | 90.0 ± 0.3 * |
| 2 | 81.5 ± 0.6 ** |
1 Data are expressed as the percentage of PEP residual activity with respect to a control without peptide (100%) and are the mean ± standard deviation (SD) of three replicates. * Significant inhibition with respect to control (p < 0.05); ** Significant inhibition with respect to control (p < 0.01) (Student’s t-test on absolute values of PEP activity).
Figure 1.
The concentration-dependent effect of lactoferrin hydrolysate (LFH) generated by proteinase K on prolyl endopeptidase (PEP) residual activity. Data are expressed as the mean ± SD of three replicates. * Significantly different from the control (p < 0.01; one-way ANOVA followed by a Dunnett multiple comparison test).
2.2. Lactoferrin-Derived Peptides from Proteinase K Hydrolysate Are Prolyl Endopeptidase Inhibitors
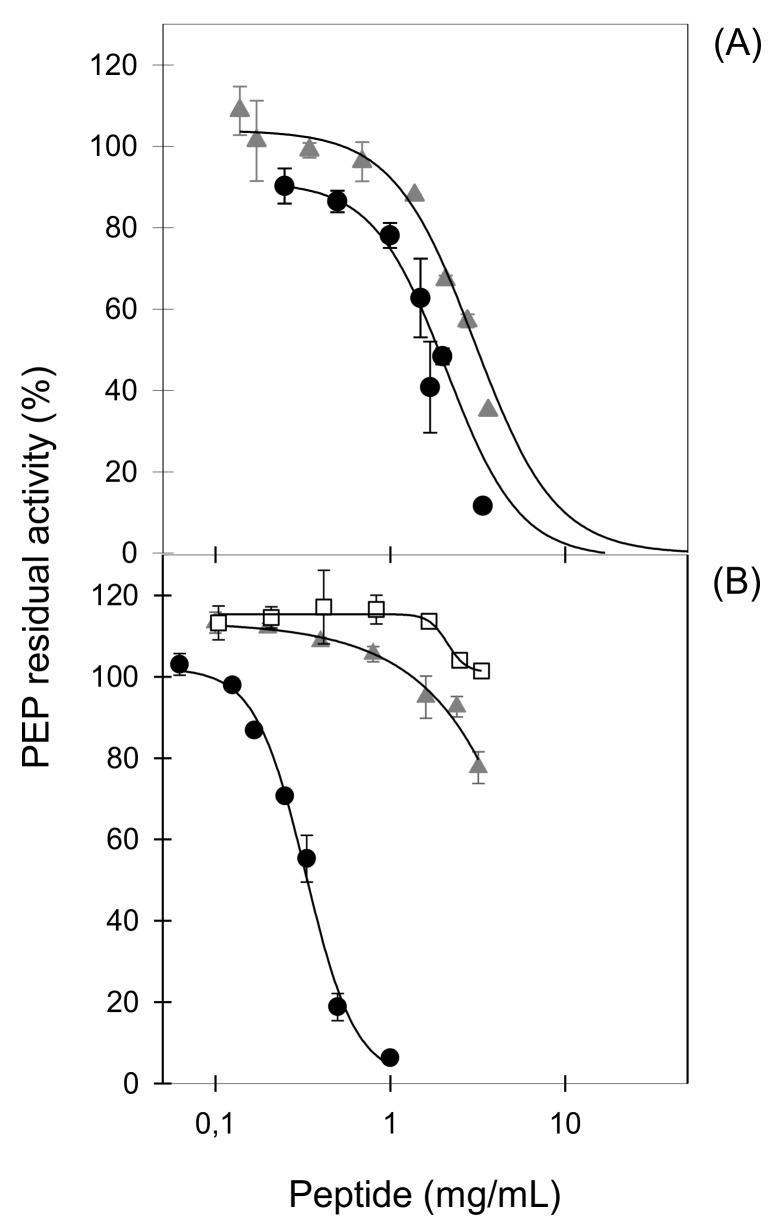
Proteinase K LFH is a complex hydrolysate from which 37 peptides were identified by high performance liquid chromatography tandem mass spectrometry (HPLC MS/MS) [38]. Eleven of the most abundant peptides identified in the hydrolysate were chemically synthesized and their in vitro PEP-inhibitory activity was tested (Table 2). Six out of eleven peptides (PKH3, PKH4, PKH5, PKH6, PKH9, and PKH10) included one P residue in the sequence but as summarized in Table 2, all of them failed to inhibit PEP. Only two sequences, the hexapeptide PKH8 (NEGLTW) and the decapeptide PKH11 (SVDGKEDLIW), which share a W residue at the C-terminal end, exhibited significant inhibitory activities of 12% and 22%, respectively, in the conditions tested. Further experiments were carried out to determine the inhibitory potency of the most promising PEP-inhibitory peptide, PKH11, which showed an IC50 value of 2.1 ± 0.2 mg/mL (Figure 2A).
Table 2.
Effects of peptides isolated from proteinase K lactoferrin hydrolysate (LFH) on prolyl endopeptidase (PEP) activity.
| Peptide (1 mg/mL) | Sequence | PEP Residual Activity (%) 1 |
|---|---|---|
| PKH1 | DRDQY | 102.7 ± 7.0 |
| PKH2 | VVKKGSNF | 115.0 ± 1.0 ** |
| PKH3 | ENLPEKA | 97.1 ± 0.1 |
| PKH4 | RIPSKV | 109.6 ± 2.2 * |
| PKH5 | GGRPTYEEY | 96.9 ± 2.1 |
| PKH6 | GILRPY | 97.2 ± 4.0 |
| PKH7 | DRDQYELL | 115.3 ± 4.2 ** |
| PKH8 | NEGLTW | 87.1 ± 1.0 ** |
| PKH9 | NIPMGL | 95.5 ± 3.4 |
| PKH10 | GILRPYL | 103.9 ± 0.8 |
| PKH11 | SVDGKEDLIW | 78.0 ± 3.1 ** |
1 Data are expressed as the percentage of PEP residual activity with respect to a control without peptide (100%) and are the mean ± SD of three replicates. * Significant inhibition with respect to control (p < 0.05). ** Significant inhibition with respect to control (p < 0.01) (Student’s t-test on absolute values of PEP activity).
Figure 2.
The concentration-dependent effect on prolyl endopeptidase (PEP) residual activity of peptides PKH11 and PACEI50L and their sequence variants obtained by W replacement with A. (A) PKH11 (SVDGKEDLIW; black circles) and its sequence variant PKH11-v1 (SVDGKEDLIA; grey triangles); (B) PACEI50L (RKWHFLW; black circles), and its two sequence variants PACEI50L-v1 (RKWHFLA; grey triangles) and PACEI50L-v2 (RKAHFLA; white squares).
2.3. Tryptophan-Containing Synthetic Heptapeptides Inhibit Prolyl Endopeptidase
The potential role of a W residue at the C-terminal end of LF-derived PEP-inhibitory peptides has also been pointed out in angiotensin-converting enzyme (ACE) inhibitory peptides, in which W at the C-terminus is associated with high inhibitory potency [20]. This prompted us to evaluate a set of six synthetic heptapeptides, which share two W residues at positions 3 and 7 (Table 3) and previously described as ACE inhibitors [39]. The inhibitory effects of 1 mg/mL heptapeptides on PEP activity are summarized in Table 3. The six heptapeptides showed significant enzyme inhibition with values ranging from 29% to 94%. Remarkably, PACEI50L exhibited a potent PEP-inhibitory activity with an IC50 value as low as 0.33 ± 0.02 mg/mL (Figure 2B).
Table 3.
Effects of heptapeptides (PACEI) on prolyl endopeptidase (PEP) activity.
| Peptide (1 mg/mL) | Sequence 1 | PEP Residual Activity (%) 2 |
|---|---|---|
| PACEI48L | Ac-RKWFHLW-NH2 | 61.4 ± 4.4 ** (a) |
| PACEI49L | Ac-RKWFLHW-NH2 | 46.8 ± 9.1 ** (b) |
| PACEI50L | Ac-RKWHFLW-NH2 | 6.4 ± 2.3 ** (c) |
| PACEI51L | Ac-RKWHLFW-NH2 | 71.6 ± 4.9 ** (a) |
| PACEI52L | Ac-RKWLFHW-NH2 | 65.9 ± 3.6 ** (a) |
| PACEI53L | Ac-RKWLHFW-NH2 | 71.5 ± 4.5 ** (a) |
1 All the peptides were acetylated at the N-terminus (Ac) and amidated at the C-terminus (NH2). 2 Data are expressed as the percentage of PEP residual activity with respect to a control without peptide (100%) and are the mean ± SD of three replicates. ** Significant inhibition with respect to control (p < 0.01) (Student’s t-test on absolute values of PEP activity). Data with the same letter do not differ at the 95% level of confidence (Tukey’s honestly significant difference (HSD) procedure).
2.4. Tryptophan Residues Are Important for Prolyl Endopeptidase Inhibition in PACEI50L and PKH11
To characterize the role of the C-terminal W in PEP-inhibitory activity, this amino acid residue was exchanged to alanine (A) in the selected sequences PACEI50L and PKH11 and thus the sequence variants RKWHFLA (PACEI50L-v1) and SVDGKEDLIA (PKH11-v1) were generated. A second PACEI50L derivative (PACEI50L-v2; RKAHFLA) was obtained by replacing both W residues at position 3 and 7 with A. We next analyzed the impact of the amino acid exchanges on PEP inhibition (Figure 2). The exchange of W to A moderately reduced PEP inhibition by PKH11-v1, in which the inhibitory potency (IC50 = 3.0 ± 0.3 mg/mL) was significantly lower than the one showed by the natural peptide PKH11 (IC50 = 2.1 ± 0.2 mg/mL), as can be seen in Figure 2A. By contrast, a severe reduction of the inhibitory activity of both PACEI50L variants was observed (Figure 2B). At the highest concentration tested (3.3 mg/mL), PACEI50L-v1 inhibited PEP by 20% while PACEI50L-v2 did not exhibit a clear inhibitory effect (Figure 2B).
2.5. Prolyl Endopeptidase-Inhibitory Peptides Alleviate Amyloid β Peptide-Induced Paralysis in the Transgenic C. elegans
To study the functional effect of PEP-inhibitory peptides in an in vivo AD model, we used the transgenic C. elegans strain CL4176 engineered to express human Aβ1–42. The active peptides PACEI50L and PKH11 were assayed at doses of 0.5, 0.1, and 0.02 µg/mL. Statistical analysis showed significant differences among paralysis curves of control and treated worms at doses of 0.5 and 0.1 µg/mL, thus, indicating a delay in paralysis (Table 4). The addition of PACEI50L and PKH11 at 0.5 and 0.1 µg/mL to the C. elegans medium provoked a reduction in the percentage of paralyzed worms at the end of the assay (49 h) and, in some cases, also a delay in the onset paralysis (Table 4). The dose of 0.02 µg/mL did not provide any effect (data not shown). Moreover, the most effective dose was 0.1 µg/mL for both PACEI50L and PKH11 peptides (Trial 2, PACEI50L 0.1 versus PACEI50L 0.5, p = 0.004; Trial 2, PKH11 0.1 versus pKH11 0.5, p = 0.006; paired log Rank survival test).
Table 4.
Effects of peptide treatments on the CL4176 paralysis and statistical analyses of the paralysis curves a.
| Treatment | Dose | Trial | Strain | Nematodes (n) | Onset Paralysis b | Paralyzed Worms c | Log Rank Χ2 | p-Value |
|---|---|---|---|---|---|---|---|---|
| PACEI50L | 0.5 | 1 | NGM | 110 | 41 (98.2%) | 70.0% | ||
| NGM + PACEI50L | 92 | 45 (96.7%) | 41.3% | 16.65 | <0.0001 | |||
| 2 | NGM | 113 | 43 (94.7%) | 68.1% | ||||
| NGM + PACEI50L | 120 | 43 (99.2%) | 41.7% | 16.94 | <0.0001 | |||
| 0.1 | 1 | NGM | 181 | 43 (96.1%) | 68.5% | |||
| NGM + PACEI50L | 189 | 41 (98.4%) | 48.7% | 15.24 | <0.0001 | |||
| 2 | NGM | 113 | 43 (94.7%) | 68.1% | ||||
| NGM + PACEI50L | 105 | 43 (99.1%) | 21.9% | 40.92 | <0.0001 | |||
| 3 | NGM | 102 | 41 (95.1%) | 65.7% | ||||
| NGM + PACEI50L | 80 | 43 (97.5%) | 56.2% | 4.386 | 0.0362 | |||
| PKH11 | 0.5 | 1 | NGM | 110 | 41 (98.2%) | 70.0% | ||
| NGM + PKH11 | 127 | 43 (96.9%) | 59.1% | 3.845 | 0.0499 | |||
| 2 | NGM | 113 | 43 (94.7%) | 68.1% | ||||
| NGM + PKH11 | 109 | 43 (97.2%) | 55.1% | 5.692 | 0.0170 | |||
| 0.1 | 1 | NGM | 113 | 41 (98.2%) | 68.8% | |||
| NGM + PKH11 | 110 | 43 (98.2%) | 32.7% | 7.643 | 0.0057 | |||
| 2 | NGM | 113 | 43 (94.7%) | 68.1% | ||||
| NGM + PKH11 | 123 | 43 (99.2%) | 30.9% | 32.09 | <0.0001 | |||
| 3 | NGM | 106 | 41 (98.1%) | 76.6% | ||||
| NGM + PKH11 | 102 | 43 (95.1%) | 64.7% | 32.71 | <0.0001 | |||
| 4 | NGM | 104 | 41 (98.1%) | 76.6% | ||||
| NGM + PKH11 | 99 | 43 (98.0%) | 61.6% | 39.88 | <0.0001 | |||
| PACEI50L-v1 | 0.1 | 1 | NGM | 181 | 43 (96.1%) | 68.5% | ||
| NGM + PACEI50L-v1 | 200 | 41 (98.0%) | 62.5% | 0.5108 | 0.4748 | |||
| 2 | NGM | 102 | 41 (95.1%) | 65.7% | ||||
| NGM + PACEI50L-v1 | 121 | 41 (91.7%) | 71.9% | 1.7 × 10−7 | 0.9997 | |||
| 3 | NGM | 113 | 41 (98.2%) | 48.7% | ||||
| NGM + PACEI50L-v1 | 134 | 41 (93.3%) | 53.7% | 0.5045 | 0.4775 | |||
| PACEI50L-v2 | 0.1 | 1 | NGM | 181 | 43 (96.1%) | 68.5% | ||
| NGM + PACEI50L-v2 | 195 | 41 (91.3%) | 74.9% | 7.489 | 0.0062 | |||
| 2 | NGM | 102 | 41 (95.1%) | 65.7% | ||||
| NGM + PACEI50L-v2 | 108 | 41 (90.7%) | 75.9% | 0.0979 | 0.7543 | |||
| 3 | NGM | 113 | 41 (98.2%) | 68.8% | ||||
| NGM + PACEI50L-v2 | 114 | 41 (94.7%) | 68.4% | 7.296 | 0.0069 | |||
| PKH11-v1 | 0.1 | 1 | NGM | 106 | 41 (98.1%) | 76.6% | ||
| NGM + PKH11-v1 | 100 | 43 (98.0%) | 52.0% | 42.14 | <0.0001 | |||
| 2 | NGM | 104 | 41 (98.1%) | 76.6% | ||||
| NGM + PKH11-v1 | 100 | 43 (98.9%) | 64.0% | 23.16 | <0.0001 |
a Analyses were performed with a paired log Rank survival test (survival curves are significantly different with respect to nematode growth medium (NGM) when p < 0.05). b Time (hours) of onset paralysis and % of non-paralyzed worms in brackets. c Percentage of paralyzed worms at 49 h.
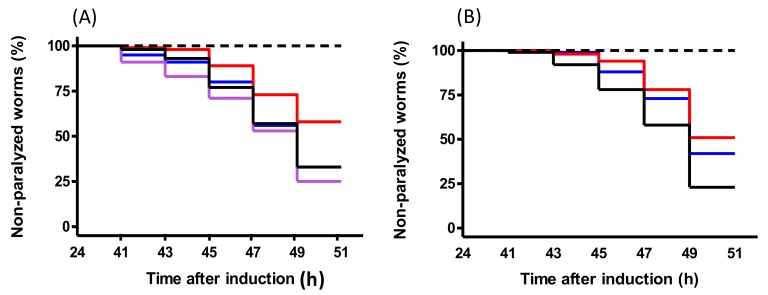
The average paralysis curves of 0.1 µg/mL PACEI50L and PKH11 treatments are shown in Figure 3, and the statistical analyses of the curves are summarized in Table 5 and Table 6. In the case of peptide PACEI50L, a decrease of 25.2% of paralyzed worms compared with control fed nematodes at 49 h was observed (Figure 3A and Table 5) whereas a delay in the onset paralysis and a reduction of 20% of final paralyzed worms were noticed for PKH11 treatment (Figure 3B and Table 6).
Figure 3.
The body paralysis of C. elegans CL4176 measured after the temperature up-shift in nematodes treated with or without peptides; worms without temperature induction were included as the negative control (dashed line). Time refers to the hours after Aβ1–42 expression induced by the temperature up-shift. (A) Non-treated worms (black line); treated-worms with 0.1 µg/mL of PACEI50L (red line), 0.1 µg/mL PACEI50L-v1 (blue line), and 0.1 µg/mL PACEI50L-v2 (purple line); data are the average of three independent trials. (B) Non-treated worms (black line); treated-worms with 0.1 µg/ml of PKH11 (red line) and 0.1 µg/mL PKH11-v1.
Table 5.
Effects of the PACEI50L, PACEI50L-v1, and PACEI50L-v2 treatments at 0.1 µg/mL on CL4176 paralysis and statistical analyses of the paralysis curves a.
| Treatment | Onset Paralysis b | Paralyzed Worms c | Log Rank X2 | p-Value (vs. NGM) | p-Value (vs. PACEI50L) |
|---|---|---|---|---|---|
| NGM | 41 (98%) | 67.5 | |||
| NGM + PACEI50L | 41 (99%) | 42.3 | 9.419 | 0.0021 | |
| NGM + PACEI50L-v1 | 41 (94%) | 62.8 | 4.5 × 10−5 | 0.9947 | 0.0022 |
| NGM + PACEI50L-v2 | 41 (92%) | 73.1 | 2.245 | 0.1341 | <0.0001 |
a Analyses were performed with a paired log Rank survival test (survival curves are significantly different with respect to control NGM or PACEI50L when p < 0.05). b Time (hours) of onset paralysis and % of non-paralyzed worms in brackets. c Percentage of paralyzed worms at 49 h.
Table 6.
Effects of the PKH11 and PKH11-v1 treatments at 0.1 µg/mL on CL4176 paralysis and statistical analyses of paralysis curves a.
| Treatment | Onset Paralysis b | Paralyzed Worms c | Log Rank X2 | p-Value (vs. NGM) | p-Value (vs. PKH11) |
|---|---|---|---|---|---|
| NGM | 41 (99%) | 67.5 | |||
| NGM + PKH11 | 43 (98%) | 47.5 | 19.26 | <0.0001 | |
| NGM + PKH11-v1 | 43 (99%) | 58.0 | 9.665 | 0.0019 | 0.1808 |
a Analyses were performed with a paired log Rank survival test (survival curves are significantly different with respect to control NGM or PKH11 when p < 0.05). b Time (hours) of onset paralysis and % of non-paralyzed worms in brackets. c Percentage of paralyzed worms at 49 h.
We also evaluated the activity of both peptide variants derived from PACEI50L at the effective dose of 0.1 µg/mL. Both modified peptides significantly changed their activity when compared with the original PACEI50L peptide, and no protective effect upon paralysis was detected (Table 4 and Table 5, and Figure 3A). In the case of PACEI50L-v2, the final percentage of non-paralyzed worms was even 5.6% lower than the control conditions. Finally, the analysis of the modified peptide PKH11-v1 (0.1 µg/mL) resulted in a partial reduction of the efficacy upon paralysis (Table 4 and Table 6, and Figure 3B), in correlation with the in vitro data of PEP inhibition.
2.6. Peptides Do Not Protect C. elegans from Oxidative Stress
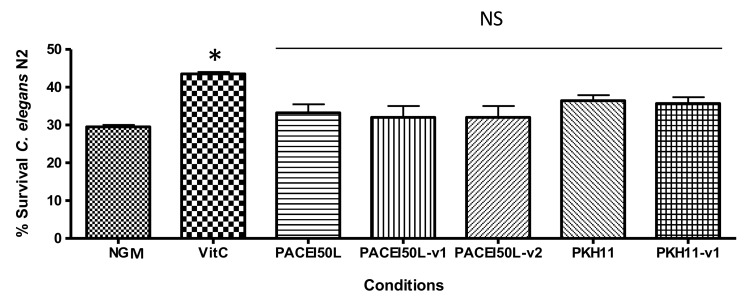
The protective effect upon oxidative stress in the wild-type strain N2 of C. elegans was assayed with PACEI50L, PKH11, and the corresponding peptide variants at a concentration of 0.1 µg/mL. Figure 4 shows that none of them was able to increase the worm survival after induction of oxidative stress with 2 mM H2O2, in contrast to the significant protection caused by vitamin C. These results indicate that peptides tested at 0.1 µg/mL, which is the most effective dose reducing C. elegans paralysis, did not show any antioxidant effect.
Figure 4.
The percentage of C. elegans N2 survival after an acute oxidative stress with H2O2. Nematodes were cultured in control conditions (NGM), with vitamin C (10 µg/mL) and with peptides (0.1 µg/mL). * Significantly different from the control (p < 0.05; one-way ANOVA followed by Tukey’s HSD); NS: not significant (p > 0.05).
2.7. Molecular Docking Analyses Reveal Different Intermolecular Interactions with Aβ1–42 Depending on the Peptide Sequence
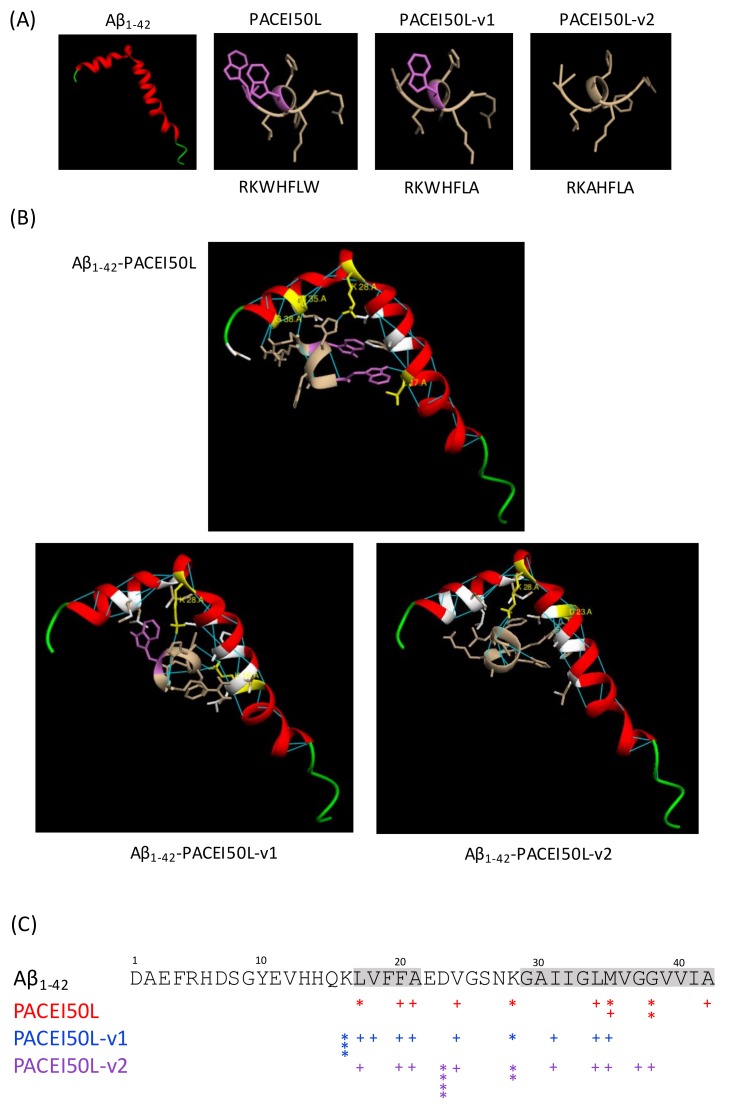
To examine the interaction between the peptides and Aβ1–42, molecular docking simulations were performed. Since the W replacement with A in the PACEI50L sequence abolished the protective effect in C. elegans, PACEI50L and its two inactive variants, PACEI50L-v1 and PACEI50L-v2, were chosen to gain insight into how peptides might interact with Aβ1–42. Peptide and Aβ1–42 monomer (ID: 1IYT) structures are shown in Figure 5A, and the simulated interactions obtained from the docking analyses are shown in Figure 5B. Figure 5C summarizes the molecular interactions in the three complexes. In the most probable model, the complex Aβ1–42-PACEI50L is stabilized by five intermolecular H-bonds: two H-bonds between R1 of PACEI50L and G38 of Aβ1–42 (R1-G38), one H-bond R1-M35, one H-bond H4-K28, and one H-bond W7-L17. PACEI50L also interacts through hydrophobic interactions with other residues of Aβ1–42 (F20/A21/V24/L34/M35/G38/A42). The interaction between Aβ1–42 and the two inactive variants is qualitatively different. The interaction with PACEI50L-v1 is stabilized by H-bonds involving K16 and K28 residues from Aβ1–42: one H-bond H4-K28, one H-bond L6-K16, and two H-bonds A7-K16. In this complex, hydrophobic interactions include residues L17/V18/F20/A21/V24/I31/L34/M35. PACEI50L-v2 docked to Aβ1–42 revealed 6 H-bonds via D23 and K28: four H-bonds R1-D23 and two H-bonds L6-K28. The binding is also stabilized by hydrophobic interactions through L17/F20/A21/V24/I31/L34/M35/G37/G38. Moreover, both inactive variants of PACEI50L change their positioning relative to Aβ1–42 when compared to the parental peptide.
Figure 5.
The molecular docking model of the Aβ1–42 monomer binding to PACEI50L, PACEI50L-v1, and PACEI50L-v2. (A) The structure of Aβ1–42 (PDB, ID:1IYT), PACEI50L, PACEI50L-v1, and PACEI50L-v2; W residues are in purple. (B) PACEI50L is bound to Aβ1–42 via five intermolecular H-bonds with L17, K28, M35, and G38. PACEI50L-v1 is bound to Aβ1–42 via four H-bonds with K16 and K28. PACEI50L-v2 is bound via six intermolecular H-bonds with D23 and K28; Aβ1–42 residues participating in molecular interactions are in yellow (H bonds) and white (hydrophobic interactions); H-bonds are in blue. (C) A schematic representation of molecular interactions between Aβ1–42 and PACEI50L (red), PACEI50L-v1 (blue), and PACEI50L-v2 (purple); * intermolecular H-bond, + hydrophobic interaction, hydrophobic domains in Aβ1–42 are highlighted in grey.
3. Discussion
Currently, peptides have a wide range of applications in medicine and biotechnology. Moreover, multifunctionality is a common trait of many peptides, which might function as polypharmacological sequences. The present study characterizes new dual neuroprotective peptides that show in vitro PEP-inhibitory properties and C. elegans protection against Aβ1–42-associated toxicity in vivo. The neuroprotective peptides include natural sequences derived from the milk protein LF and sequence-related rationally-designed synthetic heptapeptides.
LFHs obtained with three different proteases showed a moderate ability to inhibit PEP that was not shown by non-hydrolysed LF, suggesting that LF-derived peptides possess the inhibitory activity. The hydrolysates generated with three different proteases showed different PEP-inhibitory activities pointing out the importance of peptide sequences for enzyme inhibiting activity. In our study, the most potent LFH was generated using proteinase K, which exhibits specificity for peptide bonds adjacent to the carboxylic group of aliphatic and aromatic amino acids [40]. Moreover, the inhibitory activity of the hydrolysate was comparable to the previously reported IC50 values of a sodium caseinate hydrolysate (0.77 mg/mL) [29] and different fish protein hydrolysates (1.10–4.21 mg/mL) [23]. LFHs obtained with proteinase K were previously described as in vitro inhibitors of the endothelin-converting enzyme (ECE) and ex vivo as inhibitors of ECE-dependent vasoconstriction [38]. Our results describe a new biological activity related to neuroprotection for LF-derived peptides.
PEP inhibitors developed as potential therapeutic drugs are substrate-like compounds containing one P or one P-analogue residue [10]. Most of the PEP-inhibitory peptides derived from natural sources described to date contain at least one and up to six P residues in their sequences, and from 3 to 18 amino acid residues in length [22,27,29]. In spite of the fact that six of the eleven LF-derived peptides evaluated here contain one P residue in their sequences, none of them displayed PEP-inhibitory activity, as shown for some P-containing peptides derived from collagen or corn γ-zein [27]. By contrast, the only two LF-derived peptides that provoked a modest in vitro enzyme inhibition (PKH8, NEGLTW; PKH11, SVDGKEDLIW) do not contain any P residue in their sequence, suggesting that both LF-derived peptides do not act as substrate-type inhibitors. Non-P-containing peptides derived from α-casein were also described as PEP inhibitors [30].
Since the two LF-derived peptides with PEP-inhibitory activity described here shared a C-terminal W, we hypothesized its potential key contribution to enzyme inhibition. Our hypothesis was confirmed by the ability of the six sequence-related synthetic heptapeptides (PACEI48L to PACEI53L) to inhibit PEP. PACEI heptapeptides are the second generation of angiotensin converting enzyme inhibitory peptides based on two hexapeptides leads, PACEI32L (RKWHFW) and PACEI34L (RKWLFW) [41]. Heptapeptides were designed by combinations of F, H, and L residues in positions 4–6, all of them share R, K, and W residues at the N-terminus and W residue at the C-terminus of a given heptapeptide (Table 3) [39]. Among these peptides, the highest inhibitory activity was recorded for PACEI50L with H, F, and L residues at positions 4, 5, and 6, respectively. The activity of this heptapeptide is sequence-specific, as demonstrated by the swapping of H and F residues (sequences PACEI50L and PACEI48L) which drastically reduced the PEP-inhibitory activity. These results confirm and extend previously reported data on how minor amino acid exchanges affect biological properties [41,42].
Further confirmation of the key role of W residue at the C-terminus was provided by the evaluation of PACEI50L and PKH11 variants that contain W to A substitutions. Our results showed that the effect of W residue on PEP-inhibitory activity is dependent on the peptide sequence since a severe reduction of the biological activity was observed for the two variants of PACEI50L while only a 40% reduction of the inhibitory potency was recorded for the PKH11 variant. Our results also pointed to the relevance of both W residues in the PACEI50L sequence since the total loss of PEP inhibition was achieved only with the double substitution of W residues at positions 3 and 7. In agreement with these results, the most potent PEP-inhibitory peptides named 13L and 9L identified from a cocoa hydrolysate contained W in their sequence (13L, DNYDNSAGKWWVT; 9L, NYDNSAGKW) and the lower IC50 value (0.19 mg/mL) corresponded to the sequence 13L [31]. Additional studies are required to explain the role of W residues in PEP-inhibitory peptides.
C. elegans is a suitable in vivo model for research on the molecular biology and genetics of different diseases as well as for drug and bioactive compound screenings [43]. C. elegans models present alternative approaches to understanding neurodegenerative diseases for which there are currently few effective therapies [6]. Here the transgenic C. elegans model of AD, which develops a paralysis phenotype, was used to study the effects of peptides. PACEI50L and PKH11 treatments ameliorated Aβ-induced paralysis suggesting a potential in vivo protection from Aβ1–42 toxicity, as described for cocoa and maize peptides [31,32] and also for a sequence purified from scorpion venom [33]. Remarkably, and as observed for PEP-inhibitory activity, peptide variants of PACEI50L completely lost paralysis suppression in C. elegans demonstrating the positive role of W residues in the context of the heptapeptide sequence evaluated.
The effect of different compounds on Aβ1–42–mediated paralysis in transgenic C. elegans has also been associated with antioxidant effects [31,32,33,44]. In contrast to other neuroprotective peptides, the reduction of Aβ-toxicity by PACEI50L and PKH11 does not seem to operate through an antioxidant mechanism since both peptides did not produce significant C. elegans protection upon oxidative stress under the conditions tested. Conversely, the antioxidant properties of maize and scorpion venom peptides might underline, at least in part, their protective effect observed in C. elegans, since they inhibited the production of reactive oxygen species [32,33]. Additionally, in a similar experiment to the one described here, the peptide 13L from cocoa provided protection against oxidative stress, suggesting that the antioxidant activity might contribute to the protection against Aβ1–42-induced damage [31]. Therefore, our results suggest alternative mechanisms for PACEI50L and PKH11.
Aβ1–42 contains hydrophobic motifs in the central (residues 17–21) and carboxy-terminal (residues 29–42) regions of the peptide [45]. Amyloidogenic peptide aggregation seems to be primarily driven by these hydrophobic domains [46]. Here, docking analysis showed that the potential hydrogen bond interactions between the active PACEI50L and Aβ1–42 are focused on amino acids within both hydrophobic motifs (L17, M35, and G38) or close to them (K28). Particularly M35 and its hydrophobic surroundings seem to be important for the oxidative, neurotoxic, and aggregation properties of the peptide [47,48] whereas G residues can stabilize amyloidogenic structures by means of hydrogen bonds [49]. The conformation adopted by Aβ1–42 seems to be an important factor in amyloid formation since the Aβ1–42 peptide with α-helical or random coil structure aggregates slowly while Aβ with β-sheet conformation aggregates rapidly [45]. L17 included in the central hydrophobic stretch 17–21 contributes to adopt a β-sheet conformation that facilitates monomeric interaction, β-sheet oligomers, and amyloid fibrils [50]. Moreover, amino acids 17–20 served as a template for designing synthetic peptides able to inhibit fibrillogenesis in a rat brain model of amyloidosis [35]. Our docking results suggest that molecular interactions through hydrogen bonding between PACEI50L and residues L17, M35, and G38 from Aβ1–42 might impede monomeric interactions and thus, Aβ oligomerization. Docking analysis revealed the loss of hydrogen bonds between non-active PACEI50L variants and these three residues in Aβ1–42. Whether PACEI50L might interfere with the folding of Aβ1–42 to form aggregates as suggested by in silico analysis requires further in vitro and in vivo research.
Our study underscores the important role of W for the bioactivity of neuroprotective peptides. W-containing peptides as those described here display several biological activities including antihypertensive, antioxidant, antidiabetic, and satiating properties [51]. Besides W is the sole precursor of serotonin, which has been reported to have an effect on the psychological/cognitive function in humans. Remarkably, increased dietary W intake reduced intra-neuronal Aβ accumulation in a mouse model of AD, suggesting a neuroprotective role of W through the increase of serotonin levels [52]. Increased W levels also extend longevity in C. elegans and protect from alpha-synuclein and polyglutamine toxicity [53,54], suggesting a protective role against proteotoxicity in aging and age-related diseases. Additionally, in the C. elegans model, supplementation with the W-containing 13L peptide upregulated the W metabolism, including genes involved in the synthesis of serotonin and other neurotransmitters [31]. It is worthwhile to note that milk proteins and, among them, LF, are particularly rich in W in comparison with other dietary proteins opening the way to the future inclusion of LF in dietary recommendations for preventing or postponing AD.
In conclusion, we have identified novel natural and rationally-designed W-containing peptides showing in vitro PEP inhibition and in vivo protection from Aβ1–42 toxicity, although further research needs to be conducted in murine models to analyze the effectiveness of the peptides. These results add a new application to the antihypertensive proteinase K LFH and the synthetic heptapeptides confirming their multifunctionality. The effect of PACEI50L on delayed paralysis in C. elegans might be mediated, at least in part by the inhibition of Aβ1–42 oligomerization and thus, amyloid deposition, while the peptide antioxidant activity does not seem to be involved in the protective effect. Our results suggest that W-containing peptides are suitable starting leads to design effective neuroprotective agents. Further improvements to increase in vivo peptide stability based on the use of D-amino acid sequences are in progress. Future efforts are currently directed to clarify the mechanisms underlying the in vivo protective effects of W-containing peptides.
4. Materials and Methods
4.1. Materials
Bovine LF was provided by FrieslandCampina Domo (Zwolle, The Netherlands). Porcine pepsin and trypsin (type II-S), the bicinchoninic acid (BCA) kit, and the PEP substrate Z-Gly-Pro-p-nitroanilide were purchased from Sigma (Madrid, Spain). PEP was supplied by the Seikagaku Corporation (Tokyo, Japan). Recombinant proteinase K was purchased from Roche (Mannheim, Germany).
4.2. Lactoferrin Hydrolysates and Peptides
Bovine LF (5% w/v) was hydrolyzed using pepsin (3% w/w), trypsin (1% w/w), or proteinase K (1% w/v) as previously described [38,55]. LFHs were subjected to ultrafiltration through a polyethersulfone membrane with a 3 kDa cut-off (Vivascience, Sartorius Stedim Biotech, Aubagne, France) and the permeates were kept at −20 °C until use.
Synthetic peptides were purchased at >95% purity from GenScript Corporation (Piscataway, NJ, USA), wherein they were synthesized by solid phase methods using N-(9-fluorenyl) methoxycarbonyl chemistry. PACEI peptides were acetylated at the N-terminus and amidated at the C-terminus. The synthetic peptide concentration was based on the dry weight and purity provided by the manufacturer.
The protein content of LFHs was estimated by the BCA method using bovine serum albumin as the standard [55].
4.3. Prolyl Endopeptidase Assay
The PEP activity was determined as previously described [31] with minor modifications. Enzyme assays were performed in 96-well microplates using Z-Gly-Pro-p-nitroanilide as a substrate and measuring the increase in absorbance at 410 nm due to the release of p-nitroaniline. Ten µL of tenfold peptide or hydrolysate solution in 100 mM sodium phosphate buffer, pH 7 (final concentrations from 0.063 to 3.6 mg/mL for peptides and from 0.35 to 5.7 mg/L for LFHs), 30 µL of a 0.1 U/mL PEP solution in 100 mM sodium phosphate buffer (pH 7) and 35 µL of the same sodium phosphate buffer were pre-incubated at 30 °C for 5 min, and the mixture incubated with 25 µL of 2 mM Z-Gly-Pro-p-nitroanilide in 40% 1,4-dioxane for 30 min at the same temperature. The reaction was finished by the addition of 100 µL 10% Triton X-100 in 1 M sodium acetate buffer (pH 4), and the absorbance measured at 410 nm in a microplate reader. Data are expressed as the percentage of PEP residual activity with respect to a control without peptide (100%).
The IC50 value of a peptide was defined as the concentration required to inhibit 50% of the PEP activity and the value for each experiment was estimated by non-linear regression of the experimental data to a four-parameter logistic curve using the software package SigmaPlot v 13.0 (SPSS Inc., Chicago, IL, USA).
4.4. C. elegans Strains and Maintenance
Wild-type Bristol strain N2 and the transgenic strain CL4176 (smg-1ts [pAF29(myo-3/Aβ1–42/let UTR) + pRF4(rol-6(su10069))]) were obtained from the Caenorhabditis Genetics Center (College of Biological Sciences, University of Minnesota, Saint Paul, MN, USA) and were used for oxidative stress experiments and paralysis assays, respectively. C. elegans strains were routinely propagated on Nematode Growth Medium (NGM) plates with Escherichia coli strain OP50 as a food source at 20 °C (N2) or at 16 °C (CL4176). Worms were synchronized by isolating eggs from gravid adults at 20 °C (N2) or at 16 °C (CL4176), and eggs were hatched overnight in NGM plates.
In the oxidative and paralysis experiments, worms were fed with the different compounds from egg to adult stages, and transferred to new plates every two days.
4.5. C. elegans Paralysis Assay
Paralysis is induced in strain CL4176 by the expression of a muscle-specific Aβ1–42 which depends on up-shifting temperature from 16 to 25 °C [7,8]. For paralysis experiments, strain CL4176 maintained at 16 °C was egg-synchronized in plates containing NGM (control medium) and NGM with peptides (0.02–5 µg/mL). Transgene expression was induced by up-shifting the temperature from 16 to 25 °C, starting 24 h after egg laying and maintained for 24 h. The worms were incubated at 20 °C until all the worms in the experiment became paralyzed. Paralysis was scored 24 h after induction. Paralysis in induced worms was compared with non-induced worms (maintained at 16 °C until the end of the paralysis assay). Experiments were carried out at least in duplicate.
4.6. C. elegans Oxidative Stress Assays
For oxidative stress experiments, strain N2 was egg-synchronized in NGM plates (control medium) and NGM supplemented with 0.1 µg/mL peptides or 0.1 mg/mL vitamin C (positive control). The viability of C. elegans was assessed after 2 mM hydrogen peroxide (H2O2)-induced oxidative stress for 5 h [56]. Experiments were carried out in triplicate.
4.7. Molecular Docking
Structural modeling of the linear peptide PACEI50L and its variants PACEI50L-v1 and PACEI50L-v2 was carried out using the PEP-FOLD3 platform (Université Paris Diderot, Paris, France) [57] currently available on the RPBS web portal (http://mobyle.rpbs.univ-paris-diderot.fr). Solution structure of Aβ1–42 was obtained from the Protein Data Bank (PDB, ID: 1IYT). The molecular docking of the peptides to Aβ1–42 was performed using the ClusPro web server (Boston University, Boston, MA, USA; https://cluspro.bu.edu) [58]. ClusPro model selection is based on the cluster size rather than on the cluster energy score. Model 0 was selected for complexes Aβ1–42-PACEI50L (cluster size, 834; cluster score, −573.1), Aβ1–42-PACEI50L-v1 (cluster size, 642; cluster score, −549.7), and Aβ1–42-PACEI50L-v2 (cluster size, 218; cluster score, −457.4). The 3D models of the peptides and their interactions were visualized by the UCSF Chimera software (University of California, San Francisco, CA, USA) [59].
4.8. Statistics
Statistical analyses were carried out using the GraphPad Prism 4 software package (GraphPad Software, La Jolla, CA, USA). PEP residual activity data are mean ± standard deviation (SD) and were subjected to either Student’s t-test or one-way ANOVA followed by Dunnett post-test or Tukey’s honestly significant difference procedure (HSD). The comparison of the C. elegans paralysis curves was performed using the log-rank survival test. The survival data of the C. elegans was assessed after oxidative stress and differences between nematodes cultured in the control and treatment conditions were evaluated by means of a one-way ANOVA followed by Tukey’s HSD. p < 0.05 was considered significant.
Acknowledgments
This work was funded by grant AEST/2015/005 from “Generalitat Valenciana” and BIO2015-68790-C2-1-R from the “Ministerio de Economía y Competitividad” (Spain) (MINECO/FEDER Funds). S.Ga was recipient of a predoctoral scholarship (FPU13/04584) within the FPU program from “Ministerio de Educación, Cultura y Deporte” (MECD, Spain). We acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
Abbreviations
| AD | Alzheimer’s disease |
| Aβ | Amyloid β peptide |
| Aβ1–42 | Amyloid β peptide 1–42 |
| ACE | Angiotensin converting enzyme |
| βAPP | β-amyloid precursor protein |
| BCA | Bicinchoninic acid |
| HPLC MS/MS | High-performance liquid chromatography tandem mass spectrometry |
| HSD | Honestly significant difference |
| LF | Lactoferrin |
| LFH | Lactoferrin hydrolysate |
| NGM | Nematode growth medium |
| PEP | Prolyl endopeptidase |
| POP | Prolyl oligopeptidase |
| SD | Standard deviation |
Author Contributions
P.Man. D.R., S.Ge., J.F.M. and P.Mar. conceived and designed the experiments; P.Man. and S.Ge. performed PEP assays; R.M. and P.Mar. performed C. elegans assays; S.Ga. and J.F.M. performed and analyzed docking results; D.R. and J.F.M. run statistical analyses; P.Man., D.R., S.Ge., J.F.M. and P.Mar. analyzed the data; P.Man. and P.Mar. wrote the paper; all authors revised and approved the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Skovronsky D.M., Lee V.M.-Y., Trojanowski J.Q. Neurodegenerative diseases: New concepts of pathogenesis and their therapeutic implications. Annu. Rev. Pathol. Mech. Dis. 2006;1:151–170. doi: 10.1146/annurev.pathol.1.110304.100113. [DOI] [PubMed] [Google Scholar]
- 2.Holtzman D.M., Morris J.C., Goate A.M. Alzheimer’s disease: The challenge of the second century. Sci. Transl. Med. 2011;3:77sr1. doi: 10.1126/scitranslmed.3002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hardy J., Selkoe D.J. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 4.Butterfield D.A., Drake J., Pocernich C., Castegna A. Evidence of oxidative damage in Alzheimer’s disease brain: Central role for amyloid β-peptide. Trends Mol. Med. 2001;7:548–554. doi: 10.1016/S1471-4914(01)02173-6. [DOI] [PubMed] [Google Scholar]
- 5.Sultana R., Ravagna A., Mohmmad-Abdul H., Calabrese V., Butterfield D.A. Ferulic acid ethyl ester protects neurons against amyloid β-peptide(1–42)-induced oxidative stress and neurotoxicity: Relationship to antioxidant activity. J. Neurochem. 2005;92:749–758. doi: 10.1111/j.1471-4159.2004.02899.x. [DOI] [PubMed] [Google Scholar]
- 6.Alexander A.G., Marfil V., Li C. Use of Caenorhabditis elegans as a model to study Alzheimer’s disease and other neurodegenerative diseases. Front. Genet. 2014;5:279. doi: 10.3389/fgene.2014.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Link C.D. Expression of human beta-amyloid peptide in transgenic Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 1995;92:9368–9372. doi: 10.1073/pnas.92.20.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Link C.D., Johnson C.J., Fonte V., Paupard M.-C., Hall D.H., Styren S., Mathis C.A., Klunk W.E. Visualization of fibrillar amyloid deposits in living, transgenic Caenorhabditis elegans animals using the sensitive amyloid dye, X-34. Neurobiol. Aging. 2001;22:217–226. doi: 10.1016/S0197-4580(00)00237-2. [DOI] [PubMed] [Google Scholar]
- 9.García-Horsman J.A., Männistö P.T., Venäläinen J.I. On the role of prolyl oligopeptidase in health and disease. Neuropeptides. 2007;41:1–24. doi: 10.1016/j.npep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Lawandi J., Gerber-Lemaire S., Juillerat-Jeanneret L., Moitessier N. Inhibitors of prolyl oligopeptidases for the therapy of human diseases: Defining diseases and inhibitors. J. Med. Chem. 2010;53:3423–3438. doi: 10.1021/jm901104g. [DOI] [PubMed] [Google Scholar]
- 11.Barelli H., Petit A., Hirsch E., Wilk S., de Nanteuil G., Morain P., Checler F. S 17092-1, a highly potent, specific and cell permeant inhibitor of human proline endopeptidase. Biochem. Biophys. Res. Commun. 1999;257:657–661. doi: 10.1006/bbrc.1999.0366. [DOI] [PubMed] [Google Scholar]
- 12.Männistö P.T., García-Horsman J.A. Mechanism of action of prolyl oligopeptidase (PREP) in degenerative brain diseases: Has peptidase activity only a modulatory role on the interactions of PREP with proteins? Front. Aging Neurosci. 2017;9:27. doi: 10.3389/fnagi.2017.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhutia S.K., Maiti T.K. Targeting tumors with peptides from natural sources. Trends Biotechnol. 2008;26:210–217. doi: 10.1016/j.tibtech.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 14.Sultan S., Huma N., Butt M.S., Aleem M., Abbas M. Therapeutic potential of dairy bioactive peptides: A contemporary perspective. Crit. Rev. Food Sci. Nutr. 2016;58:105–115. doi: 10.1080/10408398.2015.1136590. [DOI] [PubMed] [Google Scholar]
- 15.Yoshikawa M. Bioactive peptides derived from natural proteins with respect to diversity of their receptors and physiological effects. Peptides. 2015;72:208–225. doi: 10.1016/j.peptides.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 16.Caron J., Domenger D., Dhulster P., Ravallec R., Cudennec B. Protein digestion-derived peptides and the peripheral regulation of food intake. Front. Endocrinol. 2017;8:85. doi: 10.3389/fendo.2017.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cicero A.F.G., Fogacci F., Colletti A. Potential role of bioactive peptides in prevention and treatment of chronic diseases: A narrative review. Br. J. Pharmacol. 2017;174:1378–1394. doi: 10.1111/bph.13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lafarga T., Hayes M. Bioactive peptides from meat muscle and by-products: Generation, functionality and application as functional ingredients. Meat Sci. 2014;98:227–239. doi: 10.1016/j.meatsci.2014.05.036. [DOI] [PubMed] [Google Scholar]
- 19.Erdmann K., Cheung B.W., Schroder H. The possible roles of food-derived bioactive peptides in reducing the risk of cardiovascular disease. J. Nutr. Biochem. 2008;19:643–654. doi: 10.1016/j.jnutbio.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 20.Manzanares P., Salom J.B., Garcia-Tejedor A., Fernandez-Musoles R., Ruiz-Gimenez P., Gimeno-Alcañiz J.V. Unraveling the mechanisms of action of lactoferrin-derived antihypertensive peptides: ACE inhibition and beyond. Food Funct. 2015;6:2440–2452. doi: 10.1039/C5FO00580A. [DOI] [PubMed] [Google Scholar]
- 21.Fosgerau K., Hoffmann T. Peptide therapeutics: Current status and future directions. Drug Discov. Today. 2015;20:122–128. doi: 10.1016/j.drudis.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Lafarga T., O’Connor P., Hayes M. In silico methods to identify meat-derived prolyl endopeptidase inhibitors. Food Chem. 2015;175:337–343. doi: 10.1016/j.foodchem.2014.11.150. [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Alvarez O., Batista I., Ramos C., Montero P. Enhancement of ACE and prolyl oligopeptidase inhibitory potency of protein hydrolysates from sardine and tuna by-products by simulated gastrointestinal digestion. Food Funct. 2016;7:2066–2073. doi: 10.1039/C5FO01603G. [DOI] [PubMed] [Google Scholar]
- 24.Sila A., Martinez-Alvarez O., Haddar A., Gomez-Guillen M.C., Nasri M., Montero M.P., Bougatef A. Recovery, viscoelastic and functional properties of Barbel skin gelatine: Investigation of anti-DPP-IV and anti-prolyl endopeptidase activities of generated gelatine polypeptides. Food Chem. 2015;168:478–486. doi: 10.1016/j.foodchem.2014.07.086. [DOI] [PubMed] [Google Scholar]
- 25.Sorensen R., Kildal E., Stepaniak L., Pripp A.H., Sorhaug T. Screening for peptides from fish and cheese inhibitory to prolyl endopeptidase. Nahrung. 2004;48:53–56. doi: 10.1002/food.200300358. [DOI] [PubMed] [Google Scholar]
- 26.Wilson J., Hayes M., Carney B. Angiotensin-I-converting enzyme and prolyl endopeptidase inhibitory peptides from natural sources with a focus on marine processing by-products. Food Chem. 2011;129:235–244. doi: 10.1016/j.foodchem.2011.04.081. [DOI] [PubMed] [Google Scholar]
- 27.Maruyama S., Miyoshi S., Osa T., Tanaka H. Prolyl endopeptidase inhibitory activity of peptides in the repeated sequence of various proline-rich proteins. J. Ferment. Bioeng. 1992;74:145–148. doi: 10.1016/0922-338X(92)90073-4. [DOI] [Google Scholar]
- 28.Asano M., Nio N., Ariyoshi Y. Inhibition of prolyl endopeptidase by synthetic peptide fragments of human β-casein. Agric. Biol. Chem. 1991;55:825–828. doi: 10.1271/bbb1961.55.825. [DOI] [PubMed] [Google Scholar]
- 29.Hsieh C.-H., Wang T.-Y., Hung C.-C., Hsieh Y.-L., Hsu K.-C. Isolation of prolyl endopeptidase inhibitory peptides from a sodium caseinate hydrolysate. Food Funct. 2016;7:565–573. doi: 10.1039/C5FO01262G. [DOI] [PubMed] [Google Scholar]
- 30.Sistla S. Structure-activity relationships of alpha(s)-casein peptides with multifunctional biological activities. Mol. Cell. Biochem. 2013;384:29–38. doi: 10.1007/s11010-013-1778-4. [DOI] [PubMed] [Google Scholar]
- 31.Martorell P., Bataller E., Llopis S., Gonzalez N., Alvarez B., Monton F., Ortiz P., Ramon D., Genoves S. A cocoa peptide protects Caenorhabditis elegans from oxidative stress and β-amyloid peptide toxicity. PLoS ONE. 2013;8:e63283. doi: 10.1371/journal.pone.0063283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z., Ma H., Wang X., Zhao Z., Zhang Y., Zhao B., Guo Y., Xu L. A tetrapeptide from maize protects a transgenic Caenorhabditis elegans Aβ1–42 model from Aβ-induced toxicity. RSC Adv. 2016;6:56851–56858. doi: 10.1039/C6RA06130C. [DOI] [Google Scholar]
- 33.Zhang X.-G., Wang X., Zhou T.-T., Wu X.-F., Peng Y., Zhang W.-Q., Li S., Zhao J. Scorpion venom heat-resistant peptide protects transgenic Caenorhabditis elegans from β-amyloid toxicity. Front. Pharmacol. 2016;7:227. doi: 10.3389/fphar.2016.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Estrada L.D., Soto C. Disrupting β-amyloid aggregation for Alzheimer disease treatment. Curr. Top. Med. Chem. 2007;7:115–126. doi: 10.2174/156802607779318262. [DOI] [PubMed] [Google Scholar]
- 35.Soto C., Sigurdsson E.M., Morelli L., Asok Kumar R., Castano E.M., Frangione B. β-sheet breaker peptides inhibit fibrillogenesis in a rat brain model of amyloidosis: Implications for Alzheimer’s therapy. Nat. Med. 1998;4:822–826. doi: 10.1038/nm0798-822. [DOI] [PubMed] [Google Scholar]
- 36.Tomita M., Wakabayashi H., Yamauchi K., Teraguchi S., Hayasawa H. Bovine lactoferrin and lactoferricin derived from milk: Production and applications. Biochem. Cell. Biol. 2002;80:109–112. doi: 10.1139/o01-230. [DOI] [PubMed] [Google Scholar]
- 37.Martorell P., Llopis S., Gonzalez N., Ramón D., Serrano G., Torrens A., Serrano J.M., Navarro M., Genovés S. A nutritional supplement containing lactoferrin stimulates the immune system, extends lifespan, and reduces amyloid β peptide toxicity in Caenorhabditis elegans. Food Sci. Nutr. 2017;5:255–265. doi: 10.1002/fsn3.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernández-Musoles R., Salom J.B., Martínez-Maqueda D., López-Díez J.J., Recio I., Manzanares P. Antihypertensive effects of lactoferrin hydrolyzates: Inhibition of angiotensin- and endothelin-converting enzymes. Food Chem. 2013;139:994–1000. doi: 10.1016/j.foodchem.2012.12.049. [DOI] [PubMed] [Google Scholar]
- 39.Ruiz-Giménez P., Marcos J.F., Torregrosa G., Lahoz A., Fernández-Musoles R., Vallés S., Alborch E., Manzanares P., Salom J.B. Novel antihypertensive hexa- and heptapeptides with ACE-inhibiting properties: From the in vitro ACE assay to the spontaneously hypertensive rat. Peptides. 2011;32:1431–1438. doi: 10.1016/j.peptides.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 40.Ebeling W., Hennrich N., Klockow M., Metz H., Orth H.D., Lang H. Proteinase K from Tritirachium album Limber. Eur. J. Biochem. 1974;47:91–97. doi: 10.1111/j.1432-1033.1974.tb03671.x. [DOI] [PubMed] [Google Scholar]
- 41.Centeno J.M., Burguete M.C., Castello-Ruiz M., Enrique M., Valles S., Salom J.B., Torregrosa G., Marcos J.F., Alborch E., Manzanares P. Lactoferricin-related peptides with inhibitory effects on ACE-dependent vasoconstriction. J. Agric. Food Chem. 2006;54:5323–5329. doi: 10.1021/jf060482j. [DOI] [PubMed] [Google Scholar]
- 42.Lopez-Garcia B., Perez-Paya E., Marcos J.F. Identification of novel hexapeptides bioactive against phytopathogenic fungi through screening of a synthetic peptide combinatorial library. Appl. Environ. Microbiol. 2002;68:2453–2460. doi: 10.1128/AEM.68.5.2453-2460.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calvo D.R., Martorell P., Genovés S., Gosálbez L. Development of novel functional ingredients: Need for testing systems and solutions with Caenorhabditis elegans. Trends Food Sci. Technol. 2016;54:197–203. doi: 10.1016/j.tifs.2016.05.006. [DOI] [Google Scholar]
- 44.Shi Y.C., Pan T.M., Liao V.H. Monascin from Monascus-fermented products reduces oxidative stress and amyloid-β toxicity via DAF-16/FOXO in Caenorhabditis elegans. J. Agric. Food Chem. 2016;64:7114–7120. doi: 10.1021/acs.jafc.6b02779. [DOI] [PubMed] [Google Scholar]
- 45.Soto C., Castaño E.M. The conformation of Alzheimer’s β peptide determines the rate of amyloid formation and its resistance to proteolysis. Biochem. J. 1996;314:701–707. doi: 10.1042/bj3140701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cecchini M., Curcio R., Pappalardo M., Melki R., Caflisch A. A molecular dynamics approach to the structural characterization of amyloid aggregation. J. Mol. Biol. 2006;357:1306–1321. doi: 10.1016/j.jmb.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 47.Kanski J., Aksenova M., Butterfield D.A. The hydrophobic environment of Met35 of Alzheimer’s Aβ(1–42) is important for the neurotoxic and oxidative properties of the peptide. Neurotox. Res. 2002;4:219–223. doi: 10.1080/10298420290023945. [DOI] [PubMed] [Google Scholar]
- 48.Kanski J., Aksenova M., Schöneich C., Butterfield D.A. Substitution of isoleucine-31 by helical-breaking proline abolishes oxidative stress and neurotoxic properties of Alzheimer’s amyloid β-peptide (1–42) Free Radic. Biol. Med. 2002;32:1205–1211. doi: 10.1016/S0891-5849(02)00821-3. [DOI] [PubMed] [Google Scholar]
- 49.Soto C., Brañes M.C., Alvarez J., Inestrosa N.C. Structural determinants of the Alzheimer’s amyloid β-peptide. J. Neurochem. 1994;63:1191–1198. doi: 10.1046/j.1471-4159.1994.63041191.x. [DOI] [PubMed] [Google Scholar]
- 50.Soto C., Kindy M.S., Baumann M., Frangione B. Inhibition of Alzheimer’s amyloidosis by peptides that prevent β-sheet conformation. Biochem. Biophys. Res. Commun. 1996;226:672–680. doi: 10.1006/bbrc.1996.1413. [DOI] [PubMed] [Google Scholar]
- 51.Nongonierma A.B., FitzGerald R.J. Milk proteins as a source of tryptophan-containing bioactive peptides. Food Funct. 2015;6:2115–2127. doi: 10.1039/C5FO00407A. [DOI] [PubMed] [Google Scholar]
- 52.Noristani H.N., Verkhratsky A., Rodríguez J.J. High tryptophan diet reduces CA1 intraneuronal β-amyloid in the triple transgenic mouse model of Alzheimer’s disease. Aging Cell. 2012;11:810–822. doi: 10.1111/j.1474-9726.2012.00845.x. [DOI] [PubMed] [Google Scholar]
- 53.Edwards C., Canfield J., Copes N., Brito A., Rehan M., Lipps D., Brunquell J., Westerheide S.D., Bradshaw P.C. Mechanisms of amino acid-mediated lifespan extension in Caenorhabditis elegans. BMC Genet. 2015;16:8. doi: 10.1186/s12863-015-0167-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van der Goot A.T., Zhu W., Vázquez-Manrique R.P., Seinstra R.I., Dettmer K., Michels H., Farina F., Krijnen J., Melki R., Buijsman R.C., et al. Delaying aging and the aging-associated decline in protein homeostasis by inhibition of tryptophan degradation. Proc. Natl. Acad. Sci. USA. 2012;109:14912–14917. doi: 10.1073/pnas.1203083109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruiz-Giménez P., Salom J.B., Marcos J.F., Vallés S., Martínez-Maqueda D., Recio I., Torregrosa G., Alborch E., Manzanares P. Antihypertensive effect of a bovine lactoferrin pepsin hydrolysate: Identification of novel active peptides. Food Chem. 2012;131:266–273. doi: 10.1016/j.foodchem.2011.08.076. [DOI] [Google Scholar]
- 56.Martorell P., Forment J.V., de Llanos R., Montón F., Llopis S., González N., Genovés S., Cienfuegos E., Monzó H., Ramón D. Use of Saccharomyces cerevisiae and Caenorhabditis elegans as model organisms to study the effect of cocoa polyphenols in the resistance to oxidative stress. J. Agric. Food Chem. 2011;59:2077–2085. doi: 10.1021/jf104217g. [DOI] [PubMed] [Google Scholar]
- 57.Lamiable A., Thévenet P., Rey J., Vavrusa M., Derreumaux P., Tufféry P. PEP-FOLD3: Faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016;44:W449–W454. doi: 10.1093/nar/gkw329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kozakov D., Hall D.R., Xia B., Porter K.A., Padhorny D., Yueh C., Beglov D., Vajda S. The ClusPro web server for protein–protein docking. Nat. Protoc. 2017;12:255. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]