Abstract
Malignant pleural mesothelioma (MPM) is a rare malignancy, with extremely poor survival rates. At present, treatment options are limited, with no second line chemotherapy for those who fail first line therapy. Extensive efforts are ongoing in a bid to characterise the underlying molecular mechanisms of mesothelioma. Recent research has determined that between 70–90% of our genome is transcribed. As only 2% of our genome is protein coding, the roles of the remaining proportion of non-coding RNA in biological processes has many applications, including roles in carcinogenesis and epithelial–mesenchymal transition (EMT), a process thought to play important roles in MPM pathogenesis. Non-coding RNAs can be separated loosely into two subtypes, short non-coding RNAs (<200 nucleotides) or long (>200 nucleotides). A significant body of evidence has emerged for the roles of short non-coding RNAs in MPM. Less is known about the roles of long non-coding RNAs (lncRNAs) in this disease setting. LncRNAs have been shown to play diverse roles in EMT, and it has been suggested that EMT may play a role in the aggressiveness of MPM histological subsets. In this report, using both in vitro analyses on mesothelioma patient material and in silico analyses of existing RNA datasets, we posit that various lncRNAs may play important roles in EMT within MPM, and we review the current literature regarding these lncRNAs with respect to both EMT and MPM.
Keywords: malignant pleural mesothelioma, long non-coding RNAs (lncRNAs), epithelial- mesenchymal transition
1. Introduction
Malignant pleural mesothelioma (MPM) is a rare, but aggressive form of cancer, predominantly associated with prior exposure to asbestos [1]. Whilst many countries have banned the use of asbestos [2], it is still used in developing countries. A recent report based on extrapolations for asbestos use estimated global mesothelioma deaths at 38,400 per annum [3], and while there have been some recent advances in this disease, particularly with respect to immune-oncology [4,5], the current standard of care (a combination of pemetrexed/raltitrexed and cisplatin chemotherapy) [6,7] is non-curative, and results in a response rate of approximately 40% [8].
Epithelial–mesenchymal transition (EMT) is a process by which epithelial cells shed many of their epithelial traits and acquire various features observed in mesenchymal cells. During this transition, epithelial cells lose their polarity and many of their intercellular contacts, such as desmosomes, adherens junctions, and tight junctions, resulting in their disassociation from epithelial sheets. At the end of this process, cells undergoing EMT assume a variety of mesenchymal-like properties: enhanced migratory capacity, invasiveness, heightened resistance to apoptosis, and greatly increased production of extracellular matrix components [9].
Most MPMs have three main histologic subtypes, divided into epithelioid, sarcomatoid, or mixed (biphasic) [10,11]. However, multiple morphological patterns have also been described within these subtypes, and similarities in clinical presentation and histological appearance of MPM, primary lung carcinoma, pleural metastases, reactive pleural diseases, and rare pleural malignancies can pose challenges to MPM diagnosis [12]. Indeed, “The current gold standard of MPM diagnosis is a combination of two positive and two negative immune-histochemical markers in the epithelioid and biphasic type, but sarcomatous type do not have specific markers, making diagnosis more difficult.” [12]. Because MPM has a partial fibroblastic phenotype in the context of EMT, it has been postulated that this may, in part, explain the aggressiveness of this cancer conferring both high invasiveness and chemoresistance [13], and in this regard, it may be applied to the epithelioid versus sarcomatoid histotype of MPM [13]. In this regard, the epithelioid and sarcomatoid histologic variants of MPM can be considered as E- and M-parts of the EMT axis, with the biphasic histotype considered an intermediate [14]. In support of this, hierarchical clustering of transcriptomic data from MPM separates this cancer into two distinct molecular subgroups, and one subgroup (C2) with an associated EMT molecular signature has worse overall survival (OS) [15].
A significant body of work has examined the roles of other forms of non-coding RNA such as microRNAs in both EMT [16,17] and MPM [18,19], and there is some evidence that miRNAs and lncRNAs interact or cross-talk to orchestrate EMT [20]. Despite the known roles of lncRNAs in the establishment of EMT in cancer, a topic recently reviewed in detail by us and others [21,22], very few studies have specifically examined the functional roles of lncRNAs in MPM [23,24,25,26].
With the advent of high-throughput sequencing technology, transcriptomic data for MPM is emerging. Using unsupervised consensus clustering of RNA-seq-derived expression data from 211 MPM samples, Bueno et al. [27] identified four major clusters: sarcomatoid, epithelioid, biphasic-epithelioid (biphasic-E), and biphasic-sarcomatoid (biphasic-S). Of these, differential expression analysis of the sarcomatoid and epithelioid consensus clusters identified a significant number of lncRNAs which could distinguish between these, as shown in Table 1.
Table 1.
Differentially expressed long non-coding RNAs (lncRNAs) between sarcomatoid versus epithelioid samples as identified by Bueno et al. [27], and discussed in this article.
| Name | log2 Fold Change | Unadjusted p-Value | Comments |
|---|---|---|---|
| PCAT1 | −1.227580845 | 0.000168412 | |
| HOTAIR | 4.342211972 | 1.09 × 10−10 | Associates with chromatin remodelling complexes to regulate EMT [21] |
| MALAT1 | −0.902533139 | 2.72 × 10−7 | |
| NEAT1 | −0.534058107 | 0.012990525 | Identified as an lncRNA with altered (−2.8 fold) expression in MPM [26] |
| GAS5 | 0.053707959 | 0.785538121 | GAS5 shown to have altered expression in MPM |
| HULC | −0.724711448 | 0.03946186 | Known roles in EMT in other cancers [28,29,30] |
| H19 | 2.155715056 | 1.09 × 10−9 | Promotes EMT in NSCLC [31], and various other cancers [21] |
| ZFAS1 | −0.443662478 | 0.018761094 | Known regulator of EMT in other cancer settings [32,33,34,35,36] |
| PVT1 | −0.64835701 | 7.75 × 10−5 | Previously identified as an lncRNA with altered expression in MPM [24] |
| CASC2 | −1.434979397 | 5.64 × 10−12 | Overexpression shown to inhibit EMT in lung adenocarcinoma [37]. Associated with Epithelioid and Biphasic samples and high expression associated with better OS in The Cancer Genome Atlas (TCGA) dataset |
A discussion of the putative roles for these and other lncRNAs in EMT will be presented in subsequent sections.
In this manuscript, we examined the expression of a novel series of lncRNAs (Epidermal Growth Factor Receptor- antisense RNA 1 EGFR-AS1, prostate cancer associated transcript 6 PCAT6 and zinc finger E-box binding homeobox 2 antisense RNA 1 ZEB2-AS1) for altered expression in MPM. We show that all three of these lncRNAs are overexpressed in MPM, and that one of them, PCAT6, is significantly altered across all of the histological subtypes.
Subsequently, using in silico meta-analysis of existing The Cancer Genome Atlas (TCGA) and other datasets (www.cbioportal.org; http://watson.compbio.iupui.edu/chirayu/proggene/database/?url=proggene; www.oncomine.org), we review the known lncRNAs previously described by us and others in MPM (PVT1, NEAT1, PAX8-AS1, and GAS5). Finally, using in silico analyses, combined with a review of the current literature, we examine additional lncRNAs with known roles in EMT for the dysregulated expression in MPM, and show that for many of these, this dysregulated expression is often associated with the biphasic histological subtype. These results suggest that many lncRNAs may be a factor in the transition from the epithelioid to the more aggressive sarcomatoid histotype of malignant pleural mesothelioma.
2. Results
2.1. Novel LncRNAs with Altered Expression in MPM
Several lncRNAs have recently been identified by our unit as having potentially significant roles in MPM. In the following sections we describe their expression and putative roles in both EMT and MPM.
2.1.1. EGFR-AS1
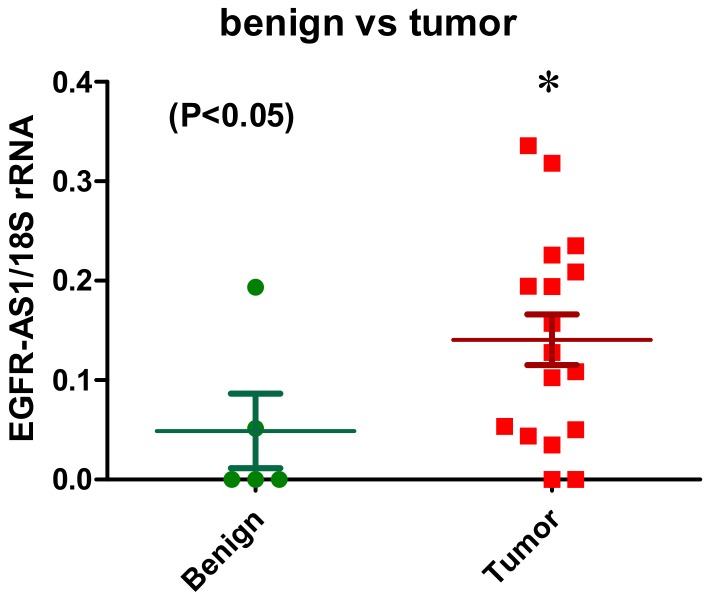
High expression of EGFR is associated with MPM [38]. However, clinical trials of EGFR tyrosine kinase inhibitors (TKIs) as single agents in MPM failed [39,40,41]. However, more recently, expression of EGFR on MPM has been used for the targeted delivery of microRNA mimics delivered by targeted bacterial minicells (TargomiRs) in a recent clinical trial in MPM [42], while most recently a patient harbouring mutations in EGFR (G719C and S768I) was successfully treated with Afatinib an EGFR TKI [43]; an lncRNA associated with EGFR called EGFR-AS1 has been identified. This lncRNA was shown to regulate EGFR expression in liver cancer [44], and most recently, expression of this lncRNA has been shown to be associated with sensitivity to EGFR TKIs in patients with head and neck SCC (HNSCC) [45]. Strikingly, knockdown of EGFR-AS1 in vitro and in vivo lead to increased sensitivity, whereas overexpression is sufficient to induce resistance to EGFR TKIs [45]. In this regard, preliminary data from our group has shown that EGFR-AS1 is significantly overexpressed in MPM (Figure 1); this may explain in part why EGFR TKIs failed as single agents in clinical trials of MPM. The role of EGFR-AS1 in EMT has as yet to be determined. However, the known role of EMT in bypassing EGFR dependence [46] suggests that this lncRNA may indeed play a role in orchestrating EMT transitions in MPM.
Figure 1.
Overexpression of EGFR-AS1 in primary malignant pleural mesothelioma (MPM). EGFR-AS1 lncRNA expression was examined by RT-PCR in a series of primary MPM (n = 17) versus benign pleura (n = 5). Semi-quantitative densitometric analysis of the results determined that EGFR-AS1 lncRNA was significantly elevated in the tumours compared to benign pleura. Statistical significance was assessed using a 1-tailed unpaired Students t-test (* p = 0.0445).
2.1.2. PCAT6
PCAT6 is a lncRNA linked to KDM5B (also known as JARID1B). This lysine demethylase has been shown to induce EMT in various cancers, including lung cancer [47,48,49]. Expression of PCAT6 has also been shown to be altered in NSCLC [50,51], and circulating levels of this lncRNA in patient blood has potential as both a diagnostic and prognostic biomarker in NSCLC [51].
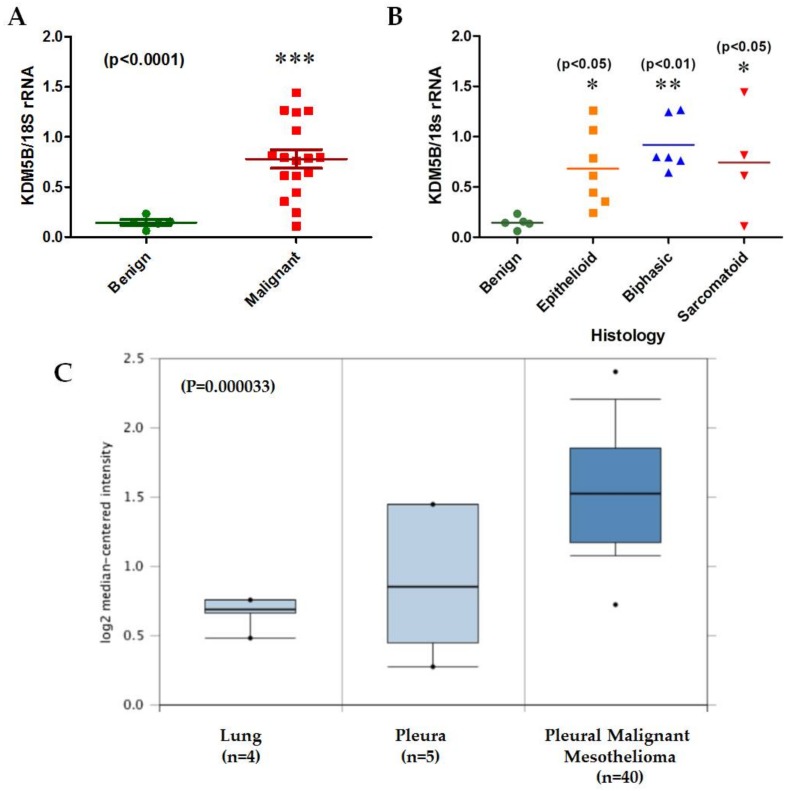
Preliminary data from our group indicates that expression of KDM5B is significantly upregulated in primary MPM (Figure 2A), remaining significant across all histological subtypes (Figure 2B). Similar significant overexpression of KDM5B is also observed in MPM samples in the Gordon et al. [52] mesothelioma dataset (Figure 2C). Across the TCGA dataset, KDM5B appears to have significant alterations in about 14% of MPM cases, including amplification of its genomic region, overexpression, or indeed downregulation of its mRNA (Figure 2D), all of which are found in either the epithelioid or biphasic subtypes (Figure 2E).
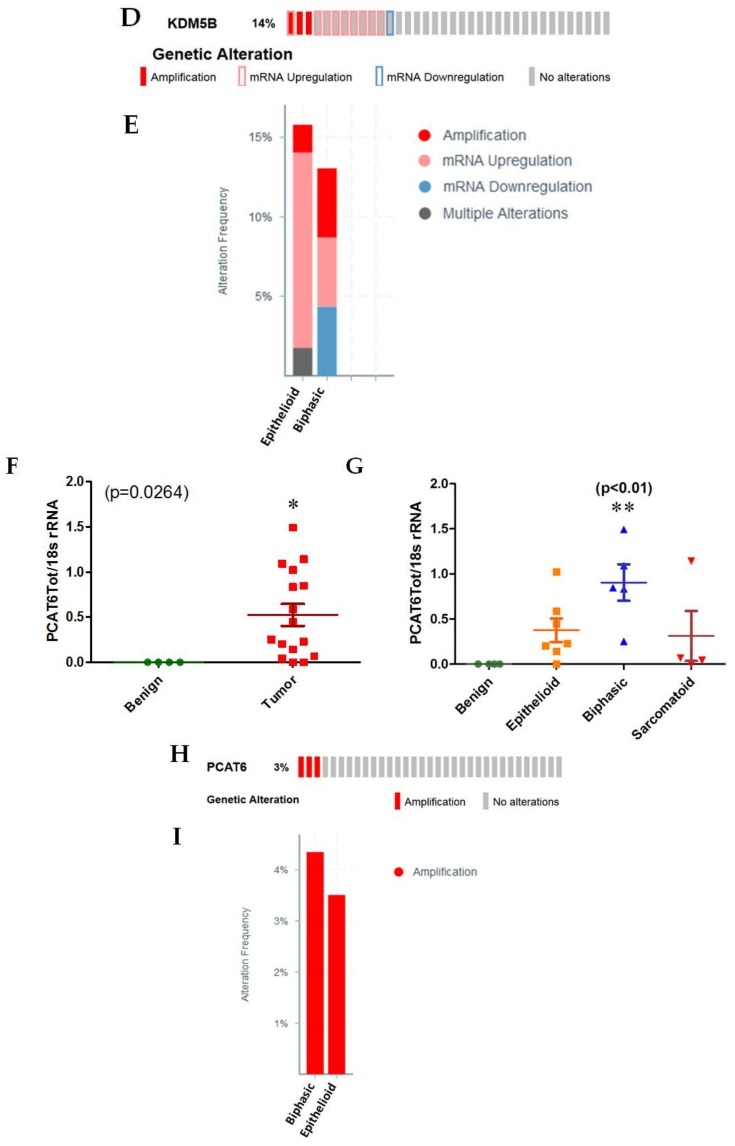
Figure 2.
An examination of KDM5B and PCAT6 expression/alterations in MPM. (A) KDM5B mRNA is significantly elevated in tumours (n = 17) compared to benign pleura (n = 5), (B) the same samples stratified by histological subtype, (C) Oncomine analysis of the Gordon mesothelioma dataset confirming significant overexpression of KDM5B, (D) in silico examination using cBioPortal reveals that 14% of samples had alterations to KDM5B, (E) when stratified by histotype, these alterations were restricted to epithelioid or biphasic subtypes, (F) total PCAT6 lncRNA is significantly elevated in tumours (n = 16(red) compared to benign pleura (n = 4—green), (G) when stratified by histological subtype (Benign = green; Epithelioid = yellow; Biphasic = blue; Sarcomatoid = red), elevated expression of total PCAT6 is significant only in the biphasic subset. Statistical significance was assessed using a Mann–Whitney t-test (* p < 0.05), or by an ANOVA using Dunnett’s Multiple Comparison Test (* p < 0.05; ** p < 0.01; *** p < 0.001), (H) in silico examination using cBioPortal reveals that 3% of samples had amplification of PCAT6, (I) when stratified by histotype, these alterations were restricted to biphasic or epithelioid subtypes.
Moreover, we have also shown that PCAT6 itself is upregulated in MPM (Figure 2F). However, when examined across histological subtypes, the upregulation observed was significant only in the biphasic subset (Figure 2G). In the TCGA dataset, expression of this lncRNA does not appear to be upregulated, although amplification of its genomic location occurs in 3% of MPM specimens (Figure 2H), again, similar to KDM5B, these are spread over the epithelioid and biphasic subsets. (Figure 2I).The functional role of this lncRNA in EMT is as yet unknown, but knockdown of this lncRNA in lung cancer is associated with inhibited cellular proliferation and metastasis [50].
2.1.3. ZEB2-AS1
ZEB2 is a known regulator of EMT [21]. Originally called ZEB2NAT, but now more often described as ZEB2-AS1, this natural anti-sense lncRNA of ZEB2 was shown to regulate ZEB2 during the process of EMT [53]. This lncRNA has been found to be upregulated in both urinary bladder cancer [54] and hepatocellular carcinoma [55], and in bladder cancer cells is partly responsible for activation of ZEB2 during EMT induction by Transforming growth factor beta (TGF-β) [54]. Furthermore, knockdown of this lncRNA in Hepatocellular Carcinoma (HCC) cells results in reduced vimentin and N-caherin expression with restoration of E-cadherin expression [55], further supporting a role for this lncRNA in the regulation of EMT.
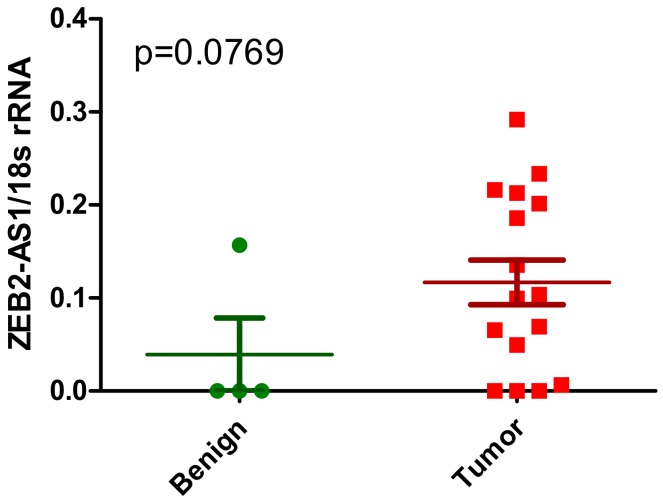
ZEB2 was found to be a significantly altered gene between the sarcomatoid vs. epithelioid clusters (unadjusted p-value: p < 2.03 × 10−26) in the analysis by Bueno et al. [27], but this has not been supported by earlier analysis in the Gordon dataset [52]. There is some suggestion that in the larger dataset by Lopez-Rios that higher expression of ZEB2 is associated with the sarcomatoid subtype (p = 0.065) [56]. Very little is known about the expression of ZEB2-AS1 in MPM. Our preliminary analysis suggests that expression of this lncRNA is potentially dysregulated in MPM (Figure 3), but further studies will be required to validate these observations.
Figure 3.
Altered expression of ZEB2-AS1 in primary MPM. ZEB2-AS1 lncRNA expression was examined by RT-PCR in a series of primary MPM (n = 16—red) vs. benign pleura (n = 4—green). Semi-quantitative densitometric analysis of the results suggests that ZEB2-AS1 lncRNA was elevated in the tumours compared to benign pleura. Statistical significance was assessed using a 1-tailed unpaired Students t-test (p = 0.0769).
2.2. Previously Published lncRNAs with Known Links to MPM
A significant body of research has shown that many short non-coding RNAs, such as microRNAs (miRNAs), have extensive alterations and diverse roles in MPM, and have been discussed by us in depth in a previous review [18]. The evidence for altered expression or roles of lncRNAs in MPM has not as yet been exhaustively analysed in MPM. In the following sections we review the current knowledge of the known lncRNAs associated with MPM, and whether or not these lncRNAs can be linked to EMT processes.
2.2.1. PVT1 and c-Myc
PVT1 is an lncRNA which has been shown to be associated with poor prognosis in many cancers [57]. Its expression has also been linked to EMT in various cancers. For example, in breast cancer, PVT1 is significantly upregulated, and directly interacts with SOX2 to drive EMT [58]. In pancreatic cancer, PVT1 has been found to promote EMT by downregulation of the cyclin-dependent kinase p21 [59]. The other ways PVT1 has been shown to elicit responses include by acting as a competitive endogenous RNA (ceRNA) for various miRNAs [60,61,62,63,64,65,66,67], or by interactions with EZH2 to epigenetically regulate genes associated with EMT [68,69,70,71,72,73,74,75].
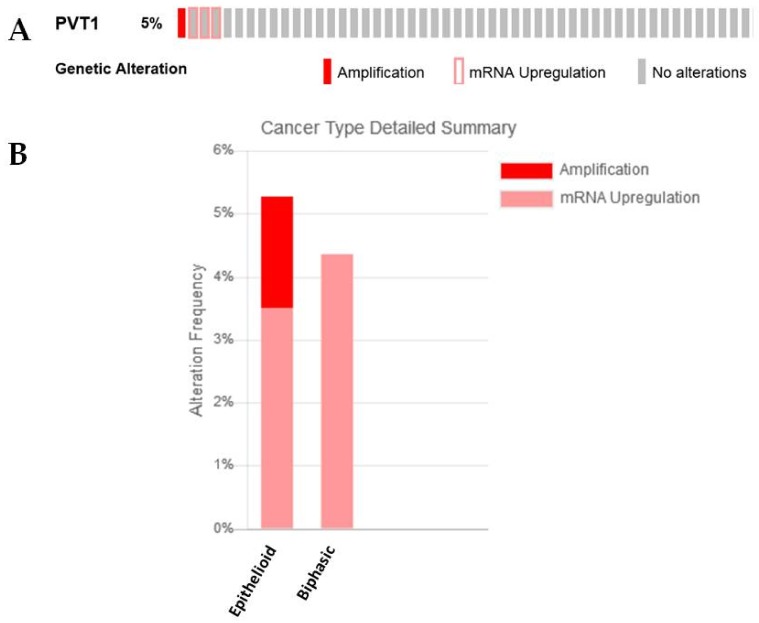
Both PVT1 and c-Myc are located at the same chromosomal location (8q24.21) and an increase in PVT1 expression is required for high MYC protein levels in 8q24-amplified human cancer cells [76]. In this regard, frequent coamplification and cooperation between c-MYC and PVT1 oncogenes have been observed to promote malignant pleural mesothelioma [24]. Next Generation Sequencing (NGS) demonstrated a downregulation of PVT1 in a sarcomatoid subset compared to epithelioid (Table 1) [27].
In silico analysis of the TCGA provisional dataset demonstrated that amplification occurred only in epithelioid samples (Figure 4A,B), which is somewhat in agreement with the observations made by Riquelme et al., where copy number gains were seen in the biphasic (6 of 26, 23%) and epithelioid (5 of 37, 13%) histotypes but not in the sarcomatoid cases [24]. In samples where PVT1 overexpression is observed it is either associated with the epithelioid or biphasic histology (Figure 4B).
Figure 4.
An examination of PVT1 expression/alterations in the TCGA dataset. (A) In silico examination using cBioPortal reveals that 5% of samples had overexpression of PVT1 RNA, (B) when stratified by histotype, only the epithelioid subtype had amplification of PVT1, whereas some patients with epithelioid and biphasic but not sarcomatoid subtypes had overexpression of this lncRNA.
2.2.2. NEAT1
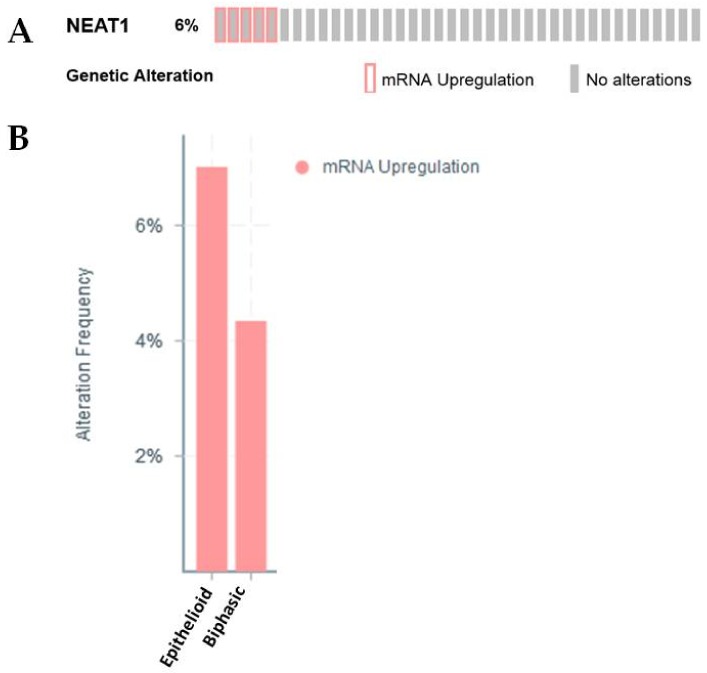
Neat1 was identified by our group as an lncRNA altered in MPM [26]. It is now well-established that this lncRNA promotes EMT [21,77], and one of the means by which it affects EMT is through regulation of EZH2 [78,79]. Most recently, the expression of NEAT1 has been shown to be BAP1 dependent [80]. Given that it is estimated that approximately 65% of mesotheliomas harbour mutations inactivating BAP1 [81], this may have implications with respect to the role of this lncRNA in MPM pathogenesis. Both our data, and that of Bueno et al. (Table 1) [27], showed an overall downregulation of this lncRNA in MPM. Further analysis of the TCGA dataset shows that a proportion of samples have upregulation of this lncRNA (Figure 5A), which when stratified by histology, is found mostly in the Epithelioid subset, with a smaller proportion in the Biphasic subset also showing elevated expression (Figure 5B).
Figure 5.
An examination of Neat1 expression/alterations in the TCGA dataset. (A) In silico examination using cBioPortal reveals that 6% of samples had overexpression of Neat1 lncRNA. (B) When stratified by histotype, the majority of samples with elevated Neat1 are found in the epithelioid subset, followed by a proportion in the biphasic subset.
2.2.3. PAX8-AS1
This lncRNA was also identified [26] as being significantly altered in MPM. The gene associated with this lncRNA, PAX8, has been shown to play important roles in the development of ovarian cancer [82], and may do this through upregulation of markers of EMT [83]; although conflicting results have emerged [84]. Interestingly, PAX8 expression is observed in peritoneal mesotheliomas [85,86], but not in pleural mesotheliomas [86]. In MPM, no significant changes in expression of PAX8 were seen in the Gordon dataset [52], whereas high expression of PAX8 was observed in 4 of 87 MPM samples (5%) in the TCGA dataset (data not shown).
Whilst our previous publication found that PAX8-AS1 was significantly altered in MPM [26], analysis of the TCGA dataset using cBioPortal found no alterations in this lncRNAs expression, suggesting that perhaps this lncRNA may not play a direct role in the regulation of EMT and/or the pathogenesis of MPM.
2.2.4. GAS5
A link between GAS5 and EGFR TKI sensitivity has also been identified. Levels of GAS5 were downregulated in the EGFR TKI resistant lung adenocarcinoma cell line A549 compared to sensitive cell lines. Moreover, restoration of GAS5 expression could greatly sensitise these cells to gefitinib treatment in xenograft mouse models [87]. In a separate study relating to prostate cancer, increased expression of GAS5 was associated with decreased Akt signalling [88]. Therefore, it could be suggested that lncRNA mediated regulation of Akt signalling seems to highly important in determining the sensitivity of NSCLC cells to EGFR TKI, such as gefitinib.
In MPM, Felley-Bosco and colleagues have shown that this lncRNA is overexpressed in malignant tumours compared to non-tumoural tissue, (* p < 0.0001 expression, Mann–Whitney test) [25]. While loss of this lncRNA is associated with a shortening of the cell-cycle in MPM cell lines, the role of this lncRNA in regulating EMT in MPM is unknown, however, studies in other cancers, such as osteosarcoma, have shown that expression of this lncRNA decreased in tumours compared to adjacent normal tissue. Furthermore, overexpression of GAS5 suppressed cellular proliferation, migration, and EMT in osteosarcoma cell lines [89].
2.3. Previously Published lncRNAs with Known Links to EMT
It is now well established that various lncRNAs play essential roles in the regulation of EMT, a subject we recently reviewed in depth [21]. Despite this, several of these key lncRNAs have not been studied in depth in MPM. In the following sections we discuss the known roles of several of these key lncRNAs, and using in silico analyses to describe the current evidence for their altered expression in mesothelioma histological subtypes.
2.3.1. HOTAIR
HOTAIR is a lncRNA transcribed from the HOXC gene cluster that promotes epigenetic silencing of target genes, including the HOXD gene cluster, through the recruitment of the PRC2 and LSD1/CoREST/REST chromatin remodelling complexes [90,91]. It is well established that HOTAIR is overexpressed in a wide variety of solid malignancies, and moreover, that this overexpression is associated with metastasis and tumour recurrence [21]. Critically, HOTAIR has been linked extensively to the promotion of EMT in solid tumours [21]. In this regard, HOTAIR has been found to regulate EMT through recruitment of PRC2 to the CDH1 promoter [92]. HOTAIR also forms a tripartite complex with Snail and EZH2, facilitating the recruitment of EZH2 to Snail binding sites at the promoters of the epithelial genes E-cadherin, Hepatocyte nuclear factor (HNF), HNF1α, and HNF4α, resulting in their epigenetic silencing [93]. HOTAIR also positively regulates the expression of JMJD3 and Snail to regulate EMT [94]. In addition, this lncRNA plays roles in the silencing of many anti-EMT regulators, such as the miRNAs miR-7, miR-34a, and miR-568 [95,96,97].
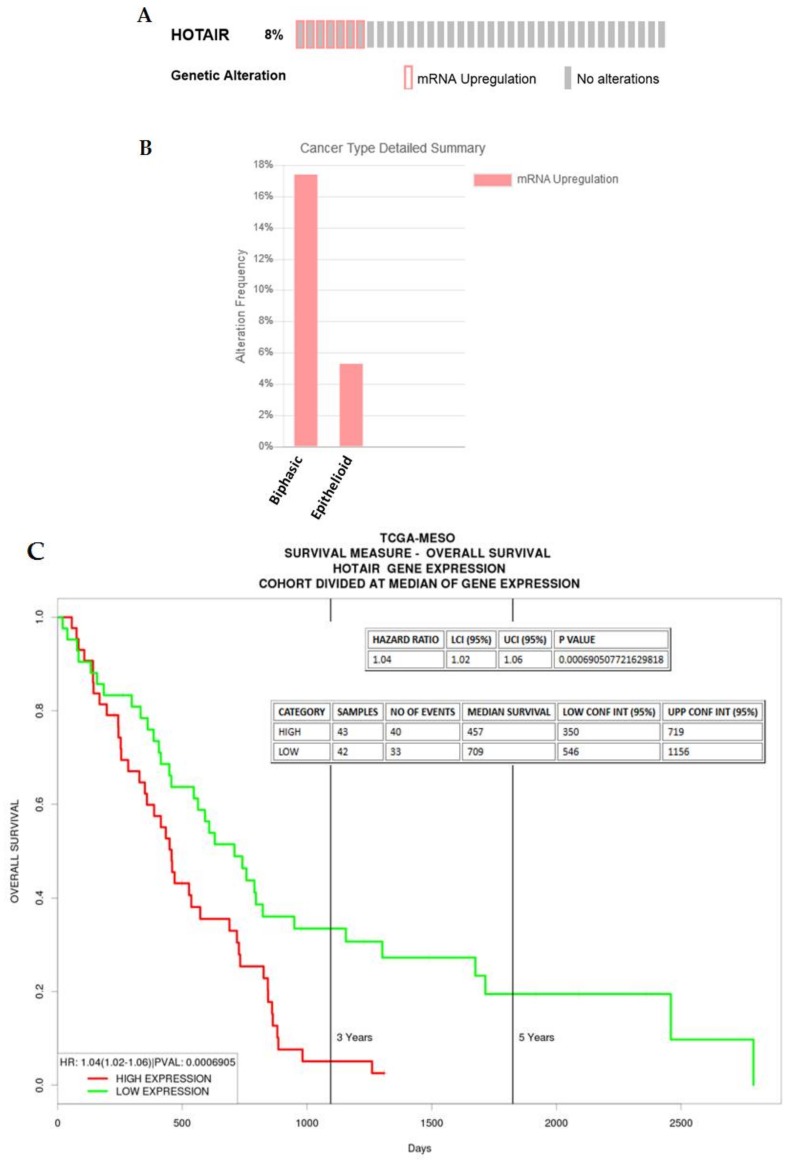
In MPM, overexpression of HOTAIR was found in the sarcomatoid subset of the Bueno NGS dataset (Table 1) [27], suggesting that HOTAIR is a lncRNA associated with the progression of MPM from the epithelioid to the sarcomatoid subtype. In silico analysis of an existing TCGA dataset shows that for those samples showing overexpression of this lncRNA the majority were biphasic (Figure 6A,B), and further analysis reveals that higher expression of HOTAIR in mesothelioma is associated with an poorer overall survival (Figure 6C).
Figure 6.
An examination of HOTAIR expression/alterations in the TCGA dataset. (A) In silico examination using cBioPortal reveals that 8% of samples had overexpression of HOTAIR RNA, (B) when stratified, the majority of these samples were associated with the Biphasic subtype, (C) when examined using ProGeneV2 (http://watson.compbio.iupui.edu/chirayu/proggene/database/?url=proggene), higher expression of HOTAIR was associated with a worse overall survival.
2.3.2. MALAT1
MALAT-1 (metastasis-associated lung adenocarcinoma transcript 1 also called NEAT2 or nuclear enriched abundant transcript 2) was first identified in NSCLC as a predictive marker associated with metastatic disease and shorter survival in early stage lung adenocarcinoma [98]. Since its initial discovery, MALAT-1 has been shown to be overexpressed and linked to the promotion of EMT in many cancers [21,99,100]. However, there are conflicting results which suggest that this lncRNA can either promote or inhibit EMT [77,101,102]. This may be in part because MALAT-1 can regulate EMT and other processes in various ways. For example, MALAT-1 can act as a competing endogenous RNA (ceRNA) for various miRNAs including miR-1, miR-200c, miRNA-204, and miR-205 resulting in the subsequent promotion of EMT [103,104,105,106]. Another mechanism by which MALAT-1 induces EMT is via the recruitment of the PRC2 components Suz12 and EZH2 to regulate E-Cadherin [105,107] and β-catenin [108,109].
MALAT-1 has been shown to activate EMT through either MAPK/ERK or PI3K/Akt signalling. MALAT-1 knockdown significantly reduced MAPK/ERK signalling in gallbladder cancer cells [110], and in glioma, MALAT-1 acts as a tumour suppressor by attenuating ERK/MAPK mediated signalling [111]. In osteosarcoma cells, downregulation of MALAT-1 inhibits PI3K/Akt signalling [112], whereas in breast and ovarian cancer cells, knockdown of MALAT-1 knockdown results in increased PI3K/Akt signalling and induction of EMT [102,113]. In this regard, in amodel of silica induced pulmonary fibrosis, MALAT-1 acts as a ceRNA for miR-503, one of whose targets is PI3K p85. By “sponging” this miRNA, MALAT-1 allows stimulation of EMT through a MALAT-1-miR-503-PI3K/Akt/mTOR/Snail pathway [114].
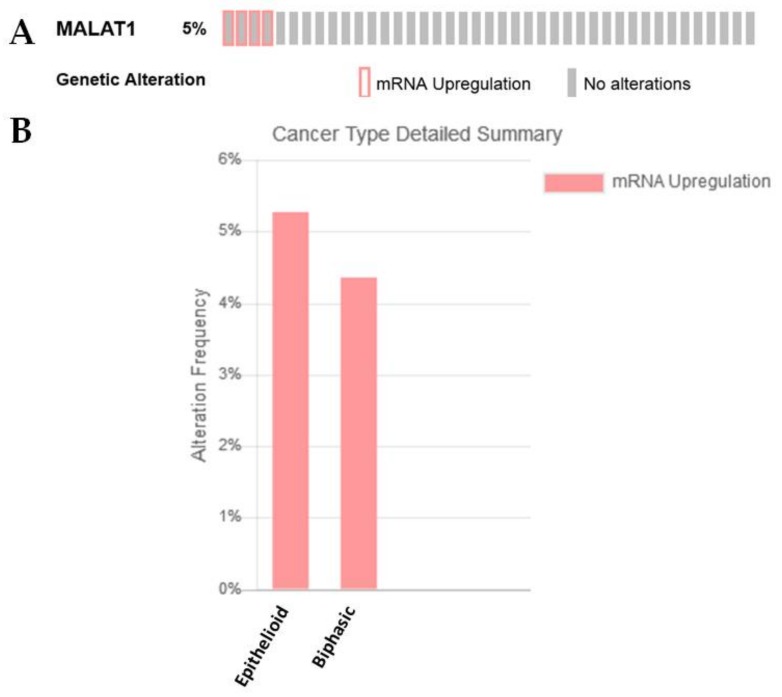
MALAT-1 is induced by TGF-β and plays a critical role during the promotion of EMT by TGF-β in bladder cancer cells [107]. TGF-β often elicits its effect through the Wnt signalling pathway, and significant evidence now suggests that lncRNAs play a major role in this process [115]. For example, MALAT-1 induces EMT in various cancers via the Wnt/β-catenin signalling pathway [116,117,118], while loss of WIF1 enhances the migratory potential of glioblastoma cells through WNT5A activation mediated by MALAT1 [119]. Intriguingly, MALAT1 expression was found to be overexpressed in the sarcomatoid subset of the Bueno NGS dataset (Table 1) [27]. In silico analysis of an existing TCGA dataset also shows that for MPM samples with overexpression of this lncRNA, the majority were epithelioid with some in the biphasic category (Figure 7A,B).
Figure 7.
An examination of MALAT1 expression/alterations in the TCGA dataset. (A) In silico examination using cBioPortal reveals that 5% of samples had overexpression of MALAT1 RNA, (B) when stratified by histotype, the majority of these samples were associated with the epithelioid subtype.
In renal cell carcinoma, a link between MALAT-1 and c-MYC, a downstream effector of Wnt/β-Catenin signalling, was found to be an element in the regulation of β-catenin and transcription factor c-Myc [116]; other lncRNAs have now been shown to play additional roles in regulating EMT via either c-Myc or n-Myc.
2.3.3. MYCNOS and N-MYC
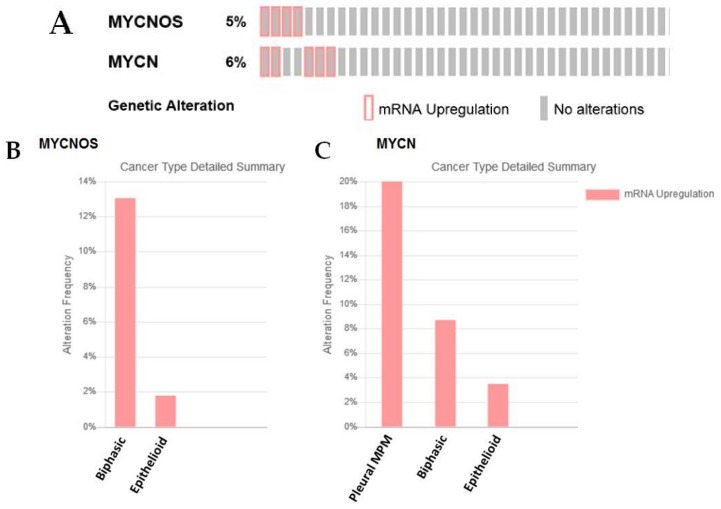
N-Myc (MYCN) belongs to the MYC family and was originally identified as being amplified in 20–30% of neuroblastoma tumours, but it is now well established that dysregulation of this transcription factor is common in many non-neuronal tumours [120]. N-Myc has also been shown to play roles in driving EMT in cancer [121]. In this regard, an lncRNA called MYCNOS has been shown to regulate the expression of N-Myc [122,123,124].
While a role for this lncRNA has not yet been identified in MPM, MYCNOS is upregulated in a proportion of MPM (5%—Figure 8A), and is mostly upregulated in the biphasic subset—Figure 8B. N-Myc also shows overexpression in a subset of MPM samples, but the majority of the samples do not fall into a defined histotype (Figure 8C). In these samples only two patients have co-overexpression of both MYCNOS and MYCN.
Figure 8.
An examination of MYCNOS and N-Myc expression/alterations in the TCGA dataset. (A) In silico examination using cBioPortal reveals that 5% of samples had overexpression of MYCNOS RNA, while 6% had overexpression of N-Myc, (B) when stratified by histotype, the majority of samples with elevated MYCNOS were found in the biphasic subset, (C) N-Myc stratification does not fall into any defined histotype.
2.3.4. H19
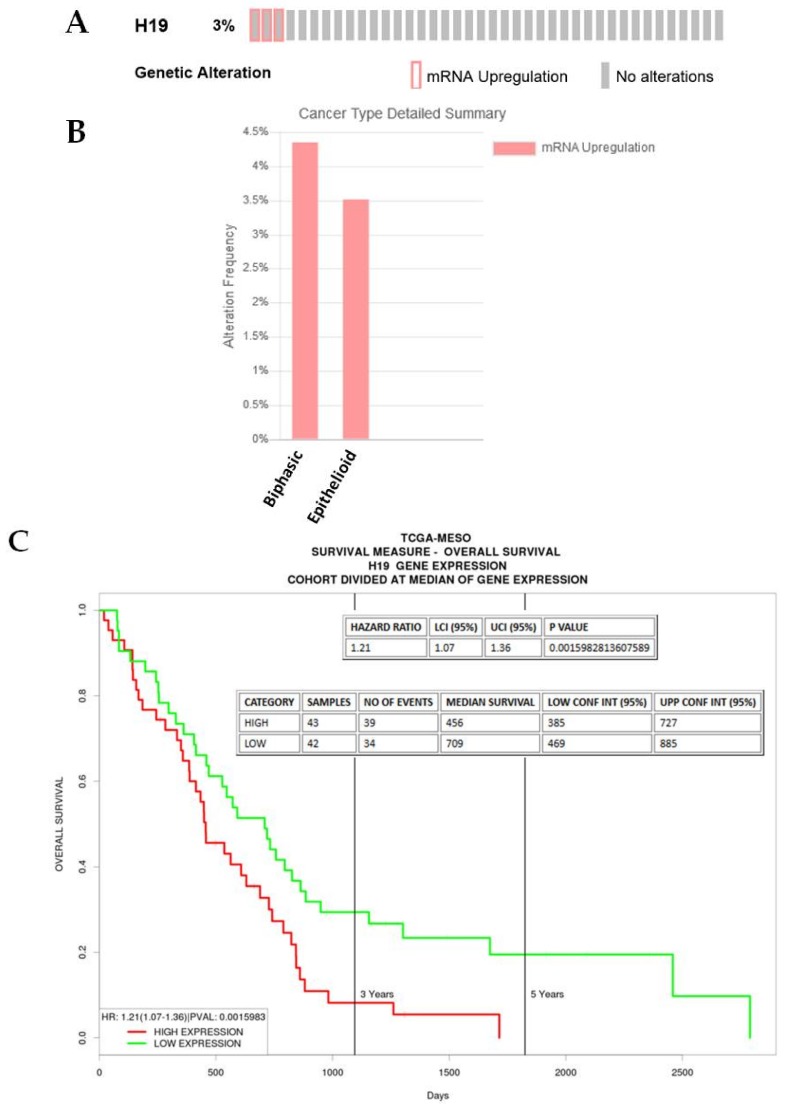
H19 is an imprinted lncRNA, and has long been identified as an aberrantly expressed non-coding RNA in a great number of cancers, and has been shown to play multi-faceted roles during the tumourigenic process [125]; and is considered to be a critical element in EMT [126]. Indeed, overexpression of this lncRNA is associated with the activation of EMT in numerous cancers, including pancreatic cancer, CRC, nasopharyngeal carcinoma, bladder cancer, gallbladder cancer, and oesophageal cancer4 [21], where it has been shown to silence E-cadherin through recruitment of EZH2 to its promoter, or functions as a ceRNA for several pro-EMT miRNAs [21].
Upregulation of this lncRNA is found in the differential analysis between the epithelioid versus sarcomatoid clusters in the analysis by Bueno et al. (Table 1) [27]. In silico analysis of the TCGA dataset suggests that a small number of samples have higher expression of H19 (Figure 9A), which are distributed between the epithelioid (n = 1) and biphasic (n = 2) (Figure 9B). However, higher median expression of H19 is associated with a worse overall survival in this dataset (Figure 9C).
Figure 9.
An examination of H19 in MPM. (A) H19 lncRNA is altered/overexpressed in a small proportion of MPM patients, as assessed using cBioPortal, (B) when separated by histology these samples fall into either the biphasic or epithelioid subsets, (C) when overall survival is assessed in this dataset, high median expression is associated with a significantly worse OS (p = 0.0016).
2.3.5. HULC
The lncRNA Highly Upregulated in Liver Cancer (HULC) was originally identified as one of the most upregulated genes in hepatocellular carcinoma (HCC) [127]; this lncRNA has now been shown to be aberrantly upregulated in several cancers [128]. Some evidence has also been reported suggesting that HULC can also act to inhibit c-Myc expression and PI3K/Akt signalling [129,130], and HULC has also been shown to cooperate with MALAT1 to promote liver cancer stem cell growth/aggressiveness [131]. Moreover, HULC has been shown to affect transcription through interaction with EZH2 [132].
A role for HULC in the regulation of EMT has been observed in HCC where it functions as a ceRNA for miRNAs (miR-122, miR-200a-3p, miR-372, and miR-488) [29,30,133,134] to mediate EMT via upregulation of Snail [135], ZEB1 [29], or ADAM9 [30], and this lncRNA has also been reported to induce EMT in gastric cancer [28].
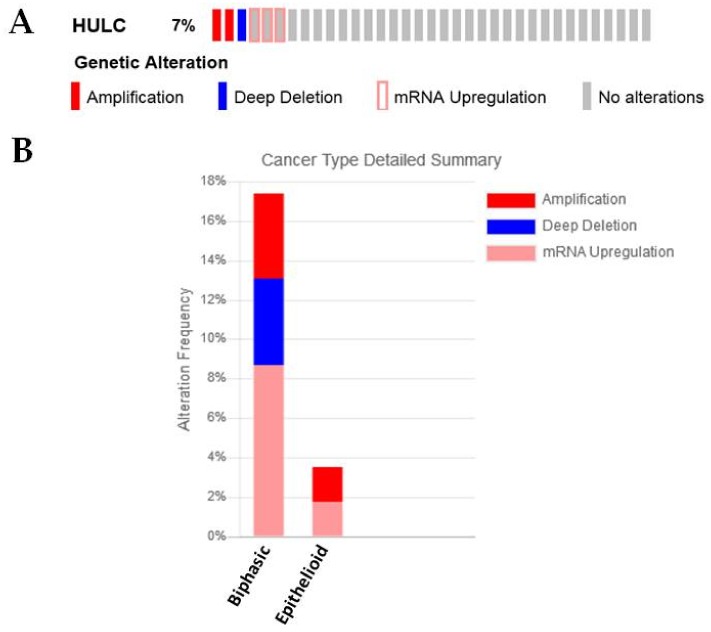
A functional role for this lncRNA in MPM has not yet been identified. However, it was observed to be significantly downregulated in the sarcomatoid compared to the epithelioid subgroup (Table 1) [27]. cBioPortal analysis of the current TCGA mesothelioma dataset finds that 7% of samples have either amplifications or deletions in HULC, or overexpress this lncRNA (Figure 10A). When separated according to histology, the majority of alterations observed were found to be of the biphasic subtype (Figure 10B).
Figure 10.
An examination of HULC in MPM. (A) HULC is either amplified/deleted or overexpressed in a small proportion (7%) of MPM patients, as assessed using cBioPortal, (B) when separated by histology the majority of these samples fall into the biphasic subgroup.
2.3.6. CASC2
In a study of NSCLC, expression of this lncRNA in the adenocarcinoma subtype was associated with inhibition of EMT through regulation of SOX4 [37]. A similar role for this lncRNA in regulating EMT in HCC has been identified, where this lncRNA has been shown to act as a ceRNA for miR-367 via a CASC2/miR-367/FBXW7 axis [136]. Furthermore, CASC2 has been shown to inhibit HCC by acting as a ceRNA for miR-362-5p, which resulted in the inhibition of the Nuclear Factor Kappa Beta (NF-κB) pathway [137].
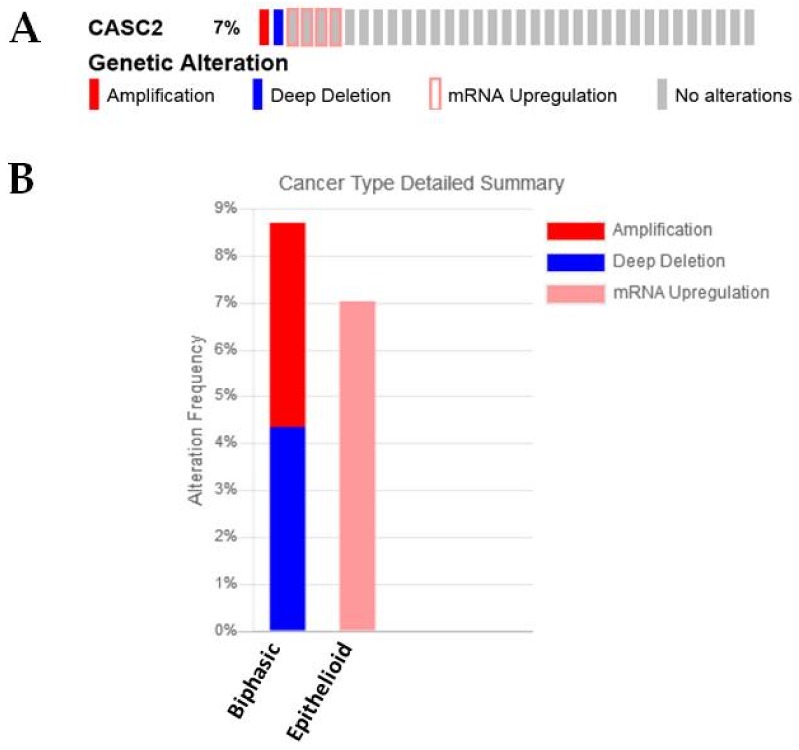
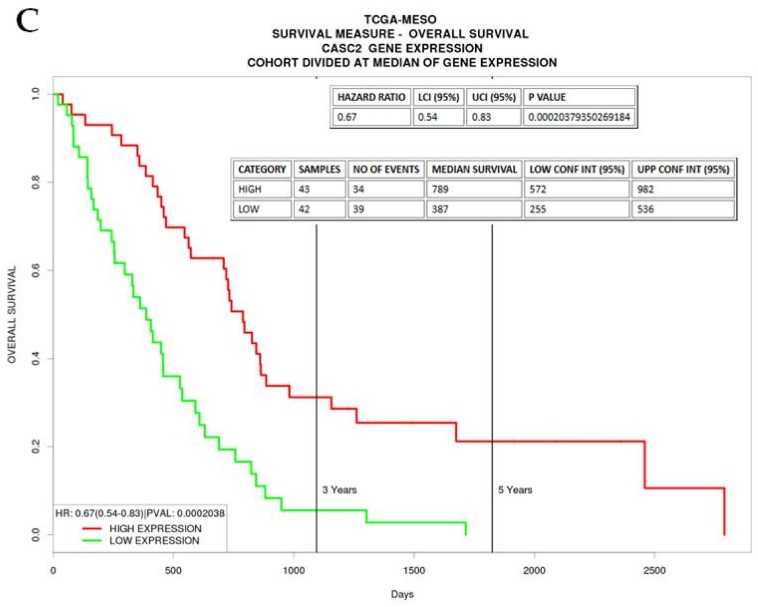
A functional role for this lncRNA in MPM has not yet been defined, but decreased expression of this lncRNA is significantly associated with the sarcomatoid subtype in the Bueno NGS samples [27] (Table 1). Moreover, analysis of the TCGA dataset in cBioPortal reveals that those samples showing high expression of this lncRNA are associated with more with epithelial and biphasic subtypes, with the majority of the overexpression being observed in the epithelioid subset, while amplifications/deletions of this lncRNA were observed in biphasic samples (Figure 11A,B). When expression of this lncRNA was examined for Overall Survival (OS) benefit using ProGeneV2, high median expression was associated with better overall survival (Figure 11C).
Figure 11.
An examination of CASC alterations and expression in MPM. (A) CASC2 mRNA is altered/overexpressed in a small proportion of MPM patients as assessed using cBioPortal, (B) when separated by histology these samples fall into either the biphasic or epithelioid subsets, (C) when overall survival is assessed in this dataset, high median expression is associated with a significantly better OS (p = 0.000203).
As CASC2 is downregulated in human HCC samples, it may therefore be of interest to examine the levels of this lncRNA in MPM to see if loss of CASC is associated with a more aggressive histological phenotype as observed in Table 1.
2.3.7. ZFAS1
ZFAS1 is a lncRNA transcribed antisense to the ZNFX1 protein-coding gene, first identified as an lncRNA involved in mammary development and subsequently found to have altered expression in breast cancer [138]. Since this initial finding, ZFAS1 has been shown to be pro-tumourigenic and promote EMT in a number of other cancers, including colon cancer, gastric carcinoma, and glioma [33,34,35,36,139,140,141,142,143,144,145,146,147,148].
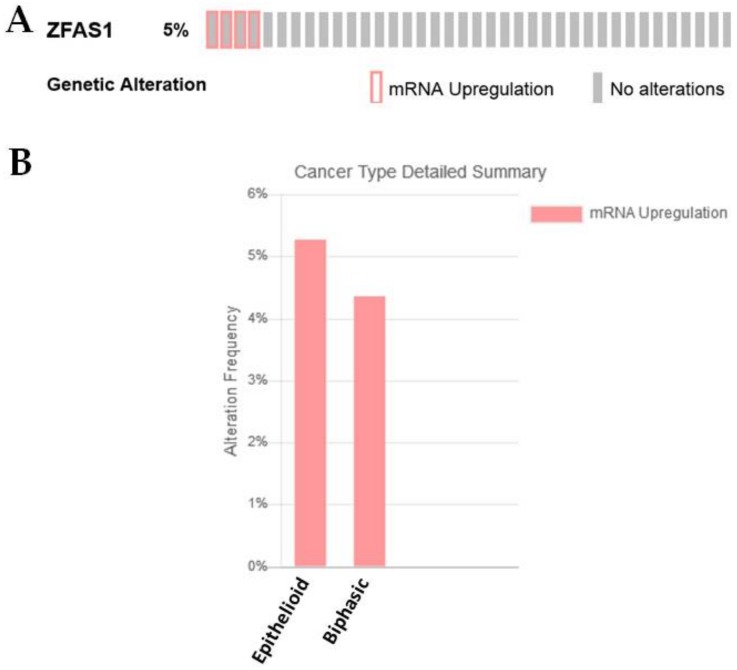
The role of this lncRNA has not yet been identified in MPM, but this lncRNA was found to be significantly altered between epithelioid versus sarcomatoid samples (Table 1) [27]. In the TCGA dataset, ZFAS1 shows overexpression in 5% of the samples; this was associated in samples with epithelioid or biphasic histologies (Figure 12A,B).
Figure 12.
ZFAS1 is altered in a subset of MPM. (A) ZFAS1 is overexpressed in a small proportion (5%) of MPM patients as assessed using cBioPortal, (B) when separated by histology the majority of these samples fall into epithelioid or biphasic subgroups.
3. Materials and Methods
3.1. Primary Tumor Samples
Surgical specimens were obtained as discarded tumour samples from patients who had undergone an extended pleuropneumonectomy at Glenfield Hospital, Leicester, UK. Benign specimens were acquired from patients never diagnosed with MPM. Informed consent was obtained from each patient, and the study was conducted after formal approval from the relevant Hospital Ethics Committee (Leicestershire Research Ethics Committee (REC) references 6742 and 6948). Samples consisted of 5 benign lesions and 17 MPM samples (epithelioid: n = 7; sarcomatoid: n = 4; biphasic: n = 6), details of which are provided in Table 2.
Table 2.
Details of pleura/mesothelioma samples used in this study.
| Sample | Pathology (Benign, Epithelial, Biphasic, Sarcomatoid) | Age | Gender |
|---|---|---|---|
| JE29 | Benign—pleural plaque | 55 | Male |
| JE30 | Benign—pleural plaque | 55 | Male |
| JE32 | Benign—pneumothorax | 30 | Male |
| JE41 | Benign—empyema | 68 | Male |
| JE48 | Benign—pleural plaque | 55 | Male |
| JE31 | Epithelioid | 62 | Male |
| JE139 | Epithelioid | 73 | Male |
| JE149 | Epithelioid | 66 | Male |
| JE155 | Epithelioid | 56 | Female |
| JE157 | Epithelioid | 52 | Male |
| JE162 | Epithelioid | 56 | Male |
| JE173 | Epithelioid | 54 | Male |
| JE86 | Biphasic | 54 | Male |
| JE89 | Biphasic | 54 | Female |
| JE136 | Biphasic | 41 | Male |
| JE150 | Biphasic | 58 | Male |
| JE151 | Biphasic | N/A | Male |
| JE160 | Biphasic | 60 | Female |
| JE106 | Sarcomatoid | 74 | Male |
| JE125 | Sarcomatoid | 64 | Male |
| JE133 | Sarcomatoid | 59 | Male |
| JE145 | Sarcomatoid (desmoplastic) | 64 | Male |
N/A—not available.
3.2. Ethics Statement
Investigations were conducted in accordance with the relevant ethical standards, the Declaration of Helsinki, national, and international guidelines, and were approved by the relevant institutional review board (041018/8804, 13 October 2004, St James’s Hospital/The Adelaide and Meath incorporating the National Childrens Hospital (SJH/AMNCH) REC).
Ethics Approval and Consent to Participate
All subjects gave their informed consent for inclusion before they participated in the study. The study was conducted in accordance with the Declaration of Helsinki.
Fresh Frozen Samples: The study was conducted after formal approval from the relevant Hospital Ethics Committee (Leicestershire REC references 6742 and 6948).
3.3. RNA Isolation and RT-PCR Amplification
Total RNA was extracted from fresh frozen patient samples using TRI reagent® (Cincinnati, OH, USA) according to manufacturer’s instructions. Prior to first strand cDNA synthesis, 200 ng of total RNA was pre-treated by digestion with amplification grade DNase (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer’s instructions. cDNA was then generated using an all-in-one cDNA Synthesis Supermix (Bimake, Houston, TX, USA) according to the manufacturer’s instructions. Patient samples were examined for the expression of various lncRNAs and 18S rRNA at the end point of PCR, using primers and annealing temperatures as outlined in Table 3. Each analysis was carried out once.
Table 3.
Primers and associated annealing temperatures.
| Gene/lncRNA | Primer Sequence | Temp | Source |
|---|---|---|---|
| EGFR-AS1 | F: 5′-CTTTGCGATCTGCACACACC-3′ | 62 | This study |
| R: 5′-GAAGCCTACGTGATGGCCAG-3′ | |||
| PCAT6 | F: 5′-CCCTAGATACACCCGCCTGGT-3′ | 64 | This study |
| R: 5′-ACATTCCAGGGCACCGAGAG-3′ | |||
| ZEB2-AS1 | F: 5′-GAGAGAGACGAGAGACCCTGAA-3′ | 60 | This Study |
| R: 5′-AAATTCATCATGCACACACCC-3′ | |||
| KDM5B (JARID1B) | F: 5′-GCTACCCCCTCCAGCTACTCAGA-3′ | 62 | This study |
| R: 5′-TCCTCCTCGACTTCCTCCTCATC-3′ | |||
| 18S rRNA | F: 5′-GATGGGCGGCGGAAAATAG-3′ | 60 | [149] |
| R: 5′-GCGTGGATTCTGCATAATGGT-3′ |
PCR cycling conditions were 1 min at 95 °C, 1 min at the appropriate annealing temperature as per Table 2, 1 min at 72 °C for 35 cycles, with a final extension of 72 °C for 10 min. RT-PCR products for each experimental gene and appropriate housekeeping genes (18S rRNA) were run on 2% agarose gels. Following image capture, product quantification was performed using TINA 2.09c (Raytest, Isotopenmeßgeräte GmbH, Straubenhardt, Germany) densitometry software. The mRNA expression was normalised to loading controls, and was expressed as a ratio of target mRNA expression: loading control expression.
3.4. Statistical Analysis
All data are expressed as mean ± SEM unless stated otherwise. Statistical analysis was performed with Prism 5.01 (GraphPad, La Jolla, CA, USA) using either t-tests or one-way analysis of variance (ANOVA) where groups in the experiment were three or more. Following ANOVA, post-test analyses utilised the Dunnett’s Multiple comparison test.
3.5. In Silico Analysis
In silico analysis was conducted on three additional mesothelioma datasets as follows:
(a) The dataset previously published by Gordon et al. [52], which was interrogated using Oncomine, (b) and an existing TCGA data set (TCGA Mesothelioma; raw data at the NCI; the dataset consists of n = 87 samples: epithelioid (57), biphasic (23), sarcomatoid (2), other mesothelioma (5).
Data-mining of the available mesothelioma datasets was conducted using Oncomine [150,151] cBioportal [152,153,154], or PROGgeneV2 [155,156], using their respective default settings.
4. Conclusions
Despite intensive efforts, the range of treatment options available to clinicians for the treatment of patients with MPM remains low. The current mainstay of treatment is a combination of cisplatin and pemetrexed (or alternatively raltitrexed), and only approximately 40% of patients will respond to this regimen. At present, no second-line strategy has been approved to date, except rechallenging the patients with long-lasting tumour control after first-line treatment with pemetrexed-based chemotherapy [157].
Across the histological subtypes of MPM, patients who have an epithelioid histology generally have the best OS. Because MPM has a partial fibroblastic phenotype in the context of EMT, it has been postulated that this may, in part, explain the aggressiveness of this cancer by conferring both its high invasiveness and chemoresistance [13]; in particular, with regard to the epithelioid rather than sarcomatoid histotype of MPM [13]. In this regard, the epithelioid and sarcomatoid histologic variants of MPM can be considered as E- and M-parts of the EMT axis, with the biphasic histotype considered an intermediate [14].
In this report, we have shown that many lncRNAs associated with EMT have predominantly altered expression, associated for the most part with the sarcomatoid histologies. Therefore, a greater understanding of the molecular mechanisms governing EMT remains imperative for the development of novel therapies that can slow or prevent metastasis, the current great unmet need of cancer therapy.
In a previous review, we discussed the role of many lncRNAs as elements associated with resistance mechanisms to cisplatin [21], and many of the lncRNAs discussed in this article such as HOTAIR or MALAT1 have well defined roles in cisplatin resistance [21].
If these lncRNAs are both associated with driving MPM from the epithelioid subtype to the more aggressive forms with poorer OS (biphasic and sarcomatoid) with resistance to cisplatin, then potentially targeting these may have therapeutic applicability. Methodologies to restore ncRNAs in MPM, such as the recently completed Phase I MesomiR 1 clinical trial [42], suggest that this technology could also be utilised or adapted to specifically target lncRNAs in MPM.
In conclusion, a large body of evidence suggests that lncRNAs associated with EMT are dysregulated in MPM, and their alteration may be associated with the more aggressive histological subtypes. More work remains to delineate how we may be able to take advantage of this clinically.
Acknowledgments
The authors wish to acknowledge all those whose research has been used in this manuscript and apologise to those whose work we may have missed.
Abbreviations
| ceRNA | competitive endogenous RNA |
| EMT | Epithelial Mesenchymal Transition |
| lncRNA | long non-coding RNA |
| NSCLC | Non-Small Cell Lung Cancer |
| miRNA | microRNA |
| MPM | Malignant Pleural Mesothelioma |
| NGS | Next Generation Sequencing |
| OS | Overall Survival |
| TCGA | The Cancer Genome Atlas |
Author Contributions
A.S.S., R.H. and S.G.G. designed the experiments; A.S.S., R.H., and~S.G.G. performed the experiments; S.G.G. wrote the paper. All authors have read and approved the final manuscript for~publication.
Funding
The research in this publication was in part supported by funding for consumables from the Masters in Translational Oncology program (TCD) for Anand S. Singh and Richard Heery.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Wagner J.C., Sleggs C.A., Marchand P. Diffuse pleural mesothelioma and asbestos exposure in the North Western Cape Province. Br. J. Ind. Med. 1960;17:260–271. doi: 10.1136/oem.17.4.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Remon J., Reguart N., Corral J., Lianes P. Malignant pleural mesothelioma: New hope in the horizon with novel therapeutic strategies. Cancer Treat. Rev. 2015;41:27–34. doi: 10.1016/j.ctrv.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Odgerel C.O., Takahashi K., Sorahan T., Driscoll T., Fitzmaurice C., Yoko O.M., Sawanyawisuth K., Furuya S., Tanaka F., Horie S., et al. Estimation of the global burden of mesothelioma deaths from incomplete national mortality data. Occup. Environ. Med. 2017;74:851–858. doi: 10.1136/oemed-2017-104298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alley E.W., Lopez J., Santoro A., Morosky A., Saraf S., Piperdi B., van Brummelen E. Clinical safety and activity of pembrolizumab in patients with malignant pleural mesothelioma (KEYNOTE-028): Preliminary results from a non-randomised, open-label, phase 1b trial. Lancet Oncol. 2017;18:623–630. doi: 10.1016/S1470-2045(17)30169-9. [DOI] [PubMed] [Google Scholar]
- 5.Quispel-Janssen J., Zago G., Schouten R., Buikhuisen W., Monkhorst K., Thunissen E., Baas P. OA13.01 A Phase II Study of Nivolumab in Malignant Pleural Mesothelioma (NivoMes): With Translational Research (TR) Biopies. J. Thorac. Oncol. 2016;12:S292–S293. doi: 10.1016/j.jtho.2016.11.300. [DOI] [Google Scholar]
- 6.Baas P., Fennell D., Kerr K.M., Van Schil P.E., Haas R.L., Peters S. Malignant pleural mesothelioma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2015;26(Suppl. 5):v31–v39. doi: 10.1093/annonc/mdv199. [DOI] [PubMed] [Google Scholar]
- 7.Kindler H.L., Ismaila N., Armato S.G., 3rd, Bueno R., Hesdorffer M., Jahan T., Jones C.M., Miettinen M., Pass H., Rimner A., et al. Treatment of Malignant Pleural Mesothelioma: American Society of Clinical Oncology Clinical Practice Guideline. J. Clin. Oncol. 2018 doi: 10.1200/JCO.2017.76.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogelzang N.J., Rusthoven J.J., Symanowski J., Denham C., Kaukel E., Ruffie P., Gatzemeier U., Boyer M., Emri S., Manegold C., et al. Phase III study of pemetrexed in combination with cisplatin versus cisplatin alone in patients with malignant pleural mesothelioma. J. Clin. Oncol. 2003;21:2636–2644. doi: 10.1200/JCO.2003.11.136. [DOI] [PubMed] [Google Scholar]
- 9.Brabletz T., Kalluri R., Nieto M.A., Weinberg R.A. EMT in cancer. Nat. Rev. Cancer. 2018;18:128–134. doi: 10.1038/nrc.2017.118. [DOI] [PubMed] [Google Scholar]
- 10.Husain A.N., Colby T.V., Ordonez N.G., Allen T.C., Attanoos R.L., Beasley M.B., Butnor K.J., Chirieac L.R., Churg A.M., Dacic S., et al. Guidelines for Pathologic Diagnosis of Malignant Mesothelioma 2017 Update of the Consensus Statement From the International Mesothelioma Interest Group. Arch. Pathol. Lab. Med. 2018;142:89–108. doi: 10.5858/arpa.2017-0124-RA. [DOI] [PubMed] [Google Scholar]
- 11.Sun H.H., Vaynblat A., Pass H.I. Diagnosis and prognosis-review of biomarkers for mesothelioma. Ann. Transl. Med. 2017;5:244. doi: 10.21037/atm.2017.06.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panou V., Vyberg M., Weinreich U.M., Meristoudis C., Falkmer U.G., Roe O.D. The established and future biomarkers of malignant pleural mesothelioma. Cancer Treat. Rev. 2015;41:486–495. doi: 10.1016/j.ctrv.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Schramm A., Opitz I., Thies S., Seifert B., Moch H., Weder W., Soltermann A. Prognostic significance of epithelial-mesenchymal transition in malignant pleural mesothelioma. Eur. J. Cardiothorac. Surg. 2010;37:566–572. doi: 10.1016/j.ejcts.2009.08.027. [DOI] [PubMed] [Google Scholar]
- 14.Thies S., Friess M., Frischknecht L., Korol D., Felley-Bosco E., Stahel R., Vrugt B., Weder W., Opitz I., Soltermann A. Expression of the Stem Cell Factor Nestin in Malignant Pleural Mesothelioma Is Associated with Poor Prognosis. PLoS ONE. 2015;10:e0139312. doi: 10.1371/journal.pone.0139312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Reynies A., Jaurand M.C., Renier A., Couchy G., Hysi I., Elarouci N., Galateau-Salle F., Copin M.C., Hofman P., Cazes A., et al. Molecular classification of malignant pleural mesothelioma: Identification of a poor prognosis subgroup linked to the epithelial-to-mesenchymal transition. Clin. Cancer Res. 2014;20:1323–1334. doi: 10.1158/1078-0432.CCR-13-2429. [DOI] [PubMed] [Google Scholar]
- 16.Legras A., Pecuchet N., Imbeaud S., Pallier K., Didelot A., Roussel H., Gibault L., Fabre E., Le Pimpec-Barthes F., Laurent-Puig P., et al. Epithelial-to-Mesenchymal Transition and MicroRNAs in Lung Cancer. Cancers. 2017;9:101. doi: 10.3390/cancers9080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jafri M.A., Al-Qahtani M.H., Shay J.W. Role of miRNAs in human cancer metastasis: Implications for therapeutic intervention. Semin. Cancer Biol. 2017;44:117–131. doi: 10.1016/j.semcancer.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 18.Quinn L., Finn S.P., Cuffe S., Gray S.G. Non-coding RNA repertoires in malignant pleural mesothelioma. Lung Cancer. 2015;90:417–426. doi: 10.1016/j.lungcan.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Rivera V., Negrete-Garcia M.C., Avila-Moreno F., Ortiz-Quintero B. Secreted and Tissue miRNAs as Diagnosis Biomarkers of Malignant Pleural Mesothelioma. Int. J. Mol. Sci. 2018;19:595. doi: 10.3390/ijms19020595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao M.X., Jiang Y.P., Tang Y.L., Liang X.H. The crosstalk between lncRNA and microRNA in cancer metastasis: Orchestrating the epithelial-mesenchymal plasticity. Oncotarget. 2017;8:12472–12483. doi: 10.18632/oncotarget.13957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heery R., Finn S.P., Cuffe S., Gray S.G. Long Non-Coding RNAs: Key Regulators of Epithelial-Mesenchymal Transition, Tumour Drug Resistance and Cancer Stem Cells. Cancers. 2017;9:38. doi: 10.3390/cancers9040038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L., Yang F., Jia L.T., Yang A.G. Missing Links in Epithelial-Mesenchymal Transition: Long Non-Coding RNAs Enter the Arena. Cell. Physiol. Biochem. 2017;44:1665–1680. doi: 10.1159/000485766. [DOI] [PubMed] [Google Scholar]
- 23.Oehl K., Kresoja-Rakic J., Opitz I., Vrugt B., Weder W., Stahel R., Wild P., Felley-Bosco E. Live-Cell Mesothelioma Biobank to Explore Mechanisms of Tumor Progression. Front. Oncol. 2018;8:40. doi: 10.3389/fonc.2018.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riquelme E., Suraokar M.B., Rodriguez J., Mino B., Lin H.Y., Rice D.C., Tsao A., Wistuba I.I. Frequent coamplification and cooperation between C-MYC and PVT1 oncogenes promote malignant pleural mesothelioma. J. Thorac. Oncol. 2014;9:998–1007. doi: 10.1097/JTO.0000000000000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Renganathan A., Kresoja-Rakic J., Echeverry N., Ziltener G., Vrugt B., Opitz I., Stahel R.A., Felley-Bosco E. GAS5 long non-coding RNA in malignant pleural mesothelioma. Mol. Cancer. 2014;13:119. doi: 10.1186/1476-4598-13-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wright C.M., Kirschner M.B., Cheng Y.Y., O’Byrne K.J., Gray S.G., Schelch K., Hoda M.A., Klebe S., McCaughan B., van Zandwijk N., et al. Long non coding RNAs (lncRNAs) are dysregulated in Malignant Pleural Mesothelioma (MPM) PLoS ONE. 2013;8:e70940. doi: 10.1371/journal.pone.0070940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bueno R., Stawiski E.W., Goldstein L.D., Durinck S., De Rienzo A., Modrusan Z., Gnad F., Nguyen T.T., Jaiswal B.S., Chirieac L.R., et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat. Genet. 2016;48:407–416. doi: 10.1038/ng.3520. [DOI] [PubMed] [Google Scholar]
- 28.Zhao Y., Guo Q., Chen J., Hu J., Wang S., Sun Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: A clinical and in vitro investigation. Oncol. Rep. 2014;31:358–364. doi: 10.3892/or.2013.2850. [DOI] [PubMed] [Google Scholar]
- 29.Li S.P., Xu H.X., Yu Y., He J.D., Wang Z., Xu Y.J., Wang C.Y., Zhang H.M., Zhang R.X., Zhang J.J., et al. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget. 2016;7:42431–42446. doi: 10.18632/oncotarget.9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu D., Shen D., Zhang M., Jiang N., Sun F., Yuan S., Wan K. MiR-488 suppresses cell proliferation and invasion by targeting ADAM9 and lncRNA HULC in hepatocellular carcinoma. Am. J. Cancer Res. 2017;7:2070–2080. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Zhang Q., Li X., Chen Z. LncRNA H19 promotes epithelial-mesenchymal transition (EMT) by targeting miR-484 in human lung cancer cells. J. Cell. Biochem. 2017 doi: 10.1002/jcb.26537. [DOI] [PubMed] [Google Scholar]
- 32.Xu W., He L., Li Y., Tan Y., Zhang F., Xu H. Silencing of lncRNA ZFAS1 inhibits malignancies by blocking Wnt/beta-catenin signaling in gastric cancer cells. Biosci. Biotechnol. Biochem. 2018;82:456–465. doi: 10.1080/09168451.2018.1431518. [DOI] [PubMed] [Google Scholar]
- 33.Zhou H., Wang F., Chen H., Tan Q., Qiu S., Chen S., Jing W., Yu M., Liang C., Ye S., et al. Increased expression of long-noncoding RNA ZFAS1 is associated with epithelial-mesenchymal transition of gastric cancer. Aging. 2016;8:2023–2038. doi: 10.18632/aging.101048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang C., Zan J., Yue B., Liu C., He C., Yan D. Long non-coding ribonucleic acid zinc finger antisense 1 promotes the progression of colonic cancer by modulating ZEB1 expression. J. Gastroenterol. Hepatol. 2017;32:1204–1211. doi: 10.1111/jgh.13646. [DOI] [PubMed] [Google Scholar]
- 35.Guo H., Wu L., Zhao P., Feng A. Overexpression of long non-coding RNA zinc finger antisense 1 in acute myeloid leukemia cell lines influences cell growth and apoptosis. Exp. Ther. Med. 2017;14:647–651. doi: 10.3892/etm.2017.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan L., Liang W., Fu M., Huang Z.H., Li X., Zhang W., Zhang P., Qian H., Jiang P.C., Xu W.R., et al. Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. J. Cancer Res. Clin. Oncol. 2017;143:991–1004. doi: 10.1007/s00432-017-2361-2. [DOI] [PubMed] [Google Scholar]
- 37.Wang D., Gao Z.M., Han L.G., Xu F., Liu K., Shen Y. Long noncoding RNA CASC2 inhibits metastasis and epithelial to mesenchymal transition of lung adenocarcinoma via suppressing SOX4. Eur. Rev. Med. Pharmacol. Sci. 2017;21:4584–4590. [PubMed] [Google Scholar]
- 38.Edwards J.G., Swinson D.E., Jones J.L., Waller D.A., O’Byrne K.J. EGFR expression: Associations with outcome and clinicopathological variables in malignant pleural mesothelioma. Lung Cancer. 2006;54:399–407. doi: 10.1016/j.lungcan.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 39.Garland L.L., Rankin C., Gandara D.R., Rivkin S.E., Scott K.M., Nagle R.B., Klein-Szanto A.J., Testa J.R., Altomare D.A., Borden E.C. Phase II study of erlotinib in patients with malignant pleural mesothelioma: A Southwest Oncology Group Study. J. Clin. Oncol. 2007;25:2406–2413. doi: 10.1200/JCO.2006.09.7634. [DOI] [PubMed] [Google Scholar]
- 40.Govindan R., Kratzke R.A., Herndon J.E., 2nd, Niehans G.A., Vollmer R., Watson D., Green M.R., Kindler H.L. Gefitinib in patients with malignant mesothelioma: A phase II study by the Cancer and Leukemia Group B. Clin. Cancer Res. 2005;11:2300–2304. doi: 10.1158/1078-0432.CCR-04-1940. [DOI] [PubMed] [Google Scholar]
- 41.Bononi A., Napolitano A., Pass H.I., Yang H., Carbone M. Latest developments in our understanding of the pathogenesis of mesothelioma and the design of targeted therapies. Expert Rev. Respir. Med. 2015;9:633–654. doi: 10.1586/17476348.2015.1081066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Zandwijk N., Pavlakis N., Kao S.C., Linton A., Boyer M.J., Clarke S., Huynh Y., Chrzanowska A., Fulham M.J., Bailey D.L., et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: A first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017;18:1386–1396. doi: 10.1016/S1470-2045(17)30621-6. [DOI] [PubMed] [Google Scholar]
- 43.Agatsuma N., Yasuda Y., Ozasa H. Malignant Pleural Mesothelioma Harboring Both G719C and S768I Mutations of EGFR Successfully Treated with Afatinib. J. Thorac. Oncol. 2017;12:e141–e143. doi: 10.1016/j.jtho.2017.04.028. [DOI] [PubMed] [Google Scholar]
- 44.Qi H.L., Li C.S., Qian C.W., Xiao Y.S., Yuan Y.F., Liu Q.Y., Liu Z.S. The long noncoding RNA, EGFR-AS1, a target of GHR, increases the expression of EGFR in hepatocellular carcinoma. Tumour Biol. 2016;37:1079–1089. doi: 10.1007/s13277-015-3887-z. [DOI] [PubMed] [Google Scholar]
- 45.Tan D.S.W., Chong F.T., Leong H.S., Toh S.Y., Lau D.P., Kwang X.L., Zhang X., Sundaram G.M., Tan G.S., Chang M.M., et al. Long noncoding RNA EGFR-AS1 mediates epidermal growth factor receptor addiction and modulates treatment response in squamous cell carcinoma. Nat. Med. 2017;23:1167–1175. doi: 10.1038/nm.4401. [DOI] [PubMed] [Google Scholar]
- 46.Barr S., Thomson S., Buck E., Russo S., Petti F., Sujka-Kwok I., Eyzaguirre A., Rosenfeld-Franklin M., Gibson N.W., Miglarese M., et al. Bypassing cellular EGF receptor dependence through epithelial-to-mesenchymal-like transitions. Clin. Exp. Metast. 2008;25:685–693. doi: 10.1007/s10585-007-9121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang B., Qi G., Tang F., Yuan S., Wang Z., Liang X., Li B., Yu S., Liu J., Huang Q., et al. JARID1B promotes metastasis and epithelial-mesenchymal transition via PTEN/AKT signaling in hepatocellular carcinoma cells. Oncotarget. 2015;6:12723–12739. doi: 10.18632/oncotarget.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haley J.A., Haughney E., Ullman E., Bean J., Haley J.D., Fink M.Y. Altered Transcriptional Control Networks with Trans-Differentiation of Isogenic Mutant-KRas NSCLC Models. Front. Oncol. 2014;4:344. doi: 10.3389/fonc.2014.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Enkhbaatar Z., Terashima M., Oktyabri D., Tange S., Ishimura A., Yano S., Suzuki T. KDM5B histone demethylase controls epithelial-mesenchymal transition of cancer cells by regulating the expression of the microRNA-200 family. Cell Cycle. 2013;12:2100–2112. doi: 10.4161/cc.25142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wan L., Zhang L., Fan K., Cheng Z.X., Sun Q.C., Wang J.J. Knockdown of Long Noncoding RNA PCAT6 Inhibits Proliferation and Invasion in Lung Cancer Cells. Oncol. Res. 2016;24:161–170. doi: 10.3727/096504016X14618564639178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wan L., Zhang L., Fan K., Wang J.J. Diagnostic significance of circulating long noncoding RNA PCAT6 in patients with non-small cell lung cancer. Onco Targets Ther. 2017;10:5695–5702. doi: 10.2147/OTT.S149314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gordon G.J., Rockwell G.N., Jensen R.V., Rheinwald J.G., Glickman J.N., Aronson J.P., Pottorf B.J., Nitz M.D., Richards W.G., Sugarbaker D.J., et al. Identification of novel candidate oncogenes and tumor suppressors in malignant pleural mesothelioma using large-scale transcriptional profiling. Am. J. Pathol. 2005;166:1827–1840. doi: 10.1016/S0002-9440(10)62492-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beltran M., Puig I., Pena C., Garcia J.M., Alvarez A.B., Pena R., Bonilla F., de Herreros A.G. A natural antisense transcript regulates ZEB2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhuang J., Lu Q., Shen B., Huang X., Shen L., Zheng X., Huang R., Yan J., Guo H. TGFbeta1 secreted by cancer-associated fibroblasts induces epithelial-mesenchymal transition of bladder cancer cells through lncRNA-ZEB2NAT. Sci. Rep. 2015;5:11924. doi: 10.1038/srep11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lan T., Chang L., Wu L., Yuan Y. Downregulation of ZEB2-AS1 decreased tumor growth and metastasis in hepatocellular carcinoma. Mol. Med. Rep. 2016;14:4606–4612. doi: 10.3892/mmr.2016.5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lopez-Rios F., Chuai S., Flores R., Shimizu S., Ohno T., Wakahara K., Illei P.B., Hussain S., Krug L., Zakowski M.F., et al. Global gene expression profiling of pleural mesotheliomas: Overexpression of aurora kinases and P16/CDKN2A deletion as prognostic factors and critical evaluation of microarray-based prognostic prediction. Cancer Res. 2006;66:2970–2979. doi: 10.1158/0008-5472.CAN-05-3907. [DOI] [PubMed] [Google Scholar]
- 57.Zhu S., Shuai P., Yang C., Zhang Y., Zhong S., Liu X., Chen K., Ran Q., Yang H., Zhou Y. Prognostic value of long non-coding RNA PVT1 as a novel biomarker in various cancers: A meta-analysis. Oncotarget. 2017;8:113174–113184. doi: 10.18632/oncotarget.22830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y., Zhou J., Wang Z., Wang P., Li S. Upregulation of SOX2 activated LncRNA PVT1 expression promotes breast cancer cell growth and invasion. Biochem. Biophys. Res. Commun. 2017;493:429–436. doi: 10.1016/j.bbrc.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 59.Wu B.Q., Jiang Y., Zhu F., Sun D.L., He X.Z. Long Noncoding RNA PVT1 Promotes EMT and Cell Proliferation and Migration through Downregulating p21 in Pancreatic Cancer Cells. Technol. Cancer Res. Treat. 2017 doi: 10.1177/1533034617700559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang T., Zhou H., Liu P., Yan L., Yao W., Chen K., Zeng J., Li H., Hu J., Xu H., et al. lncRNA PVT1 and its splicing variant function as competing endogenous RNA to regulate clear cell renal cell carcinoma progression. Oncotarget. 2017;8:85353–85367. doi: 10.18632/oncotarget.19743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H., Chen S., Liu J., Guo X., Xiang X., Dong T., Ran P., Li Q., Zhu B., Zhang X., et al. Long non-coding RNA PVT1–5 promotes cell proliferation by regulating miR-126/SLC7A5 axis in lung cancer. Biochem. Biophys. Res. Commun. 2018;495:2350–2355. doi: 10.1016/j.bbrc.2017.12.114. [DOI] [PubMed] [Google Scholar]
- 62.Wang C., Han C., Zhang Y., Liu F. LncRNA PVT1 regulate expression of HIF1alpha via functioning as ceRNA for miR199a5p in nonsmall cell lung cancer under hypoxia. Mol. Med. Rep. 2018;17:1105–1110. doi: 10.3892/mmr.2017.7962. [DOI] [PubMed] [Google Scholar]
- 63.Gao Y.L., Zhao Z.S., Zhang M.Y., Han L.J., Dong Y.J., Xu B. Long Noncoding RNA PVT1 Facilitates Cervical Cancer Progression via Negative Regulating of miR-424. Oncol. Res. 2017;25:1391–1398. doi: 10.3727/096504017X14881559833562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Conte F., Fiscon G., Chiara M., Colombo T., Farina L., Paci P. Role of the long non-coding RNA PVT1 in the dysregulation of the ceRNA-ceRNA network in human breast cancer. PLoS ONE. 2017;12:e0171661. doi: 10.1371/journal.pone.0171661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang S., Ning Q., Zhang G., Sun H., Wang Z., Li Y. Construction of differential mRNA-lncRNA crosstalk networks based on ceRNA hypothesis uncover key roles of lncRNAs implicated in esophageal squamous cell carcinoma. Oncotarget. 2016;7:85728–85740. doi: 10.18632/oncotarget.13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lan T., Yan X., Li Z., Xu X., Mao Q., Ma W., Hong Z., Chen X., Yuan Y. Long non-coding RNA PVT1 serves as a competing endogenous RNA for miR-186–5p to promote the tumorigenesis and metastasis of hepatocellular carcinoma. Tumour Biol. 2017;39 doi: 10.1177/1010428317705338. [DOI] [PubMed] [Google Scholar]
- 67.Chang Z., Cui J., Song Y. Long noncoding RNA PVT1 promotes EMT via mediating microRNA-186 targeting of Twist1 in prostate cancer. Gene. 2018;654:36–42. doi: 10.1016/j.gene.2018.02.036. [DOI] [PubMed] [Google Scholar]
- 68.Shen C.J., Cheng Y.M., Wang C.L. LncRNA PVT1 epigenetically silences miR-195 and modulates EMT and chemoresistance in cervical cancer cells. J. Drug Target. 2017;25:637–644. doi: 10.1080/1061186X.2017.1307379. [DOI] [PubMed] [Google Scholar]
- 69.Zhang S., Zhang G., Liu J. Long noncoding RNA PVT1 promotes cervical cancer progression through epigenetically silencing miR-200b. APMIS. 2016;124:649–658. doi: 10.1111/apm.12555. [DOI] [PubMed] [Google Scholar]
- 70.Wan L., Sun M., Liu G.J., Wei C.C., Zhang E.B., Kong R., Xu T.P., Huang M.D., Wang Z.X. Long Noncoding RNA PVT1 Promotes Non-Small Cell Lung Cancer Cell Proliferation through Epigenetically Regulating LATS2 Expression. Mol. Cancer Ther. 2016;15:1082–1094. doi: 10.1158/1535-7163.MCT-15-0707. [DOI] [PubMed] [Google Scholar]
- 71.Kong R., Zhang E.B., Yin D.D., You L.H., Xu T.P., Chen W.M., Xia R., Wan L., Sun M., Wang Z.X., et al. Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Mol. Cancer. 2015;14:82. doi: 10.1186/s12943-015-0355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou Q., Chen J., Feng J., Wang J. Long noncoding RNA PVT1 modulates thyroid cancer cell proliferation by recruiting EZH2 and regulating thyroid-stimulating hormone receptor (TSHR) Tumour Biol. 2016;37:3105–3113. doi: 10.1007/s13277-015-4149-9. [DOI] [PubMed] [Google Scholar]
- 73.Gou X., Zhao X., Wang Z. Long noncoding RNA PVT1 promotes hepatocellular carcinoma progression through regulating miR-214. Cancer Biomark. 2017;20:511–519. doi: 10.3233/CBM-170331. [DOI] [PubMed] [Google Scholar]
- 74.Yang A., Wang H., Yang X. Long non-coding RNA PVT1 indicates a poor prognosis of glioma and promotes cell proliferation and invasion via target EZH2. Biosci. Rep. 2017;37 doi: 10.1042/BSR20170871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen L., Ma D., Li Y., Li X., Zhao L., Zhang J., Song Y. Effect of long non-coding RNA PVT1 on cell proliferation and migration in melanoma. Int. J. Mol. Med. 2018;41:1275–1282. doi: 10.3892/ijmm.2017.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tseng Y.Y., Moriarity B.S., Gong W., Akiyama R., Tiwari A., Kawakami H., Ronning P., Reuland B., Guenther K., Beadnell T.C., et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512:82–86. doi: 10.1038/nature13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang M., Wu W.B., Wang Z.W., Wang X.H. lncRNA NEAT1 is closely related with progression of breast cancer via promoting proliferation and EMT. Eur. Rev. Med. Pharmacol. Sci. 2017;21:1020–1026. [PubMed] [Google Scholar]
- 78.Qian K., Liu G., Tang Z., Hu Y., Fang Y., Chen Z., Xu X. The long non-coding RNA NEAT1 interacted with miR-101 modulates breast cancer growth by targeting EZH2. Arch. Biochem. Biophys. 2016;615:1–9. doi: 10.1016/j.abb.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 79.Chen Q., Cai J., Wang Q., Wang Y., Liu M., Yang J., Zhou J., Kang C., Li M., Jiang C. Long Noncoding RNA NEAT1, Regulated by the EGFR Pathway, Contributes to Glioblastoma Progression Through the WNT/beta-Catenin Pathway by Scaffolding EZH2. Clin. Cancer Res. 2018;24:684–695. doi: 10.1158/1078-0432.CCR-17-0605. [DOI] [PubMed] [Google Scholar]
- 80.Parasramka M., Yan I.K., Wang X., Nguyen P., Matsuda A., Maji S., Foye C., Asmann Y., Patel T. BAP1 dependent expression of long non-coding RNA NEAT-1 contributes to sensitivity to gemcitabine in cholangiocarcinoma. Mol. Cancer. 2017;16:22. doi: 10.1186/s12943-017-0587-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yap T.A., Aerts J.G., Popat S., Fennell D.A. Novel insights into mesothelioma biology and implications for therapy. Nat. Rev. Cancer. 2017;17:475–488. doi: 10.1038/nrc.2017.42. [DOI] [PubMed] [Google Scholar]
- 82.Schaner M.E., Ross D.T., Ciaravino G., Sorlie T., Troyanskaya O., Diehn M., Wang Y.C., Duran G.E., Sikic T.L., Caldeira S., et al. Gene expression patterns in ovarian carcinomas. Mol. Biol. Cell. 2003;14:4376–4386. doi: 10.1091/mbc.E03-05-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Di Palma T., Lucci V., de Cristofaro T., Filippone M.G., Zannini M. A role for PAX8 in the tumorigenic phenotype of ovarian cancer cells. BMC Cancer. 2014;14:292. doi: 10.1186/1471-2407-14-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gardi N.L., Deshpande T.U., Kamble S.C., Budhe S.R., Bapat S.A. Discrete molecular classes of ovarian cancer suggestive of unique mechanisms of transformation and metastases. Clin. Cancer Res. 2014;20:87–99. doi: 10.1158/1078-0432.CCR-13-2063. [DOI] [PubMed] [Google Scholar]
- 85.Chapel D.B., Husain A.N., Krausz T., McGregor S.M. PAX8 Expression in a Subset of Malignant Peritoneal Mesotheliomas and Benign Mesothelium has Diagnostic Implications in the Differential Diagnosis of Ovarian Serous Carcinoma. Am. J. Surg. Pathol. 2017;41:1675–1682. doi: 10.1097/PAS.0000000000000935. [DOI] [PubMed] [Google Scholar]
- 86.Laury A.R., Hornick J.L., Perets R., Krane J.F., Corson J., Drapkin R., Hirsch M.S. PAX8 reliably distinguishes ovarian serous tumors from malignant mesothelioma. Am. J. Surg. Pathol. 2010;34:627–635. doi: 10.1097/PAS.0b013e3181da7687. [DOI] [PubMed] [Google Scholar]
- 87.Dong S., Qu X., Li W., Zhong X., Li P., Yang S., Chen X., Shao M., Zhang L. The long non-coding RNA, GAS5, enhances gefitinib-induced cell death in innate EGFR tyrosine kinase inhibitor-resistant lung adenocarcinoma cells with wide-type EGFR via downregulation of the IGF-1R expression. J. Hematol. Oncol. 2015;8:43. doi: 10.1186/s13045-015-0140-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xue D., Zhou C., Lu H., Xu R., Xu X., He X. LncRNA GAS5 inhibits proliferation and progression of prostate cancer by targeting miR-103 through AKT/mTOR signaling pathway. Tumour Biol. 2016 doi: 10.1007/s13277-016-5429-8. [DOI] [PubMed] [Google Scholar]
- 89.Ye K., Wang S., Zhang H., Han H., Ma B., Nan W. Long Noncoding RNA GAS5 Suppresses Cell Growth and Epithelial-Mesenchymal Transition in Osteosarcoma by Regulating the miR-221/ARHI Pathway. J. Cell. Biochem. 2017;118:4772–4781. doi: 10.1002/jcb.26145. [DOI] [PubMed] [Google Scholar]
- 90.Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu Y., Zhang L., Wang Y., Li H., Ren X., Wei F., Yu W., Liu T., Wang X., Zhou X., et al. Long non-coding RNA HOTAIR promotes tumor cell invasion and metastasis by recruiting EZH2 and repressing E-cadherin in oral squamous cell carcinoma. Int. J. Oncol. 2015;46:2586–2594. doi: 10.3892/ijo.2015.2976. [DOI] [PubMed] [Google Scholar]
- 93.Battistelli C., Cicchini C., Santangelo L., Tramontano A., Grassi L., Gonzalez F.J., de Nonno V., Grassi G., Amicone L., Tripodi M. The Snail repressor recruits EZH2 to specific genomic sites through the enrollment of the lncRNA HOTAIR in epithelial-to-mesenchymal transition. Oncogene. 2017;36:942–955. doi: 10.1038/onc.2016.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xia M., Yao L., Zhang Q., Wang F., Mei H., Guo X., Huang W. Long noncoding RNA HOTAIR promotes metastasis of renal cell carcinoma by up-regulating histone H3K27 demethylase JMJD3. Oncotarget. 2017;8:19795–19802. doi: 10.18632/oncotarget.15047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu Y.W., Sun M., Xia R., Zhang E.B., Liu X.H., Zhang Z.H., Xu T.P., De W., Liu B.R., Wang Z.X. LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis. 2015;6:e1802. doi: 10.1038/cddis.2015.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li J.T., Wang L.F., Zhao Y.L., Yang T., Li W., Zhao J., Yu F., Wang L., Meng Y.L., Liu N.N., et al. Nuclear factor of activated T cells 5 maintained by Hotair suppression of miR-568 upregulates S100 calcium binding protein A4 to promote breast cancer metastasis. Breast Cancer Res. 2014;16:454. doi: 10.1186/s13058-014-0454-2. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 97.Zhang H., Cai K., Wang J., Wang X., Cheng K., Shi F., Jiang L., Zhang Y., Dou J. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells. 2014;32:2858–2868. doi: 10.1002/stem.1795. [DOI] [PubMed] [Google Scholar]
- 98.Ji P., Diederichs S., Wang W., Boing S., Metzger R., Schneider P.M., Tidow N., Brandt B., Buerger H., Bulk E., et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 99.Chen Y., Xiao Z., Hu M., Luo X., Cui Z. Diagnostic efficacy of long non-coding RNA MALAT-1 in human cancers: A meta-analysis study. Oncotarget. 2017;8:102291–102300. doi: 10.18632/oncotarget.21013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cheng Y., Imanirad P., Jutooru I., Hedrick E., Jin U.H., Rodrigues Hoffman A., Leal de Araujo J., Morpurgo B., Golovko A., Safe S. Role of metastasis-associated lung adenocarcinoma transcript-1 (MALAT-1) in pancreatic cancer. PLoS ONE. 2018;13:e0192264. doi: 10.1371/journal.pone.0192264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chou J., Wang B., Zheng T., Li X., Zheng L., Hu J., Zhang Y., Xing Y., Xi T. MALAT1 induced migration and invasion of human breast cancer cells by competitively binding miR-1 with cdc42. Biochem. Biophys. Res. Commun. 2016;472:262–269. doi: 10.1016/j.bbrc.2016.02.102. [DOI] [PubMed] [Google Scholar]
- 102.Xu S., Sui S., Zhang J., Bai N., Shi Q., Zhang G., Gao S., You Z., Zhan C., Liu F., et al. Downregulation of long noncoding RNA MALAT1 induces epithelial-to-mesenchymal transition via the PI3K-AKT pathway in breast cancer. Int. J. Clin. Exp. Pathol. 2015;8:4881–4891. [PMC free article] [PubMed] [Google Scholar]
- 103.Li J., Wang J., Chen Y., Li S., Jin M., Wang H., Chen Z., Yu W. LncRNA MALAT1 exerts oncogenic functions in lung adenocarcinoma by targeting miR-204. Am. J. Cancer Res. 2016;6:1099–1107. [PMC free article] [PubMed] [Google Scholar]
- 104.Jin C., Yan B., Lu Q., Lin Y., Ma L. Reciprocal regulation of Hsa-miR-1 and long noncoding RNA MALAT1 promotes triple-negative breast cancer development. Tumour Biol. 2016;37:7383–7394. doi: 10.1007/s13277-015-4605-6. [DOI] [PubMed] [Google Scholar]
- 105.Hirata H., Hinoda Y., Shahryari V., Deng G., Nakajima K., Tabatabai Z.L., Ishii N., Dahiya R. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer Res. 2015;75:1322–1331. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li Q., Zhang C., Chen R., Xiong H., Qiu F., Liu S., Zhang M., Wang F., Wang Y., Zhou X., et al. Disrupting MALAT1/miR-200c sponge decreases invasion and migration in endometrioid endometrial carcinoma. Cancer Lett. 2016;383:28–40. doi: 10.1016/j.canlet.2016.09.019. [DOI] [PubMed] [Google Scholar]
- 107.Fan Y., Shen B., Tan M., Mu X., Qin Y., Zhang F., Liu Y. TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin. Cancer Res. 2014;20:1531–1541. doi: 10.1158/1078-0432.CCR-13-1455. [DOI] [PubMed] [Google Scholar]
- 108.Zhang Z.C., Tang C., Dong Y., Zhang J., Yuan T., Li X.L. Targeting LncRNA-MALAT1 suppresses the progression of osteosarcoma by altering the expression and localization of beta-catenin. J. Cancer. 2018;9:71–80. doi: 10.7150/jca.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang W., Zhu Y., Li S., Chen X., Jiang G., Shen Z., Qiao Y., Wang L., Zheng P., Zhang Y. Long noncoding RNA MALAT1 promotes malignant development of esophageal squamous cell carcinoma by targeting beta-catenin via Ezh2. Oncotarget. 2016;7:25668–25682. doi: 10.18632/oncotarget.8257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu X.S., Wang X.A., Wu W.G., Hu Y.P., Li M.L., Ding Q., Weng H., Shu Y.J., Liu T.Y., Jiang L., et al. MALAT1 promotes the proliferation and metastasis of gallbladder cancer cells by activating the ERK/MAPK pathway. Cancer Biol. Ther. 2014;15:806–814. doi: 10.4161/cbt.28584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Han Y., Wu Z., Wu T., Huang Y., Cheng Z., Li X., Sun T., Xie X., Zhou Y., Du Z. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death Dis. 2016;7:e2123. doi: 10.1038/cddis.2015.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dong Y., Liang G., Yuan B., Yang C., Gao R., Zhou X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol. 2015;36:1477–1486. doi: 10.1007/s13277-014-2631-4. [DOI] [PubMed] [Google Scholar]
- 113.Jin Y., Feng S.J., Qiu S., Shao N., Zheng J.H. LncRNA MALAT1 promotes proliferation and metastasis in epithelial ovarian cancer via the PI3K-AKT pathway. Eur. Rev. Med. Pharmacol. Sci. 2017;21:3176–3184. [PubMed] [Google Scholar]
- 114.Yan W., Wu Q., Yao W., Li Y., Liu Y., Yuan J., Han R., Yang J., Ji X., Ni C. MiR-503 modulates epithelial-mesenchymal transition in silica-induced pulmonary fibrosis by targeting PI3K p85 and is sponged by lncRNA MALAT1. Sci. Rep. 2017;7:11313. doi: 10.1038/s41598-017-11904-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yang G., Shen T., Yi X., Zhang Z., Tang C., Wang L., Zhou Y., Zhou W. Crosstalk between long non-coding RNAs and Wnt/beta-catenin signalling in cancer. J. Cell. Mol. Med. 2018;22:2062–2070. doi: 10.1111/jcmm.13522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liang J., Liang L., Ouyang K., Li Z., Yi X. MALAT1 induces tongue cancer cells’ EMT and inhibits apoptosis through Wnt/beta-catenin signaling pathway. J. Oral Pathol. Med. 2016;48:98–105. doi: 10.1111/jop.12466. [DOI] [PubMed] [Google Scholar]
- 117.Zhao Y., Yang Y., Trovik J., Sun K., Zhou L., Jiang P., Lau T.S., Hoivik E.A., Salvesen H.B., Sun H., et al. A novel wnt regulatory axis in endometrioid endometrial cancer. Cancer Res. 2014;74:5103–5117. doi: 10.1158/0008-5472.CAN-14-0427. [DOI] [PubMed] [Google Scholar]
- 118.Liu S., Yan G., Zhang J., Yu L. Knockdown of Long Noncoding RNA (lncRNA) Metastasis-Associated Lung Adenocarcinoma Transcript 1 (MALAT1) Inhibits Proliferation, Migration, and Invasion and Promoted Apoptosis By Targeting miR-124 in Retinoblastoma. Oncol. Res. :2017. doi: 10.3727/096504017X14953948675403. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 119.Vassallo I., Zinn P., Lai M., Rajakannu P., Hamou M.F., Hegi M.E. WIF1 re-expression in glioblastoma inhibits migration through attenuation of non-canonical WNT signaling by downregulating the lncRNA MALAT1. Oncogene. 2016;35:12–21. doi: 10.1038/onc.2015.61. [DOI] [PubMed] [Google Scholar]
- 120.Rickman D.S., Schulte J.H., Eilers M. The Expanding World of N-MYC-Driven Tumors. Cancer Discov. 2018;8:150–163. doi: 10.1158/2159-8290.CD-17-0273. [DOI] [PubMed] [Google Scholar]
- 121.Ma L., Young J., Prabhala H., Pan E., Mestdagh P., Muth D., Teruya-Feldstein J., Reinhardt F., Onder T.T., Valastyan S., et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vadie N., Saayman S., Lenox A., Ackley A., Clemson M., Burdach J., Hart J., Vogt P.K., Morris K.V. MYCNOS functions as an antisense RNA regulating MYCN. RNA Biol. 2015;12:893–899. doi: 10.1080/15476286.2015.1063773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jacobs J.F., van Bokhoven H., van Leeuwen F.N., Hulsbergen-van de Kaa C.A., de Vries I.J., Adema G.J., Hoogerbrugge P.M., de Brouwer A.P. Regulation of MYCN expression in human neuroblastoma cells. BMC Cancer. 2009;9:239. doi: 10.1186/1471-2407-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhao X., Li D., Pu J., Mei H., Yang D., Xiang X., Qu H., Huang K., Zheng L., Tong Q. CTCF cooperates with noncoding RNA MYCNOS to promote neuroblastoma progression through facilitating MYCN expression. Oncogene. 2016;35:3565–3576. doi: 10.1038/onc.2015.422. [DOI] [PubMed] [Google Scholar]
- 125.Raveh E., Matouk I.J., Gilon M., Hochberg A. The H19 Long non-coding RNA in cancer initiation, progression and metastasis—A proposed unifying theory. Mol. Cancer. 2015;14:184. doi: 10.1186/s12943-015-0458-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Matouk I.J., Halle D., Raveh E., Gilon M., Sorin V., Hochberg A. The role of the oncofetal H19 lncRNA in tumor metastasis: Orchestrating the EMT-MET decision. Oncotarget. 2016;7:3748–3765. doi: 10.18632/oncotarget.6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Panzitt K., Tschernatsch M.M., Guelly C., Moustafa T., Stradner M., Strohmaier H.M., Buck C.R., Denk H., Schroeder R., Trauner M., et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 128.Yu X., Zheng H., Chan M.T., Wu W.K. HULC: An oncogenic long non-coding RNA in human cancer. J. Cell. Mol. Med. 2017;21:410–417. doi: 10.1111/jcmm.12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lu Y., Li Y., Chai X., Kang Q., Zhao P., Xiong J., Wang J. Long noncoding RNA HULC promotes cell proliferation by regulating PI3K/AKT signaling pathway in chronic myeloid leukemia. Gene. 2017;607:41–46. doi: 10.1016/j.gene.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 130.Wang J., Ma W., Liu Y. Long non-coding RNA HULC promotes bladder cancer cells proliferation but inhibits apoptosis via regulation of ZIC2 and PI3K/AKT signaling pathway. Cancer Biomark. 2017;20:425–434. doi: 10.3233/CBM-170188. [DOI] [PubMed] [Google Scholar]
- 131.Wu M., Lin Z., Li X., Xin X., An J., Zheng Q., Yang Y., Lu D. HULC cooperates with MALAT1 to aggravate liver cancer stem cells growth through telomere repeat-binding factor 2. Sci. Rep. 2016;6:36045. doi: 10.1038/srep36045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yang X.J., Huang C.Q., Peng C.W., Hou J.X., Liu J.Y. Long noncoding RNA HULC promotes colorectal carcinoma progression through epigenetically repressing NKD2 expression. Gene. 2016;592:172–178. doi: 10.1016/j.gene.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 133.Wang J., Liu X., Wu H., Ni P., Gu Z., Qiao Y., Chen N., Sun F., Fan Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kong D., Wang Y. Knockdown of lncRNA HULC inhibits proliferation, migration, invasion, and promotes apoptosis by sponging miR-122 in osteosarcoma. J. Cell. Biochem. 2018;119:1050–1061. doi: 10.1002/jcb.26273. [DOI] [PubMed] [Google Scholar]
- 135.Zhang Y., Li Z., Zhong Q., Chen Q., Zhang L. Molecular mechanism of HEIH and HULC in the proliferation and invasion of hepatoma cells. Int. J. Clin. Exp. Med. 2015;8:12956–12962. [PMC free article] [PubMed] [Google Scholar]
- 136.Wang Y., Liu Z., Yao B., Li Q., Wang L., Wang C., Dou C., Xu M., Liu Q., Tu K. Long non-coding RNA CASC2 suppresses epithelial-mesenchymal transition of hepatocellular carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol. Cancer. 2017;16:123. doi: 10.1186/s12943-017-0702-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhao L., Zhang Y. Long noncoding RNA CASC2 regulates hepatocellular carcinoma cell oncogenesis through miR-362–5p/Nf-kappaB axis. J. Cell. Physiol. :2018. doi: 10.1002/jcp.26446. [DOI] [PubMed] [Google Scholar]
- 138.Askarian-Amiri M.E., Crawford J., French J.D., Smart C.E., Smith M.A., Clark M.B., Ru K., Mercer T.R., Thompson E.R., Lakhani S.R., et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. 2011;17:878–891. doi: 10.1261/rna.2528811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gao K., Ji Z., She K., Yang Q., Shao L. Long non-coding RNA ZFAS1 is an unfavourable prognostic factor and promotes glioma cell progression by activation of the Notch signaling pathway. Biomed. Pharmacother. 2017;87:555–560. doi: 10.1016/j.biopha.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 140.Li T., Xie J., Shen C., Cheng D., Shi Y., Wu Z., Deng X., Chen H., Shen B., Peng C., et al. Amplification of Long Noncoding RNA ZFAS1 Promotes Metastasis in Hepatocellular Carcinoma. Cancer Res. 2015;75:3181–3191. doi: 10.1158/0008-5472.CAN-14-3721. [DOI] [PubMed] [Google Scholar]
- 141.Liu G., Wang L., Han H., Li Y., Lu S., Li T., Cheng C. LncRNA ZFAS1 promotes growth and metastasis by regulating BMI1 and ZEB2 in osteosarcoma. Am. J. Cancer Res. 2017;7:1450–1462. [PMC free article] [PubMed] [Google Scholar]
- 142.Lv Q.L., Chen S.H., Zhang X., Sun B., Hu L., Qu Q., Huang Y.T., Wang G.H., Liu Y.L., Zhang Y.Y., et al. Upregulation of long noncoding RNA zinc finger antisense 1 enhances epithelial-mesenchymal transition in vitro and predicts poor prognosis in glioma. Tumour Biol. 2017;39 doi: 10.1177/1010428317695022. [DOI] [PubMed] [Google Scholar]
- 143.Nie F., Yu X., Huang M., Wang Y., Xie M., Ma H., Wang Z., De W., Sun M. Long noncoding RNA ZFAS1 promotes gastric cancer cells proliferation by epigenetically repressing KLF2 and NKD2 expression. Oncotarget. 2017;8:38227–38238. doi: 10.18632/oncotarget.9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Thorenoor N., Faltejskova-Vychytilova P., Hombach S., Mlcochova J., Kretz M., Svoboda M., Slaby O. Long non-coding RNA ZFAS1 interacts with CDK1 and is involved in p53-dependent cell cycle control and apoptosis in colorectal cancer. Oncotarget. 2016;7:622–637. doi: 10.18632/oncotarget.5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tian F.M., Meng F.Q., Wang X.B. Overexpression of long-noncoding RNA ZFAS1 decreases survival in human NSCLC patients. Eur. Rev. Med. Pharmacol. Sci. 2016;20:5126–5131. [PubMed] [Google Scholar]
- 146.Wang W., Xing C. Upregulation of long noncoding RNA ZFAS1 predicts poor prognosis and prompts invasion and metastasis in colorectal cancer. Pathol. Res. Pract. 2016;212:690–695. doi: 10.1016/j.prp.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 147.Xia B., Hou Y., Chen H., Yang S., Liu T., Lin M., Lou G. Long non-coding RNA ZFAS1 interacts with miR-150–5p to regulate Sp1 expression and ovarian cancer cell malignancy. Oncotarget. 2017;8:19534–19546. doi: 10.18632/oncotarget.14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Xie S., Ge Q., Wang X., Sun X., Kang Y. Long non-coding RNA ZFAS1 sponges miR-484 to promote cell proliferation and invasion in colorectal cancer. Cell Cycle. 2018;17:154–161. doi: 10.1080/15384101.2017.1407895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cregan S., Breslin M., Roche G., Wennstedt S., MacDonagh L., Albadri C., Gao Y., O’Byrne K.J., Cuffe S., Finn S.P. Kdm6a and Kdm6b: Altered expression in malignant pleural mesothelioma. Int. J. Oncol. 2017;50:1044–1052. doi: 10.3892/ijo.2017.3870. [DOI] [PubMed] [Google Scholar]
- 150.Rhodes D.R., Kalyana-Sundaram S., Mahavisno V., Varambally R., Yu J., Briggs B.B., Barrette T.R., Anstet M.J., Kincead-Beal C., Kulkarni P., et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Oncomine. [(accessed on 2 April 2018)]; Available online: www.oncomine.org.
- 152.cBioportal. [(accessed on 2 April 2018)]; Available online: www.cbioportal.org.
- 153.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Goswami C.P., Nakshatri H. PROGgeneV2: Enhancements on the existing database. BMC Cancer. 2014;14:970. doi: 10.1186/1471-2407-14-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.PROGgene. [(accessed on 2 April 2018)]; Available online: http://watson.compbio.iupui.edu/chirayu/proggene/database/?url=proggene.
- 157.Brosseau S., Dhalluin X., Zalcman G., Scherpereel A. Immunotherapy in relapsed mesothelioma. Immunotherapy. 2018;10:77–80. doi: 10.2217/imt-2017-0144. [DOI] [PubMed] [Google Scholar]