Abstract
Pyruvate kinase is known as the glycolytic enzyme catalyzing the final step in glycolysis. In mammals, two different forms of it exist, i.e., pyruvate kinase M1/2 (PKM) and pyruvate kinase L/R (PKLR). Also, PKM has two isoforms, i.e., PKM1 and PKM2. These genes have tissue-specific distribution. Namely, PKM1 is distributed in high-energy-demanding organs, such as brain and muscle. Also, PKM2 is distributed in various other organs, such as the colon. On the other hand, PKLR is distributed in liver and red blood cells (RBCs). Interestingly, PKM2 has been recognized as one of the essential genes for the cancer-specific energy metabolism termed the “Warburg effect”. However, the mechanism(s) underlying this fact have remained largely unclear. Recently, we found that some organ-specific microRNAs (miRNAs, MIR) regulate PKM isoform expression through direct targeting of polypyrimidine tract binding protein 1 (PTBP1), which is the splicer responsible for PKM2-dominant expression. In this study, we examined whether this machinery was conserved in the case of other PTBP1- and PKM-targeting miRNAs. We focused on the MIRs 122, 137, and 206, and investigated the expression profiles of each of these miRNAs in tissues from mouse and human organs. Also, we examined the regulatory mechanisms of PKM isoform expression by testing each of these miRNAs in human cancer cell lines. Presently, we found that brain-specific MIR137 and muscle-specific MIR206 predominantly induced PKM1 expression through direct targeting of PTBP1. Also, liver-specific MIR122 suppressed the expression of both PKM1 and PKM2, which action occurred through direct targeting of PKM to enable the expression of PKLR. Moreover, the expression levels of these miRNAs were downregulated in cancer cells that had originated from these tissues, resulting in PKM2 dominance. Our results suggest that the organ-specific distribution of miRNAs is one of the principal means by which miRNA establishes characteristics of a tissue and that dysregulation of these miRNAs results in cancer development through a change in the ratio of PKM isoform expression. Also, our results contribute to cancer diagnosis and will be useful for cancer-specific therapy for the Warburg effect in the near future.
Keywords: microRNA, PKM, PKLR, PTBP1, Warburg effect
1. Introduction
Pyruvate kinase (PK) is known as the glycolytic enzyme catalyzing the final step in glycolysis. In mammals, two different PK enzymes exist, i.e., pyruvate kinase M 1/2 (PKM) and pyruvate kinase L/R (PKLR). Also, PKM has two isoforms, i.e., PKM1/PKM2, and they are encoded by the M gene [1]. Alternative splicers, such as those in the heterogeneous nuclear ribonucleoprotein (HNRNP) family, regulate the expression of PKM1 and PKM2 isoforms through splicing of exon 9 and exon 10 [2]. Namely, the adoption of exon 9 induces PKM1; and that of exon 10, PKM2. Among HNRNP family members, polypyrimidine tract binding protein 1 (PTBP1) is one of the crucial splicers for PKM2-dominant expression, acting by repressing the expression of exon 9 [3]. Recently, PKM2 has been recognized as one of the crucial oncogenes in various cancer cells. In other words, PKM2 contributes to the establishment of the cancer-specific energy metabolism termed the “Warburg effect” [4]. Several reports have suggested that upregulation of PKM2 induces reprogramming of the Warburg effect in cancer cells. For example, PKM2 promotes aerobic glycolysis in order to acquire an advantageous environment of tumor cells by the production of lactic acid and nucleic acid [4,5]. Also, PKM2 has multiple functions for cancer growth, such as transcriptional regulation and extracellular signaling [6]. It has been thought that the PKM isoform switches from PKM1 to PKM2 to establish the Warburg effect during carcinogenesis [7]. However, our group and another have uncovered new features about this switching machinery. Namely, PKM1 is predominant only in brain, skeletal muscle, and heart among adult tissues. In these tissues, PKM switching machinery causes an expression change from PKM1 to PKM2 during cancer development. However, in other proliferative tissues, such as colon and stomach, PKM2 exists even in normal tissue, and the PKM2 ratio is further increased during cancer development [8,9].
On the other hand, PKLR is encoded by the L gene [10]. PKLR has two isoforms, i.e., PKL and PKR. PKL is predominant in the liver, and PKR in red blood cells (RBCs) [4]. PKLR acts as a feed-forward regulator of glycolysis [4,11]. Mutations in the PKLR gene cause severe congenital disease, such as nonspherocytic hemolytic anemia [12]. Thus, PKM and PKLR are clearly distributed in a tissue-specific manner. However, the mechanisms underlying their tissue distribution have been largely unknown.
MicroRNA (miRNA, MIR) negatively regulates the expression of its target gene(s) through repression of translation or induction of degradation of its target mRNA(s). MiRNAs are associated with various phenomena, including tissue differentiation [13] and carcinogenesis [14]. Individual miRNAs function as fine adjusters of gene expression, and many miRNAs modulate a given phenomenon by acting together. Recently, we found that some organ-specific miRNAs, such as brain-specific MIR124 and muscle-specific MIR-133b, determine PKM isoform expression through direct targeting of PTBP1 [8]. Also, the dysregulated expression of these miRNAs contributes to carcinogenesis through the modulation of the Warburg effect in gastrointestinal tumors [15,16]. We have assumed that some organ-specific miRNAs contribute strongly to characteristic functions and carcinogenesis of their tissue. To confirm our hypothesis, we needed to examine whether this machinery was operating even in the case of other miRNAs. Hence, in this study, we verified the universality of our hypothesis by using other PTBP1- and PKM-targeting miRNAs.
2. Results
2.1. Expression of Pyruvate Kinase (PK) Enzymes Revealed Organ Specificity
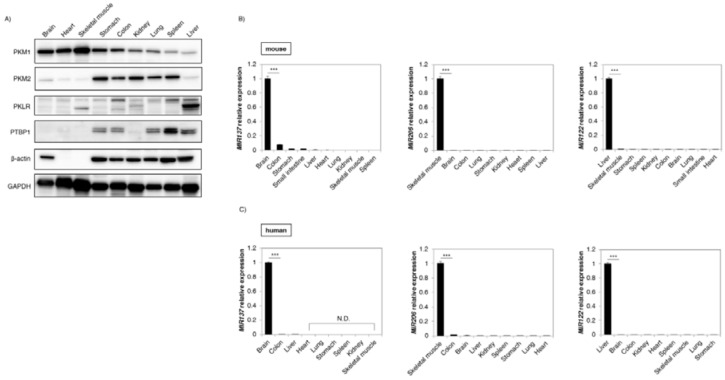
Firstly, we examined the tissue distribution of PK enzymes in the tissues from mouse organs. As shown in Figure 1A, the protein expression level of PKM1 was dominant in brain, heart, and skeletal muscle, whereas that of PKM2 expression was relatively greater in stomach, colon, kidney, lung, and spleen tissue. In the liver, PKM1 and PKM2 protein levels were not noticeably expressed as compared to the abundant expression of PKLR. Moreover, PTBP1, which is a crucial splicer for PKM2-dominant expression in PKM gene splicing, was expressed in the PKM2-dominant tissues (except kidney and liver). These findings showed that the expression profiles of PK enzymes indicated organ specificity and that PTBP1 might be associated with this specificity.
Figure 1.
Pyruvate kinase (PK) different forms and PK enzymes regulating miRNAs showed organ specificity. (A) Expression profile of PKM1, PKM2, pyruvate kinase L/R (PKLR), and PTBP1 in tissues from mouse organs. β-actin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used as internal controls; (B) Expression profile of MIR137 (Left panel), MIR206 (Middle panel), and MIR122 (Right panel) in tissues from mouse organs; (C) Profiles of the same miRNAs in tissues from human organs. The expression level is presented as a relative value compared with the tissue that had the highest expression level in each miRNA. Notably, normalization was not performed in this case because the organ specificities of these miRNAs were strong enough so as not to be affected by normalization. Human brain cortex and liver were obtained from Biochain, and others from Clontech. Results are presented as the mean ± standard deviation (SD); *** p < 0.001; N.D., not detected.
2.2. Expression of MicroRNA (MIR) 137, 206, and 122 Was Organ-Specific
Some organ-specific miRNAs, such as MIR1, 124, and 133b, which directly target PTBP1, regulate PKM1 and PKM2 expression [8,15]. The Target Scan 7.1 database (http://www.targetscan.org/) showed that MIR137 and 206 had a site that binds directly to the 3′UTR of PTBP1 and that only MIR122 had one directly binding to the 3′UTR of PKM [17]. Also, we validated the association of each miRNA and targeting gene with miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php) (Table 1). Next, we investigated the tissue distribution of these miRNAs in the tissues from mouse organs. As shown in Figure 1B, MIR137 was considerably and dominantly distributed in the brain, with MIR206 being expressed in skeletal muscle. Also, MIR122 was considerably dominant in the liver. Moreover, these findings were similar to those obtained for human tissues (Figure 1C). These findings suggested that such organ-specific expression may be associated with the PKM expression profile.
Table 1.
Validations of microRNAs (miRNAs) used in this study.
| miRNA gene name (ID) | MIR122 (406906) | MIR137 (406928) | MIR206 (406989) |
| Target gene name (ID) | PKM (5315) | PTBP1 (5725) | PTBP1 (5725) |
| Species name (ID) | Homo sapiens (9606) | Homo sapiens (9606) | Homo sapiens (9606) |
|
Genomic location of MTI
Nucleotide sequence |
5′ACACTCC | 5′AGCAATA | 5′CATTCCA |
| Location | 15:72199124–72199130 | 19:811468–811474 | 19:810871–810876 |
| Location within a part of a gene | 520–527 (location within 3′UTR) | 642–648 (location within 3′UTR) | 45–51 (location within 3′UTR) |
|
Existence of previous report (searching with miRTarBase) |
Yes | No [18] | No |
| Methods for experimental validation in this study | Luciferase reporter assay Western blot analysis |
Luciferase reporter assay Western blot analysis |
Luciferase reporter assay Western blot analysis |
| Experimental materials used in this study | HuH-7 and Hep 3B cell lines |
U-251 and U-87 cell lines |
RD and KYM-1 cell lines |
PKM = pyruvate kinase M1/2; PTBP1 = polypyrimidine tract binding protein 1; 3′UTR = 3′ untranslated region; MIR = microRNA (gene symbol); HuH-7 and Hep 3B = human hepatocellular carcinoma cell lines; U-251 and U-87 = human glioblastoma cell lines; RD and KYM-1 = human rhabdomyosarcoma cell lines; MTI = microRNA-target interaction.
2.3. MIR137 and 206 Directly Bound to Polypyrimidine Tract Binding Protein 1 (PTBP1) and Regulated Expression of the Pyruvate Kinase M 1/2 (PKM) Isoform
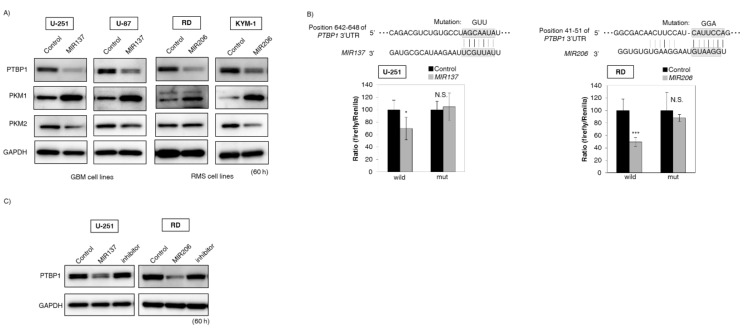
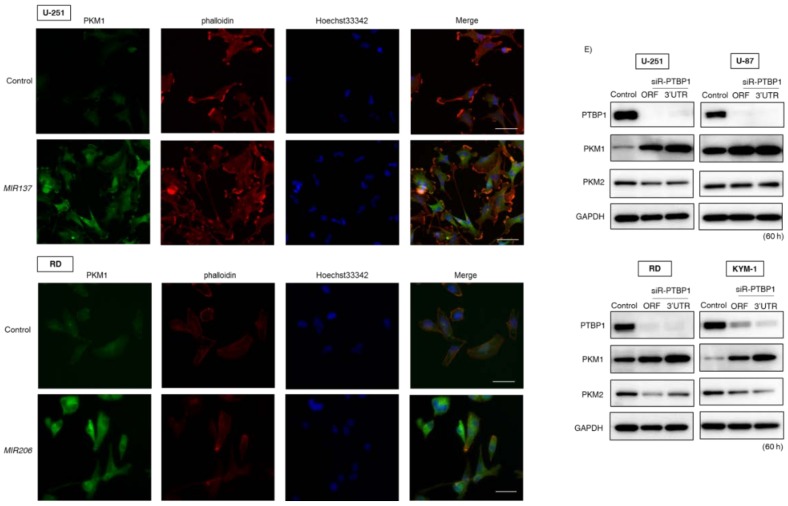
In order to further clarify the relationship between the expression of the miRNAs and PTBP1, we examined whether brain-specific MIR137 and muscle-specific MIR206 could directly bind to PTBP1. We selected glioblastoma (GBM) cell lines, i.e., U-251 and U-87, in the case of brain-specific MIR137 and rhabdomyosarcoma (RMS) RD and KYM-1 cell lines in the case of muscle-specific MIR206. As a result, the protein expression levels of PTBP1 were downregulated in these miRNA-treated cells tested. Also, the expression level of PKM1 was increased and that of PKM2 was decreased as expected (Figure 2A). The luciferase reporter activity of wild-type pMIR-PTBP1 was significantly inhibited after the introduction of brain-specific MIR137 into U-251 cells or muscle-specific MIR206 into RD cells. On the other hand, mutation of the PTBP1 3′ UTR-binding site abolished the inhibitory effect of these miRNAs (Figure 2B). In addition, treatment with an inhibitor of these miRNAs reversed the expression level of PTBP1 in both U-251 and RD cells (Figure 2C). Moreover, immunofluorescence (IFC) indicated increased signal intensity of PKM1 in these miRNAs-treated cells (Figure 2D). Furthermore, Western blotting analysis showed that knockdown of PTBP1 gave similar results; i.e., an increase in the PKM1/PKM2 ratio (by switching from PKM2 to PKM1) was observed in all cells tested (Figure 2E). These findings suggested that these organ-specific miRNAs directly bound to PTBP1 mRNA and regulated the expression of the PKM isoform.
Figure 2.
MIR137 and 206 regulated PKM isoform expression through the direct targeting of PTBP1 mRNA. (A) Protein expression levels of PTBP1, PKM1, and PKM2 at 60 h after the transfection with MIR137 or 206. Human glioblastoma (GBM) cell lines (U-251 and U-87) were transfected with brain-specific MIR137 (20 nM). Also, rhabdomyosarcoma (RMS) cell lines (RD and KYM-1) were transfected with muscle-specific MIR206 (20 nM); (B) Luciferase activities after co-transfection with control or MIR137 or 206 and wild-type or mutant-type pMIR vectors having the predicted MIR137 or 206 binding site in the 3′UTR of PTBP1. The upper panel shows the region of the 3′ UTR of human PTBP1 mRNA complementary to the mature MIR137 or 206. The box indicates the predicted binding sites for MIR137 or 206. The left panel is the case of MIR137 transfected with U-251 cells. The right panel is the case of MIR206 transfected with RD cells. Effects were assessed at 24 h after the transfection (20 nM); (C) Protein expression levels of PTBP1 in U-251 and RD cells after combined treatment with antagoMIR137/MIR137 or antagoMIR206/MIR206. Control lane: treatment with control miRNA (20 nM). MIR137 or 206 lane: treatment with MIR137 or 206 (20 nM). Inhibitor lane: treatment with MIR137 or 206 (10 nM) + antagoMIR137 or 206 (10 nM). Effects were assessed at 60 h after the transfection; (D) Immunofluorescence of PKM1 at 60 h after transfection of U-251 cells with MIR137 or transfection of RD cells with MIR206. Upper panels, treatment with control miRNA (20 nM); Lower panels, treatment with MIR137 or 206 (20 nM). PKM1 is dyed green; cell membrane, red; and nuclei, blue. Representative photographs from two experiments are shown. Scale bar = 50 µm; (E) Protein expression levels of the PKM isoform in GBM and RMS cells at 60 h after siR-PTBP1 transfection (5 nM). ORF: open reading frame. Results are presented as the mean ± SD; * p < 0.05, *** p < 0.001; N.S., not statistically significant.
2.4. MIR122 Directly Bound to PKM and Negatively Regulated PKM2 Expression
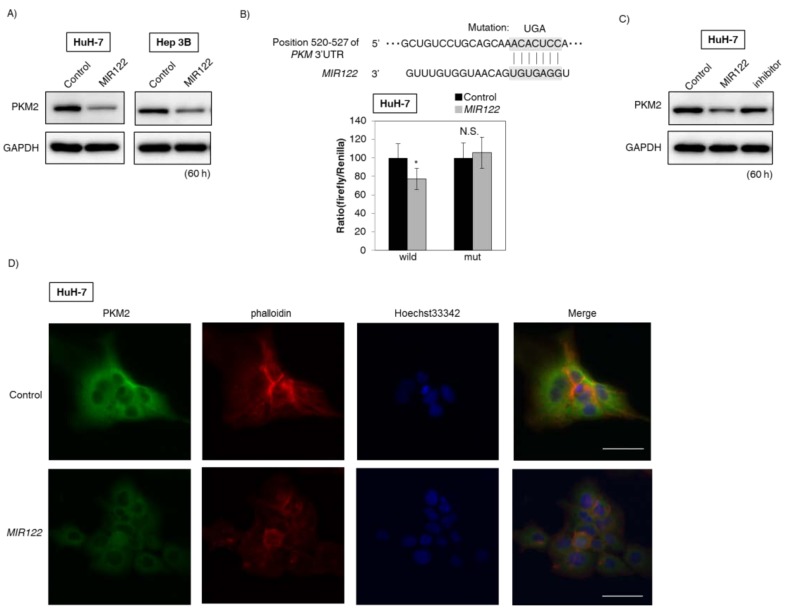
Next, we examined whether liver-specific MIR122 would directly bind to PKM. We selected hepatocellular carcinoma (HCC) cell lines for these experiments. As shown in Figure 3A, protein expression levels of PKM2 were downregulated in MIR122-treated HCC cells. In addition, the luciferase reporter activity of wild-type pMIR-PKM was significantly inhibited after the introduction of liver-specific MIR122 into HuH-7 cells. On the other hand, mutation of the PKM 3′UTR-binding site abolished the inhibitory effect of liver-specific MIR122 (Figure 3B). Moreover, treatment with an inhibitor of MIR122 reversed the expression level of PKM2 in HuH-7 cells (Figure 3C). In addition, IFC indicated that the signal intensity of PKM2 was decreased in the MIR122-treated HuH-7 cells (Figure 3D). These findings suggested that liver-specific MIR122 directly bound to PKM mRNA and negatively regulated PKM2 expression.
Figure 3.
MIR122 negatively regulated PKM expression through the direct targeting of PKM mRNA. (A) Protein expression levels of PKM2 at 60 h after the transfection of hepatocellular carcinoma (HCC) cell lines (HuH-7 and Hep 3B) with MIR122 (20 nM); (B) Luciferase activities after co-transfection with control or MIR122 and wild-type or mutant-type pMIR vectors having the predicted MIR122 binding site in the 3′-UTR of PKM. The upper panel shows the region of the 3′UTR of human PKM mRNA complementary to the mature MIR122. The box indicates the predicted binding sites for MIR122. Effects were assessed at 24 h after the transfection (20 nM); (C) Protein expression levels of PKM2 in HuH-7 cells after combined treatment with antagoMIR122/MIR122. Control lane: treatment with control miRNA (20 nM). MIR122 lane: treatment with MIR122 (20 nM). Inhibitor lane: treatment with MIR122 (10 nM) + antagoMIR122 (10 nM). Effects were assessed at 60 h after the transfection; (D) Immunofluorescence of PKM2 at 60 h after transfection of HuH-7 cells with MIR122 (20 nM). Upper panel, treatment with control miRNA; Lower panel, treatment with MIR122. PKM2 is dyed green; cell membrane, red; and nuclei, blue. Representative photographs from two experiments are shown. Scale bar = 50 µm. Results are presented as the mean ± SD; * p < 0.05; N.S., not statistically significant.
2.5. Dysregulation of These Organ-Specific miRNAs Contributed to Carcinogenesis through the Modulation of Expression of Warburg-Effect-Associated Genes
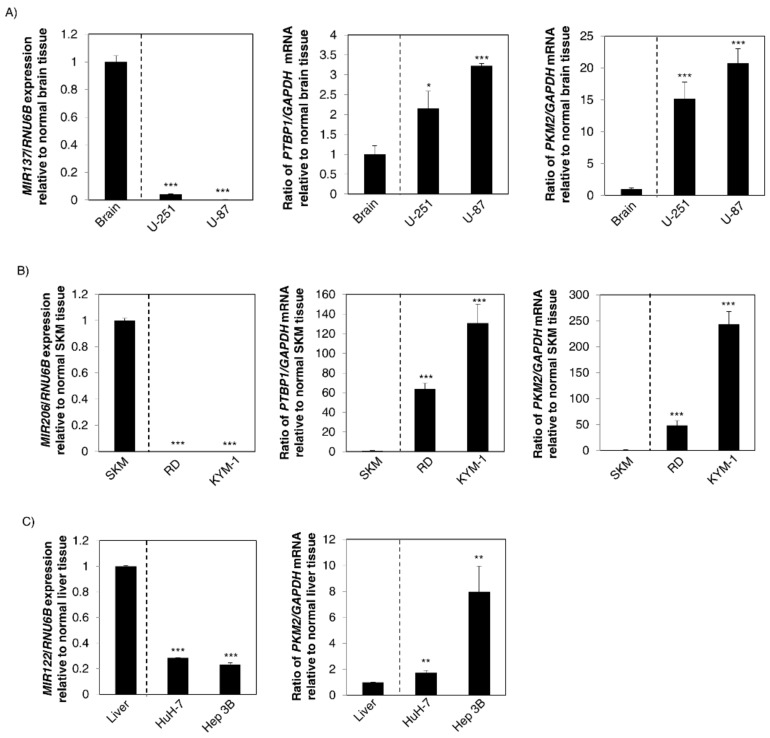
To clarify the importance of these miRNAs in carcinogenesis, we examined the association between the expression levels of these organ-specific miRNAs and the Warburg-effect-related genes, i.e., PTBP1 and PKM2. As shown in Figure 4A, brain-specific MIR137 was downregulated in GBM cell lines compared with its expression in the normal brain tissue; and mRNA expression levels of PTBP1 and PKM2 were upregulated. For MIR206, similar findings were also observed for the RMS cell lines tested (Figure 4B). On the other hand, liver-specific MIR122 was downregulated in HCC cell lines compared with its expression in normal liver tissue; and the mRNA expression level of PKM2 was increased (Figure 4C). These findings suggested that dysregulation of these organ-specific miRNAs was strongly associated with the Warburg effect through the change in PKM2 expression (Figure 5).
Figure 4.
Downregulation of these organ-specific miRNAs contributed to carcinogenesis through upregulation of Warburg-effect-related genes. (A) Left panel: Expression levels of MIR137 in the normal human brain tissues and GBM cell lines; Middle panel: mRNA expression levels of PTBP1 in the same samples; Right panel: That of PKM2 mRNA expression; (B) Left panel: Expression levels of MIR206 in the normal human skeletal muscle tissues and RMS cell lines; Middle panel: The mRNA expression levels of PTBP1 in the same samples; Right panel: That of PKM2 mRNA expression; SKM: skeletal muscle; (C) Left panel: Expression levels ofMIR122 in the normal human liver tissues and HCC cell lines; Right panel: mRNA expression levels of PKM2 in the same samples. Results are presented as the mean ± SD; * p < 0.05; ** p < 0.01; *** p < 0.001.
Figure 5.
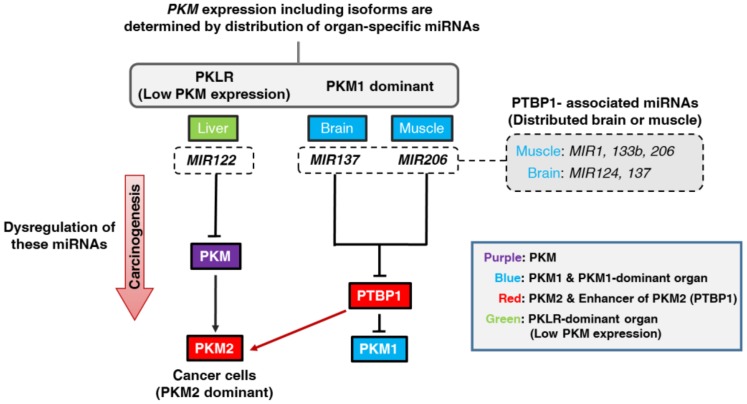
Schematic diagram of our study findings. PKLR and PKM isoforms showed organ specificity; i.e., PKLR was dominantly expressed in the liver. PKM1 was dominantly expressed in the brain and muscle. In the liver, liver-specific MIR122 suppressed the PKM expression by dominant distribution of MIR122. In the brain and muscle, brain-specific MIR137 and muscle-specific MIR206 induced the upregulation of the PKM1 expression through the suppression of the PTBP1 expression. Previously, we proved that muscle-specific MIR1 and 133b and brain-specific MIR124 had the same mechanism for PKM isoform expression. These organ-specific miRNAs, which targeted PTBP1 directly, regulate PKM isoform expression by their tissue distribution. We named these miRNAs PTBP1-associated miRNAs. Also, dysregulation of these organ-specific miRNAs strongly contributed to carcinogenesis by promoting PKM2 expression in cancer cells (Red arrow). Colors indicate the following: Purple; PKM gene, Blue; PKM1 and PKM1-dominant organ, Red; PKM2 and enhancer of PKM2 (PTBP1), Green; PKLR-dominant (low PKM expression) organ.
3. Discussion
In this study, we showed that brain-specific MIR137 and muscle-specific MIR206 enabled PKM1-dominant expression predominantly in brain and skeletal muscle tissues through the direct targeting of PTBP1 mRNA (Figure 2). Recently, we indicated that brain-specific MIR124 and muscle-specific MIR1 and 133b have the same machinery for the processing of PKM isoform expression [8,19]. Moreover, presently we showed that liver-specific MIR122 suppressed both PKM1 and PKM2 expression in normal liver tissue by binding to the 3′UTR of PKM (Figure 3). Based on these findings, we concluded that levels of PK enzymes, especially in the case of PKM expression, were strongly determined by organ-specific miRNAs. This is one of the most typical patterns of the miRNA system to determine tissue characteristics. Such organ specificities would have some biological significance for the fate or commitment of tissue differentiation. Of course, further investigations are required, because all of the miRNAs that regulate PTBP1 expression, such as MIR9, 153, and 200bc, were not examined in our studies. The association between these organ-specific miRNAs and carcinogenesis is extremely important. For example, it was reported earlier that MIR124 and 137, which are predominant in the brain, are frequently downregulated in brain tumors, such as GBM [20], and that downregulation of MIR122 induces HCC carcinogenesis [21]. Similarly, dysregulation of muscle-specific miRNAs, such as MIR1, 133 and 206, contribute to the development of sarcomas, including RMS [19,22]. Furthermore, we indicated that dysregulated expression of MIR145, which is predominant in the gastrointestinal tract, contributes significantly to the development of gastrointestinal cancer [23,24]. Namely, dysregulation of miRNAs, whose normal expression enables an organ to acquire its characteristics, most deeply contributes to carcinogenesis in these tissues. Hence, we should focus on dysregulation of these organ-specific miRNAs to better understand the mechanisms of organ-specific carcinogenesis.
Our recent study indicated that PTBP1 functions as a novel oncogene and that PTBP1 promotes the Warburg effect in various types of cancer [15,16,25]. Previously, PTBP1 was recognized as an oncogene in GBM [3,26] and as having an onco-genetic function in breast cancer cell lines and ovarian tumors [27,28]. It has been reported that c-Myc is a positive regulator of PTBP1 [3]. However, we found earlier that various organ-specific miRNAs negatively regulate PTBP1 expression both directly and indirectly [8,16,29]. The induction of switching of a PKM isoform from PKM2 to PKM1 in various cancer cells by ectopic expression of these miRNAs supports our opinion [15,18,30]. Of course, other splicers, such as HNRNPA1 and Serine and argininerich splicing factor 3 (SRSF3), work jointly with PTBP1 in the process of PKM isoform expression [3,31]. Actually, in the case of HCC cell lines, the increase in PKM2 expression in HuH-7 cells was relatively less than that of it in Hep 3B cells compared with the decrease in miR-122 expression (Figure 4C). Hence, further investigation of the association between other such splicers of the PKM gene and miRNAs will more fully clarify the mechanisms of PKM isoform expression. Understanding of the mechanism underlying the maintenance of PKM isoform expression is essential for clarifying the mechanism of carcinogenesis. Also, such understanding will contribute to clarifying the conditions for advanced-stage cancer, such as metastasis [32]. We strongly believe that our results contribute to cancer diagnosis and will be useful for cancer-specific therapy for the Warburg effect in the near future.
4. Materials and Methods
4.1. miRNA Validation and Terminology
Firstly, the Target Scan 7.1 database (http://www.targetscan.org/) was used in order to find the binding sites of each miRNA and targeting gene [17]. Also, searching in more detail for associations between them was performed with miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php). The validations are summarized in Table 1. The terminology of miRNAs was based on miRNA nomenclature guidelines [33,34].
4.2. Samples from Human and Mouse
Human RNA samples were obtained from Clontech (TAKARA BIO INC., Shiga, Japan) or Biochain (BioChain Institute Inc., Newark, CA, USA). BALB/cSLC-nu/nu (nude) mice were obtained from Japan SLC (Shizuoka, Japan). Animal experimental protocols were approved by the Committee for Animal Research and Welfare of Gifu University (11021, 10 February 2016).
4.3. Cell Culture
Human glioblastoma (GBM) cell lines U-251 and U-87, those of rhabdomyosarcoma (RMS) cell line RD and KYM-1, and those of hepatocellular carcinoma (HCC) cell line HuH-7 and Hep 3B were used in this study. Human rhabdomyosarcoma cell lines RD and KYM-1 and human hepatocellular carcinoma cell line HuH-7 were obtained from JCRB (Japanese Collection of Research Bioresources) Cell Bank. Human hepatocellular carcinoma cell line Hep 3B was a gift from Nobuhiko Tanigawa (an Emeritus Professor of our department). U-251 and U-87 were provided by Atsushi Natsume (Department of Neurosurgery Nagoya University, Graduate School of Medicine). U-251, U-87, HuH-7, and Hep B3 cells were cultured in DMEM medium. RD cells were grown in Eagle’s minimal essential medium, and KYM-1 cells in a mixture of Ham’s F12 medium and Dulbecco’s modified Eagle’s medium (1:1). Ten percent (v/v) heat-inactivated FBS (Sigma-Aldrich, St. Louis, MO, USA) and 2 mM l-glutamine were supplied to each medium. The temperature and atmosphere were maintained at 37 °C and 95% air with 5% CO2, respectively.
4.4. Western Blotting
Cells of each type were homogenized in cold-lysis buffer. The composition of it was given in previous reports of ours [8,35]. Protease Inhibitor Cocktail (1%) (Sigma-Aldrich) was added to it and reacted for 15 min on ice. Centrifugation (13,000 rpm for 20 min at 4 °C) was performed and the supernatants were collected. Protein contents were calculated with a DC Protein assay kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Six to 10 micrograms of samples were separated by SDS-PAGE using appropriate polyacrylamide gels (10.0% or 12.5%; Wako Pure Chemical Industries, Ltd., Osaka, Japan). Also, separated samples were electro-blotted onto a PVDF membrane (PerkinElmer Life Sciences, Inc., MA, USA or Bio-Rad Laboratories, Inc.). The blockage of nonspecific binding sites was performed with 5% nonfat milk (Cell Signaling Technology, Inc., Danvers, MA, USA) in PBS containing 0.1% Tween 20 (TBS-T). The membrane was incubated for the appropriate time (several hours to 1 day) at 4 °C with primary antibodies. After that, the membrane was washed with TBS-T, after which secondary antibodies were added. The immunoreaction proceeded over a 1-h period at room temperature. Finally, the immunoblots were visualized with LuminataTM Forte Western HRP Substrate (Millipore Corporation, Billerica, MA, USA). We detected protein expression by using an Image Quant Luminescent Image Analyzer LAS-4000 (Fuji Photo Film Co., Tokyo, Japan) or Fusion-FX7 (Vilber Lourmat, Marne-la-Vallée, France). Primary antibodies used were as follows: anti-PKM1, -PKM2, -PTBP1, -β-actin, and -GAPDH (Cell Signaling Technology, Inc., Danvers, MA, USA) and anti-PKLR (GeneTex, Inc., Irvine, CA, USA). Secondary antibodies (HRP-conjugated goat anti-rabbit and horse anti-mouse IgG) were obtained from Cell Signaling Technology. GAPDH or β-actin was used as an internal control according to our previous report [8].
4.5. Real-Time Reverse Transcription-PCR
MiRNA was isolated from cultured cells by using a NucleoSpin microRNA isolation kit (Takara Bio Inc., Shiga, Japan). The relative expression levels were calculated by the ΔΔCt method. Each value of ΔΔCt was determined by use of a Thermal Cycler Dice Real Time System II model TP900/960 or TP870 (Takara Bio Inc.). The primers for PTBP1, PKM1, PKM2, and GAPDH were the following: PTBP1-sense, 5′-ATCAGGCCTTCATCGAGATGCACA-3′, and PTBP1-antisense, 5′-TGTCTTGAGCTCCTTGTGGTTGGA-3′; PKM1-sense, 5′-CGAGCCTCAAGTCACTCCAC-3′, and PKM1-antisense, 5′-GTGAGCAGACCTGCCAGACT-3′; PKM2-sense, 5′-ATTATTTGAGGAACTCCGCCGCCT-3′, and PKM2-antisense, 5′-ATTCCGGGTCACAGCAATGATGG-3′; GAPDH-sense, 5′-CCACCCATGGCAAATTCCATGGCA-3′, and GAPDH-antisense, 5′-TCTAGACGGCAGGTCAGGTCCACC-3′. GAPDH and RNU6B were used as internal controls. Normalization was not performed in the case of expression profiles of organ-specific miRNAs.
4.6. Transfection Experiments
All cells were seeded in 6-well plates at a concentration of 0.5 × 105 per well (10–30% confluence) on the day before the transfection. Each transfection was performed by using LipofectamineTM RNAiMAX (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. The mature types of MIR122, 137, and 206 (mirVanaTM miRNA mimic; Ambion, CA, USA), antagoMIR122, 137, and 206 (mirVanaTM miRNA inhibitor; Ambion), or siRNA for PTBP1 (siR-PTBP1; Invitrogen, Carlsbad, CA, USA) were used for the transfection of the cells. The sequence of each miRNA or siRNA was as indicated below. The sequence of the mature type of MIR122 used in this study was 5′-UGGAGUGUGACAAUGGUGUUUG-3′; that of MIR137, 5′-UUAUUGCUUAAGAAUACGCGUAG-3′; that of MIR206, 5′-UGGAAUGUAAGGAAGUGUGUGG-3′; that of siR-PTBP1 for the open reading frame region, 5′-UGUCAUUUCCGUUUGCUGCAGAAGC-3′; and that of the 3′UTR region, 5′-AUCUCUGGUCUGCUAAGGUCACUUC-3′. The nonspecific miRNA (HSS, Hokkaido, Japan) sequence of 5′-GUAGGAGUAGUGAAAGGCC-3′ was used as a control for nonspecific effects [25,36].
4.7. Luciferase Reporter Assay
By searching the Target Scan 7.1 database (http://www.targetscan.org/) to find algorithm-based binding sites of MIR137 or 206, we found the predicted binding site to be at position 642–648 for MIR137 and at 45–51 for MIR206 in the 3′UTR of PTBP1 mRNA [17]. Also, we found the predicted binding site to be at position 520–527 for MIR122 in the 3′UTR of PKM mRNA. The sequence region containing the putative binding sequence of MIR137, 206, or 122 was inserted into a pMIR-REPORTTM Luciferase miRNA Expression Reporter Vector (Applied Biosystems Inc., Foster City, CA, USA) according to the manufacturer’s protocol. Moreover, we made other pMIR constructs, one encompassing a mutated seed sequence for MIR137 (wild-type, AGCAAUA; mutant, AGGUUUA), another for 206 (wild-type, CAUUCCAG; mutant, CAGGACAG), and another for 122 (wild-type, ACACUCC; mutant, ACUGACC) by using a PrimeSTAR® Mutagenesis Basal Kit (Takara Bio Inc.). The mutation of each vector was confirmed by sequence analysis. A pRL-TK Renilla Luciferase Reporter vector (Promega Corporation, Madison, WI, USA) was used as an internal control vector. Cells were seeded into 96-well plates at a concentration of 0.1 × 104 per well on the day before the transfection. Each cell type was co-transfected with a reporter vector (wild-type or mutated type; 0.01 μg/well) and MIR137, 206, 122, or a nonspecific non-coding siRNA (Dharmacon, Tokyo, Japan). Luciferase activities were measured by using a Dual-Glo Luciferase Assay System (Promega Corporation, Madison, WI, USA). These activities were calculated as the ratios of firefly luciferase/Renilla luciferase to define luciferase activity as in the case of our previous studies [15].
4.8. Immunofluorescence (IFC) Study
Cells of each cell type were seeded into the wells of a Lab-Tek II Chamber Slide System (Thermo Fisher Scientific Inc., Waltham, MA, USA). After transfection with a given miRNA including nonspecific miRNA (Control miRNA), the cells were immunostained with anti-PKM1 or -PKM2 antibody according to the immunofluorescence protocol of Cell Signaling Technology. The nuclei were stained with Hoechet33342 (Sigma-Aldrich), and for actin labeling the cells were incubated with a fluorescent F-actin probe, Rhodamine Phalloidin (Cytoskeleton Inc., Denver, CO, USA). The cells were observed with a BIOREVO fluorescence microscope (Keyence, Osaka, Japan).
4.9. Statistics
The statistical examinations were performed in triplicate. The two-sided Student’s t-test was used for determining the statistical significance of differences. The mean ± standard deviation was indicated in necessary cases. Statistical significance was set at a p-value < 0.05.
Acknowledgments
This work was supported in part by JSPS KAKENHI Grant Number JP16H07344 and JP15K10120 and an Osaka Medical College (OMC) Internal Research Grant. We are grateful for strong technical support by Akiko Miyamoto in our laboratory and Rintaro Oide (Division of Research Equipment and Device of OMC). Also, we are thankful for the material support by Atsushi Natsume (Department of Neurosurgery, Nagoya University School of Medicine).
Abbreviations
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| GBM | glioblastoma |
| HCC | hepatocellular carcinoma |
| HNRNP | heterogeneous nuclear ribonucleoproteins |
| IFC | immunofluorescence |
| MIR | microRNA (gene symbol in human) |
| miRNA | microRNA |
| MTI | microRNA-target interaction |
| ORF | open reading frame |
| PKLR | pyruvate kinase L/R |
| PKM | pyruvate kinase M1/2 |
| PTBP1 | polypyrimidine tract binding protein 1 |
| RMS | rhabdomyosarcoma |
| SRSF3 | Serine and arginine-rich splicing factor 3 |
| 3′UTR | 3′ untranslated region |
Author Contributions
Conception and design of the study: Kohei Taniguchi; Acquisition of data: Kohei Taniguchi and Nobuhiko Sugito. Analysis and interpretation of the data: Kohei Taniguchi, Haruka Shinohara, Yuki Kuranaga, Yosuke Inomata, Kazumasa Komura, Kazuhisa Uchiyama, and Yukihiro Akao. Material and financial support: Kazuhisa Uchiyama and Yukihiro Akao. Writing, review, and/or revision of the manuscript: Kohei Taniguchi, Yosuke Inomata, Kazumasa Komura, and Yukihiro Akao. Study supervision: Kazuhisa Uchiyama and Yukihiro Akao.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Noguchi T., Inoue H., Tanaka T. The M1- and M2-type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing. J. Biol. Chem. 1986;261:13807–13812. [PubMed] [Google Scholar]
- 2.Clower C.V., Chatterjee D., Wang Z., Cantley L.C., Heiden M.G.V., Krainer A.R. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc. Natl. Acad. Sci. USA. 2010;107:1894–1899. doi: 10.1073/pnas.0914845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.David C.J., Chen M., Assanah M., Canoll P., Manley J.L. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dayton T.L., Jacks T., Heiden M.G.V. PKM2, cancer metabolism, and the road ahead. EMBO Rep. 2016;17:1721–1730. doi: 10.15252/embr.201643300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He X., Du S., Lei T., Li X., Liu Y., Wang H., Tong R., Wang Y. PKM2 in carcinogenesis and oncotherapy. Oncotarget. 2017;8:110656–110670. doi: 10.18632/oncotarget.22529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu M.C., Hung W.C. Pyruvate kinase M2 fuels multiple aspects of cancer cells: From cellular metabolism, transcriptional regulation to extracellular signaling. Mol. Cancer. 2018;17:35. doi: 10.1186/s12943-018-0791-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen M., Zhang J., Manley J.L. Turning on a fuel switch of cancer: HnRNP proteins regulate alternative splicing of pyruvate kinase mRNA. Cancer Res. 2010;70:8977–8980. doi: 10.1158/0008-5472.CAN-10-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taniguchi K., Ito Y., Sugito N., Kumazaki M., Shinohara H., Yamada N., Nakagawa Y., Sugiyama T., Futamura M., Otsuki Y., et al. Organ-specific PTB1-associated microRNAs determine expression of pyruvate kinase isoforms. Sci. Rep. 2015;5:8647. doi: 10.1038/srep08647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bluemlein K., Gruning N.M., Feichtinger R.G., Lehrach H., Kofler B., Ralser M. No evidence for a shift in pyruvate kinase PKM1 to PKM2 expression during tumorigenesis. Oncotarget. 2011;2:393–400. doi: 10.18632/oncotarget.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noguchi T., Yamada K., Inoue H., Matsuda T., Tanaka T. The L- and R-type isozymes of rat pyruvate kinase are produced from a single gene by use of different promoters. J. Biol. Chem. 1987;262:14366–14371. [PubMed] [Google Scholar]
- 11.Dombrauckas J.D., Santarsiero B.D., Mesecar A.D. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44:9417–9429. doi: 10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- 12.Larochelle A., Magny P., Tremblay S., Medicis E.E. Erythropoiesis: Pyruvate kinase deficiency which causes nonspherocytic hemolytic anemia: The gene and its mutations. Hematology. 1999;4:77–87. doi: 10.1080/10245332.1999.11746433. [DOI] [PubMed] [Google Scholar]
- 13.Zeng Z.L., Lin X.L., Tan L.L., Liu Y.M., Qu K., Wang Z. MicroRNAs: Important regulators of induced pluripotent stem cell generation and differentiation. Stem Cell Rev. 2018;14:71–81. doi: 10.1007/s12015-017-9785-6. [DOI] [PubMed] [Google Scholar]
- 14.Fang Y., Zhang L., Li Z., Li Y., Huang C., Lu X. MicroRNAs in DNA damage response, carcinogenesis, and chemoresistance. Int. Rev. Cell Mol. Biol. 2017;333:1–49. doi: 10.1016/bs.ircmb.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Taniguchi K., Sakai M., Sugito N., Kumazaki M., Shinohara H., Yamada N., Nakayama T., Ueda H., Nakagawa Y., Ito Y., et al. PTBP1-associated microRNA-1 and -133b suppress the Warburg effect in colorectal tumors. Oncotarget. 2016;7:18940–18952. doi: 10.18632/oncotarget.8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugiyama T., Taniguchi K., Matsuhashi N., Tajirika T., Futamura M., Takai T., Akao Y., Yoshida K. MiR-133b inhibits growth of human gastric cancer cells by silencing pyruvate kinase muscle-splicer polypyrimidine tract-binding protein 1. Cancer Sci. 2016;107:1767–1775. doi: 10.1111/cas.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugito N., Taniguchi K., Kuranaga Y., Ohishi M., Soga T., Ito Y., Miyachi M., Kikuchi K., Hosoi H., Akao Y. Cancer-specific energy metabolism in rhabdomyosarcoma cells is regulated by microRNA. Nucleic Acid Ther. 2017;27:365–377. doi: 10.1089/nat.2017.0673. [DOI] [PubMed] [Google Scholar]
- 19.Sun Y., Zhao X., Zhou Y., Hu Y. miR-124, miR-137 and miR-340 regulate colorectal cancer growth via inhibition of the Warburg effect. Oncol. Rep. 2012;28:1346–1352. doi: 10.3892/or.2012.1958. [DOI] [PubMed] [Google Scholar]
- 20.Silber J., Lim D.A., Petritsch C., Persson A.I., Maunakea A.K., Yu M., Vandenberg S.R., Ginzinger D.G., James C.D., Costello J.F., et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai W.C., Hsu S.D., Hsu C.S., Lai T.C., Chen S.J., Shen R., Huang Y., Chen H.C., Lee C.H., Tsai T.F., et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J. Clin. Investig. 2012;122:2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Missiaglia E., Shepherd C.J., Patel S., Thway K., Pierron G., Pritchard-Jones K., Renard M., Sciot R., Rao P., Oberlin O., et al. MicroRNA-206 expression levels correlate with clinical behaviour of rhabdomyosarcomas. Br. J. Cancer. 2010;102:1769–1777. doi: 10.1038/sj.bjc.6605684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akao Y., Nakagawa Y., Hirata I., Iio A., Itoh T., Kojima K., Nakashima R., Kitade Y., Naoe T. Role of anti-oncomirs miR-143 and -145 in human colorectal tumors. Cancer Gene Ther. 2010;17:398–408. doi: 10.1038/cgt.2009.88. [DOI] [PubMed] [Google Scholar]
- 24.Takagi T., Iio A., Nakagawa Y., Naoe T., Tanigawa N., Akao Y. Decreased expression of microRNA-143 and -145 in human gastric cancers. Oncology. 2009;77:12–21. doi: 10.1159/000218166. [DOI] [PubMed] [Google Scholar]
- 25.Takai T., Yoshikawa Y., Inamoto T., Minami K., Taniguchi K., Sugito N., Kuranaga Y., Shinohara H., Kumazaki M., Tsujino T., et al. A novel combination RNAi toward warburg effect by replacement with miR-145 and silencing of PTBP1 induces apoptotic cell death in bladder cancer cells. Int. J. Mol. Sci. 2017;18:179. doi: 10.3390/ijms18010179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin W., McCutcheon I.E., Fuller G.N., Huang E.S., Cote G.J. Fibroblast growth factor receptor-1 alpha-exon exclusion and polypyrimidine tract-binding protein in glioblastoma multiforme tumors. Cancer Res. 2000;60:1221–1224. [PubMed] [Google Scholar]
- 27.He X., Arslan A.D., Ho T.T., Yuan C., Stampfer M.R., Beck W.T. Involvement of polypyrimidine tract-binding protein (PTBP1) in maintaining breast cancer cell growth and malignant properties. Oncogenesis. 2014;3:e84. doi: 10.1038/oncsis.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He X., Pool M., Darcy K.M., Lim S.B., Auersperg N., Coon J.S., Beck W.T. Knockdown of polypyrimidine tract-binding protein suppresses ovarian tumor cell growth and invasiveness in vitro. Oncogene. 2007;26:4961–4968. doi: 10.1038/sj.onc.1210307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minami K., Taniguchi K., Sugito N., Kuranaga Y., Inamoto T., Takahara K., Takai T., Yoshikawa Y., Kiyama S., Akao Y., et al. MiR-145 negatively regulates Warburg effect by silencing KLF4 and PTBP1 in bladder cancer cells. Oncotarget. 2017;8:33064–33077. doi: 10.18632/oncotarget.16524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taniguchi K., Sugito N., Kumazaki M., Shinohara H., Yamada N., Nakagawa Y., Ito Y., Otsuki Y., Uno B., Uchiyama K., et al. MicroRNA-124 inhibits cancer cell growth through PTB1/PKM1/PKM2 feedback cascade in colorectal cancer. Cancer Lett. 2015;363:17–27. doi: 10.1016/j.canlet.2015.03.026. [DOI] [PubMed] [Google Scholar]
- 31.Wang Z., Chatterjee D., Jeon H.Y., Akerman M., Heiden M.G.V., Cantley L.C., Krainer A.R. Exon-centric regulation of pyruvate kinase M alternative splicing via mutually exclusive exons. J. Mol. Cell Biol. 2012;4:79–87. doi: 10.1093/jmcb/mjr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fong M.Y., Zhou W., Liu L., Alontaga A.Y., Chandra M., Ashby J., Chow A., O’Connor S.T., Li S., Chin A.R., et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 2015;17:183–194. doi: 10.1038/ncb3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Desvignes T., Batzel P., Berezikov E., Eilbeck K., Eppig J.T., McAndrews M.S., Singer A., Postlethwait J.H. miRNA nomenclature: A view incorporating genetic origins, biosynthetic pathways, and sequence variants. Trends Genet. 2015;31:613–626. doi: 10.1016/j.tig.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piletic K., Kunej T. Minimal standards for reporting microRNA: Target interactions. OMICS. 2017;21:197–206. doi: 10.1089/omi.2017.0023. [DOI] [PubMed] [Google Scholar]
- 35.Taniguchi K., Iwatsuki A., Sugito N., Shinohara H., Kuranaga Y., Oshikawa Y., Tajirika T., Futamura M., Yoshida K., Uchiyama K., et al. Oncogene RNA helicase DDX6 promotes the process of c-Myc expression in gastric cancer cells. Mol. Carcinog. 2018;57:579–589. doi: 10.1002/mc.22781. [DOI] [PubMed] [Google Scholar]
- 36.Inamoto T., Taniguchi K., Takahara K., Iwatsuki A., Takai T., Komura K., Yoshikawa Y., Uchimoto T., Saito K., Tanda N., et al. Intravesical administration of exogenous microRNA-145 as a therapy for mouse orthotopic human bladder cancer xenograft. Oncotarget. 2015;6:21628–21635. doi: 10.18632/oncotarget.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]