Abstract
Colorectal cancer (CRC) is among the most common cancers. In fact, it is placed in the third place among the most diagnosed cancer in men, after lung and prostate cancer, and in the second one for the most diagnosed cancer in women, following breast cancer. Moreover, its high mortality rates classifies it among the leading causes of cancer-related death worldwide. Thus, in order to help clinicians to optimize their practice, it is crucial to introduce more effective tools that will improve not only early diagnosis, but also prediction of the most likely progression of the disease and response to chemotherapy. In that way, they will be able to decrease both morbidity and mortality of their patients. In accordance with that, colon cancer research has described numerous biomarkers for diagnostic, prognostic and predictive purposes that either alone or as part of a panel would help improve patient's clinical management. This review aims to describe the most accepted biomarkers among those proposed for use in CRC divided based on the clinical specimen that is examined (tissue, faeces or blood) along with their restrictions. Lastly, new insight in CRC monitoring will be discussed presenting promising emerging biomarkers (telomerase activity, telomere length and micronuclei frequency).
Keywords: colorectal cancer, diagnosis, prognosis, prediction, biomarkers, DNA markers, non-DNA markers
1. Introduction
Colorectal cancer (CRC) is ranked as one of the most common types of cancer. In fact, according to data from the USA, for the male population it is ranked third of the most diagnosed, after lung and prostate cancer, and for the female population, it is ranked second, following breast cancer (1). Moreover, it is considered as one of the leading causes of cancer-related death worldwide (2,3) In the USA alone, CRC is responsible for the second greatest number of cancer-related deaths. As a matter of fact, the American Cancer Society estimated that for 2013 alone, the number of first diagnosed CRC cases and that of deaths due to CRC was as high as 142,820 and about 50,830, respectively (4). However, regardless of the fact that both public and medical awareness concerning CRC has risen during the last decade, approximately 50% of the patients referred with CRC, at the time of diagnosis present distant metastases. It is clear that CRC is a rather heterogeneous disease by means of its various clinical manifestations, biological behavior and in-tumor variety of mutations (5,6) making it a true challenge, not only to detect in an early stage, but also to treat or even manage in the long term. Nowadays, it is evident that CRC is a multifactorial/polygenic disease arising both due to epigenetic as well as genetic manifestations occurring in a number of genes with an unparalleled role for the maintenance of normal cellular homeostasis. Such genes may be tumor suppressor genes, oncogenes, mismatch repair genes and cell cycle regulating genes in colon mucosal cells (7). Intense research has highlighted the importance of three major molecular pathways as the main cause of these genetic alterations that in turn result in carcinogenesis: Chromosomal instability (CIN), microsatellite instability (MSI), and the CpG island methylation phenotype (CIMP) (5,8). All these pathways attribute to the transformation of an adenoma to carcinoma, a multistep carcinogenic process known as the adenoma-carcinoma sequence (9), which is thought to be a common site for all CRCs. An illustrated form of this process is presented in Fig. 1. However, the period between those two hallmarks provides a diagnostic window for the early detection of CRC. As a result, the current research trend aims to describe new markers for diagnosis (these markers are meant to be used for risk stratification and early detection of colorectal polyps), prognosis (these markers will give an indication of the likely progression of the disease) and prediction of the biological behavior of a certain therapeutic regimen, making the clinician able to transform the knowledge of the tumor biology into a personalized decision-making process for each patient (8,10,11). Therefore, the information coming from CRC biology combined with the assessment of serum and tissue markers with prognostic and predictive value, currently constitutes, the pillars in the treatment of early-stage cases as well in the clinical management of advanced disease offering new methods to estimate the therapeutic efficacy and the overall outcome. An overview of the current and potential biomarkers used in CRC is shown in Fig. 2. In addition to CRC biomarkers, novel molecular imaging techniques using hybrid positron emission tomography (PET)/computed tomography (CT) systems enable accurate initial staging, efficient assessment of treatment response, and follow-up, facilitating individualized treatment strategies in CRC patients (12). Furthermore, novel positron emitting radiopharmaceuticals together with hybrid PET/magnetic resonance (MR) systems, which provide enhanced soft tissue resolution and incremental diagnostic information from functional MR methods such as diffusion-weighted imaging, hold promise for increased accuracy in CRC staging, restaging, early detection of recurrence, and accurate treatment planning for radiotherapy (13).
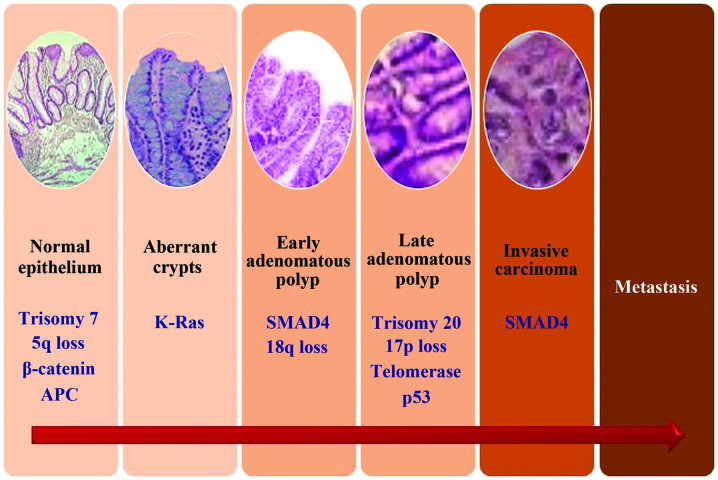
Figure 1.
Steps of CRC progression and a summarized rendering of the pathogenetic model. CRC, colorectal cancer; APC, adenomatous polyposis coli.
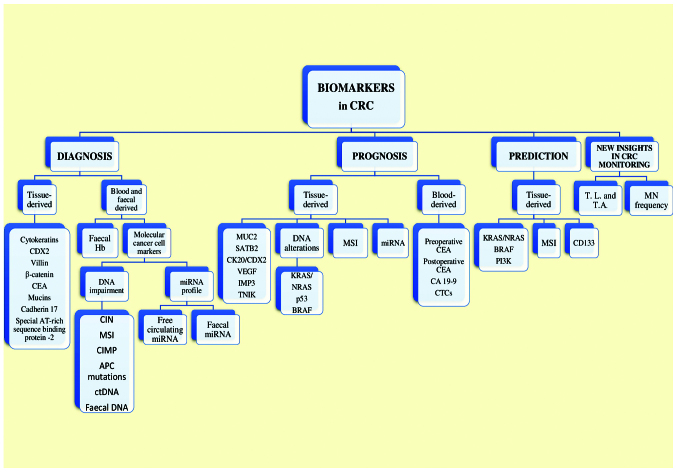
Figure 2.
A graphic overview of the current and potential biomarkers used in CRC. CRC, colorectal cancer; CDX2, caudal type homeobox 2; CEA, carcinoembryonic antigen; CIN, chromosomal instability; MSI, microsatellite instability; CIMP, CpG island methylation phenotype; APC, adenomatous polyposis coli; ctDNA, circulating tumor DNA; miRNA, microRNA; SATB2, special AT-rich sequence binding protein 2; CK, cytokeratin; VEGF, vascular endothelial growth factor; IMP3, insulin-like growth factor-II mRNA-binding protein 3; TNIK, Traf2- and Nck-interacting kinase; BRAF, B-rapidly accelerated fibro-sarcoma (proto-oncogene); MSI, microsatellite instability; CA 19-9, cancer antigen 19-9; CTCs, circulating tumor cells; PI3K, phosphoinositide 3-kinase; MN, micronuclei.
2. Diagnostic markers
General
Concerning the detection of CRC in the general population, the screening methods most commonly offered until now have included faecal occult blood testing (FOBT), flexible sigmoidoscopy and colonoscopy while CT colonography is a more recent addition to the CRC screening modalities (14). Even though screening has clearly proved to decrease the risk of CRC-associated mortality, screening effectiveness is restricted by limitations of test performance, inadequate access to CRC screening tests and loose screening compliance. Consequently, a great number of patients at the time of diagnosis present with locally advanced or metastatic disease, a phenomenon that is observed even among prosperous nations, including the United States (15). In accordance to that, CRC researchers focus their research on innovative ideas to identify molecular markers for the development of highly accurate, non-invasive screening tests for CRC in the hope of increasing the compliance of the population and to decrease potential unwanted side-effects which accompany the more invasive techniques. Several molecular classes have been tested for their potential use in CRC screening: DNA (16–18), proteins (19), messenger RNA (mRNA) (20) and microRNA (miRNA) (21–24), and have all proven to be quite promising in early phase biomarker studies (25). However, until now, only two tests (faecal haemoglobin and DNA-based markers) meet the pre-clinical and clinical criteria required for their efficient transduction from the laboratory to the clinical setting. In fact, recently, a multi-target stool DNA (MT-sDNA) test has proven better sensitivity, although with lower specificity, to faecal haemoglobin by immunochemical testing for the detection of curable-stage CRC and advanced adenomas while exhibiting overall cancer detection similar to colonoscopy (26). As a result, stool DNA testing was approved in the United States by FDA for general population screening of average risk, asymptomatic individuals in 2014.
Invasive techniques and diagnostic tissue markers
The most commonly invasive technique used, colonoscopy, provides the clinician a great advantage, to evaluate in real time the presence/absence of a polyp and to resect it at the same time, if possible, thus representing the standard tool of practice for CRC evaluation (10). Resecting a polyp gives the clinician an opportunity to use immunohistochemical (IHC) staining, typically used in the diagnosis of gastrointestinal neoplasms, in order to facilitate accurate tumor classification. By doing that, two main goals are set: The first one is to exclude morphologic mimickers or to identify the closest morphologically tissue or organ of origin in cases of metastatic carcinoma of unknown origin and thus confirm the diagnosis. The second one is to help estimate the most likely prognosis and even predict response to a given chemotherapy or novel molecular-targeted therapy (15,26). Diagnostic tissue biomarkers therefore provide additional and fundamental information that complement clinical colonoscopy findings.
Cytokeratins (CKs)
CKs, proteins expressed by epithelial cells, are members of the intermediate filaments family along with vimentin, desmin, neuro-filament, and glial-filamen. Numerous studies have attempted to identify a possible expression pattern of CKs and connect it with either the organ of origin (in order to determine whether it is a primary CRC) or with tumor progression. However, as more and more studies are conducted it is becoming clear that such a connection is not likely to be identified in the near future. To begin with, CK7 and CK20 are helpful when the clinician needs to differentiate metastases from CRC, which are usually CK7−/CK20+, from other tumors (27). CK20 almost selectively stains the normal gland cells of the colonic mucosa and Merkel cells while its expression is rarely may be seen in the urothelium or other mucosas (28,29). By contrast, CK7 is usually expressed in urinary bladder and female genital tract epithelia, mesothelium, normal lung, and, rarely, it may be observed in gastric and intestinal normal glands. However, the majority of researchers agree that it is not found in normal colonic mucosa (28,30). Based on these findings, the immunophenotype CK7/CK20 is used as a routine in order to differentiate CK20-expressing metastasis of colorectal adenocarcinomas from lung, ovarian or bladder carcinomas, which are usually stained with CK7 (31). However, it is reported that non-neoplastic colonic mucosa proximal to the rectum exhibits a CK7−/CK20+ phenotype, as is the case for 90% of CRCs (32). When CK17 is included in the diagnostic panel, the efficacy of the test is improved as less than 10% of CRCs express CK17 in contrast to other carcinomas that are more often positive for CK17 (including stomach, endometrium and urine bladder). In addition, pancreatic ductal carcinomas are consistently positive and a number of carcinomas from other sites, may exhibit CK17 expression (33). Furthermore, when CRC progression is studied, CK20 and CK7 can be useful. Results indicate that advanced CRCs were more often CK20+/CK7+ compared to early-stage cancers, which were predominantly CK20+/CK7−. Thus, CK7 expression may be a differentiating marker for the progression of CRC (34).
Caudal type homeobox 2 (CDX2)
CDX2 is a transcription factor encoded by CDX2 gene, a member of the caudal subgroup of homeobox genes. Its main role is to ensure maintenance of a cellular intestinal phenotype during the in utero and ex utero life (35). CDX2 presents strong expression patterns in epithelia of the normal small intestine, appendix, colon, and rectum as well as in the pancreatic centroacinar and interacinar ductal cells (36). It is revealed that loss of CDX2 may give birth to human CRC. CRCs, beside those exhibiting MSI, are consistently CDX2-positive (37). In fact, a quite interesting research recently investigated the effect of restoration of CDX2 expression on colon cancer cell viability, colony formation, cell cycle distribution, apoptosis, invasion ability and xenograft tumor growth in nude mice (38). According to the researchers, CDX2 upregulation significantly reduced the life span and inhibited colony formation, and the invasion and migration ability of LoVo cells. Moreover, it was able to induce cell cycle arrest and apoptosis in vitro, especially under hypoxic culture conditions (38). According to data from histological studies, expression patterns of CDX2 are found in a variety of neoplastic tissues such as adenocarcinomas that exhibit intestinal-type differentiation, including adenocarcinomas of the gastroesophageal junction, bladder, urachus, small bowel, pancreas, appendix, and ovary (37).
Villin
Villin is a cytoskeletal actin-binding protein that is associated with the microfilament family. It is normally found in cells that exhibit highly specialized, brush border-type microvilli, similar to enterocytes (39). Villin is associated with elimination of polarity that the epithelium exhibits, thus altering tissue architecture (40). Thus, in CRCs containing a micropapillary pattern, villin IHC has been effective for tracking the polarity in this type of CRC (41). The specificity of villin as a marker of intestinal origin is limited, similar to CDX2, as positivity may also be marked in adenocarcinomas with intestinal differentiation arising from a wide variety of organs including stomach, lung, and ovary as well as in malignancies of other sites such as the endocervix and liver (the latter more rarely, though) (42).
β-catenin
β-catenin is a multifunctional protein that is involved both in cell adhesion and intracellular signaling, with the latter being accomplished through β-catenin's actions through the Wnt signaling pathway (43). Activation of the Wnt signaling pathway (Fig. 3) increases the cytoplasmic pool of free β-catenin and, to a smaller extent, the nuclear pool where it induces proliferation. The Wnt signaling pathway in its normal form is highly activated in the majority of CRCs as a result of mutant adenomatous polyposis coli (APC) or β-catenin. Moreover, upregulated Wnt signaling has a key role in the pathogenesis of CRC (44). In the absence of functional APC, as it often happens in CRC, nuclear β-catenin can be identified immunohistochemically (44,45). Although nuclear expression of β-catenin is not unique to CRC, it can be proven useful as part of a diagnostic panel.
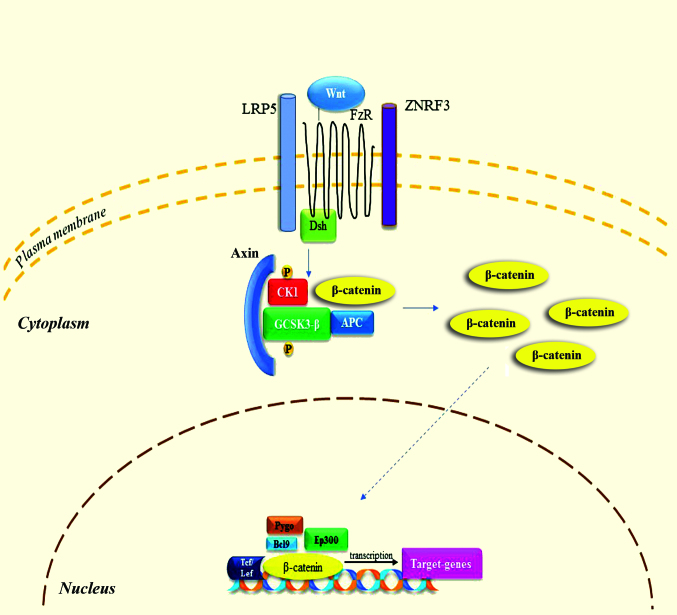
Figure 3.
Wnt/β-catenin signaling pathway. When Wnt is present, it binds to FzR and LRP5 receptor. Dsh enables the phosphorylation of GSK-3β and CK1, resulting in the binding of axin. Following to accumulation of β-catenin and translocation to the nucleus, it binds to various factors in order to modulate the transcription of target-genes. These processes lead to antiapoptotic results and promote cellular proliferation. FzR, frizzled receptor; LRP5, low-density lipoprotein receptor-related protein 5; GCSK3-β, glycogen-synthase kinase 3β; CK1, casein kinase 1; ZNRF3, zinc and ring finger 3; APC, adenomatosis polyposis coli; Pygo, pygopus; Bcl9, B-cell CLL/lymphoma 9; Ep300, E1A binding protein p300; Tcf, T-cell factor; Lef, lymphoid enhancer-binding factor 1.
Carcinoembryonic antigen (CEA)
CEA are membrane-associated glycoproteins playing a number of roles in cell adhesion or signal transduction (46). Monoclonal CEA (mCEA) may be expressed in a wide variety of adenocarcinomas, including those arising from the colon (47). Although this lack of specificity limits its value as a diagnostic marker of CRC, it remains a useful component of a broad diagnostic panel. In a meta-analysis study by Tan et al (48) based on 20 different studies, serum CEA has been demonstrated to comprise an exam of elevated specificity, although sensitivity was inadequate when tracing CRC recurrent conditions. The cut-off range varied among these studies from 3 to 15 ng/ml, suggesting that a measure of 2.2 ng/ml would be the optimal regarding sensitivity and specificity (48). Evidently, at this point circulating CEA may constitute a primary means of recording progress in patients' surgical follow-up, in accordance with the complementary tools including clinical picture, radiological response and histological results.
Mucins
Mucins are high molecular-weighted, heavily glycosylated proteins (49). Within the polypeptide of these glycoprotein molecules tandemly repeated sequences of amino acids rich in threonine and serine moieties lead to various combinations of these repeats making each mucin type unique. In colon, a mixture of neutral, sialomucin, and sulphomucin are normally met, with MUC2 being the most prominent, primarily in goblet cells. MUC4 is also abundant in the colon and its expression is found both in goblet and columnar cells, whereas MUC3 appears to be expressed primarily within enterocytes (50). MUC1, MUC5AC, and MUC6 under normal conditions are not expressed in the colonic mucosa (50). MUC2 is frequently expressed in mucinous CRC as well as mucinous carcinomas of the ovary, breast, and pancreas (50). Gastric mucins may also be expressed in CRC (51). MUC5AC expression is associated with mucinous differentiation and MSI, while most mucinous carcinomas exhibit a MUC2+/MUC5AC+ phenotype (52). Additionally, MUC21 has recently proven to be expressed in CRCs (53). Notably, although a correlation between mucin expression and clinical characteristics was observed by Wang et al (54) an increased expression of MUC5 was found to be associated with poor cellular differentiation, lymph node metastasis, advanced tumor stage and a poor overall prognosis in CRC. By contrast, a decreased expression of MUC2 was reported in cases with lymph node metastasis, poor cellular differentiation and an advanced tumor stage in CRC. These results suggest that MUC2 and MUC5 levels could be associated with tumor progression and even be used in order to facilitate the early diagnosis and clinical characterization of CRC (54).
Cadherin 17 (CDH17)
CDH17 was first described in the liver and intestine of rats (55). Later on, the research in humans revealed that its expression is limited to the intestine (both small and large) and in a part of the pancreatic duct. As for its function, it serves as an intestinal peptide transporter (56). However, its clinical applicability in GI tumors diagnosis was only recently recognized (57). Recent data indicate that positive CDH17 immuno-reactivity is frequently seen in colorectal adenocarcinomas up to 96% and to a significant portion of gastric (58), pancreatic, and biliary adenocarcinomas (25–50%) (59). By contrast, it is rarely found in adenocarcinomas arising from extra-GI tract (1–10%). Notably, even though CDH17 is transcriptionally regulated by CDX2, it is more sensitive and specific than CDX2 for the identification of primary and metastatic colorectal adenocarcinoma (57–59). CDH17 also seems to be useful in diagnosing CRC variant with poorly differentiated or even undifferentiated morphology, such as medullary carcinoma, which characteristically lacks the expression of conventional intestinal differential markers, such as CK20 and CDX2 (57). In a study where the effect of the downregulation of CDH17 upon human colon cancer cells was examined, the researchers found that the downregulation of extrinsic CDH17 gene can conspicuously promote apoptosis-inducing effects of noscapine on specific human colon cancer cell lines, which may indicate a novel strategy to improve the chemotherapeutic effects on colon cancer (60).
Special AT-rich sequence binding protein 2 (SATB2)
SATB2 is a member of the nuclear matrix-associated transcription factors family whose role is to serve as epigenetic regulators of gene expression with high tissue selectivity (61). Research has shown that SATB2 has a variety of biologic functions. However, the exact role of SATB2 in the GI tract is still unknown. Recently, Magnusson et al (61) suggested that SATB2 immunoreactivity was mainly encountered in the glandular lining cells of the lower GI tract, including appendix, colon, and rectum. In fact, SATB2 is a highly sensitive and specific marker for adenocarcinomas of the colon and rectum, with a diagnostic sensitivity of 97% (121 of 125 cases) in CRCs and of 81% in CRC metastases (61). Moreover, Bian et al (59) found that the combination of CDH17 and SATB2 serves as potential optimal markers for the differential diagnosis of pulmonary enteric adenocarcinoma, a rare type of non-small cell lung cancer exhibiting similar histological and immunohistochemical morphology to colorectal adenocarcinoma, and metastatic colorectal adenocarcinoma (59).
Non-invasive techniques and diagnostic blood-derived and faecal markers
Faecal heamoglobin detection tests
Until recently, non-invasive techniques used for diagnostic purposes were represented mainly by the guaiac faecal occult blood test (gFOBT) and faecal immunochemical test (FIT). The gFOBT detects blood loss from peptic ulcer and gastrointestinal cancer. However, its validity in terms of CRC diagnosis, is rather restricted as gFOBT is, not only unable to distinguish bleeding between the upper and the lower GI tract, but also lacks the ability to specifically distinguish human from non-human haeme (8,10). Moreover, gFOBTs are not sensitive to small bleedings, while specificity can be affected by diet or drugs and they have a fixed hemoglobin concentration cut-off determining positivity. As a result, gFOBT exhibits low sensitivity in terms of the detection of cancerous and preneoplastic lesions (30–40%) (62), whereas the sensitivity to detect an advanced adenoma (polyp, >9 mm; villous features or high grade dysplasia) is only 18% (63). On the other hand, FIT is able to detect human globin by means of an antibody-based assay providing a qualitative or quantitative result (depending on the kit used) expressed as faecal haemoglobin concentration per gram of faeces. FITs are analytically more specific, capable of quantitation and hence provide flexibility to adjust cut-off concentration for positivity and the balance between sensitivity and specificity. FITs are clinically more sensitive for cancers and advanced adenomas, and because they are easier to use, acceptance rates are higher (64). However, FIT can only detect bleeding from colonic pre-neoplastic lesions. Thus, when used for CRC screening, the sensitivity FITs demonstrates in detecting adenomas >1 cm in diameter is only 20–30%, while for advanced neoplasia or large adenomas sensitivity ranges between 20 and 67%, which is comparable or superior to the sensitivity of gFBOT (64,65). Another very important aspect that limits occult blood testing is the fact that it detects significantly more in the left- than right-sited lesions in the colon (66), which is a significant issue given the increased incidence of right-sided CRCs that has developed over the last two decades (67). In other words, it is evident that those two non-invasive diagnostic techniques are limited by their intrinsic lack of sensitivity and specificity (with the latter being the case for gFBOT) making them unable to stand on their own as diagnostic tools. Table I presents a summarization of bibliographical references regarding FOBT sensitivity and specificity.
Table I.
A summarization of bibliographical references to FOBT sensitivity and specificity.
| Author/(Refs.) | Faecal occult blood testing |
|---|---|
| Kronborg et al (185), Scholefield et al (186) | Reduction in CRC mortality with gFOBT biannually [relative risk reductions of 13% (UK trial) and 16% (Danish trial)] |
| No significant reduction in overall mortality | |
| gFOBT: Low sensitivity for CRC detection (UK trial, 45%; Danish trial, 54%) | |
| True-positive rate: 50% (UK and Danish trials) | |
| False-positive rate: 5–10% (UK and Danish trials) | |
| True-negative rate: 90–95% (UK and Danish trials) | |
| False-negative rate 50% (UK and Danish trials) | |
| Medical Advisory Secretariat (187), Dancourt et al (188), Faivre et al (189) | iFOBT sensitivity superior to those of gFOBT for CRC detection: Two studies showed sensitivity for gFOBT, 13 and 25%, respectively; pooled iFOBT sensitivity, 81% iFOBT and gFOBT: Lower sensitivities for adenoma detection than for CRC detection: Rehydrated gFOBT, 22%; pooled iFOBT, 28% |
| Lin et al (190) | FIT sensitivity, 73.8% (95% CI, 62.3 to 83.3) for quantitative (n=9,989) test categories; 78.6% (95% CI, 61.0 to 90.5) for qualitative (n=18,296) test categories |
| Koo et al (191), Moss et al (192) | Positive predictive value of FIT > positive predictive value gFOBT for advanced adenoma (1.73 vs. 0.35%) and all neoplasias (3.74 vs. 0.76%) FIT detects twice more CRCs and advanced adenomas |
| Gonzalez-Pons and Cruz-Correa (8) | gFOBT: ↓ ability to define bleeding between upper/lower GI tract |
| Kuipers et al (10) | gFOBT: ↓ ability to distinguish human haeme |
| Valori et al (62) | gFOBT: Νot sensitive in small bleedings |
| gFOBT ↓ sensitivity in detecting cancerous/preneoplastic lesions | |
| gFOBT: Specificity affected by diet/drugs | |
| Lieberman et al (63) | gFOBT: 18% sensitivity in detecting advanced adenomas |
| Whitlock et al (65), Young et al (64) | FITs sensitivity for advanced adenomas: ~20–67% (↑ than FOBT) |
| Dancourt et al (188) | FIT detects more CRC and advanced neoplasia than gFBOT (similar positive predictive value) |
| Imperiale et al (25) | |
| Rozen et al (193) | Comparative performance of gFOBT and FIT depends on the number of samples and threshold chosen for the quantitative FIT |
| Hoffman et al (194) | Screening adherence with FIT was higher than with gFOBT (61.4 vs. 50.5%) |
| Brenner and Tao (195) | Sensitivity of FITs for detecting CRC/any advanced neoplasm/any neoplasm: 2–3 times higher than gFBOT |
| Increased levels of FITs specificity vs. gFOBT | |
| Fitzpatrick-Lewis et al (196) | iFOBT vs. gFOBT on mortality outcomes: iFOBT has higher sensitivity and comparable specificity (insufficient evidence from RCTs) |
| Murphy et al (197) | Total financial burden: Lower for FIT at any threshold (expressed in µg Hb/g faeces) than for gFOBT, and this difference increases as the FIT threshold is decreased (Cohort-based Markov state transition model) |
| Lee et al (198) | FIT sensitivity, 79%; FIT specificity, 94% |
| Morikawa et al (66) | gFOBT detect notably more lesions in the left (compared to the right colon) |
FOBT, faecal occult blood test; CRC, colorectal cancer; gFOBT, guaiac faecal occult blood test; FIT, faecal immunochemical test; RCT, randomized controlled trials.
Blood-derived and faecal molecular cancer cell markers
Our improved knowledge of the molecular pathogenesis that conditions the polyp→adenocarcinoma progression sequence has made clear that the molecular changes found in polyps and adenocarcinomas have the potential to serve as neoplasm-specific molecular markers for these lesions. The concept of using these molecular markers for CRC screening is indeed the next step for a well-established non-invasive detection method for CRC. In contrast to heme detecting tests, tests focusing on molecular markers derived from neoplastic cells of the colon can prove to be more accurate. CRCs are known to bear distinguishable genetic and epigenetic changes as they develop and progress, which forms the rational of stool-based DNA and RNA testing.
DNA impairment
Approximately 90% of CRCs develop sporadically, and only a few cases (<10%) are hereditary, with familial adenomatous polyposis, hereditary non-polyposis colorectal cancer (HNPCC), MUTYH-associated polyposis, Peutz-Jeghers syndrome (PJS) and serrated polyposis syndrome being the main representatives of hereditary causes of CRC. Currently, three major paths for CRC development have been described, with CIN being the most common accounting for 70–80% of CRCs, the MSI pathway positioned in the second place accounting for 5–20% of tumors and in the third place the CIMP, which represents approximately 15% of CRCs (68). These pathogenetic pathways can be examined either in serum samples (CIN, MSI, CIMP, APC) using immunohistochemical techniques or in faeces (CIMP, APC).
a) CIN
CIN is the hallmark characteristic of most CRC cases (80–85%), and its main characteristic is the extensive abnormality in chromosome number (aneuploidy) and loss of heterozygosity. CIN can be observed in several forms, including chromosomal numerical abnormalities, small sequence modifications such as base deletions or insertions, chromosomal rearrangements and gene amplification (5).
b) MSI
Microsatellites (MSs) are short tandem-repeated base pairs of 1–6 scattered all over the genome. For the normal human genome the number of MSs is approximately half a million. Genome studies revealed that MSs are prone to duplication errors. However these errors are usually corrected by the MMR system (5,69,70). Consequently it is logical to assume that a defective MMR system would result in the accumulation of DNA mistakes and thus MSI. Indeed, MSI arises by the inhibition of MMR system either via defective methylation of MLH1 in CpG island or point mutation of any MMR genes (hMLH1, hMSH2, hMSH6, PMS1 and PMS2) especially hypermethylation of hMLH1 promoter (5). It is estimated that about 15–20% of CRC patients present MSI with a small fraction of which 2–4% are related to HNPCC (71). In order to estimate MS status, Bethesda panel was agreed in which five MS loci were included (BAT25, BAT26, D5S346, D2S123, and D17S250) (72). However, some researchers suggested an expanded Bethesda panel include 10 loci. Based on this panel, MSI can be divided into three groups: MSI-high (MSI-H), defined as having ≥30% unstable loci using mononucleotide or dinucleotide markers (8,73); MSI-low (MSI-L), with 10–30% unstable loci; and microsatellite stable (MSS), with <10% unstable loci (73). MSI status varies according to a given CRC stage: Stage II CRC exhibits high prevalence of MSI (20%) while in stage IV CRC MSI is approximately 4% (74,75). Moreover, differences based on the MSI status are found when prognosis is examined. For example, cases with MSI-H CRC share a better prognosis than that with MSS CRC (5,76).
c) CIMP
It is well accepted that DNA methylation is a key process for the normal growth of eukaryotes. If occurs in cytosine number 5 within CpG island which is present in 50% of tumor suppressor gene promoters (77). CpG island accounts for >70% of CG sequences that extend to 0.4 kB on the genome (78). Even though hypermethylation of CpG island cytosine represents a hallmark for cancer progression, both hypomethylation and hypermethylation may lead to the transformation of normal mucosa to adenoma and subsequently to the development of CRC (79). Disturbance of epigenetic programming (epigenetic modification including DNA methylation, histone modification and post-transcriptional gene regulation) is closely related to the development of CRC (80). It is reported that a wide spectrum of aberrant methylated genes in CRC, regulates crucial functions in the normal cell regarding proliferation and maintenance of genome stability (77). In the clinical setting, abnormal DNA methylation patterns can be detected in patient's blood or faeces samples from which CRC cells are examined. A number of abnormal methylated genes with variable level of specificity and sensitivity can serve as diagnostic biomarkers in CRC patients. These genes include WiF-1, AIX4, PGR, FBNI, P53, TIMP3, SEPT9, MGMT, Vimentin and GATA4 (77,80). A number of studies have indicated that instead of looking for a single gene methylation pattern, using a panel of several methylated genes is better in terms of specificity and sensitivity. Lind et al (81) tested a panel of epigenetic biomarkers for use as CRC and adenoma screening. In this study, approximately 523 human samples were examined using a gene panel consisting of: CNRIP1, FBN1, INA, MAL, SNCA and SPG20. According to their study, high level of sensitivity and specificity was achieved. Hence, those authors suggested that a combination of the six genes may serve as a non-invasive biomarker with high specificity and sensitivity for early diagnosis of CRC (81).
d) APC mutations
APC gene regulates the Wnt signaling pathway via encoding a multifunctional protein. Specifically, APC regulates Wnt pathway through the destruction of the transcription factor β-catenin, which enhances the activity of Wnt pathway. Hence, APC conversely organizes Wnt signaling (82). In addition, APC gene is involved in cell cycle regulation, cytoskeleton stabilization, intracellular adhesion, as well as apoptosis. In an attempt to determine the exact role of APC gene in CRC, Dow et al (83) investigated whether APC mutation is essential for CRC protection. For their study a CRC mouse model with inhibited APC was used. According to their findings, inhibiting APC gives rise to adenomas in colon and small intestine (83). In general, about 90% of CRC patients demonstrate APC gene mutation, which highlights its applicability as a molecular biomarker for CRC diagnosis (8). Liang et al (84) conducted a meta-analysis study to correlate APC polymorphism (D1822V, E1317Q, I1307K) and CRC risk. They concluded that E1317Q significantly increased adenoma risk. However, I1307K is associated with high risk of CRC (84). CRC-related tumor suppressor genes are thought to be altered in the early phase of cancer development, and APC mutation is the first step in the translation of normal mucosa to neoplastic tissue, leading to the activation of the WNT pathway. Subsequent mutations that occur in genes, such as KRAS, TP53, SMAD4 and type II TGF-β receptor (TGFBR2), lead to the progression from polyp to cancer similar to the process that takes place in other gastrointestinal carcinomas (85).
e) Circulating tumor DNA (ctDNA)
Circulating free DNA (cfDNA) is thought to be a natural phenomenon, existing as a result of cellular apoptosis and necrosis of both normal and cancer cells, with secretion from cancer cells being an additional source that remains to be elucidated (86). ctDNA, a subclass of a patient's cfDNA, is the material used for testing in order to identify potential DNA impairments that have been associated with CRC. Owing to the circulating DNAases, the half life ctDNA is restricted to a few hours allowing clinicians to have a more accurate look at a patient's cfDNA profile. Findings have shown that the cfDNA quantity is quite higher in patients with cancer than that in healthy individuals. Moreover, cfDNA quantity seems to be positively correlated with the cancer stage (87). Of note, benign tumors or non-neoplastic lesions in general do not lead to ctDNA avoiding, therefore, a potential pitfall of false positives (88). To date, the main DNA defect that is studied is the hypermethylated SEPT9 gene in the plasma. However, following the initial attempts to set a diagnostic tool based solely on SEPT9, researchers realized that combining multiple gene loci in a single panel in order to achieve better sensitivity and specificity is optimal. For example, Tänzer et al (89) showed that the combination of SEPT9 with ALX4 achieved a sensitivity of 71% and a specificity of 95% for advanced adenomas, thus supporting SEPT9/ALX4 as a biomarker for precancerous lesions (89). In parallel to that, apart from SEPT9, a number of genes have been investigated including HJC1, CYCD2, PAX5, RB1, SRBC, NPY, PENK, WIF1, ALX4, HLFT, HPP1, MLH1, APC, CDKN2A/P16h, TMEFF2, NGFR, FRP2, NEUROG1 and RUNX3 (90).
f) Faecal DNA
As with circulating DNA, stool DNA is examined for the presence of abnormalities in specific genes. Stool-based assays have proven to be the most efficient assay type for a number of reasons. Direct histologic observations show that CRCs and polyps exfoliate neoplastic cells and their debris into the mucocellular layer of the colonic lumen at a steady continuous rate (91). However, at first sight, one would wonder how exfoliated cells arising from a right-sided polyp or CRC could survive in the intraluminal environment in order to be detected. Indeed, most of these cells go through lysis, preserving however to a certain extent their DNA content thus allowing a DNA analysis to be conducted. To date, along with mutant KRAS a variety of hypermethylated genes including APC, ATM, BMP3, CDKN2A, SFRP2, GATA4, GSTP1, HLTF, MLH1, MGMT, NDRG4, RASSF2A, SFRP2, TFPI2, VIM and WIF1 have been analyzed in faecal DNA for the early detection of CRC, with SFRP2 and VIM proving to be the most promising ones (92,93). When these tests for both mutant and methylated DNA markers and FIT, collectively known as MT-sDNA, are combined, it is proven that they demonstrate the best clinical performance of CRC molecular marker screening assays to date.
Blood and faecal miRNA profile
miRNAs are single-stranded small non-coding RNAs that are 18–25 nucleotides in length. Their role remained unknown until 1993 when it was recognized that they act as negative post-transcriptional regulators in Caenorhabditis elegans (94,95). Until 1993, miRNA (as any other member of the non-coding RNA family) was considered to be a useless RNA product of ‘junk DNA’. However, it was shown that this is not the case as it was proven that it is able to regulate gene expression at a post-transcriptional level, either by blocking mRNA translation or by inducing their degradation. By binding miRNA to its target mRNA, miRNA can trigger the degradation of its target or otherwise inhibit its translation into protein, with the degree of sequence complementarity between the miRNA and mRNA determining which mechanism is employed (96–98). Over the last few years, research in human cancer focusing on the potential of miRNA to serve as a biomarker has increased dramatically mainly due to their unique properties. First of all, miRNAs are very stable under a variety of conditions both in the experimental and laboratory setting. Moreover, RNases cannot degrade them owing to their small size and the hairpin-loop structure (99). In addition to that, cell-free miRNAs occur in large numbers as they are packed in high density lipoprotein particles, apoptotic bodies, microvesicles, exosomes or by their binding to argonaute-2, properties that synergistically result in an increased stability (100,101). Thus, it is easy to isolate them from different forms of clinical specimens (99). Lastly, miRNAs are actively secreted by cancer cells into the circulatory system and digestive tract (102). Hence, in the clinical setting, circulating cell-free miRNAs and faecal miRNA are the main forms of RNA used as diagnostic biomarkers.
a) Blood-derived (circulating) cell-free miRNA
Various studies have shown that a series of miRNAs are pathologically excreted in CRC plasma or serum samples. However, due to the fact that standardized techniques for miRNA extraction, normalization and quantification are yet to be found, one needs to bear in mind that most of the available results in literature are not easily reproducible thus making sometimes controversial. Circulating cell-free miRNA was first evaluated in a more comprehensive and systematic way in patients with CRC by Ng et al (103) in 2009 who found an altered miRNA expression profile in tissue and plasma samples from CRC patients and healthy subjects. However, a very important observation was that two miRNAs, miR-92a and miR-17-3p, were able to indicate patients with CRC, differentiating from healthy subjects, based on their high expression profile (sensitivity: 64 89%, specificity: 70, 70% for each miRNA, respectively) (103). Since then, numerous miRNAs have been identified as indicators of CRC. However, even though a few single miRNAs have proven to be enough to make the distinction with increased sensitivity and specificity, the addition of even more dysregulated miRNAs into a single panel usually achieved better diagnostic results. Indicatively, some of the dysregulated miRNAs and their testing samples are described: miR-17-3p plasma, miR-18 plasma, miR-21 plasma, miR-21 serum, miR-21 serum (exosome), miR-29a plasma, miR-92a plasma, miR-92a serum, miR-155 serum, miR-200c plasma, miR-221 plasma, miR-21, miR-31, miR-92a, miR-181b, miR-203, let-7g (panel), miR-7, miR-93 and miR-409-3p (panel) (104). However, only three upregulated miRNAs, miR-19a-3p, miR-21 and miR-92, were identified as promising diagnostic biomarkers by more than one study (105,106).
b) Faecal miRNA
Identification of CRC-specific miRNAs is also feasible in stools. Link et al (107), using RT-PCR and microarray analysis proved that faeces from patients with CRC and colorectal adenoma contained higher levels of miR-21 and miR-106a as opposed to that from healthy controls (107). A subsequent study that assessed faecal miR-21 and miR-92a levels revealed that faecal miR-92a expression was able to differentiate patients with CRC or adenoma from healthy subjects or even those with lower-risk polyps (22). More interesting data came from the study of Zhu et al (108) as they found that stool miR-29 was significantly present in patients with rectal cancer than in those with cancer in the rest of the large intestine. Thus, the use of a panel of miRNA expression patterns can create a form of a cancer fingerprint (108). A sum of the useful stool-derived dysregulated miRNAs for diagnosis is miR-17-92 cluster, miR-20a, miR-21 up miR-135 miR-144 miR-29a, miR-223, miR-221, miR-92a and miR-224 (109).
3. Prognostic markers
General
Once CRC is diagnosed, the clinician has to take the next step in the clinical management, that is to evaluate the prognosis for this patient or in other words to estimate the likely progression of the cancer and the aggressiveness that it may exhibit (recurrence likelihood, progression and/or chance for metastasis despite adjuvant therapy). The current practice for prognosis assessment is based on radiological (CT, MRI) and pathological (TNM, lymphovascular, perineural and venous invasion) criteria. In fact, TNM staging remains the strongest prognostic tool (110). However, not only do none of the prognostic tools mentioned above provide clear evidence on which of these CRC cases is more prone to relapse, give metastases or are proven to be resistant to chemotherapy, but they also are not suitable for the personalized treatment of each patient. Thus, much effort has been made for the evaluation of the potential of several molecules and gene alterations to serve as prognostic biomarkers, taking us a step closer to a true personalized treatment.
Tissue-derived prognostic biomarkers
Molecular prognostic biomarkers
Some of the tissue-derived molecular biomarkers meant for diagnostic purposes, have demonstrated a promising potential for also serving as prognostic markers. Such markers are mucins (MUC2), SATB2 protein, CK20/CDX2, VEGF, insulin-like growth factor-II mRNA-binding protein 3 (IMP3) and Traf2- and Nck-interacting kinase (TNIK) expression. Even though some MUC2 expression profile studies have yielded mixed results, it is evident that MUC2 loss of expression may be an indicator of poor prognosis in both mis-match repair protein (MMR)-proficient and MLH1-negative CRC (111). On the other hand, studies on the SATB2 expression profile showed that an upregulated SATB2 was connected with good prognosis in CRC and could even increase sensitivity to chemotherapy and radiation, whereas a downregulated SATB2 in cases of colorectal adenocarcinomas was associated with poor prognosis, as tumor invasion, infiltrated lymph nodes and distant metastases were observed (112,113). Studies on CDX2 revealed that loss of CDX2 expression is associated with proximal origin, infiltrative characteristics and advanced TNM stage. Moreover, it was found to be an independent poor prognostic factor of progression-free survival (PFS) and overall survival (OS) (114). Vascular endothelial growth factor or VEGF is one of the main angiogenic factors in CRC as it is expressed in approximately 50% of the cases Thus, VEGF-1 expression is correlated with a worse prognosis (115). Fig. 4 shows the connection between chronic inflammation and the development of CRC. Moreover, studies on IMP3 found an increased expression in colon cancer compared to normal colonic mucosa. Of note is the fact that its expression was found to be higher in cases with lymph node infiltration with cancer cells (93%) than in primary colon cancer (65%) or normal mucosa (3.9%) (116). Lastly, research on TNIK reported that high levels of TNIK protein in primary tumors could indicate distant metastasis after surgery of stage II and III CRC patients as well as invasive characteristics of CRC (117).
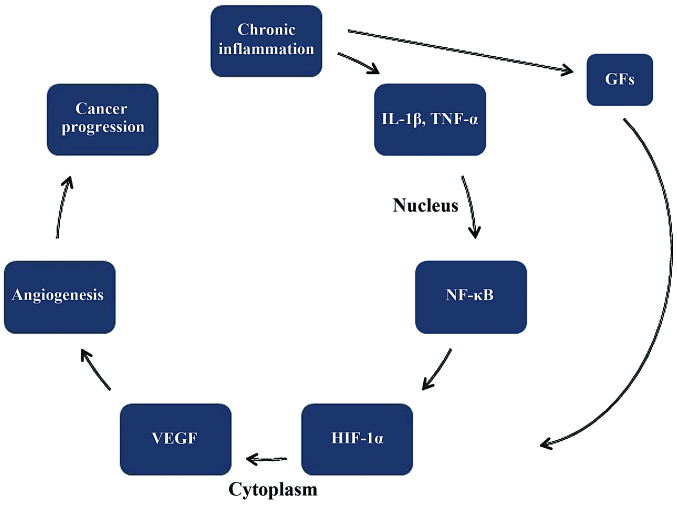
Figure 4.
The connection between chronic inflammation and development of CRC. CRC, colorectal cancer; GFs, growth factors; Il-1β, interleukin 1β; NF-κΒ, nuclear factor κB; HIF-1α, hypoxia inducible factor-1α; VEGF, vascular endothelial growth factor.
DNA alterations with prognostic value
KRAS and NRAS
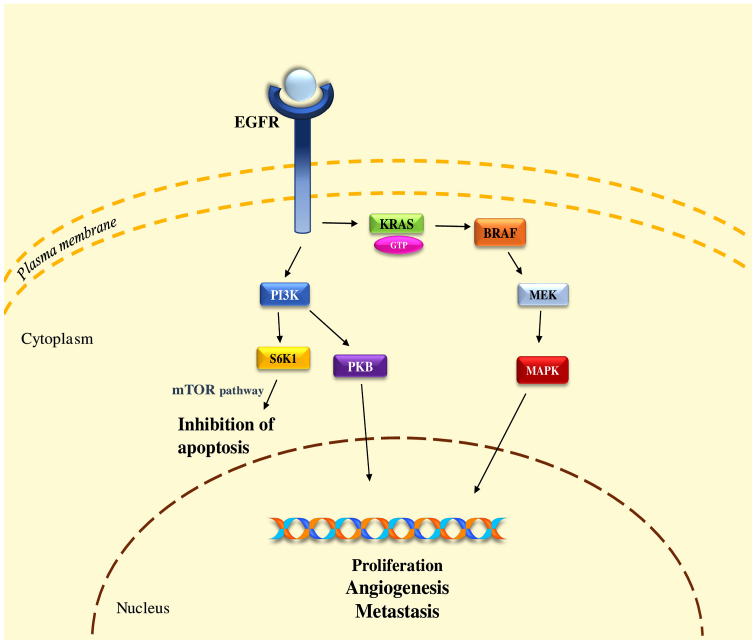
The family of RAS proteins (H-, K-, and N-RAS) is located in the intracellular side of the cell membrane involved in G-protein mediated signal transduction. Activation of the epidermal growth factor receptor (EGFR) from its ligand (e.g., EGF, TGF-α, amphiregulin) results in a change from GDP- to GTP-form of the KRAS, leading to increased concentrations of B-rapidly accelerated fibrosarcoma (proto-oncogene) (BRAF) to the plasma membrane (Fig. 5). BRAF activates the mitogen-activated protein kinases (MAPK) signaling pathway that results in the expression of proteins involved in several pathways with a crucial role including cell proliferation, differentiation, survival, angiogenesis, and cell motility. Mutant KRAS proteins present as locked in the active form as a result of an impaired GTPase activity, leading to an increased proliferating rate that is unregulated and thus resulting in an overall malignant transformation of the cells. Therefore, it is logical to assume that mutation of these oncogenes poses a great threat for carcinogenesis (118). Indeed, it is well documented that alterations of KRAS gene can act as the first step towards carcinogenesis in approximately 50% of the CRC cases (119). However, it is not that clear whether it could serve as a prognostic marker in the clinical setting. Recently, a meta-analysis of seven studies failed to associate KRAS mutation status with prognosis (however, a common limitation of such studies is the small pool of patients) (120). On the other hand, two large multicenter studies, demonstrated that only one mutation of KRAS, of codon 12, could be linked with a more aggressive progression of cancer cells. However, according to their data, KRAS mutations failed to be associated with tumor location or stage and recurrence of disease (121,122). On the other hand, the possible prognostic value of NRAS mutations is less examined, even though they appear to act in a similar way with KRAS as for the degree of negative prognostic significance (123).
Figure 5.
Intracellular signals for CRC manifestation via EGFR. CRC, colorectal cancer; EGFR, epidermal growth factor receptor; BRAF, B-rapidly accelerated fibrosarcoma (proto-oncogene); MAPK, mitogen-activated protein kinase; PI3K, phosphoinositide 3-kinase; S6K1, ribosomal protein S6 kinase β-1; PKB, protein kinase B; mTOR, mechanistic target of rapamycin.
p53
p53 is a transcription factor that participates in a variety of cellular reactions to several stress situations such as mutagenic DNA damage, oncogene activation, hypoxia and telomere shortening. In contrast to other mutations, p53 mutation seems to occur late in the development of CRC since few or even no cases of mutations are described in precancerous lesions from sporadic adenomas and polyps. Interestingly, data from one study showed that mutations of p53 in CRC exhibits a dependence on the primary tumor site thus suggesting a prognostic value of this marker. In more detail, patients with a primary tumor site in the proximal colon (caecum, ascending colon) and mutant p53 gene exhibited better survival when treated with a combination of chemotherapy and surgical removal compared to those treated by surgery alone (124).
BRAF
BRAF is serine-threonine protein kinase that is recognized downstream in the KRAS signaling cascade. BRAF mutations are linked with a poor outcome thus proving their clinical applicability as a prognostic marker in the adjuvant setting. Another indication of a poor outcome is found in stage II and III CRC cases where BRAF mutations were associated with worse OS (125). A rather interesting finding is that BRAF V600E mutation is able to predict a poor prognosis in right-sided MSS CRC (126,127). Further examination of the BRAF prognostic value was carried with a study investigating the correlation of BRAF mutation with MSI. The existence of BRAF wild-type and MSI-H exhibited favorable outcomes. Furthermore, BRAF-wild/MSS and BRAF-mutated/MSI-H exhibited intermediate outcomes (128).
MSI
MSI status is a well-studied diagnostic marker for CRC as mentioned above. However, its clinical relevance does not stop in the diagnostic setting but continues in the prognostic as well. MSI-H is associated with better survival rates than both MSI-L and MSS, not only in HNPCC, but also in sporadic cases (129,130).
miRNA with prognostic value
During the last few years, a wide variety of miRNAs has been evaluated in order to identify their prognostic value in CRC. Indeed, numerous miRNAs, among which are miR-101, the let7 family, miR-133b, miR-126, miR-337, miR-944, miR-646, miR-497 and miR-142-3p have been identified to behave as tumor suppressors (tumor-suppressive miRNAs) (131–137) while others, including miR-7, miR-20a, miR-21, miR-29a, miR-92a, miR-130b, miR-155 and miR-552, were characterized as key players in creating a favorable microenvironment for cancer cells. These miRNAs are known as oncomirs (oncogenic miRNAs) (138–141). miR-20a-5p was found to promote tumor invasion and metastasis via downregulation of SMAD4 (142). Moreover, elevated miR-21 levels correlate with CRC cell proliferation, invasion, lymph node metastases, advanced clinical stage, poor overall and disease-free survival in the different Duke stages (143,144). Increased expression of miR-29a is found to strongly correlate with metastases and especially liver metastases as, if present, it can serve as an early detector of liver metastases (145,146). High preoperative miR-155 closely correlates with advanced stage and metastasis while persistent postoperative expression correlates with recurrence and metastasis. Thus, miR-155 could be considered a prognostic marker for overall and disease-free survival (147). In a recent study, Zhao et al (148) presented that miR-411 functions as a tumor-suppressive miRNA directly targeting PIK3R3 and indirectly regulating the AKT/mTOR signaling pathway. In other words, an increase of miR-411 would result in the downregulation of PIK3R3 directly and AKT/mTOR indirectly. Additionally, they observed that a decreased expression of miR-411 was correlated with worse findings such as lymph node metastasis, distant metastasis and worse TNM stage (148).
Blood-derived prognostic biomarkers
Preoperative CEA levels
CEA may be tested in the preoperative setting in patients with CRC in order to assist staging and surgical treatment planning. Previous findings showed that increased preoperative CEA (>5 ng/ml) correlates with poorer prognosis. In fact, a study with 2,230 patients proved that pre-operative CEA levels was an important independent prognostic factor when outcome prediction was encountered (149). Similarly, another study with 1,146 patients with CRC found that, following use of a multivariate analysis, preoperative CEA levels proved a highly significant prognostic factor even when stage and grade were introduced in the model (150). In addition, elevated preoperative CEA in stage III and IV CRC are considered to be a potent independent risk factor as far as local relapse, short disease-free survival and OS are concerned (151).
Postoperative CEA levels
After surgical removal of the tumor, CEA levels should be checked as it is shown that persistent by elevated CEA levels suggest further evaluation for metastatic disease. In addition, elevated CEA is very efficient for revealing recurrence in asymptomatic patients and is the most sensitive detector for liver metastases (152).
Cancer antigen 19-9 (CA 19-9) levels
CA 19-9, also termed sialyl Lewis a, is a documented marker with prognostic value for CRC. It is shown that cases with increased CA 19-9 present more frequently metastases thus making it a marker of poor prognosis (152). Similarly, a recent study with stage IV CRC proved that the preoperative serum CA 19-9 level can be a promising marker of tumor recurrence and prognosis in cases submitted to curative resection. In detail, high levels of CA 19-9 were connected with worse 3-year relapse-free survival and 3-year overall than that in the normal CA 19-9 group (153).
Circulating tumor cells (CTCs)
Detection of CTC has demonstrated prognostic significance in predicting patient outcome in cases of metastatic CRC. Recently, two groups investigated the prognostic significance of CTCs in CRC (154,155). Both of them found that patients with high CTC count were more likely to experience worse PFS and OS in contrast to those patients with low CTC count.
4. Predictive markers
General
The final step in the clinical management of a patient with cancer, is the determination of the most suitable therapeutic regimen. It is well known that patients with cancer, exhibit different responses to a certain therapy and some patients can benefit the most from the use of a biologic factor. Thus, it is of paramount importance for us to be able to know beforehand which patient will receive each therapy or in other words to predict the most likely response to a given therapy. For this reason, predictive markers are used.
Tissue-derived predictive markers
DNA alterations
KRAS and NRAS
Mutations of KRAS gene have proven their clinical use as a predictive biomarker in response to the EGFR inhibitors clinically used, as various mutations of KRAS present resistance to therapy with EGFR receptor monoclonal antibody blockers such as cetuximab (156). In detail, anti-EGFR therapy on cases of metastatic CRC who display KRAS mutations either of the codon 12 or 13, present no benefit (157). However, a mutant KRAS does not always mean that it is a sign of bad response. For example, De Roock et al (158) have found that patients with metastatic CRC with P.G13D KRAS mutation treated with cetuximab exhibited better results than other KRAS mutations (158). Under the light of these findings, evaluation of an extended panel of RAS mutations including mutations in KRAS exon 2, 3 and 4 and NRAS exons 2, 3, and 4 can better discriminate which patients are not good candidates for treatment with anti-EGFR therapy (159).
BRAF
BRAF mutations and especially BRAF V600E is among the most common mutations found in CRC cases as it is present in approximately 8–10% of all cases (160). BRAF mutation is often used as a discrimination factor between familial and sporadic CRC as the existence of BRAF V600E mutation in MSI CRCs can virtually exclude Lynch syndrome (161). However, numerous studies have shown that tumors that exhibit BRAF mutation are resistant to anti-EGFR therapy (162).
Phosphoinositide 3-kinase (PI3K)
PI3K is a downstream mediator of the EGFR signaling cascade along with AKT and PTEN. An interesting finding regarding PI3K mutations lies on the use of cetuximab in patients with CRCs who display mutations of PI3K and especially at the PIK3CA exon 20 (which is the kinase domain) in contrast to patients with PIK3CA-wild-type CRCs. Patients with mutant PI3K exhibit much worse results (163). However, the mutation of a different exon, PIK3CA exon 9 (which is the helical domain), cannot serve as a predictive marker for anti-EGFR therapy, a fact that reflects the high complexity of the effects of specific mutations on different functions of the mutant kinases (163).
MSI status
Recent studies have investigated the application of level of MSI status as a potential predictive marker of adjuvant therapy. While there is enough evidence supporting that MSI-H may predict response to 5-fluorouracil (5-FU)-based adjuvant therapy in stage III colon cancer, numerous recent studies demonstrated that there is no significant difference between patients with MSI-H and MSS tumors when 5-FU-based adjuvant therapy is used (164–166). This finding is very important for the group with stage II disease, in which adjuvant chemotherapy (5-FU alone) is reported to improve survival by approximately 3%, allowing some investigators to suggest that stage II colon tumors should be analyzed for MSI status as well in order to assist guide decisions on the use of adjuvant therapy (165).
CD133
CD133 is a surface protein that has been associated with tumor angiogenesis and recurrence and especially with VEGF cascade (167). Previous findings have shown that the expression of CD133 was associated with worse survival rates in patients treated with surgery as a monotherapy and in patients treated with 5-FU-based chemotherapy. By contrast, CD133 expression in patients with stage III tumors, was able to determine that the absence of CD133 could benefit more from 5-FU treatment as expressed by their survival rates but not those with present CD133 expression. Positive expression of CD133 was also associated with worse clinical response to chemotherapy in stage IV patients (168).
5. New insights in CRC monitoring
General
Going through the list of the markers used for diagnostic, prognostic and predictive purposes, it is evident that even though a plethora of markers are available, the clinician still cannot achieve the best possible monitoring of his patient's disease, even when a wide panel of markers is used. For this reason, great effort is made by researchers worldwide in order to identify new markers that will be able to lead to a better, if not the best, disease monitoring. A number of these markers seem to be quite promising. We will discuss three of these markers [telomere length (TL), telomerase activity and micronuclei (MN) frequency].
TL and telomerase activity
Telomeres are DNA-protein complexes that are enrolled to guard the ends of eukaryotic chromosomes and composed of a repetitive nucleotide sequence (5′-TTAGGG-3′) which is added on by the telomerase. Telomerase is an enzyme complex that consists of two subunits, the reverse transcriptase protein human telomerase reverse transcriptase (hTERT), which is the catalytic subunit, and the telomerase RNA component (TERC), an RNA template-hTR (human telomere RNA), which serves as a template for directing the appropriate telomeric sequences onto the 3′ end of a telomeric primer (169). When telomeres become too short, cells may be unable proliferate, a situation that has been linked to the development of a variety of age-related diseases, such as cancer, cardiovascular diseases, diabetes and psychiatric disorders (170–172). A number of studies have examined the role of telomeres and telomerase in CRC in generating CIN. In CRC telomere, shortening is an initial event that directly reflects pathologic cell proliferation because of telomere's shortening in CRCs and in well-differentiated tumors (173,174). Telomeres and telomerase have been proposed as potential prognostic and diagnostic biomarkers in CRC. Telomeres and telomerase activity have universal changes along the CRC process (175). Fernández-Marcelo et al (176) examined the ratio of TL in cancer to non-cancer tissue, telomerase activity and TERT levels and their role as prognostic markers. In tumors, TL was shorter than that in non-tumor tissues and more than 80% of CRCs displayed telomerase activity and thus the use of telomere status as a prognostic factor has been suggested (176). Gertler et al (177) analyzed TL and hTERT expression in matched cancer and adjacent non-cancer mucosa samples and found that telomeres in CRC tissues were significantly shorter compared with adjacent normal mucosa samples. Another study comparing peripheral blood cell TL in CRC patients and healthy subjects, TL has an anomalous behavior. We can conclude that telomeres and telomerase emerge as useful diagnostic and prognostic markers in the clinical management of CRC (5).
MN frequency
MN are extra-nuclear bodies recognized in dividing cells, that contain chromosome fragments and/or whole chromosomes that failed to incorporate into the nucleus after cell division. It is well established that MN formation can be induced by defects in the DNA repairing system, leading to the accumulation of DNA damages and chromosomal aberrations (178). As MN formation is a result of CIN and indicative of a malfunctioning cell, it is reasonable to assume that MN could, under certain circumstances, be linked with cancer development (179). Indeed, high MN frequency is recognized in a number of cancer types. Specifically, in lung cancer MN frequency has become a useful marker for the identification of small and non-small cell lung cancer using peripheral blood samples (180). As far as CRC is concerned, data from various studies using peripheral blood samples indicate that MN frequency is a promising biomarker for the early detection of CRC while it could also be used as a prognostic biomarker (181–184). However, more studies are needed in order to describe with certainty the true potential of this biomarker.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- CRC
colorectal cancer
- CIN
chromosomal instability
- MSI
microsatellite instability
- CIMP
CpG island methylation phenotype
- CEA
carcinoembryonic antigen
- ctDNA
circulating tumor DNA
- CA 19-9
cancer antigen 19-9
- CTCs
circulating tumor cells
- PI3K
phosphoinositide 3-kinase
- CKs
cytokeratins
- APC
adenomatous polyposis coli
- CDH17
cadherin 17
- PFS
progression-free survival
- OS
overall survival
- hTERT
human telomerase reverse transcriptase
- MN
micronuclei
- TERC
telomerase RNA component
Funding
Not applicable.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
JT conceived and designed the study. TKN and LV researched the literature, performed analysis of data and drafted the manuscript. PF and GP drafted the manuscript. JT, TMS, DAS and AMT critically revised the article for important intellectual content.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Demetrios A. Spandidos is the Editor-in-Chief for the journal, but had no personal involvement in the reviewing process, or any influence in terms of adjudicating on the final decision, for this article.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Haggar FA, Boushey RP. Colorectal cancer epidemiology: Incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg. 2009;22:191–197. doi: 10.1055/s-0029-1242458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dušek L, Mužík J, Malúšková D, Šnajdrová L. Institute of Biostatistics and Analyses. Masaryk University; Brno, Czech Republic: Epidemiology of colorectal cancer: International comparison. [Google Scholar]
- 4.Bardhan K, Liu K. Epigenetics and colorectal cancer pathogenesis. Cancers (Basel) 2013;5:676–713. doi: 10.3390/cancers5020676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsiaoussis J, Vassilopoulou L, Nikolouzakis T, Rakitskii VN, Vakonaki E, Fragkiadaki P, Stivaktakis P, Tsatsakis AM. Biomolecular profile of colorectal cancer - the role of telomerase as a potent biomarker. Farmacia. 2017;65:643–659. [Google Scholar]
- 6.Souglakos J, Philips J, Wang R, Marwah S, Silver M, Tzardi M, Silver J, Ogino S, Hooshmand S, Kwak E, et al. Prognostic and predictive value of common mutations for treatment response and survival in patients with metastatic colorectal cancer. Br J Cancer. 2009;101:465–472. doi: 10.1038/sj.bjc.6605164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Migliore L, Migheli F, Spisni R, Coppedè F. Genetics, cytogenetics, and epigenetics of colorectal cancer. J Biomed Biotechnol. 2011;2011:792362. doi: 10.1155/2011/792362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez-Pons M, Cruz-Correa M. Colorectal cancer biomarkers: Where are we now? BioMed Res Int. 2015;2015:149014. doi: 10.1155/2015/149014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, Starling N. Colorectal cancer. Lancet. 2010;375:1030–1047. doi: 10.1016/S0140-6736(10)60353-4. [DOI] [PubMed] [Google Scholar]
- 10.Kuipers EJ, Rösch T, Bretthauer M. Colorectal cancer screening - optimizing current strategies and new directions. Nat Rev Clin Oncol. 2013;10:130–142. doi: 10.1038/nrclinonc.2013.12. [DOI] [PubMed] [Google Scholar]
- 11.Labianca R, Merelli B. Screening and diagnosis for colorectal cancer: Present and future. Tumori. 2010;96:889–901. doi: 10.1177/548.6506. [DOI] [PubMed] [Google Scholar]
- 12.Mahmud A, Poon R, Jonker D. PET imaging in anal canal cancer: A systematic review and meta-analysis. Br J Radiol. 2017;90:20170370. doi: 10.1259/bjr.20170370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paspulati RM, Gupta A. PET/MR imaging in cancers of the gastrointestinal tract. PET Clin. 2016;11:403–423. doi: 10.1016/j.cpet.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 14.Bond JH. Fecal occult blood test screening for colorectal cancer. Gastrointest Endosc Clin N Am. 2002;12:11–21. doi: 10.1016/S1052-5157(03)00054-0. [DOI] [PubMed] [Google Scholar]
- 15.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 16.Ahlquist DA. Molecular detection of colorectal neoplasia. Gastroenterology. 2010;138:2127–2139. doi: 10.1053/j.gastro.2010.01.055. [DOI] [PubMed] [Google Scholar]
- 17.Shah R, Jones E, Vidart V, Kuppen PJ, Conti JA, Francis NK. Biomarkers for early detection of colorectal cancer and polyps: Systematic review. Cancer Epidemiol Biomarkers Prev. 2014;23:1712–1728. doi: 10.1158/1055-9965.EPI-14-0412. [DOI] [PubMed] [Google Scholar]
- 18.Alix-Panabières C, Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016;6:479–491. doi: 10.1158/2159-8290.CD-15-1483. [DOI] [PubMed] [Google Scholar]
- 19.Shastri YM, Loitsch S, Hoepffner N, Povse N, Hanisch E, Rösch W, Mössner J, Stein JM. Comparison of an established simple office-based immunological FOBT with fecal tumor pyruvate kinase type M2 (M2-PK) for colorectal cancer screening: Prospective multicenter study. Am J Gastroenterol. 2008;103:1496–1504. doi: 10.1111/j.1572-0241.2008.01824.x. [DOI] [PubMed] [Google Scholar]
- 20.Takai T, Kanaoka S, Yoshida K, Hamaya Y, Ikuma M, Miura N, Sugimura H, Kajimura M, Hishida A. Fecal cyclooxygenase 2 plus matrix metalloproteinase 7 mRNA assays as a marker for colorectal cancer screening. Cancer Epidemiol Biomarkers Prev. 2009;18:1888–1893. doi: 10.1158/1055-9965.EPI-08-0937. [DOI] [PubMed] [Google Scholar]
- 21.Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118–126. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 22.Wu CW, Ng SS, Dong YJ, Ng SC, Leung WW, Lee CW, Wong YN, Chan FK, Yu J, Sung JJ. Detection of miR-92a and miR-21 in stool samples as potential screening biomarkers for colorectal cancer and polyps. Gut. 2012;61:739–745. doi: 10.1136/gut.2011.239236. [DOI] [PubMed] [Google Scholar]
- 23.Pan C, Yan X, Li H, Huang L, Yin M, Yang Y, Gao R, Hong L, Ma Y, Shi C, et al. Systematic literature review and clinical validation of circulating microRNAs as diagnostic biomarkers for colorectal cancer. Oncotarget. 2017;8:68317–68328. doi: 10.18632/oncotarget.19344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanaan Z, Roberts H, Eichenberger MR, Billeter A, Ocheretner G, Pan J, Rai SN, Jorden J, Williford A, Galandiuk S. A plasma microRNA panel for detection of colorectal adenomas: A step toward more precise screening for colorectal cancer. Ann Surg. 2013;258:400–408. doi: 10.1097/SLA.0b013e3182a15bcc. [DOI] [PubMed] [Google Scholar]
- 25.Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, Ahlquist DA, Berger BM. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287–1297. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 26.Imperiale TF, Ransohoff DF, Itzkowitz SH, Turnbull BA, Ross ME. Colorectal Cancer Study Group: Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med. 2004;351:2704–2714. doi: 10.1056/NEJMoa033403. [DOI] [PubMed] [Google Scholar]
- 27.Bayrak R, Yenidünya S, Haltas H. Cytokeratin 7 and cytokeratin 20 expression in colorectal adenocarcinomas. Pathol Res Pract. 2011;207:156–160. doi: 10.1016/j.prp.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 28.Righi A, Betts CM, Marchetti C, Marucci G, Montebugnoli L, Prati C, Eusebi LH, Muzzi L, Ragazzini T, Foschini MP. Merkel cells in the oral mucosa. Int J Surg Pathol. 2006;14:206–211. doi: 10.1177/1066896906290053. [DOI] [PubMed] [Google Scholar]
- 29.Stenling R, Lindberg J, Rutegård J, Palmqvist R. Altered expression of CK7 and CK20 in preneoplastic and neoplastic lesions in ulcerative colitis. APMIS. 2007;115:1219–1226. doi: 10.1111/j.1600-0643.2007.00664.x. [DOI] [PubMed] [Google Scholar]
- 30.Radović S, Selak I, Babić M, Vukobrat-Bijedić Z, Knezević Z. Anti-cytokeratin 7: A positive marker for epithelial dysplasia in flat bowel mucosa. Bosn J Basic Med Sci. 2004;4:24–30. doi: 10.17305/bjbms.2004.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gurzu S, Jung I. Aberrant pattern of the cytokeratin 7/cytokeratin 20 immunophenotype in colorectal adenocarcinomas with BRAF mutations. Pathol Res Pract. 2012;208:163–166. doi: 10.1016/j.prp.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 32.Chu P, Wu E, Weiss LM. Cytokeratin 7 and cytokeratin 20 expression in epithelial neoplasms: A survey of 435 cases. Mod Pathol. 2000;13:962–972. doi: 10.1038/modpathol.3880175. [DOI] [PubMed] [Google Scholar]
- 33.Miettinen M, Nobel MP, Tuma BT, Kovatich AJ. Keratin 17: Immunohistochemical mapping of its distribution in human epithelial tumors and its potential applications. Appl Immunohistochem. 1997;5:152–159. doi: 10.1097/00022744-199709000-00003. [DOI] [Google Scholar]
- 34.Hernandez BY, Frierson HF, Jr, Moskaluk CA, Li YJ, Clegg L, Cote TR, McCusker ME, Hankey BF, Edwards BK, Goodman MT. CK20 and CK7 protein expression in colorectal cancer: Demonstration of the utility of a population-based tissue microarray. Hum Pathol. 2005;36:275–281. doi: 10.1016/j.humpath.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 35.Silberg DG, Swain GP, Suh ER, Traber PG. Cdx1 and cdx2 expression during intestinal development. Gastroenterology. 2000;119:961–971. doi: 10.1053/gast.2000.18142. [DOI] [PubMed] [Google Scholar]
- 36.Moskaluk CA, Zhang H, Powell SM, Cerilli LA, Hampton GM, Frierson HF., Jr Cdx2 protein expression in normal and malignant human tissues: An immunohistochemical survey using tissue microarrays. Mod Pathol. 2003;16:913–919. doi: 10.1097/01.MP.0000086073.92773.55. [DOI] [PubMed] [Google Scholar]
- 37.Werling RW, Yaziji H, Bacchi CE, Gown AM. CDX2, a highly sensitive and specific marker of adenocarcinomas of intestinal origin: An immunohistochemical survey of 476 primary and metastatic carcinomas. Am J Surg Pathol. 2003;27:303–310. doi: 10.1097/00000478-200303000-00003. [DOI] [PubMed] [Google Scholar]
- 38.Zheng J, He S, Qi J, Wang X, Yu J, Wu Y, Gao Q, Wang K, Sun X. Targeted CDX2 expression inhibits aggressive phenotypes of colon cancer cells in vitro and in vivo. Int J Oncol. 2017;51:478–488. doi: 10.3892/ijo.2017.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bretscher A, Weber K. Villin: The major microfilament-associated protein of the intestinal microvillus. Proc Natl Acad Sci USA. 1979;76:2321–2325. doi: 10.1073/pnas.76.5.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patnaik S, George SP, Pham E, Roy S, Singh K, Mariadason JM, Khurana S. By moonlighting in the nucleus, villin regulates epithelial plasticity. Mol Biol Cell. 2016;27:535–548. doi: 10.1091/mbc.E15-06-0453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuroda N, Yorita K. Colon cancer with micropapillary carcinoma component: A clinopathologic study of 9 cases. Pol J Pathol. 2017;68:102–108. doi: 10.5114/pjp.2017.69685. [DOI] [PubMed] [Google Scholar]
- 42.Bacchi CE, Gown AM. Distribution and pattern of expression of villin, a gastrointestinal-associated cytoskeletal protein, in human carcinomas: A study employing paraffin-embedded tissue. Lab Invest. 1991;64:418–424. [PubMed] [Google Scholar]
- 43.Willert K, Nusse R. Beta-catenin: A key mediator of Wnt signaling. Curr Opin Genet Dev. 1998;8:95–102. doi: 10.1016/S0959-437X(98)80068-3. [DOI] [PubMed] [Google Scholar]
- 44.Wang JL, Qi Z, Li YH, Zhao HM, Chen YG, Fu W. TGFβ induced factor homeobox 1 promotes colorectal cancer development through activating Wnt/β-catenin signaling. Oncotarget. 2017;8:70214–70225. doi: 10.18632/oncotarget.19603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 46.Sheahan K, O'Brien MJ, Burke B, Dervan PA, O'Keane JC, Gottlieb LS, Zamcheck N. Differential reactivities of carcinoembryonic antigen (CEA) and CEA-related monoclonal and polyclonal antibodies in common epithelial malignancies. Am J Clin Pathol. 1990;94:157–164. doi: 10.1093/ajcp/94.2.157. [DOI] [PubMed] [Google Scholar]
- 47.Zhou M, Chinnaiyan AM, Kleer CG, Lucas PC, Rubin MA. Alpha-methylacyl-CoA racemase: A novel tumor marker over-expressed in several human cancers and their precursor lesions. Am J Surg Pathol. 2002;26:926–931. doi: 10.1097/00000478-200207000-00012. [DOI] [PubMed] [Google Scholar]
- 48.Tan E, Gouvas N, Nicholls RJ, Ziprin P, Xynos E, Tekkis PP. Diagnostic precision of carcinoembryonic antigen in the detection of recurrence of colorectal cancer. Surg Oncol. 2009;18:15–24. doi: 10.1016/j.suronc.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 49.Andrianifahanana M, Moniaux N, Batra SK. Regulation of mucin expression: Mechanistic aspects and implications for cancer and inflammatory diseases. Biochim Biophys Acta. 2006;1765:189–222. doi: 10.1016/j.bbcan.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 50.Hanski C, Hofmeier M, Schmitt-Gräff A, Riede E, Hanski ML, Borchard F, Sieber E, Niedobitek F, Foss HD, Stein H, Riecken EO. Overexpression or ectopic expression of MUC2 is the common property of mucinous carcinomas of the colon, pancreas, breast, and ovary. J Pathol. 1997;182:385–391. doi: 10.1002/(SICI)1096-9896(199708)182:4<385::AID-PATH861>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 51.Zlatian OM, Comănescu MV, Roşu AF, Roşu L, Cruce M, Găman AE, Călina CD, Sfredel V. Histochemical and immunohistochemical evidence of tumor heterogeneity in colorectal cancer. Rom J Morphol Embryol. 2015;56:175–181. [PubMed] [Google Scholar]
- 52.Park SY, Lee HS, Choe G, Chung JH, Kim WH. Clinicopathological characteristics, microsatellite instability, and expression of mucin core proteins and p53 in colorectal mucinous adenocarcinomas in relation to location. Virchows Arch. 2006;449:40–47. doi: 10.1007/s00428-006-0212-7. [DOI] [PubMed] [Google Scholar]
- 53.King RJ, Yu F, Singh PK. Genomic alterations in mucins across cancers. Oncotarget. 2017;8:67152–67168. doi: 10.18632/oncotarget.17934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang H, Jin S, Lu H, Mi S, Shao W, Zuo X, Yin H, Zeng S, Shimamoto F, Qi G. Expression of survivin, MUC2 and MUC5 in colorectal cancer and their association with clinicopathological characteristics. Oncol Lett. 2017;14:1011–1016. doi: 10.3892/ol.2017.6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dantzig AH, Hoskins JA, Tabas LB, Bright S, Shepard RL, Jenkins IL, Duckworth DC, Sportsman JR, Mackensen D, Rosteck PR, Jr, et al. Association of intestinal peptide transport with a protein related to the cadherin superfamily. Science. 1994;264:430–433. doi: 10.1126/science.8153632. [DOI] [PubMed] [Google Scholar]
- 56.Su MC, Yuan RH, Lin CY, Jeng YM. Cadherin-17 is a useful diagnostic marker for adenocarcinomas of the digestive system. Mod Pathol. 2008;21:1379–1386. doi: 10.1038/modpathol.2008.107. [DOI] [PubMed] [Google Scholar]
- 57.Panarelli NC, Yantiss RK, Yeh MM, Liu Y, Chen YT. Tissue-specific cadherin CDH17 is a useful marker of gastrointestinal adenocarcinomas with higher sensitivity than CDX2. Am J Clin Pathol. 2012;138:211–222. doi: 10.1309/AJCPKSHXI3XEHW1J. [DOI] [PubMed] [Google Scholar]
- 58.Stănculescu D, Mărgăritescu C, Stepan A, Mitruţ AO. E-cadherin in gastric carcinomas related to histological prognostic parameters. Rom J Morphol Embryol. 2011;52(Suppl):1107–1112. [PubMed] [Google Scholar]
- 59.Bian T, Zhao J, Feng J, Zhang Q, Qian L, Liu J, Jiang D, Liu Y, Zhang J. Combination of cadherin-17 and SATB homeobox 2 serves as potential optimal makers for the differential diagnosis of pulmonary enteric adenocarcinoma and metastatic colorectal adenocarcinoma. Oncotarget. 2017;8:63442–63452. doi: 10.18632/oncotarget.18828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tian X, Liu M, Zhu Q, Tan J, Liu W, Wang Y, Chen W, Zou Y, Cai Y, Han Z, Huang X. Down-regulation of liver-intestine cadherin enhances noscapine-induced apoptosis in human colon cancer cells. Expert Rev Anticancer Ther. 2017;17:857–863. doi: 10.1080/14737140.2017.1344097. [DOI] [PubMed] [Google Scholar]
- 61.Magnusson K, de Wit M, Brennan DJ, Johnson LB, McGee SF, Lundberg E, Naicker K, Klinger R, Kampf C, Asplund A, et al. SATB2 in combination with cytokeratin 20 identifies over 95% of all colorectal carcinomas. Am J Surg Pathol. 2011;35:937–948. doi: 10.1097/PAS.0b013e31821c3dae. [DOI] [PubMed] [Google Scholar]
- 62.Valori R, Rey JF, Atkin WS, Bretthauer M, Senore C, Hoff G, Kuipers EJ, Altenhofen L, Lambert R, Minoli G. International Agency for Research on Cancer: European guidelines for quality assurance in colorectal cancer screening and diagnosis First Edition - quality assurance in endoscopy in colorectal cancer screening and diagnosis. Endoscopy. 2012;44(Suppl 3):SE88–SE105. doi: 10.1055/s-0032-1309795. [DOI] [PubMed] [Google Scholar]
- 63.Lieberman DA, Weiss DG. Veterans Affairs Cooperative Study Group 380: One-time screening for colorectal cancer with combined fecal occult-blood testing and examination of the distal colon. N Engl J Med. 2001;345:555–560. doi: 10.1056/NEJMoa010328. [DOI] [PubMed] [Google Scholar]
- 64.Young GP, Symonds EL, Allison JE, Cole SR, Fraser CG, Halloran SP, Kuipers EJ, Seaman HE. Advances in fecal occult blood tests: The FIT revolution. Dig Dis Sci. 2015;60:609–622. doi: 10.1007/s10620-014-3445-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whitlock EP, Lin J, Liles E, Beil T, Fu R, O'Connor E, Thompson RN, Cardenas T. Agency for Healthcare Research and Quality. Rockville, MD: 2008. Screening for Colorectal Cancer: An Updated Systematic Review. [PubMed] [Google Scholar]
- 66.Morikawa T, Kato J, Yamaji Y, Wada R, Mitsushima T, Shiratori Y. A comparison of the immunochemical fecal occult blood test and total colonoscopy in the asymptomatic population. Gastroenterology. 2005;129:422–428. doi: 10.1053/j.gastro.2005.05.056. [DOI] [PubMed] [Google Scholar]
- 67.Gupta AK, Melton LJ, III, Petersen GM, Timmons LJ, Vege SS, Harmsen WS, Diehl NN, Zinsmeister AR, Ahlquist DA. Changing trends in the incidence, stage, survival, and screen-detection of colorectal cancer: A population-based study. Clin Gastroenterol Hepatol. 2005;3:150–158. doi: 10.1016/S1542-3565(04)00664-0. [DOI] [PubMed] [Google Scholar]
- 68.Puccini A, Berger MD, Naseem M, Tokunaga R, Battaglin F, Cao S, Hanna DL, McSkane M, Soni S, Zhang W, Lenz HJ. Colorectal cancer: Epigenetic alterations and their clinical implications. Biochim Biophys Acta. 2017;1868:439–448. doi: 10.1016/j.bbcan.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Losso GM, Moraes RS, Gentili AC, Messias-Reason IT. Microsatellite instability - MSI markers (BAT26, BAT25, D2S123, D5S346, D17S250) in rectal cancer. Arq Bras Cir Dig. 2012;25:240–244. doi: 10.1590/S0102-67202012000400006. [DOI] [PubMed] [Google Scholar]
- 70.de la Chapelle A, Hampel H. Clinical relevance of microsatellite instability in colorectal cancer. J Clin Oncol. 2010;28:3380–3387. doi: 10.1200/JCO.2009.27.0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Merok MA, Ahlquist T, Røyrvik EC, Tufteland KF, Hektoen M, Sjo OH, Mala T, Svindland A, Lothe RA, Nesbakken A. Microsatellite instability has a positive prognostic impact on stage II colorectal cancer after complete resection: Results from a large, consecutive Norwegian series. Ann Oncol. 2013;24:1274–1282. doi: 10.1093/annonc/mds614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Rüschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Laghi L, Bianchi P, Malesci A. Differences and evolution of the methods for the assessment of microsatellite instability. Oncogene. 2008;27:6313–6321. doi: 10.1038/onc.2008.217. [DOI] [PubMed] [Google Scholar]
- 74.Roth AD, Tejpar S, Yan P, Fiocca R, Dietrich D, Delorenzi M, Labianca R, Cunningham D, Van Cutsem E, Bosman F. Stage-specific prognostic value of molecular markers in colon cancer: Results of the translational study on the PETACC 3-EORTC 40993-SAKK 60–00 trial. J Clin Oncol. 2009;27(15 Suppl):4002. [Google Scholar]
- 75.Koopman M, Kortman GA, Mekenkamp L, Ligtenberg MJ, Hoogerbrugge N, Antonini NF, Punt CJ, van Krieken JH. Deficient mismatch repair system in patients with sporadic advanced colorectal cancer. Br J Cancer. 2009;100:266–273. doi: 10.1038/sj.bjc.6604867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ulamec M, Krušlin B. Colorectal cancer, novel biomarkers and immunohistochemistry - an overview. Rad Med Sci. 2014;520:41–49. [Google Scholar]
- 77.Hashimoto Y, Zumwalt TJ, Goel A. DNA methylation patterns as noninvasive biomarkers and targets of epigenetic therapies in colorectal cancer. Epigenomics. 2016;8:685–703. doi: 10.2217/epi-2015-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jair KW, Bachman KE, Suzuki H, Ting AH, Rhee I, Yen RW, Baylin SB, Schuebel KE. De novo CpG island methylation in human cancer cells. Cancer Res. 2006;66:682–692. doi: 10.1158/0008-5472.CAN-05-1980. [DOI] [PubMed] [Google Scholar]
- 79.Beggs AD, Jones A, El-Bahrawy M, Abulafi M, Hodgson SV, Tomlinson IP. Whole-genome methylation analysis of benign and malignant colorectal tumours. J Pathol. 2013;229:697–704. doi: 10.1002/path.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Toiyama Y, Okugawa Y, Goel A. DNA methylation and microRNA biomarkers for noninvasive detection of gastric and colorectal cancer. Biochem Biophys Res Commun. 2014;455:43–57. doi: 10.1016/j.bbrc.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lind GE, Danielsen SA, Ahlquist T, Merok MA, Andresen K, Skotheim RI, Hektoen M, Rognum TO, Meling GI, Hoff G, et al. Identification of an epigenetic biomarker panel with high sensitivity and specificity for colorectal cancer and adenomas. Mol Cancer. 2011;10:85. doi: 10.1186/1476-4598-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cheung AF, Carter AM, Kostova KK, Woodruff JF, Crowley D, Bronson RT, Haigis KM, Jacks T. Complete deletion of Apc results in severe polyposis in mice. Oncogene. 2010;29:1857–1864. doi: 10.1038/onc.2009.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dow LE, O'Rourke KP, Simon J, Tschaharganeh DF, van Es JH, Clevers H, Lowe SW. Apc restoration promotes cellular differentiation and reestablishes crypt homeostasis in colorectal cancer. Cell. 2015;161:1539–1552. doi: 10.1016/j.cell.2015.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liang J, Lin C, Hu F, Wang F, Zhu L, Yao X, Wang Y, Zhao Y. APC polymorphisms and the risk of colorectal neoplasia: A HuGE review and meta-analysis. Am J Epidemiol. 2013;177:1169–1179. doi: 10.1093/aje/kws382. [DOI] [PubMed] [Google Scholar]
- 85.Docea AO, Mitruţ P, Grigore D, Pirici D, Călina DC, Gofiţă E. Immunohistochemical expression of TGF beta (TGF-β), TGF beta receptor 1 (TGFBR1), and Ki67 in intestinal variant of gastric adenocarcinomas. Rom J Morphol Embryol. 2012;53(Suppl):683–692. [PubMed] [Google Scholar]
- 86.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–1665. [PubMed] [Google Scholar]
- 87.Frattini M, Gallino G, Signoroni S, Balestra D, Lusa L, Battaglia L, Sozzi G, Bertario L, Leo E, Pilotti S, Pierotti MA. Quantitative and qualitative characterization of plasma DNA identifies primary and recurrent colorectal cancer. Cancer Lett. 2008;263:170–181. doi: 10.1016/j.canlet.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 88.Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, Diaz LA, Jr, Goodman SN, David KA, Juhl H, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci USA. 2005;102:16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tänzer M, Balluff B, Distler J, Hale K, Leodolter A, Röcken C, Molnar B, Schmid R, Lofton-Day C, Schuster T, Ebert MP. Performance of epigenetic markers SEPT9 and ALX4 in plasma for detection of colorectal precancerous lesions. PLoS One. 2010;5:e9061. doi: 10.1371/journal.pone.0009061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Danese E, Montagnana M. Epigenetics of colorectal cancer: Emerging circulating diagnostic and prognostic biomarkers. Ann Transl Med. 2017;5:279. doi: 10.21037/atm.2017.04.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ahlquist DA, Harrington JJ, Burgart LJ, Roche PC. Morphometric analysis of the ‘mucocellular layer’ overlying colorectal cancer and normal mucosa: Relevance to exfoliation and stool screening. Hum Pathol. 2000;31:51–57. doi: 10.1016/S0046-8177(00)80198-7. [DOI] [PubMed] [Google Scholar]
- 92.Chen WD, Han ZJ, Skoletsky J, Olson J, Sah J, Myeroff L, Platzer P, Lu S, Dawson D, Willis J, et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst. 2005;97:1124–1132. doi: 10.1093/jnci/dji204. [DOI] [PubMed] [Google Scholar]
- 93.Itzkowitz S, Brand R, Jandorf L, Durkee K, Millholland J, Rabeneck L, Schroy PC, III, Sontag S, Johnson D, Markowitz S, et al. A simplified, noninvasive stool DNA test for colorectal cancer detection. Am J Gastroenterol. 2008;103:2862–2870. doi: 10.1111/j.1572-0241.2008.02088.x. [DOI] [PubMed] [Google Scholar]
- 94.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 95.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 96.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 97.Mendell JT. MicroRNAs: Critical regulators of development, cellular physiology and malignancy. Cell Cycle. 2005;4:1179–1184. doi: 10.4161/cc.4.9.2032. [DOI] [PubMed] [Google Scholar]
- 98.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: MicroRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 99.Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: Novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110:483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 100.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105:849–859. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ. Differential expression of microRNAs in plasma of patients with colorectal cancer: A potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 104.Carter JV, Galbraith NJ, Yang D, Burton JF, Walker SP, Galandiuk S. Blood-based microRNAs as biomarkers for the diagnosis of colorectal cancer: A systematic review and meta-analysis. Br J Cancer. 2017;116:762–774. doi: 10.1038/bjc.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Okugawa, Grady WM, Goel A. Epigenetic alterations in colorectal cancer: Emerging biomarkers. Gastroenterology. 2015;149:1204–1225.e12. doi: 10.1053/j.gastro.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen M, Lin M, Wang X. Over expression of miR-19a inhibits colorectal cancer angiogenesis by suppressing KRAS expression. Oncol Rep. 2018;39:619–626. doi: 10.3892/or.2017.6110. [DOI] [PubMed] [Google Scholar]
- 107.Link A, Balaguer F, Shen Y, Nagasaka T, Lozano JJ, Boland CR, Goel A. Fecal microRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol Biomarkers Prev. 2010;19:1766–1774. doi: 10.1158/1055-9965.EPI-10-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhu Y, Xu A, Li J, Fu J, Wang G, Yang Y, Cui L, Sun J. Fecal miR-29a and miR-224 as the noninvasive biomarkers for colorectal cancer. Cancer Biomark. 2016;16:259–264. doi: 10.3233/CBM-150563. [DOI] [PubMed] [Google Scholar]
- 109.Masuda T, Hayashi N, Kuroda Y, Ito S, Eguchi H, Mimori K. MicroRNAs as biomarkers in colorectal cancer. Cancers (Basel) 2017;9:124. doi: 10.3390/cancers9090124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Edge SB, Compton CC. The American Joint Committee on Cancer: The 7th edition of the AJCC Cancer Staging Manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 111.Perez RO, Bresciani BH, Bresciani C, Proscurshim I, Kiss D, Gama-Rodrigues J, Pereira DD, Rawet V, Cecconnello I, Habr-Gama A. Mucinous colorectal adenocarcinoma: Influence of mucin expression (Muc1, 2 and 5) on clinico-pathological features and prognosis. Int J Colorectal Dis. 2008;23:757–765. doi: 10.1007/s00384-008-0486-0. [DOI] [PubMed] [Google Scholar]
- 112.Wang S, Zhou J, Wang XY, Hao JM, Chen JZ, Zhang XM, Jin H, Liu L, Zhang YF, Liu J, et al. Down-regulated expression of SATB2 is associated with metastasis and poor prognosis in colorectal cancer. J Pathol. 2009;219:114–122. doi: 10.1002/path.2575. [DOI] [PubMed] [Google Scholar]
- 113.Eberhard J, Gaber A, Wangefjord S, Nodin B, Uhlén M, Lindquist Ericson K, Jirström K. A cohort study of the prognostic and treatment predictive value of SATB2 expression in colorectal cancer. Br J Cancer. 2012;106:931–938. doi: 10.1038/bjc.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bae JM, Lee TH, Cho NY, Kim TY, Kang GH. Loss of CDX2 expression is associated with poor prognosis in colorectal cancer patients. World J Gastroenterol. 2015;21:1457–1467. doi: 10.3748/wjg.v21.i5.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Falchook GS, Kurzrock R. VEGF and dual-EGFR inhibition in colorectal cancer. Cell Cycle. 2015;14:1129–1130. doi: 10.1080/15384101.2015.1022071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li D, Yan D, Tang H, Zhou C, Fan J, Li S, Wang X, Xia J, Huang F, Qiu G, Peng Z. IMP3 is a novel prognostic marker that correlates with colon cancer progression and pathogenesis. Ann Surg Oncol. 2009;16:3499–3506. doi: 10.1245/s10434-009-0648-5. [DOI] [PubMed] [Google Scholar]
- 117.Takahashi H, Ishikawa T, Ishiguro M, Okazaki S, Mogushi K, Kobayashi H, Iida S, Mizushima H, Tanaka H, Uetake H, Sugihara K. Prognostic significance of Traf2- and Nck-interacting kinase (TNIK) in colorectal cancer. BMC Cancer. 2015;15:794. doi: 10.1186/s12885-015-1783-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Spandidos DA, Glarakis IS, Kotsinas A, Ergazaki M, Kiaris H. Ras oncogene activation in benign and malignant colorectal tumours. Tumori. 1995;81(Suppl):7–11. [PubMed] [Google Scholar]
- 119.Kiaris H, Spandidos D. Mutations of ras genes in human tumors (Review) Int J Oncol. 1995;7:413–421. [PubMed] [Google Scholar]
- 120.Rui YY, Zhang D, Zhou ZG, Wang C, Yang L, Yu YY, Chen HN. Can K-ras gene mutation be utilized as prognostic biomarker for colorectal cancer patients receiving chemotherapy? A meta-analysis and systematic review. PLoS One. 2013;8:e77901. doi: 10.1371/journal.pone.0077901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Andreyev HJN, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten ras mutations in patients with colorectal cancer: The multicenter ‘RASCAL’ study. J Natl Cancer Inst. 1998;90:675–684. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 122.Andreyev HJN, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, Young J, Walsh T, Ward R, Hawkins N, et al. Kirsten ras mutations in patients with colorectal cancer: The ‘RASCAL II’ study. Br J Cancer. 2001;85:692–696. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang Y, Velho S, Vakiani E, Peng S, Bass AJ, Chu GC, Gierut J, Bugni JM, Der CJ, Philips M, et al. Mutant N-RAS protects colorectal cancer cells from stress-induced apoptosis and contributes to cancer development and progression. Cancer Discov. 2013;3:294–307. doi: 10.1158/2159-8290.CD-12-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Russo A, Bazan V, Iacopetta B, Kerr D, Soussi T, Gebbia N. TP53-CRC Collaborative Study Group: The TP53 colorectal cancer international collaborative study on the prognostic and predictive significance of p53 mutation: Influence of tumor site, type of mutation, and adjuvant treatment. J Clin Oncol. 2005;23:7518–7528. doi: 10.1200/JCO.2005.00.471. [DOI] [PubMed] [Google Scholar]
- 125.French AJ, Sargent DJ, Burgart LJ, Foster NR, Kabat BF, Goldberg R, Shepherd L, Windschitl HE, Thibodeau SN. Prognostic significance of defective mismatch repair and BRAF V600E in patients with colon cancer. Clin Cancer Res. 2008;14:3408–3415. doi: 10.1158/1078-0432.CCR-07-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Toon CW, Chou A, DeSilva K, Chan J, Patterson J, Clarkson A, Sioson L, Jankova L, Gill AJ. BRAFV600E immunohistochemistry in conjunction with mismatch repair status predicts survival in patients with colorectal cancer. Mod Pathol. 2014;27:644–650. doi: 10.1038/modpathol.2013.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zlobec I, Bihl MP, Schwarb H, Terracciano L, Lugli A. Clinicopathological and protein characterization of BRAF- and K-RAS-mutated colorectal cancer and implications for prognosis. Int J Cancer. 2010;127:367–380. doi: 10.1002/ijc.25042. [DOI] [PubMed] [Google Scholar]
- 128.Ogino S, Shima K, Meyerhardt JA, McCleary NJ, Ng K, Hollis D, Saltz LB, Mayer RJ, Schaefer P, Whittom R, et al. Predictive and prognostic roles of BRAF mutation in stage III colon cancer: Results from intergroup trial CALGB 89803. Clin Cancer Res. 2012;18:890–900. doi: 10.1158/1078-0432.CCR-11-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gryfe R, Kim H, Hsieh ET, Aronson MD, Holowaty EJ, Bull SB, Redston M, Gallinger S. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med. 2000;342:69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- 130.Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R, Shepherd LE, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schee K, Boye K, Abrahamsen TW, Fodstad Ø, Flatmark K. Clinical relevance of microRNA miR-21, miR-31, miR-92a, miR-101, miR-106a and miR-145 in colorectal cancer. BMC Cancer. 2012;12:505. doi: 10.1186/1471-2407-12-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Xuan Y, Yang H, Zhao L, Lau WB, Lau B, Ren N, Hu Y, Yi T, Zhao X, Zhou S, Wei Y. MicroRNAs in colorectal cancer: Small molecules with big functions. Cancer Lett. 2015;360:89–105. doi: 10.1016/j.canlet.2014.11.051. [DOI] [PubMed] [Google Scholar]
- 133.Shen WW, Zeng Z, Zhu WX, Fu GH. MiR-142-3p functions as a tumor suppressor by targeting CD133, ABCG2, and Lgr5 in colon cancer cells. J Mol Med (Berl) 2013;91:989–1000. doi: 10.1007/s00109-013-1037-x. [DOI] [PubMed] [Google Scholar]
- 134.Liu X, Wang Y, Zhao J. MicroRNA-337 inhibits colorectal cancer progression by directly targeting KRAS and suppressing the AKT and ERK pathways. Oncol Rep. 2017;38:3187–3196. doi: 10.3892/or.2017.5997. [DOI] [PubMed] [Google Scholar]
- 135.Wen L, Li Y, Jiang Z, Zhang Y, Yang B, Han F. miR-944 inhibits cell migration and invasion by targeting MACC1 in colorectal cancer. Oncol Rep. 2017;37:3415–3422. doi: 10.3892/or.2017.5611. [DOI] [PubMed] [Google Scholar]
- 136.Dai H, Hou K, Cai Z, Zhou Q, Zhu S. Low-level miR-646 in colorectal cancer inhibits cell proliferation and migration by targeting NOB1 expression. Oncol Lett. 2017;14:6708–6714. doi: 10.3892/ol.2017.7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Xu Y, Chen J, Gao C, Zhu D, Xu X, Wu C, Jiang J. MicroRNA-497 inhibits tumor growth through targeting insulin receptor substrate 1 in colorectal cancer. Oncol Lett. 2017;14:6379–6386. doi: 10.3892/ol.2017.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yang IP, Tsai HL, Miao ZF, Huang CW, Kuo CH, Wu JY, Wang WM, Juo SH, Wang JY. Development of a deregulating microRNA panel for the detection of early relapse in postoperative colorectal cancer patients. J Transl Med. 2016;14:108. doi: 10.1186/s12967-016-0856-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bovell LC, Shanmugam C, Putcha BD, Katkoori VR, Zhang B, Bae S, Singh KP, Grizzle WE, Manne U. The prognostic value of microRNAs varies with patient race/ethnicity and stage of colorectal cancer. Clin Cancer Res. 2013;19:3955–3965. doi: 10.1158/1078-0432.CCR-12-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Peng Q, Zhang X, Min M, Zou L, Shen P, Zhu Y. The clinical role of microRNA-21 as a promising biomarker in the diagnosis and prognosis of colorectal cancer: A systematic review and meta-analysis. Oncotarget. 2017;8:44893–44909. doi: 10.18632/oncotarget.16488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cao J, Yan XR, Liu T, Han XB, Yu JJ, Liu SH, Wang LB. MicroRNA-552 promotes tumor cell proliferation and migration by directly targeting DACH1 via the Wnt/β-catenin signaling pathway in colorectal cancer. Oncol Lett. 2017;14:3795–3802. doi: 10.3892/ol.2017.6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cheng D, Zhao S, Tang H, Zhang D, Sun H, Yu F, Jiang W, Yue B, Wang J, Zhang M, et al. MicroRNA-20a-5p promotes colorectal cancer invasion and metastasis by downregulating Smad4. Oncotarget. 2016;7:45199–45213. doi: 10.18632/oncotarget.9900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fukushima Y, Iinuma H, Tsukamoto M, Matsuda K, Hashiguchi Y. Clinical significance of microRNA-21 as a biomarker in each Dukes' stage of colorectal cancer. Oncol Rep. 2015;33:573–582. doi: 10.3892/or.2014.3614. [DOI] [PubMed] [Google Scholar]
- 144.Shibuya H, Iinuma H, Shimada R, Horiuchi A, Watanabe T. Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology. 2010;79:313–320. doi: 10.1159/000323283. [DOI] [PubMed] [Google Scholar]
- 145.Wang LG, Gu J. Serum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasis. Cancer Epidemiol. 2012;36:e61–e67. doi: 10.1016/j.canep.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 146.He PY, Yip WK, Chai BL, Chai BY, Jabar MF, Dusa N, Mohtarrudin N, Seow HF. Inhibition of cell migration and invasion by miR 29a 3p in a colorectal cancer cell line through suppression of CDC42BPA mRNA expression. Oncol Rep. 2017;38:3554–3566. doi: 10.3892/or.2017.6037. [DOI] [PubMed] [Google Scholar]
- 147.Lv ZC, Fan YS, Chen HB, Zhao DW. Investigation of microRNA-155 as a serum diagnostic and prognostic biomarker for colorectal cancer. Tumour Biol. 2015;36:1619–1625. doi: 10.1007/s13277-014-2760-9. [DOI] [PubMed] [Google Scholar]
- 148.Zhao J, Xu J, Zhang R. MicroRNA-411 inhibits malignant biological behaviours of colorectal cancer cells by directly targeting PIK3R3. Oncol Rep. 2018;39:633–642. doi: 10.3892/or.2017.6135. [DOI] [PubMed] [Google Scholar]
- 149.Park YJ, Park KJ, Park JG, Lee KU, Choe KJ, Kim JP. Prognostic factors in 2230 Korean colorectal cancer patients: Analysis of consecutively operated cases. World J Surg. 1999;23:721–726. doi: 10.1007/PL00012376. [DOI] [PubMed] [Google Scholar]
- 150.Park YJ, Youk EG, Choi HS, Han SU, Park KJ, Lee KU, Choe KJ, Park JG. Experience of 1446 rectal cancer patients in Korea and analysis of prognostic factors. Int J Colorectal Dis. 1999;14:101–106. doi: 10.1007/s003840050193. [DOI] [PubMed] [Google Scholar]
- 151.Wu ZY, Wan J, Zhao G, Peng L, Du JL, Yao Y, Liu QF, Lin HH. Risk factors for local recurrence of middle and lower rectal carcinoma after curative resection. World J Gastroenterol. 2008;14:4805–4809. doi: 10.3748/wjg.14.4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS, Somerfield MR, Hayes DF, Bast RC., Jr ASCO: ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313–5327. doi: 10.1200/JCO.2006.08.2644. [DOI] [PubMed] [Google Scholar]
- 153.Ozawa T, Ishihara S, Kawai K, Nozawa H, Yamaguchi H, Kitayama J, Watanabe T. Prognostic significance of preoperative serum carbohydrate antigen 19-9 in patients with stage IV colorectal cancer. Clin Colorectal Cancer. 2016;15:e157–e163. doi: 10.1016/j.clcc.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 154.Tol J, Koopman M, Miller MC, Tibbe A, Cats A, Creemers GJ, Vos AH, Nagtegaal ID, Terstappen LW, Punt CJ. Circulating tumour cells early predict progression-free and overall survival in advanced colorectal cancer patients treated with chemotherapy and targeted agents. Ann Oncol. 2010;21:1006–1012. doi: 10.1093/annonc/mdp463. [DOI] [PubMed] [Google Scholar]
- 155.Sastre J, Maestro ML, Gómez-España A, Rivera F, Valladares M, Massuti B, Benavides M, Gallén M, Marcuello E, Abad A, et al. Circulating tumor cell count is a prognostic factor in metastatic colorectal cancer patients receiving first-line chemotherapy plus bevacizumab: A Spanish Cooperative Group for the Treatment of Digestive Tumors study. Oncologist. 2012;17:947–955. doi: 10.1634/theoncologist.2012-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dinu D, Dobre M, Panaitescu E, Bîrlă R, Iosif C, Hoara P, Caragui A, Boeriu M, Constantinoiu S, Ardeleanu C. Prognostic significance of KRAS gene mutations in colorectal cancer - preliminary study. J Med Life. 2014;7:581–587. [PMC free article] [PubMed] [Google Scholar]
- 157.Peeters M, Douillard JY, Van Cutsem E, Siena S, Zhang K, Williams R, Wiezorek J. Mutant KRAS codon 12 and 13 alleles in patients with metastatic colorectal cancer: Assessment as prognostic and predictive biomarkers of response to panitumumab. J Clin Oncol. 2013;31:759–765. doi: 10.1200/JCO.2012.45.1492. [DOI] [PubMed] [Google Scholar]
- 158.De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, Siena S, Lamba S, Arena S, Frattini M, Piessevaux H, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010;304:1812–1820. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 159.Hecht JR, Douillard JY, Schwartzberg L, Grothey A, Kopetz S, Rong A, Oliner KS, Sidhu R. Extended RAS analysis for anti-epidermal growth factor therapy in patients with metastatic colorectal cancer. Cancer Treat Rev. 2015;41:653–659. doi: 10.1016/j.ctrv.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 160.Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 161.Toon CW, Walsh MD, Chou A, Capper D, Clarkson A, Sioson L, Clarke S, Mead S, Walters RJ, Clendenning M, et al. BRAFV600E immunohistochemistry facilitates universal screening of colorectal cancers for Lynch syndrome. Am J Surg Pathol. 2013;37:1592–1602. doi: 10.1097/PAS.0b013e31828f233d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kuan SF, Navina S, Cressman KL, Pai RK. Immunohistochemical detection of BRAF V600E mutant protein using the VE1 antibody in colorectal carcinoma is highly concordant with molecular testing but requires rigorous antibody optimization. Hum Pathol. 2014;45:464–472. doi: 10.1016/j.humpath.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 163.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: A retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 164.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 165.Sargent DJ, Marsoni S, Thibodeau SN, Labianca R, Hamilton SR, Torri V, Monges G, Ribic C, Grothey A, Gallinger S. Confirmation of deficient mismatch repair (dMMR) as a predictive marker for lack of benefit from 5-FU based chemotherapy in stage II and III colon cancer (CC): A pooled molecular reanalysis of randomized chemotherapy trials. J Clin Oncol. 2008;26(15 suppl):4008. doi: 10.1200/jco.2008.26.15_suppl.4008. [DOI] [Google Scholar]
- 166.Jover R, Zapater P, Castells A, Llor X, Andreu M, Cubiella J, Balaguer F, Sempere L, Xicola RM, Bujanda L, et al. Gastrointestinal Oncology Group of the Spanish Gastroenterological Association: The efficacy of adjuvant chemotherapy with 5-fluorouracil in colorectal cancer depends on the mismatch repair status. Eur J Cancer. 2009;45:365–373. doi: 10.1016/j.ejca.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 167.Pohl A, El-Khoueiry A, Yang D, Zhang W, Lurje G, Ning Y, Winder T, Hu-Lieskoven S, Iqbal S, Danenberg KD, et al. Pharmacogenetic profiling of CD133 is associated with response rate (RR) and progression-free survival (PFS) in patients with metastatic colorectal cancer (mCRC), treated with bevacizumab-based chemotherapy. Pharmacogenomics J. 2013;13:173–180. doi: 10.1038/tpj.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ong CW, Kim LG, Kong HH, Low LY, Iacopetta B, Soong R, Salto-Tellez M. CD133 expression predicts for non-response to chemotherapy in colorectal cancer. Mod Pathol. 2010;23:450–457. doi: 10.1038/modpathol.2009.181. [DOI] [PubMed] [Google Scholar]
- 169.Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 170.Wu X, Amos CI, Zhu Y, Zhao H, Grossman BH, Shay JW, Luo S, Hong WK, Spitz MR. Telomere dysfunction: a potential cancer predisposition factor. J Natl Cancer Inst. 2003;95:1211–1218. doi: 10.1093/jnci/djg011. [DOI] [PubMed] [Google Scholar]
- 171.Willeit P, Willeit J, Mayr A, Weger S, Oberhollenzer F, Brandstätter A, Kronenberg F, Kiechl S. Telomere length and risk of incident cancer and cancer mortality. JAMA. 2010;304:69–75. doi: 10.1001/jama.2010.897. [DOI] [PubMed] [Google Scholar]
- 172.Calado RT, Young NS. Telomere diseases. N Engl J Med. 2009;361:2353–2365. doi: 10.1056/NEJMra0903373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.O'Sullivan J, Risques RA, Mandelson MT, Chen L, Brentnall TA, Bronner MP, Macmillan MP, Feng Z, Siebert JR, Potter JD, Rabinovitch PS. Telomere length in the colon declines with age: A relation to colorectal cancer? Cancer Epidemiol Biomarkers Prev. 2006;15:573–577. doi: 10.1158/1055-9965.EPI-05-0542. [DOI] [PubMed] [Google Scholar]
- 174.Raynaud CM, Jang SJ, Nuciforo P, Lantuejoul S, Brambilla E, Mounier N, Olaussen KA, André F, Morat L, Sabatier L, Soria JC. Telomere shortening is correlated with the DNA damage response and telomeric protein down-regulation in colorectal preneoplastic lesions. Ann Oncol. 2008;19:1875–1881. doi: 10.1093/annonc/mdn405. [DOI] [PubMed] [Google Scholar]
- 175.Aghagolzadeh P, Radpour R. New trends in molecular and cellular biomarker discovery for colorectal cancer. World J Gastroenterol. 2016;22:5678–5693. doi: 10.3748/wjg.v22.i25.5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Fernández-Marcelo T, Sánchez-Pernaute A, Pascua I, De Juan C, Head J, Torres-García A-J, Iniesta P. Clinical relevance of telomere status and telomerase activity in colorectal cancer. PLoS One. 2016;11:e0149626. doi: 10.1371/journal.pone.0149626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gertler R, Rosenberg R, Stricker D, Friederichs J, Hoos A, Werner M, Ulm K, Holzmann B, Nekarda H, Siewert JR. Telomere length and human telomerase reverse transcriptase expression as markers for progression and prognosis of colorectal carcinoma. J Clin Oncol. 2004;22:1807–1814. doi: 10.1200/JCO.2004.09.160. [DOI] [PubMed] [Google Scholar]
- 178.Norppa H, Falck GC. What do human micronuclei contain? Mutagenesis. 2003;18:221–233. doi: 10.1093/mutage/18.3.221. [DOI] [PubMed] [Google Scholar]
- 179.Farhadi S, Mohamadi M, Mohamadi M. Repair index in examination of nuclear changes in the buccal mucosa of smokers: a useful method for screening of oral cancer. Asian Pac J Cancer Prev. 2017;18:3087–3090. doi: 10.22034/APJCP.2017.18.11.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.El-Zein RA, Abdel-Rahman S, Santee KJ, Yu R, Shete S. Identification of small and non-small cell lung cancer markers in peripheral blood using cytokinesis-blocked micronucleus and spectral karyotyping assays. Cytogenet Genome Res. 2017;152:122–131. doi: 10.1159/000479809. [DOI] [PubMed] [Google Scholar]
- 181.Maffei F, Moraga Zolezzi JM, Angelini S, Zenesini C, Musti M, Festi D, Cantelli-Forti G, Hrelia P. Micronucleus frequency in human peripheral blood lymphocytes as a biomarker for the early detection of colorectal cancer risk. Mutagenesis. 2014;29:221–225. doi: 10.1093/mutage/geu007. [DOI] [PubMed] [Google Scholar]
- 182.Karaman A, Binici DN, Kabalar ME, Calikuşu Z. Micronucleus analysis in patients with colorectal adenocarcinoma and colorectal polyps. World J Gastroenterol. 2008;14:6835–6839. doi: 10.3748/wjg.14.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ravegnini G, Moraga Zolezzi JM, Maffei F, Musti M, Zenesini C, Simeon V, Sammarini G, Festi D, Hrelia P, Angelini S. Simultaneous analysis of SEPT9 promoter methylation status, micronuclei frequency, and folate-related gene polymorphisms: The potential for a novel blood-based colorectal cancer biomarker. Int J Mol Sci. 2015;16:28486–284897. doi: 10.3390/ijms161226113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ionescu EM, Nicolaie T, Ionescu MA, Becheanu G, Andrei F, Diculescu M, Ciocirlan M. Predictive cytogenetic biomarkers for colorectal neoplasia in medium risk patients. J Med Life. 2015;8:398–403. [PMC free article] [PubMed] [Google Scholar]
- 185.Kronborg O, Jørgensen OD, Fenger C, Rasmussen M. Randomized study of biennial screening with a faecal occult blood test: Results after nine screening rounds. Scand J Gastroenterol. 2004;39:846–851. doi: 10.1080/00365520410003182. [DOI] [PubMed] [Google Scholar]
- 186.Scholefield JH, Moss S, Sufi F, Mangham CM, Hardcastle JD. Effect of faecal occult blood screening on mortality from colorectal cancer: Results from a randomised controlled trial. Gut. 2002;50:840–844. doi: 10.1136/gut.50.6.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Health Quality Ontario, corp-author. Fecal occult blood test for colorectal cancer screening: An evidence-based analysis. Ont Health Technol Assess Ser. 2009;9:1–40. [PMC free article] [PubMed] [Google Scholar]
- 188.Dancourt V, Lejeune C, Lepage C, Gailliard MC, Meny B, Faivre J. Immunochemical faecal occult blood tests are superior to guaiac-based tests for the detection of colorectal neoplasms. Eur J Cancer. 2008;44:2254–2258. doi: 10.1016/j.ejca.2008.06.041. [DOI] [PubMed] [Google Scholar]
- 189.Faivre J, Dancourt V, Denis B, Dorval E, Piette C, Perrin P, Bidan JM, Jard C, Jung S, Levillain R, et al. Comparison between a guaiac and three immunochemical faecal occult blood tests in screening for colorectal cancer. Eur J Cancer. 2012;48:2969–2976. doi: 10.1016/j.ejca.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 190.Lin JS, Piper MA, Perdue LA, Rutter CM, Webber EM, O'Connor E, Smith N, Whitlock EP. Screening for colorectal cancer: Updated evidence report and systematic review for the US Preventive Services Task Force. JAMA. 2016;315:2576–2594. doi: 10.1001/jama.2016.3332. [DOI] [PubMed] [Google Scholar]
- 191.Koo S, Neilson LJ, Von Wagner C, Rees CJ. The NHS bowel cancer screening program: Current perspectives on strategies for improvement. Risk Manag Healthc Policy. 2017;10:177–187. doi: 10.2147/RMHP.S109116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Moss S, Mathews C, Day TJ, Smith S, Seaman HE, Snowball J, Halloran SP. Increased uptake and improved outcomes of bowel cancer screening with a faecal immunochemical test: Results from a pilot study within the national screening programme in England. Gut. 2017;66:1631–1644. doi: 10.1136/gutjnl-2015-310691. [DOI] [PubMed] [Google Scholar]
- 193.Rozen P, Levi Z, Hazazi R, Waked A, Vilkin A, Maoz E, Birkenfeld S, Niv Y. Quantitative colonoscopic evaluation of relative efficiencies of an immunochemical faecal occult blood test and a sensitive guaiac test for detecting significant colorectal neoplasms. Aliment Pharmacol Ther. 2009;29:450–457. doi: 10.1111/j.1365-2036.2008.03898.x. [DOI] [PubMed] [Google Scholar]
- 194.Hoffman RM, Steel S, Yee EFT, Massie L, Schrader RM, Murata GH. Colorectal cancer screening adherence is higher with fecal immunochemical tests than guaiac-based fecal occult blood tests: A randomized, controlled trial. Prev Med. 2010;50:297–299. doi: 10.1016/j.ypmed.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 195.Brenner H, Tao S. Superior diagnostic performance of faecal immunochemical tests for haemoglobin in a head-to-head comparison with guaiac based faecal occult blood test among 2235 participants of screening colonoscopy. Eur J Cancer. 2013;49:3049–3054. doi: 10.1016/j.ejca.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 196.Fitzpatrick-Lewis D, Ali MU, Warren R, Kenny M, Sherifali D, Raina P. Screening for colorectal cancer: A systematic review and meta-analysis. Clin Colorectal Cancer. 2016;15:298–313. doi: 10.1016/j.clcc.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 197.Murphy J, Halloran S, Gray A. Cost-effectiveness of the faecal immunochemical test at a range of positivity thresholds compared with the guaiac faecal occult blood test in the NHS Bowel Cancer Screening Programme in England. BMJ Open. 2017;7:e017186. doi: 10.1136/bmjopen-2017-017186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lee JK, Liles EG, Bent S, Levin TR, Corley DA. Accuracy of fecal immunochemical tests for colorectal cancer: Systematic review and meta-analysis. Ann Intern Med. 2014;160:171. doi: 10.7326/M13-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.