Abstract
Public transcriptome databases provide a valuable resource for genome-wide co-expression network analysis and investigation of the molecular mechanisms that underlie pathogenesis. To discover genes that may affect patient survival, a large-scale analysis of human colorectal cancer (CRC) datasets that were retrieved from the NCBI Gene Expression Omnibus was performed. A gene co-expression network was constructed using weighted gene co-expression network analysis (WGCNA). A total of 18 co-expressed gene modules were identified, of which two genes corresponded to cell migration and the cell cycle, two genes were involved in immune responses, two genes corresponded to mitochondrial function, and one gene corresponded to RNA splicing. A total of eight hub genes in the cell migration/extracellular matrix module were associated with poor prognosis in CRC, and the P-value for collagen type VI α3 chain (COL6A3) was the lowest. In silico analysis of cell type-specific gene expression and COL6A3 knockout experiments indicated the clinical relevance of COL6A3 in the development of CRC. In summary, the present analysis provides a basis for understanding the molecular characterization of CRC at the transcription level. COL6A3 may be a promising biomarker or target for the prognosis and treatment of CRC.
Keywords: colorectal cancer, collagen type VI α3 chain, gene co-expression network, microarray, transcriptome, proliferation, invasion
Introduction
Colorectal cancer (CRC) is a major disease worldwide. Histopathology has been a traditional method for the diagnosis of CRC (1). However, recent genome-wide molecular analysis indicated disease heterogeneity in patients with similar pathology (2). With the advent and lower cost of microarray and next-generation sequencing (NGS) technologies, genome-wide screening of candidate genes is possible. To date, tens of millions of transcriptome datasets have been generated and deposited in public databases (3). Therefore, it is a great challenge for researchers to extract meaningful information from these huge datasets.
Many scientists reanalyze transcriptomes using publically available data to promote the understanding of CRC. For example, ColoGuideEx is a 13-gene expression classifier for prognosis prediction specific to patients with stage II CRC (4). ColoGuideEx is derived from 315 CRC transcriptomes, and its robustness was shown across patient series, populations and different microarrays (4). A 54 gene-set metastasis-prone signature was proposed for patients with early-stage mismatch-repair proficient sporadic colorectal cancer (5). Gene Set Enrichment Analysis was applied to mucosal and adenoma transcriptomes to identify gene signatures that are associated with colon carcinogenesis (6). Genome-wide association studies (GWAS) have identified several loci with weak predictive value in CRC (7). Pathway-based analyses have enhanced the interpretation of GWAS data, and transcriptome data have confirmed the pathways (7). In addition, NGS has been applied to CRC transcriptome analysis. For example, a tumor-restricted gene fusion, PRTEN-NOTCH2, was detected and experimentally confirmed, which provides deeper insights into the complexity of regulatory changes during tumorigenesis (8). There are also numerous CRC transcriptome analyses that characterize gene expression at the genome level (1).
With the development of cell separation methods, cell type-specific analyses have also been performed. For instance, cluster of differentiation (CD) 133 is an important biomarker of CRC stem cells. Transcriptome analysis of CD133+ stem cells indicated the prognostic value of survivin in CRC (9). Different cell types in the CRC niche, including cancer cells and stromal cells, were separated by flow cytometry. Transcriptome analyses have identified stromal transforming growth factor-β gene signatures that predict recurrence (10). Transcriptome analyses have also identified a stem/serrated/mesenchymal (SSM) transcriptional subtype of CRC, which is associated with poor diagnosis. The upregulated genes in the SSM subtype are prominently expressed by stromal cells, which suggest that these transcripts are derived from stromal cells instead of epithelial cancer cells (11). The involement of stromal cells in CRC has been well established (11).
In the present study, weighted gene co-expression network analysis (WGCNA) of the CRC transcriptome was performed and 18 gene co-expression modules were identified. Highly connected genes were screened, and the clinical relevance of these genes was validated in additional datasets containing clinical parameters. The expression of collagen type VI α3 chain (COL6A3) was discovered to be fibroblast-specific and associated with stromal cancer. The role of COL6A3 was experimentally verified in CRC cell line, SW480.
Materials and methods
CRC transcriptome data collection and preprocessing
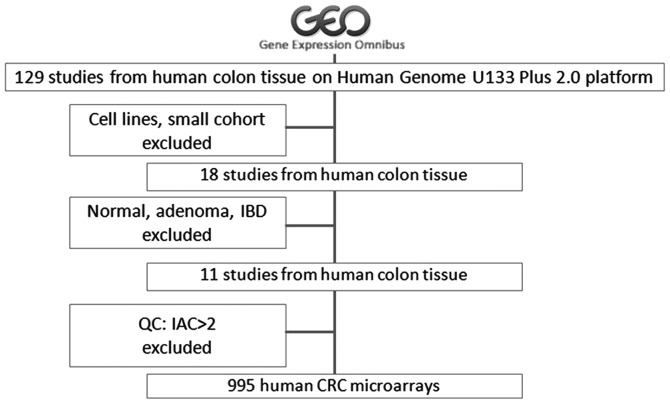
CRC transcriptome data were downloaded from the NCBI GEO database (12) using the query terms ‘(colon OR colorectal) AND GPL570’ to obtain all datasets describing microarray experiments involving CRC using the Affymetrix HG_U133_2 platform. After filtering the studies that used cell lines or normal mucosa, or involved small patient cohorts, a total of 1,045 chips from 11 studies were included for analysis (Fig. 1). The raw data were processed using the Affymetrix Expression Console software (version 1.2; Affymetrix, Inc., Santa Clara, CA, USA) (www.thermofisher.com/cn/zh/home/global/forms/life-science/download-tac-software.html). Gene expression values were generated using the MAS5 algorithm. Gene co-expression analyses are particularly sensitive to the presence of outlier samples and systematic biases in microarray data. To make the analysis more meaningful, strict quality control on chip data was performed prior to further analysis. A custom CDF file (13) was used, and non-specific and mis-targeted microarray probes were masked prior to the generation of expression values. Unlike standard CDF files, this custom file produces gene-level instead of probe set-level expression values. Scaled expression values were imported into R (version 2.13.0) for the detection and removal of outliers (14). Inter-array correlations (IACs) were averaged for each array and compared with the resulting distribution of IACs for the dataset. In general, samples with a mean IAC <2.0 standard deviations below the mean IAC for the dataset were removed. Samples were also hierarchically clustered using average linkage and 1-IAC as a distance metric to identify outliers. This procedure was repeated until no outliers were evident. This approach constitutes an unbiased method for the identification and removal of samples with aberrant global gene expression. Finally, 995 raw files were retained. Expression values were further normalized using the quantile method. Genes that were present in <30% of the samples were excluded from further analysis. Batch effects were commonly observed across multiple batches of microarray experiments or between different labs. To eliminate batch effects, additional normalization was performed using the R package ‘COMBAT’ (15). Datasets GSE17536 and GSE41258 were used for survival and metastasis analysis. GSE39397 was used for cell type specific COL6A3 expression analysis.
Figure 1.
Colorectal cancer microarray expression data used in analysis. All human microarray data from the Gene Expression Omnibus database were queried with the terms: ‘(colon OR colorectal)’ AND ‘GPL570’. After filtering datasets from studies that employed cell lines, normal samples, adenoma and inflammatory bowel disease, 11 studies were retained. These data were further filtered by expression data quality control and 995 microarrays were included.
WGCNA and module annotation
The networks were constructed from the weighted correlation matrices following the WGCNA protocols (14,16,17). Gene ontology enrichment and Kyoto Encyclopedia of Genes and Genomes pathway analysis for network modules were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (18–20). Association of the modules with genomic dysfunction was detected using DAVID on the basis of overrepresentation of genes encoded at neighboring chromosomal locations. In DAVID, an overrepresentation of a term was defined as a modified Fisher's exact P-value with an adjustment for multiple tests using the Benjamini method. For genes that were not characterized by DAVID, a PubMed literature search was performed.
Visualization
To visualize the pairwise associations between the genes, Cytoscape was used (21). A total of 150 pairs of genes with the highest intramodular topological overlap matrix (TOM) value were depicted. The lines link nodes that correspond to TOM values between the connected nodes. Kaplan-Meier survival analyses for censored data were plotted using SPSS (version 17.0; SPSS, Inc., Chicago, IL, USA) For each analysis, survival curves were constructed, and the log-rank test was used to assess the presence of significant differences between the curves for any two groups being compared.
Generation of a clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 COL6A3 knockout cell line
The SW480 cell line was purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA) was used in the present study. The cell line was cultured with Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) that was supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), streptomycin (100 µg/ml) and penicillin (100 U/ml). CRISPR/Cas9 COL6A3 knockout (KO) plasmids were constructed by Shanghai GenePharma Co., Ltd. (Shanghai, China). Briefly, the target sequence of the COL6A3 active fragment, endotrophin (5′-CGAAAGACGAAGGAACTTGC-3′) was used for the design of single guide RNA. The two sequences Y2854-S (5′-caccgCGAAAGACGAAGGAACTTGC-3′) and Y2854-A (5′-aaacGCAAGTTCCTTCGTCTTTCGc-3′) were synthesized and annealed. The sequences were inserted into the pU6-gRNA-Cas9-EGFP vector using T4 DNA ligase. Competent cells were prepared via CaCl2 treatment and used for heat-shock transformation. The sequencing of plasmids extracted by QIAGEN Plasmid Midi Kit (Qiagen, Shanghai, China) from one positive clone confirmed the target sequence. Positive colonies were then amplified to extract plasmid DNA.
A total of two knockout cell lines, SW480-20 and SW480-28, were generated by Shanghai GenePharma Co., Ltd. Briefly, SW480 cells were transfected with 0.6 µg CRISPR/Cas9 COL6A3 KO plasmids using Lipofectamine© 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) in 24-well plate. GFP-positive cells were subsequently selected by fluorescence-activated cell sorting analysis after 48 h. Single cell colonies were selected, and tested by polymerase chain reaction and sequencing. PCR was performed in a 50 µl volume reaction mixture containing 25 µl of 2X Phanta buffer, 2 µl of each primer (20 µmol l−1), 3 µl cDNA and 2 µl Phanta Max Super-Fidelity DNA polymerase (Vazyme Biotech Co., Ltd., Nanjing, China). The PCR profile employed for all primer sets consisted of an initial denaturation at 95°C for 3 min followed by 42 cycles of 95°C for 15 sec, 60°C for 15 sec, 72°C for 20 sec, and a final extension for 5 min at 72°C.
Cell proliferation, invasion and migration analyses
For the proliferation assay, the cells (density, 3×103) were seeded in a 96-well plate with five replicates for every group. The cells were then incubated in 10% Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) at 37°C. After 0, 24, 48, 72 and 96 h, proliferation rates were determined by detecting the absorbance at 450 nm.
For the invasion assay, Transwell Matrigel invasion chambers in two 24-well plates (pore size, 8 µm; BD Biosciences, San Jose, CA, USA) were used according to the manufacturer's instructions. Briefly, the cells were serum-starved for 6 h in DMEM containing 0.1% FBS. Serum-starved cells were trypsinized and resuspended in DMEM containing 0.1% FBS, and 200 µl serum-free medium containing 3×105 cells from each subgroup were added to the upper chamber of each well coated with 50 mg/l Matrigel (BD Biosciences). A volume of 0.6 ml 15% FBS-containing medium was then added to the lower chamber as a chemoattractant. After 24 h at 37°C, the cells on the upper membrane surface were removed with a cotton swab. The inserts were fixed by treatment with 95% ethanol for 30 min and stained with 0.1% crystal violet solution (Beyotime Institute of Biotechnology, Shanghai, China) at 37°C for 30 min. The cells on the bottom of the membrane were counted from three different light microscopic fields, and the mean number of cells was calculated.
For the scratch wound-healing assay, 3×105 cells were seeded into a 6-well tissue culture plate, and after 48 h the cell monolayer reached 80% confluence. Then, the monolayer was gently scratched with a 20-µl pipette tip, where a straight line was scratched in one direction. The well was gently washed twice with PBS to remove the detached cells. Then, the medium in the wells was replaced with fresh medium. The cells were grown for additional 24 and 48 h at 37°C. Images of the monolayer were captured using a light microscope. The same microscope configuration was set for capturing images at three different fields. The blank area was quantitatively evaluated using ImageJ (National Institutes of Health, Bethesda, Maryland, USA).
Flow cytometric analysis
For cell cycle analysis, the cells were harvested by trypsinization, fixed with 70% ethanol at −20°C and stored at 4°C overnight. The cells were subjected to propidium iodide (PI; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) staining and flow cytometric analysis. For apoptosis analysis, the Annexin V/PI assay was performed according to the manufacturer's instructions (M3031; MB-CHEM Corporation, Maharashtra 400009, India). Briefly, the cells were washed and resuspended in 400 µl binding buffer (M3036; MB-CHEM Corporation, Maharashtra 400009, India) and 5 µl Annexin V-fluorescein isothiocyanate, followed by incubation for 5 min at 4°C in the dark.
Statistical analysis
Differences between two groups were assessed using unpaired two-tailed t-tests. For association analysis between gene expression and patient survival, the univariate Cox model was used. When clinical parameters were also considered, the multivariate Cox model was used. Analysis of variance (ANOVA) was used to compare differences in COL6A3 expression between different types of CRC cells. The survival time statistics were calculated by log-rank and visualized in Kaplan-Meier survival curves. P<0.05 was considered to indicate a statistically significant difference. Stromal and immune scores were calculated by the ESTIMATE package (bioinformatics.mdanderson.org/estimate/rpackage.html) in R (version 2.15.3).
Results
Successful construction of a gene co-expression network for CRC
The term ‘(colon OR colorectal) AND GPL570’ was used to search the NCBI GEO database to retrieve CRC transcriptome datasets. Data from cell lines or studies with a small sample size, normal tissues, adenoma tissues or inflammatory bowel disease were excluded. Consequently, 11 GSE datasets were retained. The Affymetrix Expression Console software was used, and low-quality probe sets were filtered prior to gene expression calling. Gene co-expression network analysis is sensitive to abnormal samples. To further eliminate these outliers, the samples were evaluated by inter-array correlation analysis. As a result, data from 995 CRC samples were obtained for downstream gene network construction (Fig. 1).
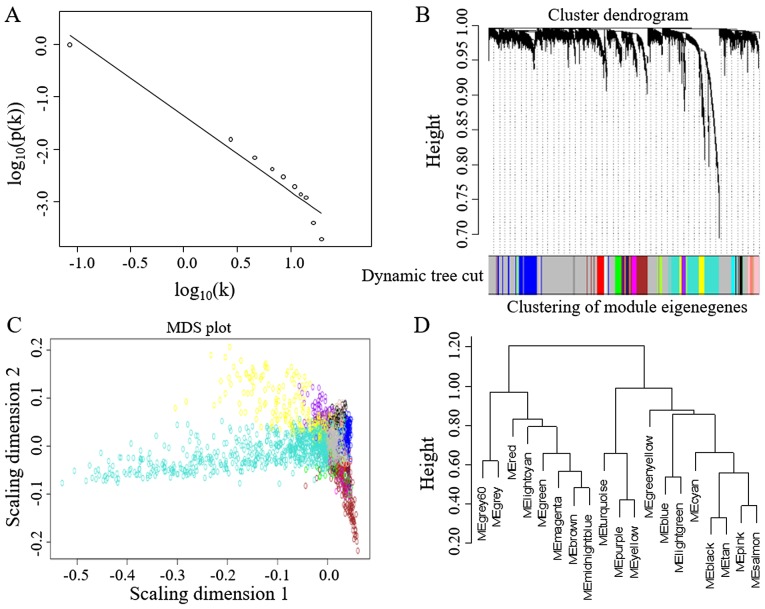
The robustness of the WGCNA method has been well established given its high citation rate in the literature (17). WGCNA could effectively identify gene modules with similar expression patterns and hub genes with high connectivity. The most variable 5,000 genes by standard deviation/average were used for gene co-expression network construction. Network statistics such as power, cluster dendrogram and module eigengenes are shown in Fig. 2. A total of 18 modules of co-expressed genes were identified (Table I). These modules could be classified into two main categories. One class of modules was enriched with chromosomal gene expression, including chromosomes 7, 8, 13, 20 and X. The other class of modules was associated with various biological processes, including mitochondrion function, extracellular matrix (ECM) function, immune responses, carbohydrate metabolism, protein localization and the cell cycle (Table I).
Figure 2.
Network analysis of gene expression in CRC identifies modules of co-expressed genes. (A) The constructed networks obey the power-law. When a power of 9 was selected, the curve corresponded to the regression line with an index of R2=0.94. The CRC network exhibits a scale-free topology. (B) Dendrograms produced by average linkage hierarchical clustering of 5,000 genes, which is based on topological overlap matrix (TOM). The modules were assigned colors as indicated in the horizontal bar beneath the liver dendrogram. (C) Classical multidimensional scaling plots in two dimensions (color-coded as in B) depict the relative size of the modules. (D) The modules were hierarchically clustered on the basis of correlations between their eigengenes (MEs). CRC, colorectal cancer.
Table I.
Functional annotation of CRC modules.
| Modulea | Annotationb | KEGG pathwaysb | Hub genes |
|---|---|---|---|
| Midnight blue (56) | Mitochondrial part (4.9E-3) 7 (3.2E-71) | C7ORF30, EIF3B, MRPS24 | |
| Tan (74) | CD55, DUSP4, LOC100507649 | ||
| Black (110) | Generation of precursor metabolites and energy (2.5E-3) | HSPA4L, LOC100507455, C1QBP | |
| Mitochondrion (7.7E-10)18 (2.0E-23) | |||
| Green (125) | X (6.2E-175)Xq28 (3.8E-17) | UBE2A, PHF16, NKRF | |
| Pink (99) | O-Glycan biosynthesis (2.7E-2) | ST6GALNAC1, SPINK4, REG4 | |
| Brown (370) | 20 (9.8E-57)20q13.13 (8.3E-5) | STAU1, DYNLRB1, DDX27 | |
| Light green (42) | 15 (5.0E-61)15q14 (2.0E-5) | COPS2, RSL24D1, MFAP1 | |
| Cyan (65) | 14 (7.7E-97)14q11.2 (5.6E-6) | C14ORF166, TMX1, TMED10 | |
| Turquoise (723) | Cell motion (4.0E-9) | Focal adhesion (1.8E-11) | SPARC, COL5A2, TIMP2 |
| Extracellular matrix (5.0E-23) | ECM-receptor interaction (2.8E-9) | ||
| Yellow (208) | Immune response (6.3E-32) | Antigen processing and presentation (3.4E-7) | CD53, LAPTM5, FCER1G |
| MHC class II protein complex (7.7E-9) | Intestinal immune network for IgA production (1.1E-6) | ||
| Grey (42) | RNA splicing (1.4E-7) | Spliceosome (4.0E-3) | NCRNA00201, NKTR, PNISR |
| Nuclear speck (4.8E-6) | |||
| Salmon (73) | Carbonate dehydratase (6.5E-3) | ZG16, CA1, CA4 | |
| Green yellow (77) | Regulation of protein localization (4.6E-2) | Pathways in cancer (2.5E-3) | DDX3X, PTPN11, G3BP2 |
| Membrane fraction (2.9E-2) | |||
| Light cyan (49) | 20 (4.5E-73)20q13 (8.9E-21) | PSMF1, MKKS, SNRPB | |
| Magenta (97) | Nucleoplasm (3.8E-3) 13 (1.8E-153)13q34 (5.8E-17) | CUL4A, IPO5, UCHL3 | |
| Red (120) | 8 (7.8E-165)8q24.3 (4.1E-18) | DCAF13, SLC25A32, ARMC1 | |
| Blue (455) | Mitotic cell cycle (5.5E-34) | Proteasome (7.8E-10) | BUB1B, OIP5, PRC1 |
| Nuclear lumen (1.9E-23) | Spliceosome (1.4E-8) | ||
| DNA replication (6.5E-8) | |||
| Purple (78) | Immune response (1.2E-17) | Antigen processing and presentation (3.9E-4) | IFIT3, CMPK2, IFIT1 |
| MHC class I protein complex (9.5E-5) |
Number of genes in the given module (the no. of genes is presented in the parentheses).
Annotation includes GO biological process, cellular component, molecular function, and chromosome. Representative functional terms overrepresented in the given module (Fisher's exact test P-values are presented in parentheses).
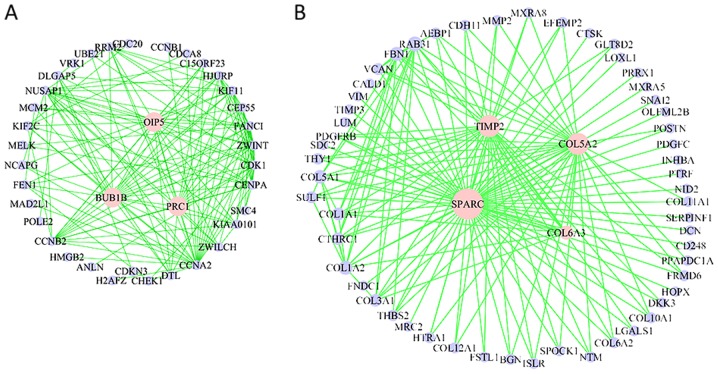
The hub genes may be important for the survival of patients with CRC (22). WGCNA results also provide the connectivity of each gene within a module. A number of these hub genes have roles in cancer development. For example, in the cell cycle module, the roles of Opa interacting protein 5, protein regulator of cytokinesis 1 and BUB1 mitotic checkpoint serine/threonine kinase B in cancer have been previously demonstrated (23). A number of the hub genes have well-known functions in regulating the DNA damage checkpoint, genome stability and cell cycle arrest, including checkpoint kinase 1, cyclin B2, cyclin A2 and nucleolar and spindle-associated protein 1 (24). In the cell migration/ECM module, there are many collagen encoding genes, including collagen type V α2 chain (COL5A2), collagen type VI α2 chain, COL6A3, TIMP metallopeptidase inhibitor 2 (TIMP2) and collagen type X α1 chain (Fig. 3A and B).
Figure 3.
Visualization of modules was performed using VisAnt, where 150 strongest connections were constructed within each module. The green lines denote the correlation between the two nodes. The pink node indicates its relative high strength of correlation. (A) The network of module blue. (B) The network of module turquoise.
Clinical implications of the hub genes in the ECM module
ECM contributes to tumor invasion and metastasis, which is a vital factor underlying patient mortality (25). ECM can act as a scaffold for cell migration, a reservoir for cytokines and growth factors, and it can transmit signals by binding with receptors (26). In the present study, the top 150 connections from the ECM module were extracted, and a co-expression network was visualized (Fig. 3B). Many of these genes have been previously implicated in cancer. For example, the frequency of the TIMP-2 rs81799090 genotype G/G was higher in patients with metastasis compared with those without metastasis (27). Cysteine-rich protein is predominantly expressed by stromal cells in CRC and is able to inhibit invasion and metastasis (28).
The ECM module is enriched with genes that are associated with cell migration, which is an important factor in metastasis. The top 15 genes with high connectivity were analyzed using the univariate Cox model, and eight of these genes were associated with overall survival, including collagen type I α1 chain (P=0.017), collagen type III α1 chain (P=0.011), sulfatase 1 (P=0.011), collagen triple helix repeat containing 1 (P=0.016), collagen type V α1 chain (P=0.029), COL6A3 (P=0.007), TIMP2 (P=0.029) and COL5A2 (P=0.014). Among these eight genes, COL6A3 was the most significant signature. After taking sex, age and American Joint Committee on Cancer (AJCC) stage (29) into consideration, COL6A3 remained associated with prognosis (multivariate Cox model; P=0.004). These results revealed that COL6A3 is an independent prognosis factor for the survival of patients with CRC.
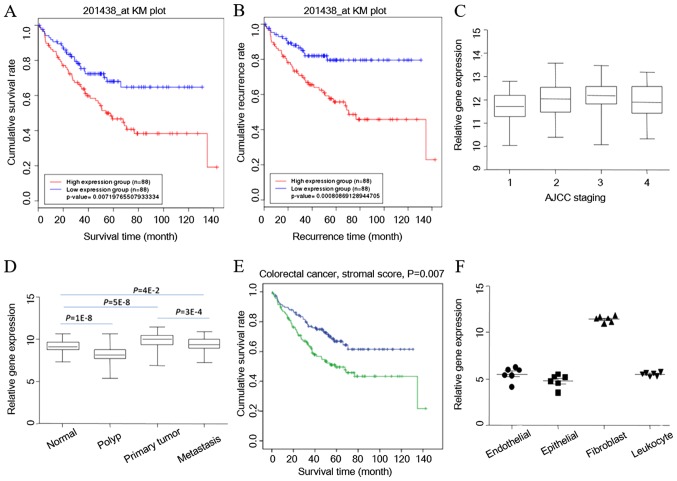
Furthermore, a Kaplan-Meier survival curve was plotted according to the survival and recurrence information in the GSE17536 dataset. COL6A3 was able to separate the patients into two groups, and was associated with survival (Fig. 4A) and recurrence (Fig. 4B). COL6A3 expression was significantly higher in patients with AJCC stage III compared with those with AJCC stage I. COL6A3 expression was also higher in patients of AJCC stage II compared with AJCC stage I patients (Fig. 4C). These findings were replicated in an additional dataset (GSE41258), which contains data on the entire disease progression spectrum. Using the GSE41258 dataset, COL6A3 expression in normal mucosa, and patients with adenoma, primary colon adenocarcinoma and metastatic CRC was compared. COL6A3 expression was significantly different in mucosa compared with cancer tissues, with the highest expression in primary CRC and the lowest expression in adenoma (P<0.05) (Fig. 4D). COL6A3 expression also increased as the stage of cancer increased.
Figure 4.
Clinical relevance of COL6A3 in CRC. (A) Survival curves indicate that COL6A3 gene expression can separate patients into two group with different survival times. (B) Survival curves indicate that COL6A3 gene expression can separate patients into two group with different recurrence times. (C) COL6A3 expression status at different American Joint Committee on Cancer stages. (D) COL6A3 is relatively highly expressed in primary CRC. (E) The stromal score of CRC tissue can separate patients with short and long survival. (F) COL6A3 expression is the highest in cancer-associated fibroblasts in CRC. COL6A3, collagen type VI α3 chain; CRC, colorectal cancer.
Cancer-associated fibroblasts are a key determinant in the malignant progression of cancer, as demonstrated by a previous study (30). Besides tumor cells, malignant solid tumor tissues consist of tumor-associated stromal cells, immune cells and vascular cells (31). The transcriptome data were analyzed using the ESTIMATE bioinformatics tool (31), and it was indicated that the stroma contributes to the survival of patients with CRC (Fig. 4E) (accepted but unpublished). To further determine COL6A3 gene expression status in cells that are present in the cancer microenvironment, the GSE39397 dataset was reanalyzed. The GSE39397 dataset contains transcriptomes of purified human CRC epithelial tumor cells, leukocytes, endothelial cells and fibroblasts (10). The results of the present study indicated that stromal cancer-associated fibroblasts are the main contributor for COL6A3 expression in CRC tissues (P=1×10−14; one-way ANOVA) (Fig. 4F).
COL6A3 knockout decreases proliferation and invasion, and increases apoptosis in vitro
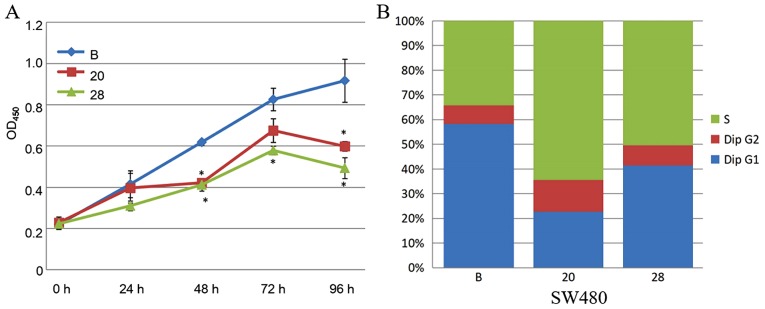
Since COL6A3 is significantly upregulated in CRC, COL6A3 was knocked out in the present study to determine whether it has any roles in the SW480 cell line, which is derived from a patient with Dukes' type B colorectal adenocarcinoma (32). The C5 terminal of COL6A3 is the active fragment (33). The fragment-encoding gene was mutated using the CRISPR-Cas9 system (GenePharma Inc.). A total of 2 COL6A3-knockout cell lines were constructed (SW480-20 and SW480-28). The CCK8 assay indicated that the proliferation rate was significantly decreased in COL6A3 knockout-cell lines (SW480-20 and SW480-28) compared to the wild-type cell line (P<0.05) (Fig. 5A).
Figure 5.
COL6A3 knockout decreases proliferation, invasion and migration in SW480 cells. (A) Cell growth of SW480 with or without COL6A3 knockout were monitored every 24 h (n=5). (B) Cell cycle arrest in SW480 cells following COL6A3 knockout as assessed by flow cytometry (n=3). (C) Representative images of the Transwell assay (left) indicate decreased invasive capacity (right) compared to the wild-type cell line (n=3). (D) Representative graphs for the scratch wound-healing assay (left) and wound healing rate in SW480, SW480-20 and SW480-28 cells (right) (n=3). *P<0.01 compared with the control. Bars, 100 µm. COL6A3, collagen type VI α3 chain.
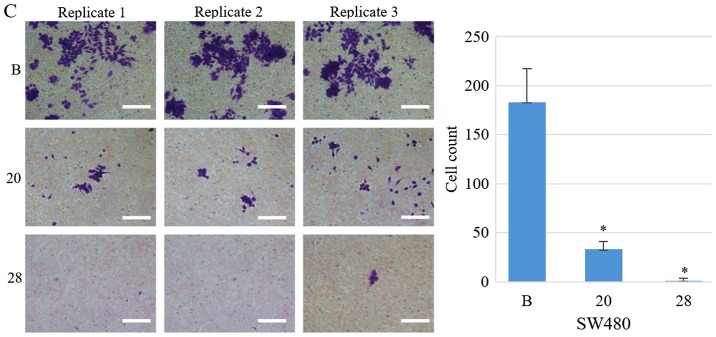
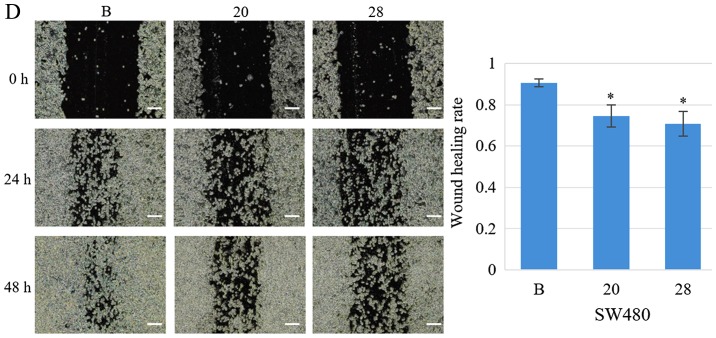
The proportion of cells in the S phase significantly increased from 34.2 to 64.5% (SW480-20, P=1.3×10−6) and to 50.5% (SW480-28, P=7.5×10−5), following COL6A3 knockout. The proportion of cells in the G1 phase decreased from 58.3 to 22.7% (SW480-20, P=4.0×10−9) and to 41.5% (SW480-28, P=6.6×10−7) following COL6A3 knockout. The proportion of cells in the G2-M phase slightly increased from 7.5 to 12.8% (SW480-20, P=0.0006) and to 8.0% (SW480-28, P>0.05) following COL6A3 knockout. Therefore, COL6A3 knockdown in SW480 cells led to cell cycle arrest in the S phase (Fig. 5B). Transwell invasion assay indicated that the COL6A3 knockout cell lines have a significantly reduced invasive capacity than the wild-type cell line (P<0.05) (Fig. 5C). Scratch wound-healing assay also indicated reduced migratory capability (Fig. 5D).
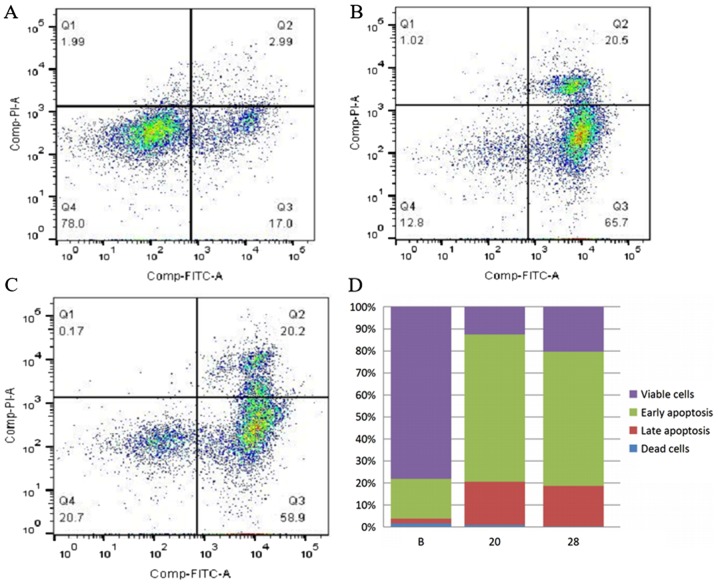
The apoptosis assay indicated that the early apoptosis rate of SW480 cells increased from 18.2 to 67.0% (SW480-20, P=2.5×10−5) and from 18.2 to 61.0% (SW480-28, P=4.2×10−5) following COL6A3 knockout. The late apoptosis rates of COL6A3 knockout cell lines (SW480-20 and SW480-28) were significantly upregulated than the wild-type cell line (P<0.01). The rate of dead cells displayed no significant difference compared to the wild-type cell line. These data indicated that the knockout of COL6A3 caused early apoptosis instead of necrosis in SW480 cells (Fig. 6A-D).
Figure 6.
COL6A3 knockout causes early apoptosis instead of necrosis in SW480 cells. Representative flow cytometry graphs for apoptosis: (A) SW480, (B), SW480-20 and (C) SW480-28 cells. (D) Proportion of dead, late apoptotic, early apoptotic and viable cells prior to and following COL6A3 knockout (n=3). Q1, dead cells; Q2, late-apoptosis; Q3, early-apoptosis; Q4, viable cells; COL6A3, collagen type VI α3 chain.
Discussion
The present study used WGCNA to construct a CRC gene co-expression network. To the best our knowledge, the present study used the largest sample size to date. The 18 identified modules were annotated, which covers many aspects of CRC, including chromosome, metabolism, cell cycle and immune response to ECM. These modules may have important roles in CRC. Considering the role of the cancer microenvironment in tumor malignancy, hub genes in the cell migration/ECM module were further analyzed for their association with patient prognosis. COL6A3, a hub gene in the cell migration/ECM module, was selected for downstream analysis. COL6A3, a factor that is predominantly expressed in stromal cancer-associated fibroblasts, was identified as an independent prognostic factor. The important role of COL6A3 in CRC malignancy was verified using an in vitro gene knockout cell experiment.
WGCNA, which was utilized in the present study, has been widely employed in the literature (14,16,17). A recent publication also applied in silico WGCNA to colon transcriptomes and indicated that a transcriptional module enriched in cell cycle processes was correlated with recurrence-free survival (22). The present authors have previously demonstrated the importance of module-based analysis in cancer transcriptome analysis (34). Compared with the study by Liu et al (22), the present study used a more stringent criterion to select datasets and included only high-quality data for final analysis. The sample size in the present study is twice the size compared with the study by Liu et al (22), which improves the robustness and confidence of the present analysis.
The emerging importance of the cancer microenvironment in cancer has been well established (25). Therefore, the present analysis focused on the ECM module and its hub genes. One of the hub genes, COL6A3, was selected for its low P-value in survival analysis. The spatial expression of COL6A3 was further analyzed, which was identified as an independent prognostic factor. We found that COL6A3 is mainly expressed in CRC-associated fibroblasts. To the best our knowledge, the clinical relevance of circulating plasma COL6A3 in CRC has only been reported by one study (35). The application of NGS has led to the identification of several mutated genes in CRC that predict survival outcomes (2,27). The COL6A3 mutation was significantly associated with improved overall survival independent of tumor-node-metastasis staging (36). These results demonstrated the validity of the present analysis.
However, there is currently no commercial human CRC-associated fibroblast cell line available. Therefore, the SW480 CRC adenocarcinoma cell line was used for the functional study of COL6A3. COL6A3 gene expression and its potential role in epithelial-mesenchymal transition in the Caco-2 human colon cancer cell line had been reported (37). Therefore, it was reasonable to examine the role of COL6A3 in the CRC cancer cell line, SW480.
In summary, the present analysis demonstrated that bioinformatics analysis is useful for identifying important candidate genes for experimental verification. The ECM module contains several hub genes that are associated with prognosis. COL6A3 is an independent prognosis factor in CRC, which is predominantly expressed in cancer-associated fibroblasts. The knockout experiments validated the role of COL6A3 in the proliferation and invasion of CRC cells. However, more insightful molecular mechanisms may be obtained in future studies. Our research may provide a framework for in-depth analysis of public transcriptome data and prioritization of candidate genes for further investigation.
Acknowledgements
Not applicable.
Funding
The present study was supported partly by the National Natural Science Foundation of China (grant nos. 31270454, 81400617 and 81502091).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
WL contributed to the design of the study, the analysis and interpretation of data, and the drafting of the manuscript. LL contributed to the acquisition of the data and analysis. HY contributed the design of the study and manuscript revising. HT collected the data and HH contributed the conception and design of the study.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Roseweir AK, McMillan DC, Horgan PG, Edwards J. Colorectal cancer subtypes: Translation to routine clinical pathology. Cancer Treat Rev. 2017;57:1–7. doi: 10.1016/j.ctrv.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Punt CJ, Koopman M, Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat Rev Clin Oncol. 2017;14:235–246. doi: 10.1038/nrclinonc.2016.171. [DOI] [PubMed] [Google Scholar]
- 3.Roychowdhury S, Chinnaiyan AM. Translating cancer genomes and transcriptomes for precision oncology. CA Cancer J Clin. 2016;66:75–88. doi: 10.3322/caac.21329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agesen TH, Sveen A, Merok MA, Lind GE, Nesbakken A, Skotheim RI, Lothe RA. ColoGuideEx: A robust gene classifier specific for stage II colorectal cancer prognosis. Gut. 2012;61:1560–1567. doi: 10.1136/gutjnl-2011-301179. [DOI] [PubMed] [Google Scholar]
- 5.Hong Y, Downey T, Eu KW, Koh PK, Cheah PY. A ‘metastasis-prone’ signature for early-stage mismatch-repair proficient sporadic colorectal cancer patients and its implications for possible therapeutics. Clin Exp Metastasis. 2010;27:83–90. doi: 10.1007/s10585-010-9305-4. [DOI] [PubMed] [Google Scholar]
- 6.Heijink DM, Fehrmann RS, de Vries EG, Koornstra JJ, Oosterhuis D, van der Zee AG, Kleibeuker JH, de Jong S. A bioinformatical and functional approach to identify novel strategies for chemoprevention of colorectal cancer. Oncogene. 2011;30:2026–2036. doi: 10.1038/onc.2010.578. [DOI] [PubMed] [Google Scholar]
- 7.Quan B, Qi X, Yu Z, Jiang Y, Liao M, Wang G, Feng R, Zhang L, Chen Z, Jiang Q, Liu G. Pathway analysis of genome-wide association study and transcriptome data highlights new biological pathways in colorectal cancer. Mol Genet Genomics. 2015;290:603–610. doi: 10.1007/s00438-014-0945-y. [DOI] [PubMed] [Google Scholar]
- 8.Wu Y, Wang X, Wu F, Huang R, Xue F, Liang G, Tao M, Cai P, Huang Y. Transcriptome profiling of the cancer, adjacent non-tumor and distant normal tissues from a colorectal cancer patient by deep sequencing. PLoS One. 2012;7:e41001. doi: 10.1371/journal.pone.0041001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim ST, Sohn I, DO IG, Jang J, Kim SH, Jung IH, Park JO, Park YS, Talasaz A, Lee J, Kim HC. Transcriptome analysis of CD133-positive stem cells and prognostic value of survivin in colorectal cancer. Cancer Genomics Proteomics. 2014;11:259–266. [PubMed] [Google Scholar]
- 10.Calon A, Espinet E, Palomo-Ponce S, Tauriello DV, Iglesias M, Céspedes MV, Sevillano M, Nadal C, Jung P, Zhang XH, et al. Dependency of colorectal cancer on a TGF-beta-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22:571–584. doi: 10.1016/j.ccr.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isella C, Terrasi A, Bellomo SE, Petti C, Galatola G, Muratore A, Mellano A, Senetta R, Cassenti A, Sonetto C, et al. Stromal contribution to the colorectal cancer transcriptome. Nat Genet. 2015;47:312–319. doi: 10.1038/ng.3224. [DOI] [PubMed] [Google Scholar]
- 12.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, et al. NCBI GEO: Archive for functional genomics data sets-10 years on. Nucleic Acids Res. 2011;39:D1005–1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 16.Miller JA, Horvath S, Geschwind DH. Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc Natl Acad Sci USA. 2010;107:12698–12703. doi: 10.1073/pnas.0914257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 19.da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu R, Zhang W, Liu ZQ, Zhou HH. Associating transcriptional modules with colon cancer survival through weighted gene co-expression network analysis. BMC Genomics. 2017;18:361. doi: 10.1186/s12864-017-3761-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lanchbury J, Gutin A, Flake D. Cancer prognosis signatures. Google Patents. 2016 [Google Scholar]
- 24.Scott RE, Ghule PN, Stein JL, Stein GS. Cell cycle gene expression networks discovered using systems biology: Significance in carcinogenesis. J Cell Physiol. 2015;230:2533–2542. doi: 10.1002/jcp.24990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedl P, Alexander S. Cancer invasion and the microenvironment: Plasticity and reciprocity. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 26.Iijima J, Konno K, Itano N. Inflammatory alterations of the extracellular matrix in the tumor microenvironment. Cancers (Basel) 2011;3:3189–3205. doi: 10.3390/cancers3033189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horvat M, Potocnik U, Repnik K, Kavalar R, Stabuc B. Single nucleotide polymorphisms as prognostic and predictive factors of adjuvant chemotherapy in colorectal cancer of stages I and II. Gastroenterol Res Pract. 2016;2016:2139489. doi: 10.1155/2016/2139489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conti J, Thomas G. The role of tumour stroma in colorectal cancer invasion and metastasis. Cancers (Basel) 2011;3:2160–2168. doi: 10.3390/cancers3022160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edge SB, Compton CC. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 30.Ostman A, Augsten M. Cancer-associated fibroblasts and tumor growth-bystanders turning into key players. Curr Opin Genet Dev. 2009;19:67–73. doi: 10.1016/j.gde.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 31.Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW, Levine DA, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomita N, Jiang W, Hibshoosh H, Warburton D, Kahn SM, Weinstein IB. Isolation and characterization of a highly malignant variant of the SW480 human colon cancer cell line. Cancer Res. 1992;52:6840–6847. [PubMed] [Google Scholar]
- 33.Park J, Scherer PE. Adipocyte-derived endotrophin promotes malignant tumor progression. J Clin Invest. 2012;122:4243–4256. doi: 10.1172/JCI63930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu W, Li L, Li W. Gene co-expression analysis identifies common modules related to prognosis and drug resistance in cancer cell lines. Int J Cancer. 2014;135:2795–2803. doi: 10.1002/ijc.28935. [DOI] [PubMed] [Google Scholar]
- 35.Qiao J, Fang CY, Chen SX, Wang XQ, Cui SJ, Liu XH, Jiang YH, Wang J, Zhang Y, Yang PY, Liu F. Stroma derived COL6A3 is a potential prognosis marker of colorectal carcinoma revealed by quantitative proteomics. Oncotarget. 2015;6:29929–29946. doi: 10.18632/oncotarget.4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu J, Wu WK, Li X, He J, Li XX, Ng SS, Yu C, Gao Z, Yang J, Li M, et al. Novel recurrently mutated genes and a prognostic mutation signature in colorectal cancer. Gut. 2015;64:636–645. doi: 10.1136/gutjnl-2013-306620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joyce T, Cantarella D, Isella C, Medico E, Pintzas A. A molecular signature for Epithelial to Mesenchymal transition in a human colon cancer cell system is revealed by large-scale microarray analysis. Clin Exp Metastasis. 2009;26:569–587. doi: 10.1007/s10585-009-9256-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.