Abstract
Melanoma is the most aggressive cutaneous cancer due to its propensity to metastasise and proliferate. Melanoma accounts for 80–90% of skin-cancer related deaths worldwide. Alhough numerous published studies have attempted to define the markers of diagnosis and prognosis of melanoma, a sensitive and specific biomarker for melanoma remains unknown. Recently, ubiquitin-like with PHD and ring finger domains 1 (UHRF1) has attracted attention due to its role in cell proliferation and it has been deemed as a potential therapeutic target for cancer. The aim of the present study was to investigate the role and the clinical significance of UHRF1 in melanoma. Immunohistochemical analysis was performed with tissue microarray (TMA) to examine the expression of UHRF1 and Ki-67, and the role of UHRF1 in cell proliferation was determined through CCK-8, colony formation and flow cytometry by interfering with the expression of UHRF1. Subsequently, the relationship among the expression of UHRF1 and several major clinical characteristics of melanoma were analysed to evaluate the role of UHRF1 in the progression of melanoma. Finally, the clinical significance of UHRF1 was estimated in 56 melanoma patients. It was observed that the expression of UHRF1 was significantly upregulated in melanoma compared with benign nevi tissues (P<0.05). In addition, the downregulation of the expression of UHRF1 significantly decreased cell proliferation. Furthermore, the level of UHRF1 was positively correlated with the expression of Ki-67 in melanoma cells, as well as in melanoma tissues. Clinically, a high level of UHRF1 was prone to be related to a high TNM classification (P=0.017) and Breslow's thickness (P=0.034) of melanoma. Furthermore, a high level of UHRF1 was positively associated with a shorter overall survival of melanoma patients. Importantly, the Cox regression model analysis demonstrated that the expression of UHRF1 was an independent prognostic factor for the overall survival of melanoma patients. In conclusion, the elevated expression of UHRF1 plays an important role in melanoma cell proliferation and progression, and it can be used as a prognostic biomarker for melanoma.
Keywords: UHRF1, melanoma, overexpression, proliferation, prognosis
Introduction
Melanoma is a highly malignant tumour originated from neural crest-derived melanocytes. Melanoma can occur in the skin, eye, digestive tract, reproductive systems and in other organs, but it mostly occurs in cutaneous tissues. In general, melanoma constitutes only 5% of all skin cancers. However, this disease has the highest malignancy and mortality rate due to its propensity for abnormal proliferation and early lymphatic and haematogenous metastasis (1). Currently, the incidence of melanoma is increasing at a rate of 4.1% per year, which is higher than that of many other malignancies, and there are over 76,000 new cases of melanoma and 10,000 melanoma-related deaths in the United States each year (2). Despite the advances and breakthroughs in the field of melanoma treatment, an effective diagnostic and therapeutic method is urgently needed. Thus, the mechanisms of proliferation and metastasis of melanoma need to be further studied.
Ubiquitin-like with PHD and ring finger domains 1 (UHRF1), which consists of 793 amino acids and binds to the inverted CCAAT box (ICB2) in the promoter of topoisomerase Iiα (3), acts as an epigenetic integrator that possesses five functional domains, including ubiquitin-like domain, tandem tudor domain, plant homeodomain, SET and RING associated domain as well as an interesting new gene domain (4). UHRF1 was reported to cooperatively regulate DNA methylation and histone modification, which epigenetically silenced tumour suppressor genes (TSGs) (5) and played a positive role in cell proliferation and tumour progression (6). A number of studies have demonstrated that aberrations in the expression of UHRF1 were a reliable biomarker for the diagnosis and prognostic prediction of lung (7) breast (8) bladder (9) and hepatocellular carcinoma (10) and that the knockdown or knockout of its expression led to weakened cell proliferation and increased cell apoptosis, which made it a potential anticancer drug target (11,12). Additionally, the elevated expression of UHRF1was recently revealed to be associated with tumour cell resistance to antitumour drugs, whereas the interference of UHRF1 sensitised cancer cells to chemotherapies or radiotherapy. Obviously, UHRF1 is considered to be a powerful diagnostic and prognostic biomarker to predict the therapeutic response and assess the risk of tumour progression and recurrence. Therefore, further investigation into the molecular mechanisms of the action of UHRF1 would help develop and improve cancer therapies.
At present, the expression and roles of UHRF1 in melanoma remain largely unknown. In the present study, the expression of UHRF1 in melanoma and the correlation between UHRF1 and the major pathological parameters of melanoma were investigated. Additionally, the prognosis of the expression of UHRF1 in melanoma was analysed.
Materials and methods
Bioinformatic analysis of the expression of UHRF1 in melanoma
To determine the expression of UHRF1 in melanoma, we used the Oncomine database (https://www.oncomine.org/resource/login.html) where we inputted the keyword ‘UHRF1’ and chose ‘melanoma, please check cancer vs. normal analysis’ as our target cancer and analysis type. Subsequently, we selected the GSE4587 and GSE7553 datasets to analyse the expression of UHRF1 mRNA between melanoma and benign nevi, respectively. In order to understand the relationship between UHRF1 and Ki-67, we searched for available data on mRNA expression in the public datasets of TCGA (https://cancergenome.nih.gov/), which contains data of 471 melanoma patients. Subsequently, the data were visualised in a heatmap using the RStudio software (RStudio, Inc., Boston, MA, USA). For analysing the correlation between the prognosis of patients with melanoma and the mRNA expression level of UHRF1 in melanoma, we inputted the keyword of ‘UHRF1’ in OncoLnc (http://www.oncolnc.org/) and chose the survival curve of skin cutaneous melanoma (SKCM) patients. Patients were sorted based on the expression of UHRF1, and we compared the bottom third vs. the top third as recommended. We obtained the survival curve and detailed survival time of the low- and high-expression groups (from TCGA).
Patients and follow-up
The present study enrolled a total of 56 melanoma patients and 16 benign nevus patients. Tumor samples and matched adjacent normal control tissues were obtained from 20 of those melanoma patients. All patients were subjected to a complete excision followed by tissue verification through pathological examination at Zhongshan Hospital Affiliated to Fudan University (Shanghai, China) from January 1, 2012 to December 31, 2016. Prior to the surgery, none of the patients had received any form of radiotherapy or chemotherapy and detailed clinicopathological and follow-up data had been obtained from them. The clinical stage of patients was evaluated by the TNM staging system of the American Joint Committee on Cancer (AJCC) and IUCC (7th edition) (13) and it was evaluated at the time of the formal pathology report. Ethical approval of the study was obtained by The Ethics Committee of the Zhongshan Hospital Biomedical Research and written informed consent was obtained from each patient. The follow-up of patients ended in December 31, 2016, and the average observation time was 25 months.
Cell culture and transfection
The melanoma cell lines, including A375, A2058, A875, M14 and MV3, were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and they were grown based on the recommended media and conditions. Knockdown of UHRF1 was performed with the specific shRNAs delivered by a lentiviral system purchased from Shanghai GeneChem Co. Ltd., (Shanghai, China) according to the manufacturer's instructions and pLenti-shRNA-Mock was used as a negative control. Subsequently, the cells with suiTable fluorescent expression were screened with puromycin at a concentration of 3 µg/ml. The transfection efficiency was verified by western blotting and RT-qPCR. Each experiment was performed in triplicate.
Tissue microarray (TMA) construction, immunohistochemical and immunofluorescence staining
TMA was constructed according to a previous study (14). Briefly, the above mentioned specimens of melanoma and benign nevus were stained with hematoxylin and eosin (H&E), and representative blocks were sampled for the TMA blocks. Subsequently, they were punched by a tissue cylinder with a 3-mm diameter and then were taken from the paraffin blocks. Finally, sections of 4-µm thickness were placed on the slides.
TMA was stained with H&E for verifying the presence of tumour tissue and stained with mouse anti-human monoclonal UHRF1 (1:100; cat. no. 612264; BD Biosciences, Franklin Lakes, NJ, USA) as well as rabbit anti-human monoclonal Ki-67 (1:100; cat. no. ab16667; Abcam, Cambridge, UK) to evaluate the two protein expression levels by immunohistochemistry (IHC). Briefly, the slides were deparaffinised and rehydrated following the manufacturer's instructions. After incubation in 0.3% H2O2 to abolish the activity of endogenous peroxidase, antigen retrieval was conducted in citrate buffer. Subsequently, the primary antibodies (UHRF1 and Ki-67) were incubated overnight, and then the immunohistochemistry kits (cat. no. GK500705; Gene Tech Co., Shanghai, China) were used for 1 h. The sections were counterstained with H&E. The TMA was viewed at ×40 and ×200 magnification and images were captured from each area using a standard Olympus microscope (Olympus Corp., Tokyo, Japan). For the IHC quantification, IPP (version 6.0; Media Cybernetics, Inc., Rockville, MD, USA) was used for digital photograph analysis of the antigen expression. The total area stained with brown was assessed in pixels, and the integrated optical density (IOD) values of each block were exported. The TMA was then examined independently by two pathologists who were unaware of the patient clinical information, and disagreements were solved by reaching to a consensus. The score for nuclear UHRF1 staining was determined based on a combination of staining percentage and intensity as previously described (15). The staining intensity was scored as follows: 0 (negative), 1 (low), 2 (moderate), or 3 (high), and the staining percentage was scored as follows: 1 (0–25%), 2 (>25–50%), 3 (>50–75%) or 4 (>75–100%). The sum was used for evaluating the expression level of UHRF1 and it was classified into two grades: low (0–3) or high (4–7).
For immunofluorescence staining, A375 and A2058 cells were seeded onto diagnostic glass slides and cultivated for 24 h with a suiTable degree of fusion. Subsequently, they were fixed with 4% paraformaldehyde (cat. no. 30314ES76; Yeasen, Shanghai, China), incubated with Triton X-100 (cat. no. 20107ES76; Yeasen) and blocked with 5% bovine serum albumin (BSA) (cat. no. 36101ES25; Yeasen) orderly. Specific primary antibodies UHRF1 and/or Ki-67 were incubated in the same conditions as IHC. After being washed with phosphate-buffered saline (PBS), they were incubated with Alexa Fluor 488-labeled anti-rabbit IgG antibody (1:100; cat. no. 33106ES60; Yeasen) and/or DyLight 594-labeled anti-mouse antibody (cat. no. 33212ES60; Yeasen) for 60 min and then, nuclear counterstaining was performed with DAPI for 15 min. The fluorescence intensities were analysed by fluorescence microscopy (Olympus Corp.).
Real-time reverse transcription-PCR (RT-qPCR) and western blot analysis
Total RNA was extracted using TRIzol reagent and reverse-transcribed using the cDNA Synthesis kit (Takara Bio, Inc., Otsu, Japan). The primer sequences used for PCR are as follows: UHRF1: 5′-ACAACGTGTGCAAGGACTGC-3′ (forward) and 5′-GAGCTGGTTGAGGACGGTCT-3′ (reverse); GAPDH: 5′-CCTGCACCACCAACTGCTTA-3′ (forward and 5′-GGCCATCCACAGTCTTCTGG-3′ (reverse). The RNA quantity and density were verified using a spectrophotometer (DeNovix Inc., Wilmington, DE, USA). RT-qPCR was performed using the SYBR-Green Master Mix kit (Takara Bio, Inc., Otsu, Japan) and the conditions were as follows: 95°C for 1 min, 35 cycles of 95°C for 15 sec, 60°C for 15 sec, 72°C for 30 sec and a final extension at 95, 65 and 30°C for 1 min. The assay was performed in triplicate using the ABI PRISM 7900HT RT-PCR system, and the relative gene expression level was determined using the 2−ΔΔCt method (16).
For total protein extraction, cell lysates were obtained using the RIPA buffer supplemented with phosphatase inhibitors. Total protein (15 µg) was injected into Bis-Tris SDS/PAGE gel and transferred to polyvinylidene difluoride (PVDF) membranes. After blocking with 5% BSA for 60 min, the membranes were incubated with mouse anti-human monoclonal UHRF1 (1:500; cat. no. 612264; BD Biosciences, Franklin Lakes, NJ, USA) and mouse monoclonal GAPDH (1:10,000; cat. no. ab8245; Abcam, Cambridge, UK) overnight at 4°C. The membranes were then exposed to the secondary antibody (1:3,000; cat. no. A0216; Beyotime Institute of Biotechnology, Shanghai, China) for 60 min. The bands were incubated with a DAB kit and analysed with an imaging system. Densitometric analysis of UHRF1 and GAPDH in every sample and cell line was performed using Adobe Photoshop CS6 (Adobe, San Jose, CA, USA).
Flow cytometric analysis, CCK-8 and colony formation assays
Flow cytometric analysis was used for detecting the cell cycle and evaluating cell proliferation. The cells were washed with PBS and fixed in 70% ethanol overnight at 4°C. Subsequently, propidium iodide (cat. no. 550825; BD Biosciences, Franklin Lakes, NJ, USA) was used for staining the cells for 30 min. Stained cells were detected by a flow cytometer and analysed using FlowJo software version 10 (BD Biosciences, Franklin Lakes, NJ, USA).
UHRF1 shRNA or negative control shRNA cells were inoculated into 96-well plates (1,000 cells/well). At each time-point (24, 48, 72 and 96 h), 10 µl of CCK-8 solution was added into the sextuplicate wells. The wells were incubated for 3 h and the absorbance of each individual well was determined at 490 nm. The data obtained are presented as a line chart. Each experiment was performed in triplicate.
The colony formation assay was also used to investigate the cell proliferation capacity. The cells were digested into a single cell suspension and seeded in a 6-well plate (1,000 cells/well). The appropriate complete medium was added in each well and then the wells were placed into an incubator with the culture medium refreshed every three days for two weeks. Following the two-week period, the cells were washed with PBS, fixed with 4% paraformaldehyde and stained with 0.4% crystal violet for 15 min. The number of colonies containing >10 cells was counted manually and averaged from the duplicate wells.
Statistical analysis
The data were analysed using SPSS statistical software package (v.20.0; IBM Corp., Armonk, NY, USA). Qualitative variables of two groups were compared by the independent or paired Student's t-test, multiple groups were compared by the one-way ANOVA and post hoc Dunnett t-test and the median UHRF1 expression values among three groups were compared by the Kruskal-Wallis test followed by a Mann-Whitney U post hoc test with Bonferroni's correction. Pearson's correlation test was adopted to analyse the relationship between the expression of UHRF1 and Ki-67 by GraphPad version 6.0 (GraphPad Software, Inc., San Diego, CA, USA). A Chi-squared test was used to analyse the association between the expression of UHRF1 and the clinical parameters of melanoma patients. Overall survival (OS) was evaluated using the Kaplan-Meier method and analysed by the log-rank test. For the univariate and multivariate analysis, a Cox proportional hazard regression model was used. A P-value of <0.05 was considered to indicate a statistically significant difference.
Results
The expression of UHRF1 is significantly upregulated in melanoma
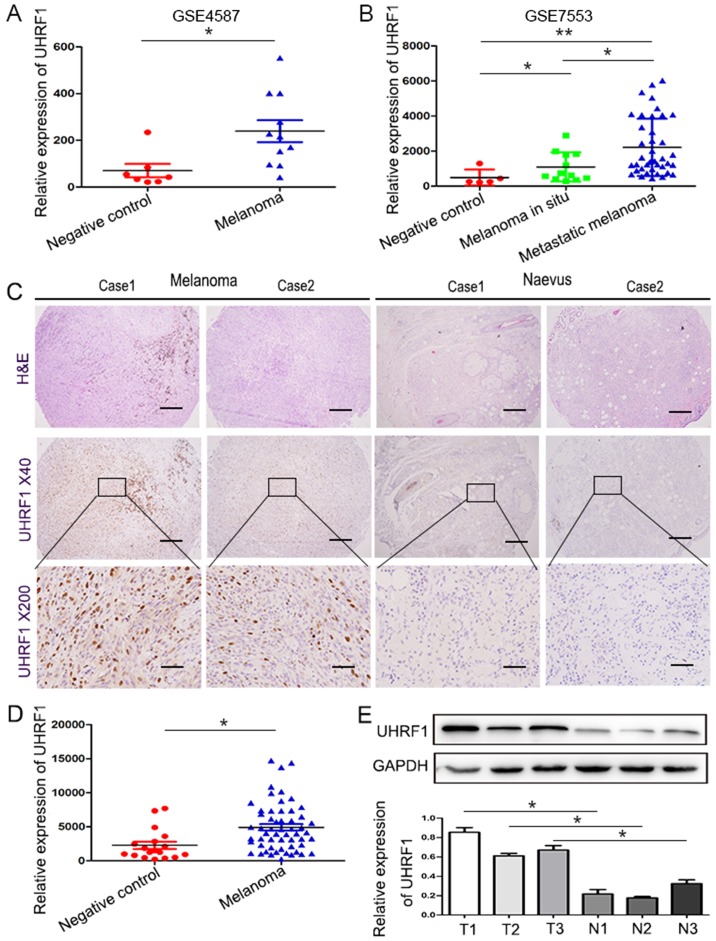
From the GSE4587 dataset, the expression of UHRF1 mRNA in benign nevus and cutaneous melanoma tissues was analysed (Fig. 1A). We revealed that the expression of UHRF1 in melanoma was significantly upregulated compared with benign nevus tissues (Fig. 1A, P=0.0112), and the average expression in the melanoma group was 5.2-fold higher than that in the benign nevus tissue group. To confirm these results, the GSE7553 dataset was further analysed (Fig. 1B) and the findings were consistent with the above mentioned results. UHRF1 mRNA expression in pre-metastatic and metastatic melanoma was obviously higher than that in benign nevus tissues (Fig. 1B, P=0.05 and P=0.0032, respectively). Additionally, the expression of UHRF1 in metastatic melanoma was found to be higher than that in pre-metastatic melanoma tissues (Fig. 1B, P=0.0146).
Figure 1.
UHRF1 is significantly upregulated in melanoma. (A and B) The expression of UHRF1 in the melanoma group is markedly upregulated compared with that of the control group by reexamining the GSE4587 and GSE7553 datasets. (C) Representative images of TMA stained with H&E and IHC for UHRF1. Scale bar, 50 µm. (D) The comparative expression of UHRF1 between the melanoma and negative control was analysed by IPP. (E) UHRF1 expression in melanoma (T1-T3) and benign nevi (N1-N3) was assessed by western blot analysis, and GAPDH expression was used as an internal reference (upper panel). Representative blots shown here were from experiments repeated three times with similar results (lower panel). *P<0.05; **P<0.01. T for tumor, N for normal tissues.
Subsequently, we attempted to investigate the expression of UHRF1 in our well-characterised melanoma TMA. H&E staining was used to verify the presence of melanoma cells, and then IHC was performed to detect the expression of the UHRF1 protein. As displayed in Fig. 1C, representative images revealed that the UHRF1 protein was mainly distributed in the nucleus. Through quantification using Image-Pro Plus 6.0, it was observed that the expression of UHRF1 in the melanoma sections was obviously upregulated compared with the benign naevus sections (P=0.0243, Fig. 1D). Further analysis using western blotting confirmed that the expression of UHRF1 protein in melanoma tissues was significantly higher compared with that of the non-cancerous tissues (Fig. 1E).
UHRF1 promotes cell proliferation and the expression of UHRF1 is positively correlated to the expression of Ki-67
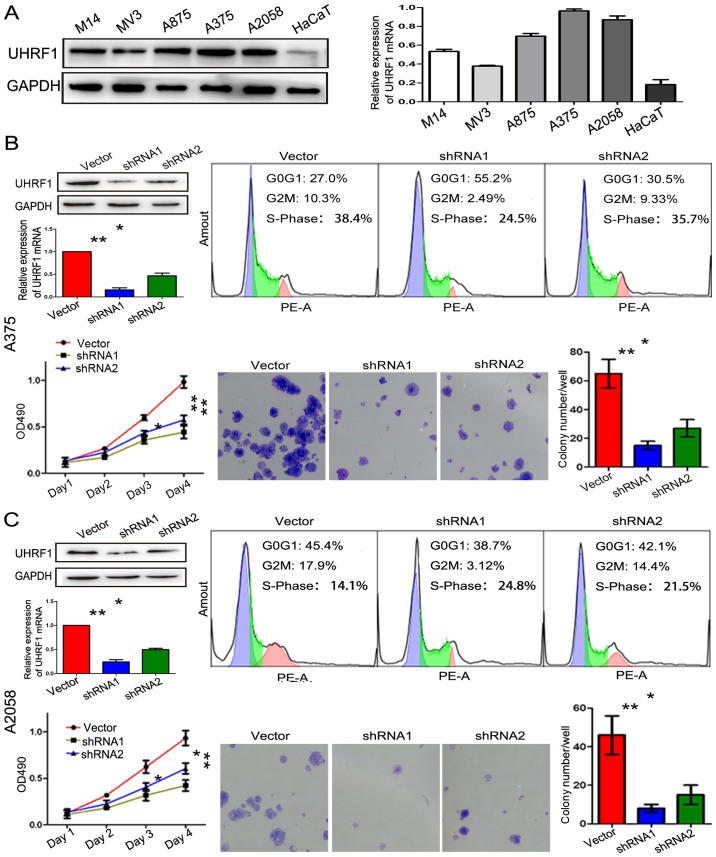
As an epigenetic integrator, UHRF1 plays a crucial role in cell proliferation in many types of cancer. As displayed in Fig. 2A, the expression of UHRF1 in five melanoma cells lines (A375, A2058, A875, M14 and MV3) and a normal skin cell line (HaCaT) was detected by western blot analysis and RT-qPCR. UHRF1 mRNA and protein were shown to be obviously upregulated in the melanoma cell lines compared with the HaCaT cell line, especially in the A375 and A2058 cell lines. In order to reveal its role in melanoma, the expression of UHRF1 in A375 and A2058 cells was downregulated by shRNAs and the downregulation was verified by RT-qPCR and western blot analysis (Figs. 2B, C and 3D). The UHRF1-shRNA significantly decreased the cell proliferation assessed by flow cytometric assay. Furthermore, the colony formation and CCK-8 assay also confirmed that the above mentioned result was statistically significant.
Figure 2.
Knockdown of the expression of UHRF1 decreased cell proliferation. (A) Analysis of the expression of UHRF1 in melanoma (M14, MV3, A875, A375 and A2058) and normal skin cell lines (HaCaT) was performed by western blot analysis and RT-qPCR. (B) The interference of UHRF1 in A375 cells inhibited cell proliferation as determined by flow cytometric analysis, CCK-8 and colony formation assay. (C) The interference of UHRF1 in A2058 cells inhibited cell proliferation as assessed by flow cytometric analysis, CCK-8 and colony formation assay. *P<0.05; **P<0.01.
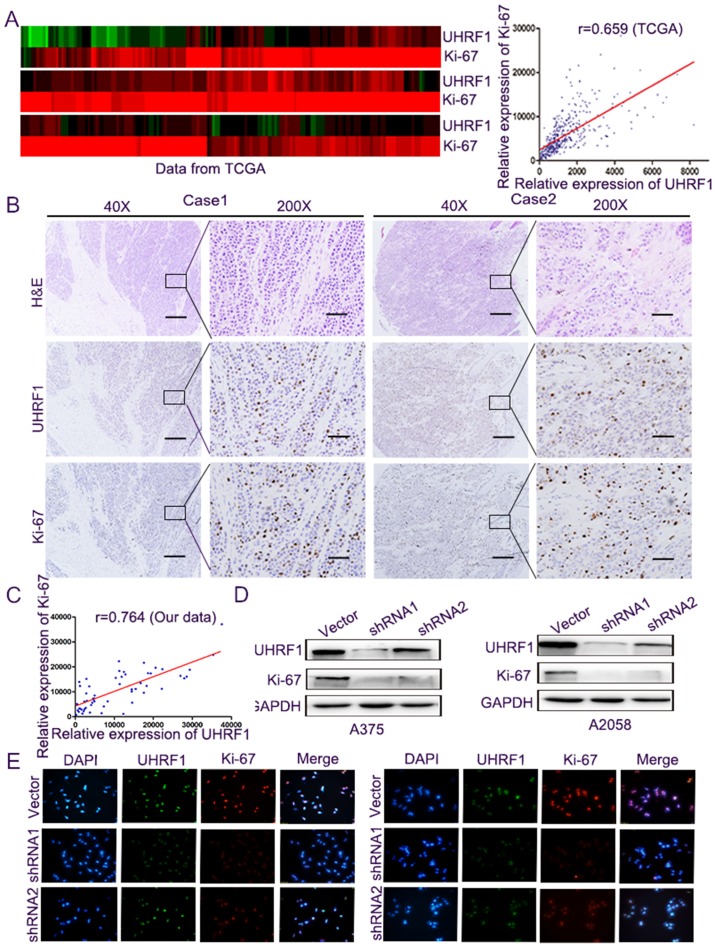
Figure 3.
The expression of UHRF1 is closely related to Ki-67 protein. (A) The TCGA data of UHRF1 and Ki-67 mRNA expression were presented through the heatmap (left panel) and the expression of UHRF1 and Ki-67 mRNA was represented by a spectrum of brightness of red (high expression) and green (low expression). Then, the relationship was analysed by GraphPad (right panel). (B) Tissue microarray was stained with UHRF1 and Ki-67 antibodies and H&E, and representative images are shown. Scale bar, 50 µm. (C) The relationship between UHRF1 and Ki-67 expression was investigated by GraphPad software. (D) Western blots were performed to detect the expression of UHRF1 and Ki-67 protein in A375-shUHRF1 and A2058-shUHRF1 cells and their control cells. (E) Immunofluorescence staining was employed to detect the expression of UHRF1 and Ki-67 protein in A375-shUHRF1 cells (left panel), A2058-shUHRF1 cells (right panel), and their control cells.
Subsequently, the relationship between the level of UHRF1 and Ki-67, a well-known biomarker of cell proliferation (17), was analysed to further determine the role of UHRF1 in cell proliferation. Firstly, we compared the mRNA expression through the public TCGA datasets, and the expression levels were converted into a heatmap where they were more easily presented. Subsequently, the correlation between them was performed, and it demonstrated a markedly positive correlation between the mRNA expression of UHRF1 and Ki-67 (Fig. 3A, r=0.659, P<0.001). Secondly, we performed immunohistochemical staining (Fig. 3B) and we calculated the comparative expression of UHRF1 and Ki-67 in TMA. The results revealed a significantly positive correlation between the expression of UHRF1 and Ki-67 (Fig. 3C, r=0.764, P<0.001). Western blot analysis and immunofluorescence staining in the A375 and A2058 melanoma cells indicated that UHRF1 and Ki-67 proteins were co-expressed in the nucleus and the knockdown of the expression of UHRF1 was accompanied by a decrease in Ki-67 protein (Fig. 3D and E).
The expression of UHRF1 is clinically relevant to melanoma
In the TMA, the blocks were divided into two groups according to the expression of UHRF1. Twenty-nine samples were found to have high expression of UHRF1 and 27 cases had low expression of UHRF1. The relationship between the expression of UHRF1 and the clinical parameters of melanoma patients is displayed in Table I. The results revealed that the expression of UHRF1 was positively correlated with TNM classification (P=0.017) and Breslow's thickness (P=0.034). However, UHRF1 did not correlate with sex (P=0.186), age (P=0.189), lymph node metastasis (P=0.299) or distant metastasis (P=0.672).
Table I.
Association of the expression of UHRF1 with the clinical parameters of 56 patients with melanoma.
| Clinical characteristics | UHRF1Low No. | UHRF1High No. | P-value |
|---|---|---|---|
| Age (years) | 0.189 | ||
| ≥55 | 14 | 10 | |
| <55 | 13 | 19 | |
| Sex | 0.186 | ||
| Male | 12 | 18 | |
| Female | 15 | 11 | |
| Lymph nodes metastasis | 0.299 | ||
| No | 13 | 10 | |
| Yes | 14 | 19 | |
| Distant metastasis | 0.672 | ||
| No | 20 | 20 | |
| Yes | 7 | 9 | |
| Breslow's depth (mm) | 0.034a | ||
| ≤5 | 16 | 9 | |
| >5 | 11 | 20 | |
| Clinical stage | 0.017a | ||
| I–II | 16 | 8 | |
| III–IV | 11 | 21 |
A Chi-square test was used for comparisons between the low and high UHRF1 expression groups.
Statistically significant.
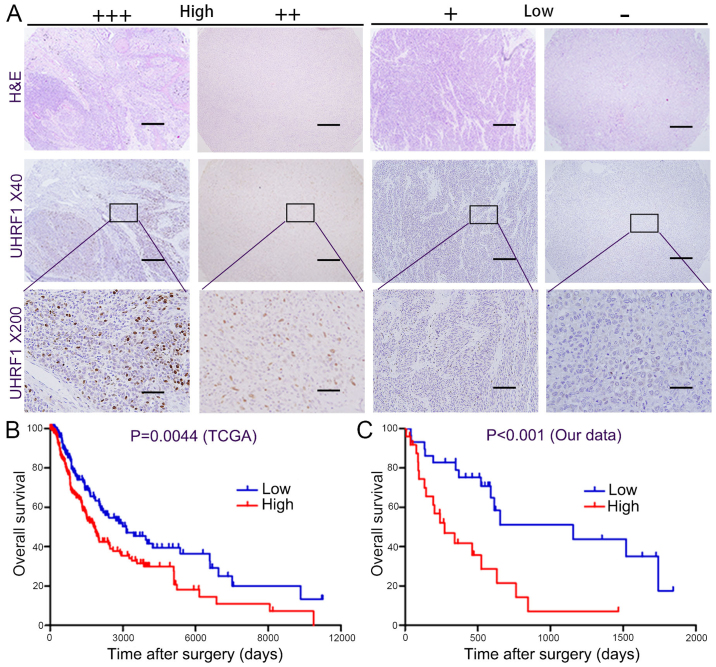
The overall survival (OS) time of 302 melanoma patients was analysed from the dataset on OncoLnc. Significantly higher survival rates were found in the low UHRF1 expression group compared with those in the high UHRF1 expression group (representive figures shown in Fig. 4A), which are displayed in Fig. 4B (P=0.0044). In our melanoma cohort, the survival rate was also found to be different between the high- and low-UHRF1 expression groups (P<0.001), and high UHRF1 expression predicted poorer prognosis (Fig. 4C). One-and two-year OS was 84.6 and 52.2% for the low UHRF1 expression group, and only 37.1 and 15.9% for the high UHRF1 expression group, respectively.
Figure 4.
The expression of UHRF1 is correlated with overall survival time. (A) Representative images of UHRF1 expression are shown; +++/++ were classified as high expression and +/- were classified as low expression. Scale bar, 50 µm. (B) Patients with a high level of UHRF1 had a lower OS than those with low UHRF1 expression by reexamining the TCGA dataset. (C) Patients with a high level of UHRF1 had a lower OS than those with low UHRF1 expression in our melanoma cohort. OS, overall survival.
The univariate analysis is displayed in Table II. We identified that lymph node metastasis, distant metastasis, Ki-67 expression, UHRF1 expression and TNM stage were correlated with the OS of melanoma patients. In particular, the expression of UHRF1 had a great impact on OS time, and the average OS for the low- and high-UHRF1 expression patients was 37 and 13.5 months, respectively.
Table II.
Univariate and multivariate analyses of factors associated with survival.
| Univariate analysis | Multivariate analysis | ||||||
|---|---|---|---|---|---|---|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value | |
| Age (years) | <55 vs. ≥55 | 1.409 | 0.658–3.017 | 0.378 | NA | ||
| Sex | Male vs. female | 0.575 | 0.274–1.206 | 0.143 | NA | ||
| Lymph nodes metastasis | No vs. yes | 0.296 | 0.129–0.680 | 0.004a | 0.253 | 0.088–0.728 | 0.011a |
| Distant metastasis | No vs. yes | 2.707 | 1.296–5.657 | 0.008a | 2.616 | 1.034–6.619 | 0.04 a |
| Breslow's depth (mm) | ≤5 vs. >5 | 2.105 | 0.979–4.529 | 0.057 | NA | ||
| Clinical stage | I–II vs. III–IV | 0.234 | 0.099–0.551 | 0.001a | 1.239 | 0.365–4.200 | 0.731 |
| UHRF1 | Low vs. high | 3.889 | 1.718–8.801 | 0.001a | 3.520 | 1.312–9.442 | 0.012a |
| Ki-67 | Low vs. high | 3.023 | 1.378–6.631 | 0.006a | 1.239 | 0.754–4.157 | 0.189 |
Univatiate and multivariate analysis, Cox proportional hazard regression model. OS, overall survival time; HR, hazard ratio; CI, confidence interval; NA, not applicable.
Statistically significant.
Subsequently, we synthesised all the factors that were statistically significant in a univariate analysis to verify the role of the expression of UHRF1 in prognosis, and then a multivariate analysis was performed. In conclusion, a high expression of UHRF1 represented a promising and independent prognostic variable for the prediction of melanoma pathogenesis (P=0.012). Other factors, including lymph node metastasis and distant metastasis, were also assessed and the results are presented in Table II.
Discussion
Recently, ubiquitin-like with PHD and ring finger domains 1 (UHRF1) has been regarded as a hub protein that participates in various activities ranging from embryonic formation to cell and tissue development. Generally, UHRF1 is highly expressed in proliferative tissues, while it is hardly expressed in low proliferative tissues (3). In addition, the expression of UHRF1 changes through the cell cycle; it peaks during late G1 and G2/M stages in normal cells and remains at a high level in tumour cells (18). Indeed, the expression of UHRF1 is markedly upregulated in multiple types of cancer, such as lung, liver and breast cancer (19–21). Overexpressed UHRF1 plays a crucial role in epigenetic changes (DNA methylation and histone modification), and several tumour suppressor genes (including p16INK4A, BRCA1, RB1, CDH13, SHP1, SOCS3 and CDX2) were reported to be related to UHRF1 epigenetic silencing, which allows cancer cells to escape apoptosis and promote tumour progression (22). Furthermore, UHRF1 plays a vital role in repairing damaged DNA and in G1/S-phase transition through its five structural constituents (6). At present, in vitro and in vivo studies have demonstrated that UHRF1 plays a vital role in tumour formation, indicating that UHRF1 is a promising candidate for cancer therapy (23).
High levels of UHRF1 act as a promoter of cell proliferation and tumour progression. In the present study, the interference of the expression of UHRF1 in A375 and A2058 melanoma cells markedly weakened the ability of cell proliferation, which is in line with the role of UHRF1 in tumour growth (24). In addition, we demonstrated that the expression of UHRF1 is positively related to the expression of Ki-67, which has been widely accepted as a biomarker of the actively proliferating cells and plays an important role in cell proliferation. Furthermore, combined research from other studies indicated that the upregulated expression of UHRF1 may be a better index in predicting tumour growth, since it is maintained throughout the cell cycle in cancer cells, but not in normal cells, while Ki-67 is overexpressed in all kinds of proliferating cells. Previous research has revealed several multi-markers that aid in the diagnosis of melanoma. For example, the co-expression of ARPC2, FN1, RGS1, SPP1 and WNT2 was found to correctly diagnose a high percentage of melanomas arising in a nevus (25), and the increasing multi-marker, NCOA3, SPP1 and RGS1, and positively immunostaining for c-Kit were reported to be helpful in predicting disease-specific survival time for melanomas. Allowing for the dynamic tumour heterogeneity of melanoma, our findings were still important in predicting the degree of melanoma malignancy.
Clinically, the information presented in our survival curve revealed that melanoma patients with a high level of UHRF1 had poorer prognosis than those with low levels of UHRF1 and that the expression of UHRF1 could be an independent prognostic factor for melanoma patients through both univariate and multivariate analyses. From the above mentioned results, it can be hypothesized that UHRF1 may be an ideal target for cancer treatment. Indeed, several studies confirmed that downregulating the expression of UHRF1 may have promising therapeutic effects. Firstly, it has been reported that many natural anticancer drugs involve an overexpression of tumour suppressor genes, accompanied with downregulation of the expression of UHRF1 (26,27). For instance, epigallocatechin-3-gallate (EGCG) has been demonstrated to decrease the proliferation of cancer cells by downregulating the expression of UHRF1 (26). Secondly, UHRF1 can inhibit tumour cell apoptosis by silencing the tumour suppressor genes (27). A relevant study demonstrated that inhibiting the expression of UHRF1 can induce apoptosis and enhance chemosensitivity in breast cancer, however it had no effect on normal cells (28). The above mentioned studies strongly support the view that UHRF1 targeting has several advantages, including specific targeting of cancer cells and enhancing chemosensitivity of cancer cells.
Currently, differential diagnosis of melanoma and benign nevi is mainly based on the H&E-stained sections and researchers have attempted to find specific markers that may distinguish melanoma from benign nevi and the progression stages (29,30). There is a number of studies on the potential biomarkers for diagnosis and prognosis of melanoma, but their sensitivity and specificity remain insufficient. C-Kit has been proposed to discriminate metastatic from non-metastatic melanoma in patients, but it cannot differentiate Spitz nevus from malignant melanoma (31). Overexpressed WT1 has been reported as an indicator of melanoma cells, however as a single immunohistochemical marker, WT1 is not able to distinguish melanoma from benign nevi (32). Other indicators, such as S100A6 (33), Melan-A (34) and HMB-45 (35), have been indicated to contribute to the differential diagnosis of melanocytic lesions. However, these markers are unlikely to be useful diagnostic tools for distinguishing malignant from benign cells because they have no differential expression between nevi and melanoma (36). Recently, a study reported a panel of five overexpressed markers (WNT2, ARPC2, RGS1, SPP1 and FN1) obtained by transcriptome analysis, and this five marker assay achieved 95% specificity and 97% sensitivity in the diagnosis of melanoma (25). However, due to the obvious heterogeneity of melanoma and the significant differences between patients, not every patient expresses common markers through transcriptome analysis in a particular population.
The expression of UHRF1 was significantly upregulated, which made it easy to distinguish it from benign nevi, and this process can be completely realized by immunohistochemistry. Furthermore, UHRF1 plays an important role in epigenetic code, and is altered during the course of tumour formation. Therefore, it is no surprising that UHRF1 may be a potential biomarker for cancer. A study has reported that UHRF1 may be an effective biomarker for differential diagnosis of pancreatic adenocarcinoma (37), and its expression is closely related to patient clinicopathological parameters (38). Similarly, it has been proposed that UHRF1 is a promising biomarker for the diagnosis and prognosis of bladder cancer, and it has been demonstrated that the expression of UHRF1 was significantly upregulated and correlated with the malignancy of bladder cancer (39).
In conclusion, the present study demonstrated that the expression of UHRF1 was markedly upregulated in melanoma, and that the downregulated expression of UHRF1 significantly decreased cell proliferation. Clinically, overexpressed UHRF1 was related to high TNM classification and Breslow's thickness. Furthermore, high UHRF1 was positively associated with shorter overall survival of melanoma patients and the expression of UHRF1 was an independent prognostic factor for the overall survival of melanoma patients.
Acknowledgements
The authors would like to thank the Pathology Department of Zhongshan Hospital for providing tumour samples.
Glossary
Abbreviations
- UHRF1
ubiquitin-like with PHD and ring finger domains 1
- TSGs
tumour suppressor genes
- GEO
Gene Expression Omnibus
- CCK-8
Cell Counting Kit-8
- AJCC
American Joint Committee on Cancer
- IUCC
International Union of Cancer Control
- IHC
immunohistochemistry
- BSA
bovine serum albumin
- TCGA
The Cancer Genome Atlas
- H&E
hematoxylin and eosin
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- DAPI
4′,6-diamidino-2-phenylindole
- PBS
phosphate-buffered saline
- RT-qPCR
real-time quantitative PCR
- TMA
tissue microarray
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
JG and FQ conceived and designed the study. CW, NL, LW and YZ performed the experiments. CW and ZF wrote the manuscript. CW, YY and NL reviewed and edited the manuscript. All authors read and approved the manuscript and agree to be accounTable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Ethical approval of the study was obtained by The Ethics Committee of the Zhongshan Hospital Biomedical Research and written informed consent was obtained from each patient.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Rajabi P, Bagheri M, Hani M. Expression of estrogen receptor alpha in malignant melanoma. Adv Biomed Res. 2017;6:14. doi: 10.4103/2277-9175.200789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Girotti MR, Gremel G, Lee R, Galvani E, Rothwell D, Viros A, Mandal AK, Lim KH, Saturno G, Furney SJ, et al. Application of sequencing, liquid biopsies, and patient-derived xenografts for personalized medicine in melanoma. Cancer Discov. 2016;6:286–299. doi: 10.1158/2159-8290.CD-15-1336. [DOI] [PubMed] [Google Scholar]
- 3.Hopfner R, Mousli M, Jeltsch JM, Voulgaris A, Lutz Y, Marin C, Bellocq JP, Oudet P, Bronner C. ICBP90, a novel human CCAAT binding protein, involved in the regulation of topoisomerase IIalpha expression. Cancer Res. 2000;60:121–128. [PubMed] [Google Scholar]
- 4.Bronner C, Krifa M, Mousli M. Increasing role of UHRF1 in the reading and inheritance of the epigenetic code as well as in tumorogenesis. Biochem Pharmacol. 2013;86:1643–1649. doi: 10.1016/j.bcp.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Kim JK, Estève PO, Jacobsen SE, Pradhan S. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res. 2009;37:493–505. doi: 10.1093/nar/gkn961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bronner C, Fuhrmann G, Chédin FL, Macaluso M, Dhe-Paganon S. UHRF1 links the histone code and DNA methylation to ensure faithful epigenetic memory inheritance. Genet Epigenet. 2010;2009:29–36. [PMC free article] [PubMed] [Google Scholar]
- 7.Unoki M, Daigo Y, Koinuma J, Tsuchiya E, Hamamoto R, Nakamura Y. UHRF1 is a novel diagnostic marker of lung cancer. Br J Cancer. 2010;103:217–222. doi: 10.1038/sj.bjc.6605717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Unoki M, Nishidate T, Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23:7601–7610. doi: 10.1038/sj.onc.1208053. [DOI] [PubMed] [Google Scholar]
- 9.Yang GL, Zhang LH, Bo JJ, Chen HG, Cao M, Liu DM, Huang YR. UHRF1 is associated with tumor recurrence in non-muscle-invasive bladder cancer. Med Oncol. 2012;29:842–847. doi: 10.1007/s12032-011-9983-z. [DOI] [PubMed] [Google Scholar]
- 10.Liang D, Xue H, Yu Y, Lv F, You W, Zhang B. Elevated expression of UHRF1 predicts unfavorable prognosis for patients with hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8:9416–9421. [PMC free article] [PubMed] [Google Scholar]
- 11.Yan F, Tan XY, Geng Y, Ju HX, Gao YF, Zhu MC. Inhibition effect of siRNA-downregulated UHRF1 on breast cancer growth. Cancer Biother Radiopharm. 2011;26:183–189. doi: 10.1089/cbr.2010.0886. [DOI] [PubMed] [Google Scholar]
- 12.Yan F, Wang X, Shao L, Ge M, Hu X. Analysis of UHRF1 expression in human ovarian cancer tissues and its regulation in cancer cell growth. Tumour Biol. 2015;36:8887–8893. doi: 10.1007/s13277-015-3638-1. [DOI] [PubMed] [Google Scholar]
- 13.Edge SB, Compton CC. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 14.Kallioniemi OP, Wagner U, Kononen J, Sauter G. Tissue microarray technology for high-throughput molecular profiling of cancer. Hum Mol Genet. 2001;10:657–662. doi: 10.1093/hmg/10.7.657. [DOI] [PubMed] [Google Scholar]
- 15.Jiang JH, Gao Q, Ke AW, Yu Y, Shi GM, Fan J, Zhou J, Huang XW. Prognostic significance of nuclear RNA export factor 3 in hepatocellular carcinoma. Oncol Lett. 2014;7:641–646. doi: 10.3892/ol.2014.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Li LT, Jiang G, Chen Q, Zheng JN. Ki67 is a promising molecular target in the diagnosis of cancer (Review) Mol Med Rep. 2015;11:1566–1572. doi: 10.3892/mmr.2014.2914. [DOI] [PubMed] [Google Scholar]
- 18.Mousli M, Hopfner R, Abbady AQ, Monté D, Jeanblanc M, Oudet P, Louis B, Bronner C. ICBP90 belongs to a new family of proteins with an expression that is deregulated in cancer cells. Br J Cancer. 2003;89:120–127. doi: 10.1038/sj.bjc.6601068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang P, Cheng CL, Chang YH, Liu CH, Hsu YC, Chen JS, Chang GC, Ho BC, Su KY, Chen HY, Yu SL. Molecular gene signature and prognosis of non-small cell lung cancer. Oncotarget. 2016;7:51898–51907. doi: 10.18632/oncotarget.10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X, Ou H, Xiang L, Li X, Huang Y, Yang D. Elevated UHRF1 expression contributes to poor prognosis by promoting cell proliferation and metastasis in hepatocellular carcinoma. Oncotarget. 2017;8:10510–10522. doi: 10.18632/oncotarget.14446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geng Y, Gao Y, Ju H, Yan F. Diagnostic and prognostic value of plasma and tissue ubiquitin-like, containing PHD and RING finger domains 1 in breast cancer patients. Cancer Sci. 2013;104:194–199. doi: 10.1111/cas.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashraf W, Ibrahim A, Alhosin M, Zaayter L, Ouararhni K, Papin C, Ahmad T, Hamiche A, Mély Y, Bronner C, Mousli M. The epigenetic integrator UHRF1: On the road to become a universal biomarker for cancer. Oncotarget. 2017;8:51946–51962. doi: 10.18632/oncotarget.17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bronner C, Achour M, Arima Y, Chataigneau T, Saya H, Schini-Kerth VB. The UHRF family: Oncogenes that are drugable targets for cancer therapy in the near future? Pharmacol Ther. 2007;115:419–434. doi: 10.1016/j.pharmthera.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 24.Wang F, Yang YZ, Shi CZ, Zhang P, Moyer MP, Zhang HZ, Zou Y, Qin HL. UHRF1 promotes cell growth and metastasis through repression of p16ink4a in colorectal cancer. Ann Surg Oncol. 2012;19:2753–2762. doi: 10.1245/s10434-011-2194-1. [DOI] [PubMed] [Google Scholar]
- 25.Kashani-Sabet M, Rangel J, Torabian S, Nosrati M, Simko J, Jablons DM, Moore DH, Haqq C, Miller JR, III, Sagebiel RW. A multi-marker assay to distinguish malignant melanomas from benign nevi. Proc Natl Acad Sci USA. 2009;106:6268–6272. doi: 10.1073/pnas.0901185106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Achour M, Mousli M, Alhosin M, Ibrahim A, Peluso J, Muller CD, Schini-Kerth VB, Hamiche A, Dhe-Paganon S, Bronner C. Epigallocatechin-3-gallate up-regulates tumor suppressor gene expression via a reactive oxygen species-dependent down-regulation of UHRF1. Biochem Biophys Res Commun. 2013;430:208–212. doi: 10.1016/j.bbrc.2012.11.087. [DOI] [PubMed] [Google Scholar]
- 27.Alhosin M, Sharif T, Mousli M, Etienne-Selloum N, Fuhrmann G, Schini-Kerth VB, Bronner C. Down-regulation of UHRF1, associated with re-expression of tumor suppressor genes, is a common feature of natural compounds exhibiting anti-cancer properties. J Exp Clin Cancer Res. 2011;30:41. doi: 10.1186/1756-9966-30-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang L, Shanqu L, Ping G, Ting H, Xi W, Ke D, Min L, Junxia W, Huizhong Z. Gene therapy with RNAi targeting UHRF1 driven by tumor-specific promoter inhibits tumor growth and enhances the sensitivity of chemotherapeutic drug in breast cancer in vitro and in vivo. Cancer Chemother Pharmacol. 2012;69:1079–1087. doi: 10.1007/s00280-011-1801-y. [DOI] [PubMed] [Google Scholar]
- 29.Guerriere-Kovach PM, Hunt EL, Patterson JW, Glembocki DJ, English JC, III, Wick MR. Primary melanoma of the skin and cutaneous melanomatous metastases: Comparative histologic features and immunophenotypes. Am J Clin Pathol. 2004;122:70–77. doi: 10.1309/R8N1CN04KKFV6M7T. [DOI] [PubMed] [Google Scholar]
- 30.van Kempen LC, van den Oord JJ, van Muijen GN, Weidle UH, Bloemers HP, Swart GW. Activated leukocyte cell adhesion molecule/CD166, a marker of tumor progression in primary malignant melanoma of the skin. Am J Pathol. 2000;156:769–774. doi: 10.1016/S0002-9440(10)64943-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Isabel Y, Fitzpatrick JE. Expression of c-kit (CD117) in Spitz nevus and malignant melanoma. J Cutan Pathol. 2006;33:33–37. doi: 10.1111/j.0303-6987.2006.00420.x. [DOI] [PubMed] [Google Scholar]
- 32.Rosner K, Mehregan DR, Moussai D, Abrams J, Tromp G, Mehregan DA. WT1 marker is not sufficient for distinguishing between melanoma and melanocytic nevi. J Cutan Pathol. 2009;36:1077–1082. doi: 10.1111/j.1600-0560.2009.01224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fullen DR, Reed JA, Finnerty B, McNutt NS. S100A6 preferentially labels type C nevus cells and nevic corpuscles: Additional support for Schwannian differentiation of intradermal nevi. J Cutan Pathol. 2001;28:393–399. doi: 10.1034/j.1600-0560.2001.028008393.x. [DOI] [PubMed] [Google Scholar]
- 34.Busam KJ, Chen YT, Old LJ, Stockert E, Iversen K, Coplan KA, Rosai J, Barnhill RL, Jungbluth AA. Expression of melan-A (MART1) in benign melanocytic nevi and primary cutaneous malignant melanoma. Am J Surg Pathol. 1998;22:976–982. doi: 10.1097/00000478-199808000-00007. [DOI] [PubMed] [Google Scholar]
- 35.Kucher C, Zhang PJ, Pasha T, Elenitsas R, Wu H, Ming ME, Elder DE, Xu X. Expression of Melan-A and Ki-67 in desmoplastic melanoma and desmoplastic nevi. Am J Dermatopathol. 2004;26:452–457. doi: 10.1097/00000372-200412000-00002. [DOI] [PubMed] [Google Scholar]
- 36.Haqq C, Nosrati M, Sudilovsky D, Crothers J, Khodabakhsh D, Pulliam BL, Federman S, Miller JR, III, Allen RE, Singer MI, et al. The gene expression signatures of melanoma progression. Proc Natl Acad Sci USA. 2005;102:6092–6097. doi: 10.1073/pnas.0501564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crnogorac-Jurcevic T, Gangeswaran R, Bhakta V, Capurso G, Lattimore S, Akada M, Sunamura M, Prime W, Campbell F, Brentnall TA, et al. Proteomic analysis of chronic pancreatitis and pancreatic adenocarcinoma. Gastroenterology. 2005;129:1454–1463. doi: 10.1053/j.gastro.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 38.Sabatino L, Fucci A, Pancione M, Carafa V, Nebbioso A, Pistore C, Babbio F, Votino C, Laudanna C, Ceccarelli M, et al. UHRF1 coordinates peroxisome proliferator activated receptor gamma (PPARG) epigenetic silencing and mediates colorectal cancer progression. Oncogene. 2012;31:5061–5072. doi: 10.1038/onc.2012.3. [DOI] [PubMed] [Google Scholar]
- 39.Unoki M, Kelly JD, Neal DE, Ponder BA, Nakamura Y, Hamamoto R. UHRF1 is a novel molecular marker for diagnosis and the prognosis of bladder cancer. Br J Cancer. 2009;101:98–105. doi: 10.1038/sj.bjc.6605123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.