Abstract
Breast cancer is the leading cause of cancer-related deaths in females worldwide. Triple-negative breast cancer (TNBC) accounts for 15% of all breast cancer cases and has a poorer prognosis than other subtypes. Moreover, the treatment for breast cancer, especially for TNBC, remains unsatisfactory. Therefore, novel therapies are urgently needed. Microribonucleic acids (miRNAs) are a class of biomarkers and therapeutic targets in many types of cancers. In the present study, the expression of miR-384 was explored in GSE58606 and in fresh breast cancer tissues by qPCR. The results showed that miR-384 was decreased in breast cancer, especially in TNBC. The results of MTT, colony formation, soft agar, Transwell migration, wound healing and the tumorigenesis assay demonstranted that overexpression of miR-384 inhibited the proliferation and migration of breast cancer in vitro and in vivo; knockdown of miR-384 enhanced the proliferation and migration of breast cancer. In addition, luciferase assay showed that Activin A receptor type 1 (ACVR1) was a direct target of miR-384 and is involved in the inhibitory effects of miR-384 on breast cancer progression. Furthermore, this study indicated that ACVR1 activated the Wnt/β-catenin signaling pathway in breast cancer. In conclusion, our findings revealed functional and mechanistic links between miR-384 and ACVR1 in the progression of breast cancer. miR-384 not only plays an important role in the progression of breast cancer, but has promise as a potential therapeutic target for breast cancer especially for TNBC.
Keywords: breast cancer, miR-384, proliferation, migration, ACVR1, Wnt/β-catenin
Introduction
Breast cancer is the second most commonly diagnosed cancer and the leading cause of cancer-related death in women worldwide (1). The prognosis of human breast cancer depends on such pathologic characteristics such as pathological markers, lymph node metastasis and tumor size. Breast cancer is divided into many subtypes according to molecular phenotypes (2,3). Of all molecular subtypes, triple-negative breast cancer (TNBC) accounts for 15% of cases and is associated with a poorer prognosis than other subtypes. TNBC refers to breast cancer which lacks expression of estrogen receptor (ER), progesterone receptor (PR) and hormone epidermal growth factor receptor-2 (HER2). This makes it more aggressive and difficult to treat. Therefore, it is of great clinical value to identify new effective molecules as diagnostic biomarkers and therapeutic targets of breast cancer, especially TNBC. Activin A receptor type 1 (ACVR1) is an important receptor of bone morphogenetic proteins (BMPs) (4). It has been reported that ACVR1 functions as an important regulator of the BMP/Wnt signaling pathway and promotes the proliferation and metastasis of many cancers (5,6). Microribonucleic acids (miRNAs), which are small non-coding RNAs, have been verified to be an important class of regulators in many cancer types (7–9). It has been reported that miRNAs negatively regulate gene expression by directly targeting the 3′-untranslated regions (3′-UTR) of their target mRNAs. Identification and characterization of miRNAs involved in breast cancer will facilitate targeting miRNAs for possible therapy (10). It has been found that the loss of various miRNAs leads to malignancy and non-response to chemotherapy, demonstrating their role as tumor suppressors (11). There are also miRNAs that promote tumor development and metastasis (12). The role of miRNAs as molecular probes as diagnostic and therapeutic targets has become the focus of research. Identification of therapeutic miRNAs for breast cancer would be of great clinical value.
Several studies have shown that miR-384 is downregulated in various human tumors (13–16). However, little is known concerning the clinical pathological correlations and biological functions of miR-384 in breast cancer. In this study, we set out to delineate the role of miR-384 in breast cancer and explore a new therapeutic target for breast cancer.
Materials and methods
Tissue specimens and cell culture
A public database GSE58606 (https://www.ncbi.nlm) was used to explore the expression of miR-384 in this study (17). Twenty-four cases of fresh breast cancer tissues and their matched adjacent normal tissues, as well as another 26 cases of fresh breast cancer tissues were collected at the Department of Pathology, Third Affiliated Hospital of Xinxiang Medical University (Xinxiang, China) from January 2010 to March 2012. All patients were female and did not receive chemotherapy, radiotherapy and immunotherapy prior to surgery. The clinicopathological information, including age, lymph node metastasis status, ER status, PR status and HER-2 status, were collected (Table I). All tissue biopsies were freshly frozen in liquid nitrogen until further use. Informed consent was obtained from all patients before surgery. Prior approval for the study was obtained from the Xinxiang Medical University Institutional Board (Xinxiang, China).
Table I.
Correlation of miR-384 expression and the clinicopathological characteristics in the 50 breast cancer cases.
| miR-384 expression | |||
|---|---|---|---|
| Clinicopathological variables | Low | High | P-value |
| Age (years) | 0.063 | ||
| ≤50 | 8 | 9 | |
| >50 | 18 | 15 | |
| Tumor size (cm) | 0.003 | ||
| ≤2 | 3 | 14 | |
| >2 | 23 | 10 | |
| Her-2 status | 0.248 | ||
| + | 10 | 20 | |
| − | 16 | 4 | |
| ER status | 0.385 | ||
| + | 2 | 13 | |
| − | 24 | 11 | |
| PR status | 0.026 | ||
| + | 5 | 14 | |
| − | 21 | 10 | |
| TNBC | 0.006 | ||
| Yes | 16 | 0 | |
| No | 10 | 24 | |
| LN metastasis | <0.001 | ||
| Yes | 22 | 7 | |
| No | 4 | 17 | |
+, positive; -, negative; ER, estrogen receptor; PR, progesterone; TNBC, triple-negative breast cancer; LN, lymph node.
The human breast cancer cell lines MCF-7 and MDA-MB-231 obtained from Cell Bank of Chinese Academy of Sciences (Shanghai, China) were cultured in DMEM medium (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.).
RNA extraction and real-time PCR (qPCR)
Total RNA was extracted from cells and tissues with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Then cDNA was synthesized from 2 µg of total RNA and the quantification of miR-384 was performed using the All-in-One™ miRNA real-time PCR Detection kit (Guangzhou GeneCopoeia, Guangzhou, China). Real-time PCR was performed via the Applied Biosystems 7500 Sequence Detection system, using iQTM SYBR-Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA) containing 5 ng cDNA and 10 pM of each primer. The cycling conditions consisted of: one cycle at 94°C for 5 min; 40 cycles at 95°C for 30 sec; 56°C for 30 sec. Melting curve analysis was conducted for each PCR reaction to confirm the specificity of amplification. The concentration of miR-384 was calculated based on the threshold cycle (CT), and the relative expression levels were calculated as 2−∆∆CT [∆∆CT = (CTmiR-384 - CTU6)T - (CTmiR-384 - CTU6)N) or 2−∆CT(∆CT = CTmiR-384 - CTU6) after normalization with reference to the quantification of U6 small nuclear RNA expression.
As for the target genes, RT was conducted with the SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. qRT-PCR was conducted by SYBR-Green I (Applied BioSystems). The data were normalized to the geometric mean of the housekeeping gene GAPDH and calculated as 2−∆∆CT. The primers were: β-catenin (F, TGC CAAGTGGGTGGTATA and R, ACGGTTCACCCACCA TAT); cyclin D1 (F, GCGAGGAACAGAAGTGCG and R, GCATCTACACCGACAACTCCA); MMP7 (F, GGAACA GGCTCAGGACTA and R, ACTTACCGCATATTACAGTG); ACVR1 (F, GGCTGCTTCCAGGTTTAT and R, AACCA AGAACGCCTCAAT); GAPDH (F, GACTCATGACCACA GTCCATGC and R, AGAGGCAGGGATGATGTTCTG).
Plasmid construction and transfection
The miR-384 binding site in the ACVR1 is located at 2394–2340 bp, whose full length of 3′UTR is 1,101 bp. The region of human ACVR1 3′UTR at 1,961-2,580 bp was PCR-amplified and inverted into the XhoI/NotI sites of the psiCHECK-2 luciferase reporter plasmid (Promega, Madison, WI, USA). The forward primer and reverse primers used to construct the plasmid were CCGCTCGAGCATTTTCATAGTGTCAAGAA and AAAT ATGCGGCCGCTTCGGCATCATTGTAAACAT, respectively. The ACVR1 construct was generated by cloning PCR-amplified full-length human ACVR1 into Psin-EF-2. miR-384 mimics, inhibitor and their control oligos, the lentiviral vectors and their paired control lentiviral vector, were purchased from Guangzhou GeneCopoeia. The sequences were the same as previously described (16). The constructed plasmids were transfected into 293FT cells to produce the lentiviral particles. Then the cells were infected with the retroviral production or purchased relevant miR-384 lentiviral vectors. Retrovirus-infected cells were selected by Puro (Sigma-Aldrich, St. Louis, MO, USA) or Hygro (Roche Diagnostics, Shanghai, China) until all of the relative uninfected cells were dead. Real-time PCR and western blot analysis were performed to confirm the stable expression.
Western blot analysis
The cells were harvested and lysed using cell lysis buffer [with phenylmethylsulfonyl fluoride (PMSF)]. Then the lysates were repeatedly pumped with 1-ml injectors and sonicated with 3–4 bursts of 5–10 sec each. Finally, the protein lysates were mixed with β-mercaptoethanol and heated in a boiling water bath for 10 min. Next, the protein lysates were subjected to SDS-PAGE, transferred to PVDF membranes, and then blotted according to standard methods with anti-ACVR1 (1:800; cat. no. 4398; Cell Signaling Technology, Inc., Danvers, MA, USA), anti-p-β-catenin (Ser657) (1:500; cat. no. 4176; Cell Signaling Technology), anti-β-catenin (1:300; cat. no. 610153; BD Biosciences, San Diego, CA, USA), anti-cyclin D1 (1:500; cat. no. 60186; ProteinTech Group, Inc., Chicago, IL, USA), anti-MMP7 (1:200; cat. no. 10374; ProteinTech Group). Anti-α-tubulin monoclonal antibody (Sigma-Aldrich) served as a loading control. All the membranes were incubated with the HRP-conjugated secondary antibody (1:2,000; anti-mouse IgG; cat. no. 7076; 1:2,000; anti-rabbit IgG; cat. no. 7074; CST Shanghai Biological Reagent, Co., Ltd., Shanghai, China). At last, the protein bands were detected by enhanced electrochemiluminescence (Tanon Science and Technology, Co., Ltd., Shanghai, China) with SuperSignal West Pico (Thermo Fisher Scientific, Inc.).
MTT assay, colony formation assay, soft agar assay, Transwell migration assay, sound healing assay and luciferase assay
The psiCHECK-2-luciferase reporter gene plasmids psiCHECK-2-ACVR1-3′-UTR, the control-luciferase plasmid, miR-384-mimics or miR-384-inhibitor were transfected into the cells using Lipofectamine 2000 reagent. The details of the MTT assay, colony formation assay, soft agar assay, Transwell migration assay, wound healing assay and luciferase assay were conducted as previously described (16,18).
Animal studies
Female BABL/c nude mice which were 4–5 weeks of age (weight, 15–18 g) were purchased from the Center of Laboratory Animal Science of Guangdong (Guangzhou, China). The mice were kept in a plastic cage with sealed air filter at 27°C, with ad libitum feeding and 10 h of light and 14 h of dark daily were maintained. All animal experiments were conducted in conformity with current Chinese regulations and standards regarding the use of laboratory animals, and all animal procedures were approved by the Xinxiang Medical University Institutional Animal Care and Use Committee. A total of 2×106 stable cells of MDA-MB-231/miR-384, MCF-7/miR-384-in and their control cells were injected subcutaneously in the hind limbs of each mouse (n=5 for each group). Then, in the following 3 weeks, the size of each tumor was measured by a slide caliper twice weekly and the tumor volume (V) was calculated as V = length × width × height. All of the mice were euthanized by cervical dislocation after 3 weeks, the tumors were excised, fixed in 10% neutral buffered formalin and embedded in paraffin. Finally, 4-µm sections were prepared and stained with hematoxylin and eosin (H&E) for immunohistochemistry (IHC). Mouse anti-Ki-67 was purchased from Fuzhou Maixin Biotech. Co., Ltd. (Fuzhou, China) to detect the proliferation activity.
Statistical analyses
All statistical analyses were carried out by SPSS 20.0 for Windows (IBM Corp., Armonk, NY, USA). The data are expressed as means ± standard deviations (SD) from at least three independent experiments. The two-tailed paired Student's t-test was conducted for the analysis of two groups. The Mann-Whitney U test was carried out to analyze the relationship between miR-384 expression and the clinicopathological features of the breast cancer cases. P<0.05 was considered to indicate a statistically significant difference. Statistically significant data are indicated by asterisks (*P<0.05 and **P<0.01) in the figures.
Results
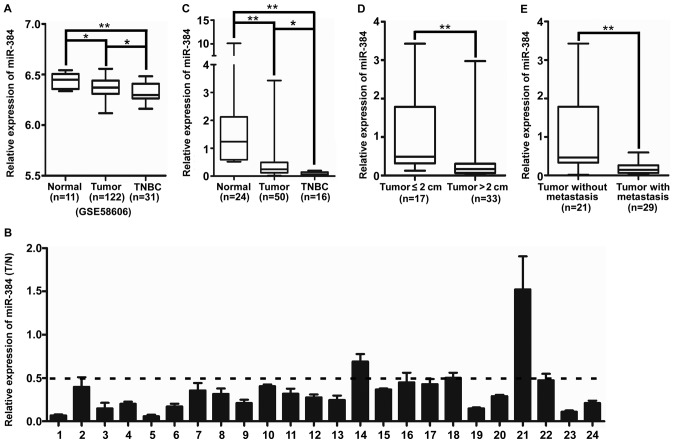
miR-384 is downregulated in breast cancer especially in TNBC compared with normal breast tissue
The expression of miR-384 was downregulated in breast cancer tissues samples, especially in TNBC, compared with that noted in the normal breast tissues in GSE58606 (Fig. 1A). Then we detected miR-384 expression in 24 cases of fresh primary breast cancer biopsies and their matched adjacent normal tissues by qPCR. The results showed that miR-384 was downregulated in 95.8% (23/24) of the breast cancer tissues; a 2-fold difference (T/N <0.5) was noted for 87.5% of the paired tissues (21/24) (Fig. 1B). In addition, the expression of miR-384 was investigated in an additional 26 cases of fresh primary breast cancer biopsies. The results showed that the expression of miR-384 was downregulated in breast cancer tissue samples, especially in TNBC, compared with that noted in the normal breast tissues (Fig. 1C and Table I). Moreover, we investigated the correlation of miR-384 expression with various clinicopathological parameters of the breast cancer cases. The median relative expression level of miR-384 in 50 cases of breast cancer sample tissues was taken as the cut-off point to separate the tumors with low expression of miR-384 from those with high expression of miR-384. The results of statistical analyses demonstrated that the expression of miR-384 was significantly associated with tumor size and lymph node metastasis (Fig. 1D and E and Table I).
Figure 1.
miR-384 is downregulated in breast cancer tissues, particularly in TNBC, compared with normal breast tissues. (A) Mean expression of miR-384 in breast cancer tissues from GEO58606 (*P<0.05, **P<0.01). (B) Expression of miR-384 in 24 cases of fresh human breast cancer tissues and their adjacent normal tissues by qPCR; miR-384 expression was normalized to U6 and expressed relative to the matched adjacent normal tissues (2−∆∆CT) (T/N, ratio of tumor/normal tissue). (C) Mean expression of miR-384 in normal breast tissues, breast cancer tissue samples and TNBC samples by qPCR (2−∆CT). (D) Mean expression of miR-384 by qPCR according to tumor size (2−∆CT). (E) Mean expression of miR-384 by qPCR according to lymph node metastasis (2−∆CT). Boundaries of boxes stand for the lower and upper quartile. Lines within the boxes and whiskers represent the median and extrema (maximum and minimum). *P<0.05, **P<0.01. TNBC, triple-negative breast cancer.
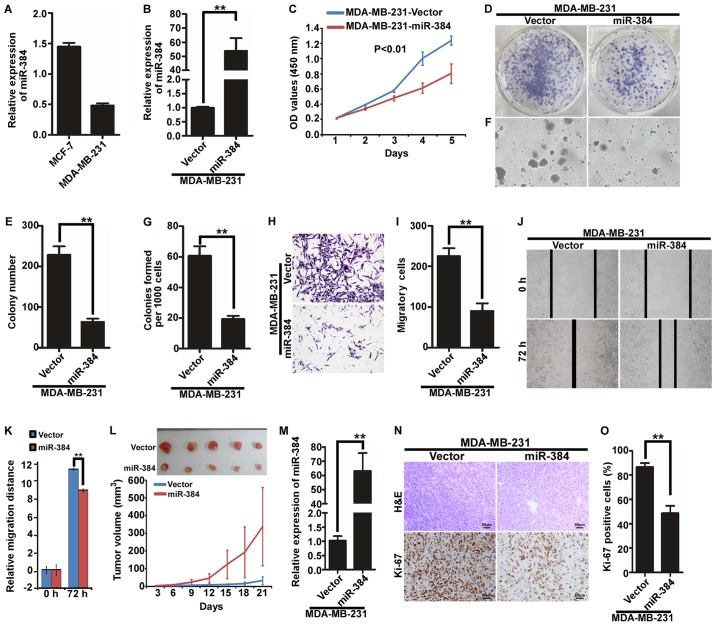
Ectopic expression of miR-384 inhibits the proliferation and migration of MDA-MB-231 cells in vitro and in vivo
We next examined the expression of miR-384 in MDA-MB-231 and MCF-7 by qPCR and found that the expression of miR-384 was relatively lower in the TNBC cell line MDA-MB-231 than that in MCF-7 cells (Fig. 2A). To evaluate the possible functions of miR-384 in breast cancer progression, we transfected hsa-miR-384 mimics into MDA-MB-231 cells and obtained cells with overexpression miR-384 (Fig. 2B). Then, we observed the effects of miR-384 on the proliferation and migration of breast cancer cells by MTT assay, colony formation assay, soft agar assay, Transwell migration assay and wound healing assay. We revealed that overexpression of miR-384 inhibited the proliferation and migration abilities of the breast cancer cells in vitro (Fig. 2C-K). To further observe the effects of miR-384 on the inhibition of the proliferation of breast cancer cells in vivo, we performed a tumorigenesis assay in nude mice by using MDA-MB-231 cells with stable miR-384 overexpression. The tumors in the MDA-MB-231/miR-384 group grew much slower than those in the MDA-MB-231/Vector group (Fig. 2L). Moreover, we detected the expression of miR-384 in the tumor tissues by qPCR and verified that miR-384 expression in the MDA-MB-231/miR-384 group was significantly higher than that in the MDA-MB-231/Vector group (Fig. 2M). In addition, H&E staining and IHC were performed. The results of IHC showed that the tumors of the MDA-MB-231/miR-384 group showed much lower Ki-67 indices than those in the control group (Fig. 2N and O).
Figure 2.
Ectopic expression of miR-384 inhibits the proliferation and migration of MDA-MB-231 cells in vitro and in vivo. (A) Expression of miR-384 in breast cancer cell lines MDA-MB-231 and MCF-7 was detected by qCR. (B) Overexpression of miR-384 in MDA-MB-231 cells was validated by qPCR. (C) Cell growth analyzed by MTT assays. (D and E) Colony formation assay. Representative images (D) and quantification (E) of colony formation; only cell colonies containing more than 50 cells were counted. (F and G) Soft agar assay. Representative micrographs are shown (F) and only cell colonies >0.1 mm in diameter were counted (G). (H and I) Transwell migration assay. Representative images (H) and quantification (I) of migrated cells across a Transwell chamber. (J and K) Wound-healing assay. Representative images (J). Histograms represent the average migrated distances at the indicated times (K). (L) MDA-MB-231/miR-384 and MDA-MB-231/Vector cells were injected into the hind limbs of nude mice (n=5). Tumor volumes were measured on the indicated days. The tumor volume data are presented as the mean ± SD. (M) miR-384 expression in the resected mouse tumor tissues derived from MDA-MB-231/miR-384 and MDA-MB-231/Vector cells by qPCR analysis. (N and O) Histopathological analyses of xenograft tumors. The tumor sections were stained with H&E or subjected to IHC staining using an antibody against Ki-67. Error bars represent mean ± SD from three independent experiments. **P<0.01.
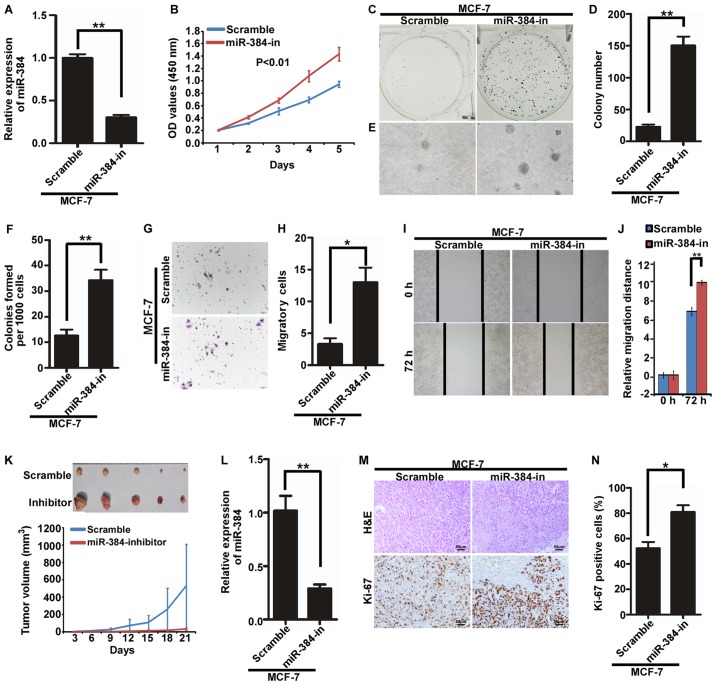
Inhibition of endogenous miR-384 promotes the proliferation and migration of MCF-7 cells in vitro and in vivo
The endogenous expression of miR-384 was suppressed by miR-384 inhibitors in MCF-7 cells (Fig. 3A). Then, we detected the proliferation and migration abilities of MCF-7/miR-384-in and MCF-7/Scramble cells by MTT assay, colony formation assay, soft agar assay, Transwell migration assay and wound healing assay. The results demonstrated that the suppression of miR-384 significantly increased the proliferation and migration abilities of the MCF-7 cells compared with the control cells (Fig. 3B-J). In order to further observe the inhibitory effects of miR-384 on tumor growth in vivo, we performed the tumorigenesis assay in nude mice by using MCF-7 cells with stable miR-384 inhibition. The results showed that the tumors in the MCF-7/miR-384-in group grew much faster than those in MCF-7/Scramble group (Fig. 3K). The expression of miR-384 was significantly lower in the MCF-7/miR-384-in group than that in the MCF-7/Scramble group (Fig. 3L). Additionally, H&E staining and IHC were performed. The results of IHC showed that the tumors from the MCF-7/miR-384-in group showed much higher Ki-67 indices than those in the control group (Fig. 3M and N).
Figure 3.
Inhibition of endogenous miR-384 promotes the proliferation and migration of MCF-7 cells in vitro and in vivo. (A) Expression of miR-384 in MCF-7 cells transfected with inhibitor (miR-384-in) or the negative control lentiviral scramble (Scramble) was detected by qPCR. (B) Cell growth analyzed by MTT assays. (C and D) Colony formation assay. Representative images (C) and quantification (D) of colony formation; only cell colonies containing more than 50 cells were counted. (E and F) Soft agar assay. Representative micrographs are shown (E) and only cell colonies >0.1mm in diameter were counted (F). (G and H) Transwell migration assay. Representative images (G) and quantification (H) of migrated cells across a Transwell chamber. (I and J) Wound-healing assay. Representative images (I). Histograms represent the average migrated distances at the indicated times (J). (K) MCF-7/miR-384-in and MCF-7/Scramble cells were injected into the hind limbs of nude mice (n=5). Tumor volumes were measured on the indicated days. The tumor volume data are presented as the mean ± SD. (L) miR-384 expression in the resected mouse tumor tissues derived from MCF-7/miR-384-in and MCF-7/Scramble cells by qPCR analysis. (M and N) Histopathological analyses of xenograft tumors. The tumor sections were stained with H&E or subjected to IHC staining using an antibody against Ki-67. Error bars represent mean ± SD from three independent experiments. *P<0.05, **P<0.01.
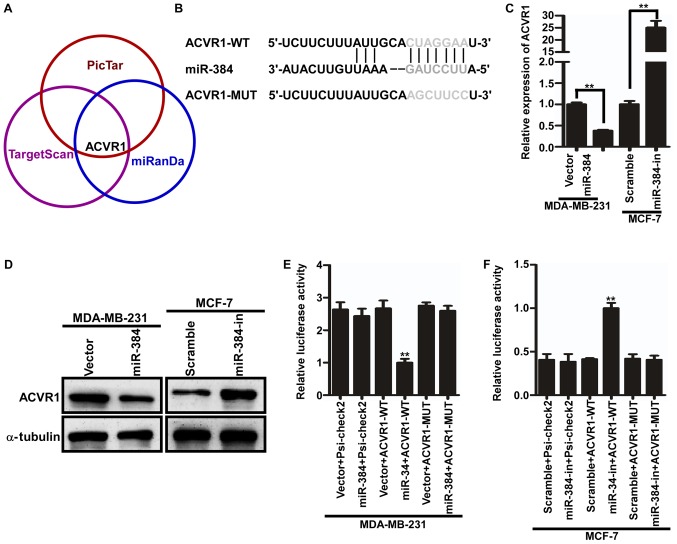
miR-384 decreases ACVR1 expression by directly binding to its 3′UTR in breast cancer
In order to further evaluate the function and mechanism of miR-384, three publicly available bioinformatic algorithms (TargetScan, Pictar and miRanda) were used to explore the target gene of miR-384. The results indicated that the 3′-UTR of ACVR1 contained the putative target sequence of miR-384 (Fig. 4A and B). To further determine that ACVR1 is a target of miR-384, qPCR and western blot analyses were performed. The results showed that the ACVR1 mRNA and protein levels were significantly downregulated in the miR-384-overexpressing cells, whereas these levels were upregulated in the miR-384-silenced cells (Fig. 4C and D). In subsequent experiments, we subcloned the 3′-UTR fragment of ACVR1 containing miR-384 binding site and the mutant fragment into psi-CHECK2 luciferase reporter vectors. Then the dual-luciferase assay analyses demonstrated that the co-expression of miR-384 mimics or inhibitors markedly inhibited or promoted the Renilla luciferase reporter activity of the wild-type ACVR1 3′UTR, but did not change the activity of the mutant 3′UTR constructs and their scramble vectors (Fig. 4E and F). Therefore, miR-384 decreased ACVR1 expression by directly binding to its 3′UTR in breast cancer cells.
Figure 4.
miR-384 decreases ACVR1 expression by directly binding to its 3′UTR in breast cancer. (A) ACVR1 is a target of miR-384 according to publicly available bioinformatic algorithms. (B) Predicted miR-384 target sequences in the 3′UTRs of ACVR1, and the mutant containing altered nucleotides in the 3′UTRs. (C) qPCR analysis of ACVR1 mRNA expression in the indicated cells. (D) Western blot analysis of ACVR1 in the indicated cells. (E and F) Luciferase assay analyses of the indicated cells transfected with the indicated reporters. Error bars represent mean ± SD from three independent experiments. **P<0.01. Vector, MDA-MB-231 cells transfected the vector control; miR-384, MDA-MB-231 cells transfected with miR-384-overexpression plasmid; Scramble, MCF-7 cells transfected with negative control lentiviral scramble; miR-384-in, MCF-7 cells transfected with inhibitor.
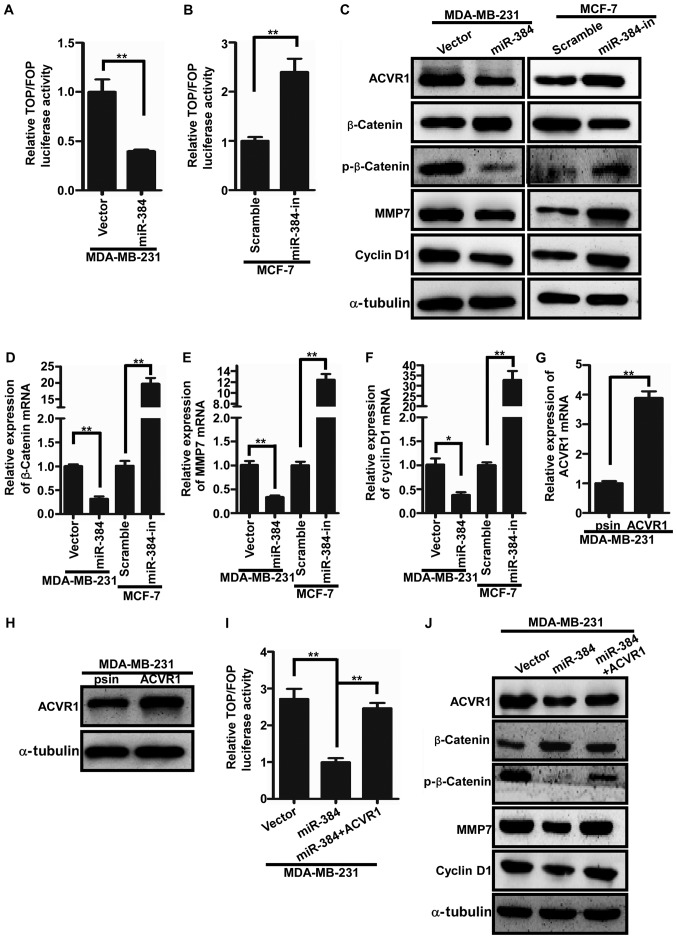
miR-384 negatively regulates the Wnt/β-catenin signaling pathway by targeting ACVR1 in breast cancer cells
It has been reported that ACVR1 is a key regulator of Wnt signaling by regulating the circuit of ACVR1/BMP/Wnt. Consequently, we speculated whether miR-384 would regulate the activity of Wnt/β-catenin signaling in breast cancer cells by ACVR1. The TOP/FOP luciferase assay was firstly performed to detect the Wnt/β-catenin signaling activity. The results showed that the activity of Wnt/β-catenin signaling was significantly increased in the miR-384-overexpressing breast cancer cells and significantly decreased in the miR-384-silenced cells (Fig. 5A and B). Furthermore, qPCR and western blotting were conducted to examine the expression levels of downstream genes of the Wnt/β-catenin signaling pathway. We found that β-catenin, cyclin D1 and MMP7 were significantly downregulated in breast cancer cells with miR-384 overexpression and upregulated in the miR-384-silenced breast cancer cells (Fig. 5C-F). To further verify whether miR-384 negatively regulates the Wnt/β-catenin signaling pathway by targeting ACVR1 in breast cancer cells, we cloned ACVR1 ORF into psin-EF-2 and transfected them into MDA-MB-231/miR-384 cells to restore ACVR1 expression (Fig. 5G and H). It was found that the activity of Wnt/β-catenin signaling was markedly increased in the MDA-MB-231/miR-384 cells with the restoration of ACVR1 expression (Fig. 5I and J). In conclusion, miR-384 was found to negatively regulate the Wnt/β-catenin signaling pathway in breast cancer cells.
Figure 5.
miR-384 negatively reguates the Wnt/β-catenin signaling pathway by targeting ACVR1 in breast cancer cells. (A and B) TOP/FOP luciferase assay revealed that Wnt/β-catenin activity was negatively correlated with miR-384 expression. (C-F) The detection of crucial downstream molecules of the Wnt/β-catenin signaling pathway by western blot and qPCR analyses. Error bars represent mean ± SD from three independent experiments. (G and H) ACVR1 overexpression in MDA-MB-231 cells by qPCR analysis or western blotting. (I and J) Restoration of Wnt/β-catenin activity with ACVR1 expression in MDA-MB-231-miR-384 cells. **P<0.01. Vector, MDA-MB-231 cells transfected the vector control; miR-384, MDA-MB-231 cells transfected with miR-384-overexpression plasmid; Scramble, MCF-7 cells transfected with negative control lentiviral scramble; miR-384-in, MCF-7 cells transfected with inhibitor; psin, MDA-MB-231/miR-384 cells transfected with psin-EF-2 without ACVR1; ACVR1, MDA-MB-231/miR-384 cells overexpressing ACVR1.
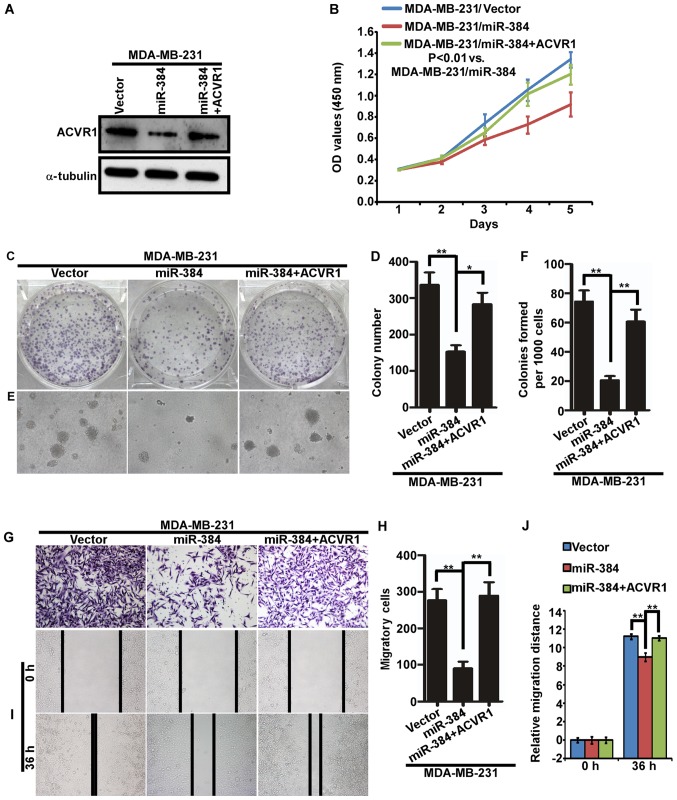
Suppression of ACVR1 plays important roles in the miR-384-silenced mediated phenotype of breast cancer cells
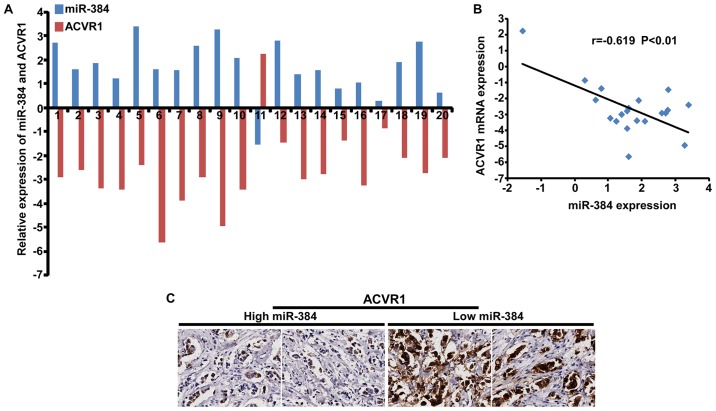
To further verify the biological function of miR-384 in the progression of breast cancer, a series of functional experiments were performed in MDA-MB-231-miR-384 cells with ACVR1 restoration (Fig. 6A). The results demonstrated that the abilities of proliferation and migration of MDA-MB-231/miR-384 cells were significantly increased with the restoration of ACVR1 (Fig. 6B-J). Moreover, we detected the expression of ACVR1 in 20 freshly collected breast cancer biopsies and explored the correlation between the expression of miR-384 and ACVR1. The results showed that there was a negative correlation between the expression of miR-384 and the expression of ACVR1 mRNA (Fig. 7A and B). IHC results detected in the same breast cancer biopsies showed that ACVR1 was increased with the downregulation of miR-384 (Fig. 7C). The results confirmed that miR-384 suppressed the aggressive phenotype of breast cancer cells by targeting.
Figure 6.
Suppression of ACVR1 plays important roles in miR-384-silenced mediated aggressive phenotype of breast cancer cells. (A) ACVR1 overexpression in MDA-MB-231/miR-384 cells. (B) Cell growth analyzed by MTT assays. (C-J) Colony formation assay, soft agar assay, Transwell migration assay and wound healing assay. Representative images (C, E, G and I) and quantification (D, F, H and J). *P<0.05, **P<0.01. Vector, MDA-MB-231 cells transfected the vector control; miR-384, MDA-MB-231 cells transfected with miR-384-overexpression plasmid; miR-384+ACVR1, cell overexpressing miR-384 and ACVR1.
Figure 7.
Correlation between the expression of miR-384 and ACVR1 in breast cancer tissues. (A) The expression of miR-384 and ACVR1 was detected by qPCR (ΔCT, n=20). (B) Spearman correlation analyses of miR-384 and ACVR1 mRNA expression. (C) The expression of ACVR1 in breast cancer tissues with high and low miR-384 expression by IHC (magnification, ×400).
Discussion
miRNAs are a class of small non-coding single stranded RNA species. They regulate the expression of target genes at the post-transcriptional levels by binding to specific sites of their mRNAs (19–21). It has been reported that miRNAs play crucial roles in the development and progression of many types of cancers and effect the outcome of therapies (22–24). However, further insights into the roles and molecular mechanisms of miRNAs during breast cancer progression are needed. To date, the deregulation of miR-384 has only been observed in a few tumor types (13–16). For example, miR-384 was found to play an essential role in melanoma metastasis by targeting HDAC3. In addition, it was recently reported that miR-384 exerts tumor-suppressive functions in colorectal cancer, glioma and hepatocellular carcinoma. However, it is still unknown whether dysregulation of miR-384 is associated with the carcinogenesis and development of breast cancer, especially TNBC. In the present study, we found that miR-384 was significantly downregulated in breast cancer, especially in TNBC tissues compared with that in normal breast tissues. Moreover, the expression of miR-384 was markedly lower in TNBC cells than that in non-TNBC cells. We inferred that miR-384 may play important roles in the carcinogenesis and development of breast cancer. Therefore, in the following study, a series of functional experiments demonstrated that the growth and migration of breast cancer cells were inhibited by miR-384 overexpression. Conversely, the inhibition of miR-384 significantly promoted the growth and migration of breast cancer cells. Xenograft experiments further demonstrated that miR-384 could suppress the proliferation of breast cancer cells in vivo. Collectively, these studies verified that miR-384 functions as a tumor suppressor in breast cancer. To explore the molecular mechanism of miR-384 in inhibiting breast cancer progression, we used three publicly available bioinformatic algorithms to analyze the target gene of miR-384. The results showed that ACVR1 (bone morphogenic protein receptor kinase activin A receptor, type I) may be an important target gene of miR-384. A dual-luciferase reporter system assay confirmed that ACVR1 is a direct target of miR-384. ACVR1 (also known as ALK2) is a key receptor of BMP7 (bone morphogenetic protein 7) and an important member of the bone morphogenetic protein (BMP) signaling pathway (25). In addition, recent studies have demonstrated that ACVR1 is not only a critical receptor of BMP7 but a key regulator of the Wnt signaling pathway and plays important roles in the occurrence and development of many diseases including breast cancer (6,26,27).
It is well known that the accumulation and nuclear localization of β-catenin is one of the hallmarks of the activation of the Wnt signaling pathway (28,29). That is to say, as the Wnt signaling pathway is activated, β-catenin is discharged from the degradation of the complex and results in the translocation of β-catenin into the nucleus, where it interacts with TCF/LEF (T-cell factor/lymphoid enhancer factor) and finally regulates the expression of specific Wnt target genes (30,31). Meanwhile, evidence has shown that miRNAs are crucial modulators of the Wnt/β-catenin signaling pathway (6,32–35). Therefore, in the present study, we aimed to ascertain whether miR-384 inhibits the proliferation of breast cancer by regulating the activation of the Wnt/β-catenin signaling pathway by targeting ACVR1. The results of TOP/FOP luciferase assays firstly demonstrated that the activity of the Wnt/β-catenin signaling pathway could be regulated by the expression of miR-384. The activity of the Wnt/β-catenin signaling pathway was obviously decreased in the miR-384-overexpressing MDA-MB-231 cells, but increased in the miR-384-silenced MCF-7 cells. Furthermore, qPCR and western blotting results verified that there were positive correlations between the expression of miR-384 and the downstream molecules of Wnt signaling and the activity of β-catenin. In subsequent experiments, we found that the repression of ACVR1 in MDA-MB-231 cells significantly decreased the activity of Wnt signaling by TOP/FOP luciferase assays. In addition, the results of the western blot analysis further demonstrated that ACVR1 restoration apparently increased the expression of the downstream molecules of Wnt signaling in the MDA-MB-231/miR-384 cells. Moreover, the proliferation and migration abilities of the MDA-MB-231/miR-384 cells were rescued following the restoration of ACVR1. Finally, there was a significant negative correlations between the expression of miR-384 and the expression of ACVR1 in 20 cases of clinical breast cancer tissues. All of these results supported that miR-384 inhibited the progression of breast cancer by impacting Wnt signaling activity by targeting ACVR1.
Since ACVR1 activates Wnt signaling, which plays an essential role in cancer stem cells, there has been extensive efforts to develop an inhibitor for ACVR1 (26,36). However, no effective ACVR1 inhibitor has become available to date. Yet miRNAs have been considered as a promising new class of therapeutic tools for cancer treatment since they are relatively stable and are naturally secreted and taken up by cells. In this study, we confirmed that miR-384 regulates the expression of ACVR1 protein by directly targeting the 3′UTR its mRNA in breast cancer. Restoration of miR-384 could inhibit the progression of breast cancer, especially TNBC.
Although our results are promising, there are still some limitations to this study. First, the results were confirmed only in MDA-MB-231 and MCF-7 cells. Furthermore, the small sample size might be another limitation of this study. We will further verify the conclusion in more cancer cell lines and expand the patient cohort in our future study.
In summary, this study confirmed that miR-384 is downregulated in breast cancer, especially in TNBC. Meanwhile, miR-384 functions as a tumor suppressor in breast cancer by targeting ACVR1 and impacting ACVR1/Wnt signaling. In addition, ACVR1 restoration could reverse the inhibition of the aggressive phenotype of breast cancer cells induced by miR-384. Furthermore, miR-384 expression was inversely correlated with the expression of ACVR1. Therefore, this study discovered a new role for miR-384 in modulating Wnt/β-catenin signaling in breast cancer tumorigenesis and further indicates that miR-384 may serve as a diagnostic and therapeutic target for breast cancer, especially for intractable TNBC.
Acknowledgements
We would like to gratefully acknowledge all people who provided the authors with considerable assistance.
Funding
The present study was supported by the Science and Technology Key Project of Henan Province Office Education of China (no. 14A310004) and the Scientific Research Fund of Xinxiang Medical University (nos. ZD200959 and ZD2011-21).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
YW designed the experiments. YW and ZZ conducted experiments and wrote the manuscript. JW provided the research materials and methods and analyzed the data. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
All participants provided written informed consent to participate and the tissue acquisition protocol was approved by the Ethic Institutional Board of Xinxiang Medical University.
Consent for publication
The relevant patients were informed and agreed for publication.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Denny L, de Sanjose S, Mutebi M, Anderson BO, Kim J, Jeronimo J, Herrero R, Yeates K, Ginsburg O, Sankaranarayanan R. Interventions to close the divide for women with breast and cervical cancer between low-income and middle-income countries and high-income countries. Lancet. 2017;389:861–870. doi: 10.1016/S0140-6736(16)31795-0. [DOI] [PubMed] [Google Scholar]
- 2.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua SA, Savage MI, Osborne CK, Hilsenbeck SG, Chang JC, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015;21:1688–1698. doi: 10.1158/1078-0432.CCR-14-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cappato S, Tonachini L, Giacopelli F, Tirone M, Galietta LJ, Sormani M, Giovenzana A, Spinelli AE, Canciani B, Brunelli S, et al. High-throughput screening for modulators of ACVR1 transcription: Discovery of potential therapeutics for fibrodysplasia ossificans progressiva. Dis Model Mech. 2016;9:685–696. doi: 10.1242/dmm.023929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang K, Sun X, Feng HL, Fei C, Zhang Y. DNALK2 inhibits the proliferation and invasiveness of breast cancer MDA-MB-231 cells through the Smad-dependent pathway. Oncol Rep. 2017;37:879–886. doi: 10.3892/or.2016.5343. [DOI] [PubMed] [Google Scholar]
- 6.Li L, Liu Y, Guo Y, Liu B, Zhao Y, Li P, Song F, Zheng H, Yu J, Song T, et al. Regulatory MiR-148a-ACVR1/BMP circuit defines a cancer stem cell-like aggressive subtype of hepatocellular carcinoma. Hepatology. 2015;61:574–584. doi: 10.1002/hep.27543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pencheva N, Tavazoie SF. Control of metastatic progression by microRNA regulatory networks. Nat Cell Biol. 2013;15:546–554. doi: 10.1038/ncb2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pencheva N, Tran H, Buss C, Huh D, Drobnjak M, Busam K, Tavazoie SF. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell. 2012;151:1068–1082. doi: 10.1016/j.cell.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jena MK. MicroRNAs in the development and neoplasia of the mammary gland. F1000 Res. 2017;6:1018. doi: 10.12688/f1000research.12005.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez-Barrueco R, Nekritz EA, Bertucci F, Yu J, Sanchez-Garcia F, Zeleke TZ, Gorbatenko A, Birnbaum D, Ezhkova E, Cordon-Cardo C, et al. miR-424(322)/503 is a breast cancer tumor suppressor whose loss promotes resistance to chemotherapy. Genes Dev. 2017;31:553–566. doi: 10.1101/gad.292318.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen Y, Ye YF, Ruan LW, Bao L, Wu MW, Zhou Y. Inhibition of miR-660-5p expression suppresses tumor development and metastasis in human breast cancer. Genet Mol Res. 2017;16 doi: 10.4238/gmr16019479. [DOI] [PubMed] [Google Scholar]
- 13.Eom S, Kim Y, Park D, Lee H, Lee YS, Choe J, Kim YM, Jeoung D. Histone deacetylase-3 mediates positive feedback relationship between anaphylaxis and tumor metastasis. J Biol Chem. 2014;289:12126–12144. doi: 10.1074/jbc.M113.521245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai YY, Shen F, Cai WS, Chen JW, Feng JH, Cao J, Xiao HQ, Zhu GH, Xu B. MiR-384 regulated IRS1 expression and suppressed cell proliferation of human hepatocellular carcinoma. Tumour Biol. 2016;37:14165–14171. doi: 10.1007/s13277-016-5233-5. [DOI] [PubMed] [Google Scholar]
- 15.Zheng J, Liu X, Wang P, Xue Y, Ma J, Qu C, Liu Y. CRNDE promotes malignant progression of glioma by attenuating miR-384/PIWIL4/STAT3 axis. Mol Ther. 2016;24:1199–1215. doi: 10.1038/mt.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Wang YX, Chen YR, Liu SS, Ye YP, Jiao HL, Wang SY, Xiao ZY, Wei WT, Qiu JF, Liang L, et al. MiR-384 inhibits human colorectal cancer metastasis by targeting KRAS and CDC42. Oncotarget. 2016;7:84826–84838. doi: 10.18632/oncotarget.12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matamala N, Vargas MT, González-Cámpora R, Miñambres R, Arias JI, Menéndez P, Andrés-León E, Gómez-López G, Yanowsky K, Calvete-Candenas J, et al. Tumor microRNA expression profiling identifies circulating microRNAs for early breast cancer detection. Clin Chem. 2015;61:1098–1106. doi: 10.1373/clinchem.2015.238691. [DOI] [PubMed] [Google Scholar]
- 18.Ye YP, Wu P, Gu CC, Deng DL, Jiao HL, Li TT, Wang SY, Wang YX, Xiao ZY, Wei WT, et al. miR-450b-5p induced by oncogenic KRAS is required for colorectal cancer progression. Oncotarget. 2016;7:61312–61324. doi: 10.18632/oncotarget.11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 20.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 21.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 22.Shi C, Yang Y, Xia Y, Okugawa Y, Yang J, Liang Y, Chen H, Zhang P, Wang F, Han H, et al. Novel evidence for an oncogenic role of microRNA-21 in colitis-associated colorectal cancer. Gut. 2016;65:1470–1481. doi: 10.1136/gutjnl-2014-308455. [DOI] [PubMed] [Google Scholar]
- 23.Liu M, Huang F, Zhang D, Ju J, Wu XB, Wang Y, Wang Y, Wu Y, Nie M, Li Z, et al. Heterochromatin protein HP1γ promotes colorectal cancer progression and is regulated by miR-30a. Cancer Res. 2015;75:4593–4604. doi: 10.1158/0008-5472.CAN-14-3735. [DOI] [PubMed] [Google Scholar]
- 24.Loo JM, Scherl A, Nguyen A, Man FY, Weinberg E, Zeng Z, Saltz L, Paty PB, Tavazoie SF. Extracellular metabolic energetics can promote cancer progression. Cell. 2015;160:393–406. doi: 10.1016/j.cell.2014.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Massagué J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 26.Kamiya N, Kaartinen VM, Mishina Y. Loss-of-function of ACVR1 in osteoblasts increases bone mass and activates canonical Wnt signaling through suppression of Wnt inhibitors SOST and DKK1. Biochem Biophys Res Commun. 2011;414:326–330. doi: 10.1016/j.bbrc.2011.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slattery ML, John EM, Torres-Mejia G, Herrick JS, Giuliano AR, Baumgartner KB, Hines LM, Wolff RK. Genetic variation in bone morphogenetic proteins and breast cancer risk in hispanic and non-hispanic white women: The breast cancer health disparities study. Int J Cancer. 2013;132:2928–2939. doi: 10.1002/ijc.27960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J Cell Sci. 2003;116:2627–2634. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- 29.Reed KR, Athineos D, Meniel VS, Wilkins JA, Ridgway RA, Burke ZD, Muncan V, Clarke AR, Sansom OJ. B-catenin deficiency, but not Myc deletion, suppresses the immediate phenotypes of APC loss in the liver. Proc Natl Acad Sci USA. 2008;105:18919–18923. doi: 10.1073/pnas.0805778105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arce L, Yokoyama NN, Waterman ML. Diversity of LEF/TCF action in development and disease. Oncogene. 2006;25:7492–7504. doi: 10.1038/sj.onc.1210056. [DOI] [PubMed] [Google Scholar]
- 31.Lu FI, Sun YH, Wei CY, Thisse C, Thisse B. Tissue-specific derepression of TCF/LEF controls the activity of the Wnt/β-catenin pathway. Nat Commun. 2014;5:5368. doi: 10.1038/ncomms6368. [DOI] [PubMed] [Google Scholar]
- 32.Yamada N, Noguchi S, Mori T, Naoe T, Maruo K, Akao Y. Tumor-suppressive microRNA-145 targets catenin δ-1 to regulate Wnt/β-catenin signaling in human colon cancer cells. Cancer Lett. 2013;335:332–342. doi: 10.1016/j.canlet.2013.02.060. [DOI] [PubMed] [Google Scholar]
- 33.Zhou AD, Diao LT, Xu H, Xiao ZD, Li JH, Zhou H, Qu LH. β-Catenin/LEF1 transactivates the microRNA-371-373 cluster that modulates the Wnt/β-catenin-signaling pathway. Oncogene. 2012;31:2968–2978. doi: 10.1038/onc.2011.461. [DOI] [PubMed] [Google Scholar]
- 34.Ji S, Ye G, Zhang J, Wang L, Wang T, Wang Z, Zhang T, Wang G, Guo Z, Luo Y, et al. miR-574-5p negatively regulates Qki6/7 to impact β-catenin/Wnt signalling and the development of colorectal cancer. Gut. 2013;62:716–726. doi: 10.1136/gutjnl-2011-301083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai J, Guan H, Fang L, Yang Y, Zhu X, Yuan J, Wu J, Li M. MicroRNA-374a activates Wnt/β-catenin signaling to promote breast cancer metastasis. J Clin Invest. 2013;123:566–579. doi: 10.1172/JCI65871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malanchi I, Peinado H, Kassen D, Hussenet T, Metzger D, Chambon P, Huber M, Hohl D, Cano A, Birchmeier W, Huelsken J. Cutaneous cancer stem cell maintenance is dependent on betacatenin signalling. Nature. 2008;452:650–653. doi: 10.1038/nature06835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.