Abstract
Esophageal squamous cell carcinoma (ESCC) is one of the most common malignancies in China, and is associated with high morbidity and mortality. However, the molecular mechanisms that control ESCC tumorigenicity and metastasis remain unclear. Here, we report that the RNA splicing factor, NONO, is an important regulator of ESCC growth, apoptosis and invasion. NONO protein levels were dramatically upregulated in ESCC when compared with that in adjacent benign esophageal squamous epithelium. Particularly, NONO expression was statistically higher in tumors with greater tumor invasion depth. Using multiple ESCC cell models, we further showed that NONO depletion using siRNA significantly inhibited proliferation, invasion, and promoted apoptosis of ESCC cells. In addition we found that knockdown of NONO could reduce protein levels of phosphorylated Akt and Erk1/2. Our findings suggest that NONO plays a potent role in multiple biological aspects of ESCC through activation of the Akt and Erk1/2 signaling pathways. Taken together, our findings suggest that NONO might play an important role in promoting tumorigenesis of ESCC. It may provide a promising approach to prevent the progress of ESCC.
Keywords: NONO, esophageal squamous cell carcinoma, apoptosis, migration, invasion
Introduction
Esophageal cancer is one of the most aggressive cancers worldwide. There are two main types of esophageal cancer: esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC) (1). In Asia, ESCC is the predominant type and EAC remains rare (2). Treatment of ESCC has greatly relied upon classical therapeutic means including surgery, radiotherapy or chemotherapy. Although multimodality therapy can apparently improve the prognosis of patients with ESCC, the overall 5-year survival rate remains unsatisfactory. Therefore, seeking new potential targets for the therapy of ESCC will help to improve the clinical outcome of patients with ESCC (3), which is reflecting our limited understanding on carcinogenesis and metastasis of ESCC at the molecular and cellular levels. It is urgent to find sensitive and specific early biomarkers for diagnosis and prognosis of ESCC, as well as novel therapeutic strategies.
The non-POU-domain-containing octamer (NONO) binding protein, also called NONO, is involved in a variety of biological processes, including regulating gene expression, DNA synthesis and repair processes (4–6). Originally identified as an RNA-binding protein (7), NONO interacts with double-stranded DNA, single-stranded DNA and RNA and is involved in multiple steps of gene transcription, RNA splicing and even non-homologous DNA end joining (8). Due to its roles in gene transcription, RNA processing and DNA repairing, NONO may be implicated in cancer progression. It has been demonstrated that NONO is associated with the initiation and progression of human cancer. NONO protein expression is strongly correlated with vascular invasions and poor patient survival of bladder cancers (9). In fact, increased NONO expression was reported in prostate cancer and malignant melanoma (10,11). Knockdown NONO reduced melanoma cell proliferation, promoted apoptosis and decreased mobility capacity. Overexpression of NONO increased human umbilical vein endothelial cells invasion (12). Chromosomal translocation of NONO and TFE3 genes are frequently detected in papillary rental cell carcinoma (13). Additionally, a previous study indicates that NONO is an independent prognostic factor for human neuroblastoma (14). One recent study has identified that the nuclear protein NONO is highly expressed in breast cancer tissues as compared with the adjacent normal tissues in human patients. It is also demonstrated that NONO-mediated regulation of breast cancer cell growth is dependent on the presence of and interaction with SREBP-1 (15). Moreover, gene microarray studies also reported that NONO mRNA levels are increased in ESCC (16,17). However, validation of NONO protein expression in human ESCC tissues has not been performed. The protein expression and functional significance of NONO in ESCC are still unclear.
Materials and methods
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Collection of all tissues and data involving human participants were approved by institutional ethics committee of Beijing Friendship Hospital on human research.
Human tissue specimens and cell lines
Human ESCC and adjacent normal esophageal tissues were collected from 42 patients undergoing surgery of ESCC at the Beijing Friendship Hospital (Beijing, China) between 2008 and 2012. No patients had been treated with chemo- or radiation therapy before surgery. Informed consent was obtained from each patient. All tissues and data were collected in accordance with institutional guidelines and approved by institutional ethics committee of Beijing Friendship Hospital on human research. The pathological tumor stage was assessed using the TNM system according to the American Joint Committee on Cancer (AJCC) staging manual (seventh edition) (18,19). Human ESCC cell lines (TE-1, TE-2, TE-10, KYSE30, KYSE70, KYSE140, KYSE450, EC109, EC9706) were generously provided by Cancer Institute and Hospital, Chinese Academy of Medical Sciences and cell authentication were reported (20,21). Cells were maintained in RPMI-1640 medium containing 10% fetal bovine serum, at 37°C in a 95% humidified incubator containing 5% CO2.
Immunohistochemistry and pathological scoring
Immunohistochemistry (IHC) followed the standard protocol with NONO antibody (BD Biosciences, Bedford, MA, USA). Slides were also counterstained with hematoxylin and mounted on coverslips with glycerol-gelatin. Pathological scoring of NONO was performed by the quickscore method (22). The percentage of stained cells (0–4, 5–19, 20–35, 40–59, 60–79 and 80–100%) was scored as 1–6, while the staining intensity (no staining, low, moderate, and high) was scored as 0–3. The quickscore of each tissue sample was calculated by multiplying the score of percentage of positive cells with the score of the staining intensity. Tumor samples with quickscore >12 were recorded as strong, moderate (quickscore <12 and >6), weak positive/negative (quickscore<6). Two doctors with no prior knowledge of each patient's clinical information evaluated all specimens independently.
siRNA and transfection
TE-1 and KYSE70 cells were plated at a density of 4×105 cells/well in 6-well plates. After 24 h of culture, the cells were transfected with siRNA using Lipofectamine® 2000 (Life Technologies, Grand Island, NY, USA) according to the manufacturer's protocol as we previously described. Small interfering RNA (siRNA) for NONO (siNONO) and control siRNA (siCTRL) were purchased from GenePharma (Shanghai, China). The siRNA sequences against NONO are 5′-CAGAGAAGCUGGUUAUAAAT-3′ and 5′-UUUAUAACCAGCUUCUCUGTT-3′. The siRNA sequences against non-specific are 5′-UUCUCCGAACGUGUCACGUTT-3′ and 5′-ACGUGACACGUUCGGAGAATT-3′.
Real-time PCR
Total RNA was extracted from ESCC cell lines using TRIzol (Life Technologies) as we previously described (23). Two micrograms of total RNA was subjected to a random-primed reverse transcription using SuperScript II reverse transcriptase (Life Technologies). Real-time qPCR was conducted in triplicates using Applied Biosystems 7500 Fast with 5 ng of cDNA, 1 µM of each primer pair and SYBR Green PCR master mix (Roche). The primers used were NONO forward, 5′-TTGTGGGAAATCTTCCTCCCGACA-3′ and reverse, 5′-GGGTTTCCAAGCGGATAAAGCCAA-3′. GAPDH forward, 5′-GGACCTGACCTGCCGTCTAGAA-3′ and reverse, 5′-GGTGTCGCTGTTGAAGTCAGAG-3′. Relative mRNA levels were normalized to GAPDH.
Western blot analysis
Antibodies against Akt, phosphorylated Akt (p-Akt) (Ser473), Erk, p-Erk1/2 (Thr202/Tyr204), caspase-3 and cleaved PARP-1 were purchased from Cell Signaling Technology (Beverly, MA, USA). The antibody against β-actin was purchased from Sigma-Aldrich. Protein lysates were extracted by lysis buffer [50 mM Tris-HCl, pH 7.4; 10 mM EDTA; 5 mM EGTA; 0.5% NP40; 1% Triton X-100 plus protease inhibitor (Roche)]. Fifty micrograms of total protein were fractionated by SDS-polyacrylamide gel electrophoresis and then transferred to PVDF membranes (Millipore, Billerica, MA, USA). The membranes were probed with primary antibodies overnight at 4°C and then incubated for 1 h with secondary peroxidase-conjugated antibodies at room temperature, followed by detection with an ECL plus system (Beyotime, Jiangsu, China). β-actin was used as the loading control.
Cell proliferation assay
Cells were plated onto a 96-well plate at a density of 1×103 cells per well. Viability of the cells was measured using a CellTiter 96® Aqueous One Solution Cell Proliferation assay (Promega, Madison, WI, USA) as described previously with modifications (20). After 0, 24, 48 and 72 h, the optical density at 492 nm was measured on a microplate reader (Smartspec Model 450; Bio-Rad Laboratories, Inc., Hercules, CA, USA) at 490 nm. Three independent experiments were performed in triplicate.
Cell apoptosis analysis
Apoptosis rates of cells were measured using an Annexin V-FITC/PI apoptosis kit (Becton Dickinson, Franklin Lakes, NJ, USA) following the manufacturer's instructions. Briefly, cells were harvested, washed with cold PBS, and resuspended in binding buffer. Then, 5 µl Annexin V-FITC and 5 µl PI were added to each sample containing 1×105 cells/100 µl. The samples were incubated at 25°C in the dark for 15 min, followed by addition of 400 µl binding buffer. Within 1 h of preparation, the samples were analyzed by a flow cytometer (Becton Dickinson). The experiments were repeated three times independently.
Cell migration and invasion assays
In cell migration assays, cells were grown on 6-well plates until 100% confluent. A 20-µl pipette tip was used to scratch and create a wound in the confluent monolayer at the center of culture plates. After washes with PBS buffer, cells were replenished with culture medium and cell migration was captured under a microscope (Olympus Corp., Tokyo, Japan) at 0 and 24 h post wound scratch. Migration rate was determined using the ImageJ software as an average closed area of the wound relative to the initial wound area at 24 h after wounding. Experiments were performed in triplicate with three independent repeats. In cell invasion assays, 10×104 of TE-1 cells, 12×104 of KYSE70 cells were suspended in serum-free RPMI-1640 medium and seeded in BD Matrigel invasion chamber (Becton Dickinson). The lower chamber contained 0.75 ml of RPMI-1640 medium plus 10% FBS. After incubation for 30 h, non-invading cells in the upper chamber were gently removed by cotton swabs. Cells that invaded through the Matrigel and reached to the lower chamber were fixed, stained with mounting medium containing DAPI (Vector Laboratories, Inc., Burlingame, CA, USA) and photographed by fluorescence microscope (Olympus). Invaded cell numbers were counted by the ImageJ software. Experiments were performed in triplicate with three independent repeats.
Immunofluorescence assays
ESCC cell lines were grown on glass coverslips (Nest Biotechnology Co., Ltd., Wuxi, China) to 50% confluence and then fixed in 4% paraformaldehyde for 15 min at room temperature. The cells were permeabilized with 0.3% Triton X-100 for 20 min, blocked with 5% bovine serum albumin for 30 min, and then incubated with the mouse monoclonal anti-NONO antibody (Becton Dickinson) at 4°C overnight, followed by an Alexa Fluor 488-conjugated secondary antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) for 45 min at 37°C. The cells were rinsed in PBS between incubation steps and mounted with medium containing 4′-6-diamidino-2-phenylindole (DAPI). Immunofluorescence images were acquired using a fluorescence microscope (Olympus).
Statistical analysis
The Wilcoxon signed rank test was used for comparison of quickscores between paired ESCC and adjacent benign tissue (Fig. 1B). Comparison of Quickscore distribution between benign and ESCC was performed by χ2 test (Fig. 1C). The associations of pathological features with NONO quickscores were evaluated by Fisher's exact test in Table I. Data from Figs. 2–4 are presented as the mean ± SEM that was calculated from at least three independent experiments. Statistical analysis was carried out using GraphPad Prism (version 4) (GraphPad Software, Inc., La Jolla, CA, USA) with the level of significance set at P<0.05, P<0.01 and P<0.001. Paired Student's t-test was used to compare two groups when the means follow the normal distribution.
Figure 1.
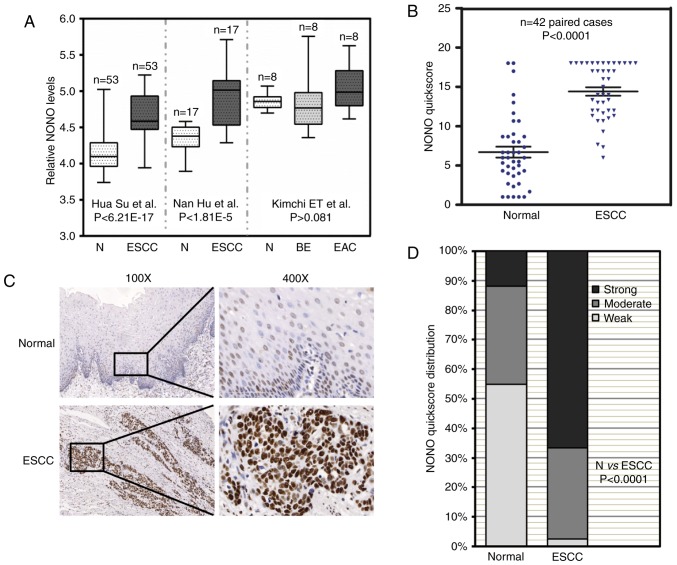
Increased expression of NONO in ESCC tissue samples. (A) Three independent microarray data from the Oncomine database showed that NONO mRNA levels were increased in ESCC patient tissue samples. Comparison of NONO mRNA levels in normal tissue (N), esophageal squamous cell carcinoma (ESCC), and Barrett's esophagus (BE), esophageal adenocarcinoma (EAC) are presented together with P-values. (B) Immunohistochemistry signal of NONO from paired normal and ESCC tissue samples were recorded by quickscore method. Comparison of quickscore distribution between adjacent normal and ESCC samples was performed by χ2 test. (C) Representative immunohistochemistry (IHC) images of NONO in paired normal and ESCC tissue samples. NONO levels were lower in adjacent normal esophageal epithelium, but were higher in ESCC. (D) NONO quickscores were further divided into three groups: strong, moderate and weak. The percentage of each group in normal and ESCC tissue samples were plotted.
Table I.
Pathological information of ESCC patients and NONO expression.
| NONO expression level | ||||
|---|---|---|---|---|
| Variables | Weak + moderate (%) | Strong (%) | No. | P-value |
| Age | ||||
| >60 | 15 | 4 (28.6) | 11 (39.3) | 0.734 |
| <60 | 27 | 10 (71.4) | 17 (60.7) | |
| Sex | ||||
| Male | 32 | 11 (78.6) | 21 (75.0) | 1.000 |
| Female | 10 | 3 (21.4) | 7 (25.0) | |
| Tumor depth | ||||
| T1-2 | 6 | 5 (35.7) | 1 (3.6) | 0.011a |
| T3-4 | 36 | 9 (64.3) | 27 (96.4) | |
| LN metastasis | ||||
| N0 | 36 | 11 (78.6) | 25 (89.3) | 0.652 |
| N1/N2/N3 | 6 | 3 (21.4) | 3 (10.7) | |
| Pathological differentiation | ||||
| Well | 7 | 2 (14.2) | 5 (17.9) | 0.651 |
| Moderate | 21 | 6 (42.9) | 15 (53.6) | |
| Poor | 14 | 6 (42.9) | 8 (28.5) | |
| AJCC stage | ||||
| I | 5 | 2 (14.3) | 3 (10.7) | 0.860 |
| II | 26 | 9 (64.3) | 17 (60.7) | |
| III–IV | 11 | 3 (21.4) | 8 (28.6) | |
AJCC, American Joint Commission on Cancer. ESCC, esophageal squamous cell carcinoma. The associations of pathological information of ESCC patients with NONO expression are summarized and statistically analyzed by Fisher's exact test.
P<0.05, significant difference.
Figure 2.
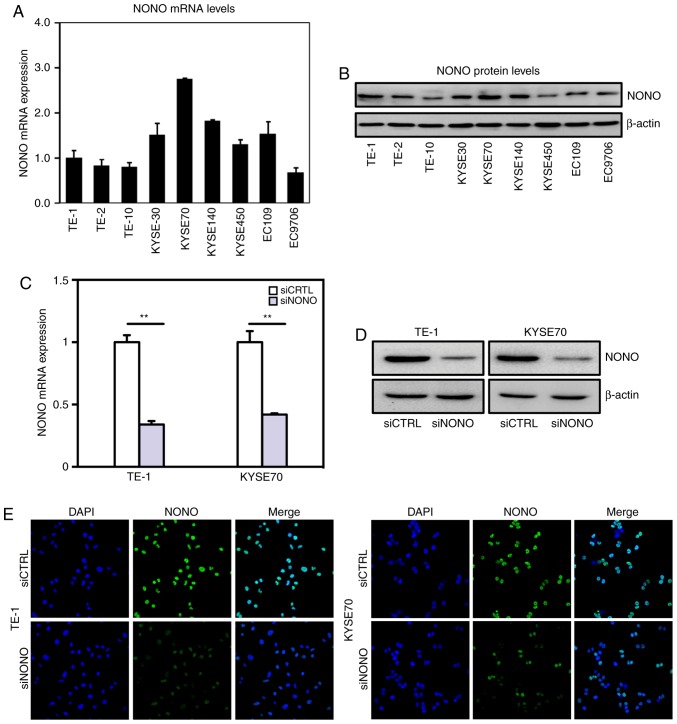
Real-time PCR and western blot analysis of NONO expression in esophageal squamous cell carcinoma (ESCC) cells. (A) Real-time PCR measured relative NONO mRNA levels to GAPDH in 9 ESCC cell lines. (B) Western blot analysis of endogenous expression of NONO in all 9 ESCC cell lines. β-actin served as a loading control. (C and D) Transfection efficiencies of NONO siRNA knockdown in TE-1, KYSE70 cells were measured by real-time PCR and western blotting. (E) Immunofluorescence staining detected expression and nuclear localization of NONO protein in human ESCC cell lines. NONO expression is reduced after NONO siRNA knockdown in TE-1, KYSE70 cells.
Figure 4.
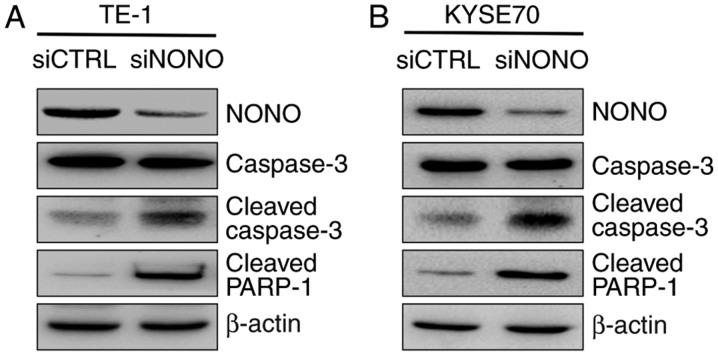
Examination of NONO targets in esophageal squamous cell carcinoma (ESCC) cells. (A) The cleavages of caspase-3, and PARP-1 were compared between siRNA control and siRNA NONO-transfected cells by western blotting at the indicated time-points. β-actin served as a loading control. NONO depletion upregulated caspase-3 and PARP-1 cleavage in TE-1 and KYSE70 ESCC cell lines.
Results
NONO is upregulated in human ESCC tissue samples
Three independent microarray studies reported that higher mRNA levels of NONO were detected in ESCC tissues (Fig. 1A) (16,17,24). By contrast, there were no significant differences of NONO mRNA levels in Barrett's esophagus, EAC and adjacent normal esophagus. In order to determine NONO protein expression in ESCC, we collected 42 paired patient samples containing ESCC and adjacent benign tissue for IHC. IHC signal of NONO was localized in the nuclei of esophageal epithelial cells as well as a sub-group of stromal and muscle cells (Fig. 1B). NONO expression was high in ESCC compared with that in adjacent benign tissue samples (P<0.0001) (Fig. 1B). Representative IHC images are presented in Fig. 1C. Of the ESCC tissue samples 66.7% showed strong NONO signal whereas only 11.9% of benign tissue samples showed strong NONO staining (Fig. 1D). The relationship between pathological background and NONO expression was further analyzed (Table I). While there was no correlation between NONO protein levels with patient age, gender, lymph node metastasis, pathological differentiation and TNM stage, strong NONO staining was detected in ESCC with greater tumor invasion depth (P=0.011). These results suggested a potential role of NONO in ESCC.
NONO expression in ESCC cell lines
Real-time PCR and western blot assays indicated that NONO is widely expressed in multiple ESCC cell lines (Fig. 2A and B). However, NONO mRNA levels varies among these cell lines with relatively higher levels in KYSE30 and KYSE70 cell lines and lower levels in TE-1, TE-2 and TE-10 cell lines. TE-1 was derived from well differentiated ESCC, whereas KYSE70 was derived from poorly differentiated tumors, respectively (25). We chose TE-1 and KYSE70 cell lines for further functional analyses of NONO. Knockdown NONO was achieved by transient transfection with efficiencies shown in Fig. 2C-E.
Knockdown NONO inhibits ESCC cell proliferation and induces cell apoptosis
Given the elevated expression of NONO in ESCC tissues and cell lines, we examined whether NONO regulated ESCC cell growth. As shown in Fig. 3A and B, NONO knockdown by specific siRNA significantly inhibited growth of TE-1 and KYSE70 cells.
Figure 3.
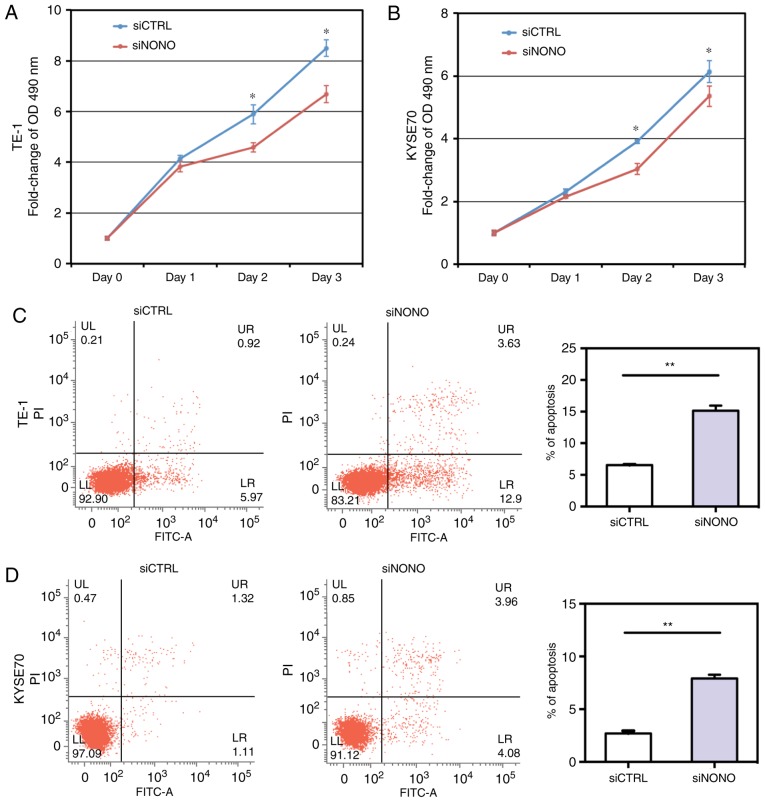
NONO modulates cell proliferation and apoptosis. (A and B) MTS assay showed that NONO knockdown inhibited proliferation in TE-1 and KYSE70 ESCC cell lines. (C and D) Apoptotic cell death was determined by flow cytometric analysis with Annexin V and PI staining. NONO knockdown increased the level of apoptosis in TE-1 and KYSE70 cell lines. Results shown are representative of three independent experiments with Student's t-test. Statistical significance *P<0.05 and **P<0.01.
To determine whether NONO could regulate apoptosis of TE-1 and KYSE70 cells, we performed an Annexin V-FITC and PI assay to detect apoptosis. Knockdown of NONO significantly induced apoptosis of TE-1 and KYSE70 cells. After NONO knockdown, the percentages of both early apoptotic (Annexin V-positive/PI-negative) and late apoptotic (Annexin V-positive/PI-positive) cells were significantly increased in NONO-knockdown cells, compared with control cells (P<0.05, Fig. 3C and D).
Because apoptosis is often mediated by the activation of caspase-3 that leads to PARP binding to fragmented DNA, western blot analysis was then used to detect caspase-3 activation. The result showed that cleavages of caspase-3 and PARP-1 were dramatically increased in NONO-knockdown cells, compared with control cells (Fig. 4).
NONO regulates ESCC cell migration and invasion in vitro
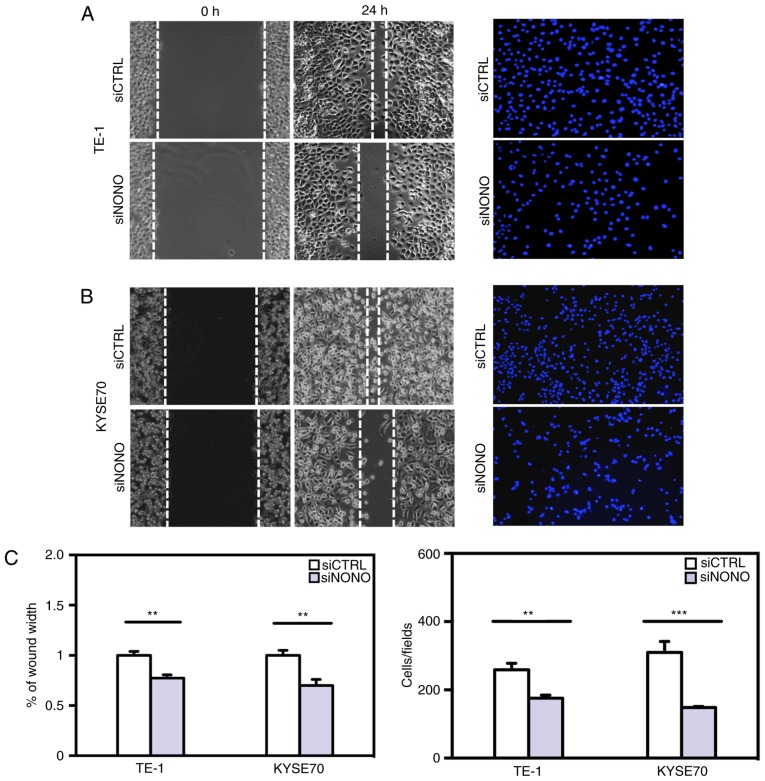
Since higher NONO protein expression was observed in tumors with greater local invasion (Table I), we further studied whether NONO is a regulator of ESCC cell migration and invasion in vitro. Knockdown of endogenous NONO expression significantly reduced TE-1 and KYSE70 cell mobility in passing through the surface of culturing plates or penetrating through Matrigel (Fig. 5). These results indicated that the expression levels of NONO are important for ESCC cell migration and invasion.
Figure 5.
NONO knockdown decreases esophageal squamous cell carcinoma (ESCC) cell migration and Matrigel invasion. TE-1 and KYSE70 cells were transfected with NONO or control siRNA for 24 h. (A and B) Wound healing assay showed that cell motility was inhibited by NONO knockdown. Representative images at time 0 and 24 h after scratching (left). Cells penetrated through the Matrigel were captured by microscope and representative images are shown (right). The numbers of cells invaded through Matrigel in each condition were counted and plotted. (C) Results shown are representative of three independent experiments with Student's t-test, the statistical significance was *P<0.05, **P<0.01 and ***P<0.001.
Knockdown NONO negatively regulates p-Erk1/2 and p-AKT expression
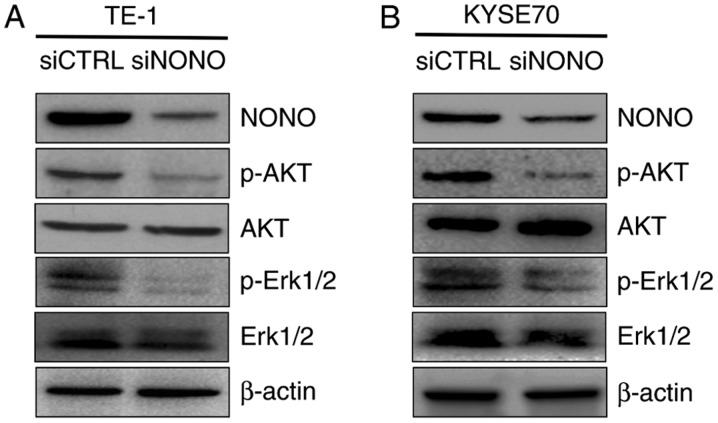
As in vitro assays showed that NONO is overexpressed in ESCC tissues and cells, and that knockdown NONO inhibits proliferation, induces apoptosis and reduces mobility of TE-1 and KYSE70 cells, we then explored the molecular mechanism underlying the effects. Both Erk1/2/MAPK and PI3K/AKT activation are frequent events in human cancer including ESCC (26,27). Since Erk1/2 and AKT are activated through phosphorylation, we detected the expression of the Thr202/Tyr204 phosphorylated form of Erk1/2 and Ser473 phosphorylated form of AKT by western blotting. TE-1 and KYSE70 cells were treated with siRNA targeting NONO or control siRNA as a negative. We found that the expression levels of phosphorylated Erk1/2 and AKT were dramatically decreased in both TE-1 and KYSE70 NONO-knockdown cells (Fig. 6). These data suggested that Erk1/2 and PI3K/AKT pathway is required for the NONO-regulated growth, apoptosis and invasion of ESCC cells.
Figure 6.
Akt and Erk1/2 are likely the downstream targets of NONO-mediated signaling. p-Akt, Akt, p-Erk1/2 and Erk1/2 were compared between TE-1 and KYSE70 cell lines treated with control siRNA and NONO siRNA by western blot analysis. β-actin served as a loading control.
Discussion
Being one of the most common causes of cancer-related deaths in China, the average 5-year survival rate of ESCC is still low. Several mechanisms promote ESCC progression and provide with independence from normal regulation of cell control (28). Treatment of ESCC especially in advanced stages still remains a clinical challenge. Therefore, new therapeutic targets remain to be elucidated. The nuclear NONO protein is an RNA-binding molecule containing two RNA recognition motifs. It is able to bind double-stranded DNA, single-stranded DNA and RNA (29). Previous studies showed that it retains hyper-edited messenger RNA in the nucleus and therefore influences gene expression and differentiation of stem cells (30). Recently, it has been shown that NONO is strongly expressed in several types of cancer. In addition, an increasing number of studies have reported that NONO modulates various pivotal intermediate molecules and ultimately regulates cell proliferation, apoptosis and mobility in different types of human carcinomas. However, the expression pattern and function of NONO in the tumorigenesis of ESCC remain unknown. Our contribution in this study is for the first time to demonstrate that NONO protein expression is upregulated in ESCC and that higher NONO protein levels are associated with greater tumor invasion depth. In addition, we demonstrated that NONO plays direct regulatory roles in ESCC cell proliferation, apoptosis, migration and invasion involving Erk1/2 and Akt signaling pathways.
Firstly, we observed a significantly elevated NONO expression in most ESCC tissues compared with paired normal esophageal epithelial tissues. Higher NONO expression was shown to be associated with deeper local invasion, but no differences between tumors with different degree of lymph node metastasis, supporting a potential significance in early stage of ESCC progression. However, the correlation of NONO expression and the poor overall survival of ESCC patients need further study.
Secondly, we observed different oncogenic effects in two ESCC cell lines following downregulation of NONO expression. Targeted NONO silencing using siRNA inhibited cell growth in TE-1 and KYSE70 ESCC cell lines. Moreover, NONO silencing increased the percentage of apoptotic cells in those cell lines, in which this oncogene is endogenously overexpressed. In addition, we examined the expression of cleaved caspase-3 and cleaved PARP-1 by western blotting. We showed that knockdown of NONO could inhibit caspase-3 and PARP-1 cleavage, which was according to its role of apoptosis inhibitor. Taken together, our data suggest that NONO may represent a promising and effective target for antitumor therapy of ESCC.
Recent studies have also shown that the in vitro proliferation, apoptosis and invasion of ESCC cell lines were regulated by inhibition of PI3K or MAPK (26,31). PI3K is a lipid kinase that generates second messengers involved in regulation of a wide spectrum of cellular functions including proliferation, survival and invasion (32). The effects of PI3K on tumor growth and progression are thought to be mediated mainly by Akt. The MAPK family includes the ERKs and the stress-activated protein kinases (SAPKs), p38 and JNK. Erk/MAPK signaling is frequently activated and promotes cancer cell proliferation, cell survival and metastasis (33,34). Some studies observed multiple biological aspects on NONO in tumor cells, such as NONO affecting cell proliferation, apoptosis, and cell cycle of melanomas, which is related to cx-43 (10). Besides, NONO stimulates SREBP-1a-dependent cell proliferation and tumor growth of breast cancer cells in vitro and in vivo (15). This study is the first to report that NONO silencing downregulated the expression level of p-Akt and p-Erk1/2 protein without affecting the total Akt and Erk1/2 expression level in ESCC cell lines, which suggested that NONO might play an important role in growth, apoptosis and metastasis of ESCC by activating both the PI3K/Akt and MAPK/Erk signaling pathways. However, this study was performed only at the cellular level. Further experiments to verify the influence of NONO on ESCC will be done in animal models. More comprehensive investigations are required to determine the associations between NONO, Akt, Erk and tumorigenesis.
Taken together, our results demonstrated that knockdown of NONO could inhibit proliferation and invasion, and promote apoptosis through the PI3K/Akt and MAPK/Erk signaling pathways in ESCC cancer cells. Our data may provide new evidence to improve the understanding of carcinogenesis of ESCC and may help to develop new therapeutic strategies for the treatment of ESCC.
Acknowledgements
This study was supported by Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (no. ZY201308), and Beijing Natural Science Foundation (no. 7152043).
References
- 1.Holmes RS, Vaughan TL. Epidemiology and pathogenesis of esophageal cancer. Semin Radiat Oncol. 2007;17:2–9. doi: 10.1016/j.semradonc.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Wang X, Fan JC, Wang AR, Leng Y, Li J, Bao Y, Wang Y, Yang QF, Ren Y. Epidemiology of esophageal cancer in Yanting - regional report of a national screening programme in China. Asian Pac J Cancer Prev. 2013;14:2429–2432. doi: 10.7314/APJCP.2013.14.4.2429. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 4.Shav-Tal Y, Zipori D. PSF and p54 (nrb)/NonO - multi-functional nuclear proteins. FEBS Lett. 2002;531:109–114. doi: 10.1016/S0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- 5.Patton JG, Porro EB, Galceran J, Tempst P, Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- 6.Li S, Kuhne WW, Kulharya A, Hudson FZ, Ha K, Cao Z, Dynan WS. Involvement of p54 (nrb), a PSF partner protein, in DNA double-strand break repair and radioresistance. Nucleic Acids Res. 2009;37:6746–6753. doi: 10.1093/nar/gkp741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong B, Horowitz DS, Kobayashi R, Krainer AR. Purification and cDNA cloning of HeLa cell p54nrb, a nuclear protein with two RNA recognition motifs and extensive homology to human splicing factor PSF and Drosophila NONA/BJ6. Nucleic Acids Res. 1993;21:4085–4092. doi: 10.1093/nar/21.17.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito T, Watanabe H, Yamamichi N, Kondo S, Tando T, Haraguchi T, Mizutani T, Sakurai K, Fujita S, Izumi T, et al. Brm transactivates the telomerase reverse transcriptase (TERT) gene and modulates the splicing patterns of its transcripts in concert with p54 (nrb) Biochem J. 2008;411:201–209. doi: 10.1042/BJ20071075. [DOI] [PubMed] [Google Scholar]
- 9.Barboro P, Rubagotti A, Orecchia P, Spina B, Truini M, Repaci E, Carmignani G, Romagnoli A, Introini C, Boccardo F, et al. Differential proteomic analysis of nuclear matrix in muscle-invasive bladder cancer: Potential to improve diagnosis and prognosis. Cell Oncol. 2008;30:13–26. doi: 10.1155/2008/686940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schiffner S, Zimara N, Schmid R, Bosserhoff AK. p54nrb is a new regulator of progression of malignant melanoma. Carcinogenesis. 2011;32:1176–1182. doi: 10.1093/carcin/bgr103. [DOI] [PubMed] [Google Scholar]
- 11.Ishiguro H, Uemura H, Fujinami K, Ikeda N, Ohta S, Kubota Y. 55 kDa nuclear matrix protein (nmt55) mRNA is expressed in human prostate cancer tissue and is associated with the androgen receptor. Int J Cancer. 2003;105:26–32. doi: 10.1002/ijc.11021. [DOI] [PubMed] [Google Scholar]
- 12.Kim YM, Seo J, Kim YH, Jeong J, Joo HJ, Lee DH, Koh GY, Lee KJ. Systemic analysis of tyrosine phosphorylated proteins in angiopoietin-1 induced signaling pathway of endothelial cells. J Proteome Res. 2007;6:3278–3290. doi: 10.1021/pr070168k. [DOI] [PubMed] [Google Scholar]
- 13.Clark J, Lu YJ, Sidhar SK, Parker C, Gill S, Smedley D, Hamoudi R, Linehan WM, Shipley J, Cooper CS. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene. 1997;15:2233–2239. doi: 10.1038/sj.onc.1201394. [DOI] [PubMed] [Google Scholar]
- 14.Liu PY, Erriquez D, Marshall GM, Tee AE, Polly P, Wong M, Liu B, Bell JL, Zhang XD, Milazzo G, et al. Effects of a novel long noncoding RNA, lncUSMycN, on N-Myc expression and neuroblastoma progression. J Natl Cancer Inst. 2014;106:106. doi: 10.1093/jnci/dju113. [DOI] [PubMed] [Google Scholar]
- 15.Zhu Z, Zhao X, Zhao L, Yang H, Liu L, Li J, Wu J, Yang F, Huang G, Liu J. p54 (nrb)/NONO regulates lipid metabolism and breast cancer growth through SREBP-1A. Oncogene. 2016;35:1399–1410. doi: 10.1038/onc.2015.197. [DOI] [PubMed] [Google Scholar]
- 16.Hu N, Clifford RJ, Yang HH, Wang C, Goldstein AM, Ding T, Taylor PR, Lee MP. Genome wide analysis of DNA copy number neutral loss of heterozygosity (CNNLOH) and its relation to gene expression in esophageal squamous cell carcinoma. BMC Genomics. 2010;11:576. doi: 10.1186/1471-2164-11-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su H, Hu N, Yang HH, Wang C, Takikita M, Wang QH, Giffen C, Clifford R, Hewitt SM, Shou JZ, et al. Global gene expression profiling and validation in esophageal squamous cell carcinoma and its association with clinical phenotypes. Clin Cancer Res. 2011;17:2955–2966. doi: 10.1158/1078-0432.CCR-10-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edge SBBD, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A, editors. American Joint Committee on Cancer Staging Manual. 7th editon. Springer; New York, NY: 2009. [Google Scholar]
- 19.Sobin LH, Compton CC. TNM seventh edition: what's new, what's changed: communication from the International Union Against Cancer and the American Joint Committee on Cancer. Cancer. 2010;116:5336–5339. doi: 10.1002/cncr.25537. [DOI] [PubMed] [Google Scholar]
- 20.Sun X, Qiu JJ, Zhu S, Cao B, Sun L, Li S, Li P, Zhang S, Dong S. Oncogenic features of PHF8 histone demethylase in esophageal squamous cell carcinoma. PLoS One. 2013;8:e77353. doi: 10.1371/journal.pone.0077353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meng XY, Zhu ST, Zhou QZ, Li P, Wang YJ, Zhang ST. Promoter methylation regulates cigarette smoke-stimulated cyclooxygenase-2 expression in esophageal squamous cell carcinoma. J Dig Dis. 2012;13:208–213. doi: 10.1111/j.1751-2980.2012.00578.x. [DOI] [PubMed] [Google Scholar]
- 22.Detre S, Saclani Jotti G, Dowsett M. A ‘quickscore’ method for immunohistochemical semiquantitation: Validation for oestrogen receptor in breast carcinomas. J Clin Pathol. 1995;48:876–878. doi: 10.1136/jcp.48.9.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang N, Sun X, Sun M, Zhu S, Wang L, Ma D, Wang Y, Zhang S, Li P. 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone promotes esophageal squamous cell carcinoma growth via beta-adrenoceptors in vitro and in vivo. PLoS One. 2015;10:e0118845. doi: 10.1371/journal.pone.0118845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimchi ET, Posner MC, Park JO, Darga TE, Kocherginsky M, Karrison T, Hart J, Smith KD, Mezhir JJ, Weichselbaum RR, et al. Progression of Barrett's metaplasia to adenocarcinoma is associated with the suppression of the transcriptional programs of epidermal differentiation. Cancer Res. 2005;65:3146–3154. doi: 10.1158/0008-5472.CAN-04-2490. [DOI] [PubMed] [Google Scholar]
- 25.Nishihira T, Hashimoto Y, Katayama M, Mori S, Kuroki T. Molecular and cellular features of esophageal cancer cells. J Cancer Res Clin Oncol. 1993;119:441–449. doi: 10.1007/BF01215923. [DOI] [PubMed] [Google Scholar]
- 26.Liu F, Zheng S, Liu T, Liu Q, Liang M, Li X, Sheyhidin I, Lu X, Liu W. MicroRNA-21 promotes the proliferation and inhibits apoptosis in Eca109 via activating ERK1/2/MAPK pathway. Mol Cell Biochem. 2013;381:115–125. doi: 10.1007/s11010-013-1693-8. [DOI] [PubMed] [Google Scholar]
- 27.Grille SJ, Bellacosa A, Upson J, Klein-Szanto AJ, van Roy F, Lee-Kwon W, Donowitz M, Tsichlis PN, Larue L. The protein kinase Akt induces epithelial mesenchymal transition and promotes enhanced motility and invasiveness of squamous cell carcinoma lines. Cancer Res. 2003;63:2172–2178. [PubMed] [Google Scholar]
- 28.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 29.Basu A, Dong B, Krainer AR, Howe CC. The intracisternal A-particle proximal enhancer-binding protein activates transcription and is identical to the RNA- and DNA-binding protein p54nrb/NonO. Mol Cell Biol. 1997;17:677–686. doi: 10.1128/MCB.17.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bond CS, Fox AH. Paraspeckles: Nuclear bodies built on long noncoding RNA. J Cell Biol. 2009;186:637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li B, Tsao SW, Li YY, Wang X, Ling MT, Wong YC, He QY, Cheung AL. Id-1 promotes tumorigenicity and metastasis of human esophageal cancer cells through activation of PI3K/AKT signaling pathway. Int J Cancer. 2009;125:2576–2585. doi: 10.1002/ijc.24675. [DOI] [PubMed] [Google Scholar]
- 32.Vara Fresno JA, Casado E, de Castro J, Cejas P, Belda-Iniesta C, González-Barón M. PI3K/Akt signalling pathway and cancer. Cancer Treat Rev. 2004;30:193–204. doi: 10.1016/j.ctrv.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 33.Wada T, Penninger JM. Mitogen-activated protein kinases in apoptosis regulation. Oncogene. 2004;23:2838–2849. doi: 10.1038/sj.onc.1207556. [DOI] [PubMed] [Google Scholar]
- 34.Chan-Hui PY, Weaver R. Human mitogen-activated protein kinase kinase kinase mediates the stress-induced activation of mitogen-activated protein kinase cascades. Biochem J. 1998;336:599–609. doi: 10.1042/bj3360599. [DOI] [PMC free article] [PubMed] [Google Scholar]