Abstract
Serous ovarian cancer (SOC) accounts for >50% of all epithelial ovarian cancers. However, patients with SOC present with various degrees of response to platinum-based chemotherapy and, thus, their survival may differ. The present study aimed to identify the candidate genes involved in the carcinogenesis and drug resistance of SOC by analyzing the microarray datasets GDS1381 and GDS3592. GDS1381 and GDS3592 were downloaded from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/gds/). A total of 219 differentially expressed genes (DEGs) were identified. Potential genes that may predict the response to carboplatin and, thus, the prognosis of SOC were analyzed. The enriched functions and pathways of DEGs included extracellular region, extracellular space and extracellular exosome, among others. Upon screening the upregulated and downregulated genes on the connectivity map, 10 small-molecule drugs were identified that may be helpful in improving drug sensitivity in patients with ovarian cancer. A total of 30 hub genes were screened for further analysis after constructing the protein-to-protein interaction network. Through survival analysis, comparison of genes across numerous analyses, and immunohistochemistry, GNAI1, non-structural maintenance of chromosomes (non-SMC) condensin I complex subunit H (NCAPH), matrix metallopeptidase 9 (MMP9), aurora kinase A (AURKA) and enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) were identified as the key molecules that may be involved in the carcinogenesis and carboplatin resistance of SOC. In conclusion, GNAI1, NCAPH, MMP9, AURKA and EZH2 should be examined in further studies for the possibility of their participation in the carcinogenesis and carboplatin response of SOC.
Keywords: differentially expressed genes, drug resistance reversal agents, platinum chemoresistance, potential targets, serous ovarian cancer
Introduction
Ovarian cancer is the most lethal gynecological cancer (1). Among all ovarian cancer cases, the histological type in >50% of cases is serous adenocarcinoma. Although patients with serous ovarian cancer (SOC) usually exhibit a fairly good response to paclitaxel and carboplatin at diagnosis, the majority of the patients relapse over time (2). The 5-year survival rate of SOC patients has remained extremely low for decades due to the high rate of delayed diagnosis, bulky residual foci following primary surgery and secondary chemoresistance (2). Recent studies on the use of poly (ADP-ribose) polymerase (PARP) inhibitors indicated their utility in the treatment of ovarian cancer, due to most patients carrying a pathogenic mutation of BRCA1/2 (3–5). However, these patients accounted for only a small proportion of all SOC cases. Thus, a profound understanding of the molecular mechanisms underlying the progression of ovarian cancer may uncover new pathways with high clinical relevance.
Cisplatin (CDDP) and carboplatin have been the main drugs used for the therapy of ovarian cancer for decades. Unfortunately, ovarian cancer cells, with their unstable genomes, are initially sensitive to these drugs, but invariably become resistant (6). Thus, novel therapeutic strategies are urgently needed (7). Investigating and identifying the genetic aberrations that participate in the basic molecular mechanisms of cancer progression and drug resistance in ovarian cancer may provide innovative therapeutic choices with novel agents (8).
The identification of novel biomarkers is currently a promising approach to designing new diagnostic and therapeutic strategies (9). Various technologies have been adopted to identify biomarkers. Furthermore, the use of bioinformatics is growing rapidly in cancer biology (6). Therefore, the present study used two gene chips downloaded from the meta-database Gene Expression Omnibus (GEO). GDS3592 was designed for comparing normal ovarian tissue and ovarian cancer and GDS1381 for comparing carboplatin-resistant and carboplatin-sensitive ovarian cancers. The present study was designed to identify differentially expressed genes (DEGs) that may affect ovarian cancer development and platinum sensitivity.
Materials and methods
Data collection
The GEO database is a public functional genomics dataset that helps users download experiments and create gene expression profiles. In the present study, the key words ‘ovarian cancer’ and ‘carboplatin resistance’ were searched in the GEO datasets (http://www.ncbi.nlm.nih.gov/geo/). After filtering out repeated experiments, relevant studies were reviewed to screen the comparison data of control and treated groups. The expression matrix or CEL file was downloaded for subsequent analysis.
Data pretreatment and identification of DEGs
All data were processed using the R software (software for statistical computing and graphics) (https://www.r-project.org/). The logarithm (base 2) of the expression value was considered for the microarray expression data. The Affy package (http://www.bioconductor.org/packages/release/bioc/html/affy.html) was used to read the expression data for CEL files. The limma package (http://www.bioconductor.org/packages/release/bioc/html/limma.html) for standardization was used for experimental data. After the limma analysis, the genes were sorted according to the log fold-change values. Subsequently, the RANK analysis (adjusted P-value <0.05 using the Bonferroni correction method) was conducted. The null hypothesis of the rank method was that each gene was randomly sorted in every experiment; the smaller the P-value of the gene, the higher ranking it would have. Secondly, the pheatmap package was used to draw the top 20 genes.
Gene Ontology (GO) analysis
The Gene Ontology Consortium aimed to produce a dynamic, controlled vocabulary of gene roles in cells (10). The GO enrichment analysis was performed using the DAVID database (https://david.ncifcrf.gov/). The F-value of the GO terms of DEGs and the FDR value of the P-value (Q-value) were calculated through statistical analysis of GO terms, positioning the most possible GO terms of DEGs. Subsequently, the GOplot R package was used to draw the chord plot.
Kyoto encyclopedia of genes and genomes (KEGG) analysis
KEGG is a database resource for understanding the interactions between molecular biology and the underlying chemical elements, allowing for analysis of high-level functions and uses of the biological system (11) (http://www.kegg.jp/). The KOBAS online tool (http://kobas.cbi.pku.edu.cn/) was used for gene annotation and KEGG pathway analyses. A P-value of <0.05 was considered to indicate statistically significance differences. Subsequently, the clusterProfiler R package was used to draw the barplot and dotplot.
Protein-protein interaction (PPI) network
PPIs are the physical contacts among protein molecules. Exploring the interactions between proteins is crucial to achieve a detailed description of their mechanisms and functions in living organisms (12). Currently, PPIs are annotated at a number of online resources, including the STRING database (http://string-db.org/), providing a comprehensive perspective for several organisms. The PPI network of DEGs was constructed using the STRING database (https://string-db.org/). Cytoscape (version 3.6.0) is a bioinformatics software platform for visualizing molecular interaction networks (13). The PPI networks were drawn by Cytoscape.
Connectivity map (CMAP) analysis
CMAP is a gene expression database based at the Broad Institute of MIT and Harvard in Cambridge (MA, USA). It currently contains data on >1,000 small molecules. CMAP uses the gene expression differences after treating human cells with small molecules to establish a biological application database that links small molecule drugs, gene expression and disease (14). This database suggests gene, disease and drug relevance, thereby helping in drug research and development. DEGs were uploaded to the CMAP to identify the relevance between genes and small-molecule drugs.
Kaplan-Meier survival analysis
The Kaplan-Meier estimator, a non-parametric statistic, is used to estimate the survival function from lifelong data. In medical research, it is used to measure a certain amount of time after patient treatment (15). Each hub gene was entered into the online tool (http://kmplot.com/analysis/) to assess the overall and progression-free survival of patients with ovarian cancer for the Kaplan-Meier curve. This tool was built using gene expression and survival data of 1,287 patients with ovarian cancer, which were downloaded from GEO and The Cancer Genome Atlas (Affymetrix HG-U133A, HG-U133A 2.0 and HG-U133 plus 2.0 microarrays) (16). As every gene mapped to a different Affy ID, each Affy ID was entered into the online tool to obtain the survival curves. One was selected for the experiment, while others were presented as supplementary data.
Immunohistochemistry (IHC) analysis
An online tool, ‘Human Tissue Proteomes’ (http://www.proteinatlas.org/), was used to assess the gene expression in the ovaries of patients with ovarian cancer. The ‘Human Tissue Proteomes’ is a database that can provide protein expression of all protein-coding genes in all major tissues and organs of the human body (17).
Results
Characteristics of the datasets
GDS1381 and GDS3592 were obtained from the GEO database. GDS1381 and GDS3592 were based on the GPL8300 and GPL570 platforms, respectively. GDS1381 included 9 carboplatin-sensitive and 9 carboplatin-resistant ovarian cancer cells from 6 cases with carboplatin-sensitive (n=3) and carboplatin-resistant (n=3) ovarian cancers, respectively, each of which included the primary ovarian cancer cells and metastatic ovarian cancer cells to the liver and omentum, respectively. Cancer cells were prepared from primary cultures. The results of GDS1381 provided insight into potential markers associated with carboplatin-resistant ovarian cancer. GDS3592 contained 12 normal ovarian surface epithelial (OSE) cells and 12 ovarian cancer epithelial cells (CEPIs). CEPIs were isolated from serous papillary ovarian adenocarcinomas (Table I).
Table I.
Characteristics of the two datasets of the present analysis.
Identification of DEGs
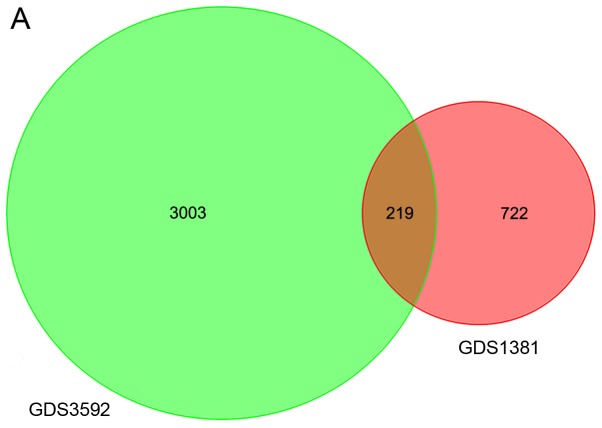
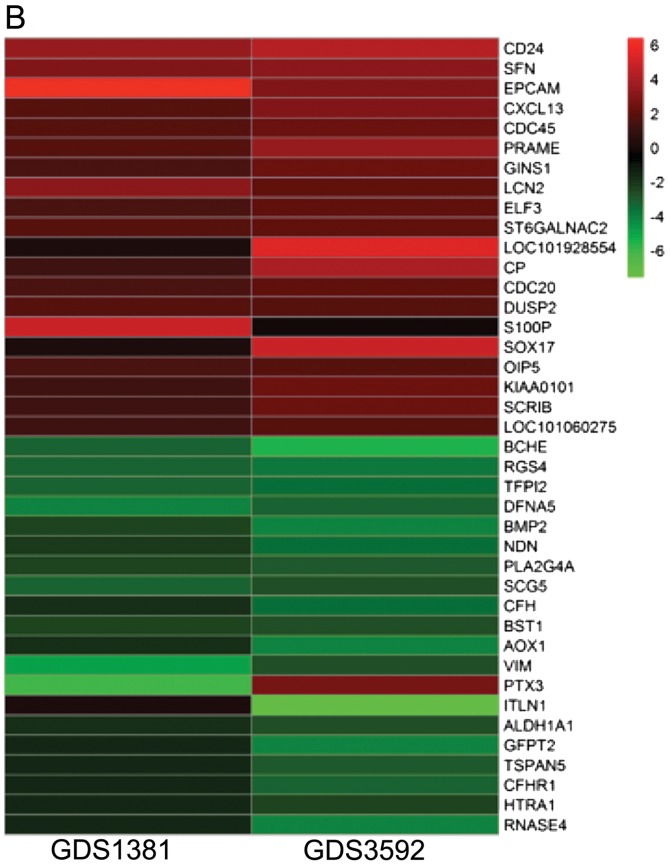
After normalization, 722 DEGs in GDS1381 and 3,003 DEGs in GDS3592 were identified. The intersection of the two datasets contained 219 genes (Fig. 1A). The heat map for DEG expression (top 15 upregulated and downregulated genes) is displayed in Fig. 1B.
Figure 1.
Venn diagram and heat map. (A) Venn diagram depicts the DEGs of GEO chips relatively and the overlapping genes. DEGs were selected through fold change >2.0 and P-value <0.05 within both GEO chips by R software. The number on the image shows the quantity of DEGs of each and overlap. (B) Heat map representation of top 40 DEGs (20 upregulated and 20 downregulated). Red modules denote upregulation, green modules denote downregulation. DEGs, differentially expressed genes; GEO, Gene Expression Omnibus.
GO analysis of DEGs
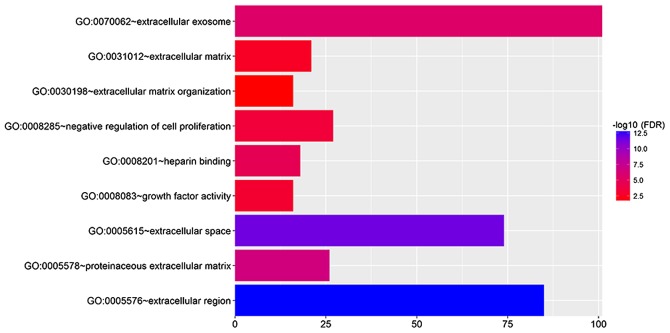
DEGs were imported into the online software DAVID to identify GO categories. According to the result, these DEGs were found to be mainly enriched in the extracellular region, extracellular space, extracellular exosome, proteinaceous extracellular matrix, heparin binding, negative regulation of cell proliferation, growth factor activity, extracellular matrix and extracellular matrix organization (Fig. 2).
Figure 2.
GO analysis of DEGs. Each gene and their correspondent GO terms are linked by different color lines on the image. GO, Gene Ontology; DEGs, differentially expressed genes.
KEGG pathways of DEGs
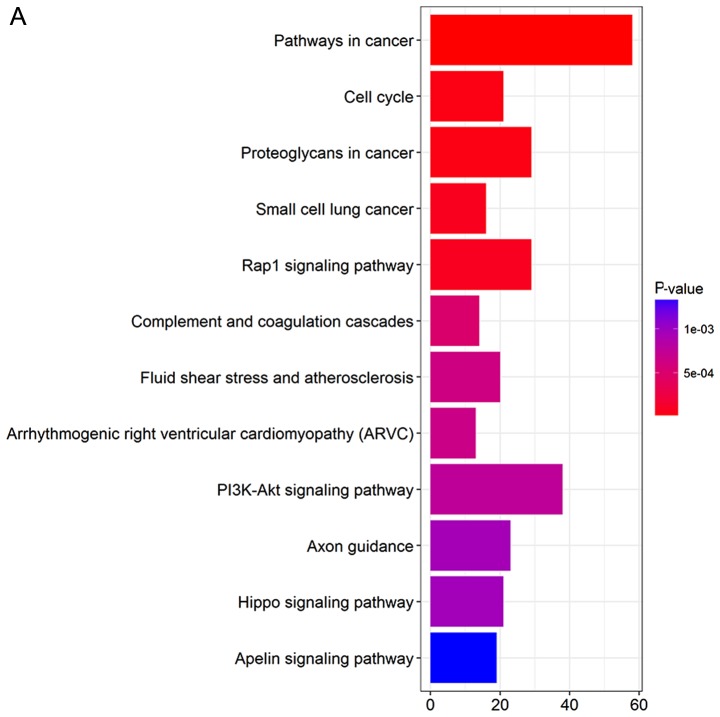
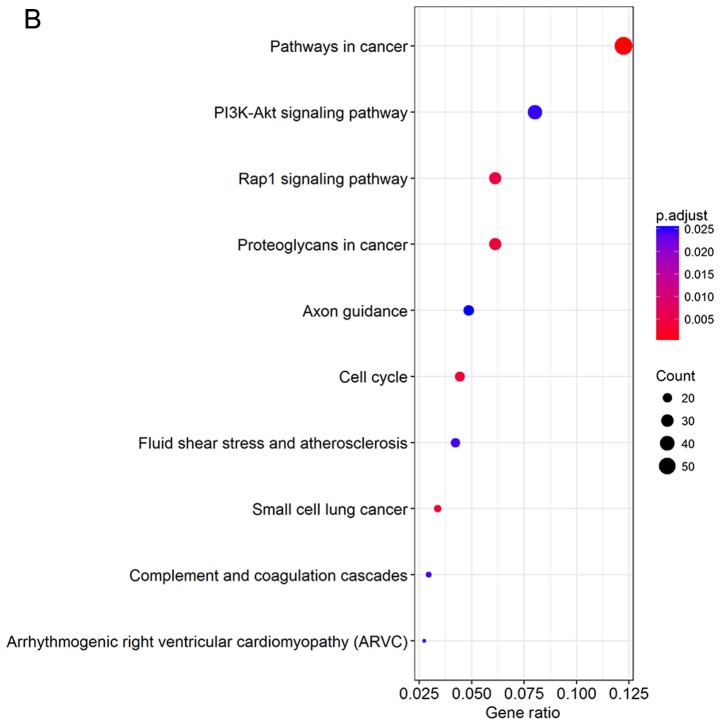
DEGs were uploaded into the KEGG database to conduct the KEGG analysis. The significantly enriched KEGG terms were pathways in cancer, phosphatidylinositol 3′-kinase (PI3K)-Akt signaling pathway, Rap1 signaling pathway, proteoglycans in cancer, axon guidance, cell cycle, fluid shear stress and atherosclerosis, small-cell lung cancer, complement and coagulation cascades, and arrhythmogenic right ventricular cardiomyopathy (Fig. 3).
Figure 3.
KEGG pathway analysis of DEGs. (A) Barplot and (B) dotplot were performed by R software. KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, differentially expressed genes.
PPI network of DEGs
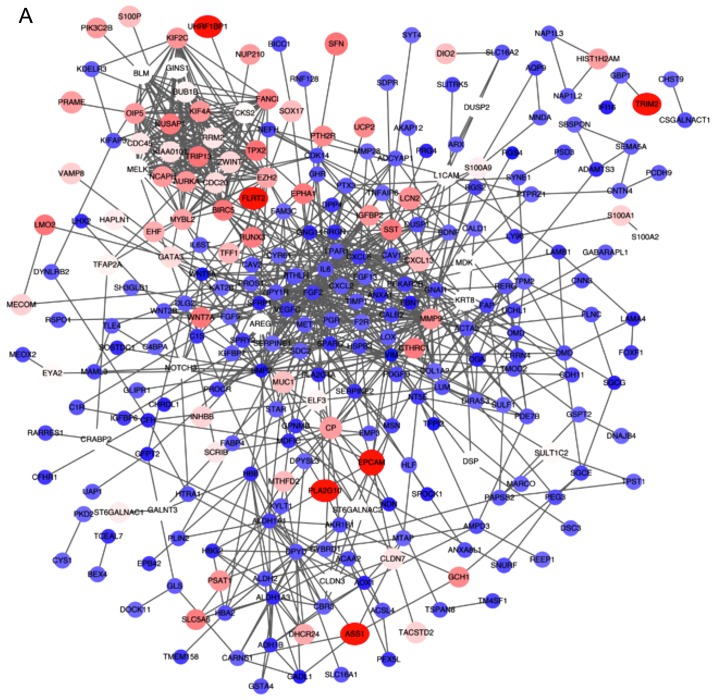
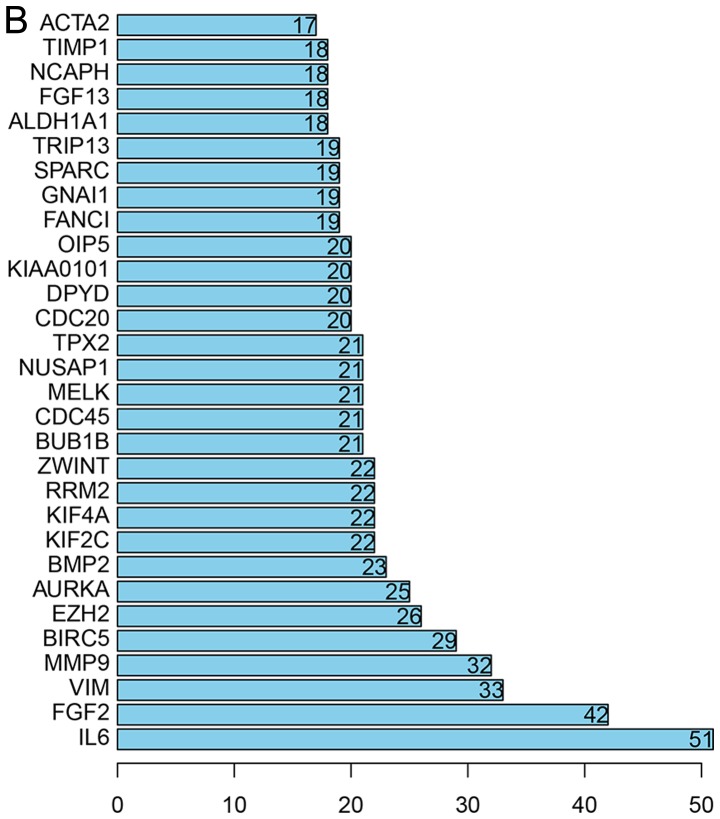
All the DEGs were screened using the STRING database to further investigate the property of these distinct genes and the internal link of DEGs. The PPI network of DEGs was constructed by Cytoscape (Fig. 4A). The number of strings for every single gene was counted to identify the hub genes. The top 30 genes are displayed in the bar chart in Fig. 4B, which helped identify the most important genes among DEGs.
Figure 4.
PPI network of DEGs. (A) The gene co-expression network of DEGs. (B) Top 30 genes with most connection nodes. DEGs, differentially expressed genes.
Related small-molecule drugs
The CMAP is a web-based tool collecting genome-wide transcriptional expression data that provides insight into functional connections between drugs, genes and diseases through the transitory feature of common gene expression changes. Through comparing microarray data from 1,300 small molecules with selected genes in a pattern-matching algorithm, those compounds with a gene expression pattern highly correlated with the selected genes would be screened, with the aim to identify a novel, potentially effective treatment for a disease (7). The present study screened the upregulated and downregulated genes on the CMAP and found 10 related small-molecule drugs. These small molecules may prove to be beneficial in terms of drug sensitivity in patients with ovarian cancer (Table II).
Table II.
Top 10 relevant small-molecule drugs.
| Rank | Cmap name | Mean | n | Enrichment | P-value | Specificity | Percent non-null |
|---|---|---|---|---|---|---|---|
| 1 | Thioridazine | −0.48 | 20 | −0.555 | 0 | 0.0645 | 90 |
| 2 | Tanespimycin | −0.576 | 62 | −0.486 | 0 | 0.0672 | 87 |
| 3 | Trichostatin A | −0.5 | 182 | −0.468 | 0 | 0.2475 | 87 |
| 4 | LY-294002 | −0.344 | 61 | −0.301 | 0.00004 | 0.3436 | 67 |
| 5 | Etiocholanolone | 0.452 | 6 | 0.817 | 0.00008 | 0 | 100 |
| 6 | Thioperamide | 0.489 | 5 | 0.83 | 0.0003 | 0 | 100 |
| 7 | Geldanamycin | −0.568 | 15 | −0.512 | 0.00034 | 0.0859 | 86 |
| 8 | Carbenoxolone | 0.457 | 4 | 0.87 | 0.0004 | 0 | 100 |
| 9 | Cloxacillin | 0.538 | 4 | 0.856 | 0.00056 | 0 | 100 |
| 10 | 8-Azaguanine | −0.7 | 4 | −0.857 | 0.00072 | 0.007 | 100 |
Upregulated and downregulated genes were screened on the Cmap and 10 related small-molecule drugs were found. Cmap, connectivity map.
Comparison of hub genes across numerous analyses
An Oncomine analysis of cancer vs. normal tissue of hub genes in ovarian cancer was performed. The results revealed that GANI1, non-structural maintenance of chromosomes (non-SMC) condensin I complex subunit H (NCAPH), fibroblast growth factor 2 (FGF2), matrix metallopeptidase 9 (MMP9), aurora kinase A (AURKA), enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) and BUB1 mitotic checkpoint serine/threonine kinase B (BUB1B) were upregulated among different analysis datasets (Fig. 5).
Figure 5.
Gene expression within ovarian cancer across multiple datasets.
Kaplan-Meier survival analysis
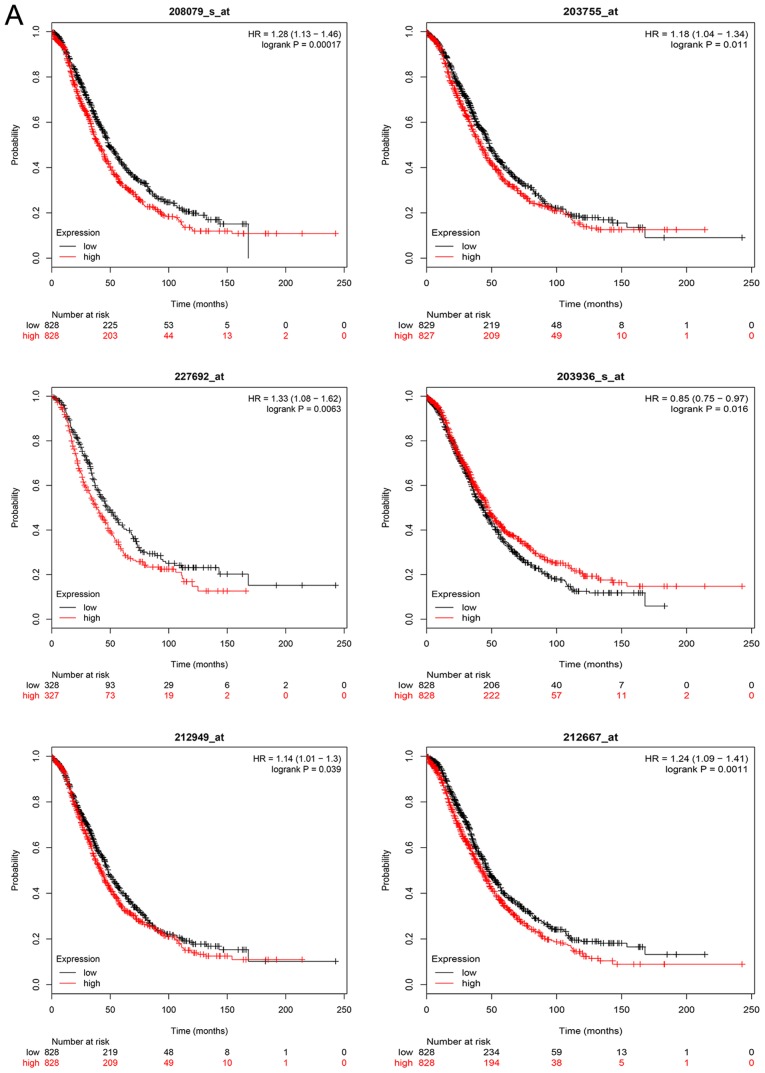
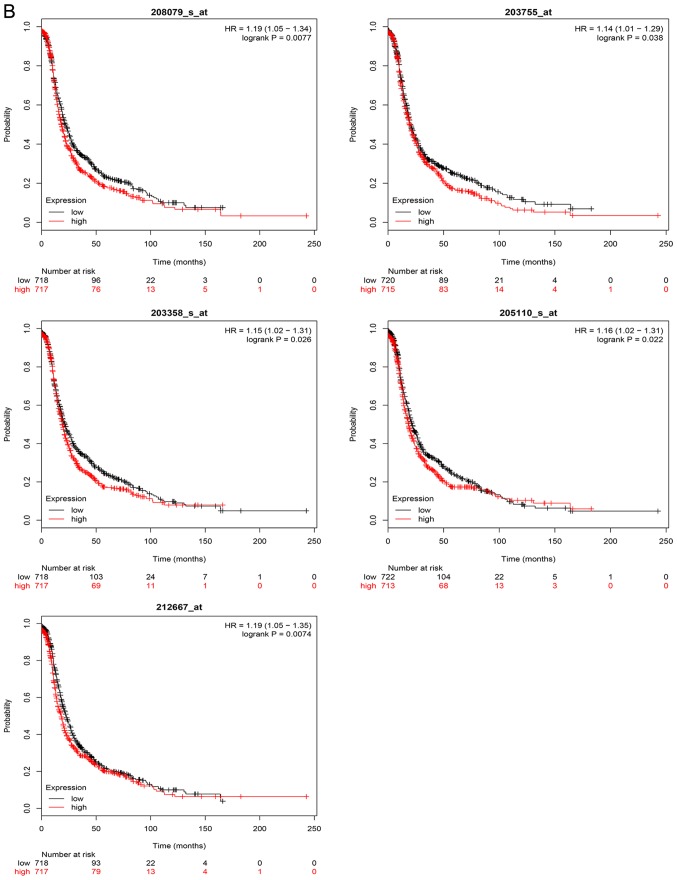
The overall survival analysis of hub genes was performed using the Kaplan-Meier curve. The results demonstrated that high expression levels of GNAI1, SPARC, NCAPH, MMP9, FGF2, AURKA, EZH2 and BUB1B were associated with worse survival of ovarian cancer patients. The overall and progression-free survival analyses of hub genes are displayed in Fig. 6A and B, respectively.
Figure 6.
Kaplan Meier survival curve of hub genes. (A) Overall survival analyses. (B) Progression-free survival analyses were performed using Kaplan Meier-plotter.
IHC
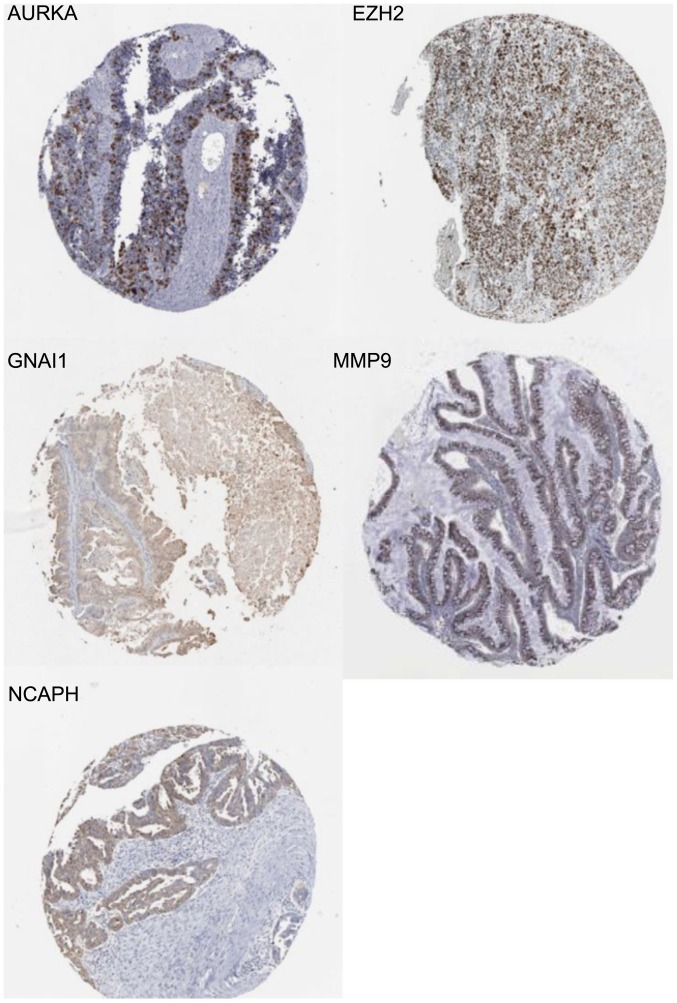
An online website (http://www.proteinatlas.org/) was used to search for the IHC results. Immunostaining of certain hub genes yielded positive results in ovarian cancer tissue. The staining intensity of GNAI1 in 12 patients with ovarian cancer included 3 weak and 9 negative staining cases using antibody CAB022449 (antibody dilution:1:1,000) (18), 1 low and 10 negative staining cases using antibody HPA042141 (antibody dilution:1:75) (18,19). The staining intensity of NCAPH in 12 ovarian cancer samples included 4 moderate, 3 weak and 5 negative staining cases using antibody HPA003008 (antibody dilution:1:150) (18), whereas 3 moderate, 3 weak and 5 negative staining cases in 11 ovarian cancer samples using antibody HPA002647 (antibody dilution: 1:100) (18,20). The staining intensity of MMP9 in 12 patients with ovarian cancer included 3 strong and 9 negative staining cases using antibody CAB000348 (antibody dilution:1:1,000) (18), whereas all were negative staining using other antibodies (21). The staining intensity of AURKA in 24 ovarian cancer samples included 1 strong, 1 moderate, 3 weak and 7 negative staining cases using antibody CAB001454 (antibody dilution:1:25) (18), 1 moderate, 3 weak and 8 negative staining cases using antibody HPA002636 (antibody dilution:1:100) (18,22). The staining intensity of EZH2 in 10 ovarian cancer samples included 4 strong, 5 moderate and 1 weak staining cases using antibody CAB009589 (antibody dilution:1:100) (18,23). IHC analysis further demonstrated the unknown connection between hub genes and ovarian cancer. An illustration of the weak staining of GNAI1 (19), strong staining of EZH2 (23), moderate staining of NCAPH (20), strong staining of MMP9 (21) and strong staining of AURKA (22) are displayed in Fig. 7.
Figure 7.
Representative images of immunostaining of certain hub genes. Immunohistochemistry was used to identify gene expression in ovarian cancer. Typical examples were selected to show. The case displays positive staining for GANI1, NCAPH, MMP9, AURKA and EZH2.
Discussion
Microarray analysis of gene expression profiles is widely used to identify genes and biological pathways associated with multifactorial diseases. However, such studies on ovarian cancer have been scarce to date. Thus, in the present study, integrated analyses were performed and 219 DEGs common between OSE and CEPIs, as well as genes common between carboplatin-sensitive and carboplatin-resistant ovarian cancer cells, were detected to identify the key platinum-resistant genes in SOC, which is the most common type of epithelial ovarian cancer. The results provided evidence that the 219 DEGs were probably involved in cancer development and the molecular mechanism underlying carboplatin resistance, which were enriched in the PI3K-Akt signaling pathway, Rap1 signaling pathway, proteoglycans in cancer, axon guidance, cell cycle, fluid shear stress and atherosclerosis, small-cell lung cancer, and complement and coagulation cascades.
Overall, 30 DEGs were identified as hub genes with high node degrees. Among these hub genes, interleukin (IL)6 exhibited the highest node degree with 51. IL6 belongs to the IL6 cytokine family and was recently reported to promote lung and pancreatic tumorigenesis (24–26). Systemic and pulmonary production of IL6 was commonly elevated in patients with lung adenocarcinoma and was correlated with poor survival (27–30). A mouse model revealed specific targeting of IL6 trans-signaling, which suppressed the pathogenesis of lung adenocarcinoma (31). Subsequently, the screened genes that may be closely associated with ovarian cancer were further analyzed. The present study identified five genes, namely GANI1, NCAPH, MMP9, AURKA and EZH2, that were closely associated with ovarian cancer. NCAPH encodes a member of the Barr gene and a regulatory subunit of the condensin complex that contributes to the conversion of interphase chromatin into condensed chromosomes (32). NCAPH is one of the three non-SMC subunits in condensin I that is a superfamily of proteins termed kleisins (16) and it was found to be crucial for condensin complex stability and resolution of sister chromatids. It promotes colonic cancerous cell proliferation and migration, affects cell cycle transition, and inhibits cellular apoptosis. Knockdown of NCAPH decreased the xenograft tumor growth of HCT116 in vivo (33). MMP9 is one of the members of the MMP family that participates in the breakdown of extracellular matrix in normal physiological processes. The majority of MMPs are converted to their active forms from inactive protein precursors after being cleaved by extracellular proteinases (32). MMP9 was found to be involved in the metastasis of lung cancer, hepatocellular carcinoma (HCC), prostate and breast cancer (34–37). MMP9 was also found to be involved in cancer progression and was identified in patients with ovarian cancer. Therefore, MMP9 may be a candidate biomarker for high-grade ovarian cancer (38). AURKA, also referred to as aurora kinase A, is a cell cycle-regulated kinase that participates in microtubule formation and stabilization at the spindle pole during chromosome segregation (32). Aurora A mapped to chromosomal region 20q13.2, which was amplified in a number of cancer cell lines and primary tumors (39–41). Previous studies demonstrated that AURKA was involved in the susceptibility to hepatitis B virus-related HCC and the progression of head and neck and gastric cancer. High expression of AURKA promoted cisplatin resistance by activating p-eIF4E, c-MYC and HDM2 (42,43). AURKA was found to be amplified in >15–25% of ovarian cancer cell lines and primary tumors (40,41). The expression of BRCA1/2 may be increased by AURKA in ovarian carcinoma cells (44), and several studies revealed a physical and functional association between AURKA and BRCA1/2 (45–47). Clinical data demonstrated that patients with BRCA1 and BRCA2 mutations exhibited a higher response rate to cisplatin (48,49). Thus, AURKA was hypothesized to exert a synergistic effect with BRCA1/2 in platinum resistance. EZH2 encoded one of the polycomb-group family members. The protein EZH2 encoding was involved in the hematopoietic and central nervous systems (32). Mutation or overexpression of EZH2 was reported to be associated with a wide variety of cancers. As in ovarian cancer, an increased expression of EZH2 was reported to promote cancer cell metastasis and migration (50,51), whereas inhibitors of EZH2 were assessed in clinical trials as potential therapeutic targets (52).
GANI1 and the interaction between ovarian cancer and NCAPH have not been widely reported and, hence, need further investigation.
The present study integrated two gene expression datasets to lower the false-positive rate of the single microarray analysis. This process also stabilized the effects of the study with a small sample size. Finally, five DEGs were identified, namely GNAI1, NCAPH, MMP9, AURKA and EZH2, which may be considered as potential novel targets for ovarian cancer therapy and drug resistance reversal agents. However, further experiments in vivo and in vitro are needed to explicate the biological function of these genes and the complex molecular processes underlying the platinum resistance of ovarian cancer require further investigation.
Acknowledgements
The authors thank all the data of all the websites involved.
Funding
The present study was supported by grants from the National Natural Science Foundation of China (nos. C81072120 and C81572562).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
Shijie Zhan conceived and designed the study and wrote the paper. Hua Linghu and Bin Liu reviewed and edited the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflicts of interest.
References
- 1. [International Agency for Research on Cancer];http://globocan.iarc.fr/Pages/fact_sheets_population.aspx Global cancer statistics. [Google Scholar]
- 2.De Las Rivas J, Fontanillo C. Protein-protein interactions essentials: Key concepts to building and analyzing interactome networks. PLoS Comput Biol. 2010;6:e1000807. doi: 10.1371/journal.pcbi.1000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 4.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 5.Wiggans AJ, Cass GK, Bryant A, Lawrie TA, Morrison J. Poly(ADP-ribose) polymerase (PARP) inhibitors for the treatment of ovarian cancer. Cochrane Database Syst Rev: CD007929. 2015 doi: 10.1002/14651858.CD007929.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olopade OI, Wei M. FANCF methylation contributes to chemoselectivity in ovarian cancer. Cancer Cell. 2003;3:417–420. doi: 10.1016/S1535-6108(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 7.Malati T. Tumour markers: An overview. Indian J Clin Biochem. 2007;22:17–31. doi: 10.1007/BF02913308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurman RJ, Shih Ie M. Pathogenesis of ovarian cancer: Lessons from morphology and molecular biology and their clinical implications. Int J Gynecol Pathol. 2008;27:151–160. doi: 10.1097/PGP.0b013e318161e4f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee E, Moon A. Identification of biomarkers for breast cancer using databases. J Cancer Prev. 2016;21:235–242. doi: 10.15430/JCP.2016.21.4.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin CM, Feng W. Microarray and synchronization of neuronal differentiation with pathway changes in the Kyoto Encyclopedia of Genes and Genomes (KEGG) databank in nerve growth factor-treated PC12 cells. Curr Neurovasc Res. 2012;9:222–229. doi: 10.2174/156720212801618983. [DOI] [PubMed] [Google Scholar]
- 12.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.https://portals.broadinstitute.org/cmap Connectivity Map. [Google Scholar]
- 15.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Statist Assoc. 1958;53:457–481. doi: 10.1080/01621459.1958.10501452. [DOI] [Google Scholar]
- 16.Gyorffy B, Lánczky A, Szállási Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 17.https://www.proteinatlas.org/about The Human Protein Atlas. [Google Scholar]
- 18.https://www.proteinatlas.org/about/releases Protein Atlas version 18. [Google Scholar]
- 19.https://www.proteinatlas.org/ENSG00000127955-GNAI1/pathology/tissue/ovarian+cancer GNAI1/pathology/tissue/ovarian+cancer. [Google Scholar]
- 20.https://www.proteinatlas.org/ENSG00000121152-NCAPH/pathology/tissue/ovarian+cancer NCAPH/pathology/tissue/ovarian+cancer. [Google Scholar]
- 21.https://www.proteinatlas.org/ENSG00000100985-MMP9/pathology/tissue/ovarian+cancer MMP9/pathology/tissue/ovarian+cancer. [Google Scholar]
- 22.https://www.proteinatlas.org/ENSG00000087586-AURKA/pathology/tissue/ovarian+cancer AURKA/pathology/tissue/ovarian+cancer. [Google Scholar]
- 23.https://www.proteinatlas.org/ENSG00000106462-EZH2/pathology/tissue/ovarian+cancer EZH2/pathology/tissue/ovarian+cancer. [Google Scholar]
- 24.Ochoa CE, Mirabolfathinejad SG, Ruiz VA, Evans SE, Gagea M, Evans CM, Dickey BF, Moghaddam SJ. Interleukin 6, but not T helper 2 cytokines, promotes lung carcinogenesis. Cancer Prev Res. 2011;4:51–64. doi: 10.1158/1940-6207.CAPR-10-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lesina M, Kurkowski MU, Ludes K, Rose-John S, Treiber M, Klöppel G, Yoshimura A, Reindl W, Sipos B, Akira S, et al. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell. 2011;19:456–469. doi: 10.1016/j.ccr.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 26.Tan X, Carretero J, Chen Z, Zhang J, Wang Y, Chen J, Li X, Ye H, Tang C, Cheng X, et al. Loss of p53 attenuates the contribution of IL-6 deletion on suppressed tumor progression and extended survival in Kras-driven murine lung cancer. PLoS One. 2013;8:e80885. doi: 10.1371/journal.pone.0080885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yanagawa H, Sone S, Takahashi Y, Haku T, Yano S, Shinohara T, Ogura T. Serum levels of interleukin 6 in patients with lung cancer. Br J Cancer. 1995;71:1095–1098. doi: 10.1038/bjc.1995.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeh HH, Lai WW, Chen HH, Liu HS, Su WC. Autocrine IL-6-induced Stat3 activation contributes to the pathogenesis of lung adenocarcinoma and malignant pleural effusion. Oncogene. 2006;25:4300–4309. doi: 10.1038/sj.onc.1209464. [DOI] [PubMed] [Google Scholar]
- 29.Gao SP, Mark KG, Leslie K, Pao W, Motoi N, Gerald WL, Travis WD, Bornmann W, Veach D, Clarkson B, Bromberg JF. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117:3846–3856. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haura EB, Livingston S, Coppola D. Autocrine interleukin-6/interleukin-6 receptor stimulation in non-small-cell lung cancer. Clin Lung Cancer. 2006;7:273–275. doi: 10.3816/CLC.2006.n.006. [DOI] [PubMed] [Google Scholar]
- 31.Brooks GD, McLeod L, Alhayyani S, Miller A, Russell PA, Ferlin W, Rose-John S, Ruwanpura S, Jenkins BJ. IL6 trans-signaling promotes KRAS-driven lung carcinogenesis. Cancer Res. 2016;76:866–876. doi: 10.1158/0008-5472.CAN-15-2388. [DOI] [PubMed] [Google Scholar]
- 32.https://www.ncbi.nlm.nih.gov/gene/ GeneSupport Center of NCBI. [Google Scholar]
- 33.Neuwald AF, Hirano T. HEAT repeats associated with condensins, cohesins, and other complexes involved in chromosome-related functions. Genome Res. 2000;10:1445–1452. doi: 10.1101/gr.147400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J, Wu Y, Guo J, Fei X, Yu L, Ma S. Adipocyte-derived exosomes promote lung cancer metastasis by increasing MMP9 activity via transferring MMP3 to lung cancer cells. Oncotarget. 2017;8:81880–81891. doi: 10.18632/oncotarget.18737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Q, Yin D, Zhang Y, Yu L, Li XD, Zhou ZJ, Zhou SL, Gao DM, Hu J, Jin C, et al. MicroRNA-29a induces loss of 5-hydroxymethylcytosine and promotes metastasis of hepatocellular carcinoma through a TET-SOCS1-MMP9 signaling axis. Cell Death Dis. 2017;8:e2906. doi: 10.1038/cddis.2017.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie H, Li L, Zhu G, Dang Q, Ma Z, He D, Chang L, Song W, Chang HC, Krolewski JJ, et al. Correction: Infiltrated pre-adipocytes increase prostate cancer metastasis via modulation of the miR-301a/androgen receptor (AR)/TGF-β1/Smad/MMP9 signals. Oncotarget. 2016;7:83829–83830. doi: 10.18632/oncotarget.13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Limoge M, Safina A, Beattie A, Kapus L, Truskinovsky AM, Bakin AV. Tumor-fibroblast interactions stimulate tumor vascularization by enhancing cytokine-driven production of MMP9 by tumor cells. Oncotarget. 2017;8:35592–35608. doi: 10.18632/oncotarget.16022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reiner AT, Tan S, Agreiter C, Auer K, Bachmayr-Heyda A, Aust S, Pecha N, Mandorfer M, Pils D, Brisson AR, et al. EV-associated MMP9 in high-grade serous ovarian cancer is preferentially localized to Annexin V-Binding EVs. Dis Markers. 2017;2017:9653194. doi: 10.1155/2017/9653194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, Schryver B, Flanagan P, Clairvoyant F, Ginther C, et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, Brinkley BR, Sen S. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 41.Tanner MM, Grenman S, Koul A, Johannsson O, Meltzer P, Pejovic T, Borg A, Isola JJ. Frequent amplification of chromosomal region 20q12-q13 in ovarian cancer. Clin Cancer Res. 2000;6:1833–1839. [PubMed] [Google Scholar]
- 42.Bao Z, Lu L, Liu X, Guo B, Zhai Y, Li Y, Wang Y, Xie B, Ren Q, Cao P, et al. Association between the functional polymorphism Ile31Phe in the AURKA gene and susceptibility of hepatocellular carcinoma in chronic hepatitis B virus carriers. Oncotarget. 2017;8:54904–54912. doi: 10.18632/oncotarget.18613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L, Arras J, Katsha A, Hamdan S, Belkhiri A, Ecsedy J, El-Rifai W. Cisplatin-resistant cancer cells are sensitive to Aurora kinase A inhibition by alisertib. Mol Oncol. 2017;11:981–995. doi: 10.1002/1878-0261.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Do TV, Hirst J, Hyter S, Roby KF, Godwin AK. Aurora A kinase regulates non-homologous end-joining and poly(ADP-ribose) polymerase function in ovarian carcinoma cells. Oncotarget. 2017;8:50376–50392. doi: 10.18632/oncotarget.18970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang G, Chang B, Yang F, Guo X, Cai KQ, Xiao XS, Wang H, Sen S, Hung MC, Mills GB, et al. Aurora kinase A promotes ovarian tumorigenesis through dysregulation of the cell cycle and suppression of BRCA2. Clin Cancer Res. 2010;16:3171–3181. doi: 10.1158/1078-0432.CCR-09-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Wang Z, Qi Z, Yin S, Zhang N, Liu Y, Liu M, Meng J, Zang R, Zhang Z, Yang G. The negative interplay between Aurora A/B and BRCA1/2 controls cancer cell growth and tumorigenesis via distinct regulation of cell cycle progression, cytokinesis, and tetraploidy. Mol Cancer. 2014;13:94. doi: 10.1186/1476-4598-13-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brodie KM, Henderson BR. Characterization of BRCA1 protein targeting, dynamics, and function at the centrosome: A role for the nuclear export signal, CRM1, and Aurora A kinase. J Biol Chem. 2012;287:7701–7716. doi: 10.1074/jbc.M111.327296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dann RB, DeLoia JA, Timms KM, Zorn KK, Potter J, Flake DD, II, Lanchbury JS, Krivak TC. BRCA1/2 mutations and expression: Response to platinum chemotherapy in patients with advanced stage epithelial ovarian cancer. Gynecol Oncol. 2012;125:677–682. doi: 10.1016/j.ygyno.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 49.Muggia F, Safra T. ‘BRCAness’ and its implications for platinum action in gynecologic cancer. Anticancer Res. 2014;34:551–556. [PMC free article] [PubMed] [Google Scholar]
- 50.Rao ZY, Cai MY, Yang GF, He LR, Mai SJ, Hua WF, Liao YJ, Deng HX, Chen YC, Guan XY, et al. EZH2 supports ovarian carcinoma cell invasion and/or metastasis via regulation of TGF-beta1 and is a predictor of outcome in ovarian carcinoma patients. Carcinogenesis. 2010;31:1576–1583. doi: 10.1093/carcin/bgq150. [DOI] [PubMed] [Google Scholar]
- 51.Gao J, Zhu Y, Nilsson M, Sundfeldt K. TGF-β isoforms induce EMT independent migration of ovarian cancer cells. Cancer Cell Int. 2014;14:72. doi: 10.1186/s12935-014-0072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Villanueva MT. Anticancer drugs: All roads lead to EZH2 inhibition. Nat Rev Drug Discov. 2017;16:239. doi: 10.1038/nrd.2017.60. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.