Abstract
Pancreatic cancer remains one of the most malignant tumors with a poor prognosis. Despite advances in diagnosis and treatment, no reliable biomarkers are available for clinical practice. Circular RNAs (circRNAs) are a novel class of endogenous non-coding RNA, which are abundant, stable and conserved, and serve crucial roles in disease, particularly in cancer. The purpose of the present study was to investigate the expression profile of circRNAs in 20 pancreatic cancer tissues and corresponding paracancerous tissues using arraystar human circRNA array analysis, high-throughput circRNA microarray, bioinformatic analysis and reverse transcription-quantitative polymerase chain reaction. It was revealed that the circRNAs expression profile was significantly different between pancreatic cancer tissue and paracancerous tissue, which indicates a potential role in pancreatic cancer. It was predicted that circRNAs may act as a micro RNA sponge to modulate gene expression in pancreatic cancer. Additionally, microarray expression analysis data was submitted to the Gene Expression Omnibus under accession no. GSE79634. The present study revealed that circRNAs expression was visibly diverse in pancreatic cancer compared with paracancerous tissue and provides more reliable biomarkers and new insights into the mechanisms of pancreatic cancer.
Keywords: circular RNAs, non-coding RNA, expression, progression, pancreatic cancer
Introduction
Pancreatic cancer continues to be the fourth leading cause of cancer-related death worldwide. It is characterized by late clinical presentation, malignant behavior and absence of effective therapeutic approaches, the 5-year overall survival rate is still less than 7% (1–3). Despite significant advance in diagnosis and treatment, the treatment efficacy is still unsatisfactory (4,5). Hence, the development of reliable methods for early detection and new therapeutic strategies to improve the outcome for patients with pancreatic cancer is urgently needed.
Circular RNAs (circRNAs) are a novel class of endogenous non-coding RNA, which are first discovered in RNA viruses as early as the 1970s (6). Unlike linear RNAs, circRNAs are terminated with 5′ caps and 3′ tails, circRNAs form covalently closed loop structures with neither 5′-3′ polarities nor polyadenylated tails. Accumulating studies have demonstrated that circRNAs are an abundant, stable, diverse and conserved class of RNA molecules (7–10). Originally, circRNAs gained little attention from researchers and the related studies were still in infancy. However, emerging evidence showed that circRNAs were implicated in multiple biological processes, including proliferation, differentiation, apoptosis and angiogenesis. Recent reports have showed the dysregulated circRNAs expression in diverse cancers, indicating crucial roles of circRNAs in carcinogenesis and progression of human cancer (11–13). With the development of biotechnology (e.g., high throughput RNA sequencing technology and bioinformatics), the characteristics of circRNAs are gradually revealed. Recent studies showed that circRNAs could regulate alternative splicing and targeted gene expression via microRNA (miRNA) sponge (14,15). Meanwhile, circRNAs dysregulation was engaged in carcinogenesis and cancer progression, including breast cancer, colorectal cancer, liver cancer and gastric cancer (16–19). The above findings suggest that circRNAs as a novel type of endogenous non-coding RNA become a new hotspot in non-coding RNA field and may play very important role in the progression of malignant tumors.
In this study, we investigated circRNAs expression profile in 20 pancreatic cancer tissues and corresponding paracancerous tissues. We found that circRNAs expression profile was significantly different between pancreatic cancer tissue and paracancerous tissue, and circRNAs could act as a miRNA sponge to modulate gene expression in pancreatic cancer. Our data indicated that circRNAs dysregulation may be associated with initiation and progression of pancreatic cancer and provide more potential biomarkers and new insights for pancreatic cancer.
Materials and methods
Samples
The Ethics Review Board of Southwest Hospital, the Third Military Medical University approved this study (no. 2016 Scientific Research no. 13), and the methods were carried out in accordance with the approved guidelines. All patients provided written informed consent. Twenty fresh-frozen pancreatic cancer tissues and corresponding paracancerous tissues were collected from Institute of Hepatopancreatobiliary Surgery, Southwest Hospital, the Third Military Medical University from April 2014 to November 2014 (clinical information of pancreatic cancer patients in Table I). All patients didn't undergo preoperative chemotherapy or radiotherapy. The pathologists from Department of Pathology, Southwest Hospital performed pathological diagnosis. According to tumor-node-metastasis (TNM) staging criteria from Union for International Cancer Control (UICC), tumor clinical stages were performed. All samples were stored in liquid nitrogen.
Table I.
Clinical characteristics of pancreatic cancer patients.
| No. | Sex | Age (years) | Diabetes | Jaundice | Histological grade | TNM stage |
|---|---|---|---|---|---|---|
| 1 | Male | 67 | Yes | Yes | G2 | T3N1M0 |
| 2 | Female | 50 | No | No | G1 | T2N0M0 |
| 3 | Male | 50 | No | No | G3 | T4N1M0 |
| 4 | Male | 62 | Yes | No | G2 | T4N1M0 |
| 5 | Male | 46 | No | No | G2 | T3N1M0 |
| 6 | Female | 71 | Yes | No | G2 | T3N1M0 |
| 7 | Male | 49 | No | No | G2 | T4N1M0 |
| 8 | Female | 47 | No | No | G2 | T3N1M0 |
| 9 | Female | 71 | No | No | G2 | T4N1M0 |
| 10 | Female | 72 | No | No | G2 | T3N0M0 |
| 11 | Male | 64 | Yes | No | G2 | T3N0M0 |
| 12 | Female | 61 | No | Yes | G1 | T3N0M0 |
| 13 | Male | 60 | No | No | G3 | T3N1M0 |
| 14 | Male | 73 | No | No | G1 | T3N1M0 |
| 15 | Female | 52 | No | No | G2 | T3N1M0 |
| 16 | Male | 61 | No | No | G2 | T3N1M0 |
| 17 | Male | 36 | No | No | G2 | T3N0M0 |
| 18 | Male | 54 | Yes | No | G2 | T3N0M0 |
| 19 | Female | 60 | No | Yes | G2 | T3N0M0 |
| 20 | Male | 60 | No | No | G2 | T3N0M0 |
TNM, tumor-node-metastasis stage.
RNA isolation
Total RNA from pancreatic cancer tissues and paracancerous tissues was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The RNA extractions were stored at −80°C.
RNA sample quality control (QC)
The purity and concentration of total RNA from each sample were quantified using a NanoDrop ND-1000. The integrity of RNA was assessed by electrophoresis on a denaturing agarose gel. Sample preparation and microarray hybridization were performed based on Arraystar's standard protocols.
RNA labeling and hybridization
Briefly, total RNA was digested with RNase R (Epicentre; Illumina, Inc., San Diego, CA, USA) to remove linear RNAs and enrich for circRNAs. The enriched circRNAs were amplified and transcribed into fluorescent cRNA utilizing a random priming method (Arraystar Super RNA Labeling kit; Arraystar, Inc., Rockville, MD, USA). The labeled cRNAs were purified by RNeasy Mini kit (Qiagen GmbH, Hilden, Germany). The concentration and specific activity of the labeled cRNAs (pmol Cy3/µg cRNA) were measured by NanoDrop ND-1000. Each labeled cRNA (1 µg) was fragmented by adding 5 µl 10X Blocking Agent and 1 µl of 25X fragmentation buffer, and then heated the mixture at 60°C for 30 min. Finally, 25 µl 2X Hybridization buffer was added to dilute the labeled cRNA. 50 µl of hybridization solution was dispensed into the gasket slide and assembled to the circRNA expression microarray slide. The labeled cRNAs were hybridized into an Arraystar Human circRNA array (8×15K; Arraystar, Inc.) and incubated for 17 h at 65°C in an Agilent Hybridization Oven (Agilent Technologies, Inc., Santa Clara, CA, USA). After the slides were washed, the arrays were scanned using an Agilent Scanner G2505C.
Microarray analysis and quality control
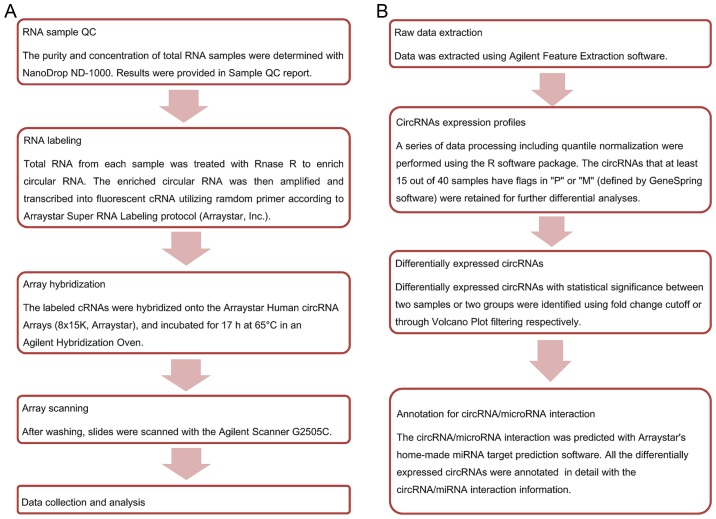
Agilent Feature Extraction software (version 11.0.1.1) was used to analyze acquired raw array images. Quantile normalization and subsequent data processing were performed using the R software package (R version 3.1.2). After quantile normalization of the raw data, low intensity filtering was performed, and circRNAs that at least 15 out of 40 samples have flags in ‘P’ or ‘M’ (‘All Targets Value’) were retained for further analyses. When comparing two groups of profile differences (such as disease vs. control), the ‘fold change’ (i.e., the ratio of the group averages) between the groups for each circRNA is computed. The statistical significance of the difference may be conveniently estimated by t-test. CircRNAs having fold changes ≥2 and P-values ≤0.05 are selected as significantly different expression. According to fold change, P-value, one can filter the analysis outputs and rank the differentially expressed circRNAs using Microsoft Excel's Data/Sort and Filter functionalities. The Statistical differences in circRNA expression between the two groups were determined with volcano plot filtering. Differentially expressed circRNAs between the two sample sets were identified by fold change filtering. Hierarchical clustering is one of the simplest and widely used clustering techniques for analysis of gene expression data. Cluster analysis arranges samples into groups based on their expression levels, which allows us to hypothesize about the relationships among samples. The dendrogram shows the relationships among the expression levels of samples. Here, hierarchical clustering was performed based on ‘All Targets Value-CircRNAs’. Our experiment consists of 40 different samples, and hierarchical clustering was performed to show a distinguishable circRNA genes expression profile among samples. The experiment workflow and data analysis flowchart are shown in Fig. 1.
Figure 1.
The experiment workflow and data analysis flowchart. (A) The experimental workflow of microarray expression profile of circRNAs. (B) The data analysis flowchart of microarray expression profile of circRNAs. circRNA, circular RNA.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) validation assay for circRNAs
Total RNA from 10 pairs human pancreatic cancer tissue and corresponding paracancerous tissue was isolated using TRIzol reagent (Invitrogen). The cDNA synthesis of RNA was performed using SuperScript™ III Reverse Transcriptase (Invitrogen). 10 differentially expressed circRNAs were respectively measured by qRT-PCR using 2X PCR Master Mix (Arraystar, Inc.) in pancreatic cancer tissue and corresponding paracancerous tissue. The reaction condition was as follows: 95°C for 10 min, 40 cycles of 95°C for 10 sec, 60°C for 60 sec, 95°C for 15 sec. The RNA levels were normalized to β-actin. All of the quantitative PCR reactions were conducted in triplicate. The related information of primers is shown in Table II. The specificity of PCR primers is validated using BLAST.
Table II.
Primers used for reverse transcription-quantitative polymerase chain reaction analysis of circRNAs, miR-15a and miR-506.
| ID | Primer sequence (5′-3′) | AT (°C) |
|---|---|---|
| β-actin (Human) | F: GTGGCCGAGGACTTTGATTG | 60 |
| R: CCTGTAACAACGCATCTCATATT | ||
| hsa_circRNA_102619 | F: GGGCATCTATTACATTCCATTCT | 60 |
| R: ATTATTCTCCGCAGCATCAGT | ||
| hsa_circRNA_102049 | F: GAAGCATTTCATCAATAACCCTC | 60 |
| R: CAAAGCCACAGTCCATCACAG | ||
| hsa_circRNA_000167 | F: TGAGCTTCGGGGAGGTGAGT | 60 |
| R: CAAGGGACATGGGAGTGGAGT | ||
| hsa_circRNA_100433 | F: CTTACCCATTCAGCCCATTCC | 60 |
| R: CGTGGCAAGGCTCTTTCTTCT | ||
| hsa_circRNA_103390 | F: CTGGGCTGACTACCTCAAACA | 60 |
| R: ACTCCACTGCACAGGGAAGAT | ||
| hsa_circRNA_101717 | F: GTCCTGTTTCTCAGATCGCTCAC | 60 |
| R: GAAGTCGGGGTTGCTGGTATT | ||
| hsa_circRNA_103076 | F: TCCTCCGTACAGCACATTCATTA | 60 |
| R: CACTCGATCGGCTTCACAAA | ||
| hsa_circRNA_104084 | F: GAGTGAGGATCAATGGGAAGAA | 60 |
| R: GGATGACTTGGTGACGGAAA | ||
| hsa_circRNA_102051 | F: CCTGAAACAAGCAGAGGAAGC | 60 |
| R: CACTCCTCCTTGGTCTTGGTG | ||
| hsa_circRNA_104270 | F: CGACAGCGGGTCTACTCACTCT | 60 |
| R: GGTACTTCCCACAAATCCTTGC | ||
| miR-506 | F: TAAGGCACCCTTCTGAGTAGA | 60 |
| R: GCGAGCACAGAATTAATACGAC | ||
| miR-15a | F: GGGGTAGCTTATCAGACTG | 60 |
| R: AGTGCGTGTCGTGGAGTC |
AT, annealing temperature; F, forward; R, reverse; circRNA, circular RNA; miR, micro RNA.
Quantitative real-time PCR for miRNAs
Total RNA from pancreatic cancer tissues was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. miRNA-specific primers were purchased from Ambion; Thermo Fisher Scientific, Inc. (Waltham, MA, USA). For miR-15a and miR-506 detection, cDNA synthesis was performed using the High Capacity cDNA Synthesis kit (Applied Biosystems, Foster City, CA, USA). Real-time qRT-PCR was carried out with miRNA-specific primers using the TaqMan Gene Expression Assay (Applied Biosystems). The reaction condition was as follows: 95°C for 10 min, 40 cycles of 95°C for 15 sec, 60°C for 60 sec, 4°C forever. β-actin was used as an endogenous control. The 2−ΔΔCt method was used to calculate expression relative to the endogenous control. The primer sequences of miR-15a and miR-506 were listed in Table II.
Statistical analysis
Data was extracted using Agilent Feature Extraction software. A series of data processing including quantile normalization were performed using the R software package. The circRNAs that at least 15 out of 40 samples have flags in ‘P’ or ‘M’ (defined by GeneSpring software) and were retained for further differential analyses. Differentially expressed circRNAs with statistical significance between two samples or two groups were identified using fold change. The statistical significance of the difference was conveniently estimated by paired t-test. CircRNAs having fold changes ≥2 and P-values ≤0.05 were selected as the significantly differentially expressed. The expression level of each circRNA was represented as fold change by 2−ΔΔCq method. The circRNAs/microRNA interaction was predicted with Arraystar's home-made miRNA target prediction software. All differentially expressed circRNAs were annotated in detail with the circRNAs/miRNA interaction information.
Results
Summary of circRNAs expression profile
The expression profile of circRNAs was performed in 20 pancreatic cancer tissues and corresponding paracancerous tissues using Arraystar Human CircRNA Array Analysis. The box plot is a convenient way to quickly visualize the distribution of a dataset. Here, it is used to compare the distributions of expression values for the samples in an experiment after normalization (Fig. 2A). The scatter-plot is a visualization method used for assessing circRNA expression variation (or reproducibility) between two compared samples or two compared groups of samples. The green lines are fold change lines. The data indicated that the expressions of circRNAs above the top green line and below the bottom green line are more than 2.0 fold changes between two compared samples (Fig. 2B). The volcano plot is useful tool for visualizing differential expression between two different conditions. The vertical lines respectively correspond to 2.0-fold up and down, and the horizontal line represents a P-value of 0.05. The red point in the plot represents the differentially expressed circRNAs with statistical significance (Fig. 2C). Hierarchical clustering is one of the simplest and widely used clustering techniques for analysis of gene expression data. Hierarchical clustering analysis shows a distinguishable circRNA expression profile among samples (Fig. 2D). Our results showed that there was a distinctly distinguishable circRNAs expression profile between pancreatic cancer tissue and paracancerous tissue. Meanwhile, we also indentified the characteristics of top 10 differently expressed circRNAs and 12 circRNAs associated with miR-15a and miR-506 (Table III). Among our results, 128 circRNAs were upregulated and 161 circRNAs were downregulated compared to paracancerous tissue (fold change ≥2.0 and P<0.05). Additionally, we validated the category of dysregulated circRNAs. Among these, upregulated circRNAs included 111 exonic, 9 intronic, 5 intragenic and 3 antisense, and downregulated circRNAs contained 122 exonic, 25 intronic, 13 intragenic and 1 antisense. We had submitted the microarray data to National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO). The GEO accession no. is GSE79634 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=cvyzseqoflahdmv&acc=GSE79634).
Figure 2.
Summary of circRNAs expression profile. (A) The box plot is a convenient way to compare the distributions of expression values for pancreatic cancer tissues and corresponding paracancerous tissues after normalization. (B) The scatter-plot is used for assessing circRNAs expression variation between pancreatic cancer tissues and corresponding paracancerous tissues. The circRNAs above the top green line and below the bottom green line indicated more than 2.0 fold change of circRNAs between two compared samples. (C) The volcano plots are useful tools for visualizing differential expression between two different conditions. The red point in the plot represents the differentially expressed circRNAs with statistical significance. (D) The Hierarchical clustering is performed for analysis of gene expression. The data shows that a distinct circRNAs expression profile is among samples. circRNA, circular RNA.
Table III.
The characteristics of top 10 differently expressed circRNAs and 12 circRNAs associated with miR-15a and miR-506.
| circRNA | P-value | FDR | FC | Regulation | circRNA type | Chrom | Strand | Gene symbol | miRNA binding sites |
|---|---|---|---|---|---|---|---|---|---|
| hsa_circRNA_102051 | 0.00000011389 | 0.00000181 | 78.901931 | Up | Exonic | chr17 | + | TADA2A | miR-302c-3p/miR-520d-3p |
| miR-302b-3p/miR-302d-3p/miR-197-5p | |||||||||
| has_circRNA_102619 | 0.00000060084 | 0.000006232 | 71.3388052 | Up | Exonic | chr2 | − | NOL10 | miR-452-5p/miR-874-3p |
| miR-218-1-3p/miR-767-5p/miR-218-2-3p | |||||||||
| hsa_circRNA_104270 | 0.00000010672 | 0.000001743 | 53.0782362 | Up | Exonic | chr6 | + | FAM120B | miR-450a-1-3p/miR-661 |
| miR-493-5p/miR-208a-5p/miR-612 | |||||||||
| hsa_circRNA_102049 | 0.00000018328 | 0.000002511 | 52.0619239 | Up | Exonic | chr17 | + | TADA2A | miR-103a-2-5p/miR-455-3p |
| miR-302c-3p/miR-520d-3p/miR-302-3p | |||||||||
| hsa_circRNA_104227 | 0.00001350199 | 0.00006792 | 10.96655 | Up | Exonic | chr6 | + | MTHFD1L | miR-329-3p/miR-593-5p |
| miR-641/miR-103a-2-5p/miR-19b-1-5p | |||||||||
| hsa_circRNA_000167 | 0.00000001124 | 0.000000389 | 10.010543 | Down | Intragenic | chr14 | − | RPPH1 | miR-330-5p/miR-328-3p |
| miR-127-5p/miR-326/miR-1296-5p | |||||||||
| hsa_circRNA_103809 | 0.00000741477 | 0.000042676 | 7.6595391 | Down | Exonic | chr5 | − | ZFR | miR-511-5p/miR-130b-5p |
| miR-642a-5p/miR-532-3p/miR-329-5p | |||||||||
| hsa_circRNA_104700 | 0.00000727569 | 0.000042007 | 7.4515712 | Down | Exonic | chr8 | − | PTK2 | miR-141-5p/miR-500a-5p |
| miR509-3p/miR-619-3p/miR578 | |||||||||
| hsa_circRNA_001846 | 0.00000006353 | 0.000001136 | 6.2821101 | Down | Intragenic | chr14 | − | RPPH1 | miR-328-3p/miR-1296-5p |
| miR-146b-3p/miR-330-5p/miR-608 | |||||||||
| hsa_circRNA_102728 | 0.00001617324 | 0.000076928 | 5.7878202 | Down | Exonic | chr2 | − | NBAS | miR-105-5p/miR-18a-5p |
| miR-455-3p/miR-624-5p/miR-7-2-3p | |||||||||
| hsa_circRNA_103076 | 0.00000004087 | 0.000000904 | 2.6239863 | Up | Exonic | chr20 | − | CSE1L | miR-330-3p/miR-155-5p |
| miR-330-5p/miR-146-3p/miR-15a-3p | |||||||||
| hsa_circRNA_100435 | 0.00000048773 | 0.000005267 | 2.1392264 | Up | Exonic | chr1 | − | DSTYK | miR-424-5p/miR-15a-5p |
| miR-10b-3p/miR-650/miR-15b-5p | |||||||||
| hsa_circRNA_103309 | 0.00122306572 | 0.002970302 | 2.127291 | Up | Exonic | chr3 | + | RBMS3 | miR-330-5p/miR-15a-5p |
| miR-429/miR-424-5p/miR-200b-3p | |||||||||
| hsa_circRNA_000780 | 0.00841633735 | 0.015881187 | 2.1957639 | Down | Intronic | chr10 | − | FAM107B | miR-651-3p/miR-15a-3p |
| miR-381-3p/miR-522-3p/miR-300 | |||||||||
| hsa_circRNA_101252 | 0.00007537652 | 0.000272424 | 2.3359115 | Down | Exonic | chr13 | − | FOXO1 | miR-654-3p/miR-15a-3p |
| miR-762/miR-1301-3p/miR-145-5p | |||||||||
| hsa_circRNA_102374 | 0.00258652534 | 0.005756195 | 2.8567112 | Down | Exonic | chr18 | + | WDR7 | miR-503-5p/miR-15a-5p |
| miR-16-5p/miR-424-5p/miR-497-5p | |||||||||
| hsa_circRNA_104433 | 0.00051721594 | 0.001427983 | 2.0563844 | Down | Exonic | chr7 | + | ARPC1B | miR-545-3p/miR-15a-5p |
| miR-636/miR-335-3p/miR-497-5p | |||||||||
| hsa_circRNA_104882 | 0.00005024961 | 0.000192181 | 2.3947053 | Down | Exonic | chr9 | + | HSDL2 | miR-16-5p/miR-15a-5p |
| miR-135b-5p/miR-424-5p/miR-135a-5p | |||||||||
| hsa_circRNA_101717 | 0.0000322857 | 0.00013679 | 2.2591213 | Up | Exonic | chr16 | + | ABCC1 | let-7e-5p/miR-124-3p |
| miR-877-5p/miR-506-3p/let-7c-5p | |||||||||
| hsa_circRNA_104084 | 0.00028685355 | 0.000860561 | 2.7046096 | Up | Exonic | chr6 | + | LINC00340 | miR-644a/miR-605-3p |
| miR-124-3p/miR-506-3p/miR-1224-3p | |||||||||
| hsa_circRNA_100646 | 0.00000451139 | 0.000029215 | 3.0830469 | Up | Exonic | chr10 | − | CPEB3 | miR-34a-5p/miR-499a-5p |
| miR-146a-5p/miR-506-5p/miR-146b-5p | |||||||||
| hsa_circRNA_102213 | 0.0000001515 | 0.000002296 | 2.214269 | Up | Exonic | chr17 | − | USP36 | miR-873-5p/miR-544a |
| miR-25-5p/miR-506-5p/miR-152-5p |
FDR, false discover rate; FC, fold change; Chrom, chromosome; miR, micro RNA; circRNA, circular RNA.
Verification for circRNAs expression
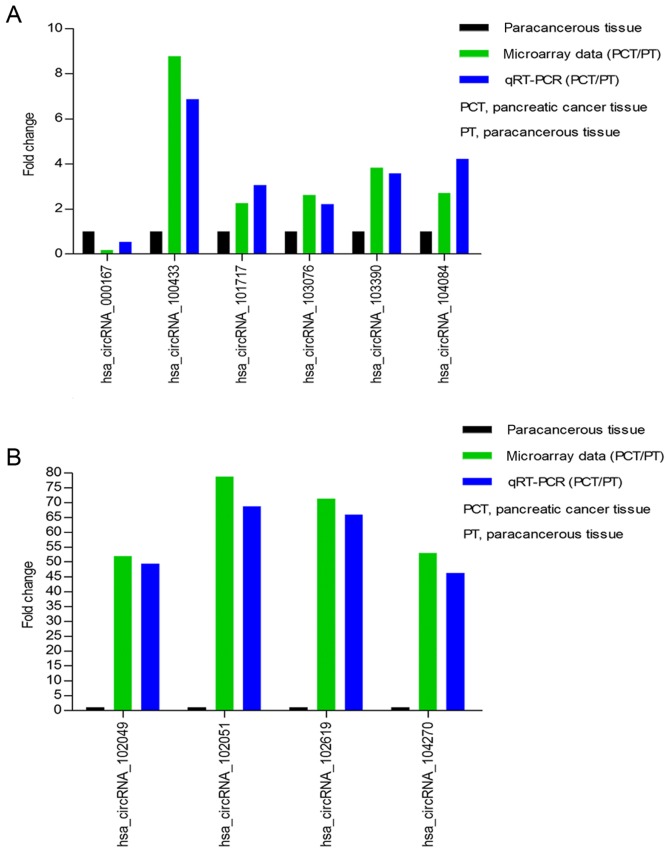
Our circRNAs expression profile revealed that the expression of circRNAs was significantly different in pancreatic cancer tissue compared to paracancerous tissue. To validate our microarray data, we randomly selected 10 differently expressed circRNAs, including one downregulated circRNA (circRNA_000167) and nine upregulated circRNAs (circRNA_100433, circRNA_101717, circRNA_102049, circRNA_102051, circRNA_102619, circRNA_103076, circRNA_103390, circRNA_104084, circRNA_104270). We respectively verified their expression levels using qRT-PCR in 10 pancreatic cancer tissues and corresponding paracancerous tissues. The qRT-PCR results showed that the expression levels of circRNAs were significantly different in pancreatic cancer tissue compared with corresponding paracancerous tissue. The log2 fold-changes were calculated for microarray data and qRT-PCR results. Our results illustrated that microarray data was consistent with the qRT-PCR results upon the expression levels of the ten circRNAs (Fig. 3A and B).
Figure 3.
Validation for circRNAs expression. (A and B) The expression levels of ten circRNAs are validated by RT-qPCR in 10 pancreatic cancer tissues and corresponding paracancerous tissues. The data are presented as mean ± SD. The comparison is performed between microarray data and RT-qPCR results. The y-axis in the diagram represents the mean of fold-change (log2 transformed) of each circRNA measured by RT-qPCR and microarray respectively. The RT-qPCR results are consistent with microarray data. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; circRNA, circular RNA; PCT, pancreatic cancer tissue; PT, paracancerous tissue.
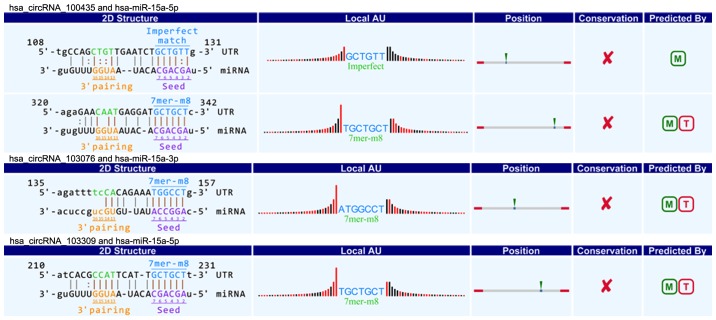
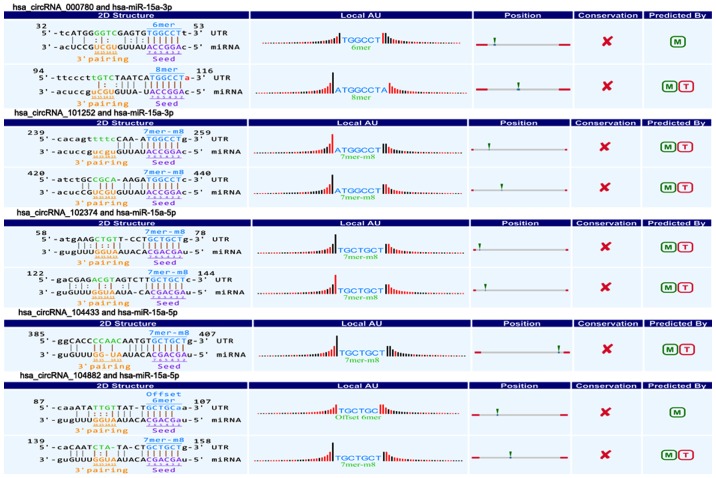
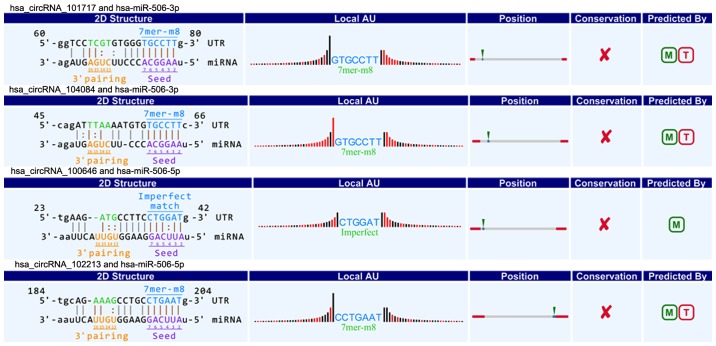
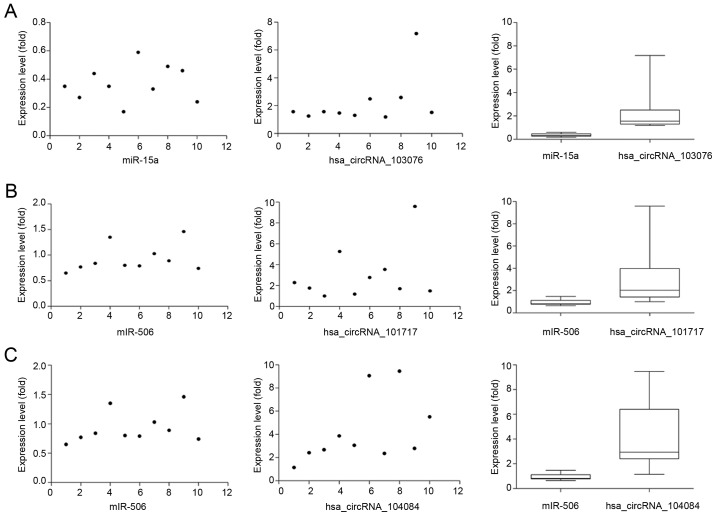
CircRNAs could act as a miRNA sponge to modulate gene expression in pancreatic cancer. Recent reports demonstrated that circRNAs played very vital role in regulating gene expression via miRNAs sponge, and could bind some miRNAs associated with disease to affect disease initiation and progression (20). For instance, circRNA, ciRS-7, contains multiple, tandem miRNA-7 binding sites, and regulates miRNA-7 functions via an endogenous miRNA sponge (21). CiRS-7 as a critical factor probably is engaged in modulating the function of neuron as well as a responsible candidate in neurological disorder and tumor development (12,22). To explore the association between circRNAs and miRNAs in pancreatic cancer, we predicted circRNA/miRNA interaction using Arraystar's home-made miRNA target prediction software based on TargetScan and miRanda (23,24). The differentially expressed circRNAs within all the comparisons were annotated in detail with the circRNA/miRNA interaction information. What's more, combined with our previous studies (25,26), we specially focused on the interaction between circRNAs and miR-15a/miR-506. Among our data, we found that eight circRNAs including three upregulated circRNAs (circRNA_100435, circRNA_103076, circRNA_103309; Fig. 4) and five downregulated circRNAs (circRNA_000780, circRNA_101252, circRNA_102374, circRNA_104433, circRNA_104882; Fig. 5) were respectively related with miR-15a. Four upregulated circRNAs (circRNA_101717, circRNA_104084, circRNA_100646, circRNA_102213) were associated with miR-506 (Fig. 6). The above circRNAs respectively had complementary sequence with miR-15a/miR-506 and diverse binding sites at the seed region, while the characteristics and differences were shown in Table III. It indicated that these circRNAs could modulate miR-15a/miR-506 expression via miRNA sponge in pancreatic cancer. To verify our predicted result, we selected three differently expressed circRNAs interacted with miR-15a and miR-506 from our prediction result, and then respectively performed qRT-PCR to quantify miR-15a and miR-506 expression levels in pancreatic cancer tissues showing altered expression of circRNAs. Our result showed that the above three circRNAs respesctively were negatively correlated with miR-15a and miR-506 (Fig. 7). Taken together, we speculated that circRNAs could regulate gene expression via miRNA sponge and play very significant role in the progression of pancreatic cancer.
Figure 4.
Three upregulated circRNAs (circRNA_100435, circRNA_103076 and circRNA_103309) contain complementary sequence and diverse binding sites with miR-15a. circRNA, circular RNA, miR, microRNA.
Figure 5.
Five downregulated circRNAs (circRNA_000780, circRNA_101252, circRNA_102374, circRNA_104433 and circRNA_104882) contain complementary sequence and diverse binding sites with miR-15a. circRNA, circular RNA; miR, microRNA.
Figure 6.
Four upregulated circRNAs (circRNA_101717, circRNA_104084, circRNA_100646 and circRNA_102213) contain complementary sequence and diverse binding sites with miR-506. circRNA, circular RNA; miR, microRNA.
Figure 7.
Verification for the interaction between circRNAs and miR-15a/miR-506. Quantifying miR-15a and miR-506 expression levels via RT-qPCR. Three circRNAs respectively are negatively correlated with miR-15a and miR-506 (P<0.05). (A) The expression of miR-15a is significantly decreased and negatively correlated with circRNA_103076. (B and C) The expression of miR-506 is significantly decreased and negatively correlated with circRNA_101717 and circRNA_104084. circRNA, circular RNA; miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Discussion
In recent years, the relationship has been extensively explored between non-coding RNA and cancer, such as miRNAs, lncRNAs and circRNAs. Emerging studies have clarified that non-coding RNA plays very significant role in the carcinogenesis and cancer progression (27–29). However, the role of circRNAs and molecular mechanism remain to be investigated in pancreatic cancer. With the development of biotechnology (e.g., high throughput RNA sequencing and bioinformatics), accumulating evidence showed that circRNAs as a novel type of endogenous non-coding RNA were an abundant, stable and conserved class of RNA molecules (7–10). What is more, recent studies revealed that circRNAs may regulate diverse biological processes and be engaged in multiple diseases, especially malignant tumors including hepatocellular carcinoma, colorectal cancer, gastric cancer, non-small cell lung cancer (14–19). Therefore, these findings indicate that circRNAs may play very important role in carcinogenesis and cancer progression.
In this study, we systematically explored the expression profile of circRNAs in 20 pancreatic cancer tissues and corresponding paracancerous tissues. Our results indicated that circRNAs expression profile was significantly different between pancreatic cancer tissue and paracancerous tissue. The expression profile showed that 289 circRNAs expression were aberrantly expressed in pancreatic cancer compared to paracancerous tissue. circRNAs (161) were downregulated and 128 circRNAs were upregulated. We also indentified the circRNAs type, chromosome location, gene symbol, and sequence. In addition, we confirmed that our microarray data was consistent with qRT-PCR result, illustrating that our microarray data was available.
Recently, some studies have demonstrated that circRNAs can play very crucial role in biological process and progression of disease via miRNA sponge (20,21). Furthermore, our previous studies have confirmed that miR-15a can inhibit pancreatic cancer cell proliferation and EMT by downregulated BMI-1, and miR-506 can suppress the progression and chemoresistance (25,26). To identify whether there was the interaction between circRNAs and miR-15a/miR-506, we performed Arraystar's home-made miRNA target prediction based on TargetScan and miRanda, and differentially expressed circRNAs were annotated in detail with circRNA/miRNA interaction information. We found that multiple circRNAs had complementary sequence with miR-15a/miR-506 and diverse miRNA binding sites, and our qRT-PCR result also validated this point. Altogether, we speculated that circRNAs could interact with miR-15a/miR-506 via miRNA sponge to modulate the progression of pancreatic cancer. However, we should perform further in-depth functional studies to indicate whether circRNAs acting as miRNA sponge play very important role in progression of pancreatic cancer in future.
In conclusion, we demonstrated that circRNAs expression profile was significantly different in pancreatic cancer tissue compared to paracancerous tissue. We also validated that dysregulated circRNAs expression in pancreatic cancer tissue using qRT-PCR, which was consistent with our microarray data. Additionally, combined with our previous studies, we successfully predicted that multiple circRNAs had complementary sequence with miR-15a/miR-506 and diverse miRNA binding sites at the seed region. We speculated that circRNAs may play very crucial role in modulating the progression of pancreatic cancer via miRNA sponge, and then we validated this point using qRT-PCR. Therefore, these findings may provide more potential biomarkers and new treatment strategies for pancreatic cancer. However, in future, we need to perform further studies to explore circRNAs' function and molecular mechanism in pancreatic cancer.
Acknowledgements
The present study was supported by grants from National Natural Science Foundation of China (grant no. 81502550), National Natural Science Foundation of China (grant no. 81672382) and Southwest Hospital Research Project (grant no. SWH2016JCYB-45).
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378:607–620. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Zeng H, Zhang S. The incidence and mortality of major cancers in China, 2012. Chin J Cancer. 2016;35:73. doi: 10.1186/s40880-016-0137-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartwig W, Werner J, Jäger D, Debus J, Büchler MW. Improvement of surgical results for pancreatic cancer. Lancet Oncol. 2013;14:e476–e485. doi: 10.1016/S1470-2045(13)70172-4. [DOI] [PubMed] [Google Scholar]
- 5.Garrido-Laguna I, Hidalgo M. Pancreatic cancer: From state-of-the-art treatments to promising novel therapies. Nat Rev Clin Oncol. 2015;12:319–334. doi: 10.1038/nrclinonc.2015.53. [DOI] [PubMed] [Google Scholar]
- 6.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/annotation/f782282b-eefa-4c8d-985c-b1484e845855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. CircularRNAs are abundant, conserved and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 11.You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 13.Boeckel JN, Jaé N, Heumüller AW, Chen W, Boon RA, Stellos K, Zeiher AM, John D, Uchida S, Dimmeler S. Identification and characterization of hypoxia-regulated endothelial circular RNA. Circ Res. 2015;117:884–890. doi: 10.1161/CIRCRESAHA.115.306319. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 15.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 16.Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bcahleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D. Correlation of circular RNA abundance with proliferation-exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis and normal human tissues. Sci Rep. 2015;5:8057. doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Genome-wide analysis of Drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, Wong DT, Xiao X. The landscape of microRNA, Piwi-interacting RNA and circular RNA in human saliva. Clin Chem. 2015;61:221–230. doi: 10.1373/clinchem.2014.230433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 20.Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: A comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. 2013;4:283. doi: 10.3389/fgene.2013.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lukiw WJ. Circular RNA (circRNA) in Alzheimer's disease (AD) Front Genet. 2013;4:307. doi: 10.3389/fgene.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 23.Enright AJ, John B, Gaul U, TuschI T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pasquinelli AE. MicroRNAs and their targets: Recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 25.Guo S, Xu X, Tang Y, Zhang C, Li J, Ouyang Y, Ju J, Bie P, Wang H. miR-15a inhibits cell proliferation and epithelial to mesenchymal transition in pancreatic cancer by down-regulating Bmi-1 expression. Cancer Lett. 2014;344:40–46. doi: 10.1016/j.canlet.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 26.Li J, Wu H, Li W, Yin L, Guo S, Xu X, Ouyang Y, Zhao Z, Liu S, Tian Y, et al. Downregulated miR-506 expression facilitates pancreatic cancer progression and chemoresistance via SPHK1/Akt/NF-κB signaling. Oncogene. 2016;35:5501–5514. doi: 10.1038/onc.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiong G, Feng M, Yang G, Zheng S, Song X, Cao Z, You L, Zheng L, Hu Y, Zhang T, Zhao Y. The underlying mechanisms of non-coding RNAs in the chemoresistance of pancreatic cancer. Cancer Lett. 2017;397:94–102. doi: 10.1016/j.canlet.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 28.Xu YF, Hannafon BN, Ding WQ. microRNA regulation of human pancreatic cancer stem cells. Stem Cell Investig. 2017;4:5. doi: 10.21037/sci.2017.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupta Chandra S, Tripathi Nandan Y. Potential of long non-coding RNAs in cancer patients: From biomarkers to therapeutic targets. Int J Cancer. 2017;140:1955–1967. doi: 10.1002/ijc.30546. [DOI] [PubMed] [Google Scholar]