Abstract
The current study aimed to identify novel long non-coding RNAs (lncRNAs) associated with gastric cancer (GC). Transcriptome sequencing of the lncRNAs and mRNAs from GC tissues and normal adjacent tissues was performed. The data were analyzed using bioinformatics analysis, specifically analysis of differentially expressed lncRNAs and mRNA, target gene prediction and functional enrichment analysis. A total of 1,181 differentially expressed mRNA and 390 differentially expressed lncRNAs were identified. The targets of upregulated lncRNAs were significantly enriched in functions associated with collagen fibril organization, whereas the downregulated lncRNA were significantly associated with ion transmembrane transport and regulation of membrane potential. A total of 7 lncRNAs were verified by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Following RT-qPCR validation, AC016735.2, AP001626.1, RP11-400N13.3 and RP11-243M5.2 were considered to be consistent with the prediction of the bioinformatics analysis. Transcriptome sequencing and RT-qPCR experiments identified 4 lncRNAs, including AC016735.2, AP001626.1, RP11-400N13.3 and RP11-243M5.2 to have an important role in the carcinogenesis of GC.
Keywords: lncRNA, gastric cancer, differential expression, transcriptome sequencing, bioinformatics analysis, RT-qPCR
Introduction
Gastric cancer (GC), developing from the lining of the stomach, is one of the most common cancers worldwide, particularly in East Asia and China (1). It is the third leading cause of death from cancer and accounts for 9% of mortality worldwide (2). If untreated, tumor cells often metastasize to other parts of the body, particularly the lungs, liver, bone, and lymph nodes; therefore, the prognosis of GC is generally unfavorable (3). The 5-year survival rate for GC is reported to be <10% (4). In China, the majority of patients with GC are diagnosed at a late stage and the prognosis is unfavorable (1). Therefore, understanding of the molecular mechanisms and identification of the key biomarkers associated with GC progression is essential for the diagnosis and therapy of GC.
The conventional view of gene regulation in biology is primarily concentrated on the protein-coding genes. However, the human genome project suggested that ~1.2% of the mammalian genome encodes proteins (5,6) and most of the genome is transcribed into long non-coding RNAs (lncRNAs) (7,8). LncRNAs are RNA molecules >200 nucleotides in length. Dysregulated lncRNAs have been demonstrated to have an important role in tumorigenesis and cancer metastasis (9–11). The association between aberrant expression of lncRNAs and GC has been previously investigated. For example lncRNA-HMlincRNA717 was determined to have a crucial role during GC occurrence and progression (12). Song et al (13) performed lncRNA microarray analysis and identified 135 differentially expressed lncRNAs between GC and normal tissues (13). However, numerous lncRNAs have been identified they are not sufficient for the treatment of GC.
In the present study, the lncRNA sequencing for GC tissues was performed using a transcriptome sequencing technique. The differentially expressed lncRNAs between GC and normal adjacent tissues were identified. The bioinformatics analysis included prediction of target genes and function enrichment analysis. Finally, the lncRNAs predicted by the present study were verified by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The current study aimed to investigate the additional lncRNAs associated with GC, which may be used as potential markers for the diagnosis and treatment of GC.
Materials and methods
Tissue samples
Between October 2015 and January 2016, a total of 3 male patients with GC (aged 65–76 years old) were included in the current study, whose diagnoses were pathologically confirmed. The cancer tissues and the normal adjacent tissues were obtained from clinically ongoing surgical specimens, were snap frozen with liquid nitrogen and subsequently stored at −80°C until RNA extraction.
All patients have provided written informed consent prior to participating in the present study. The procedures in the current study were approved by the Protection of Human Ethics Committee of Shanghai Shuguang Hospital Affiliated with Shanghai University of TCM (Shanghai, China).
Transcriptome sequencing
Total RNAs from gastric cancer tissues (3 samples) and normal adjacent tissues (3 samples) were extracted using the RNAiso Plus (Takara Biotechnology Co., Ltd., Dalian, China). Evaluation of the quality and integrity of the total RNA was performed using 1% agarose gel electrophoresis (visualized using ethidium bromide), and an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA). Following this, a cDNA library was established using the NEBNext Ultra RNA Library Prep kit (New England Biolabs, Inc., Ipswich, MA, USA) prior to Illumina sequencing. The transcriptome sequencing of mRNA and lncRNA was performed on an Illumina gene analyzer (Illumina, Inc., San Diego, USA).
The data have been deposited at National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database under accession number: SRP092509.
Quality control of the sequencing data was performed to identify the clean reads using the FASTX-toolkit (version 0.0.13) (14). The obtained clean reads were aligned to the human reference genome hg19 using TopHat software (version 2.10) (15). Then based on the mRNA and lncRNA annotation information provided by gencode version 24 (mapped to GRCh37) (16) database, the Fragments Per Kilobase of transcript per Million mapped reads values of mRNA and lncRNA and the reads number of lncRNA were identified using StringTie tool (version 1.2.3) (17).
Bioinformatics analysis of sequencing data
The differentially expressed lncRNAs and genes (DEGs) between the cancer group and the control group were identified using the limma package (18) in R (version 3.2.5) with the following criteria: |log2 fold change (FC)|>1 and P<0.05.
The downstream target genes of the differentially expressed lncRNAs were predicted based on the co-expression associations between lncRNAs and mRNAs. The threshold values were correlation coefficient >0.8 and P<0.05. Additionally, the network of lncRNAs and their target genes was constructed using Cytoscape (version 3.0) (19).
Subsequently, DEGs and target genes of lncRNAs were used to perform Gene Ontology (GO) functional and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses with the clusterprofiler package (20) in R.
RT-qPCR verification of the expression of lncRNAs
Total RNA was extracted from tissues (3 GC tissues and 3 normal adjacent tissues) using RNAiso Plus (9109; Takara Biotechnology Co., Ltd.). The concentration and purity of the isolated RNA was determined using TECAN infinite M100 PRO Biotek microplate reader (Tecan Group, Ltd., Mannedorf, Switzerland) and reverse transcription (37°C for 15 min and 85°C for 5 sec) was performed according to the PrimeScript RT Master mix RR036A (Takara Biotechnology Co., Ltd.). qPCR was performed using SYBRGreen kit (cat no. 4367659; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The reaction procedures were as follows: 50°C for 3 min, 95°C for 3 min, 95°C for 10 sec, and 60°C for 30 sec, for 40 cycles, the melting process was 60 to 95°C (increments of 0.5°C for 10 sec). According to the results of bioinformatics analysis, the expressions of 7 lncRNAs, including AC016735.2, RP11-243M5.2, RP11-400N13.2, RP11-400N13.3, AP001626.1, LINC01139 and RP11-54H7.4, were detected with the primers presented in Table I. The expression levels were calculated using the 2−ΔΔCq method (21).
Table I.
Primer sequences of the long noncoding RNAs for reverse transcription- quantitative polymerase chain reaction.
| Gene | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| AC016735.2 | CTGCTTCTCACTGCCTCG | TTTCCCAAATGGTCCTCC |
| RP11-243M5.2 | TTGCGTGAAAGCGTATGG | GAAAGCAGCCTTGAGAACAGAG |
| RP11-400N13.2 | CCCCTGTCCTCCTGCTCTT | CGGGCAGTGTCAGTCTTCA |
| RP11-400N13.3 | GCAGATGGCAAAGGATAAAGC | GGTGATATACGATGCAACGGTG |
| AP001626.1 | AGCTGCACCAAGGAGAATC | CAAAGCCAAGGTCCACTGTT |
| LINC01139 | ACCAGTCACCCAACCAGAGC | AAGCGTAAGAATGAAGACCAGTG |
| RP11-54H7.4 | TCCACTCTAGGTTCCCACG | CCTGACATTCCTGCCTTCTT |
| GAPDH | TGACAACTTTGGTATCGTGGAAGG | AGGCAGGGATGATGTTCTGGAGAG |
Statistical analysis
Data are presented as the mean ± standard error of mean. The statistical analysis was performed by Graphpad Prism (version 5.01) using the Student's t-test (Graphpad Software, Inc., San Diego, CA, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
High-throughput sequencing
From the 6 samples, a total of 3,4290 mRNAs and 10,148 lncRNAs were identified, which were expressed in at least one sample. With the criteria of|log2FC|>1 and P<0.05, a total of 1,181 DEGs were identified, 902 were upregulated and 279 were downregulated. Additionally, 390 differentially expressed lncRNAs, including 163 upregulated and 227 upregulated lncRNAs were identified (Fig. 1). The top 10 differentially expressed lncRNA with higher FCs, including RP11-171I2.1, RP11-171I2.1 and AC016735.2 are presented in Table II.
Figure 1.
Heatmap of differentially expressed lncRNAs. Red indicates high expression and green indicates low expression. Patient 1A and Patient 1B are samples derived from the same patient; Patient 2A and Patient 2B are samples derived from the same patient; and Patient 3A and Patient 3B are derived from the same patient. A, tumor tissue; B, normal tissue.
Table II.
Top 10 differentially expressed upregulated and downregulated lncRNAs.
| A, Downregulated | ||
|---|---|---|
| ID | logFC | P-value |
| RP11-171I2.1 | −4.549491715 | 0.006324281 |
| RP5-994D16.9 | −4.73821376 | 0.002778706 |
| RP11-139H15.6 | −4.802835645 | 0.002652066 |
| RP11-637N19.1 | −4.807460785 | 0.005211747 |
| RP11-243M5.2 | −4.87029533 | 0.011518821 |
| RP11-382A20.5 | −4.909287572 | 0.002375869 |
| AC003090.1 | −5.054470484 | 0.002583845 |
| FGF14-IT1 | −5.152474325 | 0.004193094 |
| AC053503.12 | −5.200695133 | 0.045876461 |
| RP11-16P20.4 | −5.309441725 | 0.00370217 |
| B, Upregulated | ||
| ID | logFC | P-value |
| RP3-416H24.1 | 6.312295751 | 0.002010312 |
| RP5-1185I7.1 | 5.627125267 | 0.002013113 |
| LINC01087 | 5.562031916 | 0.00323185 |
| RP11-1007G5.2 | 5.506776995 | 0.000837008 |
| LINC00483 | 5.440975498 | 0.01629253 |
| LINC00618 | 5.425270941 | 0.000784189 |
| RP11-120K24.4 | 5.413330508 | 0.002061786 |
| AC016735.2 | 5.383341519 | 0.030647411 |
| AC110769.3 | 5.338202236 | 0.001217988 |
| LA16c-325D7.1 | 5.296535904 | 0.002461903 |
FC, fold-change
Bioinformatics analysis of lncRNA sequencing data
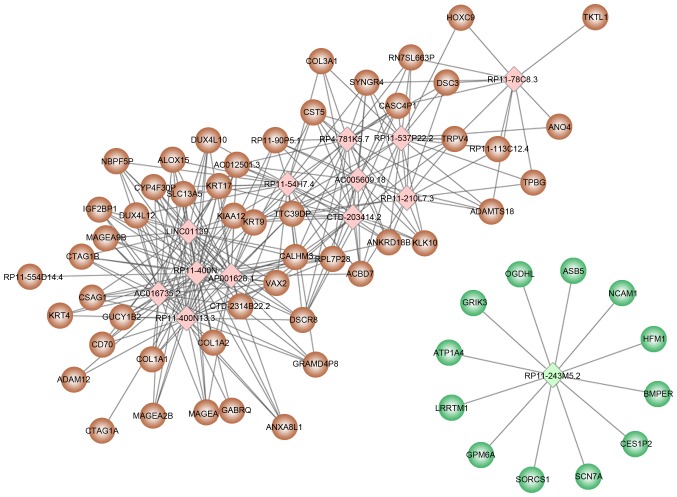
Based on the threshold values of correlation coefficient >0.8 and P<0.05, 157 differentially expressed lncRNAs were selected for target prediction and a total of 231 target genes were identified. Among the 157 differentially expressed lncRNAs, 13 lncRNAs (12 upregulated and 1 downregulated) predicted an additional 10 target genes (Table III), including RP11-400N13.2, RP11-400N13.3, AP001626.1 and RP11-54H7.4. The regulatory network constructed with 13 lncRNAs and their target genes is presented in Fig. 2.
Table III.
Long noncoding RNAs with more than 10 target genes.
| ID | No of target genes | log2FC | P-value |
|---|---|---|---|
| AC005609.18 | 16 | 5.258702066 | 0.002564097 |
| AC016735.2 | 23 | 5.383341519 | 0.030647411 |
| AP001626.1 | 30 | 3.480674967 | 0.039101426 |
| CTD-2034I4.2 | 19 | 3.225981981 | 0.039614051 |
| LINC01139 | 27 | 3.784674378 | 0.034659687 |
| RP11-210L7.3 | 15 | 3.183358792 | 0.03741905 |
| RP11-243M5.2 | 12 | −4.87029533 | 0.011518821 |
| RP11-400N13.2 | 32 | 4.411473414 | 0.015832445 |
| RP11-400N13.3 | 32 | 4.01570403 | 0.031992561 |
| RP11-537P22.2 | 19 | 3.519296946 | 0.027224444 |
| RP11-54H7.4 | 25 | 3.227024693 | 0.043767147 |
| RP11-782C8.3 | 11 | 3.386174545 | 0.029429789 |
| RP4-781K5.7 | 18 | 3.607427164 | 0.025935631 |
Figure 2.
Regulatory network constructed with 13 lncRNAs and their target genes. Brown circles indicate upregulated target genes and green circles represent downregulated target genes. Pink rhombus, upregulated lncRNA; green rhombus, downregulated lncRNA. lncRNA, long noncoding RNA.
Due to the high number of lncRNAs identified, the functions of individual lncRNAs were not analyzed. The present study focused on the functions of the 13 aforementioned lncRNAs. A total of 50 and 12 target genes were predicted for the upregulated and downregulated lncRNAs, respectively. Functional enrichment analysis of the 62 target genes determined that the upregulated lncRNAs were significantly enriched in functions associated with collagen fibril organization, whereas the downregulated lncRNA was significantly associated with ion transmembrane transport and regulation of membrane potential (Table IV).
Table IV.
Functional enrichment analysis of the target genes of the 13 lncRNAs with more than 10 target genes.
| A, Upregulated | ||||
|---|---|---|---|---|
| Ontology | ID | Function | Count | FDR |
| BP | GO:0043588 | Skin development | 5 | 0.011708982 |
| BP | GO:0030199 | Collagen fibril organization | 3 | 0.011708982 |
| BP | GO:0071230 | Cellular response to amino acid stimulus | 3 | 0.012776741 |
| BP | GO:0043589 | Skin morphogenesis | 2 | 0.012776741 |
| BP | GO:0070208 | Protein heterotrimerization | 2 | 0.016397947 |
| CC | GO:0005583 | Fibrillar collagen trimer | 3 | 0.000116529 |
| CC | GO:0098643 | Banded collagen fibril | 3 | 0.000116529 |
| CC | GO:0098644 | Complex of collagen trimers | 3 | 0.000426968 |
| CC | GO:0005581 | Collagen trimer | 3 | 0.016659231 |
| CC | GO:0044420 | Extracellular matrix component | 3 | 0.037435206 |
| MF | GO:0048407 | Platelet-derived growth factor binding | 3 | 0.000119244 |
| MF | GO:0005201 | Extracellular matrix structural constituent | 3 | 0.016290955 |
| F | GO:0005198 | Structural molecule activity | 6 | 0.036696011 |
| MF | GO:0019838 | Growth factor binding | 3 | 0.040871579 |
| KEGG | hsa04974 | Protein digestion and absorption | 3 | 0.002473586 |
| KEGG | hsa05146 | Amoebiasis | 3 | 0.002473586 |
| KEGG | hsa04933 | AGE-RAGE signaling pathway in diabetic complications | 3 | 0.002473586 |
| KEGG | hsa04611 | Platelet activation | 3 | 0.003235067 |
| KEGG | hsa04512 | ECM-receptor interaction | 2 | 0.026270733 |
| B, Downregulated | ||||
| Ontology | ID | Function | Count | FDR |
| BP | GO:0055078 | Sodium ion homeostasis | 2 | 0.164234693 |
| BP | GO:0042391 | Regulation of membrane potential | 3 | 0.164234693 |
| BP | GO:0034220 | Ion transmembrane transport | 4 | 0.164234693 |
| BP | GO:0007416 | Synapse assembly | 2 | 0.164234693 |
| BP | GO:0035725 | Sodium ion transmembrane transport | 2 | 0.164234693 |
| CC | GO:1902495 | Transmembrane transporter complex | 3 | 0.02856848 |
| CC | GO:1990351 | Transporter complex | 3 | 0.02856848 |
| CC | GO:0098794 | postsynapse | 3 | 0.02856848 |
| CC | GO:0030424 | Axon | 3 | 0.02856848 |
| CC | GO:0030426 | Growth cone | 2 | 0.041273443 |
| MF | GO:0015075 | Ion transmembrane transporter activity | 4 | 0.011272955 |
| MF | GO:0022891 | Substrate-specific transmembrane transporter activity | 4 | 0.011272955 |
| MF | GO:0022857 | Transmembrane transporter activity | 4 | 0.011272955 |
| MF | GO:0005216 | Ion channel activity | 3 | 0.011272955 |
| MF | GO:0046873 | Metal ion transmembrane transporter activity | 3 | 0.011272955 |
FDR, false discovery rate; Count, number of enriched genes; GO, gene ontology; BP, biological process; MF, molecular function; CC, cellular component; KEGG, Kyoto Encyclopedia of Genes and Genomes.
RT-qPCR verification of the expression of lncRNAs
According to the findings of the transcriptome sequencing and bioinformatics analyses, AC016735.2, RP11-243M5.2, RP11-400N13.2, RP11-400N13.3, AP001626.1, LINC01139 and RP11-54H7.4 were upregulated, and only RP11-243M5.2 was downregulated.
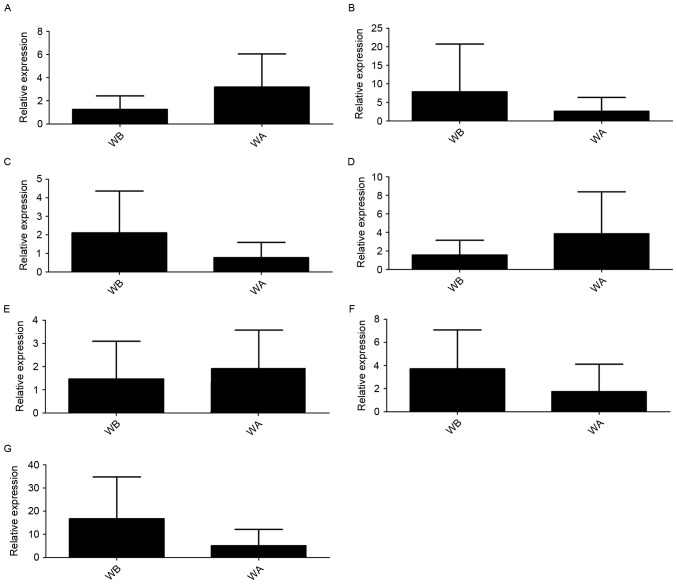
The RT-qPCR validation confirmed that AC016735.2, AP001626.1 and RP11-400N13.3 were upregulated, whereas RP11-243M5.2, RP11-400N13.2, LINC01139 and RP11-54H7.4 were downregulated in GC tissues compared with normal adjacent tissues. It is of note that although upregulation and downregulation were detected, no significant difference was identified (Fig. 3). The findings of AC016735.2, AP001626.1, RP11-400N13.3 and RP11-243M5.2 were considered to be consistent with the predicted lncRNAs in the bioinformatics analysis.
Figure 3.
LncRNA levels verified by reverse transcription-quantitative polymerase chain reaction. (A) AC016735.2, (B) RP11-243M5.2, (C) RP11-400N13.2, (D) RP11-400N13.3, (E) AP001626.1, (F) LINC01139 and (G) RP11-54H7.4. WA, cancer tissue; WB, normal adjacent tissues.
Discussion
Previous studies have identified lncRNAs to be important in the governing of fundamental biological processes, where aberrant expression may be associated with various human cancers (22,23). The present study identified 390 differentially expressed lncRNAs between GC and normal adjacent tissues via transcriptome sequencing and bioinformatics analysis. The upregulated lncRNAs were significantly enriched in functions associated with collagen fibril organization, whereas the downregulated lncRNA was significantly associated with ion transmembrane transport and regulation of membrane potential. Following RT-qPCR validation, AC016735.2, AP001626.1, RP11-400N13.3 and RP11-243M5.2 were considered to be consistent with the results of the bioinformatics prediction, suggesting that they may have a role in the tumorigenesis of GC.
RP11-400N13.3, AP001626.1 and AC016735.2 were all upregulated lncRNAs, and were predicted to regulate >10 target genes, including collagen type I a 1 (COL1A1), COL1A2, and arachidonate 15-lipoxygenase (ALOX15). It is of note that the three target genes were also DEGs. COL1A1 and COL1A2 encode type I collagen, which is the most abundant collagen of the human body that forms collagen fibers. COL1A1 and COL1A2 were identified to be significantly enriched in GO function associated with collagen fibril organization. Type I collagen has an important role in fibrosis and cancer progression (24). A previous study determined that collagen is a major contributor to diffusive hindrance in human tumors (25). Additionally, type I collagen is a prevalent component of the stromal extracellular matrix (26). The stromal extracellular matrix is a barrier to a progressing cancer cell, changes of which contribute to metastasis in cancer (27). Therefore, it is possible that the three lncRNAs may be involved in GC metastasis by regulating COL1A1 and COL1A2.
ALOX15 encoding protein is part of the lipoxygenases family. Human lipoxygenases are widely distributed in human organs, tissues and cells (28), catalyzing peroxidation of unsaturated fatty acid producing various types of eicosanoids (29). It has been previously reported that many cancers are driven by lipoxygenases and their metabolites (30,31). A previous study determined that ALOX15 is an important factor in the regulation of colorectal epithelial cell terminal differentiation and apoptosis (32). In the current study, ALOX15 was significantly enriched in engulfment of apoptotic cell (GO:0043652), which may confirm its role in apoptosis. Therefore, RP11-400N13.3, AP001626.1 and AC016735.2 may also regulate ALOX15 and have a role in the progression of GC.
RP11-243M5.2 was a downregulated lncRNA and had 12 target genes, including ATPase Na+/K+ transporting subunit a 4 (ATP1A4) and sodium voltage-gated channel a subunit 7 (SCN7A), which were significantly involved in functions associated with ion homeostasis and ion transmembrane transport. It has been previously established that cells require a balance of ions across their cell membrane in order to ensure cell survival. The homeostatic intracellular ionic environment is necessary for the correct functioning of gene expression, hormone release and cellular proteins (33,34). It is of note that Bortner and Cidlowski (35) have reported that intracellular ion homeostasis has an important role in the regulation of the cell death and changes may alter the apoptotic rate of cells. Evading apoptosis by generating genetic mutations is a key mechanism of carcinogenesis (36). Therefore, the present study suggested that RP11-243M5.2 may have a role in the carcinogenesis of GC by regulating ATP1A4 and SCN7A to participate in functions associated to ion homeostasis and ion transmembrane transport.
AC016735.2, AP001626.1, RP11-400N13.3 and RP11-243M5.2 were verified by RT-qPCR; however, the results were not statistically significant. This may be due to the heterogeneity between the samples used in the transcriptome sequencing and RT-qPCR.
In conclusion, by transcriptome sequencing and RT-qPCR experiments the present study identified 4 lncRNAs, including AC016735.2, AP001626.1, RP11-400N13.3 and RP11-243M5.2 to have an important role in the pathogenesis of GC. They may be used as potential diagnosis or treatment biomarkers of GC in the future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YW drafted the manuscript and acquired and analysed the data. JZ interpreted the data and revised the manuscript.
Ethics approval and consent to participate
The procedures in the current study were approved by the Protection of Human Ethics Committee of Shanghai Shuguang Hospital Affiliated with Shanghai University of TCM (Shanghai, China).
Consent for publication
All patients have provided written informed consent prior to participating in the present study.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Li H, Yu B, Li J, Su L, Yan M, Zhu Z, Liu B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318–2329. doi: 10.18632/oncotarget.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.IARC: World Cancer Report 2014. World Health Organization; Geneva: 2015. [Google Scholar]
- 3.Ruddon RW. Cancer Biology. Oxford Univ Pr. 2007 [Google Scholar]
- 4.Orditura M, Galizia G, Sforza V, et al. Treatment of gastric cancer. World J Gastroenterol. 2014:1635–1649. doi: 10.3748/wjg.v20.i7.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.ENCODE Project Consortium, corp-author. Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson JM, Edwards S, Shoemaker D, Schadt EE. Dark matter in the genome: Evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 2005;21:93–102. doi: 10.1016/j.tig.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 8.Furuno M, Pang KC, Ninomiya N, Fukuda S, Frith MC, Bult C, Kai C, Kawai J, Carninci P, Hayashizaki Y, Mattick JS, Suzuki H. Clusters of internally primed transcripts reveal novel long noncoding RNAs. PLoS Genet. 2006;2:e37. doi: 10.1371/journal.pgen.0020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shore AN, Herschkowitz JI, Rosen JM. Noncoding RNAs Involved in mammary gland development and tumorigenesis: There's a long way to go. J Mammary Gland Biol Neoplasia. 2012;17:43–58. doi: 10.1007/s10911-012-9247-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitehead J, Pandey GK, Kanduri C. Regulation of the mammalian epigenome by long noncoding RNAs. Biochim Biophys Acta. 2009;1790:936–947. doi: 10.1016/j.bbagen.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 12.Shao Y, Chen H, Jiang X, Chen S, Li P, Ye M, Li Q, Sun W, Guo J. Low expression of lncRNA-HMlincRNA717 in human gastric cancer and its clinical significances. Tumour Biol. 2014;35:9591–9595. doi: 10.1007/s13277-014-2243-z. [DOI] [PubMed] [Google Scholar]
- 13.Song H, Sun W, Ye G, Ding X, Liu Z, Zhang S, Xia T, Xiao B, Xi Y, Guo J. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med. 2013;11:225. doi: 10.1186/1479-5876-11-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Hauptman N, Glavač D. Long non-coding RNA in cancer. Int J Mol Sci. 2013;14:4655–4669. doi: 10.3390/ijms14034655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu MT, Hu JW, Yin R, Xu L. Long noncoding RNA: An emerging paradigm of cancer research. Tumour Biol. 2013;34:613–620. doi: 10.1007/s13277-013-0658-6. [DOI] [PubMed] [Google Scholar]
- 24.Mueller MM, Fusenig NE. Friends or foes-bipolar effects of the tumour stroma in cancer. Nat Rev Cancer. 2004;4:839–849. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- 25.Ramanujan S, Pluen A, Mckee TD, Brown EB, Boucher Y, Jain RK. Diffusion and convection in collagen gels: Implications for transport in the tumor interstitium. Biophys J. 2002;83:1650–1660. doi: 10.1016/S0006-3495(02)73933-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keely PJ, Wu JE, Santoro SA. The spatial and temporal expression of the alpha 2 beta 1 integrin and its ligands, collagen I, collagen IV, and laminin, suggest important roles in mouse mammary morphogenesis. Differentiation. 1995;59:1–13. doi: 10.1046/j.1432-0436.1995.5910001.x. [DOI] [PubMed] [Google Scholar]
- 27.Stewart DA, Cooper CR, Sikes RA. Changes in extracellular matrix (ECM) and ECM-associated proteins in the metastatic progression of prostate cancer. Reprod Biol Endocrinol. 2004;2:2. doi: 10.1186/1477-7827-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berglund L, Björling E, Oksvold P, Fagerberg L, Asplund A, Szigyarto CA, Persson A, Ottosson J, Wernérus H, Nilsson P, et al. A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol Cell Proteomics. 2008;7:2019–2027. doi: 10.1074/mcp.R800013-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Haeggström JZ, Funk CD. Lipoxygenase and leukotriene pathways: Biochemistry, biology, and roles in disease. Chem Rev. 2011;111:5866–5898. doi: 10.1021/cr200246d. [DOI] [PubMed] [Google Scholar]
- 30.Skrzypczakjankun E, Chorostowskawynimko J, Selman SH, Jankun J. Lipoxygenases-A challenging problem in enzyme inhibition and drug development. Current Enzyme Inhibition. 2007;3:119–132. doi: 10.2174/157340807780598350. [DOI] [Google Scholar]
- 31.Gohara A, Eltaki N, Sabry D, Murtagh D, Jr, Jankun J, Selman SH, Skrzypczak-Jankun E. Human 5-, 12- and 15-lipoxygenase-1 coexist in kidney but show opposite trends and their balance changes in cancer. Oncol Rep. 2012;28:1275–1282. doi: 10.3892/or.2012.1924. [DOI] [PubMed] [Google Scholar]
- 32.Shureiqi I, Wu Y, Chen D, Yang XL, Guan B, Morris JS, Yang P, Newman RA, Broaddus R, Hamilton SR, et al. Critical Role of 15-Lipoxygenase-1 in colorectal epithelial cell terminal differentiation and tumorigenesis. Cancer Res. 2011;65:11486–11492. doi: 10.1158/0008-5472.CAN-05-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lang F, Ritter M, Gamper N, Huber S, Fillon S, Tanneur V, Lepple-Wienhues A, Szabo I, Gulbins E. Cell volume in the regulation of cell proliferation and apoptotic cell death. Cell Physiol Biochem. 2000;10:417–428. doi: 10.1159/000016367. [DOI] [PubMed] [Google Scholar]
- 34.Lang F, Busch GL, Ritter M, Völkl H, Waldegger S, Gulbins E, Häussinger D. Functional significance of cell volume regulatory mechanisms. Physiol Rev. 1998;78:247–306. doi: 10.1152/physrev.1998.78.1.247. [DOI] [PubMed] [Google Scholar]
- 35.Bortner CD, Cidlowski JA. The role of apoptotic volume decrease and ionic homeostasis in the activation and repression of apoptosis. Pflugers Arch. 2004;448:313–318. doi: 10.1007/s00424-004-1266-5. [DOI] [PubMed] [Google Scholar]
- 36.Su Z, Yang Z, Xu Y, Chen Y, Yu Q. Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol Cancer. 2015;14:48. doi: 10.1186/s12943-015-0321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.