Abstract
Purpose
Keratoconus (KC) is the most common corneal ectasia. We aimed to determine the differential expression of coding and long noncoding RNAs (lncRNAs) in human corneas affected with KC.
Methods
From the corneas of 10 KC patients and 8 non-KC healthy controls, 200 ng total RNA was used to prepare sequencing libraries with the SMARTer Stranded RNA-Seq kit after ribosomal RNA depletion, followed by paired-end 50-bp sequencing with Illumina Sequencer. Differential analysis was done using TopHat/Cufflinks with a gene file from Ensembl and a lncRNA file from NONCODE. Pathway analysis was performed using WebGestalt. Using the expression level of differentially expressed coding and noncoding RNAs in each sample, we correlated their expression levels in KC and controls separately and identified significantly different correlations in KC against controls followed by visualization using Cytoscape.
Results
Using |fold change| ≥ 2 and a false discovery rate ≤ 0.05, we identified 436 coding RNAs and 584 lncRNAs with differential expression in the KC-affected corneas. Pathway analysis indicated the enrichment of genes involved in extracellular matrix, protein binding, glycosaminoglycan binding, and cell migration. Our correlation analysis identified 296 pairs of significant KC-specific correlations containing 117 coding genes enriched in functions related to cell migration/motility, extracellular space, cytokine response, and cell adhesion. Our study highlighted the potential roles of several genes (CTGF, SFRP1, AQP5, lnc-WNT4-2:1, and lnc-ALDH3A2-2:1) and pathways (TGF-β, WNT signaling, and PI3K/AKT pathways) in KC pathogenesis.
Conclusions
Our RNA-Seq–based differential expression and correlation analyses have identified many potential KC contributing coding and noncoding RNAs.
Keywords: keratoconus, RNA-seq, coding RNAs, LncRNAs, cornea
Keratoconus (KC) is the most common corneal ectatic disorder.1,2 KC is characterized by bulging and thinning cone-shaped cornea, resulting in myopia, irregular astigmatism, and, eventually, visual impairment. KC usually starts at puberty and progresses until the third or fourth decade.3,4 KC is a complex multifactorial disorder with the contribution from both genetic and environmental factors as well as biomechanical, metabolic, and hormonal factors.5–10 KC patients report significantly impaired vision-related quality of life, which worsens with time due to disease progression.11
RNA sequencing (RNA-Seq) allows the entire transcriptome of coding and/or noncoding RNAs to be surveyed in a high-throughput and quantitative manner.12–15 Long noncoding RNAs (lncRNAs) are class of noncoding RNAs that are at least 200 nucleotides long,16,17 and have been found to regulate gene expression transcriptionally and post-transcriptionally in both physiologic and pathologic conditions.17,18
Previous studies have attempted to understand processes that underlie KC using different approaches. Wentz-Hunter et al.19 have performed targeted expression analysis of the stromal layer of the cornea to identify differentially expressed transcripts in KC versus a healthy cornea. Soon afterward, microarray was used to assess RNAs isolated from either the KC-affected epithelium or keratocytes.20,21 By 2005, differentially expressed proteins in KC patients have been investigated using the whole cornea, certain layer, or tears.22–25 In 2017, Szcześniak et al.26 and Kabza et al.27 used RNA-Seq to determine differentially expressed RNAs between KC-affected and other disease affected corneas.
For the first time, we used Total RNA-Seq to identify differentially expressed coding/noncoding RNAs in KC-affected corneas versus nondiseased healthy ones, and to determine the expression correlation. We used droplet digital PCR (ddPCR) to validate the differential expression of selected RNAs. These differentially expressed RNAs may provide a better understanding of the functions of related lncRNAs and the potential pathways involved in KC pathogenesis.
Methods
Human Cornea Samples and RNA Extraction
We followed the Tenets of Declaration of Helsinki. Unaffected human cornea samples were collected from postmortem donors from North Carolina Eye Bank, and the KC-affected corneal samples were collected during corneal transplantation surgery at the Cedars-Sinai Medical Center (CSMC). The institutional research board offices at Duke University School of Medicine and CSMC approved both studies, respectively. Written informed consent was obtained from all CSMC patients. This study included 10 KC-affected corneas and eight healthy non-KC corneas (Table 1). The inclusion and exclusion criteria for KC diagnosis have been described previously.28,29 The diagnosis of KC was done by a cornea fellowship–trained ophthalmologist based on clinical examination and confirmed with videokeratography. Clinical examination included slit-lamp biomicroscopy, cycloplegic retinoscopy, and fundus evaluations. The slit-lamp biomicroscope was used to identify stromal thinning, Vogts' striae, and Fleischer ring. The retinoscopy examination was performed with a fully dilated pupil (20 minutes after phenylephrine 2.5% and cyclopentolate 1% drops had been instilled in the eye) to determine the presence or absence of retro illumination signs of KC, such as the oil droplet sign and scissoring of the red reflex. Videokeratography evaluation was performed on each eye using the Topographic Modeling System (TOMEY, Nagoya, Japan). Subjects were assigned as having KC if they had at least one clinical sign and a confirmatory videokeratography map with an asymmetric bowtie with skewed radial axis above and below the horizontal meridian (AB/SRAX) pattern.30 KC grade was assigned based on the average central keratometry readings as follows: mild: <50 diopters (D); moderate: >50 D but < 55D; moderate-severe: 55 to 60 D; severe: >60 D.
Table 1.
Clinical Phenotypes of Patients With KC and Healthy Controls
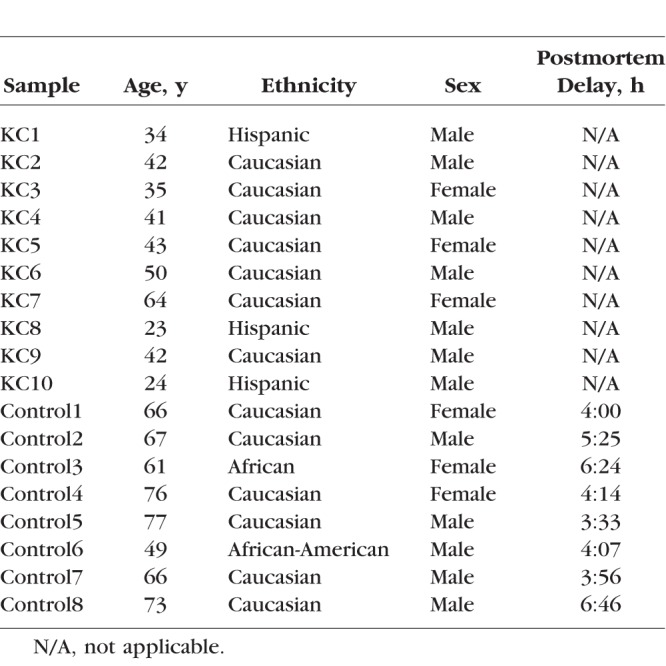
Only KC patients at the advanced stage of KC from whom all other therapeutic interventions have failed were included in this study. They were diagnosed with KC grade of above 55 D (based on the average central keratometry readings). Half of the corneal button removed from surgery was used for RNA extraction while the other half was sent for pathologic examination. The postmortem donors had no other ocular disorders affecting their cornea. These cornea samples were excluded from usage in corneal transplant mostly due to the lack or incomplete medical/health information or occasionally low number of corneal endothelial cells. The central button of the donor corneas was acquired using a 6-mm biopsy punch (Cat#33-38, Miltex, Inc., York, PA, USA) and the whole button was used for RNA isolation. The cornea was stored in RNALater solution (Qiagen, Valencia, CA, USA) to preserve RNA quality. Total RNA was extracted using mirVana miRNA isolation kit from ThermoFisher Scientific, and with NucleoSpin RNA/Protein isolation kit (MACHEREY-NAGEL, Inc., Bethlehem, PA, USA), according to the manufacturer's recommendations. RNA quality was evaluated using Agilent 2100 Bioanalyzer with Agilent's RNA 6000 Pico Kit (Agilent, Santa Clara, CA, USA). We selected RNA samples with a minimum RIN (RNA Integrity Number) score of 6 for RNA-Seq.
Stranded Total RNA-Seq
We used 200 ng total RNA from all subjects to prepare sequencing libraries with the TaKaRa SMARTer Stranded RNA-Seq kit after ribosomal RNA depletion using the RiboGone - Mammalian kit from TaKaRa (TaKaRa Bio USA, Inc., Mountain View, CA, USA). Briefly, the RiboGone-Mammalian kit removes ribosomal RNA (rRNA) and mitochondrial RNA (mtRNA) sequences from human total RNA samples based on hybridization and RNase H digestion that specifically depletes 5S, 5.8S, 18S, and 28S nuclear rRNA sequences, as well as 12S mtRNA sequences. The rRNA-depleted total RNA was reverse-transcribed to synthesize the first-strand complementary (c) DNA followed by PCR amplification using universal forward PCR primer and reverse PCR indexing primer set. Purified RNA-Seq library was validated using the Agilent 2100 Bioanalyzer with Agilent's High Sensitivity DNA Kit (Agilent, Santa Clara, CA, USA). Pooled RNA-Seq libraries were sequenced with paired-end 50-bp reads using an Illumina HiSeq 2500 sequencer (Illumina, Inc., San Diego, CA, USA) at the Georgia Cancer Center Integrated Genomics Core of Augusta University.
After quality check and control with all sequencing reads, demultiplexed reads were aligned by TopHat31 using paired-end reading with the approximation of the median library size. Counts of sequencing reads were normalized using Cufflinks in fragments per kilo bases and millions reads (FPKM).32 After normalization, we annotated the transcripts with a gene transcript file from Ensembl database at the gene isoform level. We performed differential expression analysis using the Cuffdiff package.33 We used a fold change of ≥ 2 and false discovery rate (FDR) ≤ 0.05 to identify genes with significant expression changes in KC corneas versus healthy ones. The list of differentially expressed genes was uploaded to the Web-based Gene Set Analysis Toolkit (WebGestalt; available in the public domain, www.webgestalt.org) 2013 version for gene ontology and KEGG pathway analysis.34 For lncRNAs, we annotated them using a file from the NONCODE version 5 database.35 After normalization using Cufflinks, we performed differential expression analysis using Cuffdiff. For lncRNA data, missing expression data in specific samples were replaced with a background value of 0.001 to enable the differential analysis between cases and controls. Without such replacement of missing data, the related lncRNAs would have been excluded from the differential expression. For lncRNA differential expression analysis, to avoid the possible impact of potential DNA contamination and RNA isolation bias, we required a minimal size of 300 bp for all the lncRNAs. We used |fold change| ≥ 2 and FDR ≤ 0.05 to identify lncRNAs with significant expression changes in KC corneas versus healthy ones.
Validation Using Droplet Digital PCR Technique
Twenty-one differentially expressed coding RNAs were chosen for validation with ddPCR as previously described.36,37 We selected these coding RNAs based on the following criteria: (1) reasonably high corneal expression with ≥ 10 normalized read counts; (2) relevance to corneal integrity, transparency, and cellular adhesion; (3) potential involvement in oxidative stress; and (4) related with corneal wound healing. Specific EvaGreen-based ddPCR assays were obtained from Bio-Rad Laboratories, Inc. (Hercules, CA, USA). To validate the differentially expressed lncRNAs, we selected seven lncRNAs with reasonable corneal expression (≥10 normalized read counts) and strong correlations with various other coding and lncRNAs. Specific ddPCR expression assays with specific probes were designed and ordered using the online ddPCR assay design tool with the specific RNA sequence of each selected lncRNA for copy number determination (available in the public domain, https://www.bio-rad.com/digital-assays/#/). We used RNA samples from the Total RNA-Seq experiment for validation: eight KC-affected corneas and eight control corneas. The ddPCR assays were performed using the QX200 Droplet Digital PCR system (Bio-Rad, Hercules, CA, USA) and its associated QuantaSoft Software, as previously described.28,29 We used four internal reference genes (GAPDH, PPIA, HPRT1, and RPLP0)38 to be quantified in the samples for normalization purpose. All four reference genes were stably expressed across the KC and control cornea samples and had similar performance in expression normalization, leading to almost identical successful validation of selected target genes. Thus, we decided to normalize with GAPDH for the figure purpose. Mann-Whitney U test was used to analyze the differential expression data from ddPCR for the statistical analysis.
Expression Correlation and Network Analysis
The expression levels of differentially expressed coding RNAs and lncRNAs in all 18 corneal samples were imported into RStudio Version 0.98.1062 (RStudio, Boston, MA, USA)39 from CSV raw data files. The combined expression data was structured into a table where the rows were the mapped RNA names and the columns were the case or control individual de-identified with a code name. Repeated gene names were removed and genes with no expression levels in all controls or cases were also removed. The correlation between all RNA expression levels was determined for the controls data and the cases data separately using R's built-in Pearson correlation function (cor). Using R psych package's test of significance for correlations (r.test), the P value comparing the respective mapped gene's control and case correlations were calculated. The FDR was calculated from the P values using the R's built-in Benjamini–Hochberg procedure function (p.adjust with “BH” parameter). We removed redundant correlations between each gene and itself as well as duplicated gene correlations (A-B is the same as B-A).
The list of expression correlations was filtered with the criteria of an FDR ≤ 0.05 and having at least a mean expression level ≥ 20 in the cases or the controls with one gene. The entire filtered list of correlated RNAs including lncRNAs was visualized using Cytoscape version 3.5.1 (Cytoscape Consortium, New York, NY, USA).40 Expression network hubs were defined as network nodes with multiple correlations to other RNAs. It was expected that this correlation expression analysis among coding RNAs and lncRNAs would help identify biologically important lncRNAs in human cornea. The list of correlated coding RNAs was loaded to WebGestalt 2013 version for gene ontology and KEGG pathway analyses.34
Results
Identification of Differentially Expressed Coding RNAs
Each sample generated 20 to 44 million paired reads with 100% sample index match, > 96% bases with Q30, and mean quality score > 38. Overall, 62% to 88% of sequencing reads for all RNA samples were successfully mapped to the transcriptome file from Ensembl database, containing a total of 212,368 transcript isoforms. There were a total 95,954 and 103,440 expressed transcripts in all the healthy controls and KC-affected cornea samples, respectively. The number of expressed transcripts in each sample ranged from 120,000 to 139,000. Using a cutoff of at least one count of normalized transcripts, the number of expressed transcripts in each sample ranged from 35,000 to 45,000. There is a total of 14,111 and 19,608 expressed transcripts (≥1 count) in all the healthy controls and KC-affected corneal samples, respectively. The mean expression levels of these shared transcripts (≥1 count) were 142 reads in control corneas and 63 read counts in the keratoconic corneas.
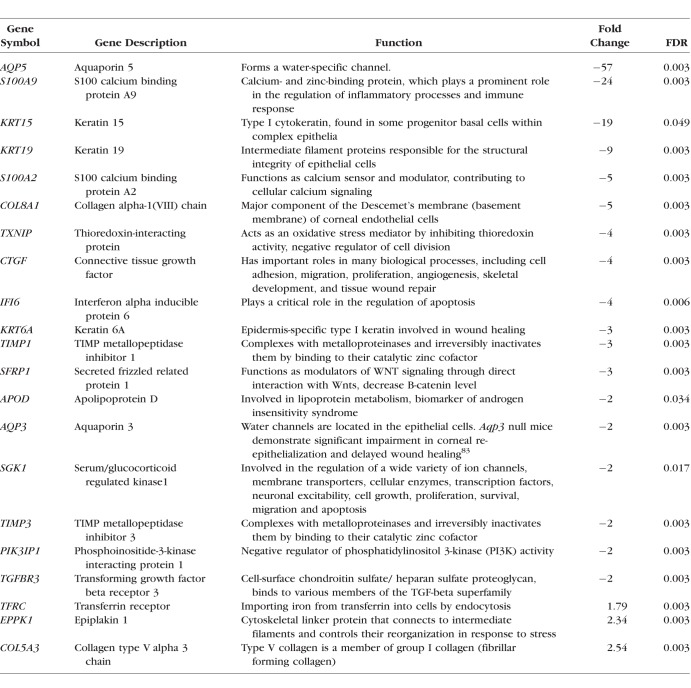
Using the cutoff with |fold change| ≥ 2 and FDR ≤ 0.05, we identified 436 differentially expressed coding RNAs (159 upregulated and 277 downregulated; Supplementary Table S1). Our analysis was consistent with previous expression studies by identifying differential expression of many known KC-related genes, including but not limited to TGFBR3, TIMP1, FBLN1, TIMP3, AQP5, and SFRP1. The top 20 up- and downregulated coding genes, showing the highest fold changes, were listed in Table 2.
Table 2.
Top Coding Genes With Significant Differential Expression in KC-Affected Corneas
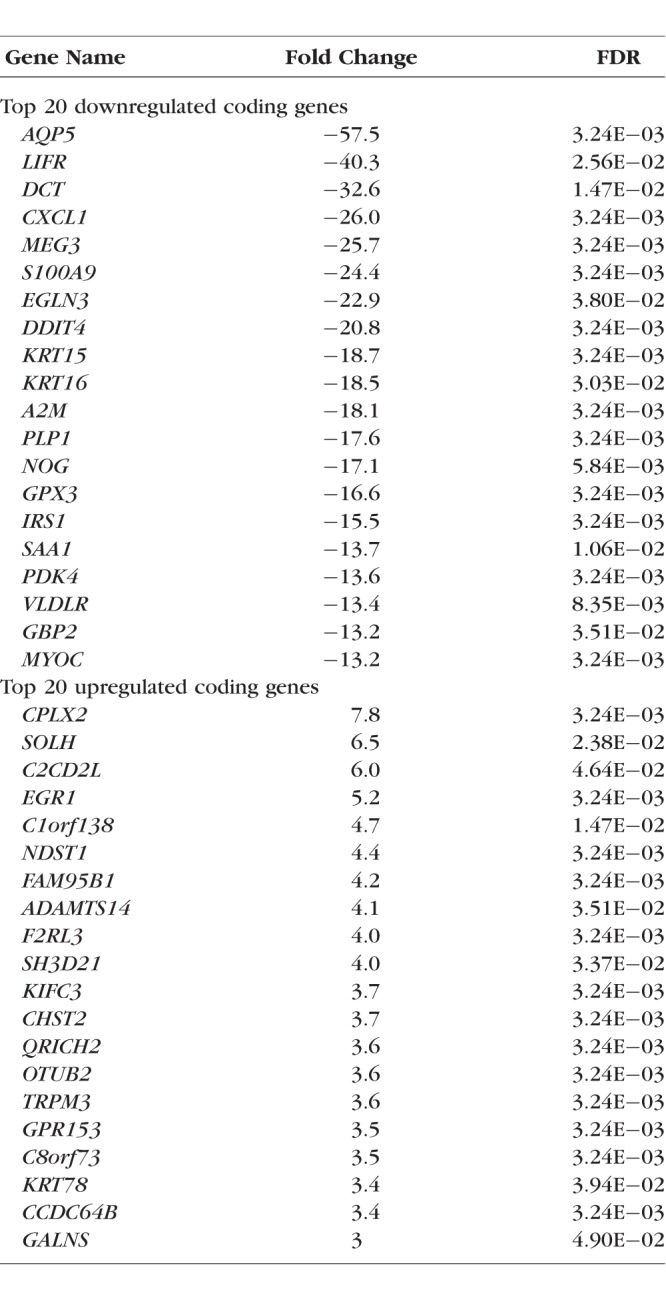
Using WebGestalt with the differentially expressed coding RNAs, our gene ontology analysis revealed disruption in different biological processes (cellular proliferation, differentiation, locomotion, migration, cellular stress, and wound response), molecular functions (glycosaminoglycan binding, iron ion binding, antioxidant activity, and insulin-like growth factor binding), and cellular components (adherens junctions, extracellular matrix region, and basement membrane) in the keratoconic corneas.
Validation of Differentially Expressed Coding RNAs
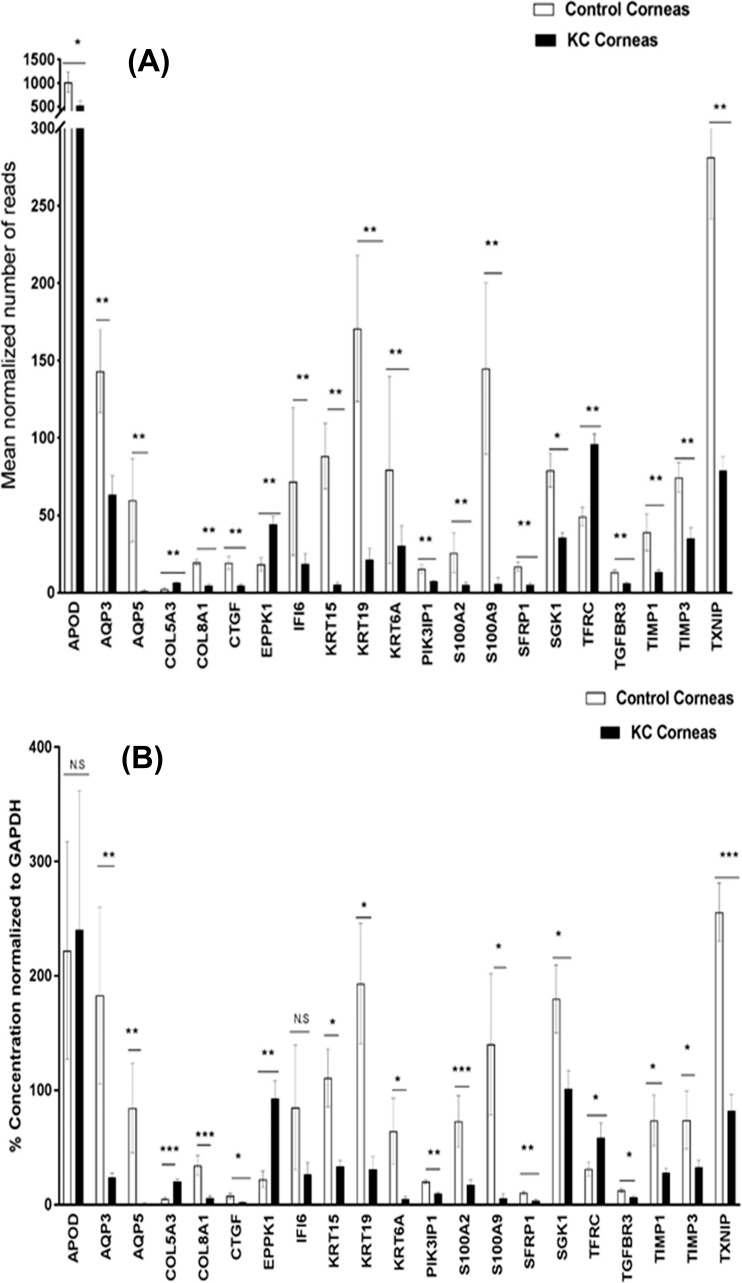
Out of 436 differentially expressed coding genes, we selected 21 genes for validation by ddPCR based on the criteria described earlier (Table 3). Of these 21 genes, 19 were confirmed to have significant and similar expression trend to the RNA-Seq results (Figs. 1A, 1B). The other two genes (APOD and IFI6) demonstrated similar expression trends but did not reach statistical significance.
Table 3.
Twenty-One Differentially Expressed Coding Genes to be Validated Using ddPCR
Figure 1.
Differentially expressed coding genes in KC-affected human corneas identified by RNA sequencing (A) and ddPCR (B). For panel A, **FDR < 0.01, *0.01< FDR < 0.05. For panel B, ***P value < 0.001, **P value < 0.01, * 0.01< P value < 0.05. Error bars: panels A and B represent standard error.
Identification of Differentially Expressed lncRNAs
A total of 15,966 lncRNAs were expressed in the control and keratoconic cornea samples. Using the cutoff with |fold change| ≥ 2 and FDR ≤ 0.05, we identified 586 differentially expressed lncRNAs (343 upregulated and 243 downregulated; Supplementary Table S2). Next, differentially expressed coding RNAs and lncRNAs were used to perform a correlation analysis.
Validation of Differentially Expressed lncRNAs
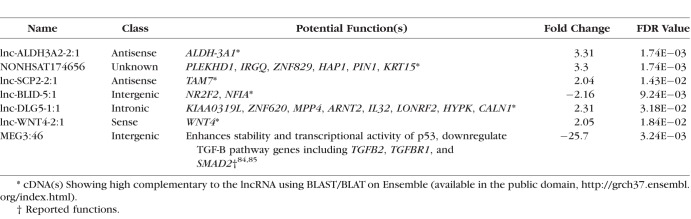
Based on their high cornea expression (≥10 normalized read counts), functional annotation in relation to KC-related pathways, and expression correlation with coding and lncRNAs, we selected seven differentially expressed lncRNAs for ddPCR validation (Table 4). Specific probe-based ddPCR assays with fluorescein dye (FAM) labeling were designed to validate the differential expression of these lncRNAs in KC-affected corneas versus the controls (Fig. 2A). The four selected internal reference genes were also used to normalize the expression of these lncRNAs. Five lncRNA showed a similar trend to RNA-Seq results (lnc-WNT4-2:1, MEG3, lnc-BLID-5:1, lnc-SCP2-2:1, and lnc-ALDH3A2-2:1). However, NONHSAT174656 and lnc-DLG-1:1 did not show any significant differential expression in KC-affected corneas versus controls (Figs. 2A, 2B).
Table 4.
Seven Differentially Expressed lncRNAs to be Validated Using ddPCR
Figure 2.
Differentially expressed lncRNAs in KC-affected human corneas identified using by RNA-Seq (A) and ddPCR (B). For panel A, **FDR < 0.01, *0.01 < FDR < 0.05. For panel B, ***P value < 0.001, **P value < 0.01, *0.01< P value < 0.05. Error bars: panels A and B represent standard error.
Correlation Analysis of Coding and Noncoding RNAs
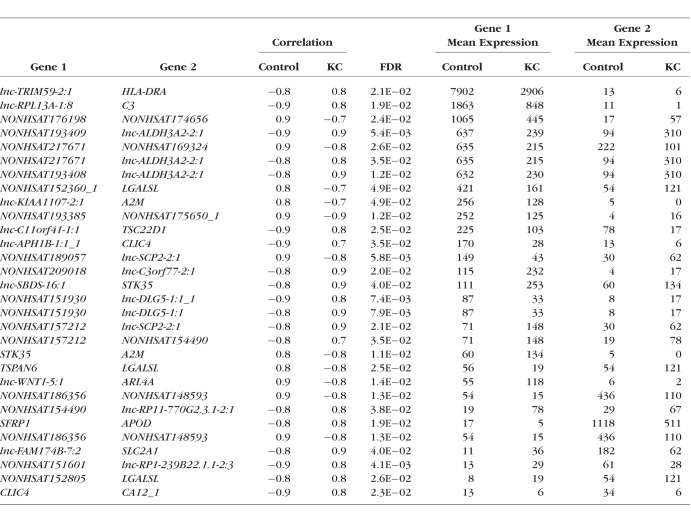
The expression correlation analysis identified a total of 492,135 pairs of RNAs with expression correlations in cases or controls. After comparing the correlations between cases and controls, a total of 352 significant differential correlations were identified with a FDR ≤ 0.05. After applying a minimum mean expression of 20 for at least one of the two paired transcripts in cases or controls, we identified 296 pairs of significant differential correlations between cases and controls (Supplementary Table S3). Many coding and noncoding RNAs were identified in multiple correlation pairs. These included A2M, APOD, MUC16, lnc-SCP2-2:1, lnc-DLG5-1:1, lnc-BLID-5:1, lnc-ALDH3A2-2:1, NONHSAT175650_1, and NONHSAT217671 (Fig. 3). We used Cytoscape to visualize the whole expression network of these 296 pairs (Supplementary Fig. S1). This network analysis revealed a number of genes with significant correlations in the keratoconic corneas versus the unaffected healthy controls (Table 5). We identified 117 coding RNAs in the significantly differential expression correlations. These 117 genes were enriched for their functions in negative regulation of cell migration and cell motility, extracellular space, cellular response to cytokine stimulus, cell surface receptor signaling pathway, and regulation of cell adhesion.
Figure 3.
Correlation networks of the differentially expressed coding RNAs and lncRNAs displayed by Cytoscape. The nodes represent mapped RNAs colored and sized by the degree of correlations with other RNAs. The edges are sized and colored based on the significant differences (i.e., FDR) between controls and cases. Correlation networks were shown for the Lnc-ALDH3A2-2:1 (A), APOD (B), CLIC4 (C), SLC2A1 (D), and A2M (E) genes.
Table 5.
Coding and lncRNAs With Significant Differences in Expression Correlation (With a Reasonable Corneal Expressions) in KC-Affected Corneas Versus the Controls
Discussion
KC is a complex disorder with both genetic and environmental factors. Epithelial and stromal layers of the cornea show the prominent histopathologic changes during KC progression. A better understanding of the key molecular players involved in KC-associated corneal pathogenesis may provide potential therapeutic targets for disease treatment. Researchers used various techniques including RNA-Seq, microarray, RT-PCR, mass spectrometry, and Western blots.20,21,25,27,41,42 Recently, Kabza et al.27 have used RNA-Seq for KC-affected corneas; however, this study was complicated by the use of other diseased corneas as controls. Compared with this study, our data have shown overlapping genes with differential expression (35 up- and 135 downregulated genes27; Supplementary Table S4).
We identified decreased expression in the cytokeratin (KRT6A, KRT13, KRT15, and KRT19) and collagen (COL8A1 and COL8A2) genes, indicating compromised integrity of KC cornea. Type VIII collagens are normally expressed in the Descemet's membrane43 and targeted inactivation of Col8a1/Col8a2 leads to corneal abnormalities in mice.44
Our study confirms the potential role of oxidative stress in KC-affected corneas by identifying differential expression of many genes involved in response to oxidative stress including GSTO1, SGK1, APOD, KLF9, GPX3, STK39, TXNIP, and lnc-ALDH3A2-2:1.45,46 The majority of these genes in defense mechanism for oxidative stress were downregulated. However, lnc-ALDH3A2-2:1 is found to have more than 3-fold increased expression and correlated to other lncRNAs (Fig. 3A). Lnc-ALDH3A2-2:1, according to Lncipedia database,47 is classified as an antisense transcript that overlaps with the protein coding gene ALDH3A2 on the opposite strand. However, using BLASTN, we found that lnc-ALDH3A2-2:1 sequence overlaps with the last exon of ALDH3A1 gene. The ALDH3A1 transcript did not show significant differential expression in our study; however, lncRNAs may alter protein level without affecting mRNA.48 Another factor impacting oxidative stress is disruption of iron homeostasis, which can lead to various corneal diseases including KC.23,24,49,50 Fleisher ring, a ring of iron deposition in the corneal cone, is a typical clinical sign for KC.51 Importantly, transferrin receptor (TFRC), a cellular receptor for iron-ferritin complex uptake,52 has been identified to have elevated expression in KC cornea in our study. This elevated expression in KC-affected corneas may result from iron deficiency.53 We also found downregulation of the Ferritin Light Chain transcript (FTL) in KC corneas. Significantly, mutations in FTL have been linked to iron accumulation in the brain, causing neurodegenerative disease.54
We observed differential expression of many genes involved in corneal wound healing, such as EPPK1, AQP3, AQP5, SLC2A1, and CTGF. Corneal wound healing is a complex process controlled by the integrated actions of multiple growth factors, cytokines, and proteases produced by epithelial cells, stromal keratocytes, and inflammatory cells.55 However, unregulated healing processes, due to genetic or epigenetic factors, can lead to impairment in cellular function.56,57 Macé et al.41 have shown antiproliferative and hyperapoptotic phenotype in KC-affected cornea, which also reveals a defect in wound healing process. Aquaporins (AQPs) are a ubiquitous family of transmembrane water channels to maintain tear film osmolarity, stromal layer thickness, and corneal transparency.58 AQP3 and AQP5, important for corneal wound healing, are highly expressed in the cornea.58 Both AQPs show reduced expressions in the KC-affected corneas (Table 2). Consistent with our results, Rabinowitz et al.59 have reported the lack of AQP5 expression in KC-affected corneas. Although Garfias et al.60 did not report this altered expression, this discrepancy could be due to their heterogeneous controls (normal corneal epithelium, sclerocorneal rims, and healthy corneal buttons). EPPK1 encodes a cytoskeletal linker protein called epiplakin, which maintains the integrity of intermediate filaments networks in epithelial cells.61,62 Migration-dependent wound healing improvement has been found in epiplakin-null epithelium.63 Upregulation of EPPK1 in our study suggests the potential involvement of EPPK1 in the impaired corneal wound healing in KC. Interestingly, solute carrier family 2 member 1 (SLC2A1) is upregulated during corneal epithelium wound healing.64 Reduced expression of SLC2A1 in KC-affected corneas may reflect a lesser glucose uptake. This is intriguing because there is a reduced risk of developing KC in diabetic patients.65–68 Moreover, it has been reported that diabetic patients have thicker cornea than healthy individuals.64
Connective tissue growth factor (CTGF) is expressed in multiple ocular tissues as a downstream signaling molecule in the TGF-β pathway.69 Importantly, loss of CTGF impairs efficient corneal wound healing.70 The decreased expression of CTGF in our study is consistent with previous studies.27,28 Dysregulation of extracellular matrix (ECM) pathway has been reported with KC progression.28,51 Previous studies have shown an upregulation of ECM degradative enzymes and/or downregulation of their inhibitors.27,45,71 Our data show significant downregulation in proteinases inhibitors (TIMP-1, TIMP-3, and A2M, and SLPI), but interestingly no significant change with ECM proteinases.
WNT signaling is essential for normal corneal development. Sequence variants in WNT7B and WNT10A genes have been associated with CCT and KC risk.72,73 A recent RNA-Seq–based study showed dysregulation in WNT signaling in KC-affected corneal epithelium.42 We found some evidence of WNT signaling disruption represented by downregulation of ANGPTL7, TGFBR3, SFRP1 and upregulation of frizzled class receptor 8 (FZD8) and early growth response 1 (EGR1). ANGPTL7 encodes angiopoietin-like 7 and is abundantly expressed in keratocytes to maintain corneal avascularity and transparency.74 SFRP1 inhibits WNT signaling by preventing the formation of the WNT-FZD complex. Iqbal et al.75 have shown an increased expression of SFRP1 in the KC-affected corneal epithelium while SFRP1 expression is reduced in tears from KC patients.76 TGFBR3 suppresses WNT signaling through binding to Wnt3a and β-catenin.77 We observed increased expression of Lnc-WNT4-2:1 in KC versus controls. Our BLASTN analysis indicated that Lnc-WNT4-2:1 is a sense transcript for its overlapping with exon 5 of WNT4 gene. Interestingly, Coding-Potential Assessment Tool (CPAT) using an alignment-free logistic regression model predicts a coding potential for this lncRNA (in the public domain, https://hg19.lncipedia.org/).47 Although no significant change in WNT4 mRNA in KC-affected corneas, this does not rule out the potential role of lnc-WNT4-2:1 in regulating WNT signaling pathway.
Another pathway in KC pathogenesis is the PI3K/AKT pathway. This pathway is involved in cell cycle regulation and cell proliferation in the cornea.78 Our data show downregulation of negative regulators of PI3K/AKT pathway, including PIK3IP1, DDIT4, IGFBP-3, -4, and -5. PIK3IP1 may suppress AKT activity by inhibiting the expression of PI3K.79 PIK3IP1 has been reported to be downregulated in KC-affected corneas.27 Insulin-like growth factor biding proteins (IGFBPs) belong to a family of six structurally similar proteins, which antagonize the binding of insulin growth factors to their receptors, thus, controlling cell survival, mitogenesis, and differentiation.80 Our results indicate that IGFBP-3, -4, and -5 have 10-, 3-, and 8-fold reduced expression in KC corneas, respectively. Our findings are consistent with previous studies in keratocytes isolated from KC-affected corneas compared to healthy corneas.20
For the first time, we have identified pairs of RNAs showing significantly different correlations in KC versus controls (Table 5). Functions of most of the identified genes are still not clear, especially in the cornea. APOD and SFRP1 showed a significant positive correlation in KC, with both being downregulated (Fig. 3B). Both genes were reported to have tumor-suppressive functions, including DNA repair, induction of apoptosis, detoxification, differentiation, and transcriptional regulation.81 Apolipoprotein D (APOD) is a stroma specific protein as lipophilic molecules carrier.82 SFRP1 is a negative regulator of WNT signaling, but it is unclear whether WNT signaling disruption is a cause for the inverse correlation between the two genes (in KC versus controls).
Despite the significant findings, our study has several limitations. First is the relatively small sample size of cases and controls. Second, age, sex, and ethnicity were not matched completely between cases and controls. It will be appropriate to study how age affects the corneal expression in controls so that we can identify KC-specific but not age-related expression alterations. The source of corneal samples (i.e., surgical versus postmortem donors) might potentially contribute to the expression profile difference between KC and controls. Third, this study used whole cornea for expression. It will be important to examine tissue-specific differential expression in the epithelium and stroma separately if possible. Fourth, our expression study is limited by transcript quantification only. Additional proteomics or protein-based studies are necessary to confirm the differential expression in KC-affected corneas.
We have not only confirmed many previous reported KC-related differential expressed genes, but also identified a number of novel coding and noncoding RNAs involved in KC pathogenesis. We have identified overabundance of genes with reduced expression, consistent with the destructive nature of corneal degradation associated with KC-related corneal thinning and with our previous RNA microarray study and another study from Macé et al.28,41
In summary, we have successfully used Total RNA-Seq to identify differentially expressed coding and noncoding RNAs in KC-affected whole-corneal tissues. These RNAs may disturb various corneal functions including cellular adhesion, oxidative stress response, proliferation, migration, apoptosis, and wound healing. Our expression correlation analysis between differentially expressed coding RNAs and lncRNAs has identified a number of potential expression regulators and provided potential functional annotation for lncRNAs with unknown functions.
Supplementary Material
Acknowledgments
The authors thank all the donors for the human corneal tissue samples. Without such samples, this study would not be possible.
Supported by National Institutes of Health (Bethesda, MD, USA) grants R01EY023242, R01EY023646, R01EY009052, and P30 EY005722.
Disclosure: M.L. Khaled, None; Y. Bykhovskaya, None; S.E.R. Yablonski, None; H. Li, None; M.D. Drewry, None; I.F. Aboobakar, None; A. Estes, None; X.R. Gao, None; W.D. Stamer, None; H. Xu, None; R.R. Allingham, None; M.A. Hauser, None; Y.S. Rabinowitz, None; Y. Liu, None
References
- 1. Kang PC,, Klintworth GK,, Kim T,, et al. Trends in the indications for penetrating keratoplasty, 1980-2001. Cornea. 2005; 24: 801–803. [DOI] [PubMed] [Google Scholar]
- 2. Ghosheh FR,, Cremona FA,, Rapuano CJ,, et al. Trends in penetrating keratoplasty in the United States 1980-2005. Int Ophthalmol. 2008; 28: 147–153. [DOI] [PubMed] [Google Scholar]
- 3. Tuft SJ,, Moodaley LC,, Gregory WM,, Davison CR,, Buckley RJ. Prognostic factors for the progression of keratoconus. Ophthalmology. 1994; 101: 439–447. [DOI] [PubMed] [Google Scholar]
- 4. Ertan A,, Muftuoglu O. Keratoconus clinical findings according to different age and gender groups. Cornea. 2008; 27: 1109–1113. [DOI] [PubMed] [Google Scholar]
- 5. Gordon-Shaag A,, Millodot M,, Shneor E,, Liu Y. The genetic and environmental factors for keratoconus. 2015; 2015: 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McKay TB,, Hjortdal J,, Sejersen H,, Karamichos D. Differential effects of hormones on cellular metabolism in keratoconus in vitro. Sci Rep. 2017; 7: 42896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McKay TB,, Hjortdal J,, Sejersen H,, Asara JM,, Wu J,, Karamichos D. Endocrine and metabolic pathways linked to keratoconus: implications for the role of hormones in the stromal microenvironment. Sci Rep. 2016; 6: 25534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Karamichos D,, Zieske JD,, Sejersen H,, Sarker-Nag A,, Asara JM,, Hjortdal J. Tear metabolite changes in keratoconus. Exp Eye Res. 2015; 132: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McKay TB,, Hjortdal J,, Priyadarsini S,, Karamichos D. Acute hypoxia influences collagen and matrix metalloproteinase expression by human keratoconus cells in vitro. PLoS One. 2017; 12: e0176017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Karamichos D,, Funderburgh ML,, Hutcheon AEK,, et al. A Role for topographic cues in the organization of collagenous matrix by corneal fibroblasts and stem cells. PLoS One. 2014; 9: e86260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kymes SM,, Walline JJ,, Zadnik K,, Gordon MO. Quality of life in keratoconus. Am J Ophthalmol. 2004; 138: 527–535. [DOI] [PubMed] [Google Scholar]
- 12. Wang Z,, Gerstein M,, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009; 10: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bradford JR,, Hey Y,, Yates T,, Li Y,, Pepper SD,, Miller CJ. A comparison of massively parallel nucleotide sequencing with oligonucleotide microarrays for global transcription profiling. BMC Genomics. 2010; 11: 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kukurba KR,, Montgomery SB. RNA sequencing and analysis. Cold Spring Harb Protoc. 2015; 2015: 951–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011; 12: 861–874. [DOI] [PubMed] [Google Scholar]
- 16. Eddy SR. Non–coding RNA genes and the modern RNA world. Nat Rev Genet. 2001; 2: 919–929. [DOI] [PubMed] [Google Scholar]
- 17. Rogoyski OM,, Pueyo JI,, Couso JP,, Newbury SF. Functions of long non-coding RNAs in human disease and their conservation in Drosophila development. Biochem Soc Trans. 2017; 45: 895–904. [DOI] [PubMed] [Google Scholar]
- 18. Harries LW. Long non-coding RNAs and human disease. Biochem Soc Trans. 2012; 40: 902–906. [DOI] [PubMed] [Google Scholar]
- 19. Wentz-Hunter K,, Cheng EL,, Ueda J,, Sugar J,, Yue BY. Keratocan expression is increased in the stroma of keratoconus corneas. Mol Med. 2001; 7: 470–477. [PMC free article] [PubMed] [Google Scholar]
- 20. Ha NT,, Nakayasu K,, Murakami A,, Ishidoh K,, Kanai A. Microarray analysis identified differentially expressed genes in keratocytes from keratoconus patients. Curr Eye Res. 2004; 28: 373–379. [DOI] [PubMed] [Google Scholar]
- 21. Nielsen K,, Birkenkamp-Demtröder K,, Ehlers N,, Orntoft TF. Identification of differentially expressed genes in keratoconus epithelium analyzed on microarrays. Invest Opthalmol Vis Sci. 2003; 44: 2466–2476. [DOI] [PubMed] [Google Scholar]
- 22. Balasubramanian SA,, Wasinger VC,, Pye DC,, Willcox MDP. Preliminary identification of differentially expressed tear proteins in keratoconus. Mol Vis. 2013; 19: 2124–2134. [PMC free article] [PubMed] [Google Scholar]
- 23. Chaerkady R,, Shao H,, Scott S-G,, Pandey A,, Jun AS,, Chakravarti S. The keratoconus corneal proteome: loss of epithelial integrity and stromal degeneration. J Proteomics. 2013; 87: 122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Joseph R,, Srivastava OP,, Pfister RR. Differential epithelial and stromal protein profiles in keratoconus and normal human corneas. Exp Eye Res. 2011; 92: 282–298. [DOI] [PubMed] [Google Scholar]
- 25. Nielsen K,, Vorum H,, Fagerholm P,, et al. Proteome profiling of corneal epithelium and identification of marker proteins for keratoconus, a pilot study. Exp Eye Res. 2006; 82: 201–209. [DOI] [PubMed] [Google Scholar]
- 26. Szcześniak MW,, Kabza M,, Karolak JA,, et al. KTCNlncDB—a first platform to investigate lncRNAs expressed in human keratoconus and non-keratoconus corneas. Database (Oxford). 2017; 2017: baw168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kabza M,, Karolak JA,, Rydzanicz M,, et al. Collagen synthesis disruption and downregulation of core elements of TGF-β, Hippo, and Wnt pathways in keratoconus corneas. Eur J Hum Genet. 2017; 25: 582–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bykhovskaya Y,, Gromova A,, Makarenkova HP,, Rabinowitz YS. Abnormal regulation of extracellular matrix and adhesion molecules in corneas of patients with keratoconus. Int J Keratoconus Ectatic Corneal Dis. 2016; 5: 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li X,, Bykhovskaya Y,, Haritunians T,, et al. A genome-wide association study identifies a potential novel gene locus for keratoconus, one of the commonest causes for corneal transplantation in developed countries. Hum Mol Genet. 2012; 21: 421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rabinowitz YS. Videokeratographic indices to aid in screening for keratoconus. J Refract Surg. 1995; 11: 371–379. [DOI] [PubMed] [Google Scholar]
- 31. Trapnell C,, Pachter L,, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009; 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Trapnell C,, Williams BA,, Pertea G,, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010; 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Trapnell C,, Roberts A,, Goff L,, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012; 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang J,, Duncan D,, Shi Z,, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013; 41: W77–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fang S,, Zhang L,, Guo J,, et al. NONCODEV5: a comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018; 46: D308–D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cai J,, Perkumas KM,, Qin X,, Hauser MA,, Stamer WD,, Liu Y. Expression profiling of human schlemm's canal endothelial cells from eyes with and without glaucoma. Invest Ophthalmol Vis Sci. 2015; 56: 6747–6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu Y,, Bailey JC,, Helwa I,, et al. A Common variant in MIR182 is associated with primary open-angle glaucoma in the NEIGHBORHOOD consortium. Invest Ophthalmol Vis Sci. 2016; 57: 3974–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kulkarni B,, Mohammed I,, Hopkinson A,, Dua HS. Validation of endogenous control genes for gene expression studies on human ocular surface epithelium. PLoS One. 2011; 6: e22301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Team Rs. RStudio: Integrated Development Environment for R. Boston, MA; 2015.
- 40. Su G,, Morris JH,, Demchak B,, Bader GD. Biological network exploration with Cytoscape 3. Curr Protoc Bioinformatics. 2014; 47: 8.13.1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Macé M,, Galiacy SD,, Erraud A,, et al. Comparative transcriptome and network biology analyses demonstrate antiproliferative and hyperapoptotic phenotypes in human keratoconus corneas. Invest Ophthalmol Vis Sci. 2011; 52: 6181–6191. [DOI] [PubMed] [Google Scholar]
- 42. You J,, Corley SM,, Wen L,, et al. RNA-Seq analysis and comparison of corneal epithelium in keratoconus and myopia patients. Sci Rep. 2018; 8: 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tamura Y,, Konomi H,, Sawada H,, Takashima S,, Nakajima A. Tissue distribution of type VIII collagen in human adult and fetal eyes. Invest Ophthalmol Vis Sci. 1991; 32: 2636–2644. [PubMed] [Google Scholar]
- 44. Hopfer U,, Fukai N,, Hopfer H,, et al. Targeted disruption of Col8a1 and Col8a2 genes in mice leads to anterior segment abnormalities in the eye. FASEB J. 2005; 19: 1232–1244. [DOI] [PubMed] [Google Scholar]
- 45. Wojcik K,, Blasiak J,, Szaflik JPJ,, Szaflik JPJ. Role of biochemical factors in the pathogenesis of keratoconus. Acta Biochim Pol. 2014; 61: 55–62. [PubMed] [Google Scholar]
- 46. Toprak I,, Kucukatay V,, Yildirim C,, Kilic-Toprak E,, Kilic-Erkek O. Increased systemic oxidative stress in patients with keratoconus. Eye. 2014; 28: 285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Volders PJ,, Verheggen K,, Menschaert G,, et al. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015; 43: 4363–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang KC,, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011; 43: 904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Loh A,, Hadziahmetovic M,, Dunaief JL. Iron homeostasis and eye disease. Biochim Biophys Acta. 2009; 1790: 637–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huelsken J,, Birchmeier W. New aspects of Wnt signaling pathways in higher vertebrates. Curr Opin Genet Dev. 2001; 11: 547–553. [DOI] [PubMed] [Google Scholar]
- 51. Khaled ML,, Helwa I,, Drewry M,, Seremwe M,, Estes A,, Liu Y. Molecular and histopathological changes associated with keratoconus. Biomed Res Int. 2017; 2017: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Goralska M,, Ferrell J,, Harned J,, et al. Iron metabolism in the eye: a review. Exp Eye Res. 2009; 88: 204–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tong X,, Kawabata H,, Koeffler HP. Iron deficiency can upregulate expression of transferrin receptor at both the mRNA and protein level. Br J Haematol. 2002; 116: 458–464. [PubMed] [Google Scholar]
- 54. Curtis ARJ,, Fey C,, Morris CM,, et al. Mutation in the gene encoding ferritin light polypeptide causes dominant adult-onset basal ganglia disease. Nat Genet. 2001; 28: 350–354. [DOI] [PubMed] [Google Scholar]
- 55. Ashby BD,, Garrett Q,, Willcox MDP. Corneal injuries and wound healing – review of processes and therapies. Austin J Clin Ophthalmol. 2014; 1: 1–25. [Google Scholar]
- 56. Guo S,, Dipietro LA. Factors affecting wound healing. J Dent Res. 2010; 89: 219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Elsharkawi-Welt K,, Hepp J,, Scharffetter-Kochanek K. Genetische Ursachen schwerer Wundheilungsstörungen. Der Hautarzt. 2008; 59: 893–903. [DOI] [PubMed] [Google Scholar]
- 58. Schey KL,, Wang Z, L. Wenke J, Qi Y. Aquaporins in the eye: expression, function, and roles in ocular disease. Biochim Biophys Acta. 2014; 1840: 1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rabinowitz YS,, Dong L,, Wistow G. Gene expression profile studies of human keratoconus cornea for NEIBank: a novel cornea-expressed gene and the absence of transcripts for aquaporin 5. Invest Ophthalmol Vis Sci. 2005; 46: 1239–1246. [DOI] [PubMed] [Google Scholar]
- 60. Garfias Y,, Navas A,, Pérez-Cano HJ,, Quevedo J,, Villalvazo L,, Zenteno JC. Comparative expression analysis of aquaporin-5 (AQP5) in keratoconic and healthy corneas. Mol Vis. 2008; 14: 756–761. [PMC free article] [PubMed] [Google Scholar]
- 61. Wang W,, Sumiyoshi H,, Yoshioka H,, Fujiwara S. Interactions between epiplakin and intermediate filaments. J Dermatol. 2006; 33: 518–527. [DOI] [PubMed] [Google Scholar]
- 62. Jang S-I,, Kalinin A,, Takahashi K,, Marekov LN,, Steinert PM. Characterization of human epiplakin: RNAi-mediated epiplakin depletion leads to the disruption of keratin and vimentin IF networks. J Cell Sci. 2005; 118: 781–793. [DOI] [PubMed] [Google Scholar]
- 63. Kokado M,, Okada Y,, Goto M,, et al. Increased fragility, impaired differentiation, and acceleration of migration of corneal epithelium of epiplakin-null mice. Invest Ophthalmol Vis Sci. 2013; 54: 3780–3789. [DOI] [PubMed] [Google Scholar]
- 64. McNamara NA,, Brand RJ,, Polse KA,, Bourne WM. Corneal function during normal and high serum glucose levels in diabetes. Invest Ophthalmol Vis Sci. 1998; 39: 3–17. [PubMed] [Google Scholar]
- 65. Woodward MA,, Blachley TS,, Stein JD. The association between sociodemographic factors, common systemic diseases, and keratoconus. Ophthalmology. 2016; 123: 457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Seiler T,, Huhle S,, Spoerl E,, Kunath H. Manifest diabetes and keratoconus: a retrospective case-control study. Graefe's Arch Clin Exp Ophthalmol. 2000; 238: 822–825. [DOI] [PubMed] [Google Scholar]
- 67. Kuo IC,, Broman A,, Pirouzmanesh A,, Melia M. Is there an association between diabetes and keratoconus? Ophthalmology. 2006; 113: 184–190. [DOI] [PubMed] [Google Scholar]
- 68. Goldich Y,, Barkana Y,, Gerber Y,, et al. Effect of diabetes mellitus on biomechanical parameters of the cornea. J Cataract Refract Surg. 2009; 35: 715–719. [DOI] [PubMed] [Google Scholar]
- 69. Sriram S,, Robinson P,, Pi L,, Lewin AS,, Schultz G. Triple combination of siRNAs targeting TGFβ1, TGFβR2, and CTGF enhances reduction of collagen I and smooth muscle actin in corneal fibroblasts. Invest Opthalmology Vis Sci. 2013; 54: 8214–8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gibson DJ,, Pi L,, Sriram S,, et al. Conditional knockout of CTGF affects corneal wound healing. Invest Ophthalmol Vis Sci. 2014; 55: 2062–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Balasubramanian SA,, Pye DC,, Willcox MDP. Are proteinases the reason for keratoconus? Curr Eye Res. 2010; 35: 185–191. [DOI] [PubMed] [Google Scholar]
- 72. Cuellar-Partida G,, Springelkamp H,, Lucas SEM,, et al. WNT10A exonic variant increases the risk of keratoconus by decreasing corneal thickness. Hum Mol Genet. 2015; 24: 5060–5068. [DOI] [PubMed] [Google Scholar]
- 73. Gao X,, Nannini DR,, Corrao K,, et al. Genome-wide association study identifies WNT7B as a novel locus for central corneal thickness in Latinos. Hum Mol Genet. 2016; 25: 5035–5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Toyono T,, Usui T,, Yokoo S,, et al. Angiopoietin-like 7 is an anti-angiogenic protein required to prevent vascularization of the cornea. PLoS One. 2015; 10: e0116838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Iqbal O,, Fisher G,, Vira S,, et al. Increased expression of secreted frizzled-related protein-1 and microtubule-associated protein light chain 3 in keratoconus. Cornea. 2013; 32: 702–707. [DOI] [PubMed] [Google Scholar]
- 76. You J,, Hodge C,, Wen L,, McAvoy JW,, Madigan MC,, Sutton G. Tear levels of SFRP1 are significantly reduced in keratoconus patients. Mol Vis. 2013; 19: 509–515. [PMC free article] [PubMed] [Google Scholar]
- 77. Jenkins LM,, Singh P,, Varadaraj A,, et al. Altering the proteoglycan state of transforming growth factor β type III receptor (TβRIII)/betaglycan modulates canonical Wnt/β-catenin signaling. J Biol Chem. 2016; 291: 25716–25728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yao B,, Wang S,, Xiao P,, Wang Q,, Hea Y,, Zhang Y. MAPK signaling pathways in eye wounds: multifunction and cooperation. Exp Cell Res. 2017; 359: 10–16. [DOI] [PubMed] [Google Scholar]
- 79. Zhu Z,, He X,, Johnson C,, et al. PI3K is negatively regulated by PIK3IP1, a novel p110 interacting protein. Biochem Biophys Res Commun. 2007; 358: 66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Clay Bunn R,, Fowlkes JL. Insulin-like growth factor binding protein proteolysis. Trends Endocrinol Metab. 2003; 14: 176–181. [DOI] [PubMed] [Google Scholar]
- 81. Lodygin D,, Epanchintsev A,, Menssen A,, Diebold J,, Hermeking H. Functional epigenomics identifies genes frequently silenced in prostate cancer. Cancer Res. 2005; 65: 4218–4227. [DOI] [PubMed] [Google Scholar]
- 82. Xuan M,, Wang S,, Liu X,, He Y,, Li Y,, Zhang Y. Proteins of the corneal stroma: importance in visual function. Cell Tissue Res. 2016; 364: 9–16. [DOI] [PubMed] [Google Scholar]
- 83. Levin MH,, Verkman AS. Aquaporin-3-dependent cell migration and proliferation during corneal re-epithelialization. Invest Ophthalmol Vis Sci. 2006; 47: 4365–4372. [DOI] [PubMed] [Google Scholar]
- 84. Zhu J,, Liu S,, Ye F,, et al. Long noncoding RNA MEG3 interacts with p53 protein and regulates partial p53 target genes in hepatoma cells. PLoS One. 2015; 10: e0139790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Mondal T,, Subhash S,, Vaid R,, et al. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat Commun. 2015; 6: 7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.