Abstract
Autism spectrum disorders (ASDs) and autistic traits in the general population may share genetic susceptibility factors. In this study, we investigated such potential overlap based on common genetic variants. We developed and validated a self-report questionnaire of autistic traits in adults. We then conducted genome-wide association studies (GWASs) of six trait scores derived from the questionnaire through exploratory factor analysis in 1981 adults from the general population. Using the results from the Psychiatric Genomics Consortium GWAS of ASDs, we observed genetic sharing between ASDs and the autistic traits 'childhood behavior', 'rigidity' and 'attention to detail'. Gene-set analysis subsequently identified 'rigidity' to be significantly associated with a network of ASD gene-encoded proteins that regulates neurite outgrowth. Gene-wide association with the well-established ASD gene MET reached significance. Taken together, our findings provide evidence for an overlapping genetic and biological etiology underlying ASDs and autistic population traits, which suggests that genetic studies in the general population may yield novel ASD genes.
Introduction
Autism spectrum disorders (ASDs) are a group of pervasive neurodevelopmental disorders that are characterized by impairments in reciprocal social interaction and communication, as well as restricted, repetitive and stereotyped patterns of behavior, interests and activities.1 The prevalence of ASDs in the general population is estimated to be ~1%,2 with approximately four times more males than females being affected by these disorders.3, 4 Family and twin studies show that ASDs are highly heritable, and ~65–90% of the phenotypic variance can be explained by genetic factors.4, 5, 6, 7 Despite this considerable heritability, the identification and replication of genetic susceptibility factors for ASDs has been challenging, as their genetic architecture is complex and highly heterogeneous, with both common and rare inherited, as well as de novo genetic variants contributing to ASD etiology.8, 9 In 2013, molecular landscape analysis by our research group, combining evidence from genome-wide assoctiation studies (GWASs) of ASDs, rare genetic variant studies (exome-sequencing and copy number variation studies) and other genetic evidence (candidate gene association, microRNA and gene expression, gene function and animal studies), showed that available genetic data for ASDs converge on protein networks regulating three biological processes, namely steroidogenesis, neurite outgrowth and (glutamatergic) synaptic function.10 Autistic traits can be defined as subthreshold deficits in social interaction and communication, as well as restricted behaviors, interests and activities that are continuously distributed in the general population.11
Population-based twin studies have shown that, like ASDs, such autistic traits are heritable, with heritability rates ranging from 36 to 87%.12, 13 There is ample evidence suggesting that ASDs represent the upper extreme on the continuous distribution of these autistic traits in the population.11, 14, 15, 16, 17, 18 Indeed, a twin study showed that autistic population traits and ASDs share genetic susceptibility factors.18 Furthermore, in a recent study, Robinson et al.19 used large genomic data sets to demonstrate that there is a genetic correlation between certain autistic traits—that is, social and communication skill impairments assessed in children from the general population—and ASDs.
The above evidence suggests that genetic studies of autistic traits in the general population could not only provide corroborating evidence for the involvement of previously identified ASD candidate genes—as shown in two previous studies20, 21—but may also aid in the identification of novel ASD susceptibility genes and loci. This could substantially ease research efforts to increase our understanding of the biology underlying ASDs. To measure autistic traits in the general population, a number of questionnaires have been developed, and of those, the Autism Spectrum Quotient (AQ) and the Social and Communication Disorders Checklist (SCDC) are commonly used in genetic studies. The AQ is a 50-item self-report questionnaire of autistic traits in adults,22, 23 the SCDC is a parent-rated questionnaire measuring social communication impairment in children.24, 25
In this article, we have further investigated the genetic overlap between ASDs and autistic traits. To this end, we first developed and validated a questionnaire of autistic traits, suitable for use in population studies. In comparison with the AQ and SCDC, our novel questionnaire captures more ASD-related features, is shorter and less theory-based. In data of 1981 adults from the Dutch general population, we then conducted GWASs of the scores for six traits, that is, the scores for ‘total autistic traits’ from our questionnaire as well as its five best-fitting constituting factors: ‘childhood behavior’, ‘rigidity’, ’social skills’, ‘attention to detail’ and ‘imagination’. Through polygenic risk score-based analysis, we observed overlap between genetic variants associated with ASDs and several of these autistic traits. Subsequent gene-set analysis identified 'rigidity' to be significantly associated with the network of ASD gene-encoded proteins that regulates neurite outgrowth (see above). In addition, the MET gene was gene-wide significantly associated with this trait.
Materials and methods
Development and validation of a questionnaire of autistic traits in adults
Based on the extensive clinical experience of authors JKB and JWM in diagnosing and treating children and adults with ASDs, we constructed a self-report questionnaire of autistic traits that consists of 18 questions/items and could be administered to healthy adults. The 18 items in the questionnaire were chosen because they comprehensively cover the three main ASD symptom clusters, that is, deficits in social interaction, deficits in communication and restricted behavior, interests and activities. In addition, some items in the questionnaire do not assess current behaviors but behaviors that occurred in childhood, and these items were included because the concept of ASDs as early onset neurodevelopmental disorders specifies the onset of symptoms in childhood. Six items are based on the Diagnostic and Statistical Manual of Mental Disorders, 4th Edition section about ASDs1 and 12 items are extracted from the AQ. The AQ is a self-report questionnaire of 50 items that was developed and validated for the purpose of quantifying ASD-related traits in adult individuals with normal intelligence in the general population.22 The AQ has been translated into the Dutch language and validated in the Dutch general population and in Dutch people with ASDs.23 Table 1 shows the 18 individual items of our self-report questionnaire and it is indicated which items were taken or derived from the AQ and the Diagnostic and Statistical Manual of Mental Disorders, 4th Edition section about ASDs, respectively. For each item, the respondents are asked to indicate on a 4-point likert scale how well that statement applies to them, with the possible answer categories ‘1=definitely agree’, ‘2=partially agree’, ‘3=partially disagree’ and ‘4=definitely disagree’. The scoring is reversed for items in which an ‘agree’ response is characteristic for ASDs, and the item scores are then summed, resulting in a minimum score of 18 (no autistic traits) and a maximum score of 72 (full endorsement of all autistic traits).
Table 1. Customized self-report questionnaire of autistic traits in the general population.
| 1 By looking at someone’s face, I find it easy to work out what he or she is thinking or feeling (1) |
| 2 I find it hard to make new friends (1)a |
| 3 I enjoy social occasions such as birthdays, receptions, and so on (1) |
| 4 I can quickly work out whether someone is fascinated by what I say (1) |
| 5 As a child, I was a late talker or I had other speech-related problems (2)a |
| 6 I don’t know how to keep a conversation going (1)a |
| 7 People tell me that I keep going on and on about the same thing (1)a |
| 8 I find making stories up easy (1) |
| 9 I often get so absorbed in one thing that I lose sight of other things (1)a |
| 10 It upsets me if my daily routine is disturbed (1)a |
| 11 I prefer to do things the same way over and over again (1)a |
| 12 I tend to notice details that others do not (1)a |
| 13 As a child, I often retreated to my own world or I rarely played with other children (2)a |
| 14 As a child, I moved in a rigid way or I tended to repeat certain movements (2)a |
| 15 As a child, I often repeated the same words or I made up new words (2)a |
| 16 As a child, I often took statements and jokes literally (2)a |
| 17 As a child, I enjoyed playing games involving pretending with other children (1) |
| 18 As a child, I frequently became upset by sudden and unexpected changes (2)a |
The 18 questionnaire items were taken or derived from the AQ (designated with (1)) and the DSM-IV section about autism spectrum disorders (designated with (2)).
Abbreviations: AQ, Autism Spectrum Quotient; DSM-IV, Diagnostic and Statistical Manual of Mental Disorders, 4th Edition.
Reverse-scored item.
To validate our questionnaire in terms of its ability to differentiate between people with less and more autistic traits within the general population, we first administered the questionnaire to a Dutch general population sample of 50 healthy adults as well as 24 Dutch adults that had been diagnosed with an ASD by an experienced psychiatrist. We used a T-test to quantify the difference between the means of the ‘total autistic traits’ scores in the healthy adult and adult ASD patient samples. Furthermore, we determined the correlation between the total scores on our novel questionnaire and the total scores on the already established AQ through also administering the complete AQ of 50 items to the same 50 Dutch general population respondents that answered our novel questionnaire. We used the corrcoef function in MATLAB (The Mathworks, Natick, MA, USA) to calculate the correlation statistics.
Measuring autistic traits in the Dutch general population and factor analysis of the questionnaire scores
After the validation step, our novel questionnaire was sent to participants from the Nijmegen Biomedical Study (NBS), a population-based survey conducted by the Department for Health Evidence and the Department of Laboratory Medicine of the Radboud University Medical Center.26 Approval to conduct the NBS was obtained from the Radboud University Medical Center Institutional Review Board (CMO 2001/055). All participants gave written informed consent.
Within the first wave of NBS 22 451 age- and sex-stratified randomly selected inhabitants of the municipality of Nijmegen received an invitation to fill out a number of questionnaires on, for example, lifestyle and medical history, and to donate an 8.5 ml blood sample in a serum separator tube and a 10 ml EDTA blood sample. The response to these questionnaires was 42% (N=9350). Of these responders, 69% (N=6468) also donated blood samples. Our novel questionnaire was part of the second wave of NBS. In this wave, 7986 individuals received an invitation, and the response rate to our autistic traits questionnaire in the NBS sample was 70% (N=5594). After further evaluation of the data, we had complete questionnaire data for 5066 NBS participants. Using the Promax method in SPSS 20.0 (SPSS Technologies, Armonk, NY, USA), we then conducted a principal component/factor analysis of the scores on the 18 individual questionnaire items of all 5066 NBS participants, to find out which combination of factors (each consisting of the added scores from two or more individual items) would explain the largest proportion of the observed variance in the total score for ‘autistic traits in the general population’. Using SPSS 20.0 and the answers of all 5066 NBS participants to the 18 questionnaire items, we also determined the internal consistency (Crohnbach’s α) of the total autistic traits score on our questionnaire.
Genome-wide association analysis
The factor analysis of the 18 individual questionnaire items revealed that a combination of five factors explained the largest proportion of the observed variance in the total autistic trait scores. Therefore, the total autistic trait scores and scores for these five factors were corrected for age and gender using information on 5066 NBS individuals. Residuals were checked for independence, equal variance and normality and log or BLOM transformation (SPSS Technologies) was applied. Residuals were saved and used for each of the six phenotypes in the genome-wide association analysis (the scores for ‘total autistic traits’ and its five constituting factors). Genome-wide genotyping data was available for 1981 of the 5066 NBS participants. For 1218 individuals, genotyping was performed using the Human 370CNV Duo Beadchip (Illumina, San Diego, CA, USA). For 763 individuals, genotyping was performed using the Human OmniExpress Beadchip (Illumina) (Supplementary Table 1). For all individuals, density of genotype data was increased by imputation using 1000 genomes Phase I version 3 as a reference and IMPUTE2 software (freely available by the University of Oxford, UK).27
Genome-wide association analysis was carried out using SNPtest version 2.528 and performed separately for individuals genotyped on the two platforms. Quality control was performed on imputation quality score (info⩾0.6), minor allele frequency (⩾0.01) and Hardy–Weinberg equilibrium (P⩾0.00001). Genome-wide results were combined in an inverse-variance-weighted meta-analysis using METAL29 accounting for genomic inflation. Final analysis included 8 610 357 autosomal single-nucleotide polymorphisms (SNPs) across the genome. Subsequently, to ascertain the extent of genetic and biological overlap between ASDs and autistic traits, we used the summary statistics from the six independent GWASs for shared genetic etiology and gene-set analyses.
Shared genetic etiology analysis
Applying the default settings of the polygenic risk score-based analysis tool PRSice,30 we determined the level of shared genetic etiology between ASDs and autistic traits, using the summary statistics data from a large GWAS of ASDs by the Psychiatric Genomics Consortium (PGC) autism group (PGC-ASD GWAS, data for 5305 ASD cases and 5305 controls—these data are publicly available at: http://www.med.unc.edu/pgc/downloads)—as 'base phenotype' sample and the summary statistics data from our six independent GWAS meta-analyses of autistic traits (see above) as 'target phenotype' samples. Before generating polygenic risk scores with PRSice clumping was performed using PLINK, to select independent index SNPs for each linkage disequilibrium (LD) block in the genome.31 The index SNPs are selected based on significance levels in the reference data set and form clumps of all other SNPs that are within 500 kb and in LD (r2>0.25). Based on the clumped summary statistics of the PGC-ASD GWAS, PRSice was then used to generate polygenic risk scores for ASDs that are the sum of genome-wide SNPs associated with ASDs weighted by their effect sizes estimated from the PGC-ASD GWAS, from which only the SNPs exceeding seven broad P-value thresholds (indicated by PT) were included, that is, the SNPs with PT<0.001, 0.05, 0.1, 0.2, 0.3, 0.4 and 0.5. As such, seven genome-wide polygenic scores were generated based on all SNPs that were associated with the base ASD phenotype at P<0.001, 0.05, 0.1 and so on. Subsequently, PRSice calculated P-values of shared genetic etiology—that is, the extent to which the combined SNPs from each of the seven PT-linked polygenic risk scores for ASD predict each of the six autistic trait phenotypes—between the base ASD phenotype on the one hand and the target phenotypes, that is, the total autistic traits score as well as the scores for each of the five autistic trait factors, on the other hand. Last, the calculated P-values of shared genetic etiology were aggregated and corrected for multiple comparisons using the false discovery rate (FDR) method, incorporating potential dependencies between P-values,32, 33 similar to the approaches used in earlier studies working with multiple traits and PRSice.34, 35 To calculate the FDR, we used the mafdr function in MATLAB (R2012a; The Mathworks) using the bootstrap selection method for the FDR parameter lambda.
Gene-set analysis
We first compiled gene sets containing all genes encoding proteins within the three protein interaction networks that regulate steroidogenesis, neurite outgrowth and (glutamatergic) synaptic function and that together constitute our 'molecular landscape' of ASDs (Supplementary Table 2).10 For each gene from the three gene sets, all SNPs within exonic, intronic and untranslated regions of the gene and within 100 kb downstream and upstream of the gene were selected. The selection of the SNPs outside each gene is based on the fact that the vast majority of expression quantitative trait loci for a given gene are located within 100 kb downstream and/or upstream of a gene36, 37, 38 and because trait-associated (GWAS) SNPs are more likely to be expression quantitative trait loci.39
As our genome-wide association analysis only included autosomal SNPs, 12 genes located on the X chromosome (ARHGAP36, DACH2, DMD, GATA1, JARID1C, MAMLD1, MSN, PLXNB3, RHFOX1, RHFOX2, SLC10A3, USP9X) were omitted before the gene-set analysis. Gene-set analysis was then performed using the Multimarker Analysis of GenoMic Annotation (MAGMA) software package (http://ctglab.nl/software/magma).40 One of the autosomal genes (MSNP1) was manually added to the genetic location file of MAGMA to be able to include it in the analysis. The single SNP P-values were transformed into a gene test-statistic by taking the mean of the χ2 statistic among the SNPs in each gene. To account for LD, the 1000 Genomes Project European sample was used as a reference to estimate the LD between SNPs within (the vicinity of) the genes (http://ctglab.nl/software/MAGMA/ref_data/g1000_ceu.zip). Gene-wide P-values were converted to z-values reflecting the strength of the association each gene has with each of the six autistic traits phenotypes.
Subsequently, we tested whether the genes in each of the three gene sets are jointly associated with each of the six autistic traits phenotypes. Two tests were performed with MAGMA. The self-contained analysis tested whether the gene set contains any association with the phenotype of interest at all. This was carried out using an intercept-only linear regression model including a subvector corresponding to the genes in the gene set, and by evaluating whether the regression coefficient of this regression was >0. The competitive analysis tested whether the difference between the association of each gene set was different from the association of the genes outside the gene set, taking into account the polygenic nature of our traits. This was carried out by expanding the regression model to include all annotated genes in the genome outside the gene set. To account for the potentially confounding factors of gene size and gene density, both gene size and gene density as well as their logarithms were included as covariates in the competitive gene-set analysis. Considering the polygenic nature of our trait, the competitive analyses have more merit. Results were considered significant if they exceeded a Bonferroni correction threshold accounting for the number of gene sets and phenotypes tested. The analysis was carried out in two steps. In step 1, the combined effect of the SNPs in (the vicinity of) all genes on the total autistic traits score as well as the scores for the five autistic trait factors was analyzed. Post hoc, in step 2, the potential effects of the genes encoding the proteins in the networks making up our molecular ASD landscape and the individual gene(s) that may drive the association were investigated, by reviewing their gene-test statistics. Genes were considered gene-wide significant if they reached the Bonferroni correction threshold adjusted for the number of genes within the molecular ASD landscape.
Results
Validation of the novel questionnaire of autistic traits in adults
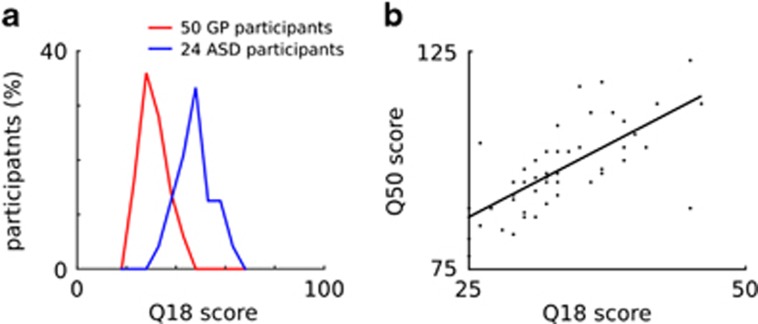
The self-report questionnaire of autistic traits consisting of 18 items that we developed was applied to 50 healthy adults and 24 adults diagnosed with an ASD by an experienced psychiatrist. Figure 1a shows the distribution of the scores for ‘total autistic traits’ in both groups. As can be derived, the ASD-diagnosed adults scored significantly higher (P<0.001) than healthy adults on the novel questionnaire.
Figure 1.
Validation of the novel questionnaire of autistic traits in adults. (a) Distributions of the total scores on the 18-item self-report questionnaire (Q18 score) of autistic traits in a Dutch general population sample of 50 adults* (General population (GP) participants, red line; total score range: 25–46; mean total score: 33 (s.d.=5.4)) and in 24 adults with a confirmed autism spectrum disorder (ASD) diagnosis (ASD participants, blue line; total score range: 37–66; mean total score: 50 (s.d.=7.5)). ASD participants scored significantly higher than the GP participants (P<0.001; two-sided T-test). (b) Correlation between Q18 score of autistic traits (total score range: 25–46; mean total score: 33 (s.d.=5.4)) and the total scores on the 50-item AQ (Q50 score; total score range: 78–123; mean total score: 98 (s.d.=10.2)) in a Dutch GP sample of 50 adults*. Each dot represents one participant. A significant correlation (correlation coefficient R=0.703; P<0.001) was observed between the total autistic traits score on our 18-item questionnaire and the total AQ score. *These are the same 50 respondents.
In addition and as shown in Figure 1b, the correlation between the total score on the new 18-item questionnaire and the total score on the 50-item AQ was 0.703 (P<0.001). Furthermore, the internal consistency of the total score on the new questionnaire was satisfactory (Cronbach’s α=0.70) and very similar to the internal consistency of the total AQ score in the Dutch general population (Cronbach’s α=0.71).23
Measuring autistic traits in the Dutch general population and factor analysis of the questionnaire scores
A total of 5066 adults from the NBS completed the questionnaire (Supplementary Table 1). Supplementary Figure 1 shows that the score for total autistic traits followed a normal distribution with a mean total score of 35.8 (s.d.=6.5). Exploratory factor analysis of the 18 individual questionnaire items revealed a five factors/subscores solution as best-fitting model to explain the variance in the total score. We named these five factors ‘childhood behavior’, ‘rigidity’, ‘social skills’, ‘attention to detail’ and ‘imagination’. In Table 2, the item content of and loadings on these five factors are shown. Taken together, the five factors explained 50.7% of the variance in the total autistic traits score.
Table 2. Item content of and loadings on the five factors that constitute the best fitting model to explain the variance in the total added score of the 18 items from our questionnaire of autistic traits that was completed by 5066 healthy adults from the NBS.
| Factor 1. Childhood behaviour | (19.33% of the variance in the total score explained) |
| Items | Factor loading |
| 5.a As a child, I was a late talker or I had other speech-related problems | 0.507 |
| 13.a As a child, I often retreated to my own world or I rarely played with other children | 0.649 |
| 14.a As a child, I moved in a rigid way or I tended to repeat certain movements | 0.712 |
| 15.a As a child, I often repeated the same words or I made up new words | 0.654 |
| 16.a As a child, I often took statements and jokes literally | 0.636 |
| 18.a As a child, I frequently became upset by sudden and unexpected changes | 0.609 |
| Factor 2. Rigidity | (11.21% of the variance in the total score explained) |
| Items | Factor loading |
| 7.a People tell me that I keep going on and on about the same thing | 0.447 |
| 9.a I often get so absorbed in one thing that I lose sight of other things | 0.599 |
| 10.a It upsets me if my daily routine is disturbed | 0.750 |
| 11.a I prefer to do things the same way over and over again | 0.689 |
| Factor 3. Social skills | (7.49% of the variance in the total score explained) |
| Items | Factor loading |
| 2.a I find it hard to make new friends | 0.633 |
| 3. I enjoy social occasions such as birthdays, receptions, etc. | 0.740 |
| 6.a I don’t know how to keep a conversation going | 0.508 |
| Factor 4. Attention to detail | (6.89% of the variance in the total score explained) |
| Items | Factor loading |
| 1. By looking at someone’s face, I find it easy to work out what he or she is thinking or feeling | 0.713 |
| 4. I can quickly work out whether someone is fascinated by what I say | 0.743 |
| 12.a I tend to notice details that others do not | −0.521 |
| Factor 5. Imagination | (5.74% of the variance in the total score explained) |
| Items | Factor loading |
| 8. I find making stories up easy | 0.723 |
| 17. As a child, I enjoyed playing games involving pretending with other children | 0.647 |
Taken together, the five factors explain 50.66% of the variance in the total score for autistic traits in the general population from our questionnaire.
Abbreviation: NBS, Nijmegen Biomedical Study.
A reverse-scored item.
Analysis of shared genetic etiology
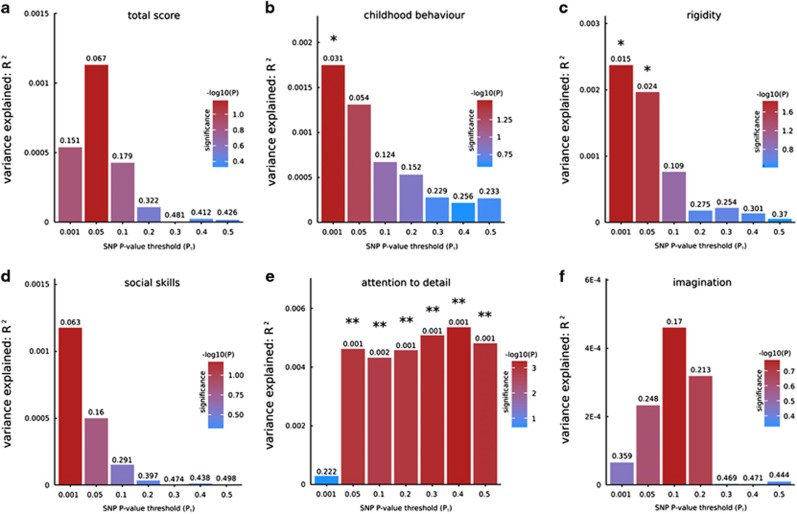
In Supplementary Figures 2 and 3, the Manhattan and QQ plots of the GWASs of the six autistic traits—that is, the scores for total autistic traits and its five constituting factors are provided. We found statistically significant evidence for overlap between genetic variants increasing the risk for ASDs derived from the publicly available data set of the Psychiatric Genomics Consortium (http://www.med.unc.edu/pgc/downloads) and genetic variants associated with three autistic traits: ‘childhood behavior’ (FDR-corrected P<0.05), ‘rigidity’ (FDR-corrected P<0.05) and ‘attention to detail’ (FDR-corrected P<0.01). The most predictive P-value thresholds (PT) were 0.001, 0.001 and 0.4, respectively (Figure 2).
Figure 2.
Bar plots from PRSice30 showing results at seven broad P-value thresholds (PT) for shared genetic etiology between autism spectrum disorders (ASDs) and the six autistic trait phenotypes (a–f) (see Materials and methods). The numbers above the bars indicate the P-values for shared genetic etiology, and these P-values were corrected using the false discovery rate (FDR) method. *FDR-corrected P<0.05, **FDR-corrected P<0.01. SNP, single-nucleotide polymorphism.
Gene-set analysis
According to the criteria described in the Materials and methods, we selected the SNPs within (100 kb from) all autosomal genes encoding proteins in the 'steroidogenesis', 'neurite outgrowth' and '(glutamatergic) synaptic function' networks that we described earlier based on published genetic studies of ASDs.10 The final data set for the gene-set analysis contained 147 unique genes and 231 002 SNPs (effective number of SNPs after adjusting for LD structure=79 993). MAGMA-based gene-set analysis38 revealed that 'rigidity' was significantly associated with the complete set of 147 genes (Pself-contained=0.036, Pcompetitive=0.005). No significant association was found for the total autistic trait score or any of the other four autistic trait factors (Table 3). Post hoc analysis of the three networks individually showed that 'rigidity' was associated with neurite outgrowth (Pself-contained=0.035, Pcompetitive=0.003), while the gene-set associations for the networks regulating steroidogenesis and (glutamatergic) synaptic function were both nonsignificant (Table 3). Within the neurite outgrowth gene-set, gene-wide association of ‘rigidity’ with the MET gene was significant after correction for multiple testing (P=0.000143), see Supplementary Figure 4.
Table 3. Association results from the autistic traits gene-set analyses with the combined gene sets are shown at the top.
| PSelf-contained | PCompetitive | |
|---|---|---|
| Autistic trait | ||
| Total score | 0.592 | 0.214 |
| Childhood behavior | 0.847 | 0.891 |
| Rigidity | 0.036 | 0.005 |
| Social skills | 0.799 | 0.506 |
| Attention to detail | 0.996 | 0.680 |
| Imagination | 0.657 | 0.602 |
| Gene set | ||
| Steroidogenesis (21 genes) | 0.076 | 0.065 |
| Neurite outgrowth (103 genes) | 0.034 | 0.003 |
| (Glutamatergic) synaptic function (43 genes) | 0.234 | 0.599 |
Results of the post hoc separate analyses for the three protein interaction networks for the rigidity trait are shown below. Significant P-values (<0.05) are in bold.
Discussion
In this study, we developed a short self-report questionnaire for the assessment of current and childhood behaviors relevant to ASDs in the adult general population, which is based on the longer AQ instrument and the Diagnostic and Statistical Manual of Mental Disorders, 4th Edition criteria for the diagnosis of ASDs. We described the successful validation of the questionnaire. After assessment of over 5000 individuals, we performed GWASs of the total questionnaire score and the subscores for five autistic traits derived through factor analysis in 1981 individuals, for whom genome-wide genotyping data was available. We subsequently used the summary statistics of the PGC-ASD GWAS to determine the genetic overlap between ASDs and autistic traits. We indeed found significant evidence for a shared genetic etiology between ASDs and three autistic traits, 'childhood behavior', 'rigidity' and 'attention to detail'.
Furthermore, gene-set analysis revealed a significant association of ‘rigidity’ with ASD-implicated genes encoding proteins interacting within a molecular network regulating neurite outgrowth. Among the neurite outgrowth-linked genes, the MET gene showed significant gene-wide association with the ‘rigidity’ trait. As rigidity is an overlapping trait of several neurodevelopmental disorders, the neurite outgrowth gene set and the MET gene could be further studied as candidates for other psychiatric disorders such as obsessive–compulsive disorder (OCD).
A number of questionnaires for autistic traits in the general population haven been developed, and of those, the AQ and the SCDC are commonly used in genetic studies. The AQ is a 50-item self-report questionnaire of autistic traits in adults,22, 23 the SCDC is a parent-rated questionnaire measuring social communication impairment in children.24, 25 Compared with the SCDC, our questionnaire has the advantage of capturing more domains relevant to autistic behavior than only social communication impairment. Advantages compared with the AQ are the brevity of the questionnaire—which makes it more readily applicable in large population studies—and the fact that its factor structure was derived in a data-driven, unbiased approach, whereas the AQ was built around five predefined, theory-based factors or subgroups of 10 questions each. The new questionnaire showed construct validity, reflected by a moderately high internal consistency of the total score of the responses to the 18 questions. In addition, it was shown to have a high discriminant validity, that is, the ability to differentiate between people with less and more autistic traits, the latter including those with a clinical ASD diagnosis. Similar to the AQ,22, 23 the total score on our questionnaire also followed a normal distribution.
Although over 5000 adults completed the questionnaire, genome-wide genotyping was only available for 1981 persons. This sample size did not lend itself to the evaluation of association results for individual genetic variants, but provided sufficient power for genetic overlap analysis with the large PGC GWAS of ASDs.
Our findings of genetic sharing between ASDs and the population traits ‘childhood behavior’, ‘rigidity’ and ‘attention to detail’ add further weight to previous reports, suggesting that the same genetic susceptibility factors underlie both ASDs and autistic traits.13, 18
Importantly, as exclusively social and communication impairments have been investigated thus far,19, 25 our results extend this concept of genetic sharing to also include traits linked to repetitive and stereotyped patterns of behavior, interests and activities observed in people with ASDs.
As shown in Figure 2, the shared genetic etiology analysis provided estimates for the proportion of the variance in autistic traits explained by common genetic variants associated with ASDs, and the highest proportions of variance explained (reaching significance) were 0.17% for ‘childhood behavior’, 0.24% for ‘rigidity’ and 0.54% for ‘attention to detail’. Although these proportions may seem quite small, they are similar to the percentages found in other studies that used polygenic risk scoring for neuropsychiatric disorders and quantitative traits. For instance, the polygenic risk derived from an attention-deficit hyperactivity disorder GWAS meta-analysis explained 0.10% of the phenotypic variance observed in an independent attention-deficit hyperactivity disorder sample,41 the polygenic risk associated with educational attainment explained 0.60% of the observed variance in adolescents' behavioral problems42 and the polygenic risk derived from a GWAS of OCD explained 0.20% of the observed variance in OCD-like symptoms in a population-based twin-family sample.43 Last, although the explained variances of polygenic risk scores are small, quantile analyses showed associations between risk scores and behavior by septiles, highlighting the potential to stratify individuals based on polygenic risk scores.42 In light of the above, the observed variance explained in the three autistic traits is likely an underestimate as the identified percentage of explained variance is dependent on the size of the 'base phenotype' sample for the generation of the polygenetic risk scores.44
In addition to the shared genetic etiology analysis, the available sample size lent itself to gene-set analysis, in which we tested the combined effect of common variants within (the vicinity of) all genes from a certain gene set. Considering multiple genetic variants within the same analysis likely increases the explained phenotypic variance, thereby boosting the power of genetic studies.45, 46, 47 Our results point toward the biological process of neurite outgrowth being implicated in both ASDs10 and the ‘rigidity’ trait, which yields evidence of a direct biological link between ASDs and a specific autistic trait. However, neurite outgrowth is not uniquely linked to ASDs and autistic traits, and genetic variance in neurite outgrowth-related genes has also been shown to be relevant to, for example, attention-deficit hyperactivity disorder,47, 48 OCD49, 50 and schizophrenia.51, 52 This reflects genetic overlap between clinically related disorders53, 54 and highlights the potential of studying traits that are shared between disorders—such as ‘rigidity’—for psychiatric genetic research.
One gene within the ‘neurite outgrowth’ gene set, MET, reached gene-wide evidence of association with ‘rigidity’ in our study. MET is a well-established ASD gene and its involvement in disease etiology has been implicated through multiple lines of evidence, including both common and rare genetic variants, gene expression studies and brain connectivity studies.55 Apart from the already established role of the MET-encoded protein in the ‘neurite outgrowth’ and ‘steroidogenesis’ networks (and hence the presence of MET in the corresponding gene sets) that form a part of our ‘molecular landscape’ of ASDs (see above),10 a recent study shows that the MET protein is also an important regulator of glutamatergic synapse development and maturation.56
The fact that both the ‘neurite outgrowth’ gene set and the established ASD gene MET are associated with a specific autistic trait in a general adult population sample provides a proof of concept for the potential of autistic trait studies in the search for novel ASD susceptibility genes and for the potential translatability of genetic findings from childhood to adulthood.
Our study should be viewed in the context of some strengths and limitations. The new and fast self-report questionnaire that we developed has the advantage that a large number of individuals can be assessed in a relatively cheap way. Although self-report data suffer from a number of limitations—such as reporter bias and misinterpretation of the questionnaire items by the respondents—we minimized these biases by validating our questionnaire in both a general population and an ASD sample and comparing it to the already well-established AQ.22, 23 The sample size of the GWASs that we conducted was too small for discovering new single genetic variant associations, but it was large enough to provide proof of concept—through shared genetic etiology and gene-set/gene-wide analyses—for sharing beyond social communication traits. Analyses of autistic traits using larger population samples can be expected to lead to the discovery of novel ASD-relevant genes, which will increase our understanding of the (altered) biology underlying ASDs. Our approach shows great potential for other psychiatric disorders that could benefit from being genetically dissected with a similar approach.
In conclusion, our findings of genetic sharing between ASDs and autistic traits, and of gene-set-based and gene-wide association of ASD-implicated genes with the autistic trait rigidity, provide evidence for a shared genetic and biological etiology underlying ASDs and autistic population traits, beyond earlier described social communication traits.
As (existing) population-based cohorts could be used in trait genetic studies, this could offer tremendous opportunities to our field, as we can increase our sample sizes considerably at relatively little cost.
Acknowledgments
The Nijmegen Biomedical Study is a population-based survey conducted at the Department for Health Evidence and the Department of Laboratory Medicine of the Radboud University Medical Center. Principal investigators of the Nijmegen Biomedical Study are LALM Kiemeney, ALM Verbeek, DW Swinkels and B Franke. The research leading to these results also received funding from the European Community’s Seventh Framework Program (FP7/2007–2013) under grant agreements no. 278948 (TACTICS) and no. 602805 (Aggressotype), from the European Community’s Horizon 2020 Programme (H2020/2014–2020) under grant agreement no. 643051 (MiND), and from the Innovative Medicines Initiative (IMI) 2 Joint Undertaking (H2020/EFPIA) under grant agreement no. 115916 (PRISM). BF is supported by a Vici grant from the Netherlands Organization for Scientific Research (NWO; Grant No. 016-130-669).
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
The authors declare no conflict of interest.
Supplementary Material
References
- American Psychiatric AssociationDiagnostic and Statistical Manual of Mental Disorders. American Psychiatric Association: Arlinton, VA, 2000. [Google Scholar]
- Baird G, Simonoff E, Pickles A, Chandler S, Loucas T, Meldrum D et al. Prevalence of disorders of the autism spectrum in a population cohort of children in South Thames: the Special Needs and Autism Project (SNAP). Lancet 2006; 368: 210–215. [DOI] [PubMed] [Google Scholar]
- Volkmar FR, Pauls D. Autism. Lancet 2003; 362: 1133–1141. [DOI] [PubMed] [Google Scholar]
- Volkmar FR, Lord C, Bailey A, Schultz RT, Klin A. Autism and pervasive developmental disorders. J Child Psychol Psychiatry 2004; 45: 135–170. [DOI] [PubMed] [Google Scholar]
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med 1995; 25: 63–77. [DOI] [PubMed] [Google Scholar]
- Freitag CM. The genetics of autistic disorders and its clinical relevance: a review of the literature. Mol Psychiatry 2007; 12: 2–22. [DOI] [PubMed] [Google Scholar]
- Tick B, Bolton P, Happe F, Rutter M, Rijsdijk F. Heritability of autism spectrum disorders: a meta-analysis of twin studies. J Child Psychol Psychiatry 2016; 57: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB et al. Most genetic risk for autism resides with common variation. Nat Genet 2014; 46: 881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm N, Turner TN, Baker C, Vives L, Mohajeri K, Witherspoon K et al. Excess of rare, inherited truncating mutations in autism. Nat Genet 2015; 47: 582–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poelmans G, Franke B, Pauls DL, Glennon JC, Buitelaar JK. AKAPs integrate genetic findings for autism spectrum disorders. Transl Psychiatry 2013; 3: e270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantino JN, Todd RD. Autistic traits in the general population: a twin study. Arch Gen Psychiatry 2003; 60: 524–530. [DOI] [PubMed] [Google Scholar]
- Ronald A, Hoekstra RA. Autism spectrum disorders and autistic traits: a decade of new twin studies. Am J Med Genet B 2011; 156B: 255–274. [DOI] [PubMed] [Google Scholar]
- Robinson EB, Koenen KC, McCormick MC, Munir K, Hallett V, Happe F et al. Evidence that autistic traits show the same etiology in the general population and at the quantitative extremes (5%, 2.5%, and 1%). Arch Gen Psychiatry 2011; 68: 1113–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillberg CL. The Emanuel Miller Memorial Lecture 1991. Autism and autistic-like conditions: subclasses among disorders of empathy. J Child Psychol Psychiatry 1992; 33: 813–842. [DOI] [PubMed] [Google Scholar]
- Piven J, Palmer P, Jacobi D, Childress D, Arndt S. Broader autism phenotype: evidence from a family history study of multiple-incidence autism families. Am J Psychiatry 1997; 154: 185–190. [DOI] [PubMed] [Google Scholar]
- Spiker D, Lotspeich LJ, Dimiceli S, Myers RM, Risch N. Behavioral phenotypic variation in autism multiplex families: evidence for a continuous severity gradient. Am J Med Genet 2002; 114: 129–136. [DOI] [PubMed] [Google Scholar]
- Ronald A, Happe F, Plomin R. The genetic relationship between individual differences in social and nonsocial behaviours characteristic of autism. Dev Sci 2005; 8: 444–458. [DOI] [PubMed] [Google Scholar]
- Lundstrom S, Chang Z, Rastam M, Gillberg C, Larsson H, Anckarsater H et al. Autism spectrum disorders and autistic like traits: similar etiology in the extreme end and the normal variation. Arch Gen Psychiatry 2012; 69: 46–52. [DOI] [PubMed] [Google Scholar]
- Robinson EB, Pourcain B St, Anttila V, Kosmicki JA, Bulik-Sullivan B, Grove J et al. Genetic risk for autism spectrum disorders and neuropsychiatric variation in the general population. Nat Genet 2016; 48: 552–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zettergren A, Jonsson L, Johansson D, Melke J, Lundstrom S, Anckarsater H et al. Associations between polymorphisms in sex steroid related genes and autistic-like traits. Psychoneuroendocrinology 2013; 38: 2575–2584. [DOI] [PubMed] [Google Scholar]
- Jones RM, Cadby G, Melton PE, Abraham LJ, Whitehouse AJ, Moses EK. Genome-wide association study of autistic-like traits in a general population study of young adults. Front Hum Neurosci 2013; 7: 658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron-Cohen S, Wheelwright S, Skinner R, Martin J, Clubley E. The autism-spectrum quotient (AQ): evidence from Asperger syndrome/high-functioning autism, males and females, scientists and mathematicians. J Autism Dev Disord 2001; 31: 5–17. [DOI] [PubMed] [Google Scholar]
- Hoekstra RA, Bartels M, Cath DC, Boomsma DI. Factor structure, reliability and criterion validity of the Autism-Spectrum Quotient (AQ): a study in Dutch population and patient groups. J Autism Dev Disord 2008; 38: 1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuse DH, Mandy WP, Scourfield J. Measuring autistic traits: heritability, reliability and validity of the Social and Communication Disorders Checklist. Br J Psychiatry 2005; 187: 568–572. [DOI] [PubMed] [Google Scholar]
- St Pourcain B, Skuse DH, Mandy WP, Wang K, Hakonarson H, Timpson NJ et al. Variability in the common genetic architecture of social-communication spectrum phenotypes during childhood and adolescence. Mol Autism 2014; 5: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galesloot TE, Vermeulen SH, Swinkels DW, de Vegt F, Franke B, den Heijer M et al. Cohort Profile: the Nijmegen Biomedical Study (NBS). Int J Epidemiol 2017. (e-pub ahead of print). [DOI] [PMC free article] [PubMed]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009; 5: e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet 2010; 11: 499–511. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26: 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euesden J, Lewis CM, O'Reilly PF. PRSice: Polygenic Risk Score software. Bioinformatics 2015; 31: 1466–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodological) 1995; 57: 289–300. [Google Scholar]
- Glickman ME, Rao SR, Schultz MR False. discovery rate control is a recommended alternative to Bonferroni-type adjustments in health studies. J Clin Epidemiol 2014; 67: 850–857. [DOI] [PubMed] [Google Scholar]
- Harris SE, Hagenaars SP, Davies G, David Hill W, Liewald DC, Ritchie SJ et al. Molecular genetic contributions to self-rated health. Int J Epidemiol 2016. (e-pub ahead of print). [DOI] [PMC free article] [PubMed]
- Hagenaars SP, Harris SE, Davies G, Hill WD, Liewald DC, Ritchie SJ et al. Shared genetic aetiology between cognitive functions and physical and mental health in UK Biobank (N=112 151) and 24 GWAS consortia. Mol Psychiatry 2016; 21: 1624–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherman A, Wang R, Avramopoulos D. Orientation, distance, regulation and function of neighbouring genes. Hum Genomics 2009; 3: 143–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 2010; 464: 768–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrieras JB, Kudaravalli S, Kim SY, Dermitzakis ET, Gilad Y, Stephens M et al. qHigh-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet 2008; 4: e1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet 2010; 6: e1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 2015; 11: e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamshere ML, Langley K, Martin J, Agha SS, Stergiakouli E, Anney RJ et al. High loading of polygenic risk for ADHD in children with comorbid aggression. Am J Psychiatry 2013; 170: 909–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapohl E, Euesden J, Zabaneh D, Pingault JB, Rimfeld K, von Stumm S et al. Phenome-wide analysis of genome-wide polygenic scores. Mol Psychiatry 2016; 21: 1188–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Braber A, Zilhao NR, Fedko IO, Hottenga JJ, Pool R, Smit DJ et al. Obsessive-compulsive symptoms in a large population-based twin-family sample are predicted by clinically based polygenic scores and by genome-wide SNPs. Transl Psychiatry 2016; 6: e731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet 2013; 9: e1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Jia P, Wolfinger RD, Chen X, Zhao Z. Gene set analysis of genome-wide association studies: methodological issues and perspectives. Genomics 2011; 98: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bralten J, Arias-Vasquez A, Makkinje R, Veltman JA, Brunner HG, Fernandez G et al. Association of the Alzheimer's gene SORL1 with hippocampal volume in young, healthy adults. Am J Psychiatry 2011; 168: 1083–1089. [DOI] [PubMed] [Google Scholar]
- Bralten J, Franke B, Waldman I, Rommelse N, Hartman C, Asherson P et al. Candidate genetic pathways for attention-deficit/hyperactivity disorder (ADHD) show association to hyperactive/impulsive symptoms in children with ADHD. J Am Acad Child Adolesc Psychiatry 2013; 52: 1204–1212. e1201. [DOI] [PubMed] [Google Scholar]
- Poelmans G, Pauls DL, Buitelaar JK, Franke B. Integrated genome-wide association study findings: identification of a neurodevelopmental network for attention deficit hyperactivity disorder. Am J Psychiatry 2011; 168: 365–377. [DOI] [PubMed] [Google Scholar]
- Ozomaro U, Cai G, Kajiwara Y, Yoon S, Makarov V, Delorme R et al. Characterization of SLITRK1 variation in obsessive-compulsive disorder. PLoS ONE 2013; 8: e70376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Vondervoort I, Poelmans G, Aschrafi A, Pauls DL, Buitelaar JK, Glennon JC et al. An integrated molecular landscape implicates the regulation of dendritic spine formation through insulin-related signalling in obsessive-compulsive disorder. J Psychiatry Neurosci 2016; 41: 140327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci 2011; 14: 285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valles A, Martens GJ, De Weerd P, Poelmans G, Aschrafi A. MicroRNA-137 regulates a glucocorticoid receptor-dependent signalling network: implications for the etiology of schizophrenia. J Psychiatry Neurosci 2014; 39: 312–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics C. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 2013; 381: 1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 2013; 45: 984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Huentelman M, Smith C, Qiu S. MET receptor tyrosine kinase as an autism genetic risk factor. Int Rev Neurobiol 2013; 113: 135–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Lu Z, Li G, Piechowicz M, Anderson M, Uddin Y et al. The autism-associated MET receptor tyrosine kinase engages early neuronal growth mechanism and controls glutamatergic circuits development in the forebrain. Mol Psychiatry 2016; 21: 925–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.