Abstract
The amygdala is a key brain region that is critically involved in the processing and expression of anxiety and fear-related signals. In parallel, a growing number of preclinical and human studies have implicated the microbiome–gut–brain in regulating anxiety and stress-related responses. However, the role of the microbiome in fear-related behaviours is unclear. To this end we investigated the importance of the host microbiome on amygdala-dependent behavioural readouts using the cued fear conditioning paradigm. We also assessed changes in neuronal transcription and post-transcriptional regulation in the amygdala of naive and stimulated germ-free (GF) mice, using a genome-wide transcriptome profiling approach. Our results reveal that GF mice display reduced freezing during the cued memory retention test. Moreover, we demonstrate that under baseline conditions, GF mice display altered transcriptional profile with a marked increase in immediate-early genes (for example, Fos, Egr2, Fosb, Arc) as well as genes implicated in neural activity, synaptic transmission and nervous system development. We also found a predicted interaction between mRNA and specific microRNAs that are differentially regulated in GF mice. Interestingly, colonized GF mice (ex-GF) were behaviourally comparable to conventionally raised (CON) mice. Together, our data demonstrates a unique transcriptional response in GF animals, likely because of already elevated levels of immediate-early gene expression and the potentially underlying neuronal hyperactivity that in turn primes the amygdala for a different transcriptional response. Thus, we demonstrate for what is to our knowledge the first time that the presence of the host microbiome is crucial for the appropriate behavioural response during amygdala-dependent memory retention.
Introduction
The neurobiology of fear critically implicates the amygdala as the key brain structure in anxiety disorders.1, 2, 3, 4 Currently, our understanding of the underlying molecular pathophysiology of such disorders is not well understood, although it is well appreciated that the amygdala plays a key role in the acquisition and expression of fear and anxiety-related behaviours.5, 6 Over the past decade, it has become clear that the host intestinal microbiota has the capacity to alter behaviours relevant to anxiety and stress responses7, 8 and to regulate relevant central nervous system molecular changes at a transcriptional level.9, 10 In addition, the use of microbiota-deficient animal models, such as germ-free (GF) mice or gut microbiota-depleted animals, has allowed in-depth assessment of the impact of an absence of the intestinal bacteria on multiple aspects of host physiology11, 12 including brain and behaviour.13, 14, 15, 16, 17, 18 These behaviours are known to be controlled, at least in part, by the amygdala, a brain region critically involved in fear learning.4, 19, 20
Recently, we have shown that the amygdala transcriptome in GF mice is widely dysregulated and shows features of neuronal hyperactivity at the gene expression level.21 Coinciding with these findings, GF animals display greater amygdalar volume with increased excitatory spine density in the basolateral nuclei,22 demonstrating that the presence of the host microbiome critically regulates the morphology and transcriptional programming within the amygdala. Attempts to understand the underlying molecular mechanisms of many psychiatric disorders have focussed attention on the role of microRNAs (miRNAs)23 that act as posttranscriptional regulators of gene expression.24, 25 It has been demonstrated that brain-specific miRNA candidates within the amygdala regulate anxiety and fear-related behaviours in mice.26, 27, 28 However, there is a paucity of information about whether the microbiome could regulate amygdala-dependent fear behaviours or modify the expression of miRNAs in the central nervous system.
To this end we investigated amygdala-dependent fear memory in GF and an additional group of GF colonized mice (ex-GF). Manipulating the timing of colonization in GF animals further supports the importance that the microbiome has during neurodevelopmental time windows that are crucial for establishing normal behavioural responses in these mice.14, 29 We used a modified cued fear conditioning paradigm in a 1-day setting to test the hypothesis that the microbiome regulates fear-related behaviours in an amygdala-dependent manner. To compliment this, we investigated, using unbiased genome-wide RNA sequencing the transcriptional and posttranscriptional landscape in the amygdala of naive GF mice and after stimulation by a memory-based recall challenge. Our data uncovered a heretofore-undescribed role of the microbiome in regulating fear memory in GF mice.
Materials and methods
Animals
C57BL/6J GF and conventionally raised (CON) breeding pairs were obtained from Taconic (Germantown, NY, USA) with F1-generation offspring used in all experiments. Male GF mice were housed in gnotobiotic flexible-film isolators (2–4 mice per cage) kept on a strict 12 h light/dark cycle. The ex-GF mice were all initially raised within the GF isolators until postnatal day 21 when they were removed and housed in a standard animal unit next to CON mice in order to allow for efficient colonization by environmental microbes.30, 31 The ex-GF mice were initially put in clean cages with dirty bedding from CON, as mice are coprophagic this will allow for efficient colonization. Male CON mice were housed in controlled conditions with regulated temperature (20–21 °C) and humidity (55–60%) with mice 2–4 per cage on the same 12 h light/dark cycle as GF mice and in the same cage type. All mice, CON, GF and ex-GF received the same autoclaved, pelleted diet (Special Diet Services, product code 801010, Essex, UK) until ∼10 weeks of life when tissue was harvested. All experiments were conducted in accordance with the European Directive 86/609/EEC. Approval by the Animal Experimentation Ethics Committee of University College Cork and Health Products Regulatory Authority (HPRA) was obtained before commencement of all animal-related experiments. Both the conventional and germ-free facility adhere to the same animal care guidelines in terms of temperature, humidity and noise levels.
Experimental design
Two cohorts of male CON, GF and ex-GF mice were used in this study. Cohort 1 was used to investigate the impact of GF status on amygdala-dependent behaviours using a modified 1-day cued fear conditioning paradigm (n=13–15/group; Figure 1a). In addition, we investigated the impact of microbiota colonization early in life on fear-related behaviours. Cohort 2 was used to confirm the memory retention deficit and assess gene expression priming and changes using both naive and fear conditioned CON and GF animals 1 h after a cued fear stimulus26, 32 (n=5–12/group; Figure 2). CON and GF cohort 2 mice were culled 1 h after the fear retention stimulus.
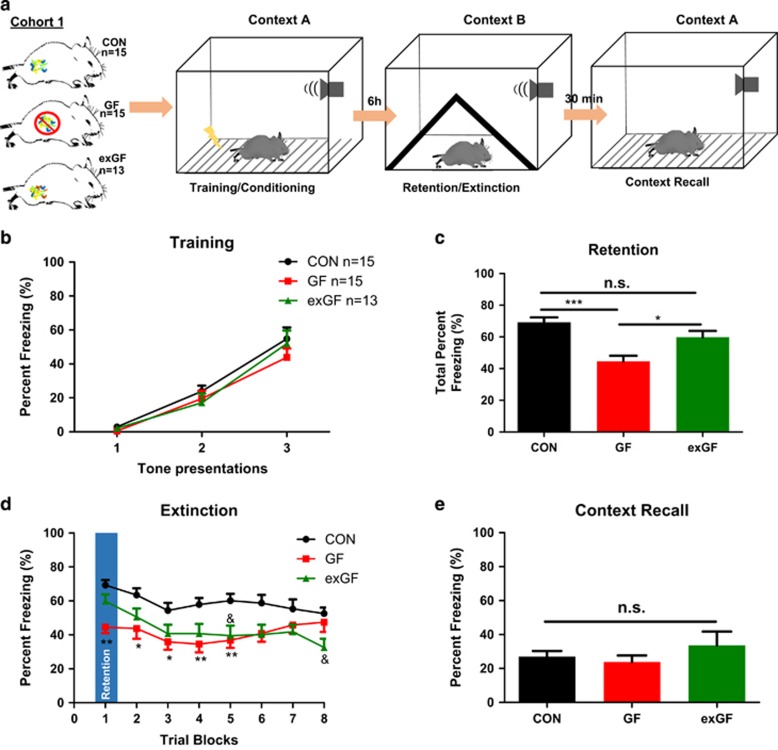
Figure 1.
Behavioural assessment to cued fear conditioning in germ-free (GF) mice. (a) Schematic representation of 1-day fear conditioning protocol. (b) Percentage freezing during training to tone (context A). (c) Total percentage freezing for the first trial block (initial 5 tones) used to demonstrate cued memory retention (context B). (d) Cued fear extinction percentage freezing during all 8 trial blocks (40 conditioned stimulus (CS) tone presentations, context B). (e) Total percentage freezing during context recall (context A) (5 min no stimulus trial). CON, conventionally raised mice. *P<0.05, **P<0.01, ***P<0.001. *Represents comparison between CON and GF; &represents comparison between CON and ex-GF.
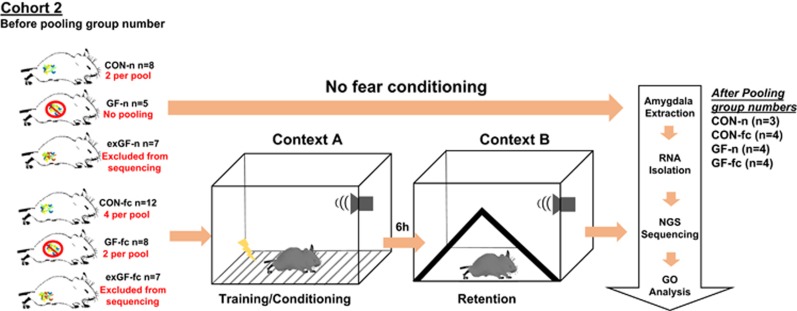
Figure 2.
Experimental approach and RNA pooling for investigating mRNA change in the amygdala. (a) Schematic depiction of experimental design including stimulated and naive mice. Schematic indicates number of mice per group. Red indicates the number of mice used to create one pool (equal volumes of RNA were used). The final number of animals per group after pooling was 4 pools per group (except CON-n, pooled to make n=3) for RNA sequencing. GF-n mice were not pooled as only 5 animals were in this group. CON, conventionally raised mice; CON-n, CON naive; GF, germ free; GF-n, GF naive.
Cued fear conditioning
In order to complete the testing protocol as quickly as possible, once animals were removed from the isolators, a modified 1-day fear conditioning was utilized. This was adapted from the protocol of Izquierdo et al.33 See Supplementary Materials and Methods for full details of adapted cued fear conditioning protocol.
Amygdala and RNA extraction
Whole mouse half brains were snap frozen in isopentane on dry ice and stored in −80 °C. Tissue was placed in a cryostat and allowed to reach −20° C. A standard adult mouse brain matrix was used and 1 mm thick brain slices were taken. Using the Palkovits punch technique34 and a standard mouse atlas as a guide, whole amygdala tissue punches were taken (1 punch per mouse, approximately bregma 1.82 mm). Amygdala punches were stored in RNAlater (Sigma-Aldrich, Wicklow, Ireland) for 24 h, removed and tissue was again stored at −80 °C until extraction. Following the manufacturer’s protocol, a mirVana miRNA kit (Ambion/Life Technologies, Dublin, Ireland) was used to extract total RNA from all animals. A Nanodrop 1000 (Thermo Scientific, UK) was used to determine RNA concentration. Only germ-free samples and conventional, both naive and fear conditioned, were sent for Illumina next-generation sequencing (Exiqon, Vedbæk, Denmark). RNA was pooled (except for GF-n, n=5 after RNA extraction) within each group by combining equal amounts of RNA from two animals resulting in a final sample group of four (except CON-n, n=3, and GF-n, GF-fc and CON-fc, n=4/group after pooling). Figure 2 depicts experimental groups and describes RNA pooling before sequencing.
mRNA sequencing
Sequencing was conducted by Exiqon on the NextSeq500 with an average of 50 million reads with a 50 bp single-end read length. Reference genome of obtained sequences was performed using the reference annotation: Mus musculus (organism), GRCm38, UCSC (University of California, Santa Cruz) Genome browser (reference genome) and Ensemble (Annotation reference). For full details see Supplementary Materials and Methods.
Differential gene expression and GO term enrichment analyses
Data analysis conducted by Exiqon is as follows
Data analysis pipeline is based on the Tuxedo software package that combines open-source software and implements peer-reviewed statistical methods. Complementary to this, Exiqon employs specialized software developed internally to interpret and improve readability of the final results. The components of next-generation RNA-sequencing pipeline include Bowtie2 (v.2.2.2), Tophat (v2.0.11) and Cufflinks (v2.2.1). Briefly, Tophat is used to align the sequencing reads to the reference genome (GRCm38, UCSC Genome browser and Ensemble (Annotation reference)). Cufflinks takes the alignment results to assemble the aligned sequence into transcripts, constructing a map of the transcriptome. Cufflinks assembles aligned reads into different transcript isoforms based on exon usage and also determines the transcriptional start sites. For comparison of groups Cufflinks is used to calculate the FPKM (number of fragments per kilobase per million mapped fragments) and tests for differential expression and regulation among the assembled transcripts across the submitted samples using the Cufflinks output. Differential expression analysis is conducted in the EdgeR statistical software package (Bioconductor, http://www.bioconductor.org/). An adjusted P-value of ⩽0.1 (Benjamini–Hochberg method)21, 35 was considered significantly differentially regulated. Differentially expressed genes were then analysed for enrichment of Gene Ontology (GO) terms using the DAVID Bioinformatics Resources (v6.8).36
MicroRNA sequencing
For full details of miRNA sequencing in CON and GF mice and predicted interaction with dysregulated mRNA see Supplementary Materials and Methods.
Statistics
Fear conditioning
The effects of GF status on freezing during cued fear conditioning behaviour was analysed using two-way repeated measures analysis of variance for trial block in the case of extinction and respective post hoc comparisons. Trial block 1 was additionally separated and analysed for deficits in retention. One-way analysis of variance with Tukey’s post hoc was conducted on fear retention data. Comparison of trial blocks 1 and 2 and 1 and 3 was conducted to determine presence of extinction learning.
mRNA sequencing
Differential expression analysis for RNA sequencing is conducted in the EdgeR statistical software package (Bioconductor, http://www.bioconductor.org/). An adjusted P-value of ⩽0.1 (Padj, Benjamini–Hochberg method) was considered significantly differentially regulated as per previously published sequencing data.21, 37 A false discovery rate cutoff of Padj<0.1 is commonly used in transcriptome-wide studies; using the DESeq2 package it is the default setting in this analysis pipeline.35 In order to further highlight the consistency of our data and test for potential statistical confounding, we have also analysed our data using an even more stringent cutoff of Padj<0.05 (Supplementary Figure 3).
Results
One-day cued fear conditioning in CON mice
We successfully established a 1-day fear conditioning protocol adapted from Izquierdo et al.33 This was necessary because of the logistical and practical constraints of GF facilities and in order to reduce potential for colonization. We subjected CON mice to cued fear conditioning, either 2 or 3 conditioned stimulus/unconditioned stimulus pairings (Supplementary Figure 1a). Following a 6 h delay, all mice were subjected to fear retention/extinction trials. Both groups of mice were able to learn the association between conditioned stimulus and unconditioned stimulus and show memory retention to tone presentations in a 1-day setting. The freezing response was quantitatively somewhat lower in our CON mice when compared with previous studies in this strain;33 however, the delay of 6 h was sufficient time to allow for memory consolidation and subsequent expression of conditioning to tone (Supplementary Figures 1b and c). Scrutinizing the individual trial blocks (each block composed of 5 tones in the absence of foot shock), they showed reduced freezing (extinction) from trial blocks 1 to 2 (Supplementary Figure 1d).
GF mice display reduced freezing in retention and extinction trial
No difference in acquisition was observed between all three experimental groups during conditioning to tone (Figure 1b). When challenged 6 h after acquisition, GF mice displayed reduced percentage freezing during retention (F(2,42)=1.3028, P<0.0001) and extinction trials (trial blocks 1–5) (housing, F(2,32)=34.08, P<0.0001 and trial blocks, F(7,32)=3.222, P<0.01) when compared with CON mice (Figures 1c and d). The ex-GF mice during the initial first trial block (fear retention) was indistinguishable from CON mice (69.30±2.957 vs 59.76±4.021, mean±s.e.m; Figure 1c). However, like GF mice, they displayed reduced percentage freezing during trial blocks 5 and 8 (Figure 1d). When mice were placed back into the initial condition context they displayed minimal percentage freezing to context with no observed differences between the groups (Figure 1e). Cohort 2 again replicated our finding with GF animals displaying reduced percentage freezing to tone during the retention test (F(2,26)=4.913, P=0.0163) with no statistically significant difference in acquisition between groups (Interaction, F(4,66)=1.0941, P=0.1139) (Supplementary Figures 2a and b). Again, ex-GF mice displayed comparable freezing to CON mice both during acquisition and retention tests (Supplementary Figures 2a and b). In addition, CON and ex-GF mice displayed reduced freezing from trial blocks 1 to 3 indicating extinction of the conditioned stimulus/unconditioned stimulus association; however, this effect was not present in GF mice. Interestingly, in our validation cohort of 1-day fear conditioning (Supplementary Figure 1c) there was a faster extinction in CON mice than test cohort (Figure 1) but both cohorts of CON mice show reduced freezing by trial block 3 during the extinction trial.
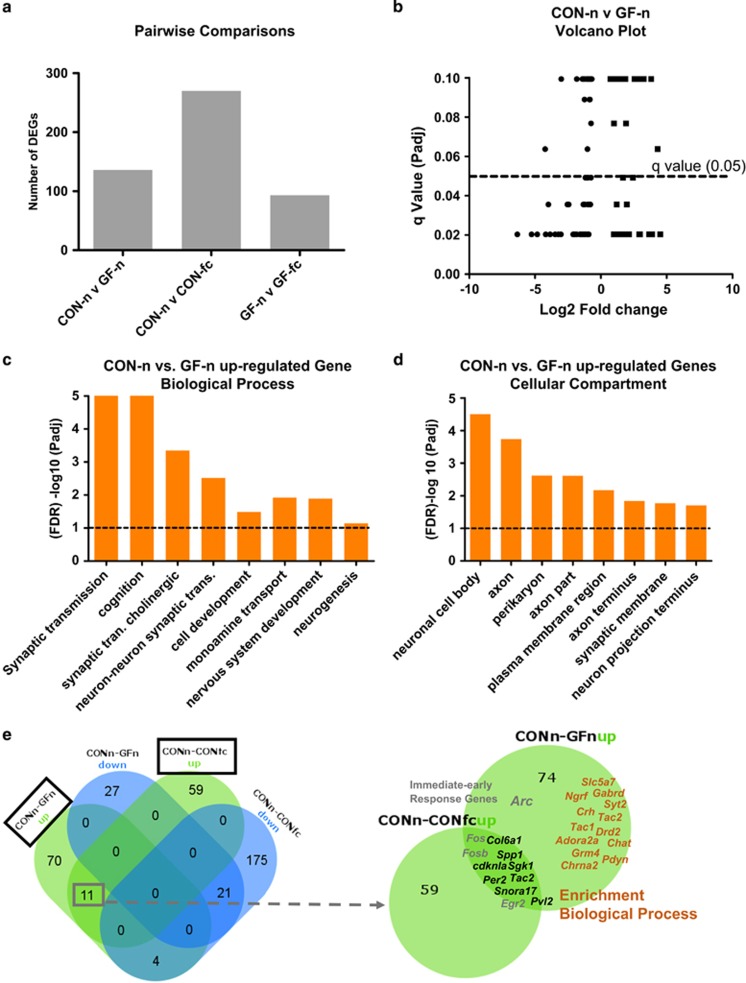
Enrichment in genes related to neuronal transmission and nervous system development in the amygdala of GF naive mice
We performed unbiased deep sequencing of mRNA in the amygdala of naive and fear conditioned GF and CON mice. Analysing gene expression among all four groups by pairwise comparisons, we found a number of differentially regulated genes across all groups when comparing naive with stimulated mice (CON-n vs CON-fc (270) and GF-n vs GF-fc (93); Figure 3a and Supplementary Table 1). First, in comparison between naive CON-n and GF-n mice, we see a total of 133 genes to be altered because of germ-free status that can be further broken down to 85 up- and 48 down-regulated genes (Figures 3a and b and Supplementary Table 1). Next, we investigated biological functions linked to the differentially expressed genes. GO analysis of up-regulated genes (CON-n vs GF-n) shows significant overrepresentation of genes for both biological process and cellular compartment categories (Figures 3c and d). Specifically, we found enrichment in GO terms such as synaptic transmission, cholinergic transmission, nervous system development and neurogenesis (Figure 3c). In addition, we see a number of immediate-early responding genes (Fos, Fosb, Egr2, Arc and Nr4a1) and genes involved in neurotransmission (Drd2, Syt2, Chat and Adora2a) to be up-regulated in naive GF mice, in line with our previous finding indicating hyperactivity in the amygdala of GF naive mice.21 To further investigate this potential hyperactive state present in naive GF amygdala, we compared differentially regulated genes under naive conditions (CON-n vs GF-n) to genes that are differentially regulated upon the fear retention test in control conditions (CON-n vs CON-fc) (Figure 3e). We note common up-regulated or down-regulated of 11 genes and 21 genes, respectively, in both comparisons. Interestingly, overlapping up-regulated genes also include several immediate-early genes such as Fos, Fosb and Egr2. Thus, a significant proportion of genes (P<1.6−13 for up-regulated genes, P<2.3−27 for down-regulated genes) differentially regulated between CON and GF mice are genes that are necessarily induced during fear memory retention. This further suggests elevated neuronal activity in the amygdala in naive GF animals, whereas no significant enrichment in GO terms was noted among down-regulated genes in GF-n mice.
Figure 3.
Altered gene expression in the amygdala of GF naive mice when compared with naive CON. (a) Number of differentially regulated genes (DEGs) between all pairwise comparisons (including fear conditioned mice). (b) Volcano plot representing the number of differentially regulated genes between CON-n and GF-n mice. Each gene is graphed with fold change (Log2) against q-value (P-value adjusted (Padj) for multiple comparisons). (c) Gene Ontology analysis for a selection of significant biological processes from the up-regulated gene between CON-n and GF-n mice. (d) Enriched terms for cellular compartment from differentially up-regulated gene between CON-n and GF-n mice. (e) Venn diagrams showing overlaps between CON-n and GF-n and CON-n and CON-fc up- and down-regulated and the gene symbols that contribute to the enrichment in biological process in orange. In grey are immediate-early response genes that are up-regulated in naive GF mice and stimulated CON mice. CON, conventionally raised mice; CON-fc, CON after fear retention; CON-n, CON naive; GF, germ free; GF-n, GF naive.
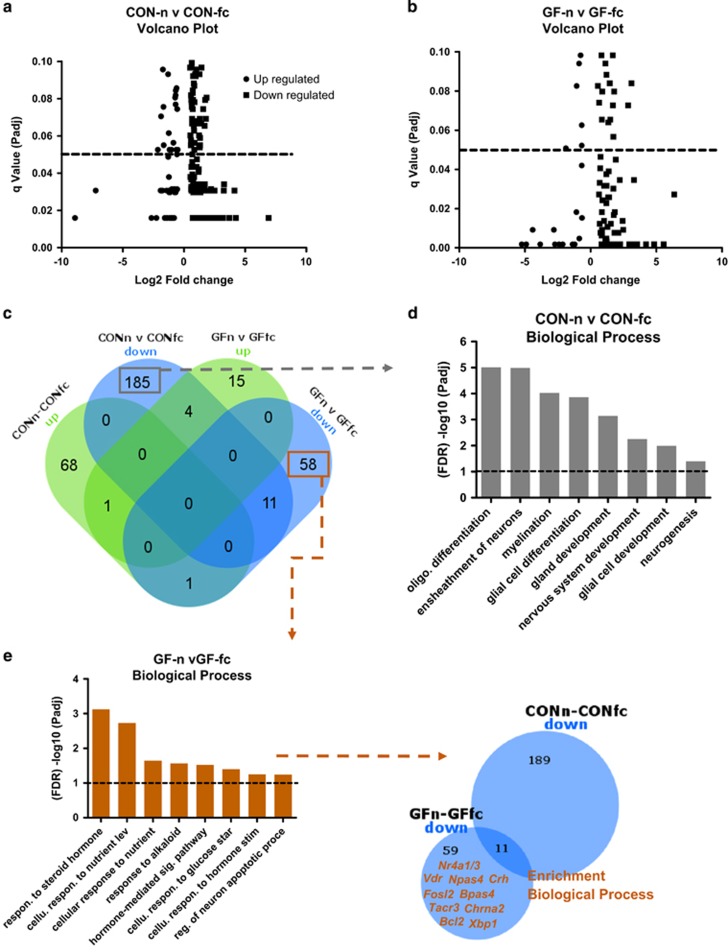
Unique transcriptional response to fear retention stimulus in CON and GF mice
We found a number of genes to be differentially regulated in both CON-fc and GF-fc 1 h after the retention test when compared with their unconditioned counterparts (Figures 4a and c). Very little overlap between CON-fc and GF-fc mice (11 down-regulated and 1 up-regulated) was present after fear retention (Col8a2, Lbp, Thbs4, Capn11, Rp17a, Foxj1, Dnahc6, Aqp1, Hbb-1b, Dynlrb2 and Ccdc135; Figure 4c). The 185 up-regulated genes and 68 down-regulated genes were unique to CON-fc mice after fear retention, with 58 and 15 up- and down-regulated genes unique in GF-fc conditioned mice respectively (Figures 4a–c). Overall, we note that CON-fc and GF-fc mice respond differently to retention testing at the level of gene expression in the amygdala. Next, to investigate the cellular response that was occurring in CON-fc mice, we conducted GO analysis on the 185 down-regulated genes that were unique to CON-fc mice that were not altered in GF-fc mice after retention testing. We found a significant overrepresentation of genes associated with GO terms such as oligodendrocyte development, glial cell development and myelination (Sox10, Sox9, Aspa, Plp1, Tlr4, Id4, Trp73, Fa2h; Figure 4d). However, some of these genes were also associated with biological processes such as gland development (Sox10, Sox9, Id4 and Fa2h). We found no significant enrichment in up-regulated genes unique to CON-fc fear conditioned mice. Enrichment on the 58 down-regulated genes unique to GF-fc mice showed significant enrichment in many GO terms in response to steroid hormones, nutrient starvation, hormone stimulation and hormone-mediated signalling (Figure 4e). In addition, a number of genes (22, overlap not shown) that were up-regulated in CON-n vsGF-n were now down-regulated in GF-n vs GF-fc (for example, Nr4a1, Crh, Fosl2, Gabre and Chrna2).
Figure 4.
Transcriptional response to fear retention in CON and GF mice. (a, b) Volcano plot of all the differentially regulated genes between CON-n and CON-fc and GF and GF-fc. (c) Venn diagram showing the overlap between up- and down-regulated genes after comparison of naive and fear conditioned CON and GF mice. (d) Enrichment among unique up-regulated genes in CON-fc mice that are not differentially regulated in GF-fc mice. (e) Enrichment of down-regulated genes that are unique to GF-fc mice. Venn diagram represents the overlap between down-regulated genes in CON-n vs CON-fc and GF-n vs GF-fc. Genes highlighted in orange directly contribute to the Gene Ontology (GO) terms enriched for biological processes in GF-fc mice. CON, conventionally raised mice; CON-fc, CON after fear retention; CON-n, CON naive; GF, germ free; GF-fc, GF after fear retention; GF-n, GF naive.
Functional enrichment and predicted interaction with dysregulated miRNAs and mRNAs in GF-n mice
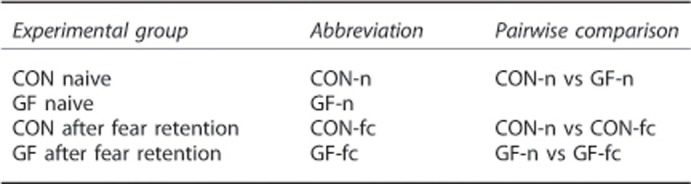
We found by pairwise comparison a total of 13 miRNAs, 7 up-regulated and 6 down-regulated, in GF-n mice when compared with CON-n (Figure 5a). Expression profile of miRNAs in GF-n compared with CON-n is represented in Supplementary Table 2. All up-regulated miRNAs have a fold change of >1.4 with strong fold reduction in down-regulated miRNAs (Supplementary Table 2). Supplementary Table 3 shows the altered miRNA/mRNA (both up and down-regulated) predicted interaction with dysregulated mRNAs in GF-n mice. As our mRNA sequencing revealed among up-regulated gene an orchestrated enrichment for GO terms for synaptic transmission, cognition, cholinergic transmission, nervous system development and neurogenesis, we investigated whether any of our miRNAs have predicted targets that are enriched for such GO terms. As only our up-regulated mRNAs showed significant enrichment we listed all the predicated targets (present in 3 prediction algorithms) of down-regulated miRNAs in GF-n mice (Figure 5b) and conducted GO enrichment analysis. We then overlapped significant GO terms from our mRNA sequencing with significant GO terms from our predicted miRNA target enrichment. We found that miR-211-5p, miR-204-5p and miR-200b-3p displayed significant enrichment for select GO terms that overlapped with those from mRNA sequencing (Figures 5c and d). Supplementary Table 3 indicates which of our observed dysregulated mRNAs in GF-n mice these miRNAs are predicted to target.
Figure 5.
Altered miRNAs in GF-n mice and their relevant Gene Ontology (GO) terms. (a) Number of dysregulated miRNAs based on raw P-value for each experimental comparison. (b) Number of predicted mRNA for individual down-regulated miRNAs in GF-n mice when compared with CON-n that occur in more than 3 prediction algorithms. (c, d) Venn diagram of the number of significant (Padj) GO terms for biological processes for each miRNA overlapped with significant GO terms from mRNA sequencing. Bar graphs represent a selection of significant terms that were common between mRNA sequencing and enriched from individual mRNAs. (e) Venn diagram showing the number of overlapping miRNAs between groups after fear retention test (CON-fc and GF-fc). CON, conventionally raised mice; CON-fc, CON after fear retention; CON-n, CON naive; GF, germ free; GF-fc, GF after fear retention; GF-n, GF naive.
Unique miRNA response to fear retention stimulus in CON and GF mice with predicted interaction with dysregulated mRNAs
We found a number of miRNAs to be differentially regulated after the retention test in both GF-fc and CON-fc when compared with the unconditioned counterparts respectively (Supplementary Tables 4 and 5). The CON-fc mice after fear retention test had altered expression of 25 miRNAs (7 up-regulated and 18 down-regulated; Figure 5e and Supplementary Table 4). After retention, GF-fc mice had 7 altered miRNAs in the amygdala (2 up-regulated and 5 down-regulated; Figure 5e and Supplementary Table 5). miR-34b-5p, miR-34c-5p and miR-34b-3p were down-regulated in both CON-fc and GF-fc mice (Figure 5e). The remaining differentially regulated miRNAs were unique to CON-fc and GF-fc. Again we listed the predicted mRNA targets of differentially regulated miRNAs between CON-n and CON-fc and GF-n and GF-fc and overlapped them with up- or down-regulated mRNAs to identify potential relevant targets of these miRNAs (Supplementary Tables 6 and 7).
Discussion
A growing body of literature implicated the host microbiome in brain and behaviour. Here we show, for what is to our knowledge the first time, that the microbiome regulates amygdala-dependent fear conditioning in GF mice. We found that after training, GF animals displayed reduced freezing to the conditioned stimulus indicative of impaired cued memory recall. Therefore, it appears that GF animals were unable to retain the association between tone and shock to the same degree as CON mice. Exposure to environmental microbes appeared to reverse the deficit in memory retention displayed by GF mice. Previous colonization studies have shown normalization of behavioural phenotypes related to reduced anxiety,14 social deficits29 and hypothalamic–pituitary–adrenal axis activity.38 Regarding extinction, there is evidence of reduced freezing during the initial 3 trial blocks in CON mice, but this was not evident in GF animals. The lack of extinction cannot be easily disentangled from the deficits in retention in GF mice and may be a consequence of the reduced recall-induced freezing response in these mice. In addition, it is worth noting that GF mice display reduced anxiety-related behaviours in a variety of approach–avoidance tasks.13, 14, 39 Thus, the relative contribution of this reduced anxiety response to the observed differences in cued fear conditioning is unclear, although it is worth noting that the neurobiological basis of learned fear and ethological anxiety responses in such rodent paradigms are quite distinct.40, 41, 42 Moreover, we did not see a difference in freezing response during the acquisition training between groups. In addition, future studies should investigate the role of the dysfunctional hypothalamic–pituitary–adrenal axis evident in GF animals38 in potentially driving the altered response to fear conditioning given the relationship between stress hormones such as glucocorticoids and memory formation.43, 44
Accompanying this impairment in fear memory recall was a marked reshaping of the transcriptional landscape in the amygdala of GF-n mice when compared with CON-n mice. The in-depth analysis of differentially up-regulated genes in the amygdala demonstrates a marked increase in the expression of immediate-early genes and genes involved in synaptic transmission with specific enrichment for genes involved in cholinergic transmission. Among others, we found an increase in Fos, Fosb, Egr2 and Arc in GF-n mice, in line with our previous finding indicating orchestrated up-regulation of these genes in the amygdala of naive GF mice.21 It is worth noting that these findings are consistently independent of host genetics/strain and method of tissue isolation and is more related to the microbiota-deficient animal model used in both studies. Activation of immediate early-response genes and CREB activation, which is essential for activity-driven neuronal gene expression, are markedly up-regulated in the amygdala following social novelty and fear memory retrieval.21, 45 Our data confirm this typical observation as CON-fc mice display increased expression of immediate-early genes, for example, Fos, Fosb and Erg2, indicating that cued fear memory recall induces activation of these early-response genes in the amygdala of CON mice. Indeed, up-regulated of these typical genes (for example, Fos, Nr4a1, Egr1) has previously been demonstrated to follow cued memory recall.46
Increased neuronal activity of the amygdala in GF-n mice is further suggested by specific up-regulated of genes related to cholinergic and dopaminergic neurotransmission (Drd2, Adora2a, Tac1, Chrnra2 and Chat) that have been shown to be critical for many aspects of amygdala-dependent conditioned learning and extinction.47, 48 Many of these genes additionally contribute to enrichment for GO terms such as nervous system development, neurogenesis and cell development with specific increases in genes such as nerve growth factor receptor (Ngfr) and early growth factor 1 and 2 (Egr1, Egr2), with the latter previously shown to regulate fear memory conditioning.49, 50 Together, these data suggest that the amygdalar circuitry of GF mice at baseline is in a hyperactive state, potentially originating from altered microglial signalling during critical phases of neurodevelopment.51, 52 Interestingly, we find no overlap in differentially regulated genes when comparing our current findings with those of a previous study that investigated changes in gene expression in other brain regions (hippocampus, striatum and frontal cortex) of GF mice using hybridization-based microarray technology.16 Despite the use of different brain regions, many of the genes seen in both studies are associated with several common neuronal processes such as synaptic transmission and nervous system development. However, we see no similarity at the individual gene level suggesting that gene expression in different brain regions are differentially affected by the microbiota.
We next tested whether this differential baseline priming of the amygdala in GF mice would translate to differential gene expression patterns in response to a recall stimulus. Surprisingly, minimal overlap existed between comparisons and large differences in gene expression were present in response to the retention test between CON-fc and GF-fc mice when compared with naive, unconditioned counterparts (CON-n and GF-n; Figure 4c). Although we saw the expected increase in expression of immediate early genes Fos, Fosb and Egr2, indicating activation of the amygdala after fear retention in CON-fc mice, a comparable increase in the expression of these genes was not observed in the GF-fc animals. This suggests altered cellular signalling pathways are associated with impaired memory retention in GF animals. This unique response pattern is likely because of already elevated levels of immediate-early gene expression in naive GF animals and the potentially underlying neuronal hyperactivity that in turn primes the amygdala for a different transcriptional response. In fact, a number of genes that were up-regulated in GF-n mice when compared with CON-n (for example, Nr4a1, Crh, Gabre and Fosl2) and that contributed to the enrichment in increased synaptic transmission-related genes were down-regulated after fear retention. GF-fc mice had decreased expression of Nr4a1 and Nr4a3 that are a class of nuclear receptor transcription factors that are important for the formation of long-lasting memory as blockade of these receptors impairs long-term but not short-term memory in the hippocampus53 and are up-regulated following cued and contextual fear conditioning.46, 53 Scrutinizing further down-regulated genes unique to GF-fc mice shows a decrease in many activity-related pathways and transcription factors that further supports that GF-n amygdala is primed for a different transcriptional response to fear retention.
Research from our group has previously demonstrated that there is marked volumetric expansion in the amygdala of GF mice.22 In addition, the basolateral amygdala nuclei GF animals displayed increased spine density, specifically in mushroom spines, that are critical for formation of glutamatergic synapses.54 This suggests that these cells receive more excitatory inputs.22 Though our transcriptomic study is not exclusive to the basolateral amygdala, hyperactivity in this specific nucleus may underlie the inability to regulate fear memory retention or in many behavioural responses that depend on this brain region. Disruption of cued fear memory is less well characterized in animal models of autism spectrum disorder. Deficits in social behaviour are more commonly observed in autism spectrum disorder models, and this is also a feature of the GF phenotype.29, 55, 56 The valproic acid model of autism spectrum disorder also showed increased amygdala volume57 with evidence of hyperactivity.58 In this model, offspring of mothers exposed to valproic acid during gestation display increased memory retention with evidence of increased fear as valproic acid-exposed rats were unable to extinguish cued fear memory.58 This could be related to the increased anxiety displayed by these animals, although this model is also associated with microbiome alterations.59 Clearly, amygdala hyperactivity and dramatic reshaping of the dendritic landscape can have detrimental effects on amygdala-dependent memory.
The most unique and surprising finding in our RNA sequencing was in relation to the impact of the retention test on downregulated gene in the amygdala of CON-fc mice. We found significant enrichment of genes related to oligodendrocyte differentiation and myelination and specific decreases in critical transcription factors (Sox9 and Sox10), gene encoding enzymes responsible for the biosynthesis of sphinogolipids of the myelin sheath (Ugt8a). However, we only found changes in one myelin structural, Plp1.60, 61 There is growing evidence for a relationship between the microbiome and brain myelination patterns.37, 58, 62 More work is required to investigate whether this type of stimulus could correspond with demyelination in the amygdala, but it must be noted that this was unique to CON-fc mice. The same orchestrated down-regulated of myelin-related genes was not seen in GF-fc mice following the retention stimulus.
Although other microbiota-mediated manipulation strategies have investigated cued fear conditioning in rodents, to our knowledge this is the first demonstration of such a behavioural deficit in GF mice. In line with this, chronic probiotic administration of Lactobacillus rhamnosus resulted in increased freezing during cued memory recall.63 Faecal microbiota transplantation from mice on a high-fat diet to microbiota-depleted donors display significant decrease in freezing behaviours when cued memory retention was assessed when compared with mice receiving microbiota form control diet feed donors. High-fat diet fed mice had significant alterations in their microbiota composition, and when transferred to a donor mouse, it resulted in amygdala-dependent memory impairment.64 Together, these data demonstrate that changes at the level of gut microbiota composition can dramatically affect amygdala-dependent learning.
As previous studies have shown the involvement of miRNAs in amygdala-dependent stress and fear-related outputs,21, 65, 66 we next investigated whether posttranscriptional regulation via miRNAs may be a contributing factor underlying impairments in fear memory learning and the observed altered transcriptional network. Indeed, in line with our mRNA sequencing, we investigated miRNA expression in the amygdala of naive CON and GF mice and 1 h after fear retention that is in line with previous studies expanding miRNA expression 30 min to 1 h after conditioning. Analysis by pairwise comparisons revealed a number of differentially regulated miRNAs. We found that 3 miRNAs, miR-211-5p, miR-204-5p and miR-200-3p, had predicted targets that were enrichment for GO terms overlapping with our RNA sequencing analysis and therefore could be contributing to enrichment at the mRNA level. As such, we found enrichment in GO terms such as synaptic transmission, cholinergic transmission and cell development (for example, Chrna2, Arc, Egr2, Crh, Fosb). Interestingly, miR-204-5p and miR-211-5p are from the same miRNA family and therefore have very similar sequence alignments that is reflected by the similarity in the mRNAs they are predicted to target (Supplementary Table 3). Currently, there are no studies to our knowledge investigating the importance of these miRNAs in neuronal function within the amygdala; however, studies have shown the importance of miR-204 family in lens and retina development.67 Further work is needed to tease apart whether the microbiota recruits these miRNAs in regulation of these altered transcriptional networks. In addition, we found a number of miRNAs that are altered after fear retention testing and that are unique to CON-fc and GF-fc that show a predicted interaction with many mRNAs altered from those comparisons (Supplementary Tables 5 and 7). Only three miRNAs were commonly down-regulated after fear retention in CON and GF mice (miR-34b-5p, miR-34c-5p and miR-34b-3p). A recent study demonstrated that targeted deletion of this miRNA family results in resilience to stress-induced anxiety and displayed reduced freezing during retention/extinction trials.68 Conversely, miR-34c has also been shown to be down-regulated in the central amygdala 30 min after acute restraint stress and lentiviral overexpression in this nucleus has an anxiolytic effect.69 The diverging behavioural impact of this miRNA following different experimental approaches in conjunction with acute stressors notwithstanding, these studies highlight the potential important of this target in regulating anxiety-like behaviours.
Conclusion
In conclusion, the present study indicates that retention of cued fear memory relies on the presence of a functional microbiota during critical windows of neurodevelopment. The in-depth analysis of the transcriptome data confirms a functional hyperactivity in the GF amygdala. Here we demonstrate a unique transcriptional response to cued fear retention testing that may, at least in part, underlie the observed impairment in retaining cued fear memory. The apparent hyperactive start of the amygdala at baseline may ultimately prime GF animals to respond differentially to external stimuli. Converging evidence of predicted interaction between miRNAs and their target genes indicates that the observed orchestrated GO enrichments in naive GF mice may be regulated by miRNAs. Future studies should focus on investigating changes in gene expression at multiple time points in order to better appraise a temporal response in the amygdala of GF mice following fear recall. Moreover, this aberrant behavioural profile can be (partially) normalized by introduction of a microbiome after weaning. It will be of interest to investigate the molecular mechanisms driving this partial reversal in future studies as it may unmask specific target genes that may be under the influence of the gut microbiota. Furthermore, given that the microbiome has now become implicated in a wide variety of brain process such as myelination,37 microglia activation/neuroimmune function,52, 70, 71 blood brain barrier function72 and dendritic morphology,22 it will be of interest to investigate the relationship between these processes and microbial regulation of fear recall in the future. Taken together, our data indicate that the microbiome may be a promising new therapeutic target for developing psychobiotic approaches73 for fear-related disorders.
Acknowledgments
We thank Patrick Fitzgerald and Frances O’Brien for technical assistance with animal husbandry. The APC Microbiome Institute is a research centre funded by Science Foundation Ireland (SFI), through the Irish Government’s National Development Plan (Grant Number 12/RC/2273). RMS is supported by the Irish Research Council (IRC) through a Government of Ireland Postdoctoral Fellowship (Grant Number GOIPD/2014/355). GF is supported by a NARSAD Young Investigator Grant from the Brain and Behavior Research Foundation (Grant Number 20771). TGD and JFC are also supported by the Irish Health Research Board, the Department of Agriculture, Food and the Marine and Enterprise Ireland.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
AEH and RMS report no biomedical financial interests or potential conflict of interest. TGD, FS and JFC are principal investigators in the APC Microbiome Institute, University College Cork. GC is a faculty member of the APC Microbiome Institute. The APC Microbiome Institute has conducted research funded by many Pharmaceutical and Food Companies. TGD has been an invited speaker at meetings organized by Servier, Lundbeck, Janssen and AstraZeneca and has received research funding from Mead Johnson, Cremo, Suntory Wellness, Nutricia and 4D Pharma. JFC has been an invited speaker at meetings organized by Mead Johnson, Yakult, Alkermes and Janssen and has received research funding from Mead Johnson, Cremo, Suntory Wellness, Nutricia and 4D Pharma. GM declares no conflict of interest.
Supplementary Material
References
- Apps R, Strata P. Neuronal circuits for fear and anxiety - the missing link. Nat Rev Neurosci 2015; 16: 642-. [DOI] [PubMed] [Google Scholar]
- Aggleton EJ, Everitt BJ, Cardinal RN, Hall J. The amygdala: a functional analysis, 2nd edn. Oxford University Press, 2000.
- Barton RA, Aggleton JP, Grenyer R. Evolutionary coherence of the mammalian amygdala. Proc R Soc Lond B Biol Sci 2003; 270: 539–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDoux J. The emotional brain, fear, and the amygdala. Cell Mol Neurobiol 2003; 23: 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvarci S, Pare D. Amygdala microcircuits controlling learned fear. Neuron 2014; 82: 966–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovote P, Fadok JP, Lüthi A. Neuronal circuits for fear and anxiety. Nat Rev Neurosci 2015; 16: 317–331. [DOI] [PubMed] [Google Scholar]
- Sherwin E, Sandhu KV, Dinan TG, Cryan JF. May the force be with you: the light and dark sides of the microbiota–gut–brain axis in neuropsychiatry. CNS Drugs 2016; 30: 1019–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster JA, Lyte M, Meyer E, Cryan JF. Gut microbiota and brain function: an evolving field in neuroscience. Int J Neuropsychopharmacol 2016; 19: pyv114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stilling R, Dinan T, Cryan J. Microbial genes, brain & behaviour–epigenetic regulation of the gut–brain axis. Genes Brain Behav 2014; 13: 69–86. [DOI] [PubMed] [Google Scholar]
- Stilling RM, Bordenstein SR, Dinan TG, Cryan JF. Friends with social benefits: host-microbe interactions as a driver of brain evolution and development? Front Cell Infect Microbiol 2014; 4: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg R, Toft MF, August B, Hansen AK, Hansen CH. Antibiotic-treated versus germ-free rodents for microbiota transplantation studies. Gut Microbes 2016; 7: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luczynski P, Neufeld K-AM, Oriach CS, Clarke G, Dinan TG, Cryan JF. Growing up in a bubble: using germ-free animals to assess the influence of the gut microbiota on brain and behaviour. Int J Neuropsychopharmacol 2016; 19: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld K-AM, Kang N, Bienenstock J, Foster JA. Effects of intestinal microbiota on anxiety-like behavior. Commun Integr Biol 2011; 4: 492–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke G, Grenham S, Scully P, Fitzgerald P, Moloney R, Shanahan F et al. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol Psychiatry 2013; 18: 666–673. [DOI] [PubMed] [Google Scholar]
- Gareau MG, Wine E, Rodrigues DM, Cho JH, Whary MT, Philpott DJ et al. Bacterial infection causes stress-induced memory dysfunction in mice. Gut 2011; 60: 307–317. [DOI] [PubMed] [Google Scholar]
- Heijtz RD, Wang S, Anuar F, Qian Y, Björkholm B, Samuelsson A et al. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci USA 2011; 108: 3047–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoban AE, Moloney RD, Golubeva AV, Neufeld KM, O’Sullivan O, Patterson E et al. Behavioural and neurochemical consequences of chronic gut microbiota depletion during adulthood in the rat. Neuroscience 2016; 339: 463–477. [DOI] [PubMed] [Google Scholar]
- Desbonnet L, Clarke G, Traplin A, O’Sullivan O, Crispie F, Moloney RD et al. Gut microbiota depletion from early adolescence in mice: Implications for brain and behaviour. Brain Behav Immun 2015; 48: 165–173. [DOI] [PubMed] [Google Scholar]
- Davidson RJ. Anxiety and affective style: role of prefrontal cortex and amygdala. Biol Psychiatry 2002; 51: 68–80. [DOI] [PubMed] [Google Scholar]
- Pare D, Duvarci S. Amygdala microcircuits mediating fear expression and extinction. Curr Opin Neurobiol 2012; 22: 717–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stilling RM, Ryan FJ, Hoban AE, Shanahan F, Clarke G, Claesson MJ et al. Microbes & neurodevelopment–absence of microbiota during early life increases activity-related transcriptional pathways in the amygdala. Brain Behav Immun 2015; 50: 209–220. [DOI] [PubMed] [Google Scholar]
- Luczynski P, Whelan SO, O'Sullivan C, Clarke G, Shanahan F, Dinan TG et al. Adult microbiota‐deficient mice have distinct dendritic morphological changes: differential effects in the amygdala and hippocampus. Eur J Neurosci 2016; 44: 2654–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alural B, Genc S, Haggarty SJ. Diagnostic and therapeutic potential of microRNAs in neuropsychiatric disorders: past, present, and future. Prog Neuropsychopharmacol Biol Psychiatry 2016; 73: 87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Carroll D, Schaefer A. General principals of miRNA biogenesis and regulation in the brain. Neuropsychopharmacology 2013; 38: 39–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet 2015; 16: 421–433. [DOI] [PubMed] [Google Scholar]
- Griggs EM, Young EJ, Rumbaugh G, Miller CA. MicroRNA-182 regulates amygdala-dependent memory formation. J Neurosci 2013; 33: 1734–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias BG, Goodman JV, Ahluwalia R, Easton AE, Andero R, Ressler KJ. Amygdala-dependent fear memory consolidation via miR-34a and Notch signaling. Neuron 2014; 83: 906–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton DE, Cooke CL, Carter BS, Akil H, Watson SJ, Thompson RC. Basal microRNA expression patterns in reward circuitry of selectively bred high-responder and low-responder rats vary by brain region and genotype. Physiol Genomics 2014; 46: 290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbonnet L, Clarke G, Shanahan F, Dinan T, Cryan J. Microbiota is essential for social development in the mouse. Mol Psychiatry 2014; 19: 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen CHF, Nielsen DS, Kverka M, Zakostelska Z, Klimesova K, Hudcovic T et al. Patterns of early gut colonization shape future immune responses of the host. PLoS ONE 2012; 7: e34043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motherway MOC, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ et al. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci USA 2011; 108: 11217–11222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Wei W, Coelho CM, Li X, Baker-Andresen D, Dudley K et al. The brain-specific microRNA miR-128b regulates the formation of fear-extinction memory. Nat Neurosci 2011; 14: 1115–1117. [DOI] [PubMed] [Google Scholar]
- Izquierdo A, Wellman CL, Holmes A. Brief uncontrollable stress causes dendritic retraction in infralimbic cortex and resistance to fear extinction in mice. J Neurosci 2006; 26: 5733–5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palkovits M. Isolated removal of hypothalamic or other brain nuclei of the rat. Brain Res 1973; 59: 449–450. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Hoban A, Stilling R, Ryan F, Shanahan F, Dinan T, Claesson M et al. Regulation of prefrontal cortex myelination by the microbiota. Transl Psychiatry 2016; 6: e774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu XN et al. Postnatal microbial colonization programs the hypothalamic–pituitary–adrenal system for stress response in mice. J Physiol 2004; 558: 263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld K, Kang N, Bienenstock J, Foster J. Reduced anxiety‐like behavior and central neurochemical change in germ‐free mice. Neurogastroenterol Motil 2011; 23: 255–e119. [DOI] [PubMed] [Google Scholar]
- Gross CT, Canteras NS. The many paths to fear. Nat Rev Neurosci 2012; 13: 651–658. [DOI] [PubMed] [Google Scholar]
- Trent NL, Menard JL. The ventral hippocampus and the lateral septum work in tandem to regulate rats' open-arm exploration in the elevated plus-maze. Physiol Behav 2010; 101: 141–152. [DOI] [PubMed] [Google Scholar]
- Engin E, Treit D. The role of hippocampus in anxiety: intracerebral infusion studies. Behav Pharmacol 2007; 18: 365–374. [DOI] [PubMed] [Google Scholar]
- Dominique J-F, Aerni A, Schelling G, Roozendaal B. Glucocorticoids and the regulation of memory in health and disease. Front Neuroendocrinol 2009; 30: 358–370. [DOI] [PubMed] [Google Scholar]
- Roozendaal B, Okuda S, De Quervain D-F, McGaugh J. Glucocorticoids interact with emotion-induced noradrenergic activation in influencing different memory functions. Neuroscience 2006; 138: 901–910. [DOI] [PubMed] [Google Scholar]
- Hall J, Thomas KL, Everitt BJ. Fear memory retrieval induces CREB phosphorylation and Fos expression within the amygdala. Eur J Neurosci 2001; 13: 1453–1458. [DOI] [PubMed] [Google Scholar]
- Keeley MB, Wood MA, Isiegas C, Stein J, Hellman K, Hannenhalli S et al. Differential transcriptional response to nonassociative and associative components of classical fear conditioning in the amygdala and hippocampus. Learn Mem 2006; 13: 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandão ML, de Oliveira AR, Muthuraju S, Colombo AC, Saito VM, Talbot T. Dual role of dopamine D2‐like receptors in the mediation of conditioned and unconditioned fear. FEBS Lett 2015; 589: 3433–3437. [DOI] [PubMed] [Google Scholar]
- Robinson L, Platt B, Riedel G. Involvement of the cholinergic system in conditioning and perceptual memory. Behav Brain Res 2011; 221: 443–465. [DOI] [PubMed] [Google Scholar]
- Ploski JE, Park KW, Ping J, Monsey MS, Schafe GE. Identification of plasticity‐associated genes regulated by Pavlovian fear conditioning in the lateral amygdala. J Neurochem 2010; 112: 636–650. [DOI] [PubMed] [Google Scholar]
- Malkani S, Wallace KJ, Donley MP, Rosen JB. An egr-1 (zif268) antisense oligodeoxynucleotide infused into the amygdala disrupts fear conditioning. Learn Mem 2004; 11: 617–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borre YE, O’Keeffe GW, Clarke G, Stanton C, Dinan TG, Cryan JF. Microbiota and neurodevelopmental windows: implications for brain disorders. Trends Mol Med 2014; 20: 509–518. [DOI] [PubMed] [Google Scholar]
- Erny D, de Angelis ALH, Jaitin D, Wieghofer P, Staszewski O, David E et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci 2015; 18: 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawk JD, Bookout AL, Poplawski SG, Bridi M, Rao AJ, Sulewski ME et al. NR4A nuclear receptors support memory enhancement by histone deacetylase inhibitors. J Clin Invest 2012; 122: 3593–3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmaat AJ, Trachtenberg JT, Wilbrecht L, Shepherd GM, Zhang X, Knott GW et al. Transient and persistent dendritic spines in the neocortex in vivo. Neuron 2005; 45: 279–291. [DOI] [PubMed] [Google Scholar]
- Buffington SA, Di Prisco GV, Auchtung TA, Ajami NJ, Petrosino JF, Costa-Mattioli M. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell 2016; 165: 1762–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arentsen T, Raith H, Qian Y, Forssberg H, Heijtz RD. Host microbiota modulates development of social preference in mice. Microb Ecol Health Dis 2015; 26: 29719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loohuis NO, Kole K, Glennon J, Karel P, Van der Borg G, Van Gemert Y et al. Elevated microRNA-181c and microRNA-30d levels in the enlarged amygdala of the valproic acid rat model of autism. Neurobiol Dis 2015; 80: 42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markram K, Rinaldi T, La Mendola D, Sandi C, Markram H. Abnormal fear conditioning and amygdala processing in an animal model of autism. Neuropsychopharmacology 2008; 33: 901–912. [DOI] [PubMed] [Google Scholar]
- Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 2013; 155: 1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn O, Tenzer S, Werner HB. Myelin proteomics: molecular anatomy of an insulating sheath. Mol Neurobiol 2009; 40: 55–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner M. A matter of identity: transcriptional control in oligodendrocytes. J Mol Neurosci 2008; 35: 3–12. [DOI] [PubMed] [Google Scholar]
- Gacias M, Gaspari S, Santos P-MG, Tamburini S, Andrade M, Zhang F et al. Microbiota-driven transcriptional changes in prefrontal cortex override genetic differences in social behavior. elife 2016; 5: e13442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG et al. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci USA 2011; 108: 16050–16055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce-Keller AJ, Salbaum JM, Luo M, Blanchard E, Taylor CM, Welsh DA et al. Obese-type gut microbiota induce neurobehavioral changes in the absence of obesity. Biol Psychiatry 2015; 77: 607–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerson A, Cacheaux L, Goosens KA, Sapolsky RM, Soreq H, Kaufer D. Changes in brain microRNAs contribute to cholinergic stress reactions. J Mol Neurosci 2010; 40: 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KA, Hoban AE, Clarke G, Moloney GM, Dinan TG, Cryan JF. Thinking small: towards microRNA-based therapeutics for anxiety disorders. Expert Opin Investig Drugs 2015; 24: 529–542. [DOI] [PubMed] [Google Scholar]
- Conte I, Carrella S, Avellino R, Karali M, Marco-Ferreres R, Bovolenta P et al. miR-204 is required for lens and retinal development via Meis2 targeting. Proc Natl Acad Sci USA 2010; 107: 15491–15496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolina D, Di Segni M, Bisicchia E, D'Alessandro F, Cestari V, Ventura A et al. Effects of lack of microRNA-34 on the neural circuitry underlying the stress response and anxiety. Neuropharmacology 2016; 107: 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramati S, Navon I, Issler O, Ezra-Nevo G, Gil S, Zwang R et al. MicroRNA as repressors of stress-induced anxiety: the case of amygdalar miR-34. J Neurosci 2011; 31: 14191–14203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung TC, Olson CA, Hsiao EY. Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci 2017; 20: 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell 2016; 167: 1469–1480 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braniste V, Al-Asmakh M, Kowal C, Anuar F, Abbaspour A, Tóth M et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci Transl Med 2014; 6: 263ra158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnet PW, Cowen PJ. Psychobiotics highlight the pathways to happiness. Biol Psychiatry 2013; 74: 708–709. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.