Abstract
Implementation of genomic medicine into clinical care continues to increase in prevalence in medical centers worldwide. As defined by the National Human Genome Research Institute, “Genomic medicine is an emerging medical discipline that involves using genomic information about an individual as part of their clinical care.…” The genomic information utilized falls broadly into two categories: 1) highly penetrant genetic disorders1 and 2) pharmacogenomics. Herein, we focus on pharmacogenomics, although the Pharmacogenomics Clinical Annotation Tool (PharmCAT) tool could be extended to include other types of genetic variation.
RATIONALE FOR PharmCAT
Pharmacogenomics (PGx) decision support and return of results is an active area of genomic medicine implementation at many healthcare organizations and academic medical centers.2 Groups around the world have established guidelines surrounding gene–drug pairs that can and should lead to prescribing modifications based on genetic variant(s) including the Royal Dutch Association for the Advancement of Pharmacy – Pharmacogenetics Working Group (DPWG),3, 4 the Canadian Pharmacogenomics Network for Drug Safety (CPNDS),5, 6 and the Clinical Pharmacogenetics Implementation Consortium (CPIC).7, 8 One of the challenges in implementing PGx is extracting genomic variants and assigning possible diplotypes (one haplotype on each chromosome, including star‐allele definitions) from genetic data derived from sequencing and genotyping technologies to apply the prescribing recommendations of these established guidelines. In a collaboration between the former Pharmacogenomics Research Network (PGRN) Statistical Analysis Resource (P‐STAR) and the Pharmacogenomics Knowledgebase (PharmGKB), with input from other groups, we are developing a software tool (PharmCAT) to extract all PGx variants, beginning with variants with CPIC guideline recommendations, from a genetic dataset resulting from sequencing or genotyping (represented as VCF data; VCF specifications can be viewed at https://github.com/samtools/hts-specs), infer diplotypes/genotypes, and generate an interpretation report containing the relevant CPIC recommendations. The PharmCAT report can then be used to inform prescribing decisions. The first release of PharmCAT will annotate VCF data using CPIC guideline recommendations, but later versions will include additional PGx associations and guidelines from other sources such as those mentioned above.
We assembled a focus group of thought leaders from the PGRN, Clinical Genome Resource (ClinGen), electronic Medical Records and Genomics network (eMERGE), and CPIC to brainstorm the issues and requirements for the software tool. We then hosted a 1‐week Hackathon at the PharmGKB to bring together computer programmers with scientific curators to implement the first version of this tool (see http://www.pharmgkb.org/page/pharmcat for a listing of early participants of PharmCAT).
UNMET NEEDS
As mentioned, one of the challenges in implementing PGx is assigning possible diplotypes (one haplotype on each chromosome, including star‐allele definitions) from genetic data derived from sequencing/genotyping technologies to apply CPIC prescribing recommendations, specifically in an automated manner. All CPIC guidelines include two tables: one describes the assignment of phenotypes based on example genotypes/diplotypes, and the second describes the dosing recommendations based on phenotypes. Diplotype assignment requires mapping genotype data to Allele Definition Tables, which supplement CPIC guidelines. With some genotyping technologies, only a subset of gene alleles are assayed, and diplotype assignments can vary depending on the alleles assayed. Complete genotyping data are necessary for defining all important alleles and determining accurate diplotypes.
Assuming that the genotype data are available to match against the allele definition tables, why is the diplotype assignment process still difficult? It is challenging for two reasons. First, the Allele Definition Tables are available as Excel spreadsheets that are difficult to compute upon. Second, many of the actionable alleles contain multiple variants, which presents a combinatorial problem when assessing variants from genotype/sequencing data from both chromosomes simultaneously. This combinatorial problem needs to be solved separately for each gene with CPIC guidelines. For an example of diplotype assignment for one of the pharmacogenes, see Figure 1.
Figure 1.
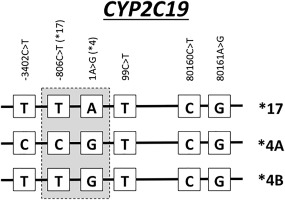
Diplotype example from CYP2C19. Three different diplotype assignments for CYP2C19 are shown: *17, *4A, and *4B. These are defined based on two specific positions (–806C>T and 1A>G).
Once the diplotype assignments have been made, they can be translated to phenotypes and then annotated with the prescribing recommendations. This process is also not easy to automate from the CPIC tables in guideline PDF files. Again, this is due to the multiple diplotype possibilities, the existence of this information in a noncomputable format, and the caveats related to some of the dosing recommendations.
Why is this process important to automate? It is critical to understand the percentage of individuals in the population that carry these variants in their genome. In the eMERGE‐PGx project, 5,000 individuals were evaluated for the presence of the genetic variants in the CPIC‐level A genes and ∼96% of individuals were found to carry one or more genetic variants from this list.9 Thus, manually annotating these genes in a large dataset will be extremely time‐consuming. If implementation of PGx is going to become available in medical centers and clinics around the country/world, standardizing the automation of diplotype assignments based on allele definition from genetic variants and marrying those with the CPIC dosing recommendations is essential. Additionally, proper haplotype/diplotype matching needs to be documented explicitly and made reproducible for all by using PharmCAT. As an open resource, involvement of the community leads to a standard. This is the motivation for PharmCAT.
PharmCAT
PharmCAT, the Pharmacogenomics Clinical Annotation Tool, was sparked by a series of conversations among researchers and clinical experts in the pharmacogenomics community. Clinical implementation of PGx variants was beginning to happen in medical centers around the world.2, 10 However, as each new group began the implementation process, they faced the same challenge: How do I take the CPIC Allele Definition Tables and a patient‐level VCF file and find out which patients in my dataset have the variants of interest? Each research or clinical team developed their own series of scripts or computer programs to perform this process, which can result in different systems generating conflicting reports. As such, the community of experts agreed that it was time to standardize this process and make it available to the entire community. Our plan is to develop PharmCAT, initially using the CPIC guidelines (with other pharmacogenomics guidelines in future versions), the PharmGKB knowledgebase, and new software to automate this annotation process. PharmCAT will provide a solution that will enable sites implementing PGx a way to more consistently and transparently interpret genomic results and link those results to published clinical guidelines. We will release PharmCAT under the Mozilla Public License (MPL 2.0) and disseminate it in GitHub for the scientific and clinical community to test, explore, and improve. It will be available to academics, nonprofits, and commercial/for‐profit entities, and any changes or improvements that they make to the software will be required to be re‐released to the entire community. The goal is to standardize the way in which the CPIC guidelines are used to identify patients who possess these variants and produce consistent guideline‐based reports regardless of where the genetic test is being performed. This license will provide the structure needed to do this effectively. Furthermore, we are assembling (and will be maintaining) the computational translation tables based on CPIC allele definitions that support the tool, which will significantly reduce the effort required to implement PGx clinically and ensure more uniform interpretation of PGx knowledge.
As shown in Figure 2, the general framework for PharmCAT has been designed. The input for PharmCAT is a VCF file including both reference and alternate allele calls for all important genetic variants within these genes: CFTR, CYP2C19, CYP2C9, CYP2D6, CYP3A5, CYP4F2, DPYD, IFNL3, SLCO1B1, TPMT, UGT1A1, VKORC1. A few additional CPIC guideline genes (such as G6PD and HLA‐B) will be implemented in a later version of PharmCAT. We incorporated CPIC prescribing recommendations and the supplemental Allele Definition Tables. These tables have been given a more precise computational data structure and integrated into the PharmCAT codebase to allow for the allele calls to be made. The Named Allele Matcher uses the allele definition data and the VCF input file to perform diplotype assignment. Due to the extremely complex nature of CYP2D6 alleles, CYP2D6 diplotypes will not be assigned by PharmCAT's Named Allele Matcher, but rather, PharmCAT uses the output of Astrolabe11 for the CYP2D6 calling. The CPIC prescribing recommendations are paired with the diplotype assignment using PharmGKB's internal representation of the CPIC guidelines (diplotype‐phenotype‐recommendation mappings) to generate the reports. With this process automated, we will ensure that the process is standardized, accurate, and comprehensive based on the CPIC diplotypes, phenotypes, and recommendations. We plan to be thorough in the reports to include all relevant content, including missing allele calls and caveats from the guidelines.
Figure 2.
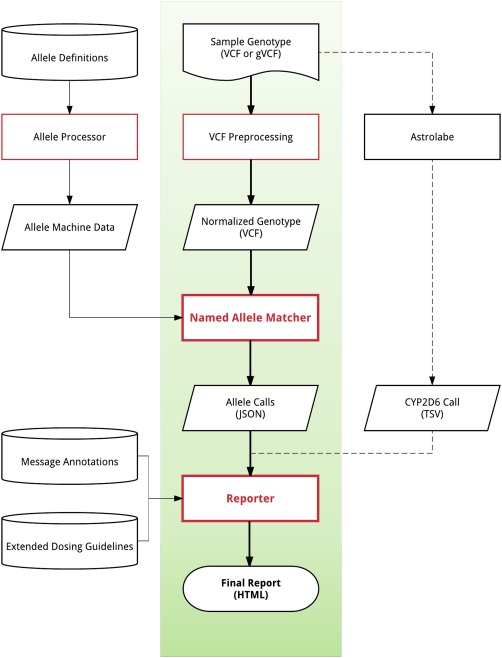
Overview of the PharmCAT tool. Data provided by the user includes the sample genotype (VCF or gVCF). The allele definitions, message annotations, and extended dosing guidelines are extracted from PGx guidelines. The elements of PharmCAT that are the core components include the allele processor, VCF processor, named allele matcher, and reporter. Astrolabe calls will be done externally and the CYP2D6 calls will integrate into the reporter for the Final Report.
REMAINING ISSUES
While we anticipate that PharmCAT will solve many of the problems related to the implementation of PGx in clinical care, there are several issues that remain. First, consideration of the genetic variant data used as input to PharmCAT. The process of sequence alignment, variant calling, and quality control processes can dramatically alter the VCF data used for input. The selection of the reference sequence is also an important decision; PharmCAT is based on the GRCh38 assembly. This means that any genetic data on a different assembly will need to be converted. The quality metrics on the sequence data are still a matter of debate; PharmCAT assumes high‐quality sequence/genotype data and will not use these metrics in the haplotype calling, although we may include those metrics along with the allele calls in the final report in future versions of PharmCAT. Second, because the CPIC, DPWG, CPNDS, and other guidelines continue to evolve and grow, it will be important for users of PharmCAT to reprocess their VCF files as new content becomes available. This will be particularly important as more international guidelines continue to emerge and include a wider array of ethnic diversity. Third, with the increasing use of whole exome sequencing and whole genome sequencing, many more rare variants are being identified that have not been observed before. These variants, by and large, are categorized as variants of unknown significance (VUS) because their function is not yet known. These VUS will not be interpretable into a guideline‐based recommendation if they are not included in one of the guidelines. Lastly, the clinical decision support tools for implementing genetics into an electronic health record (EHR) also continue to evolve and change. We will generate a PharmCAT report that is computer‐readable so that it can be digested by other tools. But until those tools and standards have been finalized, we will not know what structure will be needed.
Due to the reality that over 95% of individuals carry one or more genetic variants that are important for drug dosing recommendations,9 the ability to annotate genetic sequence information with the appropriate CPIC guidelines will be essential for precision medicine to truly be realized in medical centers across the world. Without a standard, reproducible software tool to generate these annotations, the field risks erroneous and irreproducible results. As precision medicine is moved to the forefront of clinical care, pharmacogenomics variants are likely to be an overwhelming priority. Thus, the need for PharmCAT is imminent; the need for PharmCAT is now.
ACKNOWLEDGMENTS
The authors thank Dr. Michelle Whirl‐Carrillo and Dr. Katrin Sangkuhl for their careful review of this commentary. This work is supported by NIH R24 GM61374 (to T.E.K.) and NIH U01 HL065962 (to M.D.R.).
CONFLICT OF INTEREST
The authors report no conflicts of interest.
References
- 1. Kalia, S.S. et al Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet. Med. Off. J. Am. Coll. Med. Genet. 19, 249–255 (2017). [DOI] [PubMed] [Google Scholar]
- 2. Dunnenberger, H.M. et al Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annu. Rev. Pharmacol. Toxicol. 55, 89–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Swen, J.J. et al Pharmacogenetics: from bench to byte. Clin. Pharmacol. Ther. 83, 781–787 (2008). [DOI] [PubMed] [Google Scholar]
- 4. Swen, J.J. et al Pharmacogenetics: from bench to byte—an update of guidelines. Clin. Pharmacol. Ther. 89, 662–673 (2011). [DOI] [PubMed] [Google Scholar]
- 5. Madadi, P. et al Clinical practice guideline: CYP2D6 genotyping for safe and efficacious codeine therapy. J. Popul. Ther. Clin. Pharmacol. J. Ther. Popul. Pharamcol. Clin. 20, e369–396 (2013). [PubMed] [Google Scholar]
- 6. Amstutz, U. et al Recommendations for HLA‐B*15:02 and HLA‐A*31:01 genetic testing to reduce the risk of carbamazepine‐induced hypersensitivity reactions. Epilepsia 55, 496–506 (2014). [DOI] [PubMed] [Google Scholar]
- 7. Relling, M.V. & Klein, T.E. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin. Pharmacol. Ther. 89, 464–467 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moriyama, B. et al Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for CYP2C19 and voriconazole therapy. Clin. Pharmacol. Ther. (2016) doi:10.1002/cpt.583 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bush, W.S. et al Genetic variation among 82 pharmacogenes: the PGRNseq data from the eMERGE network. Clin. Pharmacol. Ther. 100, 160–169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Relling, M.V. & Evans, W.E. Pharmacogenomics in the clinic. Nature 526, 343–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gaedigk, A. et al In vivo characterization of CYP2D6*12, *29 and *84 using dextromethorphan as a probe drug: a case report. Pharmacogenomics 18, 427–431 (2017). [DOI] [PubMed] [Google Scholar]


