Abstract
In utero exposure to vinclozolin (VIN), an antiandrogenic fungicide, is linked to multigenerational phenotypic and epigenetic effects. Mechanisms remain unclear. We assessed the role of antiandrogenic activity and DNA sequence context by comparing effects of VIN vs. M2 (metabolite with greater antiandrogenic activity) and wild-type C57BL/6 (B6) mice vs. mice carrying mutations at the previously reported VIN-responsive H19/Igf2 locus. First generation offspring from VIN-treated 8nrCG mutant dams exhibited increased body weight and decreased sperm ICR methylation. Second generation pups sired by affected males exhibited decreased neonatal body weight but only when dam was unexposed. Offspring from M2 treatments, B6 dams, 8nrCG sires or additional mutant lines were not similarly affected. Therefore, pup response to VIN over two generations detected here was an 8nrCG-specific maternal effect, independent of antiandrogenic activity. These findings demonstrate that maternal effects and crossing scheme play a major role in multigenerational response to in utero exposures.
Keywords: Epigenetic inheritance, endocrine disruptor, vinclozolin, gene-environment, maternal effect, genomic imprinting, DNA methylation
1. Introduction
Endocrine-disrupting compounds (EDCs) are environmental compounds that interfere with the homeostasis of the endocrine system. EDC exposure is of growing concern because it is linked to increased prevalence of human diseases such as cancer, diabetes, obesity, asthma, neurodegenerative disorders, and reproductive disorders [1]. Furthermore, experimental models show that exposure to EDCs in utero can negatively influence offspring health outcomes over multiple generations [2]. Vinclozolin (VIN) is a dicarboximide fungicide that is still used commercially but in limited application in the United States (US) due to adverse effects on male reproduction detected in experimental models [3–5]. In rodents, VIN exposure in utero disrupts male reproduction causing reduced anogenital distance (AGD) and sperm count, cleft phallus, hypospadias, ectopic testes, vaginal pouches, and epididymal granulomas [6–8].
The mechanism by which VIN perturbs male reproduction is not fully understood. However, VIN and two of its metabolites (M1 & M2) have been shown to compete with dihydrotestosterone (DHT) for binding the androgen receptor (AR) and therefore may act in part by disrupting genomic activity of AR [9,10]. Compared to VIN and M1, M2 seemingly has greater potential to interfere with AR activity. M2 was previously reported to have a greater ability to compete with androgen for binding to AR (DHT (Ki = 100 nM) > hydroxyflutamide (Ki = 175 nM) > testosterone (Ki = 220 nM) > M2 (Ki = 9.65 μM) > M1 (Ki = 92 μM) > VIN (Ki> 700 μM)) [11,12]. M2 was also shown to be a more potent antagonist of AR transcriptional activity, acting at a dose that was more similar to hydroxyflutamide (a similarly structured known antiandrogenic compound) [9]. VIN and its metabolites have also been reported to interact with other steroid receptors. Here also, M2 was shown to be a more potent antagonist of transcriptional activity of progesterone receptor (PR) and mineralocorticoid receptor (MR) and a more potent agonist of AR and estrogen receptors (ER) α and β [13–15].
VIN exposure in utero was the first reported model of transgenerational epigenetic inheritance. Male rats exposed to VIN in utero exhibited adverse reproductive outcomes and altered DNA methylation in sperm over 3 generations [8,16]. A separate study also reported transgenerational DNA methylation changes at imprinted loci including the H19/Igf2 imprinting control region (ICR) in mature sperm [17]. Genetic differences have been shown to influence heritable responses to VIN in reports showing that transgenerational germline epimutations could be detected in outbred CD-1 mice but not inbred 129 mice [18]. Exact sequences responsible were not investigated. The mechanism of persistence of VIN-induced phenotypic and epigenetic changes are unclear. However, recent studies show that VIN-induced methylation changes do not persist through stages of reprogramming during male germ cell development [19] suggesting an alternate mechanism of transmission.
Here, we used the VIN mouse exposure model to investigate factors that may contribute to differences in sensitivity to endocrine disruptors. We used a two-stage study design to examine the role of M2 metabolite availability (compared to VIN) and DNA sequence context (wildtype vs. mutations at the H19/Igf2 ICR) in determining the extent and heritability of epigenetic and phenotypic outcomes in response to VIN. The mutant lines used, 8nrCG, Δ2,3 and ΔIVS, carry targeted mutations at the H19/Igf2 ICR that mimic genetic mutations found at the epigenetically perturbed human locus in patients with the overgrowth disorder Beckwith-Wiedemann syndrome (BWS) [20–26]. Aberrant H19/Igf2 expression is also associated with metabolic disorders such as diabetes and obesity, Wilms tumor of the kidney, adrenocortical tumors, and various other forms of cancer [27–31]. The cause of these epimutations remains unknown, however, studies suggest they may act by disrupting epigenetic stability at the locus [22–26]. While these mutations alone in the mouse do not perturb epigenetic status [20,21], this study will investigate whether the presence of these mutations increase sensitivity to VIN-induced epimutation.
2. Materials and Methods
2.1. Animals – genetic line descriptions, sources, and housing
Animal handling was performed in accordance with the Guide for the Care and Use of Laboratory Animals under the corresponding animal use protocol at the University of North Carolina at Chapel Hill and housed at the David H. Murdock Institute Vivarium in Kannapolis, NC. Five mouse lines were used in the study including: (1) wild-type fully inbred C57BL/6 (B6) mice; (2) wild-type C7 mice homozygous for CAST/EiJ across chromosome 7 and mixed CAST/EiJ-B6 genetic background [32]; (3) mutant 8nrCG mice [20] carrying targeted mutation of 8 base pairs at 8CpGs at the H19/Igf2 locus; (4) mutant ΔIVS mice [20] carrying a targeted deletion of 0.9 kb at the H19/Igf2 imprinting control region; and (5) mutant Δ2,3 mice [21] carrying a targeted deletion of 1.3 kb at the H19/Igf2 imprinting control region. All mutant lines were previously backcrossed 6–10 generations into B6 to result in mostly (>98%) B6 genetic background as confirmed by genotyping. B6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and used directly (study 1) or purchased separately and bred in-house to generate animals for study 2. B6-Cast7 (C7), 8nrCG, Δ2,3, and ΔIVS were derived in Dr. Marisa Bartolomei’s lab at the University of Pennsylvania [32,20,21], then transferred and bred in-house at the David H. Murdoch Institute Vivarium in Kannapolis to generate animals for study 1 and study 2. Sterilized water and rodent chow were fed ad libitum. All mice were euthanized in accordance with current recommendations by the American Veterinary Medical Association (AVMA) guidelines. All tissues were collected and flash frozen in liquid nitrogen following euthanasia.
2.2. Breeding and treatments (see Figure 1 for specific cross descriptions and timelines)
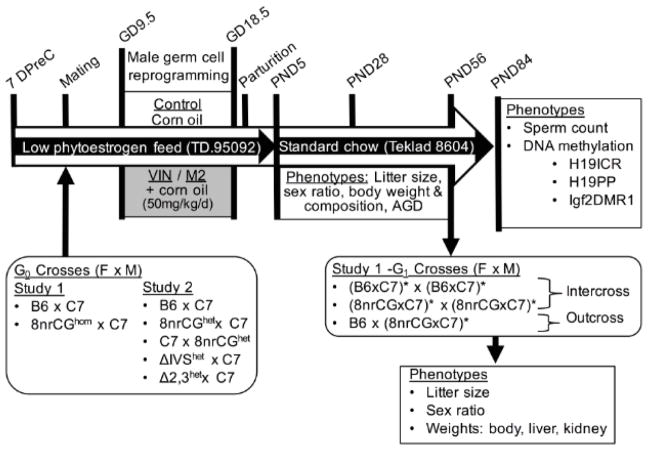
Figure 1.
Treatment Scheme. Illustrates timing of crosses, special diets, experimental treatments, and phenotypes measured. Cross descriptions are shown in order female x male. Homozygous mutant animals are indicated with “hom“, heterozygous mutant animals indicated with “het”. Asterisks (*) indicate which parental G1 animals were exposed to treatments in utero. F, female; M, male; DPreC, days precoitus.
Supplemental Table 1 lists the number of pups, males, females, litters (dams), and average number of males per litter assessed in each experimental group. For generating G1 animals, G0 sires and dams (virgin females) were group housed after weaning/purchasing and set up to breed at approximately 8–10 weeks of age. Use of G0 C7 males allowed for consistency in paternal contribution between lineages and also allowed for allelic expression or methylation analyses should the data have shown significant changes in the latter. G0 dams were bred to stud males (8–24 weeks of age) and checked for vaginal plugs every morning. The first morning of a successful plug was considered GD1 and plugged females were immediately separated from stud males and placed into treatment cages. Group housed females were separated before parturition to enable identification of each litter separately. G1 pups were counted on the morning of birth to measure litter size, sexed at PND5 to measure the number of males and females, weaned at PND28, and euthanized at PND84. For deriving the second generation of animals (G2), 10wk old G1 males and females treated in utero were intercrossed (avoiding brother sister mating) or simultaneously outcrossed to untreated inbred B6 females. G2 pups were counted on the morning of birth, sexed at PND5, and euthanized at PND5.
All G0 dams were placed on a low-phytoestrogen purified diet (Teklad, TD.95092) one-week prior to mating and throughout breeding and gestation until PND5 to standardize nutrient intake and minimize potential xenoestrogenic properties of soy based diet [33]. Pregnant females were group-housed based on treatment group and plug date and treated daily with either VIN (VIN) (analytical grade >99% purity, Chem Service Inc, PA), M2 (analytical grade >99% purity, Cayman Chemical, MI), or pure corn oil (CON) (Sigma) by intraperitoneal injection. VIN and M2 were dissolved in corn oil at room temperature, prepared fresh weekly, and injected at a dose of 50 milligrams/kilogram of body weight/day (mg/kg/d) at a volume of 5 ul per gram of body weight. To reduce potential of lineage effects confounding the treatment effects, we avoided placing sibling females in the same treatment group and stud males were used across treatment groups. Mice were treated daily from GD9.5 to GD18.5. At PND5 for G1 pups, dams and pups and were transferred to standard rodent chow (Teklad 8604) where they remained throughout subsequent breeding until the end of the study.
We performed two separate rounds of study. For both rounds of study, all methods of treatment and breeding schemes are maintained except where described (Figure 1). Study 1 compared the effects of VIN and M2 to CON between two genetic lineages (crossed dam x sire): B6xC7 and 8nrCGxC7. G0 dams for these crosses were either homozygous wild-type (B6) or homozygous mutant 8nrCG mice. Study 2 compared the effects of VIN to CON among 5 genetic crosses (dam x sire): B6xC7, 8nrCGxC7, C7x8nrCG, ΔIVSxC7, and Δ2,3xC7. G0 dams for these crosses were either homozygous wild-type (B6 and C7) or heterozygous mutant 8nrCG, Δ2,3, ΔIVS mice. See Supplemental Table 1 for litter and pup numbers.
2.3. Body weight, body composition and anogenital distance measurements
Body weight at PND5, PND28, and PND56 was measured by analytical balance. Body composition was measured by magnetic resonance imaging (EchoMRI-100) at PND28 and PND56 in the UNC Nutrition Obesity Research Center Animal Metabolism Phenotyping Core (UNC NORC AMP). Percent fat and lean mass were calculated using the ratio of fat or lean mass/(total body mass - free water mass) x 100. AGD was measured using calipers. Mice were weighed at AGD measurement and changes in AGD due to body weight changes were adjusted for using a ratio of AGD/body weight.
2.4. Sperm isolation and count
Intact cauda epididymis and vas deferens were isolated immediately after euthanasia in 1 ml of HTF buffer (pH7.5, 100 mM NaCl, 5 mM KCl, 0.368 mM KH2PO4, 0.2 mM MgSO4, 2 mM CaCl2 and 5 mg/ml BSA) at 37°C and scored in 4-well petri dishes under a dissection microscope. Scored tissues were incubated for 10 minutes at 37°C for sperm swim out. Sperm in supernatant was transferred to fresh 1.5 ml microcentrifuge tubes and 3 ul were used in a 1:50 dilution of phosphate buffered saline (PBS) while remaining cells were pelleted by centrifugation, flash frozen in liquid nitrogen and stored at −80°C. Sperm counts and purity was measured in the 1:50 dilution using a hemocytometer and inverted light microscope.
2.5. Methylation detection & genotyping
Genomic DNA from sperm, tongue, and abdominal fat was isolated using phenol-chloroform extraction [34] and from liver using the Anaprep 12, an automated magnetic-bead based nucleic acid extraction system (BioChain Institute Inc, CA). DNA quality was assessed by NanoDrop 2000 spectrometer (Thermo Scientific, DE) and double stranded DNA concentrations measured by Quant-it PicoGreen dsDNA assay (#P7589, Life technologies, NY). Genomic DNA was bisulfite-converted using the EZ DNA methylation Gold kit according to the manufacturer’s protocol (#D5006, Zymo research, CA). All pyrosequencing PCR and sequencing primers and sequenced regions were confirmed to be devoid of SNPs or deletions. Pyrosequencing was performed using Pyromark Q96MD instrument according to the manufacturers’ instructions (Qiagen, MD). Assays for Igf2DMR1 and H19PP were previously described [21].
H19ICR_Rp1 assays methylation at the H19/Igf2 ICR using primers forward 5’-GAGGTATAAGAATTTTGTAAGGAGATTATG-3’ and biotinylated reverse 5’-ACTAAACTTAAATAACCCACAACATTAC-3’ and pyrosequenced using primer 5’-ATTAGTTGTGGGGTT-3’. PCRs for each sample and locus were performed in duplicate and CpG methylation was determined by pyrosequencing. The average methylation of all CpGs combined within the assayed region are presented as mean methylation (%) and used in all downstream analysis. The number of CpGs assayed for each region was 3 (Igf2DMR1), 6 (H19Rp1), and 3 (H19PP).
Sex of G1 offspring was confirmed by SRY amplification using primers forward 5’-TTGTCTAGAGAGCATGGAGGGCCATGTCAA-3’ and reverse 5’-CCACTCCTCTGTGACACTTTAGCCCTCCGA-3’[35].
2.6. Statistical analyses
Least squares linear regression was performed separately for each genetic line with adjustment for sex and genotype where applicable using the JMP Pro software version 13.0 (SAS, NC) to compare among treatment groups for phenotypes in study 1 G1 phenotypes (litter size, % male, litter survival, PND56 AGD/body weight, PND5 body weight, PND28 body weight, PND28 % fat mass, PND28 % lean mass, liver H19ICR_Rp1, Igf2DMR1 and H19PP methylation, and abdominal fat and tongue H19ICR_Rp1 methylation); and G2 phenotypes (litter size, % male, PND5 body weight, PND5 kidney weight/body weight, and PND5 liver weight/body weight); and phenotypes in study 2 (adult body weight and sperm methylation at H19/Igf2). Treatment effects with p<0.1 were assessed further using Dunnett’s test to compare CON to either VIN or M2. Correlations between sperm H19ICR_Rp1 methylation and body weight were determined separately for each genetic line by Spearman correlation using the JMP Pro software version 13.0 (SAS, NC). Statistical analyses to compare between treatment groups for study 1 G1 phenotypes (PND56 body weight, PND56 percent fat mass, PND56 percent lean mass, sperm count, liver and sperm methylation levels at H19/Igf2 ICR) were performed using linear mixed effects models with fixed effects for treatments and random effect for litter using the nlme package in R [36,37]. For all models, we adjusted for sex where applicable and compared the difference between CON and either VIN or M2 for each line. Benjamini-Hochberg (BH) correction [38] was used to adjust p-values (adj.p) for multiple comparisons within each dataset and a false discovery rate of 0.1 was used as the significance threshold.
3. Results
3.1. Phenotypic effects of in utero VIN exposure are genetic line-dependent
To determine whether the underlying genetic sequence at the site of an epimutation influences phenotypic and epigenetic response to VIN, we first compared the effects of in utero VIN exposure in offspring from wild-type inbred B6 females to offspring from homozygous mutant 8nrCG females (both crossed to wild-type inbred C7 sires, Figure 1). Pregnant mice were treated in three separate groups with either corn oil (CON), VIN dissolved in corn oil (VIN) or M2 dissolved in corn oil (M2), from GD9.5 to GD18.5 (Figure 1). This is a critical period in which the fetus undergoes gonadal sex determination, genome wide epigenetic reprogramming, imprint establishment at ICRs, and widespread developmental changes [39–41].
3.1.1. Cross reproductive outcomes
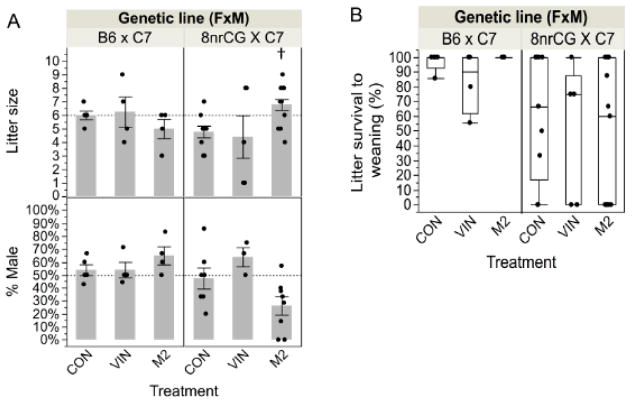
To assess the effect of treatment on reproductive outcomes, we measured litter size, sex ratio and pup survival to weaning. For B6 mice, litter size, sex ratios and pup survival to weaning were not significantly affected by treatment with either VIN nor M2 (Figure 2A–B). VIN treatment in 8nrCG mice did not significantly affect litter size or sex ratio (Figure 2A). However, M2 treatment in 8nrCG mice was linked to a significant increase in litter size (42%, p=0.044, adj.p=0.099) (Figure 2A). Interestingly, M2 had the lowest average in percent of males per litter (26.4% compared to 47.5% for CON) but this was not statistically significant (p=0.105, adj.p=0.189). We assessed further whether males were disproportionately dying perinatally or inaccurately sexed due to feminized features. We found no direct correlation between litter size and proportion of males in the 8nrCG M2 group and sex misdetermination was ruled out by SRY genotyping. Compared to B6, 8nrCG crosses exhibited a substantial difference in survival to weaning (p=0.010, adj.p=0.084) with a high rate (9/28 litters) of whole litter mortality in the 8nrCG crosses (Figure 2B). There was no significant difference in gestational weight at GD18.5 or weight gain from GD9.5-GD18.5 between B6 and 8nrCG mice (p=0.412, p=0.810, respectively) making additional prenatal lethality unlikely. Partial litter mortality in the 8nrCG mice resulted in the death of an additional 18 pups, 15 of which died before the animals were sexed at PND5. Postnatal mortality in the 8nrCG mice was similar across all treatment groups (Figure 2B), and was not correlated with male/female ratios (p=0.148) indicating that male and female offspring exhibited similar levels of postnatal mortality.
Figure 2.
G0 cross reproductive outcomes (Study 1). (A) Bar graph of average litter size at birth (top panel) and average percent of males in each litter (% male, bottom panel). Each dot represents data for one litter. Dashed horizontal lines represent expected litter size and expected proportion of males for B6 litters. (B) Box and whiskers plot of percent of pups in each litter that survived to weaning. Error bars indicate standard error of the mean (SEM). Treatment group(s) with a significant difference from CON is denoted by (†). Sample sizes for each treatment group are listed from left to right as shown in each graph (N=number of litters): litter size at birth N= 5, 4, 4, 9, 5, 13; % male at PND5 N= 5, 4, 4, 7, 3, 8; litter survival to weaning N= 5, 4, 4, 9, 5, 13). Reduction in N between litter size and % male reflects death of whole litter before pups were sexed at PND5.
3.1.2. G1 body weight & body composition outcomes
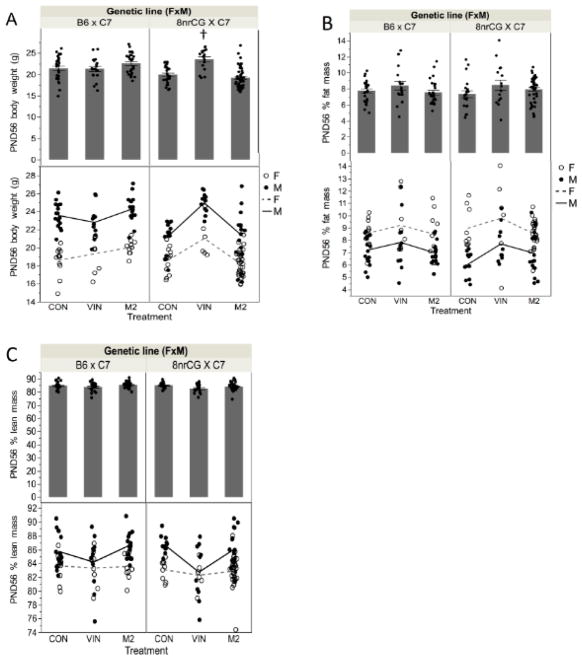
To assess the effect of in utero treatment on adult phenotypes, we measured adult (PND56) body weight and body composition as growth/developmental endpoints. B6 mice showed no significant difference in PND56 body weight for VIN or M2 treatment groups (Figure 3A). However, VIN-treated 8nrCG mice had significantly higher PND56 body weight (3.6g,18% increase on average, p=0.0008, adj.p=0.024), while M2-treated mice were unaffected (p=0.718, adj.p=0.886) (Figure 3A). Male and female adult offspring were affected similarly (Figure 3A). The increase in VIN-treated 8nrCG bodyweight was present as early as PND28 (1.8g, 12%, p=0.0018, adj.p=0.0273) but not detected at PND5 (Supplemental Figure 1A & 1B).
Figure 3.
G1 body weight and body composition outcomes (Study 1). PND56 (A) body weight; (B) percent fat mass; (C) percent lean mass. Top panels show bar graph of average body weight for both sexes combined. Bottom panel is enlarged to only show portion y-axis covered by data points. Individual (dots) weights are separated by sex, male (filled dots), female (unfilled dots); lines connect means between groups for males (solid line) or females (dashed line)). Error bars indicate standard error of the mean (SEM). F, female; M, male. Treatment group(s) with a significant difference from CON is denoted by (†). Sample sizes for each treatment group are listed from left to right as shown in each graph (n=number of mice, both sexes combined; N=number of litters): PND56 bodyweight n= 28, 19, 26, 27, 17, 50; N= 5, 4, 5, 7,3, 7; PND56 % fat mass n= 28,19, 26, 20, 16, 43; N=5, 4, 5, 5, 3, 6; PND56 % lean mass n= 28, 19, 26, 20, 16, 43; N=5, 4, 5, 5, 3, 6.
To determine whether the increase in body weight in the 8nrCG line was related to a change in body composition (disproportionate increase in fat or lean mass), we compared percent fat and lean mass (tissue mass/(body mass – free water mass)) between treatments and genetic lines. Percent fat mass was significantly increased at PND28 for VIN-treated 8nrCG mice (15% increase, p=0.019, adj.p=0.0988) (Supplemental Figure 1C). However, by PND56, percent fat mass was not significantly increased for any treatment group in either B6 or 8nrCG lineage (p=0.248, adj.p=0.623 and p=0.083, adj.p=0.313, respectively) (Figure 3B). Percent lean mass was not significantly changed for any treatment group for either lineage either at PND28 or PND56 (Figure 3C & Supplemental Figure 1D). Taken together, these data suggest that the increase in adult body weight is likely due to developmental phenotype that persisted into adulthood and though changes in adiposity may play a role, this is not the main driver of this phenotype. Since these effects were only observed in VIN-treated mice, and not M2, they likely occur independent of antiandrogenic activity.
3.1.3. G1 reproductive development outcomes
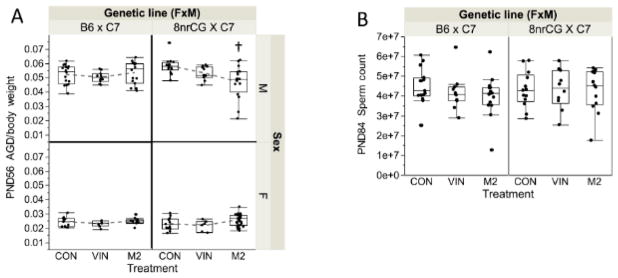
To determine whether the antiandrogenic activity of VIN and M2 reported previously, is sufficient to alter reproductive development and male fertility in this model, we compared adult AGD and sperm count between genetic lineages and treatment groups. After normalization to body weight (AGD/body weight), B6 males and females did not exhibit any significant differences in PND56 AGD in either VIN or M2 treatment groups (Figure 4A). On the other hand, 8nrCG males exhibited a significant decrease in PND56 AGD in the M2 treatment group (19% decrease, p=0.0014, adj.p=0.024) (Figure 4A), while females were not significantly affected (p=0.070, adj.p=0.298) (Figure 4A). There were no statistically significant changes in sperm count for either genetic lineage or treatment group (Figure 4B). These data show that in this model of VIN and M2 exposure, antiandrogenic effects on reproductive development were minimal and unlikely to affect fertility. M2 exposure had a greater effect on reproductive development compared to VIN (as expected based on previously reported antiandrogenic activity), and 8nrCG mice were more susceptible than B6. Importantly, it confirms that body weight changes detected in the VIN treatment group are not linked to antiandrogenic activity.
Figure 4.
G1 reproductive development outcomes (Study 1). Box and whiskers plot shown for PND56 (A) male AGD (normalized to body weight, top panel) and female AGD (normalized to body weight, bottom panel); (B) PND84 total sperm count. Error bars indicate standard error of the mean (SEM). Treatment group(s) with a significant difference from CON is denoted by (†). Sample sizes for each treatment group are listed from left to right as shown in each graph (n=number of mice, both sexes combined; N=number of litters): PND56 male AGD/bodyweight n=16, 11, 16, 14, 11, 16; N=5, 4, 5, 6, 3, 6; PND56 female AGD/bodyweight n= 12, 8, 10, 13, 6, 34; N= 5, 4, 5, 5, 3, 7; PND84 sperm count n= 15, 11, 16, 13, 10, 13; N=5, 4, 5, 5, 3, 5).
3.2. VIN-induced body weight increase is independent of methylation state at H19/Igf2
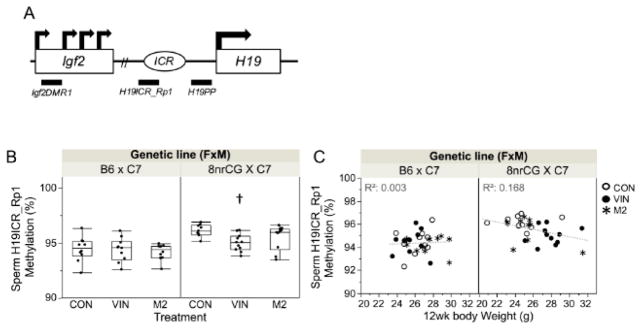
The VIN dose used in this study (50 mg/kg/d) was previously reported to alter DNA methylation state at the H19/Igf2 locus in sperm of FVB/N mice [17]. To test whether the body weight sensitivity of 8nrCG mice to VIN exposure is due to methylation perturbation at the mutated H19/Igf2 locus, we compared methylation state at 3 loci across the H19/Igf2 imprinted domain between B6 and 8nrCG lineages treated with VIN or M2. These loci were selected due to proximity to the 8nrCG mutation at the H19/Igf2 ICR (H19ICR_Rp1) and proximity to differentially methylated regions (DMR) linked to expression of Igf2 (Igf2DMR1) and H19 (H19PP). We found that methylation levels in adult male liver were not affected by genetic lineage or treatment groups for any of the 3 loci tested (Supplemental Figure 2A). A small but statistically significant decrease in methylation was detected in 8nrCG VIN sperm at the H19ICR_Rp1 (1%, p=0.012, adj.p=0.099) (Figure 5B). Sperm methylation at H19ICR_Rp1 was seemingly related to body weight in 8nrCG and not B6 mice, but this relationship was not statistically significant after correction for multiple testing (p=0.025, adj.p=0.139) (Figure 5C). Methylation at H19ICR_Rp1 in tongue (muscle) or abdominal fat in 8nrCG mice was not affected by treatment (Supplemental Figure 2B). In sum, these data suggest that individuals may have shared susceptibility to VIN-induced body weight and sperm methylation changes. However, body weight differences are likely not directly driven by methylation differences at the H19/Igf2 locus in 8nrCG mice, thus another mechanism is at play.
Figure 5.
G1 methylation outcomes at H19/Igf2 locus (Study 1). (A) Schematic of H19/Igf2 imprinted domain and loci assayed for methylation changes: Igf2DMR1, H19ICR_Rp1, and H19PP (locations indicated by horizontal bars below the domain). (B) Box and whiskers plots of percent methylation at H19ICR_Rp1 in G1 sperm samples; each dot represents the mean methylation of 6 CpGs across the region analyzed for each individual sample. Error bars indicate standard error of the mean (SEM). (C) Correlation between G1 sperm H19ICR_Rp1 methylation levels and 12wk body weight. Treatment group(s) with a significant difference from CON is denoted by (†). Sample sizes for each treatment group are listed from left to right as shown in each graph (n=number of mice, both sexes combined; N=number of litters): Sperm H19ICR_Rp1 Methylation n= 10, 10, 10, 9, 11, 10; N= 5, 4, 5, 4, 3, 5.
3.3. VIN-induced outcomes are 8nrCG specific and due to a maternal effect and not pup genotype
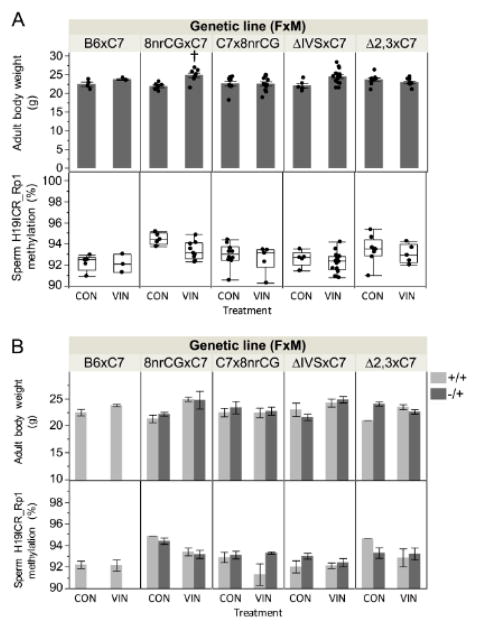
To determine whether VIN-induced phenotypic and epigenetic outcomes observed in the 8nrCG mutant mouse line are caused by genetic mutations at the H19/Igf2 ICR, we screened a panel including two additional H19/Igf2 ICR mutant lines. We compared the response of adult male offspring from heterozygous 8nrCG, Δ2,3, ΔIVS, and wild-type B6 dams (all crossed to C7 males) and offspring from the reciprocal cross C7 dams x 8nrCG males (to determine parent of origin dependence) (Figure 1).
When comparing 8–9wk old male body weights and methylation at the H19/Igf2 ICR in sperm between VIN and CON-treated mice across the panel of mice, we reproduced the 8nrCG lineage specific VIN-induced effects observed in study 1. Similar to study 1, offspring from 8nrCG but not B6 dams exhibited a significant increase in body weight in the VIN treatment group compared to CON (3g, 14% increase, p=0.002, adj.p=0.020) (Figure 6A). However, sperm methylation at the H19/Igf2 ICR in VIN-exposed offspring from 8nrCG dams showed a similar trend but was not statistically significant after correction for multiple testing, possibly due to the smaller sample size reducing power to detect such a small change (1%, p=0.031, adj.p=0.103). Although we could not statistically test due to small sample size, both 8nrCG offspring genotypes (+/+ and −/+) were on average affected similarly for body weight and methylation (Figure 6B). Neither B6 nor the other mutant mice (Δ2,3 and ΔIVS) were affected by treatment (Figure 6A). VIN-exposed offspring from 8nrCG sires (reciprocal cross, C7x8nrCG) were also unaffected (Figure 6A). We conclude that the VIN-induced methylation and body weight changes are likely due to an 8nrCG lineage specific effect that was only observed when the mother carried an 8nrCG allele and was independent of pup genotype.
Figure 6.
G1 male adult body weight and methylation outcomes across 5 genetic lines screened for VIN sensitivity (Study 2). (A) Bar graph of average adult (8–9wks) body weight (top panel) and box and whiskers plots of percent methylation at H19ICR_Rp1 in G1 sperm samples (bottom panel); (B) Bar graph of same data shown in (A) but figure is enlarged to show differences/similarities between genotypes. Error bars indicate standard error of the mean (SEM). Treatment group(s) with a significant difference from CON is denoted by (†). Sample sizes for each treatment group are listed from left to right as shown in each graph (n=number of mice, genotypes combined; N=number of litters): Adult body weight n= 4, 3, 7, 8, 9, 12, 5, 16, 9, 9; N=2, 2, 3, 3, 3, 4, 2, 4, 4, 4 ;Sperm H19ICR_Rp1 methylation n= 5, 3, 5, 9, 10, 6, 5, 16, 8, 5; N=2, 2, 2, 3, 3, 3, 2, 4, 4, 3.
3.4. Paternal VIN exposure in utero decreases pup neonatal body weight in a cross-dependent manner
VIN exposure was previously shown to result in heritable phenotypes [8,16,18]. Therefore, we determined whether paternal (G1) VIN exposure in utero alters unexposed second generation offspring body weight or epigenetic outcomes. To determine whether the exposed male germline was sufficient to transmit any VIN-induced effects, we compared offspring from study 1 G1 males generated by intercross (to treated G1 females) or outcross (to untreated B6 females) (Figure 1).
3.4.1. Cross reproductive outcomes
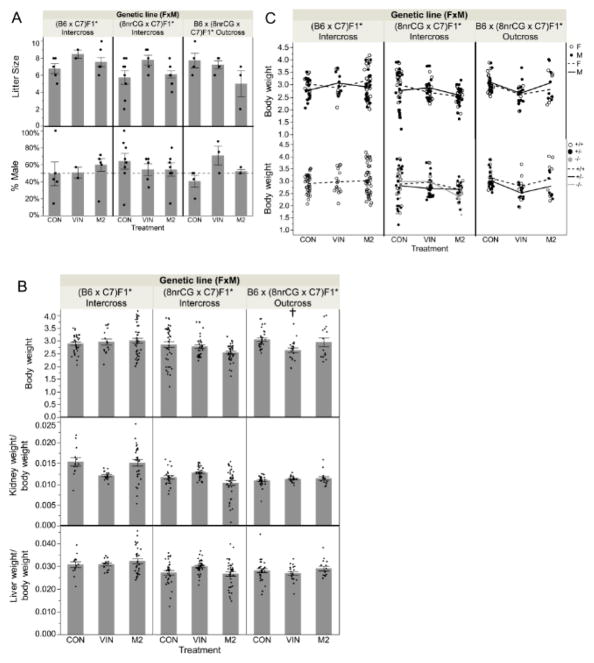
There were no statistically significant reductions detected in litter size or sex ratio associated with treatment group or genetic lineage for either intercross or outcross groups (Figure 7A). Therefore, in support of data described above, G1 male and female fertility were not substantially affected by VIN or M2 treatment in utero.
Figure 7.
G2 offspring reproductive and birth outcomes at PND5 (Study 1). Asterisks (*) indicates treated parent(s). (A) Bar graph of average litter size (top panel) and % of litter born male (bottom panel). (B) Bar graph of average body weight (top panel); kidney weight/normalized to body weight (middle panel); and liver weight/normalized to body weight (bottom panel). (C) Samples separated by sex, male (M, filled dots) and female (F, unfilled dots) (top panel); or separated by genotype, wildtype (+/+,unfilled dots), heterozygous mutant (+/−, filled dots), and homozygous mutant (−/−, grey dots) (bottom panel); lines connect means between treatment groups for sexes (top panel) or genotypes (bottom panel). Error bars indicate standard error of the mean (SEM). Treatment group(s) with a significant difference from CON is denoted by (†). Sample sizes for each treatment group are listed from left to right as shown in each graph (n=number of mice, genotypes combined; N=number of litters): Litter size N= 5, 2, 7, 8, 5, 9, 4, 4, 3; % male N= 5, 2, 7, 8, 5, 7, 4, 3, 3; PND5 Body weight n= 32, 16, 44, 40, 36, 43, 31, 20, 15; N= 5, 2, 6, 7, 5, 7, 5, 3, 3 ; PND5 Kidney weight/Body weight n= 13, 16, 32, 23, 36, 36, 31, 20, 15; N= 2, 2, 4, 5, 5, 6, 5, 3, 3; PND5 Liver weight/Body weight n= 13, 16, 32, 32, 36, 37, 31, 20, 15; N=2, 2, 4, 6, 5, 6, 5, 3, 3.
3.4.2. G2 growth/developmental outcomes
After adjustment for pup sex and genotype, we found that G2 neonatal (PND5) body weight was only significantly different for pups from VIN-treated 8nrCG outcrossed sires compared to CON sires (approx. 420 mg, 14%, p=0.0025, adj.p=0.035) (Figure 7B). Although we could not statistically test due to small sample size, both 8nrCG offspring genotypes (+/+ and −/+) and sexes were on average affected similarly for body weight (Figure 7C). Differences in body weight for 8nrCG outcross pups is likely due to differences in growth and not adiposity since kidney and liver showed similar decreases in weight that were completely normalized after correction for body weight (Figure 7B). Similar to our findings in the parental mice, VIN-related changes in G2 8nrCG neonatal (PND5) body weight did not coincide with methylation changes at the H19/Igf2 ICR (Supplemental Figure 3) as one might expect for a loss of imprinting-related growth phenotype. PND5 body weight, kidney and liver weight (after normalization to body weight) was not significantly different from CON for any other lineages or treatments (Figure 7B). Taken together, these data demonstrate that first generation in utero exposure to VIN can alter second generation offspring development when transmitted through the paternal germline in a manner seemingly independent of antiandrogenic activity but strongly influenced by genetic lineage and crossing scheme.
4. Discussion
Here, we use several genetic mouse lines carrying mutations at the H19/Igf2 ICR to assess the effect of genetic sequence differences on phenotypic and epigenetic response to VIN exposure during fetal development. We found that compared to wild-type B6 mice, the 8nrCG mutant line was more susceptible to VIN-induced changes in body weight, body composition and H19/Igf2 ICR methylation. Importantly, we reproduced the body weight finding and showed that these changes were seemingly independent of sex and pup genotype at the H19/Igf2 ICR. Furthermore, assessment of reciprocal crosses showed that a maternal effect specific to the 8nrCG line was likely responsible for VIN-induced effects since they were present when offspring from 8nrCG dams were assessed but not when 8nrCG sires were used. We cannot rule out that maternal genotype at the H19/Igf2 ICR is still responsible, but this seems unlikely since none of the other mutant lines that exhibit greater loss of imprinting at the ICR were significantly affected [20,21]. It is possible that 129/SvJ loci in the <2% of the genome that remains after backcrossing to B6 are responsible. We also showed that in utero VIN treatment in one generation altered body weight in a second generation but significant changes were only detected in the 8nrCG lineage and specifically when mice were outcrossed to an unexposed female. Thus, contrary to expectation, intercrossing schemes may actually mask intergenerational effects. Overall, these data show that even small differences in maternal genetic lineage confer significant susceptibility to phenotypic and epigenetic effects of environmental exposures in utero and therefore careful attention should be taken in study design to control for such features.
We were unable to replicate previous findings of sperm methylation changes at the H19/Igf2 ICR associated with VIN exposure [17]. This could be explained by either a strain effect or an aggregate effect of other environmental factors such as diet. We did detect a very small change (~1%) in G1 sperm methylation associated with VIN in the 8nrCG lineage. No methylation differences were detected in second generation pups. It is very unlikely that the 1% methylation change in itself is biologically relevant or causative of any of the phenotypes observed in either generation tested. However, the correlation between sperm methylation and body weight in the first generation suggests the two may be somehow physiologically linked. We propose that both sperm methylation changes and body weight differences associated with VIN are separate and indirect outcomes of perturbation of gross development that likely began in utero at the time of treatment. 8nrCG crosses exhibited increased perinatal lethality, and although it was not linked to treatment, it may allow for increased susceptibility to such developmental perturbation when exposed to VIN in utero. Further studies are required to dissect the pathways responsible.
In our assessment of the effects of VIN compared to effects of the M2 metabolite, we found that offspring exposed to M2 behaved more similarly to CON-treated animals for almost all of the phenotypes tested except those related to male reproductive development. Even then, there were only minimal antiandrogenic effects on male reproductive development. This was surprising since M2 has generally been shown to have greater antagonistic activity as a ligand for steroid receptors including AR and ER [13]. However, it does demonstrate that the effects of VIN reported here are likely independent of such activity and confirms previous studies that could not replicate VIN effects using flutamide [42]. Since our sample sizes here are somewhat modest, the lack of effect of the M2 treatment group also presents as a secondary control in this study showing that the effects we observe are likely specific to VIN and not due to random variation in the population. This is true for outcomes in both generations and genetic lineages.
5. Conclusions
These data show that VIN may alter other important developmental pathways in a manner independent of antiandrogenic function and that genetic strain sensitivity can have a significant effect on outcomes. While VIN may not be of particular concern as a toxicant due to restricted use in many countries, these data are highly relevant for similar dicarboximide fungicides such as iprodione that are still widely used in the US [43] as well as other common EDCs. Studies determining the effects of EDC exposure for safety and risk assessment purposes will benefit significantly from comparing effects on different genetic lineages.
Supplementary Material
Highlights.
Pup bodyweight and sperm methylation are altered by in utero vinclozolin exposure
Pup sensitivity to vinclozolin in utero is dependent on maternal genetic background
Pup sensitivity to vinclozolin in utero is not dependent on antiandrogenic activity
Genetic mutation at H19/Igf2 does not alter epigenetic sensitivity to vinclozolin
Uni- vs. bi-parental in utero exposure determines second generation pup response
Acknowledgments
We thank Dr. Marisa Bartolomei for generously providing the targeted H19/Igf2 mutant mouse lines and the C7 mice; the UNC Nutrition Obesity Research Core (NORC) for performing the body composition measurements; the David H. Murdock Research Institute CLAS for animal care and husbandry. This work was supported by the National Institutes of Health Transition to Independent Environmental Health Research Career Development Award from NIEHS [KES023849A] and the NIEHS sponsored University of Pennsylvania Center of Excellence in Environmental Toxicology (CEET) Mentored Scientist Transition Award [1P30 ES013508-05] to F.Y.I.; NIDDK grant [P30DK056350] to the UNC NORC AMP. KK was supported by NIH/NIEHS grant [T32ES007018]. MIL was supported by NIH/NIEHS grant [ES010126].
Abbreviated Terms
- VIN
Vinclozolin, or 3-(3,5-dichlorophenyl)-5-methyl-5-vinyloxazolidine-2,4-dione
- EDC
Endocrine disrupting compound
- M1
2-[[(3,5-dichlorophenyl)-carbamoyl]oxy]-2-methyl-3-butenoic acid
- M2
3′,5′-dichloro-2-hydroxy-2-methylbut-3-enanilide
- AR
Androgen Receptor
- ER
Estrogen Receptor
- PR
Progesterone Receptor
- GD
Gestational Day
- PND
Postnatal day
- AGD
Anogenital distance
- DHT
Dihydrotestosterone
- SRY
Sex determining region of the Y chromosome gene
- ICR
Imprinting Control Region
- BWS
Beckwith-Wiedemann syndrome
- DMR
Differentially methylated region
- G0
Generation zero, parental generation
- G1
First generation
- G2
Second generation
Footnotes
Author contributions
Study design, conception, and supervision of research was performed by FI. Animal husbandry, treatment, and tissue/cell collection was performed by KC, EP, RK, and FI. Pyrosequencing data generation analyses were performed by EP, ME, and FI. Statistical analyses were performed by KK, ML, and FI. Manuscript was drafted and revised by EP, ME and FI with input from all authors. All authors have approved this manuscript.
Data Statement
All raw data are available upon request.
Conflict of Interest
The authors declare no conflict of interest. The authors alone are responsible for the content and writing of this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Edwards TM, Myers JP. Environmental Exposures and Gene Regulation in Disease Etiology. Environ Health Perspect. 2007;115:1264–1270. doi: 10.1289/ehp.9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.USEPA Notices. Federal Register. 1998;63(30):59557–59558. [Google Scholar]
- 4.USEPA Notices. Federal Register. 1998;63(248):71553–71567. [Google Scholar]
- 5.USGS. Pesticide National Synthesis Project. [accessed September 28, 2017];Estim Annu Agric Pestic Use. 2015 https://water.usgs.gov/nawqa/pnsp/usage/maps/show_map.php?year=2015&map=VINCLOZOLIN&hilo=L&disp=Vinclozolin.
- 6.Gray LE, Ostby JS, Kelce WR. Developmental Effects of an Environmental Antiandrogen: The Fungicide Vinclozolin Alters Sex Differentiation of the Male Rat. Toxicol Appl Pharmacol. 1994;129:46–52. doi: 10.1006/taap.1994.1227. [DOI] [PubMed] [Google Scholar]
- 7.Gray LE, Ostby J, Monosson E, Kelce WR. Environmental antiandrogens: low doses of the fungicide vinclozolin alter sexual differentiation of the male rat. Toxicol Ind Health. 1999;15:48–64. doi: 10.1177/074823379901500106. [DOI] [PubMed] [Google Scholar]
- 8.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic Transgenerational Actions of Endocrine Disruptors and Male Fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong C, Kelce WR, Sar M, Wilson EM. Androgen Receptor Antagonist versus Agonist Activities of the Fungicide Vinclozolin Relative to Hydroxyflutamide. J Biol Chem. 1995;270:19998–20003. doi: 10.1074/jbc.270.34.19998. [DOI] [PubMed] [Google Scholar]
- 10.French FS, Lubahn DB, Brown TR, Simental JA, Quigley CA, Yarbrough WG, Tan JA, Sar M, Joseph DR, Evans BA. Molecular basis of androgen insensitivity. Recent Prog Horm Res. 1990;46:1–38. doi: 10.1016/b978-0-12-571146-3.50005-5. discussion 38–42. [DOI] [PubMed] [Google Scholar]
- 11.Kelce WR, Monosson E, Gamcsik MP, Laws SC, Gray LE. Environmental Hormone Disruptors: Evidence That Vinclozolin Developmental Toxicity Is Mediated by Antiandrogenic Metabolites. Toxicol Appl Pharmacol. 1994;126:276–285. doi: 10.1006/taap.1994.1117. [DOI] [PubMed] [Google Scholar]
- 12.Kemppainen JA, Lane MV, Sar M, Wilson EM. Androgen receptor phosphorylation, turnover, nuclear transport, and transcriptional activation. Specificity for steroids and antihormones. J Biol Chem. 1992;267:968–974. [PubMed] [Google Scholar]
- 13.Molina-Molina J-M, Hillenweck A, Jouanin I, Zalko D, Cravedi J-P, Fernández F, Pillon A, Nicolas J-C, Olea N, Balaguer P. Steroid receptor profiling of vinclozolin and its primary metabolites. Toxicol Appl Pharmacol. 2006;216:44–54. doi: 10.1016/j.taap.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Buckley J, Willingham E, Agras K, Baskin LS. Embryonic exposure to the fungicide vinclozolin causes virilization of females and alteration of progesterone receptor expression in vivo: an experimental study in mice. Environ Health. 2006;5:4. doi: 10.1186/1476-069X-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laws SC, Carey SA, Kelce WR, Cooper RL, Gray LE. Vinclozolin does not alter progesterone receptor (PR) function in vivo despite inhibition of PR binding by its metabolites in vitro. Toxicology. 1996;112:173–182. doi: 10.1016/0300-483x(96)03354-9. [DOI] [PubMed] [Google Scholar]
- 16.Guerrero-Bosagna C, Settles M, Lucker B, Skinner MK. Epigenetic Transgenerational Actions of Vinclozolin on Promoter Regions of the Sperm Epigenome. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stouder C, Paoloni-Giacobino A. Transgenerational effects of the endocrine disruptor vinclozolin on the methylation pattern of imprinted genes in the mouse sperm. Reproduction. 2010;139:373–379. doi: 10.1530/REP-09-0340. [DOI] [PubMed] [Google Scholar]
- 18.Guerrero-Bosagna C, Covert TR, Haque MM, Settles M, Nilsson EE, Anway MD, Skinner MK. Epigenetic Transgenerational Inheritance of Vinclozolin Induced Mouse Adult Onset Disease and Associated Sperm Epigenome Biomarkers. Reprod Toxicol Elmsford N. 2012;34:694–707. doi: 10.1016/j.reprotox.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iqbal K, Tran DA, Li AX, Warden C, Bai AY, Singh P, Wu X, Pfeifer GP, Szabó PE. Deleterious effects of endocrine disruptors are corrected in the mammalian germline by epigenome reprogramming. Genome Biol. 2015;16:59. doi: 10.1186/s13059-015-0619-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ideraabdullah FY, Abramowitz LK, Thorvaldsen JL, Krapp C, Wen SC, Engel N, Bartolomei MS. Novel cis-regulatory function in ICR-mediated imprinted repression of H19. Dev Biol. 2011;355:349–357. doi: 10.1016/j.ydbio.2011.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ideraabdullah FY, Thorvaldsen JL, Myers JA, Bartolomei MS. Tissue-specific insulator function at H19/Igf2 revealed by deletions at the imprinting control region. Hum Mol Genet. 2014;23:6246–6259. doi: 10.1093/hmg/ddu344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sparago A, Cerrato F, Vernucci M, Ferrero GB, Silengo MC, Riccio A. Microdeletions in the human H19 DMR result in loss of IGF2 imprinting and Beckwith-Wiedemann syndrome. Nat Genet. 2004;36:958–960. doi: 10.1038/ng1410. [DOI] [PubMed] [Google Scholar]
- 23.De Crescenzo A, Coppola F, Falco P, Bernardo I, Ausanio G, Cerrato F, Falco L, Riccio A. A novel microdeletion in the IGF2/H19 imprinting centre region defines a recurrent mutation mechanism in familial Beckwith-Wiedemann syndrome. Eur J Med Genet. 2011;54:e451–454. doi: 10.1016/j.ejmg.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beygo J, Citro V, Sparago A, De Crescenzo A, Cerrato F, Heitmann M, Rademacher K, Guala A, Enklaar T, Anichini C, Cirillo Silengo M, Graf N, Prawitt D, Cubellis MV, Horsthemke B, Buiting K, Riccio A. The molecular function and clinical phenotype of partial deletions of the IGF2/H19 imprinting control region depends on the spatial arrangement of the remaining CTCF-binding sites. Hum Mol Genet. 2013;22:544–557. doi: 10.1093/hmg/dds465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abi Habib W, Azzi S, Brioude F, Steunou V, Thibaud N, Das Neves C, Le Jule M, Chantot-Bastaraud S, Keren B, Lyonnet S, Michot C, Rossi M, Pasquier L, Gicquel C, Rossignol S, Le Bouc Y, Netchine I. Extensive investigation of the IGF2/H19 imprinting control region reveals novel OCT4/SOX2 binding site defects associated with specific methylation patterns in Beckwith-Wiedemann syndrome. Hum Mol Genet. 2014;23:5763–5773. doi: 10.1093/hmg/ddu290. [DOI] [PubMed] [Google Scholar]
- 26.Poole RL, Leith DJ, Docherty LE, Shmela ME, Gicquel C, Splitt M, Temple IK, Mackay DJG. Beckwith-Wiedemann syndrome caused by maternally inherited mutation of an OCT-binding motif in the IGF2/H19-imprinting control region, ICR1. Eur J Hum Genet EJHG. 2012;20:240–243. doi: 10.1038/ejhg.2011.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith FM, Garfield AS, Ward A. Regulation of growth and metabolism by imprinted genes. Cytogenet Genome Res. 2006;113:279–291. doi: 10.1159/000090843. [DOI] [PubMed] [Google Scholar]
- 28.Taniguchi T, Sullivan MJ, Ogawa O, Reeve AE. Epigenetic changes encompassing the IGF2/H19 locus associated with relaxation of IGF2 imprinting and silencing of H19 in Wilms tumor. Proc Natl Acad Sci U S A. 1995;92:2159–2163. doi: 10.1073/pnas.92.6.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao Z-H, Suppola S, Liu J, Heikkilä P, Jänne J, Voutilainen R. Association of H19 promoter methylation with the expression of H19 and IGF-II genes in adrenocortical tumors. J Clin Endocrinol Metab. 2002;87:1170–1176. doi: 10.1210/jcem.87.3.8331. [DOI] [PubMed] [Google Scholar]
- 30.Kim HT, Choi BH, Niikawa N, Lee TS, Chang SI. Frequent loss of imprinting of the H19 and IGF-II genes in ovarian tumors. Am J Med Genet. 1998;80:391–395. doi: 10.1002/(sici)1096-8628(19981204)80:4<391::aid-ajmg16>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 31.van Gurp RJ, Oosterhuis JW, Kalscheuer V, Mariman EC, Looijenga LH. Biallelic expression of the H19 and IGF2 genes in human testicular germ cell tumors. J Natl Cancer Inst. 1994;86:1070–1075. doi: 10.1093/jnci/86.14.1070. [DOI] [PubMed] [Google Scholar]
- 32.Mann MRW, Chung YG, Nolen LD, Verona RI, Latham KE, Bartolomei MS. Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol Reprod. 2003;69:902–914. doi: 10.1095/biolreprod.103.017293. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Tranguch S, Xie H, Hanley G, Das SK, Dey SK. Variation in commercial rodent diets induces disparate molecular and physiological changes in the mouse uterus. Proc Natl Acad Sci U S A. 2005;102:9960–9965. doi: 10.1073/pnas.0501632102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartolomei MS, Webber AL, Brunkow ME, Tilghman SM. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 1993;7:1663–1673. doi: 10.1101/gad.7.9.1663. [DOI] [PubMed] [Google Scholar]
- 35.Chen G, Yang L, Begum S, Xu L. GPR56 is essential for testis development and male fertility in mice. Dev Dyn. 2010;239:3358–3367. doi: 10.1002/dvdy.22468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinheiro J, Bates D, DebRoy S, Sarkar D. R Core Team, nlme: Linear and Nonlinear Mixed Effects Models. 2017 https://CRAN.R-project.org/package=nlme.
- 37.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2017. https://www.R-project.org/ [Google Scholar]
- 38.Benjamini Y, Hochberg Y. Controlling The False Discovery Rate - A Practical And Powerful Approach To Multiple Testing. J R Stat Soc Ser B. 1995;57:289–300. doi: 10.2307/2346101. [DOI] [Google Scholar]
- 39.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 40.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14(Spec No 1):R47–58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 41.Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 42.Anway MD, Rekow SS, Skinner MK. Comparative anti-androgenic actions of vinclozolin and flutamide on transgenerational adult onset disease and spermatogenesis. Reprod Toxicol. 2008;26:100–106. doi: 10.1016/j.reprotox.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.USGS. Pesticide National Synthesis Project. [accessed September 28, 2017];Estim Annu Agric Pestic Use. 2015 https://water.usgs.gov/nawqa/pnsp/usage/maps/show_map.php?year=2015&map=IPRODIONE&hilo=L&disp=Iprodione.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.