Abstract
Purified human eosinophils treated for 18-24 h with IL-3 adopt a unique activated phenotype marked by increased reactivity to aggregated immunoglobulin-G (IgG). To characterize this phenotype, we quantified protein abundance and phosphorylation by multi-plexed isobaric labeling combined with high-resolution mass spectrometry. Purified blood eosinophils of five individuals were treated with IL-3 or no cytokine for 20 hours, and comparative data were obtained on abundance of 5385 proteins and phosphorylation at 7330 sites. The 1150 proteins that were significantly up-regulated (q<0.05, pair-wise t test with Benjamini-Hoachberg correction) by IL-3 included the IL3RA and CSF2RB subunits of the IL-3 receptor, the low-affinity receptor for IgG (FCGR2B), 96 proteins involved in protein translation, and 55 proteins involved in cytoskeleton organization. Among the 703 proteins that decreased were 78 mitochondrial proteins. Dynamic regulation of protein phosphorylation was detected at 4218 sites. These included multiple serines in CSF2RB; Y694 of STAT5, a key site of activating phosphorylation downstream of IL3RA/CSF2RB; and multiple sites in RPS6KA1, RPS6, and EIF4B, which are responsible for translational initiation. We conclude that IL-3 up-regulates overall protein synthesis and targets specific proteins for up-regulation, including its own receptor.
Keywords: eosinophil, IL-3, mass spectrometry-based proteomics, phosphorylation sites
Graphical abstract
Introduction
Eosinophils (EOS) are terminally differentiated cells that mature in the bone marrow, enter the blood stream, and arrest and migrate at sites of inflammation guided by chemoattractant proteins (1). At the site of the inflammatory response, EOS become activated and release a variety of toxic proteins and mediators that damage tissues and direct the immune response (2). Tissue EOS are implicated in many diseases including asthma, eosinophilic esophagitis, and hyper-eosinophilic syndrome (3). Maturation and activation of the EOS are enhanced by cytokines such as IL-5, which is produced by T lymphocytes and innate lymphoid cells in response to an allergen, viral infection and IL-1ß (4). IL-5, along with GM-CSF and IL-3, belongs to the IL-5-family cytokines that target receptors with a specific α-chain (IL5RA, CSF2RA, or IL3RA) and a common ß-chain (CSF2RB) to initiate signaling pathways. Many other agonists acting via other receptors and signaling pathways are physiological activators of EOS. Knowledge of the consequences of activation by the different agonists as well as the responsible signaling pathways is fundamental to therapeutic targeting of EOS (5).
We have demonstrated that among the three IL-5-family cytokines, IL-3 is uniquely effective in prolonging intracellular signaling, maintaining a polarized shape, and up-regulation of the abundance of FCGR2B and SEMA7A as assessed by immuno-blotting and flow cytometry (6–8). IL-3 is relevant in allergy inasmuch as it is released by activated Th-2 lymphocytes, mast cells and basophils following IgE cross-linking (9, 10), and elevated in the airways to as much as 10 ng/ml after a segmental allergen challenge (11). In addition, IL-3 is elevated in serum in poorly controlled asthmatic patients, and airway IL-3-positive cells are increased with asthma severity (12, 13). Importantly we found that EOS chronically activated in vitro with IL-3 react vigorously to aggregated immunoglobulin-G (IgG) via activated FCGR2 and ITGAM/ITGB2 integrin receptor (8).
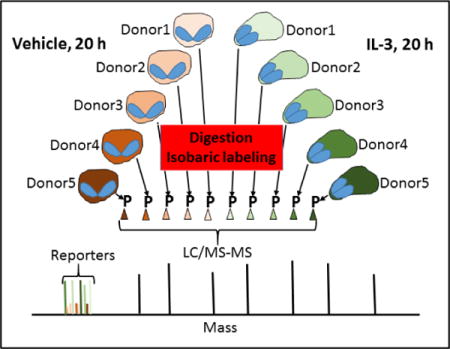
To characterize and understand more completely the EOS phenotype induced by IL-3, we quantified global changes of proteins and phospho-sites in IL-3-stimulated EOS by two-dimensional chromatography coupled to high-resolution mass spectrometry (LC/MS-MS)(14). Previously, we compared protein abundance and phosphorylation in unactivated EOS and EOS activated acutely (5 min) with IL-5 (15). Tryptic peptides were generated individually from activated or non-activated EOS of five individuals and labeled with isobaric labels, mixed together, and analyzed by LC-MS/MS. The relative amounts of a given peptide from each of the ten samples was ascertained by comparison of the respective reporter ion signals in the MS2 spectrum. Significant changes (p<0.01, two-tailed t-test) were found for 220 of 1819 localized phospho-sites (12.1%) when the two experimental conditions were compared. As would be expected given the short time of activation, differences in protein abundance were much less striking. We used the same approach to compare blood EOS incubated for 20 h with IL-3 to control EOS. We identified 5385 proteins, including 1150 proteins up-regulated by IL-3, and 7330 phosphorylated sites of which more than half were altered in IL-3-treated EOS. The results, therefore, reveal reshaping of the EOS proteome and a host of phosphorylation events that support and sustain activation.
Methods
Cell preparation
The study protocol was approved by the University of Wisconsin-Madison Health Sciences Institutional Review Board. Informed written consent was obtained from subjects prior to participation. Peripheral blood EOS were obtained from allergic subjects with rhinitis with or without mild asthma, and EOS blood counts between 229 and 308 per μl (Supplemental Table 1). Subjects with prescriptions for low doses of inhaled corticosteroids did not use their corticosteroids the day of the blood draw. Eosinophils were purified by negative selection as previously described (8). Briefly, heparinized blood was diluted 1:1 in HBSS and was overlaid above Percoll (1.090 g/ml). After centrifugation at 700 × g for 20 min at room temperature, the mononuclear cells were removed from the plasma/Percoll interface, and erythrocytes were eliminated from the cell pellet by hypotonic lysis. The remaining pellet was resuspended in 2% NCS in HBSS. Cells were then incubated with beads coupled to anti-CD16, anti-CD3, anti-CD14, and anti-glycophorin-A (Miltenyi, San Diego, CA) and processed with an AutoMACS (Miltenyi). EOS preparations with purity > 99% and viability ~98% were used the same day, ~5 h after the blood draw. EOS, 1 × 107, were cultured at 37°C in 10 mL in medium (RPMI 1640 plus 10% fetal bovine serum) without cytokine (resting) or with IL-3 (4 ng/mL; BD Biosciences, San Jose, CA, USA) for 20 h. At the end of the incubations, cells were washed with PBS, snap frozen in cold ethanol, and stored at -80°C until all ten samples (five sets of two) had been obtained.
Proteomic workflow
The samples were thawed and suspended in 100 μL of 6 M guanidine HCl and boiled for 5 minutes. Methanol was added to 90% final concentration, samples were centrifuged at 10,000 × g for 5 min, and the supernatant was discarded. The protein pellet was dissolved in 200 μL 8 M urea, 100 mM TRIS pH 8, 10 mM TCEP, and 40 mM chloroacetamide. Endoproteinase LysC was added to an estimated 50:1 (protein:enzyme) ratio, and the digestion was incubated at ambient temperature for 2 hours. Samples were diluted to 2 M urea in 100 mM TRIS, and trypsin was added at a 50:1 ratio followed by overnight incubation at ambient temperature. Samples were titrated with TFA to pH 2, followed by centrifugation, solid phase extraction of the supernatant peptides, and drying. Desalted peptides were dissolved in 100 mM TEAB and labeled with one of the ten tandem mass tags (10plexTMT10, ThermoFisher). The concentration of labeled peptides in each sample was determined, and equal amounts from each sample were combined and dried. The combined sample was dissolved in 80% acetonitrile and 6% TFA (IMAC wash solution) and enriched for phospho-peptides using Titanium(IV) immobilized metal ion affinity chromatography. In brief, the beads were washed twice with IMAC wash solution, incubated with the sample on a shaker for 30 min, washed again three times with IMAC wash solution, once with 80% acetonitrile, once with 0.5 M glycolic acid/80% acetonitrile, and once again with 80% acetonitrile. Phospho-peptides were eluted with 1% ammonium hydroxide/50% acetonitrile. Samples depleted of phospho-peptides were saved from the unbound portion following the incubation step. Both types of samples were dried and fractionated by high pH reversed phase chromatography on an Agilent 1200 HPLC with automated fraction collector using mobile phase A: 20 mM ammonium bicarbonate and mobile phase B: 80% methanol/20 mM ammonium bicarbonate. Samples were loaded onto a 4.6 × 150 mm 3.5 μm BEH C18 column and eluted with increasing mobile phase B. For the protein and phosphorylation samples, 15 and 10 fractions were collected, respectively.
LC-MS/MS analysis
Each fraction was dissolved in 0.2% formic acid and injected onto an in-house fabricated 75-μm capillary column with an embedded emitter packed to 30 cm with 1.7 μm BEH C18 particles and eluted using a Thermo Fisher RSLC nano pump and autosampler with mobile phase A: 0.2% formic acid and mobile phase B: 70% acetonitrile/0.2% formic acid. For protein samples, an estimated 1 μg was injected onto the column, for phospho-peptide samples 1/3 of total volume was analyzed each injection. Peptides were eluted off the column with increasing mobile phase B, followed by washing with 100% B, and equilibration of the column at 0% B. Eluted peptides were analyzed with a Thermo Orbitrap Fusion Lumos instrument. For these analyses, survey scans and MS/MS scans were performed at 60,000 resolving power in the Orbitrap. Peptide precursors selected for MS/MS were isolated in the quad with a 1.6-Da isolation window and fragmented in the HCD cell with a normalized collision energy of 35 prior to measurement in the Orbitrap. The AGC target was set to 200,000, and the maximum injection time was set to 118 ms. Two injection replicates were analyzed for each phospho-peptide fraction.
Data analysis
All data processing was performed with the COMPASS software suite. In brief, raw files were converted to text files and then searched with OMSSA against a target-decoy UniProt reference proteome (downloaded March 2017). Theoretical tryptic peptides with up to three missed cleavages were searched for with carbamidomethylation of cysteine set as a fixed modification and methionine oxidation set as a variable modification. Phospho-peptide searches included phosphorylation of S and T with neutral loss and phosphorylation of Y. The search tolerance was 75 ppm and 0.01 Da for MS1 and MS2 respectively. Search results were filtered to 1% FDR at the unique peptide level with a two-dimensional ppm and E-value score filtering. All resulting peptides had < 11 ppm mass error. TMT reporter ion intensity measurements were extracted from the raw files for all peptides spectral matches. Proteins were grouped according to subsumable protein grouping rules, groups were filtered to 1% FDR, and quantified. Phosphorylation sites were localized and quantified using an adapted phosphoRS algorithm. Sites with localization probability >75% were considered localized in this analysis.
Analysis of proteins
Differentially expressed genes were analyzed using DAVID Bioinformatic Resources 6.8(Beta) (National Institute of Allergy and Infectious Diseases (NIAID), NIH) (16). Phosphosites that went up by >+0.5 or down by <-0.5 were analyzed by MotifX (17, 18) using the default parameters of p<0.000001 and 20 required occurrences. All the sites were used as the background.
Results and Discussion
After a 20-hour culture of eosinophils, 5385 proteins were identified by two-dimensional liquid chromatography coupled with high-resolution mass spectrometry (Supplemental Spreadsheet 1). Intensities of reporter ions in the MS2 spectra originating from peptides derived from the proteins were determined for each of the ten samples: resting and IL-3-activated eosinophils from five donors. Compared to resting EOS (vehicle), IL-3-activated EOS significantly (q<0.05) up-regulated 1150 proteins, whereas 703 proteins were significantly down-regulated, as displayed by Volcano plot of log2 differences versus significance of differences (Figure 1, left). Because equal amounts of peptides from each sample are mixed together before analysis, if some proteins increase, others must decrease. To assess whether changes in more minor proteins would be biased by large changes in major proteins, we analyzed IL-3-induced changes in the 15 most abundant proteins that accounted for 25% of the molar mass of proteins in a prior intensity-based absolute quantitation (IBAQ) study of the EOS proteome (15), as shown in Table 1. Two of the four histones, which are considered a “proteomic ruler” because cellular histone content is proportional to DNA content (19), were increased modestly but significantly in IL-3-treated EOS. Two actin remodeling proteins and two cytoplasmic proteins also were increased modestly but significantly compared to control. The four major granule proteins were slightly decreased, but the changes did not reach significance. The 15 proteins in the IL-3 treated sample, normalized for abundance, were 104% of control samples.
Figure 1. Volcano plots of proteomic changes.
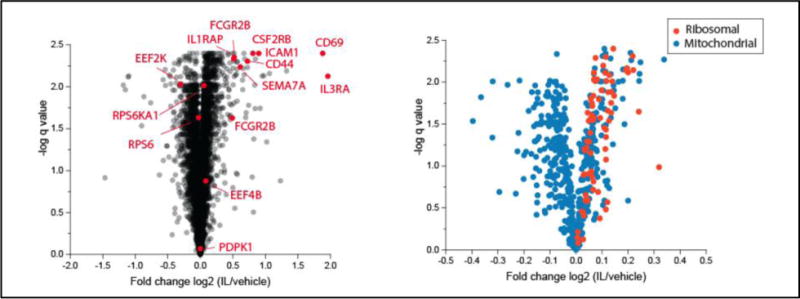
Left, all identified proteins. Right, mitochondrial and ribosomal proteins. IL/vehicle: IL-3-activated versus unactivated EOS. Note that the ranges of horizontal axes are different.
Table 1.
Changes in the 15 most abundant EOS proteins in response to IL-3
| Resting EOS (From Wilkerson)15 | IL-3-stimulated (I) versus vehicle (V) control EOS | ||||
|---|---|---|---|---|---|
| Gene | Description | IBAQ (×10−10) | Log2 FC I/V | % of Control | q−value |
| ACTB | Actin | 12.5 | 0.04 | 103 | 0.147 |
| RNASE2 | Granule protein | 11.1 | −0.03 | 98 | 0.504 |
| HIST1H4A | Histone | 11.1 | 0.10 | 107 | 0.015 |
| HIST1H2A | Histone | 9.8 | 0.07 | 105 | 0.426 |
| CLC | Cytoplasmic protein | 9.0 | −0.01 | 99 | 0.667 |
| HIST1H2B | Histone | 7.8 | 0.37 | 129 | 0.015 |
| PFN1 | Actin remodeling | 7.4 | 0.09 | 106 | 0.025 |
| PRG2 | Granule protein | 6.6 | −0.05 | 96 | 0.051 |
| RNASE3 | Granule protein | 6.4 | −0.03 | 98 | 0.483 |
| CFL1 | Actin remodeling | 4.9 | 0.12 | 108 | 0.023 |
| S100A9 | Cytoplasmic protein | 4.5 | 0.06 | 104 | 0.031 |
| PRG3 | Granule protein | 4.3 | −0.03 | 98 | 0.239 |
| ANXA1 | Cytoplasmic protein | 4.3 | 0.09 | 106 | 0.013 |
| GAPDH | Metabolic enzyme | 4.2 | 0.06 | 104 | 0.044 |
| HIST1H3A | Histone | 3.7 | −0.15 | 90 | 0.069 |
IBAQ values from Wilkerson et al.15 were compared with the fold change (FC) log2 ratios of the intensities of reporter ions from IL-3 treated (I) and vehicle control (V) EOS, the log2 ratios expressed as of % vehicle control, and q-values for differences between the treated and control EOS.
Regulated proteins were analyzed using DAVID Bioinformatic Resources 6.8 (DAVID). Of the 1150 and 703 up- and down-regulated proteins, 1097 and 650 proteins, respectively, were recognized and included in DAVID analyses. DAVID cluster analysis indicates that the proteins up-regulated by IL-3 are implicated in protein translation, cytoskeleton regulation and composition, vesicle transport, intracellular signaling, and leukocyte functions (adhesion and migration) (Table 2). Both up- and down-regulated proteins are highly associated with nucleotide binding activity and cell death as well (Tables 2 and 3). The list of the 96 up-regulated proteins associated with protein translation (Supplemental Table 2) includes many initiation and elongation factors of the protein translational machinery, and components of large and small subunits of the ribosome (Figure 1, right). Fifty-five up-regulated proteins are related to cytoskeleton organization, in large part proteins involved in actin cytoskeleton organization (Table 2 and Supplemental Table 3). Of the proteins down-regulated by IL-3, 78 are connected to the mitochondrion (Table 3 and Figure 1, right), particularly the mitochondrial membrane respiratory chain NADH dehydrogenase (ubiquinone; Complex I) (Supplemental Table 4). The change in mitochondrial proteins is intriguing given publications describing catapulted release of mitochondrial DNA from activated EOS (20, 21). In those studies, the discharge of mitochondrial DNA was followed by the release of reactive oxygen species (ROS), which was required for vacuolization, loss of membrane integrity, and release of granule proteins (21). Based on our prior finding that IL-3 primes EOS for release of granule proteins upon exposure to aggregated IgG (8), we speculate that alterations in mitochondria may be part of the priming process.
Table 2.
Cluster analysis of the proteins up-regulated by IL-3 versus vehicle (resting)
DAVID 6.8 Functional Annotation Clustering
| Cluster | Functional annotation | # of detected proteins* | # of upregulated proteins** | P value (Benjamini) (DAVID)*** | % upregulated versus detected |
|---|---|---|---|---|---|
|
| |||||
| Translation and protein processing | Translation | 153 | 96 | 2.8 ×10−31 | 62.7 |
| Protein biosynthesis | 128 | 80 | 2.1 ×10−46 | 62.5 | |
| Protein transport | 355 | 123 | 2.8 ×10−16 | 34.6 | |
| Golgi apparatus | 348 | 72 | 4.9 ×10−8 | 20.6 | |
| Cytoskeleton | Cytoskeleton | 150 | 89 | 3.4 ×10−13 | 59.3 |
| Actin cytoskeleton organization | 71 | 38 | 9.7 ×10−5 | 53.5 | |
| Cytoskeleton organization | 112 | 55 | 1.4 ×10−3 | 49.1 | |
| Nucleotide binding/RNA | Nucleotide binding | 707 | 229 | 1.2 ×10−9 | 32.4 |
| RNA binding | 395 | 122 | 3.3 ×10−19 | 30.8 | |
| Vesicle/transport | Vesicle-mediated transport | 306 | 96 | 3.9 ×10−13 | 31.4 |
| Vesicle | 280 | 86 | 3.2 ×10−7 | 30.7 | |
| Intracellular signaling | ATP binding | 608 | 140 | 9.4 ×10−4 | 23.0 |
| Kinase | 263 | 60 | 1.2 ×10−2 | 22.8 | |
| GTPase activity | 97 | 34 | 3.0 ×10−4 | 35.0 | |
| Cell death | Regulation of cell death | 330 | 92 | 2.4 ×10−4 | 27.9 |
| Positive regulation of cell death | 182 | 50 | 1.6 ×10−2 | 27.4 | |
| Leukocyte function | Leukocyte migration | 45 | 12 | 4.7 ×10−2 | 26.6 |
| Leukocyte adhesion | 11 | 8 | 6.5 ×10−2 | 72.7 | |
: number of detected proteins among the 5385 detected proteins that are part of the indicated functional annotation as determined by DAVID analysis
: number of proteins up-regulated by IL-3 (q<0.05) and part of the indicated functional annotation as determined by DAVIS analysis
: adjusted enrichment p value (Benjamini correction). The smaller, the more enrichment of the IL-3-upregulated proteins in the indicated functional annotation
%: percentage of proteins IL-3-up-regulated among the total proteins detected and present in the indicated functional annotation
Underlined are clusters for which a list of proteins is shown in Supplemental Tables 2 and 3
Table 3.
Cluster analysis of the proteins down-regulated by IL-3
DAVID 6.8 Functional Annotation Clustering
| Cluster | Functional annotation | # of detected proteins* | # of down-regulated proteins** | P value (Benjamini) (DAVID)*** | % downregulated versus detected |
|---|---|---|---|---|---|
|
| |||||
| Mitochondria | Mitochondrion | 482 | 96 | 6.8 ×10−14 | 16.1 |
| RNA/DNA | mRNA transport | 52 | 14 | 4.1 ×10−5 | 26.9 |
| Nucleotide binding | 707 | 130 | 1.0 ×10−3 | 18.3 | |
| mRNA processing | 200 | 35 | 1.5 ×10−4 | 17.5 | |
| RNA binding | 395 | 42 | 6.2 ×10−2 | 10.6 | |
| Transcription activator activity | 87 | 32 | 5.1 ×10−2 | 36.8 | |
| Cell death | Cell death | 296 | 47 | 4.9 ×10−2 | 15.8 |
: number of detected proteins among the 5385 detected proteins that are part of the indicated functional annotation as determined by DAVID analysis
: number of proteins down-regulated by IL-3 (q<0.05) and part of the indicated functional annotation as determined by DAVIS analysis
: Adjusted enrichment p value (Benjamini correction). The smaller, the more enrichment of the IL-3-downregulated proteins in the indicated functional annotation
%: percentage of proteins IL-3-down-regulated among the total proteins detected and present in the indicated functional annotation
Underlined are clusters for which a list of proteins is shown in Supplemental Table 4
The present studies were motivated by immuno-blotting and qPCR analyses demonstrating that SEMA7A and FCGR2B/C are regulated at the translational level in EOS by IL-3, but not by the other ß-chain cytokines, IL-5 or GM-CSF (6–8). SEMA7A and two splice variants (isoforms 2/5 and 1/3/4) of FCGR2B were among the top 30 proteins up-regulated by IL-3 as defined by Log2 >0.5 fold increase and q values <0.01 (Figure 1 left and Supplemental Spreadsheet 1). Because the peptides, ISALPGYPECR and ISANPTNPDEADK, that define the FCGR2B splice variants are not present in FCGR2C, these data resolve the ambiguity in previous immuno-blotting studies (8) caused by the inability of the antibody used to distinguish between FCGR2B and FCGR2C. However, because the present analysis did not detect any peptides unique to FCGR2C, directed reaction-monitored analyses using an internal standard would be needed to ascertain if FCGR2C also increases after IL-3 treatment.
Among the top 30 proteins up-regulated by IL-3, were IL3RA and CSF2RB, the α and common ß subunits of the receptor for IL-3 (Supplemental spreadsheet 1 and Figure 1, left). In contrast, CSF2RA (GM-CSF receptor α-chain) did not change significantly, and IL5RA (IL-5 receptor α-chain) was undetectable. IL-3-enhanced IL3RA is in agreement with the previous finding showing that the surface receptor for IL-3 is enhanced on EOS by a prolonged (~20 h) activation with the ß-chain cytokines (IL-5, GM-CSF, or IL-3) while the amount of surface receptors for IL-5 and GM-CSF are decreased or unchanged, respectively, in the same conditions (22, 23). The enhanced amount of both IL3RA and CSF2RB explains the unique ability for IL-3 compared to IL-5 and GM-CSF to sustain intracellular signaling in EOS (7, 24).
High among the other 26 most up-regulated proteins (Figure 1 left and Supplemental Spreadsheet 1) was the well-known leukocyte activation marker, CD69 (25). Of the other 25, only five have been previously studied in EOS. CD44 and ICAM1 are proteins involved in EOS cell-cell/matrix interaction, migration, and activation (26–28). IL1RAP is co-receptor with IL1R1 for IL1 and with IL1RL1 for IL-33 signaling. The receptor for IL-33, IL1RL1/IL1RAP is expressed on mature blood and airway EOS, and EOS progenitors (29–31). IL-33 activates EOS to release cytokines such as GM-CSF, which increases their survival in an autocrine manner (32, 33). IL-33 also potently triggers EOS degranulation and superoxide anion production (29) and up-regulates the membrane proteins CCR3 and CD69 and the adhesion molecule ITGB2 (reviewed in (34)). LMNA codes for lamin A and C, which play an important role in chromatin organization and nuclear membrane dynamics. In EOS, lamin A/C fragmentation occurs following NO-induced apoptosis in a caspase 6-dependent manner (35). Finally, increases in numbered HLA-Ds were found. HLA-Ds are class II molecules that play a central role in the immune system by presenting peptides derived from extracellular proteins. Class II molecules are expressed on antigen presenting cells (B-lymphocytes, dendritic cells, and macrophages) and are increased in vivo on airway EOS (36) and in vitro on EOS by the ß-chain cytokines (37, 38). Tissue EOS in eosinophilic esophagitis also display increased HLA-DR expression (39) and have the potential to act as an antigen-presenting cell (reviewed in (40)).
The other top proteins up-regulated by IL-3, although unstudied in EOS, have recognized cellular functions related to cytoskeleton organization, vesicle and protein transport, cell adhesion, and cell trafficking (CCPG1, DSC2, JAML, PRNP, SEMA4A, TAX1BP1, and TMF1) (41–46). Some of these proteins also possess pro-survival/anti-apoptotic features (CFLAR, PRNP, RARA, and TAX1BP1) (47–49) while others are involved in gene transcription (DDX5 and NFE2) (50, 51). P2RY14 (Supplemental Spreadsheet 1), a purinergic receptor for UDP-glucose and other UDP-sugars is an asthma risk gene (52). Finally, CDC123 (Supplemental Spreadsheet 1) is critical for the eukaryotic translation initiation factor 2 (eIF2) activity, which is required for the onset of protein synthesis (53). Conversely, EEF2K, a kinase inhibitor of protein translation elongation (54) was one of the top 30 proteins down-regulated by IL-3 (Supplemental Spreadsheet 1).
Importantly, along with EEF2K, IL-3 reduced levels of PAIP2 and PDCP4, two inhibitors of the cap-dependent protein translation initiation, blocking the interaction of PABP with the poly(A) RNA tail and the helicase activity of the EIF4 complex, respectively (55–57). Additionally, in agreement with a previous study (58), Supplemental Spreadsheet 1 shows that IL-3 significantly reduces the amount of CDKN1B, cyclin-dependent kinase inhibitor. Interestingly, and relevant to the effect of IL-3 on mitochondrial function discussed above, the mitochondrial dehydrogenase DHTKD1 was strongly decreased by IL-3. Although DHTKD1 function has not been studied in EOS, its suppression may lead to impaired mitochondrial biogenesis and increased production of ROS (59). Finally, histone deacetylase 1 (HDAC1), which is produced by EOS (15), is targeted by both TSC22D3 (aka GILZ) and DDX5. While TSC22D3 and DDX5 were, respectively, highly down- and up-regulated by IL-3 (Supplemental Spreadsheet 1), both can repress transcription via HDAC1 (60, 61).
The phospho-proteomic analysis identified 7330 phosphorylated sites, of which 4217 (57.5%) were significantly (q<0.05) either hyper- or hypo-phosphorylated by IL-3 activation (Figure 2 and Supplemental Spreadsheet 2). Changes of phosphorylation state were observed on 1765 different proteins, with separate sites in 485 proteins being both hyper- and hypo-phosphorylated. Motif-X (17, 18) identified eight motifs that were hyper-phosphorylated, and four that were hypo-phosphorylated (by >+0.5 or down by <-0.5 log2) (Figure 3). The most enriched hyper-phosphorylated motif above the background of all sites was K/R.K...S, which is best known as a site of phosphorylation by AKT (62).
Figure 2. Volcano plots of phospho-proteomic changes.
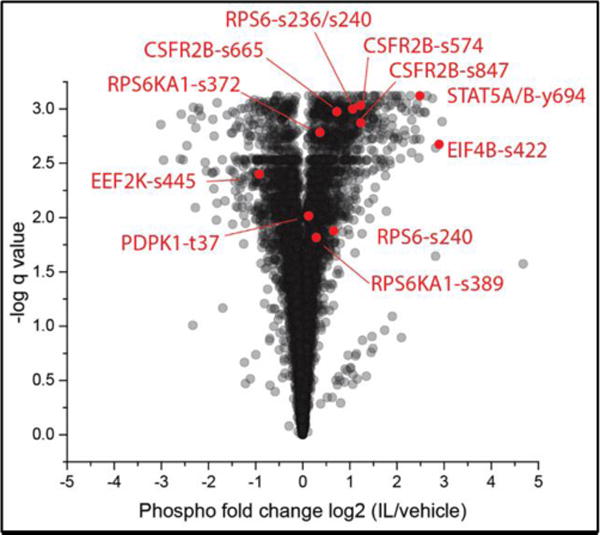
IL/vehicle: IL-3-activated versus unactivated EOS.
Figure 3. Phospho-proteome motifs over-represent among phospho-sites that changed significantly.
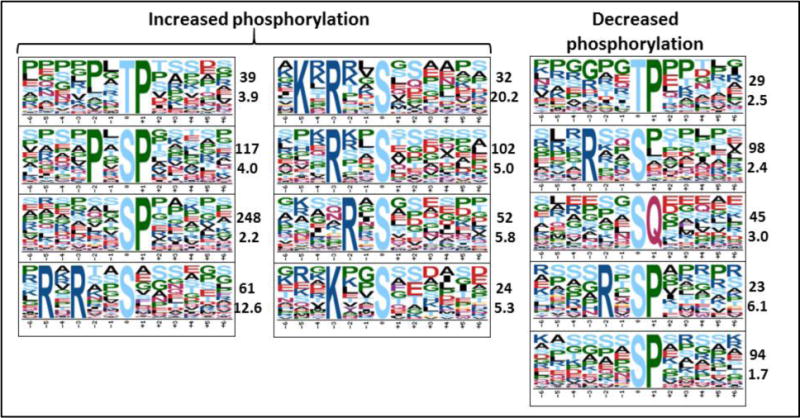
The two numbers next to each motif refer to number of times the motif was identified and fold increase above background.
The 1765 proteins hyper- or hypo-phosphorylated (Supplemental spreadsheet 2) were analyzed using DAVID. DAVID cluster analysis indicates that the proteins with a change in phosphorylation state are highly implicated in intracellular signaling, including as many as 164 kinases, while 226 proteins were also related to the cytoskeleton, and 58 proteins were involved in actin cytoskeleton organization (not shown). Other significant clusters were part of intracellular movement, gene transcription and other types of cellular responses such as cell death, adhesion, trafficking and degranulation. Another important group of proteins was linked to RNA binding and regulation of protein translation. Signaling pathways implicated in the regulation of protein translation were further analyzed (Figure 4), according to several review articles (63–65) as well as our recent review article describing the regulation of protein translation in EOS (54). Figure 4 shows up-regulation and hyper-phosphorylation of proteins that are well known to enhance mRNA translatability and protein translation. Notably, a major inhibitor of protein elongation, EEF2K was down-regulated and hypo-phosphorylated by IL-3. Moreover, our data show that EEF2K activity may be inhibited by numerous upstream kinases, including the RPS6 kinases and the MAP kinases in IL-3-activated EOS (Figure 4), thus leading to enhanced protein production.
Figure 4. Effects of IL-3 on proteins controlling initiation and elongation of protein translation.
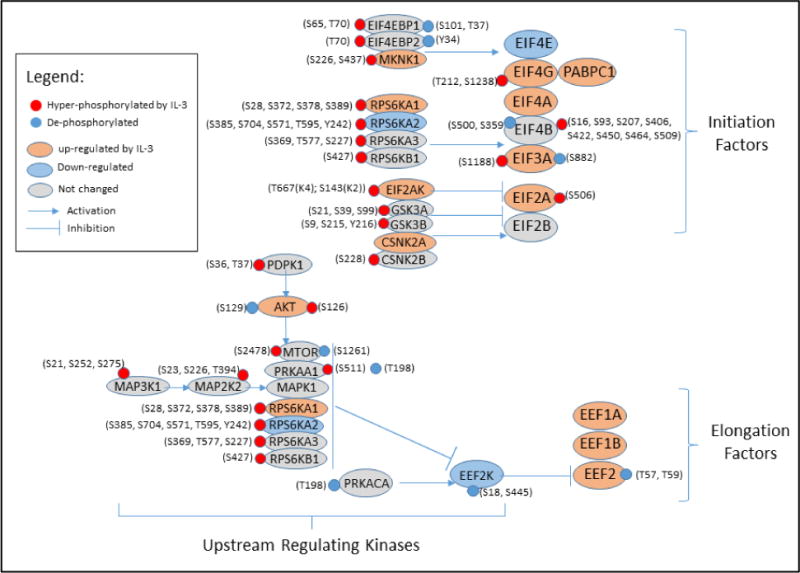
Changes in protein amount (Supplemental spreadsheet 1) and phosphorylation state (Supplemental spreadsheet 2) of initiation factors (EIF), elongation factors (EEF), and their upstream regulating kinases are indicated by color. Changed phosphorylation sites at serine (S), threonine (T), and tyrosine (Y) of each protein are shown. EIF4B interaction with EIF3 (translational initiation) is increased by RPS6KB1- and RPS6KA1/2/3- mediated phosphorylation. RPS6KB1 and RPS6KA1/2/3 along with MAPK1 and MTOR increase protein elongation by blocking EEF2K activity.
More profound changes in phosphorylation status as defined by q<0.01 and Log2 fold change > 0.25, were extracted from the 2099 sites significantly (q<0.05) hyper-phosphorylated by IL-3 activation. Using these criteria, hyper-phosphorylation was observed on 1108 sites and 700 proteins. DAVID cluster analysis indicates that the hyper-phosphorylated proteins are mostly implicated in intracellular signaling, cytoskeleton organization, RNA binding, regulation of protein translation, and intracellular movement (Table 4 and Supplemental Tables 5, 6, 7 and 8). The present phospho-proteome confirms our previous published study, in which we demonstrated continuous phosphorylation of RPS6KA1 (RSK1), RPS6, and EIF4B by a 14 h activation of EOS with IL-3 (7). Additionally, we previously found that IL-3-prolonged RPS6KA1 phosphorylation was critical for SEMA7A and FCGR2B translation and protein accumulation (7, 8), and as described above, SEMA7A and FCGR2B were both highly up-regulated by IL-3.
Table 4.
Cluster analysis of the proteins hyper-phosphorylated by IL-3 (q<0.01 and log 2 fold change > 0.25)
DAVID 6.8 Functional Annotating Clustering
| Cluster | Functional annotation |
|---|---|
| Signaling | Kinases (72; p=1.6 ×10−15) |
| Serine/threonine-protein kinase (46, p=1.2 ×10−11) | |
| Regulation of Ras protein signal transduction (25, p=3.9 ×10−4) | |
| MAPKKK cascade (21, p=3.4 ×10−3) | |
| Transmembrane receptor protein tyrosine kinase signaling pathway (21, p=2.2 ×10−2) | |
| Cytoskeleton | Cytoskeleton (96; p=1.3×10−9) |
| Actin cytoskeleton (29; p=8.9×10−6) | |
| RNA/translation | RNA-binding (41, p=5.1×10−5) |
| mRNA transport (13, p=9.6×10−5) | |
| Ubiquitin-associated/translation elongation factor EF1B (11, p=1.3×10−4) | |
| Posttranscriptional regulation of gene expression (21, p=1.2×10−2) | |
| Translation regulation (9, p=1.5×10−2) | |
| mTOR signaling pathway (9, p=3.4×10−2) | |
| Golgi apparatus (47, p=2.7×10−2) | |
| Intracellular movement | Vesicle-mediated transport (50, p=3.4×10−5) |
| Nucleocytoplasmic transport (19, p=3.6×10−3) | |
| Membrane-bounded vesicle (36, p=8.0×10−3) | |
| Other cell responses | Focal adhesion (16, p=6.0×10−5) |
| Positive regulation of apoptosis (33, p=1.5×10−2) |
In parentheses: number of proteins hyper-phosphorylated by IL-3 (q<0.01 and log 2 fold change > 0.25) for each functional annotation; p values (Benjamini correction)
Underlined are clusters for which a list of proteins is shown in Supplemental Tables 5, 6, 7 and 8
STAT5A/B phosphorylation on Y694/699 was ranked the second most phosphorylated protein by IL-3 (q<0.00001 and fold change Log2 =+2.48; Figure 2 and Supplemental Spreadsheet 2). This residue is a substrate for JAKs that bind to ligated IL3RA/CSF2RB receptor and tyrosine phosphorylate CSF2RB (66, 67). In our present study, however, no phosphorylation of tyrosine in the receptor was found. However, significantly increased phosphorylation occurred at S574, S665, and S847 (numbering specific for isoform 2) (Figure 2). Interestingly, constitutive phosphorylation of S577 in isoform 1 of CSF2RB (corresponding to S582 in isoform 2) has been identified in acute leukemia and is thought to contribute to leukemic cell survival (68).
The present study highlights new candidates that may be involved in RPS6 and RPS6KA regulation, such as PDPK1, which is a known potential activator of RPS6KA1 (69). Other kinases that regulate protein translation are also present in Supplemental Table 5, including MKNK1 and MTOR, which increase translation initiation and elongation (Figure 4 and reviewed in (54)). It is well known that protein translation is also controlled by RNA binding proteins (54). Of interest, among these RNA-binding proteins, YBX1 (YB-1), a regulator of mRNA stability in EOS (70), was both hyper-phosphorylated and up-regulated by IL-3 (Supplemental Tables 7 and 8, and Supplemental Spreadsheets 1 and 2).
Hypo-phosphorylation by IL-3 as defined by q<0.01 and Log2 fold change >0.25 was observed on 732 sites and 483 different proteins. DAVID cluster analysis indicates that the hypo-phosphorylated proteins are mostly implicated in DNA/RNA binding, mRNA transcription, intracellular signaling, and cytoskeleton organization (Table 5). While not present as a functional annotation cluster among the hyper-phosphorylated proteins (Table 4), one of the most significant clusters among the hypo-phosphorylated proteins is linked to mRNA transcription (Table 5). Hypo-phosphorylation of some of these proteins part of mRNA transcription (not shown) may lead to increased transcription while others would lose their transcriptional activity. Therefore, increased transcription may only target a subset of genes. Of interest also is both the hypo-phosphorylation (Supplemental Spreadsheet 2) and the down-regulation (Supplemental Spreadsheet 1) of EEF2K (Figure 4), suggesting strong enhanced protein translation elongation in IL-3-activated EOS (54). Of note also, while 72 kinases were hyper-phosphorylated by IL-3 (Tables 4; p=1.6 ×10−15), only 44 were hypo-phosphorylated in this same condition (Tables 5; p=1.8×10−7).
Table 5.
Cluster analysis of the proteins hypo-phosphorylated by IL-3 (q<0.01 and log 2 fold change <-0.25)
DAVID 6.8 Functional Annotating Clustering
| Cluster | Functional annotation |
|---|---|
| DNA/transcription | Chromosome (43, p=1.6×10−11) |
| Transcription (95, p=1.8×10−7) | |
| DNA binding (114, p=5.2×10−6) | |
| Signaling | GTPase regulator activity (47, p=2.9×10−12) |
| Kinases (44; p=8.2×10−7) | |
| Serine/threonine-protein kinase (28, p=1.8×10−5) | |
| Regulation of Ras protein signal transduction (19, p=1.6×10−3) | |
| Cytoskeleton | Intracellular non-membrane-bounded organelle (146, p=6.3×10−22) |
| Cytoskeleton (74; p=1.6×10−8) | |
| Actin binding (28; p=1.2×10−4) | |
| RNA | RNA-binding (n=40, p=7.3×10−3) |
| RNA processing (n=42, p=1.1×10−5) | |
| Cell response | Response to DNA damage stimulus (33, p=1.5×10−5) |
| Cellular response to stress (40, p=1.3×10−4) | |
| Establishment and maintenance of cell polarity (9, p=5.7×10−3) | |
| Cell projection (30, p=3.9×10−2) |
In parentheses: number of proteins de-phosphorylated by IL-3 (q<0.01 and log 2 fold change > 0.25) for each functional annotation; p values (Benjamini correction)
According to our previous (15) and current study, proteomic analyses of EOS demonstrated a low level of individual variation with the exception of HLA proteins with coefficients of variation >1. Therefore, despite the limited number of EOS donors included in this study, we believe our data set to be highly representative of the general allergic population with rhinitis with or without mild asthma. We, however, acknowledge that higher number of donors would allow testing potential effects of variables such as age, sex, type of diseases, and use of medications, as described in Supplemental Table 1. As another limitation, our current study did not include combination of other cytokines with IL3 as we have previously shown that the combination of TNFα with IL3 led to synergistic increase of activin A and matrix metalloproteinase 9 mRNAs and proteins. (71, 72)
Conclusion
These proteome and phospho-proteome analyses indicate that chronic activation of EOS with IL-3 leads to widespread changes in protein translation and cytoskeletal organization and possible decreases in gene transcription and mitochondrial activity. The results are consistent with and greatly extend our recently published description of the IL-3-induced phenotype (6–8) of EOS by demonstrating persistent intracellular signaling associated with translation of a subset of proteins, including those that support EOS adhesion, spreading, degranulation, and cytolysis upon exposure to aggregated IgG. The lengthy list of proteins regulated in EOS will allow generation of new hypotheses to understand the biology of this inflammatory cell and to target deleterious functions of EOS in human diseases such as asthma, eosinophilic esophagitis, and other eosinophilic syndromes. We achieved much deeper phospho-site coverage obtained here than previously (15) which we attribute to refinement of labeling methodology and use of the latest generation mass spectrometry instrumentation. The approach and data provide a point of reference for future proteomic and phospho-proteomic investigations of EOS activated with other agonists and in vivo populations of EOS in individuals with EOS-associated diseases.
Supplementary Material
Supplemental Table 1. Characteristics of the eosinophil (EOS) donors
Supplemental Table 2. Proteins up-regulated by IL-3 and associated with protein translation as indicated in Table 2
Supplemental Table 3. Proteins upregulated by IL-3 and related to cytoskeleton organization as indicated in Table 2
Supplemental Table 4. Proteins downregulated by IL-3 and associated with mitochondrion as indicated in Table 3
Supplemental Table 5. Kinases (n=72) hyper-phosphorylatated by IL-3 (q<0.01 and log 2 fold change > 0.025), as indicated in Table 4
Supplemental Table 6. Proteins hyper-phosphorylatated by IL-3 (q<0.01 and log 2 fold change > 0.025), and part of actin cytoskeleton organization (n=29) as indicated in Table 4
Supplemental Table 7. Proteins hyper-phosphorylatated by IL-3 (q<0.01 and log 2 fold change > 0.025), and having an RNA binding activity (n=41) as indicated in Table 4
Supplemental Table 8. Proteins hyper-phosphorylatated by IL-3 (q<0.01 and log 2 fold change > 0.025), and involved in posttranscriptional regulation of gene expression, translation regulation and mTOR signaling (n=26) as indicated in Table 4
The mass spectrometry data have been deposited to the Chorus project (https://chorusproject.org) identifier 1464.
Supplemental Spreadsheet 1: 5385 proteins identified by two-dimensional liquid chromatography coupled with high-resolution mass spectrometry in eosinophils cultured for 20 h
Supplemental Spreadsheet 2: 7330 phosphorylated sites identified by two-dimensional liquid chromatography coupled with high-resolution mass spectrometry in eosinophils cultured for 20 h
Acknowledgments
The authors wish to thank the subject volunteers who participated in this study, Elizabeth McKernan, BS, and Paul Fichtinger, BS of the Eosinophil Core facility (P.I., Sameer Mathur, M.D., Ph.D.) for blood eosinophil purification, the research nurse coordinators for subject recruitment and screening, and Elizabeth (Becky) Kelly, Ph.D. for her inputs in the manuscript.
Funding: National Institutes of Health grants P01HL088594 (N.N.J.) and R01 AI125390 (D.F.M.).
Footnotes
List of supporting information:
The following supporting information is available free of charge at ACS webside http://pubs.acs.org
Author Contributions: All authors made substantial contributions to conception, design, acquisition, analysis, and interpretation of data. All authors participated in drafting and revising the manuscript with critical intellectual inputs. All authors approved the final version of the manuscript.
Conflict of Interest: The authors report no conflicts of interest.
References
- 1.Blanchard C, Rothenberg ME. Biology of the eosinophil. Adv Immunol. 2009;101:81–121. doi: 10.1016/S0065-2776(08)01003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weller PF, Spencer LA. Functions of tissue-resident eosinophils. Nature reviews Immunology. 2017;17:746–760. doi: 10.1038/nri.2017.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenberg HF, Dyer KD, Foster PS. Eosinophils: changing perspectives in health and disease. Nature reviews Immunology. 2013;13:9–22. doi: 10.1038/nri3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morita H, Moro K, Koyasu S. Innate lymphoid cells in allergic and nonallergic inflammation. J Allergy Clin Immunol. 2016;138:1253–1264. doi: 10.1016/j.jaci.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 5.Soman KV, Stafford SJ, Pazdrak K, Wu Z, Luo X, White WI, Wiktorowicz JE, Calhoun WJ, Kurosky A. Activation of human peripheral blood eosinophils by cytokines in a comparative time-course proteomic/phosphoproteomic study. Journal of proteome research. 2017;16:2663–2679. doi: 10.1021/acs.jproteome.6b00367. [DOI] [PubMed] [Google Scholar]
- 6.Esnault S, Kelly EA, Johansson MW, Liu LY, Han SH, Akhtar M, Sandbo N, Mosher DF, Denlinger LC, Mathur SK, Malter JS, Jarjour NN. Semaphorin 7A is expressed on airway eosinophils and upregulated by IL-5 family cytokines. Clin Immunol. 2014;150:90–100. doi: 10.1016/j.clim.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esnault S, Kelly EA, Shen ZJ, Johansson MW, Malter JS, Jarjour NN. IL-3 maintains activation of the p90S6K/RPS6 pathway and increases translation in human eosinophils. J Immunol. 2015;195:2529–2539. doi: 10.4049/jimmunol.1500871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esnault S, Johansson MW, Kelly EA, Koenderman L, Mosher DF, Jarjour NN. IL-3 up-regulates and activates human eosinophil CD32 and alphaMbeta2 integrin causing degranulation. Clin Exp Allergy. 2017;47:488–498. doi: 10.1111/cea.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schroeder JT, Chichester KL, Bieneman AP. Human basophils secrete IL-3: evidence of autocrine priming for phenotypic and functional responses in allergic disease. J Immunol. 2009;182:2432–2438. doi: 10.4049/jimmunol.0801782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ackerman V, Marini M, Vittori E, Bellini A, Vassali G, Mattoli S. Detection of cytokines and their cell sources in bronchial biopsy specimens from asthmatic patients. Chest. 1994;105:687–696. doi: 10.1378/chest.105.3.687. [DOI] [PubMed] [Google Scholar]
- 11.Johansson MW, Kelly EA, Busse WW, Jarjour NN, Mosher DF. Up-regulation and activation of eosinophil integrins in blood and airway after segmental lung antigen challenge. J Immunol. 2008;180:7622–7635. doi: 10.4049/jimmunol.180.11.7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patil SP, Wisnivesky JP, Busse PJ, Halm EA, Li XM. Detection of immunological biomarkers correlated with asthma control and quality of life measurements in sera from chronic asthmatic patients. Ann Allergy Asthma Immunol. 2011;106:205–213. doi: 10.1016/j.anai.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson DS, Ying S, Bentley AM, Meng Q, North J, Durham SR, Kay AB, Hamid Q. Relationships among numbers of bronchoalveolar lavage cells expressing messenger-ribonucleic-acid for cytokines, asthma symptoms, and airway methacholine responsiveness in atopic asthma. J Allergy Clin Immunol. 1993;92:397–403. doi: 10.1016/0091-6749(93)90118-y. [DOI] [PubMed] [Google Scholar]
- 14.Mosher DF, Wilkerson EM, Turton KB, Hebert AS, Coon JJ. Proteomics of eosinophil activation. Frontiers in medicine. 2017;4:159. doi: 10.3389/fmed.2017.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilkerson EM, Johansson MW, Hebert AS, Westphall MS, Mathur SK, Jarjour NN, Schwantes EA, Mosher DF, Coon JJ. The peripheral blood eosinophil proteome. Journal of proteome research. 2016;15:1524–1533. doi: 10.1021/acs.jproteome.6b00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 17.Chou MF, Schwartz D. Biological sequence motif discovery using motif-x. Current protocols in bioinformatics Chapter 13. 2011:15–24. doi: 10.1002/0471250953.bi1315s35. Unit 13. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 19.Wisniewski JR, Hein MY, Cox J, Mann M. A “proteomic ruler” for protein copy number and concentration estimation without spike-in standards. Molecular & cellular proteomics : MCP. 2014;13:3497–3506. doi: 10.1074/mcp.M113.037309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yousefi S, Gold JA, Andina N, Lee JJ, Kelly AM, Kozlowski E, Schmid I, Straumann A, Reichenbach J, Gleich GJ, Simon HU. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat Med. 2008;14:949–953. doi: 10.1038/nm.1855. [DOI] [PubMed] [Google Scholar]
- 21.Radonjic-Hoesli S, Wang X, de Graauw E, Stoeckle C, Styp-Rekowska B, Hlushchuk R, Simon D, Spaeth PJ, Yousefi S, Simon HU. Adhesion-induced eosinophil cytolysis requires the receptor-interacting protein kinase 3 (RIPK3)-mixed lineage kinase-like (MLKL) signaling pathway, which is counterregulated by autophagy. J Allergy Clin Immunol. 2017;140:1632–1642. doi: 10.1016/j.jaci.2017.01.044. [DOI] [PubMed] [Google Scholar]
- 22.Gregory B, Kirchem A, Phipps S, Gevaert P, Pridgeon C, Rankin SM, Robinson DS. Differential regulation of human eosinophil IL-3, IL-5, and GM-CSF receptor alpha-chain expression by cytokines: IL-3, IL-5, and GM-CSF down-regulate IL-5 receptor alpha expression with loss of IL-5 responsiveness, but up-regulate IL-3 receptor alpha expression. J Immunol. 2003;170:5359–5366. doi: 10.4049/jimmunol.170.11.5359. [DOI] [PubMed] [Google Scholar]
- 23.Yoshimura-Uchiyama C, Yamaguchi M, Nagase H, Matsushima K, Igarashi T, Iwata T, Yamamoto K, Hirai K. Changing expression of IL-3 and IL-5 receptors in cultured human eosinophils. Biochem Biophys Res Commun. 2003;309:26–31. doi: 10.1016/s0006-291x(03)01526-2. [DOI] [PubMed] [Google Scholar]
- 24.Kampfer SS, Odermatt A, Dahinden CA, Fux M. Late IL-3-induced phenotypic and functional alterations in human basophils require continuous IL-3 receptor signaling. Journal of Leukocyte Biology. 2017;101:227–238. doi: 10.1189/jlb.2A0715-292RR. [DOI] [PubMed] [Google Scholar]
- 25.Kelly EA, Esnault S, Liu LY, Evans MD, Johansson MW, Mathur S, Mosher DF, Denlinger LC, Jarjour NN. Mepolizumab attenuates airway eosinophil numbers, but not their functional phenotype, in asthma. Am J Respir Crit Care Med. 2017;196:1385–1395. doi: 10.1164/rccm.201611-2234OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen ZJ, Esnault S, Malter JS. The peptidyl-prolyl isomerase Pin1 regulates the stability of granulocyte-macrophage colony-stimulating factor mRNA in activated eosinophils. Nat Immunol. 2005;6:1280–1287. doi: 10.1038/ni1266. [DOI] [PubMed] [Google Scholar]
- 27.Esnault S, Malter JS. Hyaluronic acid or TNF-alpha plus fibronectin triggers granulocyte macrophage-colony-stimulating factor mRNA stabilization in eosinophils yet engages differential intracellular pathways and mRNA binding proteins. J Immunol. 2003;171:6780–6787. doi: 10.4049/jimmunol.171.12.6780. [DOI] [PubMed] [Google Scholar]
- 28.Johansson MW. Activation states of blood eosinophils in asthma. Clin Exp Allergy. 2014;44:482–498. doi: 10.1111/cea.12292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cherry WB, Yoon J, Bartemes KR, Iijima K, Kita H. A novel IL-1 family cytokine, IL-33, potently activates human eosinophils. J Allergy Clin Immunol. 2008;121:1484–1490. doi: 10.1016/j.jaci.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esnault S, Kelly EA, Schwantes EA, Liu LY, Delain LP, Hauer JA, Bochkov YA, Denlinger LC, Malter JS, Mathur SK, Jarjour NN. Identification of genes expressed by human airway eosinophils after an in vivo allergen challenge. PloS one. 2013;8:e67560. doi: 10.1371/journal.pone.0067560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnston LK, Hsu CL, Krier-Burris RA, Chhiba KD, Chien KB, McKenzie A, Berdnikovs S, Bryce PJ. IL-33 Precedes IL-5 in Regulating Eosinophil Commitment and Is Required for Eosinophil Homeostasis. J Immunol. 2016;197:3445–3453. doi: 10.4049/jimmunol.1600611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esnault S, Malter JS. Minute quantities of granulocyte-macrophage colony-stimulating factor prolong eosinophil survival. J Interferon Cytokine Res. 2001;21:117–124. doi: 10.1089/107999001750069980. [DOI] [PubMed] [Google Scholar]
- 33.Willebrand R, Voehringer D. IL-33-Induced Cytokine Secretion and Survival of Mouse Eosinophils Is Promoted by Autocrine GM-CSF. PloS one. 2016;11:e0163751. doi: 10.1371/journal.pone.0163751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnston LK, Bryce PJ. Understanding Interleukin 33 and Its Roles in Eosinophil Development. Frontiers in medicine. 2017;4:51. doi: 10.3389/fmed.2017.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ilmarinen-Salo P, Moilanen E, Kankaanranta H. Nitric oxide induces apoptosis in GM-CSF-treated eosinophils via caspase-6-dependent lamin and DNA fragmentation. Pulm Pharmacol Ther. 2010;23:365–371. doi: 10.1016/j.pupt.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 36.Sedgwick JB, Calhoun WJ, Vrtis RF, Bates ME, McAllister PK, Busse WW. Comparison of airway and blood eosinophil function after in vivo antigen challenge. J Immunol. 1992;149:3710–3718. [PubMed] [Google Scholar]
- 37.Hansel TT, Braunstein JB, Walker C, Blaser K, Bruijnzeel PL, Virchow JC, Jr, Virchow C. Sputum eosinophils from asthmatics express ICAM-1 and HLA-DR. Clin Exp Immunol. 1991;86:271–277. doi: 10.1111/j.1365-2249.1991.tb05809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weller PF, Rand TH, Barrett T, Elovic A, Wong DT, Finberg RW. Accessory cell function and human eosinophils. HLA-DR-dependent, MHC-restricted antigen-presentation and IL-1 alpha expression. J Immunol. 1993;150:2554–2562. [PubMed] [Google Scholar]
- 39.Patel AJ, Fuentebella J, Gernez Y, Nguyen T, Bass D, Berquist W, Cox K, Sibley E, Kerner J, Nadeau K. Increased HLA-DR expression on tissue eosinophils in eosinophilic esophagitis. Journal of pediatric gastroenterology and nutrition. 2010;51:290–294. doi: 10.1097/MPG.0b013e3181e083e7. [DOI] [PubMed] [Google Scholar]
- 40.Akuthota P, Wang HB, Weller PF. Eosinophils as antigen-presenting cells in allergic upper airway disease. Curr Opin Allergy Clin Immunol. 2010;10:14–19. doi: 10.1097/ACI.0b013e328334f693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kostenko EV, Olabisi OO, Sahay S, Rodriguez PL, Whitehead IP. Ccpg1, a novel scaffold protein that regulates the activity of the Rho guanine nucleotide exchange factor Dbs. Mol Cell Biol. 2006;26:8964–8975. doi: 10.1128/MCB.00670-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber DA, Sumagin R, McCall IC, Leoni G, Neumann PA, Andargachew R, Brazil JC, Medina-Contreras O, Denning TL, Nusrat A, Parkos CA. Neutrophil-derived JAML inhibits repair of intestinal epithelial injury during acute inflammation. Mucosal Immunol. 2014;7:1221–1232. doi: 10.1038/mi.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Atkinson CJ, Zhang K, Munn AL, Wiegmans A, Wei MQ. Prion protein scrapie and the normal cellular prion protein. Prion. 2016;10:63–82. doi: 10.1080/19336896.2015.1110293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ito D, Kumanogoh A. The role of Sema4A in angiogenesis, immune responses, carcinogenesis, and retinal systems. Cell adhesion & migration. 2016;10:692–699. doi: 10.1080/19336918.2016.1215785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morriswood B, Ryzhakov G, Puri C, Arden SD, Roberts R, Dendrou C, Kendrick-Jones J, Buss F. T6BP and NDP52 are myosin VI binding partners with potential roles in cytokine signalling and cell adhesion. J Cell Sci. 2007;120:2574–2585. doi: 10.1242/jcs.007005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamane J, Kubo A, Nakayama K, Yuba-Kubo A, Katsuno T, Tsukita S, Tsukita S. Functional involvement of TMF/ARA160 in Rab6-dependent retrograde membrane traffic. Exp Cell Res. 2007;313:3472–3485. doi: 10.1016/j.yexcr.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 47.Roucou X, LeBlanc AC. Cellular prion protein neuroprotective function: implications in prion diseases. J Mol Med (Berl) 2005;83:3–11. doi: 10.1007/s00109-004-0605-5. [DOI] [PubMed] [Google Scholar]
- 48.Robert C, Delva L, Balitrand N, Nahajevszky S, Masszi T, Chomienne C, Papp B. Apoptosis induction by retinoids in eosinophilic leukemia cells: implication of retinoic acid receptor-alpha signaling in all-trans-retinoic acid hypersensitivity. Cancer Res. 2006;66:6336–6344. doi: 10.1158/0008-5472.CAN-06-0078. [DOI] [PubMed] [Google Scholar]
- 49.Verstrepen L, Verhelst K, Carpentier I, Beyaert R. TAX1BP1, a ubiquitin-binding adaptor protein in innate immunity and beyond. Trends Biochem Sci. 2011;36:347–354. doi: 10.1016/j.tibs.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 50.Fuller-Pace FV. The DEAD box proteins DDX5 (p68) and DDX17 (p72): multi-tasking transcriptional regulators. Biochim Biophys Acta. 2013;1829:756–763. doi: 10.1016/j.bbagrm.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 51.Toki T, Itoh J, Arai K, Kitazawa J, Yokoyama M, Igarashi K, Yamamoto M, Ito E. Abundant expression of erythroid transcription factor P45NF-E2 mRNA in human peripheral granurocytes. Biochem Biophys Res Commun. 1996;219:760–765. doi: 10.1006/bbrc.1996.0307. [DOI] [PubMed] [Google Scholar]
- 52.Ferreira MA, Jansen R, Willemsen G, Penninx B, Bain LM, Vicente CT, Revez JA, Matheson MC, Hui J, Tung JY, Baltic S, Le Souef P, Montgomery GW, Martin NG, Robertson CF, James A, Thompson PJ, Boomsma DI, Hopper JL, Hinds DA, Werder RB, Phipps S, Australian Asthma Genetics Consortium, C Gene-based analysis of regulatory variants identifies 4 putative novel asthma risk genes related to nucleotide synthesis and signaling. J Allergy Clin Immunol. 2017;139:1148–1157. doi: 10.1016/j.jaci.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perzlmaier AF, Richter F, Seufert W. Translation initiation requires cell division cycle 123 (Cdc123) to facilitate biogenesis of the eukaryotic initiation factor 2 (eIF2) J Biol Chem. 2013;288:21537–21546. doi: 10.1074/jbc.M113.472290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Esnault S, Shen ZJ, Malter JS. Protein translation and signaling in human eosinophils. Frontiers in medicine. 2017;4:150. doi: 10.3389/fmed.2017.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee SH, Oh J, Park J, Paek KY, Rho S, Jang SK, Lee JB. Poly(A) RNA and Paip2 act as allosteric regulators of poly(A)-binding protein. Nucleic Acids Res. 2014;42:2697–2707. doi: 10.1093/nar/gkt1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waters LC, Strong SL, Ferlemann E, Oka O, Muskett FW, Veverka V, Banerjee S, Schmedt T, Henry AJ, Klempnauer KH, Carr MD. Structure of the tandem MA-3 region of Pdcd4 protein and characterization of its interactions with eIF4A and eIF4G: molecular mechanisms of a tumor suppressor. J Biol Chem. 2011;286:17270–17280. doi: 10.1074/jbc.M110.166157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dennis MD, Jefferson LS, Kimball SR. Role of p70S6K1-mediated phosphorylation of eIF4B and PDCD4 proteins in the regulation of protein synthesis. J Biol Chem. 2012;287:42890–42899. doi: 10.1074/jbc.M112.404822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dijkers PF, Medema RH, Pals C, Banerji L, Thomas NS, Lam EW, Burgering BM, Raaijmakers JA, Lammers JW, Koenderman L, Coffer PJ. Forkhead transcription factor FKHR-L1 modulates cytokine-dependent transcriptional regulation of p27(KIP1) Mol Cell Biol. 2000;20:9138–9148. doi: 10.1128/mcb.20.24.9138-9148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu W, Zhu H, Gu M, Luo Q, Ding J, Yao Y, Chen F, Wang Z. DHTKD1 is essential for mitochondrial biogenesis and function maintenance. FEBS Lett. 2013;587:3587–3592. doi: 10.1016/j.febslet.2013.08.047. [DOI] [PubMed] [Google Scholar]
- 60.Wilson BJ, Bates GJ, Nicol SM, Gregory DJ, Perkins ND, Fuller-Pace FV. The p68 and p72 DEAD box RNA helicases interact with HDAC1 and repress transcription in a promoter-specific manner. BMC molecular biology. 2004;5:11. doi: 10.1186/1471-2199-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bruscoli S, Donato V, Velardi E, Di Sante M, Migliorati G, Donato R, Riccardi C. Glucocorticoid-induced leucine zipper (GILZ) and long GILZ inhibit myogenic differentiation and mediate anti-myogenic effects of glucocorticoids. J Biol Chem. 2010;285:10385–10396. doi: 10.1074/jbc.M109.070136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Obata T, Yaffe MB, Leparc GG, Piro ET, Maegawa H, Kashiwagi A, Kikkawa R, Cantley LC. Peptide and protein library screening defines optimal substrate motifs for AKT/PKB. J Biol Chem. 2000;275:36108–36115. doi: 10.1074/jbc.M005497200. [DOI] [PubMed] [Google Scholar]
- 63.Proud CG. Signalling to translation: how signal transduction pathways control the protein synthetic machinery. Biochem J. 2007;403:217–234. doi: 10.1042/BJ20070024. [DOI] [PubMed] [Google Scholar]
- 64.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hinnebusch AG, Ivanov IP, Sonenberg N. Translational control by 5'-untranslated regions of eukaryotic mRNAs. Science. 2016;352:1413–1416. doi: 10.1126/science.aad9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cornelis S, Fache I, Van der Heyden J, Guisez Y, Tavernier J, Devos R, Fiers W, Plaetinck G. Characterization of critical residues in the cytoplasmic domain of the human interleukin-5 receptor alpha chain required for growth signal transduction. Eur J Immunol. 1995;25:1857–1864. doi: 10.1002/eji.1830250710. [DOI] [PubMed] [Google Scholar]
- 67.Callus BA, Mathey-Prevot B. Interleukin-3-induced activation of the JAK/STAT pathway is prolonged by proteasome inhibitors. Blood. 1998;91:3182–3192. [PubMed] [Google Scholar]
- 68.Powell JA, Thomas D, Barry EF, Kok CH, McClure BJ, Tsykin A, To LB, Brown A, Lewis ID, Herbert K, Goodall GJ, Speed TP, Asou N, Jacob B, Osato M, Haylock DN, Nilsson SK, D’Andrea RJ, Lopez AF, Guthridge MA. Expression profiling of a hemopoietic cell survival transcriptome implicates osteopontin as a functional prognostic factor in AML. Blood. 2009;114:4859–4870. doi: 10.1182/blood-2009-02-204818. [DOI] [PubMed] [Google Scholar]
- 69.Jensen CJ, Buch MB, Krag TO, Hemmings BA, Gammeltoft S, Frodin M. 90-kDa ribosomal S6 kinase is phosphorylated and activated by 3-phosphoinositide-dependent protein kinase-1. J Biol Chem. 1999;274:27168–27176. doi: 10.1074/jbc.274.38.27168. [DOI] [PubMed] [Google Scholar]
- 70.Capowski EE, Esnault S, Bhattacharya S, Malter JS. Y box-binding factor promotes eosinophil survival by stabilizing granulocyte-macrophage colony-stimulating factor mRNA. J Immunol. 2001;167:5970–5976. doi: 10.4049/jimmunol.167.10.5970. [DOI] [PubMed] [Google Scholar]
- 71.Kelly EA, Esnault S, Johnson SH, Liu LY, Malter JS, Burnham ME, Jarjour NN. Human eosinophil activin A synthesis and mRNA stabilization are induced by the combination of IL-3 plus TNF. Immunol Cell Biol. 2016;94:701–708. doi: 10.1038/icb.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kelly EA, Liu LY, Esnault S, Quinchia Johnson BH, Jarjour NN. Potent synergistic effect of IL-3 and TNF on matrix metalloproteinase 9 generation by human eosinophils. Cytokine. 2012;58:199–206. doi: 10.1016/j.cyto.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Characteristics of the eosinophil (EOS) donors
Supplemental Table 2. Proteins up-regulated by IL-3 and associated with protein translation as indicated in Table 2
Supplemental Table 3. Proteins upregulated by IL-3 and related to cytoskeleton organization as indicated in Table 2
Supplemental Table 4. Proteins downregulated by IL-3 and associated with mitochondrion as indicated in Table 3
Supplemental Table 5. Kinases (n=72) hyper-phosphorylatated by IL-3 (q<0.01 and log 2 fold change > 0.025), as indicated in Table 4
Supplemental Table 6. Proteins hyper-phosphorylatated by IL-3 (q<0.01 and log 2 fold change > 0.025), and part of actin cytoskeleton organization (n=29) as indicated in Table 4
Supplemental Table 7. Proteins hyper-phosphorylatated by IL-3 (q<0.01 and log 2 fold change > 0.025), and having an RNA binding activity (n=41) as indicated in Table 4
Supplemental Table 8. Proteins hyper-phosphorylatated by IL-3 (q<0.01 and log 2 fold change > 0.025), and involved in posttranscriptional regulation of gene expression, translation regulation and mTOR signaling (n=26) as indicated in Table 4
The mass spectrometry data have been deposited to the Chorus project (https://chorusproject.org) identifier 1464.
Supplemental Spreadsheet 1: 5385 proteins identified by two-dimensional liquid chromatography coupled with high-resolution mass spectrometry in eosinophils cultured for 20 h
Supplemental Spreadsheet 2: 7330 phosphorylated sites identified by two-dimensional liquid chromatography coupled with high-resolution mass spectrometry in eosinophils cultured for 20 h


