Abstract
Introduction
Small renal masses (SRMs) with evidence of clear cell renal cell carcinoma (ccRCC) are understudied. Current algorithms for the management of SRMs include surgical resection, ablation and active surveillance. We sought to identify genomic biomarkers that could potentially refine the management of ccRCC in SRMs, especially in patients being evaluated for AS.
Methods
We identified patients who had SRM’s (4 cm or less) at time of surgery, had sequencing performed on their primary tumor and had a diagnosis ccRCC. Patients were selected from three publicly available cohorts, The Cancer Genome Atlas (n = 110), University of Tokyo (n = 37), The International Cancer Genome Consortium (n = 31) and from our own institutional prospective database (n = 25). Among this cohort we analyzed mutations with at least 5% of tumors, assessing for the enrichment of mutations and progression-free survival using the composite endpoint of recurrence or death of disease. Analysis was adjusted for multiple testing. A Cox regression model was used to asses clinical variables with significant mutations.
Results
203 patients were available for analysis. Median follow-up was 43.1 months among survivors. Mutations in VHL, PBRM1, SETD2, BAP1, KDM5C, and MTOR were present in more than 5% of tumors. Twenty-three patients (11.3%) had recurrence or died of their disease. Mutations in KDM5C were associated with inferior survival from either recurrence or death from disease, adjusted P-value 0.033.
Conclusions
We identified mutations in SRMs in ccRCC that are associated with recurrence and lethality. The strongest association was seen in those with KDM5C mutations. Use of these genomic biomarkers may improve stratification of patients with SRMs and for those who may be appropriate for AS. Prospective evaluation of these markers is needed.
Keywords: Small renal masses, Genomic biomarkers, KDM5C mutations, Risk stratification
1. Introduction
The detection and diagnosis of kidney cancer has steadily risen over the last few decades in the US [1] and worldwide [2]. The rise in these rates has largely been attributed to the use of more cross-sectional imaging and the identification of incidental renal masses that are concerning for malignancy. The majority of these masses are ≤ 4 cm in diameter and are commonly classified as small renal masses (SRMs). SRMs have proven to be a heterogeneous group with regard to histology [3]. The most common histology encountered in these cases, however, is clear cell renal cell carcinoma (ccRCC), followed by a significant percentage of tumors having non-clear cell histology and benign disease [4]. Once SRMs are identified, both patients and clinicians are faced with a variety of treatment decisions.
Several physician organizations have published clinical guidelines on the management of renal masses including SRMs. The American Urological Association, European Association of Urology, and National Comprehensive Cancer Network have all published guidelines to assist physicians with clinical decision-making [5,6]. The algorithm for the clinical management of localized SRMs includes surgical resection, ablation, and active surveillance (AS). The majority of patients who elect treatment, either surgery or ablation, have favorable long-term outcomes indicated by recurrence and cancer-specific survival [7]. Nonetheless, some of these patients will develop recurrence and even metastatic disease and eventually die from their disease [8].
The tumor biology of patients with SRMs destined for poor clinical outcomes at the time of diagnosis is poorly understood. Many factors believed to be associated with inferior outcomes used in clinical nomograms rely on time-dependent factors such as tumor growth and changes in radiographic appearance [9]. Changes in these factors are often difficult to predict and require long term follow up and frequent imaging. The use of preoperative biopsy can provide important information for clinical decision making but still fails to stratify long-term risk among patients within the most commonly diagnosed histology, ccRCC [10].
The use of biomarkers to assist in the management and treatment of patients with ccRCC is currently being explored by many groups. However, the evaluation of these markers in patients with SRMs has been underreported to date in the current literature. Our study aimed to better characterize and analyze the use of somatic mutations commonly encountered in ccRCC in SRMs to identify possible biomarkers in this specific cohort of patients. These biomarkers could serve to give focus to future prospective studies and to augment the management of patients with SRMs, especially those being considered for AS.
2. Methods
Our cohort consisted of patients with available genomic sequencing data from their primary tumors who had ≤ 4 cm masses and had elected to undergo surgical resection with a diagnosis of ccRCC. Patients who met these criteria were identified from the prospective database at our institution (Memorial Sloan Kettering Cancer Center, MSKCC; n = 25) and from three publicly available cohorts: The Cancer Genome Atlas (n = 110) [11], University of Tokyo (n = 37) [12], and The International Cancer Genome Consortium (n = 31) [13]. Corresponding clinical data was also required for inclusion in our study including recurrence and survival outcomes. We excluded patients with multiple kidney tumors (n = 1), a history of kidney cancer (n = 1), those with known syndromic kidney cancer diseases (i.e., Von-Hippel Lindau; n = 2), those with suspected local recurrence (n = 2), or those who did not have sufficient clinical data (n = 6).
Sequencing of patients’ tumors was done using whole genome, whole exome, or targeted deep sequencing methods. A single tumor sample from each tumor, originating from the pathologic specimen at the time or surgical resection, was used for sequencing. The details of tumor sample processing and the sequencing pipeline employed from patients’ data gathered from the public databases may be found in their original publications. Details of the MSKCC genomic pipeline have also been previously published [14]. Tumor samples from patients from our institution were analyzed using MSK-IMPACT (Integrated Mutation Profiling of Actionable Cancer Targets), a hybridization-based exon-capture assay of select introns and commonly altered oncogenes and tumor suppressor genes. The panel of targeted genes for this assay is listed in the Supplemental Materials (Supplemental List 1).
A panel of 28 genes was chosen for initial analysis based on the clinical relevance of significantly mutated genes across all cohorts in ccRCC as identified in previous studies [12,13,15]. For adequate statistical power, we restricted our survival analysis to the subset of genes that were mutated in at least 5% of the entire cohort. We felt that a 5% mutation occurrence threshold in these SRMs would offer a reasonable and practical limit that could facilitate implementation of our results in potential a clinical study.
2.1 Statistical analysis
We first sought to determine differences in the frequency of mutations in patients experiencing recurrence or death from their disease compared to patients with no such events. We constructed (for each gene) 2×2 contingency tables describing the number of patients with or without a non-silent mutation in the gene of interest across the two subsets of patients. The first subset of patients was those who either had recurrence of their disease during follow-up or who died as a result of their kidney cancer. The second subset was those patients who were alive and not found to have disease recurrence during follow-up. Fisher’s exact test was applied to these contingency tables, and the resulting P-values were adjusted for multiple hypotheses testing using the Benjamini-Hochberg procedure.
Next, we conducted survival analysis using recurrence of disease or death from kidney cancer as our primary endpoints. Follow-up time was calculated from date of surgical resection. Recurrence was determined according to previously published methods among the patients from the public cohort. Similar to those published methods, among MSKCC patients, date of recurrence was defined as date of pathologic confirmation of diagnosis—i.e., biopsy. For patients with no pathology-confirmed recurrence, we used the date of the radiologic exam used to diagnose recurrence. Kaplan-Meier curves were generated using the R statistical package version 3.2.2 (The R Foundation for Statistical Computing, Vienna, Austria). Statistical significance of differences in survival time between mutated and non-mutated (wild-type) tumors were assessed using the log-rank test.
3. Results
We had 203 patients included in our final cohort for analysis. The demographics and clinical characteristics for these patients can be found in Table 1. The frequency of mutations in all genes from our selected gene panel can be seen in Supplementary Table 1.
Table 1.
Demographics and baseline clinical characteristics
| Overall (n=203) | No Recurrence (n=180) | Recurrence (n=23) | P-value | |
|---|---|---|---|---|
| Median age at diagnosis (IQR) | 60 (52–71) | 60 (52–72) | 59 (52.5–63.5) | 0.42 |
| No. gender (%) | ||||
| Male | 132 (65) | 115 (63.9) | 17 (73.9) | 0.49 |
| Female | 71 (35) | 65 (36.1) | 6 (26.1) | |
| No. race (%) | ||||
| White | 160 (78.8) | 143 (79.4) | 17 (73.9) | 0.67 |
| Black | 3 (1.5) | 3 (1.7) | 0 (0) | |
| Asian | 2 (1) | 2 (1.1) | 0 (0) | |
| Unavailable | 38 (18.7) | 32 (17.8) | 6 (26.1) | |
| No. primary tumor laterality (%) | ||||
| Right | 111 (54.7) | 100 (55.6) | 11 (47.8) | 0.13 |
| Left | 83 (40.9) | 74 (41.1) | 9 (39.1) | |
| Unavailable | 9 (4.4) | 6 (3.3) | 3 (13) | |
| No. primary tumor grade (%) | ||||
| G1 | 12 (5.9) | 12 (6.7) | 0 (0) | 0.02 |
| G2 | 131 (64.5) | 121 (67.2) | 10 (43.5) | |
| G3 | 55 (27.1) | 43 (23.9) | 12 (52.2) | |
| G4 | 5 (2.5) | 4 (2.2) | 1 (4.3) | |
| No. T stage (%) | ||||
| T1 | 179 (88.2) | 162 (90) | 17 (73.9) | 0.05 |
| T3a | 10 (4.9) | 8 (4.4) | 2 (8.7) | |
| T3b | 6 (3) | 5 (2.8) | 1 (4.3) | |
| Unavailable | 8 (3.9) | 5 (2.8) | 3 (13) | |
| No. N stage (%) | ||||
| N0 | 82 (40.4) | 75 (41.7) | 7 (30.4) | 0.02 |
| N1 | 1 (0.5) | 1 (0.6) | 0 (0) | |
| N2 | 1 (0.5) | 0 (0) | 1 (4.3) | |
| NX | 111 (54.7) | 99 (55) | 12 (52.2) | |
| Unavailable | 8 (3.9) | 5 (2.8) | 3 (13) | |
| No. M stage (%) | 0.0005 | |||
| M0 | 150 (73.9) | 142 (78.9) | 8 (34.8) | |
| M1 | 11 (5.4) | 3 (1.7) | 8 (34.8) | |
| MX | 34 (16.8) | 30 (16.7) | 4 (17.4) | |
| Unavailable | 8 (3.9) | 5 (2.8) | 3 (13) | |
| No. AJCC stage (%) | 0.0005 | |||
| Stage I | 177 (87.2) | 164 (91.1) | 13 (56.5) | |
| Stage III | 13 (6.4) | 13 (7.2) | 0 (0) | |
| Stage IV | 13 (6.4) | 3 (1.7) | 10 (43.5) |
A total 23 patients (11.3%) had recurrence or died of their disease at the time of our analysis. Among the patients who presented with stage IV disease (13/203, 6.4 %), more than three-quarters (10/13, 76.9%) died of their disease during follow-up. The median follow-up for patients who were alive without recurrence was 43.1 months (IQR: 27.9–64.3 months).
Mutations in the following genes were found to be present in at least 5% of patients in our cohort: VHL (127/203, 62.6%), PBRM1 (67/203, 33.0%), SETD2 (20/203, 9.9%), BAP1 (15/203, 7.4%), MTOR (14/203, 6.9%), KDM5C (12/203, 5.9%). Fig. 1 provides a visual depiction of the frequency and co-occurrence of these mutations across the cohort.
Fig. 1.
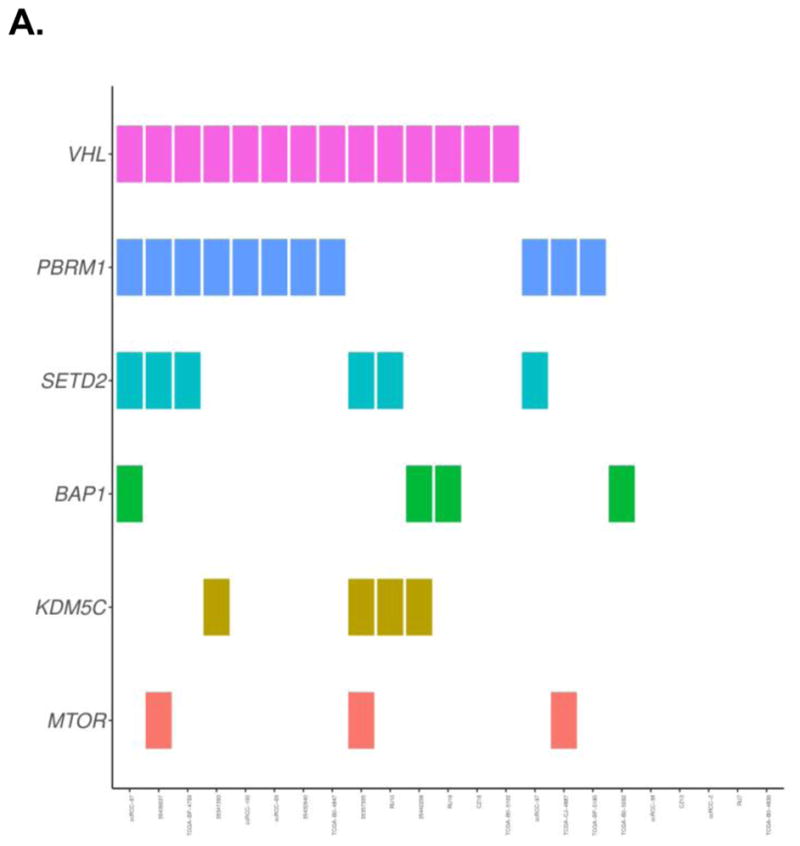
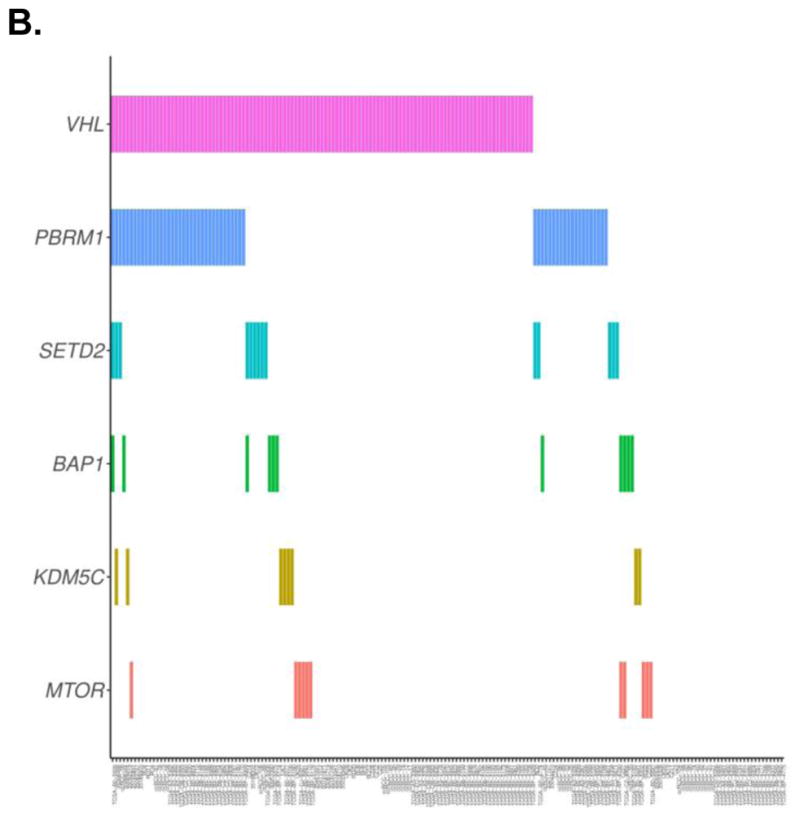
Depiction of the frequency and co-occurrence of mutations in a cohort of patients who had recurrence or died of their disease (A), and patients who were alive without recurrence (B)
We compared the frequency of the 6 mutations that occurred in at least 5% of the cohort in those patients who either had recurrence or death from their disease to those who were alive and free of recurrence (Table 2). On unadjusted analysis mutations in SETD2 (p = 0.014) and KDM5C (p = 0.033) were enriched in patients with recurrence or death of disease. On adjusted analysis these mutations are no longer found to remain significant; SETD2 adjusted P =0.087 and KDM5C adjusted P =0.101.
Table 2.
Enrichment of common mutations (≥5% frequency among cohort) comparing patients with recurrence or death of disease to those who were alive and free of recurrence and survival analysis of patients with these mutations.
| Enrichment of gene mutation in patients tumors with recurrence or death | Survival Analysis Comparing patients with the mutation to wild-type | |||
|---|---|---|---|---|
|
| ||||
| Mutation | Fischer Exact Unadjusted | Adjusted P-value | Log-Rank Unadjusted | Adjusted P-Value |
| VHL | 1 | 1 | 0.676 | 0.676 |
| PBRM1 | 0.155 | 0.233 | 0.302 | 0.422 |
| SETD2 | 0.014 | 0.087 | 0.023 | 0.071 |
| BAP1 | 0.073 | 0.146 | 0.050 | 0.100 |
| KDM5C | 0.033 | 0.101 | 0.005 | 0.033 |
| MTOR | 0.200 | 0.241 | 0.351 | 0.422 |
Survival analysis from recurrence or death of disease was conducted for each of the 6 genes found to be frequently mutated in our cohort. In this analysis (Fig. 2), KDMC5 mutations were strongly associated with either recurrence or death of disease (p = 0.005; adjusted P = 0.033). Patients with SETD2 (p = 0.023) and BAP1 (p = 0.050) mutations showed significance in our unadjusted survival analysis but became insignificant after adjustment for multiple testing; SETD2 adjusted P =0.071 and BAP1 adjusted P = 0.100.
Fig. 2.
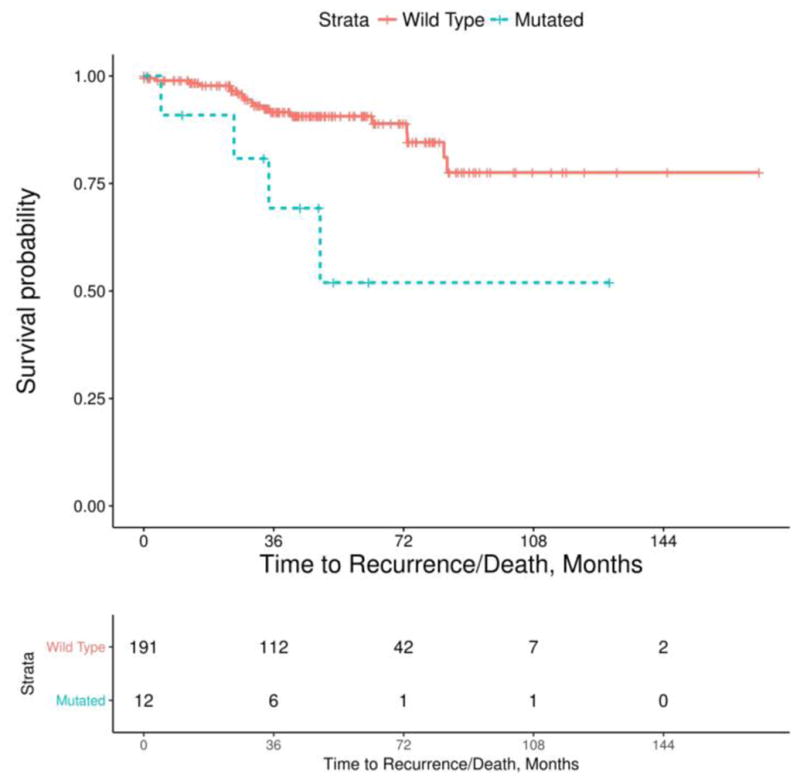
Kaplan-Meier survival plot comparing risk of recurrence or death of disease between patients with wild type and KDM5C mutations in their primary tumor
4. Discussion
Our study identified recurrently mutated genes in ccRCC that may be associated with clinically aggressive SRMs. The strongest relation (adjusted P = 0.033) was seen between KDM5C mutations and the risk of recurrence or death from disease. Mutations in other genes like SETD2 and BAP1 which have been associated with inferior clinical outcomes in other studies were not found to be statistically significant in our analysis after adjustment.
The gene KDM5C is located on the X-chromosome, therefore males only carry a single-allele making them susceptible to complete loss of the gene. The protein product of KDM5C plays a role in histone modification, and has been associated with antagonization of hypoxia-inducible factors central to the pathogenesis of ccRCC [16]. Despite having only 12 patients in our cohort with this mutation, it was persistently associated with an increased risk of recurrence or death from disease in our analysis [17,18].
While most patients who present with SRMs will have excellent clinical outcomes either with resection or possible active surveillance; there is a small but significant number or patients who with present with metastatic disease or will have disease recurrence and die from their kidney cancer [9,19]. Management decisions for most patients with SRMs are made based on estimations of malignant potential and heavily influenced by the treating surgeon’s discretion and patient preference. The clinical management of SRMs is a complex task, one that is often underestimated by both the clinician and the investigator.
Several proposed predictive nomograms for recurrence and cancer-specific survival have been published in the literature. Most of these nomograms rely solely on clinical parameters and originated to evaluate patients across the spectrum of tumor size and stage [20,21]. The quest to identify biomarkers in ccRCC has led to the evaluation of several known genomic aberrations present in this disease.
Several studies have tried to clarify the carcinogenic and clinical impacts of recurrent somatic mutations found in ccRCC [22,23]. A disproportionate number of these investigations have been tailored to those patients with more advanced or metastatic disease [23,24]. Mutations in BAP1, SETD2, and TP53 have been associated with inferior clinical results in those patients who undergo primary treatment with surgical resection [25]. In our study patients with BAP1 and SETD2 mutations appeared to have a trend for an increased risk of recurrence or death with survival analysis, however, this did not meet statistical significance (SETD2 p = 0.023, adjusted P =0.071; BAP1 p = 0.050, adjusted P = 0.100), possibly due to our small number of patients with this mutation (SETD2 = 20, 9.9%; BAP1 = 15, 7.3%). Many of these studies included patients irrespective of the tumor’s size or clinical stage [22,25]. The number of patients with SRMs included in these studies is small and limits the extrapolation of these results to our clinically distinct cohort of patients.
Identification and development of a useful and practical biomarker in ccRCC face several challenges. One of the biggest hurdles is the well-known presence of intratumoral heterogeneity (ITH) present in ccRCC tumors [26]. Our group has previously established that ITH is present even in relatively small localized tumors [27]. Even in the face of known ITH, tradeoffs between more detailed genomic data must be balanced with associated cost and feasibility if this technology is to be brought in to the clinical setting. The practicality of performing next-generation sequencing on multiple distinct tumor samples for each patient in the clinical setting is likely unrealistic, especially in SRMs. One potential solution is the use of immunohistochemistry in favor of targeted sequencing to identify these genomic alterations [23,28]. Another possible answer may be the incorporation of radiographic characteristics with genomic phenotypes found in the tumor, known as radiomics [29].
In addition to ITH, our study is also limited due to its retrospective nature, relatively small number of patients and events in our analysis and the use of multiple sequencing platforms.
We believe the recognition of KDM5C mutations and their association with inferior clinical outcomes in patients with ccRCC SRMs has the potential to impact possible future clinical studies of clinically aggressive SRMs. While relatively common among patients diagnosed with renal cancer, SRMs are understudied, with little research aimed at identifying the genomic drivers of pathogenesis for this unique cohort of patients. Our study provides a foundation for future studies among these patients, especially those with ccRCC. We hope that prospective evaluation of these and other genomic markers will improve the care and management of patients with SRMs.
5. Conclusion
In our analysis, patients presenting with SRMs and mutations in KDM5C are associated with inferior recurrence free or cancer specific survival. While clinical outcomes for managing patients with SRM are typically excellent, identification of those SRM that portend an aggressive clinical course deserve further clinical and mechanistic studies.
Supplementary Material
Highlights.
In small renal masses, KDM5C mutations were linked to poor outcomes.
These results may be used to study management strategies of patients with small renal masses.
Evaluation of genomic markers could improve management of small renal masses.
Acknowledgments
Funding
National Institutes of Health/National Cancer Institute Cancer Center Support (Grant P30 CA008748), Ruth L. Kirschstein National Research Service Award T32CA082088 (BJM and MG), Clinical and Translational Science Center at Weill Cornell Medical Center UL1TR00457 (BJM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Noone A-M, Cronin KA, Altekruse SF, Howlader N, Lewis DR, Petkov VI, et al. Cancer incidence and survival trends by subtype using data from the Surveillance Epidemiology and End Results Program, 1992–2013. Cancer Epidemiology and Prevention Biomarkers. 2016 doi: 10.1158/1055-9965.EPI-16-0520. cebp. 0520.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Znaor A, Lortet-Tieulent J, Laversanne M, Jemal A, Bray F. International variations and trends in renal cell carcinoma incidence and mortality. European urology. 2015;67:519–30. doi: 10.1016/j.eururo.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Kutikov A, Fossett LK, Ramchandani P, Tomaszewski JE, Siegelman ES, Banner MP, et al. Incidence of benign pathologic findings at partial nephrectomy for solitary renal mass presumed to be renal cell carcinoma on preoperative imaging. Urology. 2006;68:737–40. doi: 10.1016/j.urology.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 4.Remzi M, Özsoy M, Klingler H-C, Susani M, Waldert M, Seitz C, et al. Are small renal tumors harmless? Analysis of histopathological features according to tumors 4 cm or less in diameter. The Journal of urology. 2006;176:896–9. doi: 10.1016/j.juro.2006.04.047. [DOI] [PubMed] [Google Scholar]
- 5.Campbell SC, Novick AC, Belldegrun A, Blute ML, Chow GK, Derweesh IH, et al. Guideline for management of the clinical T1 renal mass. The Journal of urology. 2009;182:1271–9. doi: 10.1016/j.juro.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Network NCC. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines™) Kidney Cancer. Version 2.2017. 2016 [Google Scholar]
- 7.LEE CT, Katz J, Shi W, Thaler HT, Reuter VE, Russo P. Surgical management of renal tumors 4 cm. or less in a contemporary cohort. The Journal of urology. 2000;163:730–6. [PubMed] [Google Scholar]
- 8.Thompson RH, Atwell T, Schmit G, Lohse CM, Kurup AN, Weisbrod A, et al. Comparison of partial nephrectomy and percutaneous ablation for cT1 renal masses. European urology. 2015;67:252–9. doi: 10.1016/j.eururo.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 9.Smaldone MC, Kutikov A, Egleston BL, Canter DJ, Viterbo R, Chen DY, et al. Small renal masses progressing to metastases under active surveillance. Cancer. 2012;118:997–1006. doi: 10.1002/cncr.26369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halverson SJ, Kunju LP, Bhalla R, Gadzinski AJ, Alderman M, Miller DC, et al. Accuracy of determining small renal mass management with risk stratified biopsies: confirmation by final pathology. The Journal of urology. 2013;189:441–6. doi: 10.1016/j.juro.2012.09.032. [DOI] [PubMed] [Google Scholar]
- 11.Network CGAR. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–9. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nature genetics. 2013;45:860–7. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 13.Scelo G, Riazalhosseini Y, Greger L, Letourneau L, Gonzàlez-Porta M, Wozniak MB, et al. Variation in genomic landscape of clear cell renal cell carcinoma across Europe. Nature communications. 2014:5. doi: 10.1038/ncomms6135. [DOI] [PubMed] [Google Scholar]
- 14.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. The Journal of Molecular Diagnostics. 2015;17:251–64. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hakimi AA, Chen Y-B, Wren J, Gonen M, Abdel-Wahab O, Heguy A, et al. Clinical and pathologic impact of select chromatin-modulating tumor suppressors in clear cell renal cell carcinoma. European urology. 2013;63:848–54. doi: 10.1016/j.eururo.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niu X, Zhang T, Liao L, Zhou L, Lindner DJ, Zhou M, et al. The von Hippel–Lindau tumor suppressor protein regulates gene expression and tumor growth through histone demethylase JARID1C. Oncogene. 2012;31:776–86. doi: 10.1038/onc.2011.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hakimi AA, Ostrovnaya I, Reva B, Schultz N, Chen Y-B, Gonen M, et al. Adverse outcomes in clear cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: a report by MSKCC and the KIRC TCGA research network. Clinical Cancer Research. 2013;19:3259–67. doi: 10.1158/1078-0432.CCR-12-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu W, Fu Q, An H, Chang Y, Zhang W, Zhu Y, et al. Decreased expression of SETD2 predicts unfavorable prognosis in patients with nonmetastatic clear-cell renal cell carcinoma. Medicine. 2015:94. doi: 10.1097/MD.0000000000002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klatte T, Patard J-J, De Martino M, Bensalah K, Verhoest G, De La Taille A, et al. Tumor size does not predict risk of metastatic disease or prognosis of small renal cell carcinomas. The Journal of urology. 2008;179:1719–26. doi: 10.1016/j.juro.2008.01.018. [DOI] [PubMed] [Google Scholar]
- 20.Russo GI, Di Rosa A, Favilla V, Fragalà E, Castelli T, Privitera S, et al. Accuracy capabilities comparisons between Karakiewicz, Kattan and Cindolo nomograms in predicting outcomes for renal cancer carcinoma: A systematic review and meta-analysis. Canadian Urological Association Journal. 2015;9:E359. doi: 10.5489/cuaj.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lane BR, Babineau D, Kattan MW, Novick AC, Gill IS, Zhou M, et al. A preoperative prognostic nomogram for solid enhancing renal tumors 7 cm or less amenable to partial nephrectomy. The Journal of urology. 2007;178:429–34. doi: 10.1016/j.juro.2007.03.106. [DOI] [PubMed] [Google Scholar]
- 22.Kapur P, Peña-Llopis S, Christie A, Zhrebker L, Pavía-Jiménez A, Rathmell WK, et al. Effects on survival of BAP1 and PBRM1 mutations in sporadic clear-cell renal-cell carcinoma: a retrospective analysis with independent validation. The lancet oncology. 2013;14:159–67. doi: 10.1016/S1470-2045(12)70584-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Liu L, Qu Y, Xi W, Xia Y, Bai Q, et al. Prognostic value of SETD2 expression in patients with metastatic renal cell carcinoma treated with tyrosine kinase inhibitors. The Journal of Urology. 2016 doi: 10.1016/j.juro.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh JJ, Chen D, Wang PI, Marker M, Redzematovic A, Chen Y-B, et al. Genomic Biomarkers of a Randomized Trial Comparing First-line Everolimus and Sunitinib in Patients with Metastatic Renal Cell Carcinoma. European Urology. 2016 doi: 10.1016/j.eururo.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manley BJ, Zabor EC, Casuscelli J, Tennenbaum DM, Redzematovic A, Becerra MF, et al. Integration of Recurrent Somatic Mutations with Clinical Outcomes: A Pooled Analysis of 1049 Patients with Clear Cell Renal Cell Carcinoma. European Urology Focus. 2016 doi: 10.1016/j.euf.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. New England journal of medicine. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sankin A, Hakimi AA, Mikkilineni N, Ostrovnaya I, Silk MT, Liang Y, et al. The impact of genetic heterogeneity on biomarker development in kidney cancer assessed by multiregional sampling. Cancer medicine. 2014;3:1485–92. doi: 10.1002/cam4.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang W, Dulaimi E, Devarajan K, Parsons T, Wang Q, Liao L, et al. Immunohistochemistry Successfully Uncovers Intratumoral Heterogeneity and Widespread Co-Losses of Chromatin Regulators in Clear Cell Renal Cell Carcinoma. PloS one. 2016;11:e0164554. doi: 10.1371/journal.pone.0164554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jamshidi N, Jonasch E, Zapala M, Korn RL, Aganovic L, Zhao H, et al. The radiogenomic risk score: construction of a prognostic quantitative, noninvasive image-based molecular assay for renal cell carcinoma. Radiology. 2015;277:114–23. doi: 10.1148/radiol.2015150800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


