Summary
Background
Medulloblastoma is associated with rare hereditary cancer predisposition syndromes; however, consensus medulloblastoma predisposition genes have not been defined and screening guidelines for genetic counselling and testing for paediatric patients are not available. We aimed to assess and define these genes to provide evidence for future screening guidelines.
Methods
In this international, multicentre study, we analysed patients with medulloblastoma from retrospective cohorts (International Cancer Genome Consortium [ICGC] PedBrain, Medulloblastoma Advanced Genomics International Consortium [MAGIC], and the CEFALO series) and from prospective cohorts from four clinical studies (SJMB03, SJMB12, SJYC07, and I-HIT-MED). Whole-genome sequences and exome sequences from blood and tumour samples were analysed for rare damaging germline mutations in cancer predisposition genes. DNA methylation profiling was done to determine consensus molecular subgroups: WNT (MBWNT), SHH (MBSHH), group 3 (MBGroup3), and group 4 (MBGroup4). Medulloblastoma predisposition genes were predicted on the basis of rare variant burden tests against controls without a cancer diagnosis from the Exome Aggregation Consortium (ExAC). Previously defined somatic mutational signatures were used to further classify medulloblastoma genomes into two groups, a clock-like group (signatures 1 and 5) and a homologous recombination repair deficiency-like group (signatures 3 and 8), and chromothripsis was investigated using previously established criteria. Progression-free survival and overall survival were modelled for patients with a genetic predisposition to medulloblastoma.
Findings
We included a total of 1022 patients with medulloblastoma from the retrospective cohorts (n=673) and the four prospective studies (n=349), from whom blood samples (n=1022) and tumour samples (n=800) were analysed for germline mutations in 110 cancer predisposition genes. In our rare variant burden analysis, we compared these against 53 105 sequenced controls from ExAC and identified APC, BRCA2, PALB2, PTCH1, SUFU, and TP53 as consensus medulloblastoma predisposition genes according to our rare variant burden analysis and estimated that germline mutations accounted for 6% of medulloblastoma diagnoses in the retrospective cohort. The prevalence of genetic predispositions differed between molecular subgroups in the retrospective cohort and was highest for patients in the MBSHH subgroup (20% in the retrospective cohort). These estimates were replicated in the prospective clinical cohort (germline mutations accounted for 5% of medulloblastoma diagnoses, with the highest prevalence [14%] in the MBSHH subgroup). Patients with germline APC mutations developed MBWNT and accounted for most (five [71%] of seven) cases of MBWNT that had no somatic CTNNB1 exon 3 mutations. Patients with germline mutations in SUFU and PTCH1 mostly developed infant MBSHH. Germline TP53 mutations presented only in childhood patients in the MBSHH subgroup and explained more than half (eight [57%] of 14) of all chromothripsis events in this subgroup. Germline mutations in PALB2 and BRCA2 were observed across the MBSHH, MBGroup3, and MBGroup4 molecular subgroups and were associated with mutational signatures typical of homologous recombination repair deficiency. In patients with a genetic predisposition to medulloblastoma, 5-year progression-free survival was 52% (95% CI 40–69) and 5-year overall survival was 65% (95% CI 52–81); these survival estimates differed significantly across patients with germline mutations in different medulloblastoma predisposition genes.
Interpretation
Genetic counselling and testing should be used as a standard-of-care procedure in patients with MBWNT and MBSHH because these patients have the highest prevalence of damaging germline mutations in known cancer predisposition genes. We propose criteria for routine genetic screening for patients with medulloblastoma based on clinical and molecular tumour characteristics.
Funding
German Cancer Aid; German Federal Ministry of Education and Research; German Childhood Cancer Foundation (Deutsche Kinderkrebsstiftung); European Research Council; National Institutes of Health; Canadian Institutes for Health Research; German Cancer Research Center; St Jude Comprehensive Cancer Center; American Lebanese Syrian Associated Charities; Swiss National Science Foundation; European Molecular Biology Organization; Cancer Research UK; Hertie Foundation; Alexander and Margaret Stewart Trust; V Foundation for Cancer Research; Sontag Foundation; Musicians Against Childhood Cancer; BC Cancer Foundation; Swedish Council for Health, Working Life and Welfare; Swedish Research Council; Swedish Cancer Society; the Swedish Radiation Protection Authority; Danish Strategic Research Council; Swiss Federal Office of Public Health; Swiss Research Foundation on Mobile Communication; Masaryk University; Ministry of Health of the Czech Republic; Research Council of Norway; Genome Canada; Genome BC; Terry Fox Research Institute; Ontario Institute for Cancer Research; Pediatric Oncology Group of Ontario; The Family of Kathleen Lorette and the Clark H Smith Brain Tumour Centre; Montreal Children's Hospital Foundation; The Hospital for Sick Children: Sonia and Arthur Labatt Brain Tumour Research Centre, Chief of Research Fund, Cancer Genetics Program, Garron Family Cancer Centre, MDT's Garron Family Endowment; BC Childhood Cancer Parents Association; Cure Search Foundation; Pediatric Brain Tumor Foundation; Brainchild; and the Government of Ontario.
Introduction
Medulloblastoma is an embryonal brain tumour of the cerebellum, with an annual age-adjusted incidence ranging from 2·0 cases per 1 000 000 to 5·8 cases per 1 000 000 worldwide.1 The cause of medulloblastoma is largely unclear and most cases are presumed to arise sporadically.2 Medulloblastoma has been observed in conjunction with several rare disorders, including Gorlin syndrome (associated with mutations in SUFU and PTCH1),3, 4 Li-Fraumeni syndrome (TP53),5 APC-associated polyposis conditions,6 and Fanconi anaemia (BRCA2).7 However, the full spectrum and prevalence of genetic predisposition involving damaging germline mutations in paediatric-onset and adult-onset cancer predisposition genes8 remains unknown. Following the recognition9 of four consensus molecular subgroups (WNT [MBWNT], SHH [MBSHH], group 3 [MBGroup3], and group 4 [MBGroup4]) with distinct demographics and clinical outcomes,10 patients with germline TP53, SUFU, and PTCH1 mutations have been reported to be predisposed to develop MBSHH.11, 12 Although several case studies have reported medulloblastomas arising in patients with Fanconi anaemia, whether these predispose to specific medulloblastoma subgroups, and whether heterozygous germline mutations13 also confer an increased risk of developing medulloblastoma, remain unknown. Furthermore, whether and to what extent the presence of predisposing germline mutations affects clinical outcome is unclear. A comprehensive study14 of damaging germline mutations in cancer predisposition genes across a diverse set of paediatric cancers identified variants likely to predispose to the disease in three (8%) of 37 patients with medulloblastoma—a cohort size that is too small to allow these issues to be thoroughly addressed. Owing to these uncertainties, and since knowledge about germline mutations can be useful for clinical practice,8 assessment of larger patient series is crucial for the identification of consensus medulloblastoma predisposition genes to estimate the contribution of genetic predisposition towards consensus molecular subgroups, and to investigate whether affected patients have distinct clinical outcomes. A comprehensive understanding of molecular alterations in affected patients would further help in the development of clinical screening guidelines for genetic risk assessment in paediatric patients.
In this study, we provide a comprehensive description of genetic risk factors across 1022 patients with medulloblastoma based on a retrospective discovery cohort and validation in a prospective clinical cohort. We validated a set of six consensus medulloblastoma predisposition genes, and report associations between germline mutations and patient characteristics, molecular subgroups, somatic mutation landscapes, and clinical outcomes.
Methods
Study design and participants
This series was based on 1022 patients with medulloblastoma (491 published12, 15, 16, 17, 18 and 531 as yet unpublished cases), and comprised whole-genome and whole-exome sequencing data for the whole set of donor samples available to use from the International Cancer Genome Consortium (ICGC), the Medulloblastoma Advanced Genomics International Consortium (MAGIC), and the CEFALO series,19 and from four prospective clinical studies (SJMB03 [NCT00085202], SJMB12 [NCT01878617], SJYC07 [NCT00602667], and I-HIT-MED [NCT02417324]). We obtained biological samples from all patients, who all provided written informed consent, in accordance with institutional review board guidelines.
Research in context.
Evidence before this study
We searched PubMed for journal articles and case reports from Aug 1, 1925, up to Dec 31, 2017, with the terms “medulloblastoma” and “germline OR familial OR syndrome OR heritable OR susceptibility”, and the names of 110 genes in which germline mutations confer increased risk of paediatric and adult cancers. About 100 studies identified damaging germline mutations in nearly 20 genes, primarily in APC, BRCA2, PTCH1, SUFU, and TP53. Several studies were based on case reports and small-scale series. Additionally, which patients would benefit from genetic counselling in the context of molecular subgrouping—nowadays routinely applied in clinical trials and implemented into the revised WHO classification of CNS tumours in 2016—and whether genetic predisposition can be recognised based on familial patterns were unclear. Additionally, several paediatric cancer centres have implemented routine multigene panel analysis and whole-exome analysis of medulloblastomas; however, these centres encounter several germline mutations with uncertain clinical significance. No study has previously aimed to define a consensus set of medulloblastoma predisposition genes or has investigated under which circumstances genetic counselling and testing should be offered to patients with medulloblastoma.
Added value of this study
This study is based on an international cohort of 1022 patients with medulloblastoma, and includes detailed information about medulloblastoma subgroups (WNT [MBWNT], SHH [MBSHH], group 3 [MBGroup3], and group 4 [MBGroup4]), somatic mutation landscapes, and clinical outcomes. We defined and characterised six consensus, clinically relevant medulloblastoma predisposition genes on the basis of rare variant burden analysis (APC, BRCA2, PALB2, PTCH1, SUFU, and TP53). Half of all patients with damaging germline mutations were not recognised based on familial history of cancer; however, these patients exhibited distinct phenotypes with respect to age at diagnosis, molecular subgroups, somatic mutation patterns, and clinical outcomes. Paediatric patients with heterozygous germline PALB2 mutations exhibited mutational signatures consistent with homologous recombination repair deficiency and all patients with medulloblastoma showed no signs characteristic of Fanconi anaemia. About one in five patients in the MBSHH subgroup developed medulloblastoma in the context of a genetic predisposition, underscoring the need for a dedicated genetic screening programme and surveillance programme in this patient group. Patients in the MBWNT subgroup had an intermediate risk of damaging germline mutations, and clear genotype–phenotype associations that guide the ordering of genetic tests. Damaging germline mutations in known cancer predisposition genes were rare in paediatric patients in the MBGroup3 and MBGroup4 subgroups, which indicates conservative ordering of genetic tests in these groups. We propose clinical guidelines for genetic screening in medulloblastoma based on routinely acquired clinical and molecular tumour phenotypes.
Implications of all the available evidence
A significant prevalence of clinically important germline mutations in two of four molecular subgroups reveals that genetic counselling and testing should be established as a standard-of-care procedure in the management of patients with medulloblastoma. The proposed testing algorithm has been implemented in the International Society of Paediatric Oncology clinical trial PNET 5 MB (NCT02066220), which is currently recruiting patients.
Patient accrual for our retrospective cohort was done from 2003 until 2016. Patient accrual for the SJMB03 trial was done from Sept 9, 2003, until March 7, 2013 and for the SJYC07 trial was done from Dec 17, 2007, until March 31, 2017. The SJMB12 and I-HIT-MED trials are still accruing. The age limit for eligibility in the prospective studies was 5 years or younger (SJYC07), 3–21 years (SJMB03), and 3–39 years (SJMB12). Patients of all ages were eligible for the I-HIT-MED study. The retrospective cohort had no age limit for inclusion. Patients in the I-HIT-MED study were excluded if they were registered in another clinical trial for the same diagnosis (relapse is defined as a second diagnosis) or if a valid ethical committee approval was lacking.
Procedures
Whole-genome and whole-exome sequencing data for germline and tumour samples were generated at the German Cancer Research Centre (ICGC PedBrain; Heidelberg, Germany), Canada's Michael Smith Genome Sciences Centre (MAGIC; Vancouver, BC, Canada), Broad Institute of Massachusetts Institute of Technology and Harvard (Cambridge, MA, USA), and St Jude Children's Research Hospital (Pediatric Cancer Genome Project; Memphis, TN, USA).
To ensure standardisation of genomic data processing, we used the same uniform computational analysis workflows for all germline and tumour samples. We first unaligned samples that were provided to us as genomic alignment format (BAM) files. Subsequently, we aligned all samples using the same set of algorithms and the same human reference genome build (GRCH37, also known as hg19), based on data-processing standards defined by the Pan-Cancer Analysis of Whole Genomes (PCAWG) project.
For germline variant discovery, we used freebayes (version 1.1.0) in single-sample and paired-sample calling modes for discovery of single-nucleotide variants (SNVs), multiple nucleotide variants (MNVs), and insertions or deletions (indels) smaller than 50 base-pairs (bp; used parameters were –min-repeat-entropy 1, –report-genotype-likelihood-max, –alternate-fraction 0·2, and –no-partial-observations). Raw variant predictions were further filtered for quality (QUAL >20, QUAL/AO >2; where QUAL is −10 log10 Pr[alternative allele is wrong], and AO is alternative allele observation count), strand bias artifacts (SAF >1, SAR >1; where SAF is the number of alternate observations on the forward strand, and SAR is number of alternate observations on the reverse strand), read position artifacts (RPR >1, RPL >1; where RPR is the number of reads supporting the alternate balanced to the right [3ʹ] of the alternate allele, and RPL is the number of reads supporting the alternate balanced to the left [5ʹ] of the alternate allele), and normalised for consistent representation across patients with the variant analysis software vt (version 0.5). We used delly for germline genomic structural variant discovery with default settings for whole-genome sequencing samples and a custom read-depth-based copy-number variation (CNV)-calling pipeline for whole-exome sequencing samples. We based this custom pipeline on read quantification by bedtools in exome capture regions (MAQ >30), followed by variance stabilising transformation of count data with the vst function from the DESeq2 R package (version 1.8.2), unsupervised estimation of hidden confounders using the R package PEER (30 hidden confounders), and copy-number segmentation based on standardised residuals using circular binary segmentation (R package DNAcopy, version 1.50, default settings). Raw CNV calls were filtered further for size (to include only those >10 kb), minimum number of exons (>2), and CNV signal intensity (>4 SDs). Mosaic mutation discovery was based on freebayes and for donors with paired blood and tumour sequencing data. Any SNV, short indel, and MNV that was observed at low variant allele frequency in blood (3–15%) and high variant allele frequency in tumours (>50%) was considered a putative mosaic mutation. We restricted the analysis to reads with mapping quality of more than 30 (−10 log10 Pr[mapping position is wrong]), base quality of more than 20 (−10 log10 Pr[mapping position is wrong]), and sites with more than 30 times sequencing coverage (ie, number of reads). We made no assumptions regarding the pathogenicity of putative mosaic mutations. We excluded any variant that was present in public genetic archives and discovered in germline genomes of other patients with medulloblastoma.
For classification of damaging germline mutations, germline variants were annotated with the Ensembl Variant Effect Predictor (VEP; r81). High-impact (ie, damaging) germline mutations were defined as frameshift, stop gain, start lost, canonical splice site, exon or gene deletions, known (ClinVar; accessed Feb 16, 2017) damaging non-canonical splice site variants, and somatic mosaic mutations (defined as mutations present in a subset of normal cells). Putative damaging germline mutations were removed if the estimated minor allele frequency in at least one continental population was more than 0·1%, which we judged on the basis of 53 105 sequenced individuals that were assigned to known (control) populations and without a cancer diagnosis from the Exome Aggregation Consortium (ExAC) resource, the 1000 Genomes Project, and the National Heart, Lung, and Blood Institute GO Exome Sequencing Project. Putative gain-of-function missense variants in TP53 were further evaluated by use of information in the International Agency for Research on Cancer TP53 database and annotated as pathogenic if TP53 mutations were classified as non-functional in experimental transcriptional activity assays. Finally, all germline mutations were excluded from the analysis if annotated as (likely) benign in ClinVar. Damaging germline structural variants were defined if they overlapped at least one exon and were absent in samples from the 1000 Genomes Project. We estimated the primary population ancestry (European, African, east Asian, south Asian, and Native American) for all patients with medulloblastoma with a supervised decomposition approach20 and ancestry-informative markers that are within ExAC-defined exome capture target regions.
Identification of somatic mutations (SNVs, small indels, and copy-number alterations) was pursued in a standardised manner across all samples (matched tumour or normal genome as well as exome pairs) with the German Cancer Research Center (known as DKFZ) and European Molecular Biology Laboratory cancer genome analysis workflow of the PCAWG consortium. Somatic SNVs and indels were further stringently filtered for germline contamination using information from dbSNP and the 1000 Genomes Project. Somatic structural variant discovery was pursued in a standardised way by use of an optimised version of delly. We used a high-stringency structural variant set by additionally filtering somatic structural variants detected in at least 1% of a set of 1105 germline samples from healthy individuals belonging to phase 1 of the 1000 Genomes Project, and by requiring absence of somatic structural variants in all medulloblastoma germline samples of this study. For inference of high-stringency structural variants, we additionally required at least four supporting read pairs21 with a minimum mapping quality of 20 (−10 log10 Pr[mapping position is wrong]) and restricted valid somatic structural variant sizes from 100 bp to 500 Mb.
We quantified previously defined somatic mutational signatures22 (termed 1, 3, 5, and 8) using tumour-specific somatic point-mutation spectra and published signature probabilities. These signatures comprise combinations of somatic mutations and represent consequences of mutagenic exposures and defective DNA repair pathways. Signatures 1 and 5 (“clock-like” signatures) are associated with ageing (a clock-like accumulation of mutations) and occur in normal, non-malignant cells,22 whereas signatures 3 and 8 are associated with homologous recombination repair deficiency (HRD) and have been reported in cancer tissues. We used these mutational signatures to further classify medulloblastoma genomes into two groups, a clock-like group (signatures 1 and 5) and a HRD-like group (signatures 3 and 8). Somatic mutation spectra were quantified with the R package SomaticSignatures (version 2.6) and decomposed into contributions of known mutational signatures based on the Lawson–Hanson algorithm for non-negative least squares (nnls version 1.1, R package). The quantification error (residual sum of squares) was correlated with the total somatic mutation burden (Spearman's r=0·74; p<0·0001) and it was highest in tumours with fewer than 100 somatic SNVs. We have retained 375 whole-genome sequencing samples with more than 100 SNVs and low quantification error (<0·02) for signature analysis. Medulloblastomas were classified as homologous recombination repair deficient if most (>50%) somatic SNVs were assigned to mutational signatures 3 or 8.
To assess the burden of rare germline SNVs and indels in cancer predisposition genes, we performed case–control association testing in a subgroup-specific manner as well as across all subgroups based on a collection of 110 autosomal cancer predisposition genes8, 23 (appendix p 2). All patients with medulloblastoma in our retrospective discovery cohort were eligible for rare variant burden analysis. We used 53 105 sequenced individuals without a known cancer diagnosis from ExAC (release 0.3) as controls. Age and sex were not matched between cases and controls because of missing information from ExAC. To ensure comparability with variants found in patients with medulloblastoma, ExAC germline mutations were normalised with vt normalize, annotated with VEP (r81), and filtered for sites that passed quality controls, as defined by ExAC. Moreover, we excluded any germline mutations that were present outside ExAC-defined target regions to reduce any possible advantages derived from improved variant calling in whole-genome sequences (eg, exons not being covered in ExAC). Furthermore, we also did not include pathogenic structural variants in our rare variant burden analysis. We assumed equal effect sizes for frameshift, nonsense, splice site, and gain-of-function missense variants, as well as for variants located at any position along a gene. The total number of damaging (a) and wild-type (A) alleles were counted for each gene in cases and controls, recorded in a 2 × 2 contingency table, and evaluated for statistical independence of rows (case and control) and columns (allele a and allele A) with Fisher's exact test (to 1 df) implemented in R version 3.4 using the fisher.exact function. Unadjusted p values were recorded for each gene and adjusted for multiple testing correction with the Bonferroni method. We only considered genes for which more than four pathogenic alleles were observed in cases and controls. Allelic relative risks were estimated by the odds ratio (OR), which describes the association between medulloblastoma and damaging alleles by comparing the odds of medulloblastoma in an individual carrying a wild-type allele to the odds of medulloblastoma in an individual carrying one damaging allele. We assumed that the penetrance of monoallelic germline mutations was lower than that of biallelic germline mutations (ie, homozygous and compound heterozygous mutations).
We assessed secondary somatic gene hits in tumours on three levels: point mutations, loss-of-heterozygosity, and allele-specific gene expression. Somatic point mutations were assumed to occur in trans (ie, on a different haplotype). Loss of heterozygosity was quantified by use of genotyping germline alleles in available tumour genomes or exomes with freebayes and requiring a minimum coverage of ten reads, minimum base quality of 10 (−10 log10 Pr[base is wrong]), and minimum mapping quality of 10 (−10 log10 Pr[mapping position is wrong]). Loss of heterozygosity was inferred if the variant allele frequency was above 80% in tumour sequences. Allele-specific gene expression was quantified at heterozygous SNVs and indels and mRNA sequencing data by use of freebayes (minimum mapping quality of 10 [−10 log10 Pr(mapping position is wrong)] and minimum base quality of 10 [−10 log10 Pr(base is wrong)]) and was predicted on the basis of binomial tests, an expected ratio of 0·5, and p values lower than 0·05. When possible, multiple sites within the same gene were phased with paired-end RNA sequencing data and individual sites were merged to calculate haplotype-specific expression ratios.
We investigated chromothripsis using previously established criteria.24 These criteria distinguish chromothripsis from DNA rearrangements occurring in a stepwise fashion. First, we analysed breakpoint clustering in the entire genome based on high-confidence somatic structural variant calls and did statistical analysis for non-randomness of breakpoint distributions, under the assumption of an exponential distribution (null hypothesis). Breakpoint clustering generates highly clustered DNA breaks that might be followed by long tracts of intact DNA segments. For each sample and chromosome, a Kolmogorov–Smirnov test was used to test against the null hypothesis. Then we assessed the randomness of DNA rearrangements using a multinomial test (implemented in the R package EMT) for chromosomes with putative chromothripsis patterns. For chromosomes undergoing chromothripsis, the shattered fragments were randomly stitched together according to the original chromothripsis model. This model implies that for each DNA break, the orientation of the two joined DNA fragment ends will be random. We tested the observed distribution of rearrangement joins (tail-to-head, head-to-tail, head-to-head, and tail-to-tail) against a background model of occurrence with equal probability (ie, a probability of 0·25 for each rearrangement).
Molecular classification of medulloblastomas into consensus subgroups9 (MBWNT, MBSHH, MBGroup3, and MBGroup4) was determined by DNA methylation profiling.25
Statistical analysis
No formal sample size and power calculations were done in this study, since we made use of all germline genomes and exomes available to us. Samples with missing data were excluded from specific analysis. We used Fisher's exact and χ2 tests for associations with binary and categorical variables, and Mann-Whitney U and Kruskal-Wallis tests for associations with quantitative values. Survival analysis was based on the Kaplan-Meier estimator and log-rank tests using the R package suvival (version 2.41), and survival curves were visualised using the R package survminer (version 0.4.2). Overall survival and progression-free survival were based on definitions consistent with how they were evaluated within each respective patient cohort from each prospective trial: SJMB03, SJMB12, and SJYC07. The prospective observational patient registry I-HIT-MED had no formal endpoints and so no statistical analyses were defined. Detailed outcome measures for all four studies are available on ClinicalTrials.gov. One-sided binomial tests were used for replication analysis with the alternative hypothesis defined as a lower probability of observing germline mutations in the replication cohort.
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding authors had full access to all the data and had the final responsibility to submit for publication.
Results
We analysed germline genome-sequencing and exome-sequencing data (appendix pp 8–9) from 1022 patients with medulloblastoma, of whom 673 were in the retrospective discovery cohort and 349 were in the prospective clinical study cohort (figure 1A, appendix p 3). Patient-matched tumour genomes were sequenced for 800 samples from both cohorts (n=451 retrospective and n=349 prospective) from whole-genome sequencing (n=397) and whole-exome sequencing (n=403; figure 1A). Molecular classification of medulloblastomas into subgroups was done for 844 patients (n=496 retrospective cohort and n=348 prospective cohort). SNVs, MNVs, small (<30 bp) indels, and large structural variants were predicted across 110 paediatric-onset and adult-onset cancer predisposition genes8, 23 and classified as pathogenic based on stringent criteria.
Figure 1.
Discovery and replication of medulloblastoma predisposition genes
(A) Study design and patient characteristics. (B) Rare variant burden analysis based on 673 patients with medulloblastoma and 53 105 controls from the Exome Aggregation Consortium. Diagnosis of medulloblastoma was the outcome variable. Proportion of patients with a genetic predisposition in our (C) retrospective cohort and (D) prospective cohort. (D) p values indicate statistical differences in the proportion of germline mutation carriers in the retrospective compared with prospective cohort. Infant=age 3 years or younger. Child=age 4–17 years. Adult=age 18 years or older. RR=relative risk. *p<0·001. †p<0·01. ‡p<0·05. §Patients with damaging germline mutations in APC, PTCH1, SUFU, TP53, PALB2, and BRCA2.
We identified damaging germline mutations in 76 (11%) of 673 patients in our retrospective cohort and across 32 genes (appendix pp 10–12). Most damaging germline mutations have been previously described in the literature (in 55 [71%] of 77 mutations) and already classified as (likely) pathogenic (42 [55%] of 77) in public archives (ClinVar, LOVD; appendix pp 10–12). Discovery of consensus medulloblastoma predisposition genes was based on rare variant burden analysis using 673 patients with medulloblastoma and 53 105 ExAC controls. In total, six genes showed a significant excess of damaging germline mutations for patients with medulloblastoma—namely, APC, BRCA2, PALB2, PTCH1, SUFU, and TP53 (adjusted p value for multiple testing <0·05; figure 1B, appendix p 4). Restricting the analysis to patients and controls with European population ancestry revealed no additional candidate medulloblastoma predisposition genes (appendix p 4). Moreover, we did not identify patients with biallelic germline mutations in mismatch repair genes (ie, MSH2, MSH6, MLH1, and PMS2). However, a single patient in our discovery cohort harboured a heterozygous germline mutation in MSH6 (appendix pp 10–12).
The overall prevalence of genetic predisposition based on these six genes in the retrospective discovery cohort was 6% (40/673), with a highest prevalence of 20% (28/141) in patients in the MBSHH subgroup (figure 1C). Replication of these estimates was based on 349 patients who were enrolled in the prospective clinical studies. Key patient characteristics, such as sex, age at diagnosis, and molecular tumour subgroups, were similar between both cohorts (psex=0·68, page=0·17, psubgroup=0·12; figure 1A). The overall prevalence of genetic predisposition in our prospective cohort (17 [5%] of 349) was consistent with estimates from our retrospective cohort (p=0·24; figure 1D, appendix pp 10–12). We further replicated the high prevalence of genetic predisposition in patients in the MBSHH subgroup (figure 1D). Notably, patients in the MBWNT subgroup also had an increased prevalence of germline predisposition in both the discovery and replication cohort, albeit more modest than for patients in the MBSHH subgroup (figure 1C, D).
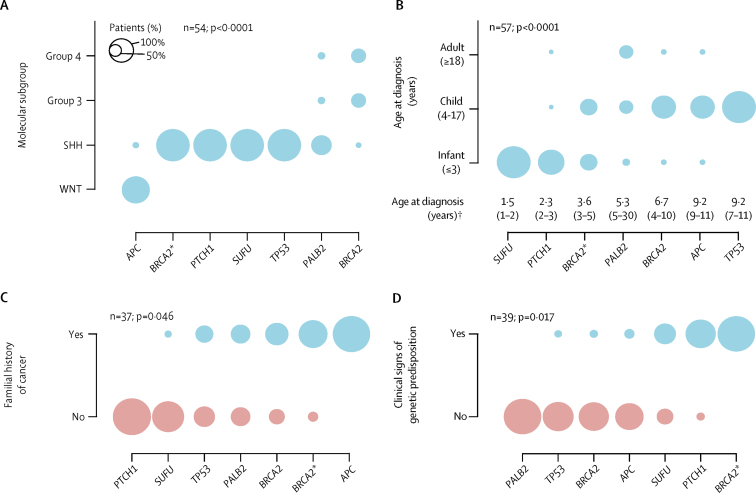
We closely analysed key demographic, clinical, and molecular characteristics of patients with a genetic predisposition in these six genes. Most patients with available subgroup information developed MBSHH (41 [76%] of 54; figure 2A) and age at diagnosis also differed significantly between patients with germline mutations in medulloblastoma predisposition genes (p<0·0001; figure 2B). Patients with germline SUFU or PTCH1 mutations were typically diagnosed as infants at a median age of 2·0 years (IQR 1·3–2·3), whereas patients with APC or TP53 mutations were diagnosed as children at a median age of 9·8 years (IQR 8·0–11). Analysis of histological subtypes in relation to genetic predisposition is presented in the appendix (p 2). Clinical signs of a genetic predisposition were noted in 16 (41%) of 39 patients and familial history of cancer in 17 (46%) of 37 patients, and both were different between patients with germline mutations in medulloblastoma predisposition genes (p=0·017 and p=0·046, respectively; figure 2C, 2D). For example, only one (9%) of all 11 PTCH1 and SUFU mutation carriers with available medical records had a family history of cancer; however, eight (67%) of 12 patients with available medical records had clinical symptoms consistent with Gorlin's syndrome (eg, macrocephaly, jaw cysts, and frontal bossing; appendix pp 10–12). By contrast, all four APC mutation carriers with available medical records had a familial history of familial adenomatous polyposis and associated cancers (eg, adrenocortical carcinoma). Of note, additional malignancies were observed in six (12%) of 50 patients with medulloblastoma and all were restricted to patients with germline APC (n=3) or TP53 (n=3) mutations (appendix pp 10–12).
Figure 2.
Clinical characteristics of patients with a genetic predisposition
(A) Molecular tumour subgroups. (B) Age at diagnosis. (C) Family history of cancer. (D) Clinical signs of a genetic predisposition. p values indicate whether there is a difference between patients with different types of genetic predisposition. *Patients with compound heterozygous germline BRCA2 mutations. †Data are median (IQR). Circle sizes indicate the proportion of patients with a genetic predisposition.
At a median follow-up of 48 months (IQR 28–78), 5-year progression-free survival for 49 patients with a genetic predisposition who were evaluable for this outcome was 52% (95% CI 40–69). 5-year overall survival in 47 evaluable patients with a genetic predisposition was 65% (95% CI 52–81). Progression-free survival (log-rank p=0·0056) and overall survival (log-rank p=0·00032) differed significantly across patients with germline mutations in different medulloblastoma predisposition genes (figure 3).
Figure 3.
Kaplan-Meier curves for survival outcomes of patients with a genetic predisposition
(A) Progression-free survival. (B) Overall survival. Log-rank p values indicate differences across all patient groups.
Germline APC mutations were found in seven (1%) of all 1022 patients with medulloblastoma and included one infant, five children, and one adult patient. Two of these seven patients harboured partial or full gene deletions (appendix p 5) and the remaining five patients had frameshift and nonsense mutations between codons 554 and 1113, a region associated with classical familial adenomatous polyposis phenotypes. Molecular subgroup information was available for six cases and showed that all five children developed MBWNT. One infant developed MBSHH (appendix p 1). All five WNT-driven medulloblastomas lost the wild-type APC allele and the SHH-driven medulloblastoma showed retention of the wild-type allele (figure 4A). Furthermore, germline APC mutations in MBWNT patients were mutually exclusive with somatic CTNNB1 exon 3 mutations (p<0·0001), the primary26 somatic driver event in MBWNT (figure 4B). Overall, germline APC mutations were identified in five (71%) of seven CTNNB1-wild-type MBWNT cases and, together with somatic CTNNB1 mutations, explained 97% (64/66) of all WNT-driven medulloblastomas. By contrast, monosomy 6—a frequent somatic chromosome aberration in MBWNT (in 55 [83%] of 66 cases)—was not mutually exclusive with germline APC mutation status (p=0·19); although we observed two patients with APC mutations and balanced chromosome 6 (figure 4B). We recorded no difference in age at diagnosis between patients with germline APC mutations and somatic CTNNB1 mutations (p=0·92; figure 4B). Overall and progression-free survival for patients with APC-MBWNT was 100% (95% CI 100–100).
Figure 4.
Molecular medulloblastoma characteristics for patients with a genetic predisposition
(A) Patterns of somatic inactivation of wild-type alleles for all patients with a germline mutation in one of the consensus medulloblastoma predisposition genes. Circle sizes indicate the proportion of patients with a genetic predisposition. (B) Somatic driver events in patients with medulloblastoma in the WNT subgroup (top panel) and age at diagnosis for all patients with germline APC mutations and patients in the WNT subgroup with somatic CTNNB1 mutations (bottom panel). (C) Association between germline PALB2 (n=5) and BRCA2 (n=7) mutation status and somatic mutational signatures 1, 3, 5, and 8. EXP=allele-specific gene expression. MUT=somatic inactivation via single nucleotide variation, insertion, or deletion. LOH=loss of heterozygosity. ND=no somatic alteration detected. NS=not significant. HRD=homologous recombination repair deficiency. *Present study and Hamilton and colleagues.5 †p<0·001. ‡p<0·01. §p<0·05.
Germline TP53 mutations were found in 14 (1%) of all 1022 patients with medulloblastoma and were only present in patients with MBSHH (13/13; data missing for one patient). Notably, germline TP53 mutations were identified in 13 (8%) of 170 paediatric MBSHH patients and 13 (20%) of 63 children aged between 5 years and 16 years with MBSHH. Only five (4%) of 11 patients with germline TP53 mutations had a family history of cancer (figure 2B). Most germline TP53 mutations (13/14) clustered within the DNA-binding domain (appendix p 6) and somatic inactivation of the wild-type TP53 allele was detected in all 13 available medulloblastoma genomes via loss of heterozygosity (figure 4A). All eight whole-genome-sequenced TP53-deficient MBSHH exhibited complex somatic genomic rearrangements consistent with chromothripsis21 and accounted for eight (57%) of all 14 chromothripsis events in the MBSHH subgroup. The remaining six patients with chromothripsis-positive MBSHH harboured somatic TP53 mutations. 5-year overall survival for patients with germline TP53 mutations was 27% (95% CI 10–72; figure 3).
Germline SUFU and PTCH1 mutations were detected in 20 (2%) of all 1022 patients with medulloblastoma, exclusively in the MBSHH subgroup (19/19; data missing for one patient), and accounted for 11% (18/170) of all paediatric patients with MBSHH and 21% (17/80) of all infant patients with MBSHH. We observed somatic loss of the SUFU or PTCH1 wild-type allele in all (n=18) sequenced MBSHH that were diagnosed in patients with germline SUFU or PTCH1 mutations. Most (15/18) SUFU-deficient or PTCH1-deficient MBSHH lost the wild-type allele via loss of heterozygosity (figure 4A). Only six of 20 germline SUFU and PTCH1 mutations have been previously described in the literature (appendix pp 10–12), suggesting either appreciable amounts of de-novo mutagenesis or poor reporting to public archives. In support of the de-novo mutagenesis theory, a de-novo germline PTCH1 mutation was observed in a patient with MBSHH from our retrospective cohort, for whom whole-exome sequences were also available for both parents. We also identified putative protein-truncating mosaic PTCH1 mutations in three patients with MBSHH (in whom variant allele frequency ranged from 4·7% to 6·7% in the blood). Clinical information was available for one patient with a mosaic PTCH1 mutation, which showed that the patient was clinically diagnosed with Gorlin's syndrome and the patient had no family history of cancer and no family history of Gorlin's syndrome. Moreover, comparison of clinical outcomes between patients with germline mutations in SUFU and PTCH1 showed no differences in progression-free survival (p=0·50, 16 patients with germline mutations) and overall survival (p=0·91, 17 patients with germline mutations). Notably, patients with germline mutations in either SUFU or PTCH1 had a better overall survival than progression-free survival (combined 5-year progression-free survival 56% for SUFU and PTCH1 mutations, 95% CI 37–87; combined 5-year overall survival 85%, 95% CI 67–100; figure 3).
Germline BRCA2 mutations were present in 11 (1%) of all 1022 patients with medulloblastoma and included ten paediatric patients and one adult (survival data not available for all patients with these mutations). The median age at diagnosis was 5·7 years (IQR 3·0–8·3). Most of these mutations (16 [89%] of 18) were previously classified as pathogenic according to ClinVar (appendix pp 10–12). We observed compound heterozygosity at BRCA2 in four (36%) of 11 patients. Clinical signs of a genetic predisposition or family history of cancer were noted in all four compound heterozygous patients and in four (80%) of five heterozygous patients with available medical records. Patients with compound heterozygous mutations at BRCA2 developed exclusively MBSHH (p=0·0060 when compared with any other subgroup; figure 2A) and exhibited worse progression-free survival (p=0·025) and overall survival (p=0·022) relative to patients with heterozygous germline BRCA2 mutations (figure 3). When compared with 53 105 controls, the burden of rare germline mutations in BRCA2 was associated with increased risk of MBSHH (relative risk [RR] 13·8 [5·4–29·4]; p<0·0001) and MBGroup3/4 (RR 4·2 [1·4–10·1]; p=0·0077). Overall and progression-free survival of heterozygous germline BRCA2 mutation carriers was 100% (95% CI 100–100) and none developed secondary malignancies. Analysis of medulloblastoma genomes revealed that all seven heterozygous BRCA2 mutation carriers retained the wild-type BRCA2 allele. Additional details about family history of cancer, parental genetic testing, and somatic mutation profiles in heterozygous and compound heterozygous BRCA2 mutation carriers are provided in the appendix (p 1).
Germline PALB2 mutations were found in five (<1%) of all 1022 patients with medulloblastoma. We identified three patients with MBSHH, one patient with MBGroup3 and one patient with MBGroup4. All five damaging germline mutations were heterozygous in patients affected with medulloblastoma and were previously reported in familial pancreatic and breast cancer studies.27 All five germline mutations were classified as pathogenic according to ClinVar (appendix pp 10–12). Although heterozygous PALB2 mutations are known8 to increase the risk of adult-onset cancers (eg, breast cancer), predisposition to paediatric malignancies has so far only been described in the context of Fanconi anaemia.28 We excluded the predisposition to Fanconi anaemia in all five cases because of an absence of additional rare germline mutations (including protein-truncating variants, missense mutations, and inframe indels). Furthermore, clinical signs of a genetic disorder were absent in both paediatric cases with available medical records (figure 2D). Analysis of medulloblastoma genomes revealed somatic inactivation of the wild-type PALB2 allele in all three paediatric cases and retention of heterozygosity in both adult cases (figure 4A). 5-year overall and progression-free survival for patients with germline PALB2 mutations was 75% (95% CI 43–100). Additional details about family history of cancer, parental genetic testing, and somatic mutation profiles available for one affected patient are presented in the appendix (p 1).
Since the clinical relevance of heterozygous germline mutations in PALB2 and BRCA2 is uncertain, we aimed to corroborate our findings through investigation of somatic mutation patterns (mutational signatures) in medulloblastoma genomes. Quantification of four previously implicated mutational signatures (signatures 1, 3, 5, and 8) across 375 medulloblastoma genomes showed a significant association between both HRD signatures (signatures 3 and 8) and germline BRCA2 and PALB2 mutation status (figure 4C, appendix p 7). Moreover, although the overall prevalence of an HRD-like mutation spectrum was modest in medulloblastoma (58 [15%] of 375), it was enriched in BRCA2-deficient and PALB2-deficient tumours as compared with tumours with other genetic mutations (OR 19·0 [95% CI 4·5–113], p<0·0001). In total, nine (75%) of 12 BRCA2-deficient and PALB2-deficient tumours showed evidence for an HRD-like mutation spectrum. Strikingly, eight (89%) of nine patients with MBSHH and an HRD-like mutation spectrum harboured germline mutations in consensus medulloblastoma predisposition genes (BRCA2 n=4, PALB2 n=2, and TP53 n=2), suggesting that HRD might serve as a biomarker for genetic predisposition in this patient group. We also observed five patients in the MBWNT subgroup with an HRD-like mutation spectrum and noticed that four (89%) of five cases were diagnosed as adults (p=0·0003 for paediatric vs adult cases). The only paediatric patient in the MBWNT subgroup with an HRD-like mutation spectrum harboured a pathogenic heterozygous germline mutation in ATM along with somatic inactivation of the wild-type ATM allele (appendix pp 10–12). Finally, we also identified heterozygous germline mutations in FANCA (n=1) and FANCQ (n=1) in patients with MBGroup3 and MBGroup4, respectively, and an HRD-like mutation spectrum. Taken together, these results corroborate our genetic findings and indicate that genetic alteration of homologous recombination genes is associated with an HRD-like mutation spectrum in medulloblastoma.
Discussion
To the best of our knowledge, this study is the largest so far on genetic predisposition in a single paediatric brain tumour entity. Our rare variant burden analyses revealed that genetic predisposition has a major role in the cause of medulloblastoma after accounting for molecular subgroups, with a high prevalence in MBWNT and MBSHH patient subgroups. Moreover, we identified that less than half of all germline cases were suspected based on medical records and that patients with genetic predispositions exhibit distinct clinical outcomes. Our data show the urgent need to establish genetic counselling and genetic testing as a standard-of-care procedure in patients with MBWNT and MBSHH. We therefore provide clinical screening guidelines based on routinely acquired patient and tumour characteristics (figure 5). Recommendations for surveillance and clinical management of individual childhood cancer predisposition syndromes have been summarised in a series of articles29 from the American Association for Cancer Research Childhood Cancer Predisposition Workshop. Our study complements these recommendations by providing additional diagnostic criteria based on clinical and molecular characteristics.
Figure 5.
Proposed clinical guidelines for genetic counselling and testing in medulloblastoma based on clinical and molecular tumour characteristics
Here, we identified heterozygous germline APC mutations in 6–8% of patients in the MBWNT molecular subgroup and showed that genetic cases were indistinguishable from sporadic cases based on age at diagnosis. Importantly, all patients with APC-deficient MBWNT did not have the hallmark somatic driver event of MBWNT—namely, somatic missense mutations in CTNNB1 (the gene encoding β-catenin). Overall, germline APC and somatic CTNNB1 mutations accounted for nearly all (97%) MBWNT diagnoses. We therefore recommend genetic counselling only for germline APC mutations in patients with MBWNT. In our series, patients with MBWNT and germline APC mutations showed favourable clinical outcomes, which mirrors the favourable outcome for patients with MBWNT with nuclear accumulation of β-catenin.30 Nevertheless, several patients with germline APC mutations had an additional malignancy, which emphasises the need to provide genetic counselling for patients with MBWNT in the future, irrespective of clinical outcomes. Notably, we also observed a patient with atypical APC who was diagnosed during infancy with MBSHH and retained the wild-type APC allele. Additional studies will be needed to assess whether or not germline APC mutations are a genetic risk factor for MBSHH.
Given the particularly high prevalence of damaging germline mutations in patients with MBSHH, we recommend that all patients with MBSHH should be counselled for genetic testing. We recommend ordering of these tests by age at diagnosis and molecular phenotypes (eg, chromothripsis or HRD signatures). Patients younger than 3 years of age should initially be tested for germline mutations in SUFU and PTCH1 (especially in view of the high prevalence of SUFU mutations in infant patients with MBSHH in our study, which is consistent with previous reports31), and children older than 3 years for germline TP53 mutations. By contrast, germline mutations in PALB2 and BRCA2 showed no strict associations with age at diagnosis (observed in infants, children, and adults). We thus recommend that patients with MBSHH should be screened for germline mutations in PALB2 and BRCA2 following a negative test result for the aforementioned genes. We further observed germline PALB2 and BRCA2 mutations in 1–2% of patients with MBGroup3/4. Based on these findings we recommend conservative ordering of genetic tests in these patient groups (eg, those with familial history of BRCA-associated cancers or if mutational signatures are suggestive of HR deficiency—specifically signatures 3 and 8, the latter of which has previously been reported to be associated with breast cancer).32
Genetic counselling and testing for germline PALB2 and BRCA2 mutations in paediatric patients has so far been pursued only in case of suspected Fanconi anaemia. By use of integrative genomic analyses, we showed that heterozygous mutations in BRCA2 and PALB2 are associated with an increased risk of medulloblastoma and of HR-deficient tumours. All identified heterozygous germline mutations are rare in the general population (minor allele frequency <0·01%) and were classified in most patients as (likely) pathogenic in ClinVar. Magnusson and colleagues33 reported that families with germline mutations in BRCA2 had an increased prevalence of childhood tumours compared with population-based control families. Medulloblastoma genomes from patients with heterozygous germline mutations in BRCA2 and PALB2 described in our study show strong signs of HRD. Many studies have sought to elucidate the mechanisms of genomic instability in heterozygous BRCA2 and PALB2 mutation carriers. For example, it was shown that cells from heterozygous PALB2 mutation carriers exhibited an aberrant DNA replication stress response and increased amounts of spontaneous genomic instability,34 and analysis of double-strand break repair outcomes35 revealed a shift towards error-prone DNA repair pathways. Furthermore, one study36 presented evidence showing that naturally occurring concentrations of formaldehyde, a product of cellular metabolism, can selectively deplete BRCA2 via proteasomal degradation and induce genomic instability. Although wild-type cells retained adequate protein concentrations in that study, cells heterozygous for truncating BRCA2 mutations showed enhanced susceptibility to formaldehyde-induced genomic instability. Future research is needed to understand whether and to what extent BRCA2 and PALB2-deficient medulloblastomas might be sensitive to aldehyde-induced DNA damage. Moreover, consequent identification of paediatric patients with heterozygous germline mutations in PALB2 and BRCA2 will be valuable to further evaluate whether HR-deficient tumours show a particularly favourable response to standard platinum-based chemotherapy and whether they might benefit from combination therapies with PARP inhibitors.
Germline mutations in TP53 accounted for the highest proportion of genetic cases among paediatric patients in the MBSHH subgroup. This patient group showed especially poor clinical outcomes as well as secondary malignancies. This finding underscores the need for a dedicated treatment protocol, which is being prepared by the International Society of Paediatric Oncology PNET 5 Medulloblastoma study group and by an international registry for this high-risk patient group.37 Additionally, clinical surveillance of germline TP53 mutation carriers has been shown to result in earlier detection of tumours and therefore improved long-term survival.38
We also observed rare and damaging germline mutations with potential clinical relevance in additional genes based on loss of heterozygosity analysis and somatic mutation patterns (eg, ATM and PTEN). The molecular and clinical evidence reported in this study was, however, not fully conclusive for these additional genes. Long-term follow-up studies and more detailed molecular analysis will be necessary to provide unambiguous recommendations for routine genetic screening of these genes. Of note, we did not identify patients with biallelic germline mutations in mismatch repair genes (MSH2, MSH6, MLH1, and PMS2). A single patient harboured a heterozygous germline MSH6 mutation in our discovery cohort; however, the meaning of this observation remains uncertain owing to the absence of tumour material for molecular analyses.
Our study does have some limitations. Because of the multiple (and in part heterogeneous) cohorts used in our study, familial history of cancer could not be obtained in a standardised form for all patients carrying a genetic predisposition. Furthermore, we did not make any effort to pursue retrospective collection of clinical information from patients without a detected genetic predisposition. Larger cohort sizes in further studies will be necessary to assess the impact of germline mutations on clinical outcomes within molecular subgroups as well as relative to patients who develop sporadic medulloblastoma due to somatic mutations in the same set of genes (eg, PTCH1, SUFU, and TP53). These larger molecular cohorts will further enable pinpointing of shared somatic driver events in patients with a genetic predisposition. Furthermore, we note that our rare variant burden analysis against ExAC was restricted to regions covered by whole-exome sequencing, was restricted to genes previously involved in cancer predisposition, and excluded pathogenic germline structural variants. Although these steps aimed to reduce potential biases from analysing heterogeneous sequencing cohorts, we cannot rule out that particular classes of germline variation might have been under-represented. Finally, it is also possible that additional genes are involved in medulloblastoma predisposition, which were not previously associated with hereditary cancer syndromes.
Acknowledgments
Acknowledgments
This project was supported by the PedBrain Tumor Project contributing to the ICGC, funded by German Cancer Aid (109252), the German Federal Ministry of Education and Research (BMBF; 01KU1201A and 01KU1201C), and additionally through BMBF grants BioTop (01EK1502A and 01EK1502B), ICGC-Data Mining (01KU1505F), MedSys (0315416C) and NGFNplus (01GS0883). Additional support came from the German Cancer Research Center-Heidelberg Center for Personalized Oncology (German Cancer Research Centre [DKFZ]-HIPO), the St Jude Comprehensive Cancer Center Core Grant (CA 21765), the American Lebanese Syrian Associated Charities, German Cancer Aid (111234), and the German Childhood Cancer Foundation (Deutsche Kinderkrebsstiftung; A2013/46 DKS2014.12). JOK was supported by a European Research Council Starting Grant (336045) and EurocanPlatform (260791) funding from the European Commission. DM was supported by the Canadian Institutes for Health Research (143234). PAN is the recipient of a Roman-Herzog Postdoctoral Fellowship (Hertie Foundation), V Foundation V Scholar Award, Sontag Foundation Distinguished Scientist Award, and is a Pew-Stewart Scholar for Cancer Research (Alexander and Margaret Stewart Trust). AG acknowledges funding from Musicians Against Childhood Cancer. MM acknowledges support from the BC Cancer Foundation and Canadian Institutes for Health Research (FDN 143288). MDT is supported by the Garron Family Chair in Childhood Cancer Research, and grants from the Cure Search Foundation, the National Institutes of Health (R01CA148699 and R01CA159859), the Pediatric Brain Tumor Foundation, the Terry Fox Research Institute, and Brainchild. This study was done with the support of the Ontario Institute for Cancer Research through funding provided by the Government of Ontario. SMW was supported by a Swiss National Science Foundation Early Postdoc.Mobility Fellowship (P2ELP3_155365) and an EMBO Long-Term Fellowship (ALTF 755-2014). SLP was supported by the NIH (2R01 CA109467, U01 CA184898, and U54 HD090255). TCA was supported by the NIH (T32 HL007901). RJG was supported by the Cancer Research UK (A17197) and the NIH (P01CA96832 and R0CA1129541). Tissue banking was supported by funds from the Faculty of Medicine, Masaryk University (Brno, Czech Republic) and KZ was supported by the project AZV 15-30657A from the Ministry of Health of the Czech Republic. CEFALO was supported by grants from the Swedish Council for Health, Working Life and Welfare (2004-0504, 2007-0224), the Swedish Research Council (K2008-70X-15366-04-3), the Swedish Cancer Society (09 0666), the Swedish Childhood Cancer Foundation (PROJ06/050, PROJ09/086), the Swedish Radiation Protection Authority (SSI P 1572), the Danish Strategic Research Council (2103-05-0006, 2064-04-0010), the Swiss Federal Office of Public Health (05.001626), the Swiss Research Foundation on Mobile Communication (A2006.18), the Swiss National Science Foundation (PDFMP3_122873), and the Research Council of Norway (175163/V40). The Medulloblastoma Advanced Genomics International Consortium (or MAGIC) project is financially supported by Genome Canada, Genome BC, Terry Fox Research Institute, Ontario Institute for Cancer Research, Pediatric Oncology Group Ontario, Funds from The Family of Kathleen Lorette and the Clark H Smith Brain Tumour Centre, Montreal Children's Hospital Foundation, Hospital for Sick Children: Sonia and Arthur Labatt Brain Tumour Research Centre, Chief of Research Fund, Cancer Genetics Program, Garron Family Cancer Centre, BRAIN Child, MDT's Garron Family Endowment, and BC Childhood Cancer Parents Association. We thank the DKFZ Genomics and Proteomics Core Facility, Andrea Wittmann (Pediatric Glioma Research Group, DKFZ, Heidelberg, Germany), Laura Sieber (Division of Pediatric Neurooncology, DKFZ), Rolf Kabbe (Division of Theoretical Bioinformatics, DKFZ; Department for Bioinformatics and Functional Genomics, Institute for Pharmacy and Molecular Biotechnology, and BioQuant, Heidelberg University, Heidelberg, Germany), Matthias Bieg (Division of Theoretical Bioinformatics, Heidelberg Center for Personalised Oncology, DKFZ), Matthias Schlesner (Bioinformatics and Omics Data Analytics, DKFZ), and Bernd Klaus (Genome Biology Unit, European Molecular Biology Laboratory [EMBL], Heidelberg, Germany) for technical support. We also acknowledge the IT facilities at EMBL and DKFZ for invaluable support. We thank Brandon Stelter (Biomedical Communications, St Jude Children's Research Hospital, Memphis, TN, USA) for assistance with artwork. This work is dedicated to Prof Dr Enno Kleihauer, who introduced molecular genetics to the field of neuro-oncology in the early 1980s, and who was one of the most influential paediatric haematologist-oncologists in Germany and beyond.
Contributors
JOK, PAN, SMP, and SMW contributed to manuscript preparation and to study design. PAN and SMP contributed to study conception. JOK contributed to study supervision. PL and RE contributed to supervision of the International Cancer Genome Consortium (ICGC) PedBrain Tumor Project. AE, ASM, BRM, CL, DSt, DTWJ, GWR, IB, JOK, JW, JZ, LC, MK, MS-W, PAN, SB, SG, SMP, SMW, TR, TZ, and XZ contributed to data analysis. MH contributed to information technology infrastructure. AEB, AG, AK, AP, BL, CEK, CJ, CMM, DAS, DCB, DM, DSa, DSt, DSu, DWF, ED, FW, GF, GM, GWR, H-KN, JAC, JOK, JS, KK, KZ, KvH, LB, LG-R, MDT, MF, MG, MM, MMC, MR, MVR, NJ, PH, PHD, PV, RC, RJG, RS, SF, SLP, SMP, SR, TCA, TE, TH, TJM, TM, TVA, VR, WG, WS, and Y-SR contributed to data collection. AG, CB, CDB, CPK, CRB, CS, DM, FMDLV, KBG, KWP, and SSS contributed to data interpretation.
Declaration of interests
SMW reports grants from the European Molecular Biology Organization (EMBO) and from the Swiss National Science Foundation, during the conduct of the study. GM reports grants from the St Jude Children's Research Hospital, during the conduct of the study. KvH reports grants from the German Childhood Cancer Foundation, during the conduct of the study. MF reports grants from the Swedish Research Council, the Swedish Council for Health, Working Life and Welfare, the Swedish Cancer Society, the Swedish Childhood Cancer Foundation, and the Swedish Radiation Protection Authority, during the conduct of the study. SLP reports grants from the National Institutes of Health, during the conduct of the study. FMDLV reports financial activities from Annai Systems Inc and financial activities from TOMA Biosciences Inc, outside the submitted work. CDB reports grants from the National Institutes of Health (NIH), during the conduct of the study; grants from the University of Miami, financial activities from the University of Chicago; University of California, Los Angeles; HUGO; New York Genome Center; University of Iowa; Rockefeller University; Alexandria; FH Foundation; Concert Genetics; National Autonomous University of Mexico; Mexican Institute of Social Security; Colorado State University; MacArthur Foundation; and Gordon Conference; and personal fees from CDB Consulting Ltd, Personalis, Inc, 23andme “Roots into the Future Project”, Ancestry.com, Liberty Biosecurity, Med-Tek, IdentifyGenomics LLC, Mars Inc, Etalon Inc, Fish & Richardson PC, Eden Roc, Hypatia, the Nicklaus Children's Hospital Research Institute, Knox Medical, Arc Bio LLC, Embark Veterinary, Digitalis Ventures, Humancode, Web Shield, and Luna DNA, outside the submitted work. All other authors declare no competing interests.
Supplementary Material
References
- 1.Johnson KJ, Cullen J, Barnholtz-Sloan JS. Childhood brain tumor epidemiology: a Brain Tumor Epidemiology Consortium review. Cancer Epidemiol Biomarkers Prev. 2014;23:2716–2736. doi: 10.1158/1055-9965.EPI-14-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Northcott PA, Jones DTW, Kool M. Medulloblastomics: the end of the beginning. Nat Rev Cancer. 2012;12:818–834. doi: 10.1038/nrc3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolter M, Reifenberger J, Sommer C, Ruzicka T, Reifenberger G. Mutations in the human homologue of the drosophila segment polarity gene patched (PTCH) in sporadic basal cell carcinomas of the skin and primitive neuroectodermal tumors of the central nervous system. Cancer Res. 1997;57:2581–2585. [PubMed] [Google Scholar]
- 4.Taylor MD, Liu L, Raffel C. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31:306–310. doi: 10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- 5.Li FP, Fraumeni JF. Rhabdomyosarcoma in children: epidemiologic study and identification of a familial cancer syndrome. J Natl Cancer Inst. 1969;43:1365–1373. [PubMed] [Google Scholar]
- 6.Hamilton SR, Liu B, Parsons RE. The molecular basis of Turcot's syndrome. N Engl J Med. 1995;332:839–847. doi: 10.1056/NEJM199503303321302. [DOI] [PubMed] [Google Scholar]
- 7.Offit K, Levran O, Mullaney B. Shared genetic susceptibility to breast cancer, brain tumors, and Fanconi anemia. J Natl Cancer Inst. 2003;95:1548–1551. doi: 10.1093/jnci/djg072. [DOI] [PubMed] [Google Scholar]
- 8.Rahman N. Realizing the promise of cancer predisposition genes. Nature. 2014;505:302–308. doi: 10.1038/nature12981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor MD, Northcott PA, Korshunov A. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123:465–472. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Northcott PA, Korshunov A, Pfister SM, Taylor MD. The clinical implications of medulloblastoma subgroups. Nat Rev Neurol. 2012;8:340–351. doi: 10.1038/nrneurol.2012.78. [DOI] [PubMed] [Google Scholar]
- 11.Zhukova N, Ramaswamy V, Remke M. Subgroup-specific prognostic implications of TP53 mutation in medulloblastoma. J Clin Oncol. 2013;31:2927–2935. doi: 10.1200/JCO.2012.48.5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kool M, Jones DT, Jager N. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell. 2014;25:393–405. doi: 10.1016/j.ccr.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu J, Margol AS, Shukla A. Disseminated medulloblastoma in a child with germline BRCA2 6174delT mutation and without Fanconi anemia. Front Oncol. 2015;5:191. doi: 10.3389/fonc.2015.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Walsh MF, Wu G. Germline mutations in predisposition genes in pediatric cancer. N Engl J Med. 2015;373:2336–2346. doi: 10.1056/NEJMoa1508054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones DTW, Jager N, Kool M. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pugh TJ, Weeraratne SD, Archer TC. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson G, Parker M, Kranenburg TA. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Northcott PA, Buchhalter I, Morrissy AS. The whole-genome landscape of medulloblastoma subtypes. Nature. 2017;547:311–317. doi: 10.1038/nature22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aydin D, Feychting M, Schüz J. Mobile phone use and brain tumors in children and adolescents: a multicenter case–control study. J Natl Cancer Inst. 2011;103:1264–1276. doi: 10.1093/jnci/djr244. [DOI] [PubMed] [Google Scholar]
- 20.Waszak SM, Tiao G, Zhu B. Germline determinants of the somatic mutation landscape in 2,642 cancer genomes. bioRxiv. 2017 DOI:10.1101/208330 published online Nov 1. [Google Scholar]
- 21.Rausch T, Jones DT, Zapatka M. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexandrov LB, Nik-Zainal S, Wedge DC. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bourdeaut F, Miquel C, Richer W. Rubinstein-Taybi syndrome predisposing to non-WNT, non-SHH, group 3 medulloblastoma. Pediatr Blood Cancer. 2014;61:383–386. doi: 10.1002/pbc.24765. [DOI] [PubMed] [Google Scholar]
- 24.Mardin BR, Drainas AP, Waszak SM. A cell-based model system links chromothripsis with hyperploidy. Mol Syst Biol. 2015;11:828. doi: 10.15252/msb.20156505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hovestadt V, Remke M, Kool M. Robust molecular subgrouping and copy-number profiling of medulloblastoma from small amounts of archival tumour material using high-density DNA methylation arrays. Acta Neuropathol. 2013;125:913–916. doi: 10.1007/s00401-013-1126-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson MC, Fuller C, Hogg TL. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24:1924–1931. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- 27.Antoniou AC, Casadei S, Heikkinen T. Breast-cancer risk in families with mutations in PALB2. N Engl J Med. 2014;371:497–506. doi: 10.1056/NEJMoa1400382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reid S, Schindler D, Hanenberg H. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 29.Clinical Cancer Research Clinical Cancer Research Pediatric Oncology Series. http://clincancerres.aacrjournals.org/pediatricseries?utm_source=landingpage&utm_medium=snippet2&utm_campaign=pediatricseries
- 30.Ellison DW, Kocak M, Dalton J. Definition of disease-risk stratification groups in childhood medulloblastoma using combined clinical, pathologic, and molecular variables. J Clin Oncol. 2011;29:1400–1407. doi: 10.1200/JCO.2010.30.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brugières L, Remenieras A, Pierron G. High frequency of germline SUFU mutations in children with desmoplastic/nodular medulloblastoma younger than 3 years of age. J Clin Oncol. 2012;30:2087–2093. doi: 10.1200/JCO.2011.38.7258. [DOI] [PubMed] [Google Scholar]
- 32.Nik-Zainal S, Davies H, Staaf J. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magnusson S, Wiebe T, Kristoffersson U, Jernström H, Olsson H. Increased incidence of childhood, prostate and breast cancers in relatives of childhood cancer patients. Fam Cancer. 2012;11:145–155. doi: 10.1007/s10689-011-9493-3. [DOI] [PubMed] [Google Scholar]
- 34.Nikkilä J, Christin Parplys A, Pylkäs K. Heterozygous mutations in PALB2 cause DNA replication and damage response defects. Nat Commun. 2013;4:2578. doi: 10.1038/ncomms3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obermeier K, Sachsenweger J, Friedl TWP, Pospiech H, Winqvist R, Wiesmuller L. Heterozygous PALB2 c.1592delT mutation channels DNA double-strand break repair into error-prone pathways in breast cancer patients. Oncogene. 2016;35:3796–3806. doi: 10.1038/onc.2015.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan SLW, Chadha S, Liu Y. A class of environmental and endogenous toxins induces BRCA2 haploinsufficiency and genome instability. Cell. 2017;169:1105. doi: 10.1016/j.cell.2017.05.010. 18.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kratz CP, Achatz MI, Brugieres L. Cancer screening recommendations for individuals with Li-Fraumeni syndrome. Clin Cancer Res. 2017:e38–e45. doi: 10.1158/1078-0432.CCR-17-0408. [DOI] [PubMed] [Google Scholar]
- 38.Villani A, Shore A, Wasserman JD. Biochemical and imaging surveillance in germline TP53 mutation carriers with Li-Fraumeni syndrome: 11 year follow-up of a prospective observational study. Lancet Oncol. 2016;17:1295–1305. doi: 10.1016/S1470-2045(16)30249-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.