Correction
Following the publication of this article [1], the authors noticed that Figs. 2, 3 and 4 were in the incorrect order and thus had incorrect captions. The images that were incorrectly published as Figs. 2, 3 and 4 should have been published as Figs. 3, 4 and 2 respectively.
The correct versions of Figs. 2, 3 and 4 with captions have been included in this Correction.
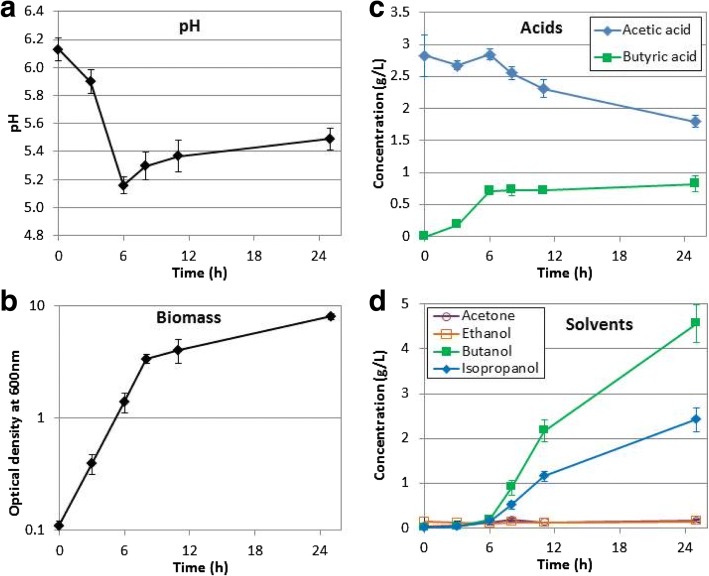
Fig. 2.
Fermentation profile of Clostridium beijerinckii DSM 6423 on glucose. C. beijerinckii DSM 6423 was cultivated in bioreactors in GAPES medium. a pH, b biomass followed by OD600, c acids and d solvents. Values are the mean and standard deviation of the 6 biological replicates. See Additional file 3 for details on the biological replicates
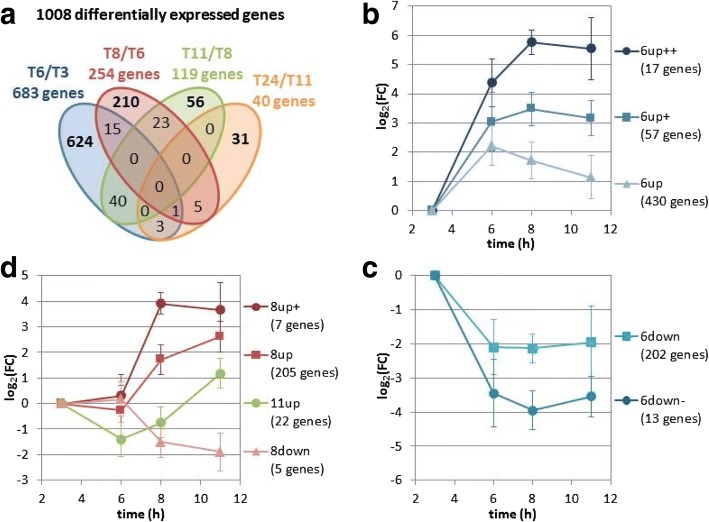
Fig. 3.
Global transcriptomic analysis of C. beijerinckii DSM6423 fermentation on glucose. a Venn Diagram showing the number of genes regulated in various physiological time points. b to d: kinetic expression profiles of various clusters of genes: genes up-regulated at 6 h b, genes down-regulated at 6 h c, and genes regulated at 8 h or 11 h d
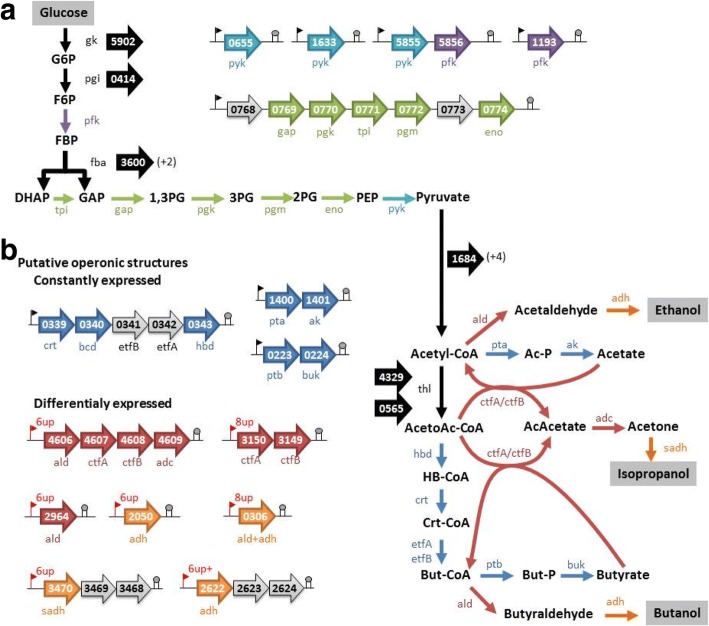
Fig. 4.
Main genes and predicted operonic structures involved in the central metabolism of in C. beijerinckii DSM6423. a glycolysis; b acids and solvents production) . Number of isozymes, predicted by Microscope tool (Genoscope, Evry, France) are indicated in brackets
The original article has been corrected.
Footnotes
The original article can be found online at 10.1186/s12864-018-4636-7
Reference
- 1.de Gérando M, et al. BMC Genomics. 2018;19:242. doi: 10.1186/s12864-018-4636-7. [DOI] [PMC free article] [PubMed] [Google Scholar]